Abstract
Background
Gliomas are the most common primary tumors in central nervous system. Despite advances in diagnosis and therapy, the prognosis of glioma remains gloomy. Autophagy is a cellular catabolic process that degrades proteins and damaged organelles, which is implicated in tumorigenesis and tumor progression. Autophagy related 4C cysteine peptidase (ATG4C) is an autophagy regulator responsible for cleaving of pro-LC3 and delipidation of LC3 II. This study was designed to investigate the role of ATG4C in glioma progression and temozolomide (TMZ) chemosensitivity.
Methods
The association between ATG4C mRNA expression and prognosis of gliomas patients was analyzed using the TCGA datasets. The role of ATG4C in proliferation, apoptosis, autophagy, and TMZ chemosensitivity were investigated by silencing ATG4C in vivo. Ectopic xenograft nude mice model was established to investigate the effects of ATG4C on glioma growth in vivo.
Results
The median overall survival (OS) time of patients with higher ATG4C expression was significantly reduced (HR: 1.48, p = 9.91 × 10− 7). ATG4C mRNA expression was evidently increased with the rising of glioma grade (p = 2.97 × 10− 8). Knockdown ATG4C suppressed glioma cells proliferation by inducing cell cycle arrest at G1 phase. ATG4C depletion suppressed autophagy and triggered apoptosis through ROS accumulation. Depletion of ATG4C suppressed TMZ-activated autophagy and promoted sensitivity of glioma cells to TMZ. Additionally, ATG4C knockdown suppressed the growth of glioma remarkably in nude mice.
Conclusion
ATG4C is a potential prognostic predictor for glioma patient. Targeting ATG4C may provide promising therapy strategies for gliomas treatment.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1287-8) contains supplementary material, which is available to authorized users.
Keywords: Glioma, Glioblastoma, ATG4C, Autophagy, Temozolomide
Background
Gliomas are the most common primary malignant tumors of central nervous system (CNS) in adults [1, 2]. According to the morphopathology features of tumor cells, gliomas are mainly classified into subtypes including astrocytoma, oligodendroglioma, ependymoma and choroid plexus papilloma [3, 4]. Based on pathological evaluation, gliomas are categorized as Grade I to IV, and higher grade indicates worse prognosis [3, 5]. The median survival time of patients with low-grade glioma (LGG, Grade II and III) is 3 to 10 years, compared with 1.5 years for patients with Grade IV (Glioblastoma) [6–10]. Temozolomide (TMZ) is considered to be the most effective drug for the treatment of glioblastoma, which can prolong the survival of glioblastoma patients by 2 to 5 months [9, 11–13]. Several molecular biomarkers, such as IDH mutation, 1p/19q co-deletion and MGMT promoter methylation, are shown to predict prognosis and/or drug responses for gliomas [3, 9, 13]. However, the overall prognosis of gliomas has not been improved effectively.
Macroautophagy (Autophagy) is an evolutionally conserved dynamic catabolic process that degrades proteins and damaged organelles [14, 15]. The role of autophagy in tumorigenesis and progression is complicated. On the one hand, autophagy deficiency promotes tumorigenesis; on the other hand, autophagy is essential for the progression of tumor [16–21]. Additionally, recent studies have suggested that TMZ-induced autophagy sustained the survival of glioblastoma cells, thereby contributing to drug resistance and recurrence [10, 22]. Blocking autophagy by inhibitor can significantly enhance TMZ cytotoxicity [23–25]. Whereas, non-specific autophagy inhibition may bring unexpected safety problems, which limits its clinical application [10, 26, 27].
The Cancer Genome Atlas (TCGA) project, aimed to depict profile of cancer genome, is widely used in tumor research. Based on TCGA data mining, we observed that expression level of autophagy related 4C cysteine peptidase (ATG4C), a member of ATG4 family, is deeply associated with prognosis of glioma patients. ATG4C is a cysteine peptidase responsible for lipidation and delipidation of LC3 during autophagy [28, 29]. Previous study has shown that ATG4C is involved in the maintenance of stem-like phenotype in breast cancer cells [30]. However, the role of ATG4C in gliomas progression and TMZ chemosensitivity remains unclear.
In this study, we tried to determine whether interference with ATG4C can affect autophagy activity and TMZ sensitivity in glioma cell lines, as well as its role in disease progression of glioma both in vitro and in vivo.
Methods
Patients samples
Glioma was diagnosed according to the 2016 WHO Classification of Tumors of the Central Nervous System. The glioma tissues and clinical data were obtained from patients who underwent surgery between 2015 June and 2018 January at Xiangya Hospital of Central South University. The informed consents were provided to all patients or their family members. This study was approved by Central South University Xiangya Hospital Medical Ethics Committee.
Bioinformatic data mining
The integrated gliomas expression profile data and corresponding clinical data were downloaded from the TCGA database (http://www.cbioportal.org/study.do?cancer_study_id=lgggbm_tcga_pub). Clinical characteristics of the patients were shown in Table 1.
Table 1.
Baseline clinical characteristics of patients
| Clinical characteristic | Prognosis analysis in gliomas | Prognosis analysis in LGG | Prognosis analysis in GBM |
|---|---|---|---|
| NO. of patients | 975 | 414 | 561 |
| Age (years) | |||
| ≥55 | 546 | 326 | 220 |
| <55 | 429 | 88 | 341 |
| Gender | |||
| Male | 573 | 227 | 346 |
| Female | 402 | 187 | 215 |
| WHO grade | |||
| II | 198 | 198 | 0 |
| III | 216 | 216 | 0 |
| IV | 561 | 0 | 561 |
| IDH status | |||
| Wild | 483 | 77 | |
| Mutant | 372 | 337 | |
| Undefined | 120 | 2 | |
| MGMT status | |||
| Unmethylated | 280 | 211 | |
| Methylated | 514 | 169 | |
| Undefined | 181 | 181 | |
Cell culture
The human glioma cell line U87-MG was purchased from Cell Bank of Chinese Academy, Shanghai Institute of Biochemistry and Cell Biology, China Academy of Science. Human glioma cell line T98G was kindly provided by Dr. Lv from Jiangxi Cancer Hospital, China. The cell lines were identified using short tandem repeat (STR) markers. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, CA, USA) in a humiliated incubator containing 5% CO2 at 37 °C.
Cell transfection
Three different small interfering RNA (siRNA) against ATG4C and negative control siRNA were synthesized by RiboBio (Guangzhou, China). Exponentially growing cells were seeded in 6 well plates and cultured overnight. The plated cell at 60–70% confluence was transfected with indicated siRNAs at a final concentration of 50 nM using Lipofectamine RNAiMAX reagent (Invitrogen, CA, USA) following the manufacturer’s instruction. The plasmid of RFP-GFP-LC3B was kindly provided by Professor Cheng, Central South University, China. Cells were transfected with 1.0 μg plasmid per well of a 6-well plate using Lipofectamine 3000 reagent (Invitrogen, CA, USA) according to the manufacturer’s instruction. Six hours after transfection, the medium was replaced with fresh medium containing 10% FBS. The transfected cells were collected for further analysis.
Lentivirus infection and stable cell lines establishment
The knockdown lentivirus vector of ATG4C (LV-ATG4C-shRNA) and the lentivirus vector carrying scramble sequences (LV-NC-shRNA) were purchased from Genechem (Shanghai, China). All the lentivirus vectors were verified by DNA sequencing. After seeded in 6-well plates for 24 h, cells were infected with ATG4C sh-RNA (sh-ATG4C) or negative control sh-RNA (sh-NC) at a multiplicity of infection (MOI) of 10 in the Opti-MEM medium. The culture medium was replaced with DMEM containing 10% serum 12 h after, and cells were continually cultured in incubator at 37 °C with 5% CO2. To establish ATG4C stable knockdown cell lines, infected cells were treated with puromycin (2 μg/ml for U87-MG, 1 μg/ml for T98G) for 7 days. The knockdown efficiency was evaluated by Western blot analysis.
Total RNA extraction and real-time qPCR
Total RNA was extracted from tissues or cells using RNAiso Plus reagent (Takara, China) and then reverse-transcribed to complementary DNA (cDNA) by PrimeScript RT Master Mix (Takara, China) according to the manufacturer’s instruction. Real-time PCR was performed with LightCycler 480 type II instrument (Roche, Switzerland). PCR were conducted using the following cycling parameters: pre-denaturation at 95 °C for 30 s, followed by 45 cycles of two step: 95 °C for 5 s and 60 °C for 20 s. GAPDH was selected to normalize genes expression levels using the ΔΔCt method. Primers used in this study were shown as Additional file 5: Table S1 (Ribobio, China).
Western blot analysis
The whole-cell lysates were produced by RIPA buffer containing PMSF. The protein concentrations were quantified using BCA Protein Assay kit (Beyotime, China). Equal amounts of proteins were added to SDS-PAGE and then transferred onto PVDF membrane. Blots were blocked in TBST buffer containing 5% (m/v) nonfat milk at room temperature for 2 h and then incubated at 4 °C overnight with primary antibodies as follows: GAPDH (Protein Tech), ATG4C (Abcam), LC3 (Cell Signaling Technology), P62 (Cell Signaling Technology), p21 (Ab clonal), p53 (AB clonal), Cyclin E (Cell Signaling Technology), Bcl-2 (Protein Tech), BAX (Protein Tech) and PARP (Protein Tech). After washing with TBST buffer, the membranes were probed with secondary antibodies (Invitrogen, USA) at room temperature for 1 h. Immunoreactive binding was detected with BeyoECL star kit (Beyotime, China) using Bio-Rad ChemiDoc XRS imaging system (Bio-Rad, USA).
Cell proliferation and viability assay
Cell proliferation and viability were detected by MTS. Briefly, cells were seeded in 96-well at a density of 1000 cells/well. After culturing for indicated time, the medium was replaced with 100 μL DMEM containing 10 μL MTS (Promega, CA, USA) and incubated at 37 °C for 1 h. Then, the optical density (OD) values were detected at 490 nm using multi-well spectrophotometer. The survival rate was calculated as AT490/CT490 × 100% (AT490 = Absorbance value of the experimental group at 490 nm; CT490 = Absorbance value of the control group at 490 nm). IC50 values were derived by GraphPad Prism 5 software through plotting the survival rates on a logarithmic curve.
Colony forming assay
After transfection, cells were dissociated and seeded in 3.5 cm dishes at a destiny of 1.0 × 103 cells/well and then cultured for 14 days. The cells were fixed with 4% (v/v) formaldehyde for 15 min, then stained with 0.1% (w/v) crystal violet for 10 min. After washing with PBS for three times, the number of colonies was counted.
Cell cycle analysis
Briefly, cells were harvested and fixed overnight in a freezer at − 20 °C with precooled 70% ethanol, then centrifuged at 1000 g for 5 min. The pellet was resuspended with 100 μL PBS containing 2 μL of RNase A (10 mg/mL), then incubated at 37 °C for 30 min. After that, 400 uL of PBS including 20 uL of PI (1 mg/mL) and 0.4 μL of Triton-X were added and incubated at 37 °C for 30 min. The cell cycle distribution was analyzed flow cytometer (Beckman Coulter, USA).
Cell apoptosis assay
Cells apoptosis was evaluated by Annevin V-FITC/PI and Hoechst 33342 staining. For Annevin V-FITC/PI staining, cells were dissociated by trypsin without EDTA and centrifuged at 1000 g for 5 min. The pellet was resuspended with staining solution containing 5 μL Annexin V-FITC and 10 μL PI, then incubated at room temperature for 20 min in dark. The apoptosis was analyzed by flow cytometer following manufacture’s instruction (Beyotime, China). For Hoechst 33342 staining, cells were fixed by 4% (v/v) formaldehyde for 10 min and washed with PBS, then staining buffer with Hoechst 33342 were added. The apoptotic cells were observed under fluorescence microscope. For Caspase-3/6/9 activity assay, cells were harvested and centrifuged at 1000 g for 5 min, and then incubated with lysis buffer for 15 min on ice. The lysate was centrifuged at 12000 g for 15 min. Then the supernatant was detected for the caspase-3/6/9 activity by multi-well spectrophotometer, according to the manufacture’s instruction (Beyotime, China).
Determination of ROS in cultured cells
After transfection, T98G cells were harvested and centrifuged at 1000 g for 5 min. The pellet was resuspended with DMEM containing DCFH-DA (10 μM) and incubated for 20 min. After that, the cell suspension was centrifuged at 1000 g for 5 min and resuspended with 400 μL PBS, then analyzed by flow cytometer.
Nude mice xenograft study
This study was performed in compliance with the guidelines of the Ethics Committee of Institutional Animal Care and Use in Central South University. The BALB/C male nude mice were obtained from Hunan SLAC Co., Ltd. The mice were housed in a specific pathogen-free facility at Department of Laboratory, Central South University. The ATG4C stable knockdown U87-MG cells were harvested and resuspended in PBS containing Matrigel matrix (BD, USA) at a density of 1 × 108 cells/ml. Then mice were injected subcutaneously with 100 μL of cells suspension (1 × 107 cells/mouse). Tumor volume and mice weight were measured after implanted for indicated time. Tumor volume (mm3) = (L × W2)/2, where L and W (L > W) are the tumor’s length and width. At the end point, tumor tissues were weighted and fixed in paraffin for further analysis.
Immunohistochemistry (IHC)
IHC was performed to determine the expression level of Ki67, LC3 and ATG4C in tumor tissue implanted in mice. Briefly, the formalin-fixed, paraffin-embedded samples were cut into 4 μm slices. Then, tissues sections were baked, dewaxed, hydrated and blocked. Afterwards, the sections were incubated with primary antibodies against Ki-67, LC3B and ATG4C. The following day, sections were incubated with secondary antibody at room temperature. After visualizing, sections were photographed using microscope.
Statistical analysis
All statistical analysis was performed using SPSS software (version 16.0). Data were presented as the mean ± standard deviation. Statistical analysis between two groups was performed using t-test. One-way ANOVA with Tamhane post-hoc was used for comparisons of different groups. Comparisons of survival rate were analyzed using Kaplan-Meier method and log-rank test. Multivariate Cox proportional hazard models were used to analyze the effect of clinical variables on patients’ overall survival. P-value of < 0.05 was considered as statistically significant in this study.
Results
Higher ATG4C mRNA expression predicted poor prognosis for glioma patients
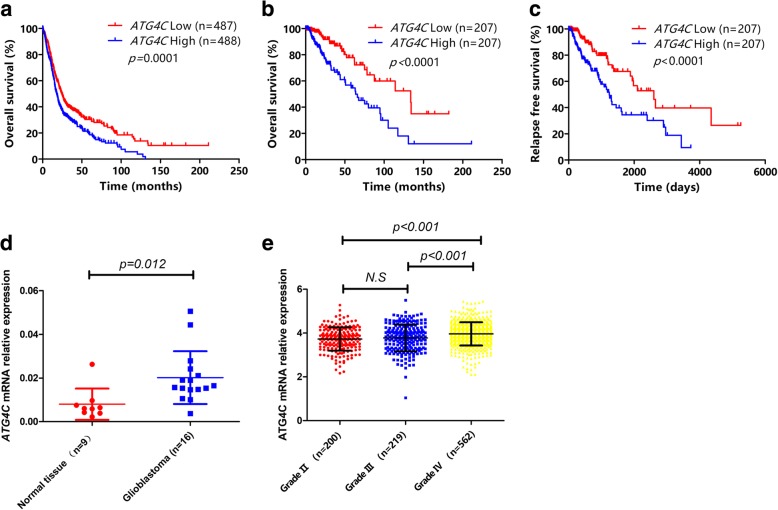
In order to investigate the role of autophagy-related genes (ATGs) in prognosis of glioma patients, we developed an analysis flowchart (Additional file 1: Figure S1). The baseline clinical characteristics of glioma patients were shown in Table 1. We observed that the mRNA expression of 13 ATGs (ATG3, ATG4A, ATG4C, ATG5, ATG7, ATG9B, ATG10, ATG12, ATG16L1, ATG2A, ATG2B, ATG9A and ATG16L2) were closely associated with the OS of glioma patients (Fig. 1a and Additional file 2: Figure S2). To rule out the potential influence of WHO grade, MGMT promoter methylation, IDH mutation and other clinical variables on prognosis of patients, Cox proportional hazards regression was further performed. The results showed that mRNA expression of ATG3 (HR = 0.63, 95% CI: 0.40–0.99, p = 0.05), ATG4C (HR = 1.54, 95% CI: 1.16–2.01, p = 3 × 10− 3) and ATG5 (HR = 0.61, 95% CI: 0.43–0.86, p = 5 × 10− 3) were independent OS prognostic factors in gliomas patients (Table 2). Given the significant influence of pathological grade on patient prognosis, these findings were further validated in low-grade glioma (LGG) patients and glioblastoma patients (GBM), respectively. We observed that only ATG4C mRNA expression was significantly associated with OS in LGG patients (Fig. 1b and Additional file 3: Figure S3). Moreover, the median relapse-free survival (RFS) time of LGG patients with low ATG4C mRNA expression was significantly longer than those with high expression (Fig. 1c). However, no significant association was observed between ATGs mRNA expression and OS in GBM patients (Additional file 3: Figure S3).
Fig. 1.
Kaplan-Meier survival analysis of glioma patients based on ATG4C mRNA expression levels. a Comparison of overall survival (OS) profiles between patients with ATG4C high and low expression levels in 975 glioma patients. b Comparison of overall survival (OS) profiles between patients with ATG4C high and low expression levels in 414 LGG patients. c Comparison of relapse free survival (RFS) time between patients with ATG4C high and low expression levels in 414 LGG patients. d Comparison of ATG4C mRNA expression between 16 glioblastoma tissues and 9 normal brain tissues. e Comparison of the mRNA expression of ATG4C in patients with different pathological grades of gliomas
Table 2.
Cox proportional hazards regression analysis of factors associated with overall survival of glioma patients
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| MGMT | 0.37 (0.30–0.46) | 6.81 × 10−19 | 0.98 (0.75–1.28) | 0.88 |
| IDH mutation vs wild | 0.13 (0.10–0.17) | 6.05 × 10−51 | 0.40 (0.26–0.63 | 6.40 × 10− 5 |
| Grade III vs Grade II | 3.20 (1.93–5.29) | 5.86 × 10−6 | 2.44 (1.44–4.12) | 9.34 × 10−4 |
| Grade IV vs Grade II | 14.28 (9.14–22.32) | 1.88 × 10− 31 | 5.02 (2.70–9.36) | 3.71 × 10−6 |
| Age | 1.06 (1.05–1.06) | 7.91 × 10−58 | 1.05 (1.03–1.06) | 3.85 × 10− 15 |
| Gender | 1.13 (0.94–1.35) | 0.19 | 1.51 (1.17–1.93) | 0.014 |
| ATG2A mRNA | 0.41 (0.35–0.48) | 2.95 × 10− 30 | 1.17 (0.79–1.73) | 0.43 |
| ATG2B mRNA | 0.47 (0.40–0.54) | 4.50 × 10−24 | 0.98 (0.75–1.27) | 0.86 |
| ATG3 mRNA | 1.42 (1.10–1.84) | 0.008 | 0.63 (0.40–0.99) | 0.05 |
| ATG4A mRNA | 2.37 (2.02–2.79) | 1.04 × 10− 25 | 0.90 (0.64–1.28) | 0.56 |
| ATG4C mRNA | 1.48 (1.27–1.73) | 9.91 × 10− 7 | 1.54 (1.16–2.01) | 3 × 10− 3 |
| ATG5 mRNA | 1.74 (1.43–2.13) | 4.32 × 10− 8 | 0.61 (0.43–0.86) | 5 × 10−3 |
| ATG7 mRNA | 2.32 (1.89–2.86) | 1.66 × 10− 15 | 1.36 (0.94–1.98) | 0.1 |
| ATG9A mRNA | 0.42 (0.34–0.51) | 7.05 × 10− 17 | 0.74 (0.49–1.13) | 0.16 |
| ATG9B mRNA | 1.44 (1.33–1.57) | 1.57 × 10− 18 | 0.97 (0.88–1.07) | 0.52 |
| ATG10 mRNA | 2.18 (1.86–2.55) | 1.08 × 10− 21 | 1.11 (0.82–1.41) | 0.50 |
| ATG12 mRNA | 2.05 (1.62–2.61) | 4.59 × 10− 9 | 0.998 (0.66–1.52) | 0.99 |
| ATG16L1 mRNA | 3.17 (2.53–3.97) | 8.53 × 10−24 | 1.01 (0.77–1.09) | 0.96 |
| ATG16L2 mRNA | 0.78 (0.70–0.88) | 7.22 × 10−5 | 0.92 (0.77–1.09) | 0.34 |
mRNA expression of ATG4C was increased with pathological grades in glioma patients
To make clear whether ATG4C was differentially expressed between glioblastoma and normal brain tissues, mRNA levels of ATG4C was detected in 16 glioblastoma and 9 normal brain tissues. As compared with normal brain tissues, the expression of ATG4C mRNA was significantly higher in glioblastoma tissues (Fig. 1d). We then compared the expression of ATG4C mRNA in different grades of gliomas using TCGA database. As shown in Fig. 1e, ATG4C mRNA expression was significantly increased with the increase in glioma grade. When the patients were divided into two groups according to ATG4C mRNA expression levels, we observed that patients with higher ATG4C mRNA expression were more likely to develop advanced grades glioma (Table 3).
Table 3.
Comparison of distribution of clinical factors between glioma patients with high and low ATG4C mRNA expression
| Clinical characteristic | NO. of patients | p value | |
|---|---|---|---|
| ATG4C low expression | ATG4C high expression | ||
| NO. of cases | 487 | 488 | |
| Age (years) | |||
| ≥ 55 | 297 (60.99%) | 249 (51.02%) | 0.002 |
| < 55 | 190 (39.01%) | 239 (48.98%) | |
| Gender | |||
| Male | 279 (57.29%) | 294 (60.25%) | 0.348 |
| Female | 208 (42.71%) | 194 (39.75%) | |
| WHO grade | |||
| II | 121 (24.85%) | 77 (15.78%) | 0.000 |
| III | 120 (24.64%) | 96 (19.67%) | |
| IV | 246 (50.51%) | 315 (64.55%) | |
| IDH status | |||
| Wild | 185 (37.99%) | 130 (26.64%) | 0.000 |
| Mutant | 242 (49.69%) | 298 (61.07%) | |
| Undefined | 60 (12.32%) | 60 (12.30%) | |
| MGMT status | |||
| Unmethylated | 110 (22.59%) | 224 (45.90%) | 0.000 |
| Methylated | 290 (59.55%) | 170 (34.84%) | |
| Undefined | 87 (17.86%) | 94 (19.26%) | |
Knockdown of ATG4C suppressed glioma cells proliferation
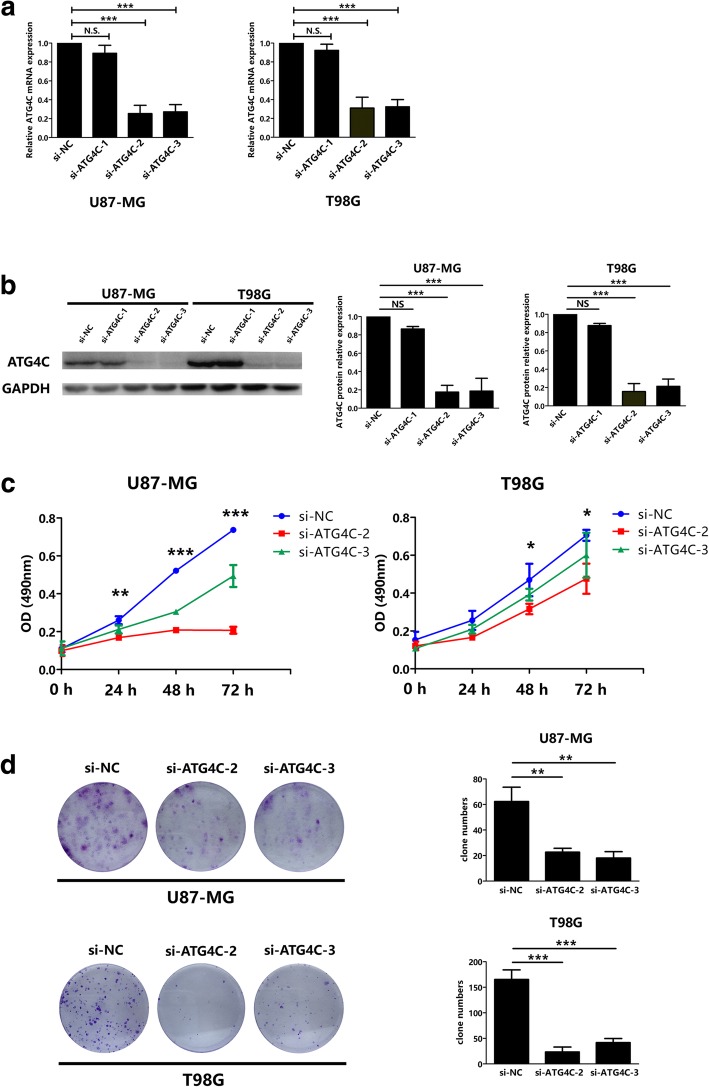
The above results indicated that ATG4C mRNA expression could predict the prognosis of glioma patients, implying that ATG4C might be involved in glioma progression and/or therapeutic response. The MTS assay and colony formation were performed to evaluate the role of ATG4C on glioma proliferation. The interference efficiency of siRNAs was validated at both mRNA and protein levels (Fig. 2a and b). Compared with control group, knockdown of ATG4C by si-RNA transfection significantly suppressed the proliferation of glioma cells indicated by proliferation curves (Fig. 2c). Results from colony-formation assay also showed that knockdown of ATG4C by si-RNA transfection reduced the number of colonies remarkably (Fig. 2d).
Fig. 2.
Knockdown of ATG4C inhibited the proliferation of glioma cells. a-b ATG4C siRNAs interference efficiency was tested at both mRNA and protein levels. c ATG4C knockdown by si-ATG4C transfection suppressed the proliferation of glioma cells in a time-dependent manner. d ATG4C knockdown by si-ATG4C transfection impaired clone formation ability of glioma cells. Left panel: representative crystal violet staining images; right panel: quantitative analysis of colony numbers. *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant
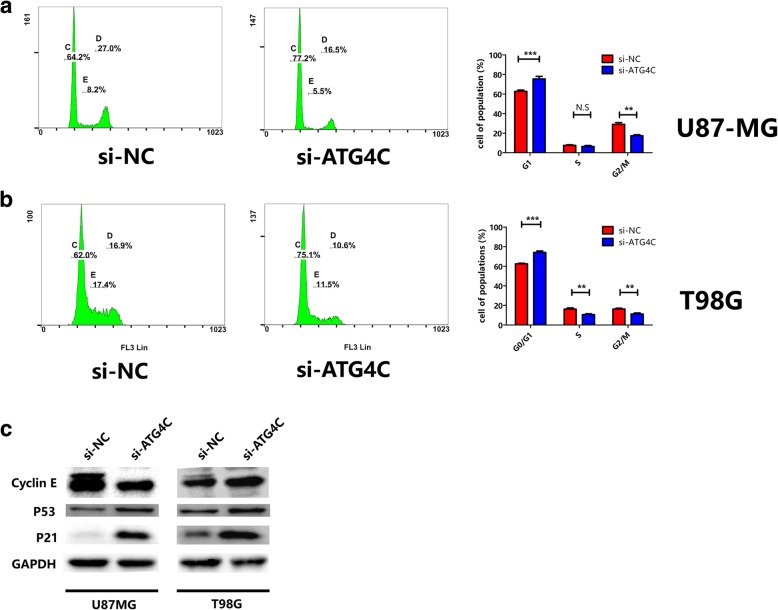
ATG4C ablation interfered with cell cycle and the expression of cell cycle related proteins in glioma cells
The impaired proliferation of glioma cell by si-ATG4C might be a result, at least in part, from unorganized cell cycle. For this reason, we analyzed the cell cycle distribution of glioma cells after ATG4C silencing. Both U87-MG and T98G cells transfected with ATG4C siRNA were accumulated in G1 phase (Fig. 3a and b). Meanwhile, knockdown of ATG4C increased the protein expression of p21 and p53 while decreased the expression of Cyclin E (Fig. 3c).
Fig. 3.
Influence of ATG4C knockdown on cell cycle and the expression of cell-cycle related proteins in glioma cells. a-b Changes in proportion of cells in G1 phase with ATG4C depletion. c Changes in protein expression of p21 and p53 by ATG4C depletion
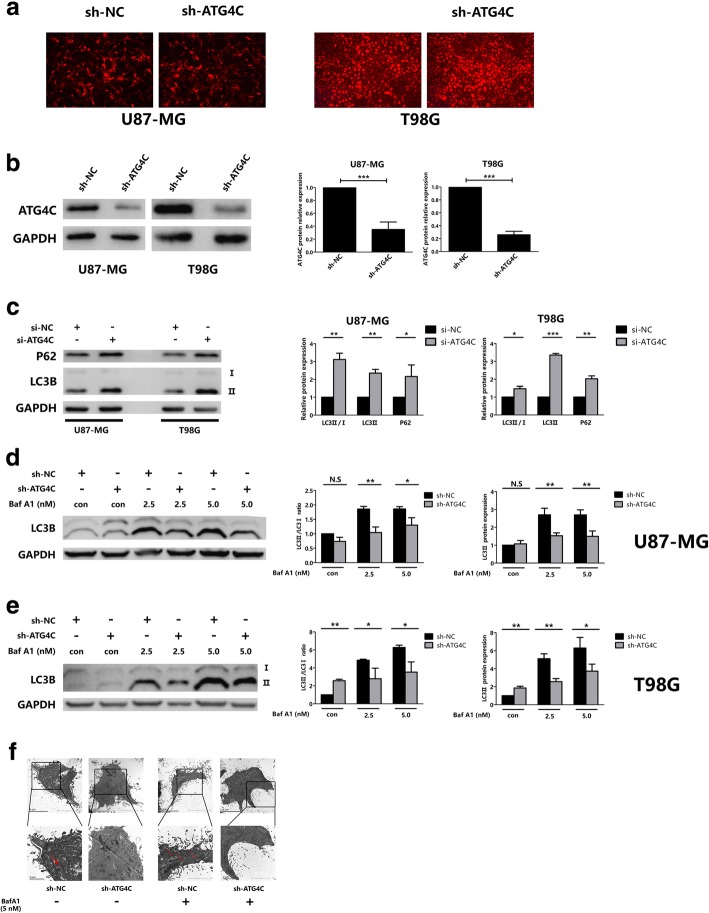
ATG4C knockdown suppressed autophagic flux in glioma cells
Previous studies have shown that ATG4C was implicated in the formation of autophagsome [31, 32]. However, the role of ATG4C in autophagy in glioma cells was unclear. ATG4C stable knockdown glioma cell lines were established (Fig. 4a and b). The LC3II/LC3-I ratio and expression of LC3-II and P62 are established indicators of autophagy. Intriguingly, we found that both of LC3-II and P62 protein levels were obviously increased in cells treated with si-ATG4C (Fig. 4c). In order to further confirm the role of ATG4C in autophagy, an autophagic flux assay was performed in ATG4C stable knockdown cells. Autophagic flux can be evaluated by LC3-II protein levels in the presence or absence of inhibitors of lysosomal degradation such as Bafilomycin A1 (BafA1). Our results showed that, in the presence of BafA1, knockdown of ATG4C reduced the protein expression of LC3-II remarkably, which suggested impaired autophagic flux (Fig. 4d and e). The analysis of transmission electron microscope (TEM) was performed to observe the influence of ATG4C on the formation of autophagy vacuoles in T98G cells. As shown in Fig. 4f, autophagic vacuoles was increased in T98G cells by BafA1 treatment. And in the presence of BafA1, knockdown of ATG4C reduced autophagic vacuoles.
Fig. 4.
Knockdown of ATG4C suppressed autophagy and autophagic flux. a, b ATG4C stable knockdown glioma cells were established by lentivirus infection, and the interference efficiency was detected by Western blot analysis. c Protein expression of LC3-II and P62 in glioma cells transfected with si-ATG4C. d, e Knockdown of ATG4C on the expression of LC3-II in the presence of BafA1. f Transmission electron microscopy observation of cellular ultrastructure tin T98G cells by ATG4C depletion. Arrows indicate autophagic vacuoles. *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant
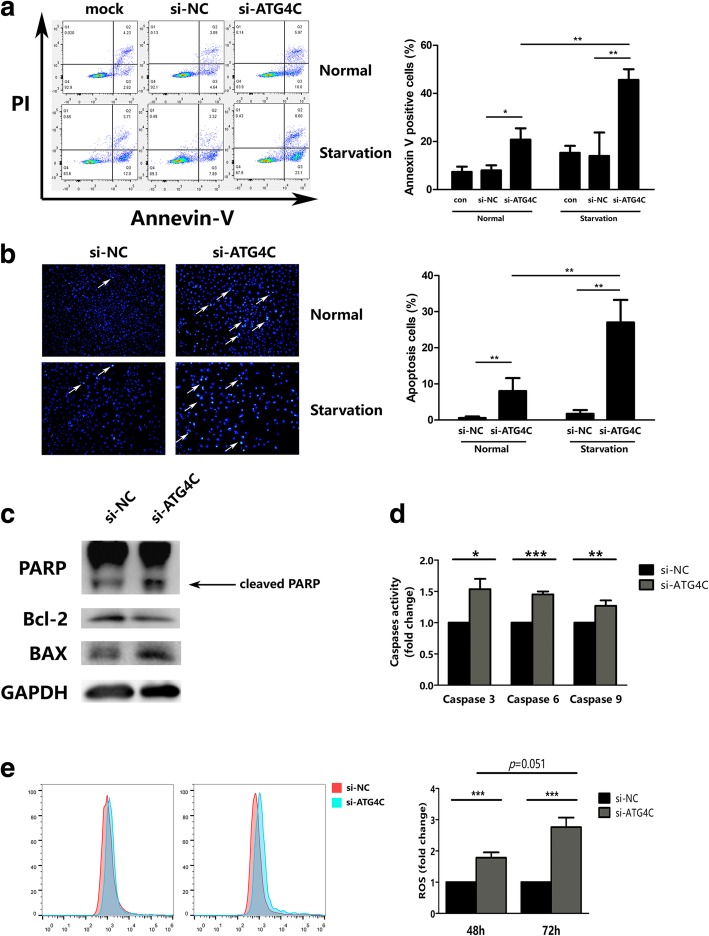
ATG4C depletion triggered apoptosis by increasing ROS
Beside immortal proliferation, decreased apoptosis is another important malignant biological behavior of gliomas. The effect of ATG4C on apoptosis was investigated in T98G using Annexin V-FITC/PI and Hoechst 33342 staining. As shown in Fig. 5a and b, knockdown of ATG4C significantly promoted apoptosis in T98G cells. And ATG4C depletion-induced apoptosis was further enhanced by starvation, a known factor for autophagy stimulation. Western blot analysis showed that ATG4C ablation remarkably increased the expression of pro-apoptosis proteins BAX and cleaved-PARP, while reduced the expression of the anti-apoptosis protein Bcl-2 (Fig. 5c). Consistently, ATG4C ablation significantly increased the activity of caspase-3/6/9 (Fig. 5d). Autophagy deficiency is reported to lead to an increase in ROS levels and then trigger apoptosis. For this reason, we further investigated the influence of ATG4C ablation on ROS production. As shown in Fig. 5e, ATG4C ablation increased the level of ROS remarkably in a time-dependent manner in T98G cells.
Fig. 5.
Knockdown of ATG4C promoted apoptosis and increased ROS accumulation in T98G cells. a-b The apoptosis in T98G cell transfected with si-ATG4C was detected by Annevin-V and Hoechst 33342 staining, respectively. c Changes in protein levels of cleaved-PARP, BAX and Bcl-2 expression in T98G cells transfected with ATG4C siRNAs. d Changes in Caspase-3/6/9 activity in T98G cells transfected with ATG4C siRNAs. e Changes in ROS levels in T98G cells transfected with ATG4C siRNAs. *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant
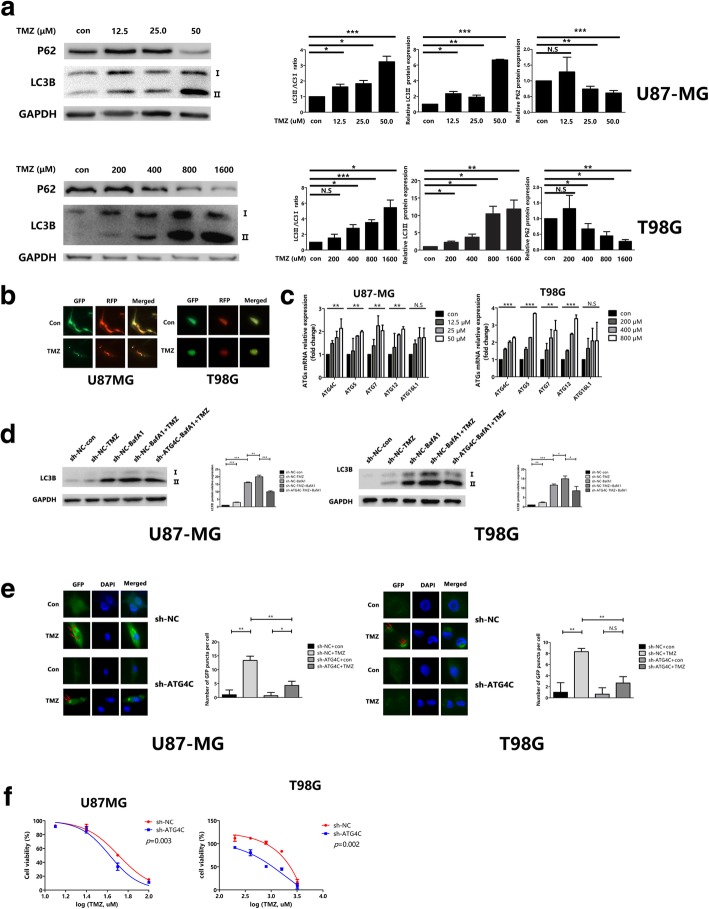
ATG4C depletion increased the sensitivity of glioma cells to TMZ
Previous studies have shown that TMZ-induced autophagy sustained the survival of tumor cells, which contributed to treatment resistance. Blocking autophagy with inhibitors can significantly increase TMZ cytotoxicity in glioma cells. However, it is unknown whether ATG4C is involved in the TMZ-activated autophagy. To validate the effect of TMZ in activating autophagy in glioma cells, U87-MG and T98G cells were treated with series concentrations of TMZ (12.5, 25, and 50 μM for U87-MG cells, and 200, 400, 800, 1600 μM for T98G cells). Western blot analysis showed that TMZ increased the LC3-II/LC3-I ratio and LC3-II protein levels in a concentration-dependent manner in both cells (Fig. 6a). TMZ treatment also decreased the protein expression of P62 significantly (Fig. 6a). The autophagy activating effect of TMZ in glioma cells was further confirmed by an increase in RFP-GFP-LC3 puncta (Fig. 6b). Additionally, RT-qPCR analysis showed a significant increase in ATG4C mRNA levels with increasing TMZ concentrations (Fig. 6c). Moreover, we observed that ATG4C ablation blocked the accumulation of LC3II in the presence of BafA1 in the glioma cells treated with TMZ (Fig. 6d). Consistently, ATG4C depletion significantly decreased the increase of GFP-LC3 dots induced by TMZ (Fig. 6e). All these results suggested that ATG4C was involved in TMZ-induced autophagy. As shown in Fig. 6f, ATG4C ablation also decreased the IC50 of TMZ in both U87MG and T98G cells obviously (Fig. 6f).
Fig. 6.
Knockdown of ATG4C enhanced TMZ cytotoxicity in glioma cells by suppressing autophagy. a Both LC3-II/LC3-I ratio and LC3-II expression were increased and P62 expression was decreased in glioma cells treated with TMZ. b The number of yellow puncta was elevated in glioma cells treated with TMZ. c The expression of ATGs mRNA were increased in glioma cells treated with TMZ. d Knockdown of ATG4C by sh-ATG4C decreased the expression of LC3-II in the presence of BafA1 in glioma cells treated with TMZ. e Knockdown of ATG4C by sh-ATG4C decreased the number of green puncta (red arrows) in glioma cells treated with TMZ. f Knockdown of ATG4C by sh-ATG4C increased TMZ cytotoxicity in glioma cells. *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant
ATG4C knockdown inhibited the growth of xenograft tumor
In order to investigate the effects of ATG4C on glioma growth in vivo, a xenograft nude mouse model was established. ATG4C stable knockdown U87-MG cells were implanted in nude mice and showed significantly slower growth than those in control group (Fig. 7a and b). Additionally, the tumors from mice injected with ATG4C depleted U87-MG cells was significantly smaller than those in control group (Fig. 7c). Moreover, ATG4C knockdown also remarkably impaired the proliferation of primary glioblastoma cells (Additionaly file 4: Figure S4). Immunohistochemical (IHC) staining was performed to detect the expression of Ki67, LC3 and ATG4C in the xenograft. As shown in Fig. 7d, the expression of Ki67, LC3 and ATG4C were decreased significantly in tumor tissues from sh-ATG4C group.
Fig. 7.
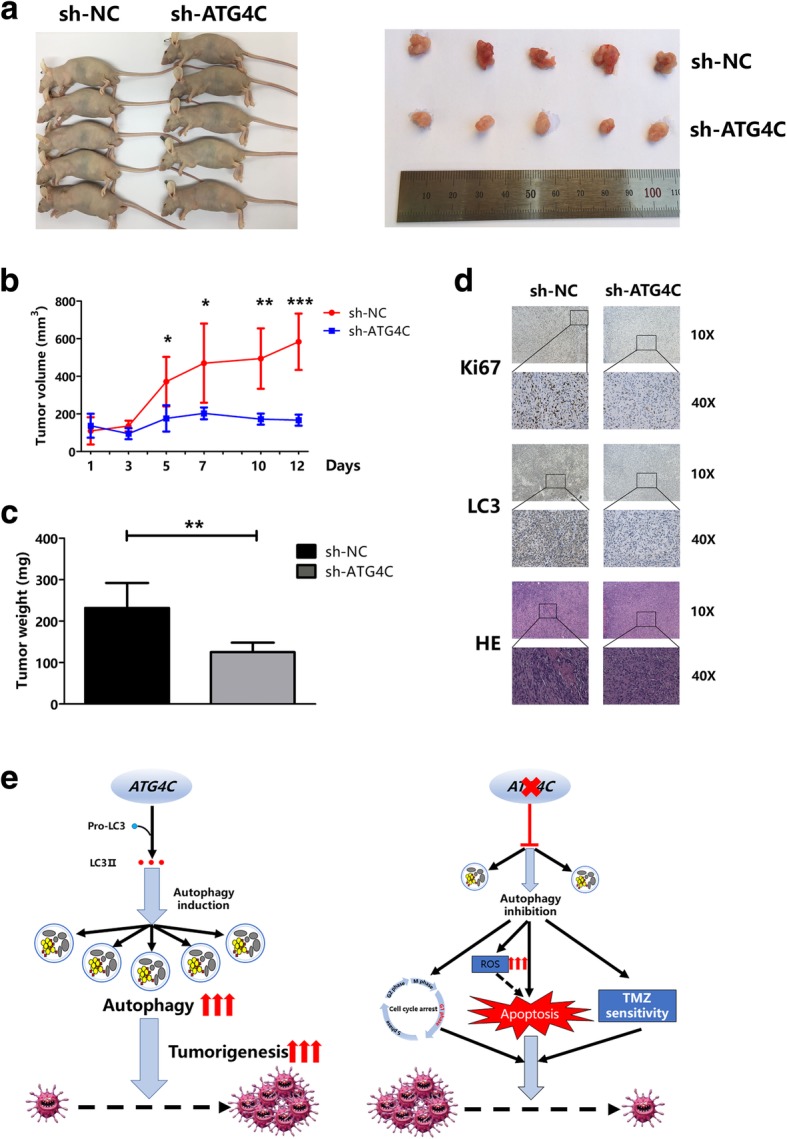
Knockdown of ATG4C by sh-ATG4C suppressed xenograft tumor and the graphic abstract. a-b The growth of glioma was restricted in nude mice by ATG4C knockdown. c Knockdown of ATG4C decreased the xenograft weights. d Depletion of ATG4C reduced the expression of Ki67, LC3 and ATG4C protein in xenograft. e Graph depicts the role of ATG4C in glioma progression and TMZ sensitivity
Discussion
In this study, we identified that the mRNA level of ATG4C was associated with worse prognosis in glioma patients. ATG4C levels was evidently elevated with the rising of glioma grade. Knockdown of ATG4C markedly suppressed the growth of glioma and promoted apoptosis in glioma cells, which was accompanied by increased ROS production. Additionally, cytotoxicity of TMZ was increased while the TMZ-induced autophagy was reduced in ATG4C depleted glioma cells.
More and more molecular biomarkers are identified and applied to improve the diagnosis and treatment of gliomas. However, the prognosis of the disease still remains gloomy [3, 33–37]. And also, it is of desperate need to find promising therapeutic targets for glioma. In this study, we observed that ATG4C was differently expressed between normal brain and glioblastoma tissues, and its expression was increased with the grade of glioma. And also, we observed that ATG4C level was an independent predictor for OS and RFS in LGG patients (Additional files 6 and 7: Tables S2 and S3). To make clear whether ATG4C acts through affecting cell growth in gliomas, we observed its role in proliferation of gliomas cells. We demonstrated that knockdown ATG4C remarkably suppressed the proliferation of glioma cell lines by inducing cell cycle arrest at G1 phase. It is obvious that cell cycle transitions in eukaryotic cells are regulated by cyclin-dependent kinases (CDKs)-cyclin complexes [38]. The CDK inhibitor P21 is an important regulator of cell growth in mammalian cells [39] as well as a universal cell-cycle inhibition controlled by P53 [40, 41]. In our current works, we observed that knockdown ATG4C induced a decrease in Cyclin E and an increase in P21 and P53 expression, which indicate obvious cell cycle arrest caused by ATG4C ablation. Additionally, we observed that knockdown ATG4C could restrain the proliferation of primarily cultured glioblastoma cells from patients (Additional file 3: Figure S3). A large body of evidence suggests that inhibition of the activity of other ATG4 family members, including ATG4A, ATG4B and ATG4D, could suppress tumor progression and enhance chemosensitivity or the efficacy of radiotherapy [42–49]. These findings consistently implied that the ATG4 family played an important role in tumorigenesis and cancer therapy. Development of ATG4C specific inhibitors may bring new promising strategy for glioma treatment.
The role of ATG4C in autophagy in glioma cells is not clear. Previous researches suggested that the miR-376 mediated ATG4C silencing could suppress autophagy in breast cancer cells and hepatocarcinoma cells [31, 32]. Recently, there was data implied that ATG4C had a limited role in autophagy [50]. In the current work, we observed that LC3-II expression was significantly reduced in the presence of BafA1 in sh-ATG4C cells, indicating impaired autophagic flux. Results from transmission electron microscope (TEM) analysis also observed a decrease in autophagic vacuoles by ATG4C silencing, in the presence of BafA1. However, we observed concomitantly increased protein expression of LC3-II and P62 in glioma cells transfected with si-ATG4C. It seems to be intriguing. According to the functions of ATG4 family members in LC3 processing, ATG4C is putative to cleavage of pro-LC3 and delipidation of LC 3 II from the membrane of the autophagsome through the cysteine peptidase activity [28, 31, 32]. Of course, the possible role of ATG4C in LC3-II degradation at late stage of autophagy could not be excluded in our study.
Autophagy is essential for the removal of damaged mitochondria, which are primary sources of intracellular ROS [16]. Autophagy deficiency can lead to an increase in ROS, which contribute to the DNA damage, genomic instability and then trigger apoptosis [51–54]. In our study, we found that ROS levels were increased remarkably in T98G cells transfected with si-ATG4C for 48 h, and a significant increase in apoptosis was observed followed by si-ATG4C transfection for 72 h. These findings implied that ATG4C ablation could suppress autophagy in glioma cells and then trigger apoptosis by increasing ROS levels. However, ROS scavengers should be used to test whether ATG4C depletion-induced apoptosis was ROS-dependent. Moreover, ectopic xenograft nude mice model was used to establish the influence of ATG4C on glioma growth in vivo. Consistent with the in vivo findings, knockdown ATG4C restrained the proliferation of glioma with significantly decreased tumor volumes and weights. In tumor tissues of mice xenograft, the expression of both Ki67 and LC3 proteins were decreased in the sh-ATG4C group. These results indicated that ATG4C ablation may impaired the proliferation of glioma through suppressing autophagy in vivo.
TMZ is the first-line chemotherapeutic drug for glioma. The drug undergoes spontaneous hydrolysis to 5-(3-methyltriazen-1-yl) imidazole-4-carboxamide (MITC) and then forms O6-methylguanine (O6-MeG) in neutral pH. O6-MeG lead to DNA mispairs during DNA replication, and that results in DNA double strand breaks through futile cycles of mismatch repair system [55, 56]. In the treatment of glioma, cytotoxicity chemotherapeutics drugs including TMZ can induce autophagy, which may promote the development of treatment resistance [23–25, 57]. In our study, we observed for the first time that ATG4C is involved in TMZ-induced autophagy, and depletion of ATG4C significantly increased the sensitivity of U87-MG and T98G cells to TMZ, which are thought to be TMZ sensitivity and resistance cells, respectively [58]. Selective and specific cytotoxicity to tumor cells is expected during glioma chemotherapy, which can increase the anti-tumor activity of TMZ with less toxic effects. However, these findings need to be further validated in vivo. Given that ATG4C expression was increased in tumor tissues of glioma, a therapy targeting ATG4C may provide a promising strategy for gliomas treatment.
Conclusion
Taken together, our results suggested that increased ATG4C expression was associated with worse prognosis in glioma patients. Knockdown of ATG4C suppressed glioma progression by inducing cell cycle arrest and promoting apoptosis of glioma cells possibly through increasing ROS production. Additionally, ATG4C ablation could promote TMZ cytotoxicity to glioma cells by inhibiting autophagy. Therefore, ATG4C might be potential target for the treatment of gliomas.
Additional files
Figure S1. Flow chart of procedures for screening of ATGs associated with outcomes of glioma patients. (TIF 90 kb)
Figure S2. Survival analysis of glioma patients based on expression of ATGs. (a-m) Kaplan-Meier analysis for OS in gliomas patients. (TIF 2897 kb)
Figure S3. Survival analysis of LGG based on expression of ATGs. (TIF 2249 kb)
Figure S4. Knockdown of ATG4C suppressed cell growth in glioblastoma primary cells. a The expression of GFAP in glioblastoma primary cells. b ATG4C knockdown suppressed the proliferation of primarily cultured glioblastoma cells. (TIF 952 kb)
Table S1. Sequences of primers used for RT-qPCR. (DOCX 14 kb)
Table S2. Cox proportional hazards regression analysis for OS in LGG patients. (DOCX 17 kb)
Table S3. Cox proportional hazards regression analysis for RFS in LGG patients. (DOCX 18 kb)
Acknowledgements
The RFP-GFP-LC3 plasmid were gift from Prof. Yan Cheng (Department of Pharmacology, School of Pharmaceutical Science, Central South University). We most cordially thank all members of the Department of Neurosurgery for assisting in samples collection of this works.
Abbreviations
- ATG4C
Autophagy Related 4C Cysteine Peptidase
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
Fetal Bovine Serum
- GBM
Glioblastoma
- IHC
Immunohistochemistry
- LGG
Low Grade Glioma
- MITC
5-(3-methyltriazen-1-yl) imidazole-4-carboxamide
- O6-MeG: ROS
O6-methylguanine; Reactive Oxygen Species
- OS
Overall Survival
- qPCR
Quantitative polymerase chain reaction
- RFS
Relapse-Fess Survival
- TCGA
The Cancer Genome Atlas
- TEM
Transmission electron microscope
- TMZ
Temozolomide
- WHO
World Health Organization
Authors’ contributions
Substantial contributions to conception and design, data acquisition and manuscript drafting: ZPW; Glioma tissues collection: YHC and JYW; TCGA data mining: QC; Transmission electron microscope analysis: JY; Drafting the article or critically revising it for important intellectual content: WJZ, HL, JY and XPC; Final approval of the version to be published: JX and XPC; Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved: HHZ and XPC. All authors read and approved to the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (NO. 81803582, NO.81703622); China Postdoctoral Science Foundation (NO. 2018 M633002); Hunan Provincial Natural Science Foundation of China (NO.2018JJ3838); Fundamental Research Funds for the Central Universities of Central South University (NO.2018zzts902).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The TCGA data set can download from: http://www.cbioportal.org/study.do?cancer_study_id=lgggbm_tcga_pub.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of the Xiangya Hospital of Central South University (No. 201803806). In this study, all procedures were performed in accordance with the Declaration of Helsinki. The animal experiment in this study was approved by the Department of Laboratory Animals of Central South University and performed in compliance with the guidelines of the Ethics Committee of Institutional Animal Care and Use in Central South University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhi-peng Wen, Email: wenzhipeng1989@sina.com.
Wen-jing Zeng, Email: wenjinganne@126.com.
Yan-hong Chen, Email: hihollychen@163.com.
He Li, Email: lh0530@csu.edu.cn.
Jie-ya Wang, Email: wangjy694@163.com.
Quan Cheng, Email: chengquan@csu.edu.cn.
Jing Yu, Email: xiaojingyu112@163.com.
Hong-hao Zhou, Email: hhzhou2003@126.com.
Zheng-zheng Liu, Phone: 86-731-84805380, Email: lzzsmile@aliyun.com.
Jian Xiao, Phone: 86-731-84805380, Email: admanoas@163.com.
Xiao-ping Chen, Phone: 86-731-84805380, Email: chenxiaoping@csu.edu.cn.
References
- 1.Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127(2):415–426. doi: 10.1172/JCI89587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Zeng WJ, Yang YL, Liu ZZ, Wen ZP, Chen YH, Hu XL, Cheng Q, Xiao J, Zhao J, Chen XP. Integrative analysis of DNA methylation and gene expression identify a three-gene signature for predicting prognosis in lower-grade gliomas. Cell Physiol Biochem. 2018;47(1):428–439. doi: 10.1159/000489954. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro-Oncology. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 8.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 10.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30(17):1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanegrungsunk D, Onchan W, Chattipakorn N, Chattipakorn SC. Current evidence of temozolomide and bevacizumab in treatment of gliomas. Neurol Res. 2015;37(2):167–183. doi: 10.1179/1743132814Y.0000000423. [DOI] [PubMed] [Google Scholar]
- 12.Wait SD, Prabhu RS, Burri SH, Atkins TG, Asher AL. Polymeric drug delivery for the treatment of glioblastoma. Neuro-Oncology. 2015;17(Suppl 2):ii9–ii23. doi: 10.1093/neuonc/nou360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 14.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. Front Oncol. 2012;2:171. doi: 10.3389/fonc.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T, Kim CK, Alvarez AA, Pangeni RP, Wan X, Song X, Shi T, Yang Y, Sastry N, Horbinski CM, et al. MST4 phosphorylation of ATG4B regulates Autophagic activity, Tumorigenicity, and Radioresistance in glioblastoma. Cancer Cell. 2017;32(6):840–855. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SW, Kim HK, Lee NH, Yi HY, Kim HS, Hong SH, Hong YK, Joe YA. The synergistic effect of combination temozolomide and chloroquine treatment is dependent on autophagy formation and p53 status in glioma cells. Cancer Lett. 2015;360(2):195–204. doi: 10.1016/j.canlet.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Lin CJ, Lee CC, Shih YL, Lin TY, Wang SH, Lin YF, Shih CM. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med. 2012;52(2):377–391. doi: 10.1016/j.freeradbiomed.2011.10.487. [DOI] [PubMed] [Google Scholar]
- 25.Zanotto-Filho A, Braganhol E, Klafke K, Figueiro F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini AM, Forcelini CM, et al. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett. 2015;358(2):220–231. doi: 10.1016/j.canlet.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 26.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Li J, Ouyang L, Liu B, Cheng Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett. 2016;373(1):19–26. doi: 10.1016/j.canlet.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286(9):7327–7338. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonelli M, Strappazzon F, Arisi I, Brandi R, D'Onofrio M, Sambucci M, Manic G, Vitale I, Barila D, Stagni V. ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget. 2017;8(13):21692–21709. doi: 10.18632/oncotarget.15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 32.Korkmaz G, Tekirdag KA, Ozturk DG, Kosar A, Sezerman OU, Gozuacik D. MIR376A is a regulator of starvation-induced autophagy. PLoS One. 2013;8(12):e82556. doi: 10.1371/journal.pone.0082556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J, Fang J, Cheng Z, Fan L, Hu W, Zhou F, Shen H. Overexpression of the Kininogen-1 inhibits proliferation and induces apoptosis of glioma cells. J Exp Clin Cancer Res. 2018;37(1):180. doi: 10.1186/s13046-018-0833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu T, Xie P, Gao YF, Huang MS, Li X, Zhang W, Zhou HH, Liu ZQ. Nucleolar and spindle-associated protein 1 is a tumor grade correlated prognosis marker for glioma patients. CNS Neurosci Ther. 2018;24(3):178–186. doi: 10.1111/cns.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang CH, Li XJ, Zhou YZ, Luo Y, Li C, Yuan XR. Expression and clinical significance of EGFL7 in malignant glioma. J Cancer Res Clin Oncol. 2010;136(11):1737–1743. doi: 10.1007/s00432-010-0832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang CM, Brat DJ. Genomic profiling of lower-grade gliomas uncovers cohesive disease groups: implications for diagnosis and treatment. Chin J Cancer. 2016;35:12. doi: 10.1186/s40880-015-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Lv QL, Huang YT, Zhang LH, Zhou HH. Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2 promotes progression of human glioma. J Exp Clin Cancer Res. 2017;36(1):105. doi: 10.1186/s13046-017-0573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9(11):910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 39.el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54(5):1169–1174. [PubMed] [Google Scholar]
- 40.El-Deiry WS. p21(WAF1) mediates cell-cycle inhibition, relevant to Cancer suppression and therapy. Cancer Res. 2016;76(18):5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 42.Akin D, Wang SK, Habibzadegah-Tari P, Law B, Ostrov D, Li M, Yin XM, Kim JS, Horenstein N, Dunn WA., Jr A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10(11):2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, Su X, Maganti A, Jiang C, Lu W, et al. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563(7732):569–573. doi: 10.1038/s41586-018-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 45.Tran E, Chow A, Goda T, Wong A, Blakely K, Rocha M, Taeb S, Hoang VC, Liu SK, Emmenegger U. Context-dependent role of ATG4B as target for autophagy inhibition in prostate cancer therapy. Biochem Biophys Res Commun. 2013;441(4):726–731. doi: 10.1016/j.bbrc.2013.10.117. [DOI] [PubMed] [Google Scholar]
- 46.Andaloussi AE, Habib S, Soylemes G, Laknaur A, Elhusseini H, Al-Hendy A, Ismail N. Defective expression of ATG4D abrogates autophagy and promotes growth in human uterine fibroids. Cell Death Discov. 2017;3:17041. doi: 10.1038/cddiscovery.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y, Hong L, Xu J, Zhong G, Gu Q, Gu Q, Guan Y, Zheng X, Dai Q, Luo X, et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy. 2018;20:1–17. [DOI] [PMC free article] [PubMed]
- 48.Liu PF, Tsai KL, Hsu CJ, Tsai WL, Cheng JS, Chang HW, Shiau CW, Goan YG, Tseng HH, Wu CH, et al. Drug repurposing screening identifies Tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes Cancer cells to chemotherapy. Theranostics. 2018;8(3):830–845. doi: 10.7150/thno.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Ni Z, Yan X, Dai X, Hu C, Zheng Y, He F, Lian J. Targeting the MIR34C-5p-ATG4B-autophagy axis enhances the sensitivity of cervical cancer cells to pirarubicin. Autophagy. 2016;12(7):1105–1117. doi: 10.1080/15548627.2016.1173798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrotis A, Pengo N, Burden JJ, Ketteler R. Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy. 2019;15(6):976–997. doi: 10.1080/15548627.2019.1569925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kongara S, Kravchuk O, Teplova I, Lozy F, Schulte J, Moore D, Barnard N, Neumann CA, White E, Karantza V. Autophagy regulates keratin 8 homeostasis in mammary epithelial cells and in breast tumors. Mol Cancer Res. 2010;8(6):873–884. doi: 10.1158/1541-7786.MCR-09-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106(8):2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanzawa T, Bedwell J, Kondo Y, Kondo S, Germano IM. Inhibition of DNA repair for sensitizing resistant glioma cells to temozolomide. J Neurosurg. 2003;99(6):1047–1052. doi: 10.3171/jns.2003.99.6.1047. [DOI] [PubMed] [Google Scholar]
- 57.Agnihotri S, Burrell K, Buczkowicz P, Remke M, Golbourn B, Chornenkyy Y, Gajadhar A, Fernandez NA, Clarke ID, Barszczyk MS, et al. ATM regulates 3-methylpurine-DNA glycosylase and promotes therapeutic resistance to alkylating agents. Cancer Discov. 2014;4(10):1198–1213. doi: 10.1158/2159-8290.CD-14-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo Dico A, Martelli C, Diceglie C, Lucignani G, Ottobrini L. Hypoxia-inducible factor-1alpha activity as a switch for glioblastoma responsiveness to Temozolomide. Front Oncol. 2018;8:249. doi: 10.3389/fonc.2018.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flow chart of procedures for screening of ATGs associated with outcomes of glioma patients. (TIF 90 kb)
Figure S2. Survival analysis of glioma patients based on expression of ATGs. (a-m) Kaplan-Meier analysis for OS in gliomas patients. (TIF 2897 kb)
Figure S3. Survival analysis of LGG based on expression of ATGs. (TIF 2249 kb)
Figure S4. Knockdown of ATG4C suppressed cell growth in glioblastoma primary cells. a The expression of GFAP in glioblastoma primary cells. b ATG4C knockdown suppressed the proliferation of primarily cultured glioblastoma cells. (TIF 952 kb)
Table S1. Sequences of primers used for RT-qPCR. (DOCX 14 kb)
Table S2. Cox proportional hazards regression analysis for OS in LGG patients. (DOCX 17 kb)
Table S3. Cox proportional hazards regression analysis for RFS in LGG patients. (DOCX 18 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The TCGA data set can download from: http://www.cbioportal.org/study.do?cancer_study_id=lgggbm_tcga_pub.