Abstract
Neurodevelopmental disorders (NDDs) represent a growing medical challenge in modern societies. Ever-increasing sophisticated diagnostic tools have been continuously revealing a remarkably complex architecture that embraces genetic mutations of distinct types (chromosomal rearrangements, copy number variants, small indels, and nucleotide substitutions) with distinct frequencies in the population (common, rare, de novo). Such a network of interacting players creates difficulties in establishing rigorous genotype-phenotype correlations. Furthermore, individual lifestyles may also contribute to the severity of the symptoms fueling a large spectrum of gene-environment interactions that have a key role on the relationships between genotypes and phenotypes.
Herein, a review of the genetic discoveries related to NDDs is presented with the aim to provide useful general information for the medical community.
Keywords: Neurodevelopmental disorders, Brain-related genes, Deleterious mutations, de novo mutations, Polymorphisms, Risk alleles, Gene interaction
Introduction
Neurodevelopment is the biological process resulting in the development and maturation of the nervous system. In humans, the process starts at the third week of embryonic growth with the formation of the neural tube [1–5]. From the ninth week onward, the brain orderly maturates and acquires its typical structure, under a tightly orchestrated chain of events that includes abundant cell proliferation, migration, and differentiation [1, 4, 5]. Any disruption to such orderly and complex chain of events may lead to dysfunctional brain development, and consequently to a neurodevelopmental phenotype. Under the designation neurodevelopmental disorders (NDDs) falls a group of complex and heterogeneous disorders showing symptoms associated to abnormal brain development that may give rise to impaired cognition, communication, adaptive behavior, and psychomotor skills [6–8]. NDDs include, for example, autism spectrum disorder, intellectual disability, attention deficit hyperactivity disorder, schizophrenia, and bipolar disorder [7, 9, 10]. The prevalence of these disorders constitutes a serious health problem in modern days. Previous reviews in distinct populations indicated a median global estimate of 62/10,000 for autism [11], 10.37/1000 for intellectual disability [12], and a median lifetime prevalence of 4/1000 for schizophrenia [13].
Multiple causes have been associated with NDDs, including genetic, environmental, infectious, and traumatic, among others, which in general do not operate alone but instead interacting between each other [6]. Importantly, the co-occurrence of distinct NDD entities has been often reported in the literature (e.g., [14]) suggesting the existence of shared underlying biological/cellular mechanisms [15, 16].
This review intends to focus on the molecular mechanisms associated with the most common neurodevelopmental illnesses, for which the precise etiology remains still largely unknown, but yet the genetic component has been increasingly deciphered with the massive sequencing of genomes of affected individuals.
Gene/variant discovery by genome/exome screenings
Although the genetic etiology of NDDs is far from being completely known, significant advances have been made in the last years, achieved hand-in-hand with progresses in ascertaining specific biological pathways underlying the molecular mechanisms of these illnesses. The current mutational spectrum of NDDs includes many hundreds of genes related to neurodevelopmental pathways such as those associated with chromatin remodeling, synaptic function, and transcriptional regulation [17–19]. There is convincing evidence for the huge genetic heterogeneity not only within but also between and across different NDDs, once it is documented a considerable overlap of genes involved in more than one NDD, and the number of known causative genes continues to increase.
Whole exome sequencing (WES) has revealed to be among the most useful approaches in the identification of novel causal mutations [20–32] in particular WES-Trio (proband and parents) studies due to be based on the comparison of the genotypes of an affected child and their parents, allowing thus the identification both of de novo mutations as inherited risk variants with variable penetrance. The success of the WES approach was clearly demonstrated in a recent study based in consanguineous families with NDDs, in which 14 new candidate genes not previously associated with NDD disorders were identified (GRM7, STX1A, CCAR2, EEF1D, GALNT2, SLC44A1, LRRIQ3, AMZ2, CLMN, SEC23IP, INIP, NARG2, FAM234B, and TRAP1) all in patients who were homozygous for truncating mutations in each of the genes [31]. Importantly, the same study allowed the identification of a de novo dominant truncating mutation at the PARD6A (p.Arg312Term), a gene never yet associated with any human disease but whose mouse homolog had been demonstrated to control glial-guided neuronal migration [33]. Although future studies still need to address whether PARD6A plays a similar functional role present in humans, this illustrates the importance of WES in revealing new candidate genes that may have a critical role in the neurodevelopment.
Intronic mutations can also be identified through WES. In 2017, Prchalova et al. [34] reported on an adult female with severe intellectual disability, epilepsy, and autistic features among other symptoms in whom the WES analysis led to the detection of an intronic mutation in the SYNGAP1 gene that was experimentally demonstrated to interfere with mRNA splicing. SYNGAP1 encodes the Ras/Rap GTP-activating protein, which has a critical role in synaptic function [35, 36] and has been associated with NDDs [37].
Along with WES, whole genome sequencing (WGS) is further revealing the role of non-coding mutations in the development of NDD phenotypes, adding an extra dimension to the already complex etiology of these disorders [38–41]. Very recently, Short et al. [41] estimated that pathogenic de novo variants in fetal brain regulatory elements account for about 1–3% of exome-negative NDD probands. Therefore, WGS should be considered whenever exome analyses do not provide evidence regarding putative causative mutations in NDD phenotypes.
Polymorphic variants and risk assessment
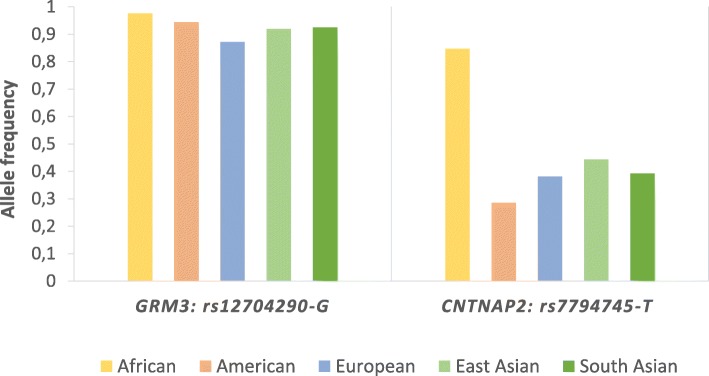
It is widely acknowledged that common genetic variations play an important role in the majority of complex disorders; actually, both rare and common alleles can contribute towards disease susceptibility [42]. Usually, variants with high frequency in the general population confer low relative risk [43, 44] while rare alleles highly penetrant may confer high risk [44]. Similarly to what is commonly found in other complex genetic disorders, the risk of developing NDDs seems to be highly influenced by the combined effect of common variants [45]. Up to now, thousands of common low-risk genetic variants that collectively can contribute to NDD susceptibility have been described [46]. Although the specific common risk alleles may differ between distinct NDDs, given their overall relevance here, we selected two single nucleotide polymorphisms (SNPs), highly polymorphic and showing replicated evidence of being associated with NDDs [47–51] to dissect their patterns of population distribution. In Fig. 1 is plotted the frequency of the assumed risk allele at each SNP across five major human populations.
Fig. 1.
Risk allele frequency in five populations for two single nucleotide polymorphisms (SNPs) found to be associated to increased risk of neurodevelopmental disorders [47–51]. Data extracted from 1000 Genomes Project Phase 3 [52]
The rs12704290 is an intronic variant located at GRM3, the gene that encodes the glutamate metabotropic receptor 3 involved in the glutamatergic neurotransmission. At this position, the assumed risk allele is rs12704290-G, which has been associated with a significant increased risk to schizophrenia [48, 50]. This allele is highly frequent across the five major human populations (Fig. 1), reaching the highest frequency in Africans (0.976) whereas the lowest is typically observed in Europeans (0.872).
The other common variant, rs7794745, is localized in the CNTNAP2 gene, which encodes a neurexin family protein involved in cell-cell adhesion [53]. The allele rs7794745-T was previously associated with an increased risk of developing autism spectrum disorder [47, 49, 51] and is highly frequent in all human populations (Fig. 1). The presence of risk alleles showing high frequencies in different human populations led to the question on whether they were ancestral or derived alleles. To find the answer, we investigated which allele was present in the homologous positions in the available orthologous primate sequences using sequences available at the Ensembl project [54]. Interestingly, the two risk alleles (GRM3 rs12704290-G and CNTNAP2 rs7794745-T) were the ancestral configurations, a finding that likely explains the worldwide high frequencies both reach. According to di Rienzo and Hudson [55], cases in which the ancestral alleles contribute to increase risk to common diseases or disease-related phenotypes, whereas the derived alleles are protective, may have an evolutionary explanation whereby the ancestral alleles mainly reflect ancient adaptations of ancient human populations, but due to the environmental and lifestyle changes suffered in modern populations, such ancestral alleles become now variants that increase the risk of common diseases.
Variants in chromatin-modifying/remodeling genes
The synaptic function may be influenced by many factors, among which are changes in chromatin dynamics caused by the disruption of a number of highly conserved genes [18]. Accordingly, chromatin-remodeling genes have been frequently reported in gene ontology analyses of data retrieved from WGS involving complex NDDs. For instance, CHD2, CHD7, and CHD8, three genes encoding chromodomain helicase DNA-binding (CHD) proteins that modulate chromatin structure, regulate gene expression, and play several other important roles, were previously linked to neurodevelopmental disorders such as intellectual disability [56]. Very recently, Kikawwa et al. [57] discussed the role played by the product of PAX6 gene—Pax6, a chromatin modulator, in autism, reinforcing the importance of chromatin alterations in NDD genes.
Clinical relevance of de novo mutations
De novo mutations are non-inherited sporadic mutations that arise either in the germline or in early embryonic development. As so, they do not conform to some rules of Mendelian inheritance, rendering more difficult to validate the prediction of their functional effect. When de novo mutations are associated with a clinical phenotype in a person without family history of a given condition, they can contribute to sporadic cases of the disease, including NDDs [58]. The de novo mutational rate of the human genome is approximately 1–3 × 10− 8 per base per generation [19, 59, 60] being well known that this rate is influenced by several factors, among which is the parental age [61]. The number of de novo mutations associated with NDDs has increased due to the strong investment in large-scale genetic screenings (exonic or genomic) of patients, which facilitate the identification of all types of molecular lesions as copy number variants (CNVs), indels, and mutations that cause gene disruption (missense, frameshift, and loss of splice site) [8, 62–68]. These spontaneous mutations were often found in candidate protein-coding genes with a high degree of haploinsufficiency or in regulatory elements involved in alternative splicing, in transcriptional regulation (enhancer and promoter), and in conserved non-coding sequences [41]. For example, in autistic patients, several de novo mutations were independently identified in the autism-associated genes ADNP, ARID1B, CHD8, and SYNGAP1 [18, 69–71] revealing a likely deleterious effect. De novo mutations have also been identified in GATAD2B [72], SCN2A [73], and FBXO11 [74] genes associated to intellectual disability, and in PTPRG, TGM5, SLC39A13, BTK, and CDKN3 linked to schizophrenia [64].
Some mutations overlap distinct neurodevelopmental disorders [14, 75]. Accordingly, a WES-Trios study with schizophrenic patients conducted by McCarthy et al. [76] suggested a shared genetic etiology between schizophrenia, autism, and intellectual disability. Although the complete set of genes involved in NDD is far being from fully characterized, the recurrent identification of de novo mutations in a shared set of genes may allow further clarification and delineation of the molecular pathways that underlie NDDs. Also, experimental/functional validation of the identified de novo mutations is essential to separate disease-causing alleles from neutral variation.
Genetic interaction
Genetic interaction (or epistasis) between genes or within the same gene is a major determinant of genotype-phenotype correlations [77–83]. The net result of distinct combinations of variants can result in distinct severities of the disease. Epistatic interactions between alleles are known for some Mendelian diseases revealing the interplay between mutations and polymorphisms which result in distinct functional outcomes [84, 85]. In what concerns the genetically heterogeneous neurodevelopmental disorders, the impact of the interaction between distinct alleles within the same locus or between interacting loci seems now to be giving its first steps. Evidence is emerging on intermolecular epistasis in autism spectrum disorders [86] regarding intramolecular and intermolecular epistasis between variants in the SHANK2 family that were very recently documented [87]. This is in accordance with previous observations on the cumulative effect of disease-associated alleles in modulating neurodevelopmental phenotypes [88].
Variants in the sodium channel gene SCN2A have been often described in cohorts of patients with NDDs [89–91]. Among them is the common rs10174400-T allele, associated to impairment of cognitive ability in schizophrenic patients [92, 93] but with an unlikely effect in healthy individuals, which points towards a pathogenic effect that is conditionally dependent on the genetic background and, therefore, on the cumulative effect of distinct alleles as mentioned above. Extending these promising results to other neurodevelopmental disorders, it is expected that more cases of allelic interaction could highlight the etiology of these diseases, further explaining the genotype-phenotype correlation and the genetic overlap often observed [94].
Conclusions
Neurodevelopmental disorders are a public health challenge due to complexity and heterogeneity of the etiology in conjugation with the high prevalence attained. Several biological pathways are disrupted in neurodevelopmental disorders, mainly at genes involved in synaptogenesis, chromatin remodeling, cell proliferation, and differentiation. Many of these genes, expressed during brain embryonic development, are intolerant to haploinsufficiency. It is important to continue the collection of information provided by WES and WGS data and focus deeply on epistatic interactions between identified mutations and polymorphic variants. In a more ambitious perspective, epigenetics may reveal itself as a promising therapeutic approach in the near future, exploiting the promise of numerous epigenome-wide association studies that are addressing neurodevelopmental disorders. Finally, it cannot be devaluated the major role that gene-environment interactions play in the outcomes of the diseases, implying that much attention should be given in the future to implement measures able to promote NDD prevention.
Acknowledgements
Not applicable.
Abbreviations
- CHD
Chromodomain helicase deoxyribonucleic acid binding
- CNV
Copy number variant
- NDD
Neurodevelopmental disorder
- SNP
Single nucleotide polymorphism
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Authors’ contributions
ARC, MLM, LA, RS, and MJP contributed to writing the first draft of the paper. ARC generated the image. ARC, CS, and MLM analyzed the data. LA, MLM, MJP, and AA revised the paper. All authors read and approved the final manuscript.
Funding
This work was financed by FEDER - Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 - Operacional Programme for Competitiveness and Internationalization (POCI), Portugal 2020, and by Portuguese funds through FCT - Fundação para a Ciência e a Tecnologia, in the framework of the project POCI-01-0145-FEDER-007274 to i3S and UID/BIM/04501/2013 and UID/BIM/04501/2019 to iBiMED, as well as by national funds (OE), through FCT, in the scope of the framework contract foreseen in the numbers 4, 5, and 6 of the article 23, of the Decree-Law 57/2016, of August 29, changed by Law 57/2017, of July 19 to RMS, and by FCT research project POCI-01-0145-FEDER-29723. ARC and CS hold FCT PhD fellowships (SFRH/BD/141702/2018-ARC and SFRH/BD/137925/2018-CS). Funders had no role in the design, collection, analysis, interpretation of the data, and writing of the manuscript.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20(4):327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310(5749):805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- 3.Bishop KM, Rubenstein JL, O'Leary DD. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J Neurosci. 2002;22(17):7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stiles J. Brain development and the nature versus nurture debate. Prog Brain Res. 2011;189:3–22. doi: 10.1016/B978-0-444-53884-0.00015-4. [DOI] [PubMed] [Google Scholar]
- 5.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacol. 2010;35(1):147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12(4):406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullin AP, Gokhale A, Moreno-De-Luca A, Sanyal S, Waddington JL, Faundez V. Neurodevelopmental disorders: mechanisms and boundary definitions from genomes, interactomes and proteomes. Transl Psychiat. 2013;3:e329. doi: 10.1038/tp.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platzer K, Sticht H, Edwards SL, Allen W, Angione KM, Bonati MT, et al. De novo variants in MAPK8IP3 cause intellectual disability with variable brain anomalies. Am J of Hum Genet. 2019;104(2):203–212. doi: 10.1016/j.ajhg.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562(7726):268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owen MJ, O'Donovan MC. Schizophrenia and the neurodevelopmental continuum:evidence from genomics. World Psychiatry. 2017;16(3):227–235. doi: 10.1002/wps.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsabbagh M, Divan G, Koh Y-J, Kim YS, Kauchali S, Marcín C, Montiel-Nava C, Patel V, Paula CS, Wang C, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5(3):160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32(2):419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Welham J, Saha S, Chant D, McGrath J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30(1):67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 14.Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49(8):1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilinc M, Creson T, Rojas C, Aceti M, Ellegood J, Vaissiere T, et al. Species-conserved SYNGAP1 phenotypes associated with neurodevelopmental disorders. Mol Cell Neurosci. 2018;91:140–150. doi: 10.1016/j.mcn.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnell Jennifer C, Van Driesche SJ, Zhang C, Hung Ka Ying S, Mele A, Fraser Claire E, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Mubarak B, Abouelhoda M, Omar A, AlDhalaan H, Aldosari M, Nester M, et al. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: a trio study from Saudi families. Sci Rep. 2017;7(1):5679. doi: 10.1038/s41598-017-06033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carneiro TN, Krepischi AC, Costa SS, Tojal da Silva I, Vianna-Morgante AM, Valieris R, et al. Utility of trio-based exome sequencing in the elucidation of the genetic basis of isolated syndromic intellectual disability: illustrative cases. Appl Clin Genet. 2018;11:93–98. doi: 10.2147/TACG.S165799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du X, Gao X, Liu X, Shen L, Wang K, Fan Y, et al. Genetic diagnostic evaluation of trio-based whole exome sequencing among children with diagnosed or suspected autism spectrum disorder. Front Genet. 2018;9:594. doi: 10.3389/fgene.2018.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagnamenta AT, Lise S, Harrison V, Stewart H, Jayawant S, Quaghebeur G, et al. Exome sequencing can detect pathogenic mosaic mutations present at low allele frequencies. J Hum Genet. 2012;57(1):70–72. doi: 10.1038/jhg.2011.128. [DOI] [PubMed] [Google Scholar]
- 24.Giacopuzzi E, Gennarelli M, Minelli A, Gardella R, Valsecchi P, Traversa M, et al. Exome sequencing in schizophrenic patients with high levels of homozygosity identifies novel and extremely rare mutations in the GABA/glutamatergic pathways. PLoS One. 2017;12(8):e0182778. doi: 10.1371/journal.pone.0182778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guipponi M, Santoni FA, Setola V, Gehrig C, Rotharmel M, Cuenca M, et al. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9(11):e112745. doi: 10.1371/journal.pone.0112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka M, Matoba N, Sawada T, Kazuno AA, Ishiwata M, Fujii K, et al. Exome sequencing for bipolar disorder points to roles of de novo loss-of-function and protein-altering mutations. Mol Psychiatry. 2016;21:885. doi: 10.1038/mp.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maaser A, Forstner AJ, Strohmaier J, Hecker J, Ludwig KU, Sivalingam S, et al. Exome sequencing in large, multiplex bipolar disorder families from Cuba. PLoS One. 2018;13(10):e0205895. doi: 10.1371/journal.pone.0205895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43(6):585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiat. 2017;74(3):293–299. doi: 10.1001/jamapsychiatry.2016.3798. [DOI] [PubMed] [Google Scholar]
- 32.Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77(2):259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6α signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 34.Prchalova D, Havlovicova M, Sterbova K, Stranecky V, Hancarova M, Sedlacek Z. Analysis of 31-year-old patient with SYNGAP1 gene defect points to importance of variants in broader splice regions and reveals developmental trajectory of SYNGAP1-associated phenotype: case report. BMC Med Genet. 2017;18(1):62. doi: 10.1186/s12881-017-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clement James P, Aceti M, Creson Thomas K, Ozkan Emin D, Shi Y, Reish Nicholas J, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151(4):709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berryer MH, Chattopadhyaya B, Xing P, Riebe I, Bosoi C, Sanon N, et al. Decrease of SYNGAP1 in GABAergic cells impairs inhibitory synapse connectivity, synaptic inhibition and cognitive function. Nat Commun. 2016;7:13340. doi: 10.1038/ncomms13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berryer MH, Hamdan FF, Klitten LL, Moller RS, Carmant L, Schwartzentruber J, et al. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum Mutat. 2013;34(2):385–394. doi: 10.1002/humu.22248. [DOI] [PubMed] [Google Scholar]
- 38.Devanna P, Chen XS, Ho J, Gajewski D, Smith SD, Gialluisi A, et al. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol Psychiatry. 2017;23:1375. doi: 10.1038/mp.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams SM, An JY, Edson J, Watts M, Murigneux V. Whitehouse AJO, et al. Mol Psychiatry: An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder; 2018. [DOI] [PubMed]
- 40.Zeledón M, Eckart N, Taub M, Vernon H, Szymanksi M, Wang R, et al. Identification and functional studies of regulatory variants responsible for the association of NRG3 with a delusion phenotype in schizophrenia. Mol neuropsychiatry. 2015;1(1):36–46. doi: 10.1159/000371518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555(7698):611–616. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemminki K, Forsti A, Bermejo JL. The ‘common disease-common variant’ hypothesis and familial risks. PLoS One. 2008;3(6):e2504. doi: 10.1371/journal.pone.0002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21(21):4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henriksen MG, Nordgaard J, Jansson LB. Genetics of schizophrenia: overview of methods, findings and limitations. Front Hum Neurosci. 2017;11:322. doi: 10.3389/fnhum.2017.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorstman JA, Anney RJ, Derks EM, Gallagher L, Gill M, de Jonge MV, et al. No evidence that common genetic risk variation is shared between schizophrenia and autism. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(1):55–60. doi: 10.1002/ajmg.b.32121. [DOI] [PubMed] [Google Scholar]
- 47.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang M, Sun L, Liu X, Sun W, Ji M, Wang Z, et al. Evaluation of relationship between GRM3 polymorphisms and cognitive function in schizophrenia of Han Chinese. Psychiatry Res. 2015;229(3):1043–1046. doi: 10.1016/j.psychres.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Nascimento PP, Bossolani-Martins AL, Rosan DB, Mattos LC, Brandao-Mattos C, Fett-Conte AC. Single nucleotide polymorphisms in the CNTNAP2 gene in Brazilian patients with autistic spectrum disorder. Genet Mol Res. 2016;15(1):gmr.15017422. [DOI] [PubMed]
- 50.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zare S, Mashayekhi F, Bidabadi E. The association of CNTNAP2 rs7794745 gene polymorphism and autism in Iranian population. J Clin Neurosci. 2017;39:189–192. doi: 10.1016/j.jocn.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, et al. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24(4):1037–1047. doi: 10.1016/S0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 54.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2017;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21(11):596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Chenier S, Yoon G, Argiropoulos B, Lauzon J, Laframboise R, Ahn JW, et al. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord. 2014;6(1):9. doi: 10.1186/1866-1955-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kikkawa T, Casingal CR, Chun SH, Shinohara H, Hiraoka K, Osumi N. The role of Pax6 in brain development and its impact on pathogenesis of autism spectrum disorder. Brain Res. 2018;1705:95. doi: 10.1016/j.brainres.2018.02.041. [DOI] [PubMed] [Google Scholar]
- 58.Ku CS, Polychronakos C, Tan EK, Naidoo N, Pawitan Y, Roukos DH, et al. A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol Psychiatry. 2013;18(2):141–153. doi: 10.1038/mp.2012.58. [DOI] [PubMed] [Google Scholar]
- 59.Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87(3):316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conrad DF, Keebler JEM, DePristo MA, Lindsay SJ, Zhang Y, Casals F, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Girard SL, Bourassa CV, Lemieux Perreault L-P, Legault M-A, Barhdadi A, Ambalavanan A, et al. Paternal age explains a major portion of de novo germline mutation rate variability in healthy individuals. Plos One. 2016;11(10):e0164212. doi: 10.1371/journal.pone.0164212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamdan FF, Srour M, Capo-Chichi J-M, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10(10):e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2011;17:142. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kranz TM, Harroch S, Manor O, Lichtenberg P, Friedlander Y, Seandel M, et al. De novo mutations from sporadic schizophrenia cases highlight important signaling genes in an independent sample. Schizophr Res. 2015;166(1):119–124. doi: 10.1016/j.schres.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy D, Ronemus M, Yamrom B, Lee Y-H, Leotta A, Kendall J, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 66.Malhotra D, McCarthy S, Michaelson Jacob J, Vacic V, Burdick Katherine E, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72(6):951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 69.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs M-O, et al. De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol Psychiatry. 2011;69(9):898–901. doi: 10.1016/j.biopsych.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 71.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ueda K, Yanagi K, Kaname T, Okamoto N. A novel mutation in the GATAD2B gene associated with severe intellectual disability. Brain and Development. 2019;41(3):276–279. doi: 10.1016/j.braindev.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 73.Baasch A-L, Hüning I, Gilissen C, Klepper J, Veltman JA, Gillessen-Kaesbach G, et al. Exome sequencing identifies a de novo SCN2A mutation in a patient with intractable seizures, severe intellectual disability, optic atrophy, muscular hypotonia, and brain abnormalities. Epilepsia. 2014;55(4):e25–e29. doi: 10.1111/epi.12554. [DOI] [PubMed] [Google Scholar]
- 74.Fritzen D, Kuechler A, Grimmel M, Becker J, Peters S, Sturm M, et al. De novo FBXO11 mutations are associated with intellectual disability and behavioural anomalies. Hum Genet. 2018;137(5):401–411. doi: 10.1007/s00439-018-1892-1. [DOI] [PubMed] [Google Scholar]
- 75.Autism Spectrum Disorders Working Group of The Psychiatric Genomics C Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19(6):652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Azevedo L, Carneiro J, van Asch B, Moleirinho A, Pereira F, Amorim A. Epistatic interactions modulate the evolution of mammalian mitochondrial respiratory complex components. BMC Genomics. 2009;10(1):266. doi: 10.1186/1471-2164-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Azevedo L, Mort M, Costa AC, Silva RM, Quelhas D, Amorim A, et al. Improving the in silico assessment of pathogenicity for compensated variants. Eur J Hum Genet. 2016;25:2. doi: 10.1038/ejhg.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azevedo L, Suriano G, van Asch B, Harding RM, Amorim A. Epistatic interactions: how strong in disease and evolution? Trends Genet. 2006;22(11):581–585. doi: 10.1016/j.tig.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 80.Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27(8):323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Okser S, Pahikkala T, Aittokallio T. Genetic variants and their interactions in disease risk prediction - machine learning and network perspectives. BioData Min. 2013;6(1):5–5. doi: 10.1186/1756-0381-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suriano G, Azevedo L, Novais M, Boscolo B, Seruca R, Amorim A, et al. In vitro demonstration of intra-locus compensation using the ornithine transcarbamylase protein as model. Hum Mol Genet. 2007;16(18):2209–2214. doi: 10.1093/hmg/ddm172. [DOI] [PubMed] [Google Scholar]
- 83.Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85(3):309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lage MD, Pittman AMC, Roncador A, Cellini B, Tucker CL. Allele-specific characterization of alanine: glyoxylate aminotransferase variants associated with primary yperoxaluria. PLoS One. 2014;9(4):e94338. doi: 10.1371/journal.pone.0094338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longley MJ, Chan SSL, Copeland WC. Modulation of the W748S mutation in DNA polymerase γ by the E1143G polymorphismin mitochondrial disorders. Hum Mol Genet. 2006;15(23):3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitra I, Lavillaureix A, Yeh E, Traglia M, Tsang K, Bearden CE, et al. Reverse pathway genetic approach identifies epistasis in autism spectrum disorders. PLoS Genet. 2017;13(1):e1006516. doi: 10.1371/journal.pgen.1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiu S, Li Y, Bai Y, Shi J, Cui H, Gu Y, et al. SHANK1 polymorphisms and SNP–SNP interactions among SHANK family: a possible cue for recognition to autism spectrum disorder in infant age. Autism Res. 2019;12(3):375–383. doi: 10.1002/aur.2065. [DOI] [PubMed] [Google Scholar]
- 88.Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med. 2018;21:816. doi: 10.1038/s41436-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Imbrici P, Conte Camerino D, Tricarico D. Major channels involved in neuropsychiatric disorders and therapeutic perspectives. Front Genet. 2013;4:76. doi: 10.3389/fgene.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weiss LA, Escayg A, Kearney JA, Trudeau M, MacDonald BT, Mori M, et al. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry. 2003;8:186. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- 91.Yokoi T, Enomoto Y, Tsurusaki Y, Naruto T, Kurosawa K. Nonsyndromic intellectual disability with novel heterozygous SCN2A mutation and epilepsy. Hum Genome Var. 2018;5(1):20. doi: 10.1038/s41439-018-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dickinson D, Straub RE, Trampush JW, Gao Y, Feng N, Xie B, Shin JH, Lim HK, Ursini G, Bigos KL, et al. Differential effects of common variants in SCN2A on general cognitive ability, brain physiology, and messenger RNA expression in schizophrenia cases and control individuals. JAMA Psychiatry. 2014;71(6):647–656. doi: 10.1001/jamapsychiatry.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scult MA, Trampush JW, Zheng F, Conley ED, Lencz T, Malhotra AK, Dickinson D, Weinberger DR, Hariri AR. A common polymorphism in SCN2A predicts general cognitive ability through effects on PFC physiology. J Cogn Neurosci. 2015;27(9):1766–1774. doi: 10.1162/jocn_a_00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009;1(10):102. doi: 10.1186/gm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.