Abstract
Generation of glial cell diversity in the developing spinal cord is known to depend on spatio‐temporal patterning programs. In particular, expression of the transcription factor Olig2 in neural progenitors of the pMN domain is recognized as critical to their fate choice decision to form oligodendrocyte precursor cells (OPCs) instead of astrocyte precursors (APs). However, generating some confusion, lineage‐tracing studies of Olig2 progenitors in the spinal cord provided evidence that these progenitors also generate some astrocytes. Here, we addressed the role of the heparan sulfate‐editing enzyme Sulf2 in the control of gliogenesis and found an unanticipated function for this enzyme. At initiation of gliogenesis in mouse, Sulf2 is expressed in ventral neural progenitors of the embryonic spinal cord, including in Olig2‐expressing cells of the pMN domain. We found that sulf2 deletion, while not affecting OPC production, impairs generation of a previously unknown Olig2‐expressing pMN‐derived cell subtype that, in contrast to OPCs, does not upregulate Sox10, PDGFRα or Olig1. Instead, these cells activate expression of AP identity genes, including aldh1L1 and fgfr3 and, of note, retain Olig2 expression as they populate the spinal parenchyma at embryonic stages but also as they differentiate into mature astrocytes at postnatal stages. Thus, our study, by revealing the existence of Olig2‐expressing APs that segregate early from pMN cells under the influence of Sulf2, supports the existence of a common source of APs and OPCs in the ventral spinal cord and highlights divergent regulatory mechanism for the development of pMN‐derived OPCs and APs.
Keywords: astrocyte, gliogenesis, olig2, oligodendrocyte, spinal cord, sulfatase 2
1. INTRODUCTION
Glial cells are recognized for serving critical functions in the central nervous system (CNS). Among them, astrocytes and oligodendrocytes, namely glial cells of neural origin, populate all regions of the mature CNS. Astrocytes are active regulators of synaptogenesis and neurotransmission and are also critical for formation of the blood–brain barrier. The mostly known feature of oligodendrocytes is formation of myelin sheaths that provides insulation of neuronal axons.
Acquisition of these two distinct glial cell fates is acknowledged to result from spatial segregation of neural progenitors that is established early, during patterning of the neural tube. A good understanding of organizing signals and target genes involved in this process was acquired studying the development of the embryonic spinal cord (Dessaud, McMahon, & Briscoe, 2008). Under the influence of patterning signals, progenitor cells of the ventral neural tube activate or repress sets of transcription factors that subsequently subdivide this structure into distinct progenitor domains, each dedicated to first generate specific neuronal subtypes, followed by glial cells (Ben Haim & Rowitch, 2017). Among them, the pMN domain, populated by progenitor cells expressing the transcription factor Olig2, first generates all motor neurons (MNs) and change with time to produce most oligodendrocyte precursor cells (OPCs) of the spinal cord. Meanwhile, the ventrally and dorsally located p1 to p3 domains initially produce V1 to V3 interneurons and then change their fate to generate three distinct ventral astrocyte precursor cell (AP) subtypes (VA1‐3). Thus, the origins of OPCs and APs are considered to be more closely connected to neuron subtype progenitor cells than with each other. The mechanistic parallel between neuron and glial cell development is well exemplified by functional analysis of the transcription factor Olig2, expressed in pMN cells. In Olig2 null spinal cords, pMN cells fail to generate both MNs and OPCs (Lu et al., 2002; Takebayashi, Nabeshima, Yoshida, Chisaka, & Ikenaka, 2002; Zhou & Anderson, 2002). Instead, in these mutants, supernumerary V2 interneurons and APs are produced as a consequence of transformation of the presumptive pMN domain into a p2 domain. Therefore, spinal OPCs and APs have been proposed to develop along mutually exclusive paths, Olig2 being viewed as a repressor of a “pro‐astrocytic” program (Rowitch & Kriegstein, 2010). Olig2 is also considered as a key determinant of OPC specification, maturation, and differentiation and it continues to be present in differentiated oligodendrocytes of the adult spinal cord (Meijer et al., 2012). However, contrasting with the notion that Olig2 represses the astrocytic fate, lineage‐tracing studies brought evidence that Olig2‐expressing progenitors of the pMN domain generate some astrocytes (Masahira et al., 2006; Tsai et al., 2012).
Beyond spatial control, a temporal component is important to initiate gliogenesis. Although timing mechanism regulating AP commitment remains elusive, OPC commitment from pMN cells is known to rely on a temporal rise of sonic hedgehog (Shh) signaling activity (Danesin & Soula, 2017). The heparan sulfate–editing enzyme Sulf1, known to modulate the sulfation state of heparan sulfate proteoglycans (HSPGs) at the cell surface (Lamanna et al., 2007), is a key player in this process. This enzyme is expressed specifically by Shh‐secreting cells of the floor plate immediately prior to OPC specification. Sulf1, by eliminating 6O sulfate groups on HS chains, locally lowers Shh/HSPG interaction and thereby promotes Shh release at the right time to trigger OPC specification from Olig2‐expressing pMN cells (Al Oustah et al., 2014; Touahri et al., 2012). Jiang et al. (2017) more recently reported that Sulf2, the second member of the Sulf protein family (El Masri, Seffouh, Lortat‐Jacob, & Vivès, 2017), plays a similar role as Sulf1 in triggering the MN to OPC fate change. In the present work, we also investigated the function of Sulf2 in the control of gliogenesis during spinal cord development and came to the different conclusion that Sulf2 is dispensable for OPC production. Instead, we found that Sulf2, which in contrast to Sulf1 is expressed in pMN cells, promotes generation of APs. Furthermore, we showed that APs generated under the influence of Sulf2 are distinguishable from other APs by the expression of Olig2 from initial stages of their generation in the ventral spinal cord to their final maturation at postnatal stages. These results therefore reveal a previously uncharacterized spinal cord AP subtype characterized by expression of Olig2. By highlighting the role of Sulf2 in this glial subtype development, our data also reveal distinct developmental mechanisms for generation of OPC and AP from progenitor cells of the pMN domain.
2. MATERIALS AND METHODS
2.1. Mouse strains
All procedures were performed in agreement with the European Community guiding principles on the care and use of animals (Scientific Procedures) Act, 1987 and approved by the national Animal Care and Ethics Committee (APAFIS#01331.02) following Directive 2010/63/EU. Sulf2 mutant and floxed mice have been generated by Ai et al. (2007). Sulf2 mutant carry deletion of the second coding exon (exon 2) of sulf2, which encodes essential amino acids in the enzymatic domain of the protein (Dhoot et al., 2001). Sulf2 floxed mice carry LoxP sites flanking the exon2 of sulf2. Sulf2 mutant and floxed mice were genotyped as previously reported (Ai et al., 2007; Tran, Shi, Zaia, & Ai, 2012). Parental olig2‐cre +/−;sulf2 fl/+ ;R26R‐tomato mice were generated by crossing heterozygous knock‐in olig2‐cre line (Dessaud et al., 2007) with mice carrying one floxed sulf2 allele and the R26R‐tomato reporter (sulf2 fl/+; R26R‐tomato). Olig2‐cre +/−; sulf2 fl/+ ; R26R‐tomato mice were crossed and olig2‐cre +/−; sulf2 fl/fl ; R26R‐tomato and olig2‐cre +/−; sulf2 fl/+ ; R26R‐tomato littermate embryos were selected by genotyping. GFAP‐GFP and Aldh1L1‐GFP transgenic mice were genotyped as previously reported (Gong et al., 2003; Heintz, 2004; Nolte et al., 2001). Parental sulf2 +/− ; Aldh1L1‐GFP mice were generated by crossing heterozygous sulf2 mutant mouse to an Aldh1L1‐GFP transgenic mouse. Then, sulf2 +/− ; Aldh1L1‐GFP mice were crossed and sulf2 +/+ ; Aldh1L1‐GFP and sulf2 −/− ; Aldh1L1‐GFP littermate embryos were selected by genotyping. All mice were maintained on a C57BL/6 background.
2.2. Thymidine analogue labeling
For proliferation assays, the timed pups (P0) were injected intraperitoneally with 5‐ethylyl‐2‐deoxyuridine (EdU, Invitrogen) at 50 μg/g body weight once a day from postnatal Day 0 to postnatal Day 2. The animals were then sacrificed 2 hr after the last injection. EdU incorporation was detected using the AlexaFluor‐647 Click‐iT Imaging Kit (Invitrogen).
2.3. Tissue processing
Spinal cords at the brachial level were isolated from E12.5 to E18.5 mouse embryos and fixed in 4% paraformaldehyde (PFA, Sigma) in phosphate‐buffered saline (PBS) overnight at 4 °C. Adult mice were perfused intracardially with 4% PFA (Sigma) in PBS. Adult spinal cords were dissected and postfixed in 4% PFA in PBS overnight at 4 °C. Tissues were then sectioned either at 60–80 μm using a vibratome (Microm) or at 15 to 25 μm using a cryostat (Leica CM1950) after cryoprotection in 20% sucrose (Sigma) in PBS and freezing in OCT media on the surface of dry ice.
2.4. In situ RNA hybridization and immunofluorescent staining
Simple or double in situ hybridization and immunofluorescent staining were performed on transverse sections as previously reported (Touahri et al., 2012; Ventéo et al., 2012). Digoxigenin‐ or Fluorescein‐labeled antisens RNA probes for sulf2, aldh1L1, fgfr3, sox10, and tenascin‐C were synthesized using DIG‐ or Fluorescein‐labelling kit (Roche) according to the manufacturer's instructions and revealed using either NBT/BCIP (Roche), INT/BCIP (Roche) or Alexa‐fluor 488 Tyramide (Molecular Probes). Antibodies used in this study were as follows: goat anti‐AldoC (Santa Cruz Biotechnology), rabbit and mouse anti‐GFAP (DAKO), rabbit anti‐NFIA (Active Motif), rabbit and goat anti‐Olig2 (Millipore and R&D Systems), goat anti‐Olig1 (R&D Systems), goat anti‐Sox10 (Santa Cruz Biotechnology), rat anti‐PDGFRα (BD Pharmingen), rabbit anti‐Zeb1 (Novus), mouse anti‐Nkx6.1 (Hybridoma Bank), mouse anti‐Cx43 (BD Biosciences) and rabbit anti‐Cx30 (Thermo Fisher Scientific). Alexa Fluor‐594‐ or Alexa Fluor‐488 or Alexa Fluor‐647‐conjugated secondary antibodies (Thermo Fisher Scientific) were used.
2.5. Imaging, cell counting, and statistical analysis
Confocal images were acquired from tissue sections using Leica SP5 or SP8 confocal microscopes and were always represented as single optical plane sections. In some instances, Z‐stacks were acquired at high magnification (×63 objective) and images along the Z‐axis were obtained using the Reslice feature of ImageJ software along a straight or broken line drawn in the X/Y plane. Images of simple or double bright‐field ISHs were collected with Nikon digital camera DXM1200C on a Nikon Eclipse 80i microscope. Double ISH stainings and double ISH‐immunofluorescence staining were imaged on Leica SP5 confocal microscope according to the method of Trinh et al. (2007) which allows acquiring in the same optical plane a high resolution confocal image of NBT/BCIP stain in the near infrared range together with the immunofluorescence or TSA signals. Images were processed (size adjustment, luminosity and contrast, and merging or separating layers) using Adobe Photoshop CS6 software (Adobe). Provided data are the average of four to eight individuals per condition. Numbers of individuals (n) are indicated in figure legends. Cell counts were performed using ImageJ software and obtained from at least five tissue sections of brachial spinal cord (anterior limb level). For each 60–80 μm tissue slice, at least four optical sections were acquired at 6 μm intervals and cells were counted in each optical section. The same approach was applied for each 15–25 μm tissue slices except that only two optical sections were used. For regional cell number quantification at embryonic and postnatal stages, distribution of cells in the ventral spinal cord was obtained by drawing a straight line through the midline dividing the spinal cord into two equal regions. Cell counts on adult spinal cord sections were limited to the ventral horns. Quantifications are expressed as the mean number of cells (mean ± SEM) in an optical section of hemi‐ventral spinal cord sections. Statistical analyses were performed using the Mann–Whitney U test on Prism software (GraphPad). Significance was determined at p < .05. p values are indicated in figure legends.
3. RESULTS
3.1. Sulf2 controls generation of glial precursor cells marked by Olig2 expression but distinct from OPCs
In the mouse developing spinal cord, Olig2‐expressing (Olig2+) pMN cells start generating OPCs from 12.5 days of development (E12.5). At this initial stage of gliogenesis, sulf2 is expressed in ventral progenitor cells, including in Olig2+ cells of the pMN domain (Figure 1a). Noticeably, variable levels of sulf2 mRNA staining were observed in this domain (Figure 1a′), indicating heterogeneous levels of sulf2 expression within the population of Olig2+ pMN cells. Later on, sulf2 expression is maintained in ventral progenitor cells but also in few cells streaming away in the spinal parenchyma (Figure 1b). At E14.5, a subset of sulf2+ parenchymal cells retains expression of Olig2 (Figure 1b′). Based on these data, Sulf2 appeared to be a reasonable candidate to play a role in OPC development. To test this possibility, we analyzed OPC generation in sulf2 knocked‐out (sulf2−/−) embryos using Olig2 together with Sox10, a reliable marker of oligodendroglial cells which is upregulated in Olig2+ pMN cells as soon as they commit to the OPC fate (Kuhlbrodt, Herbarth, Sock, Hermans‐Borgmeyer, & Wegner, 1998; Zhou, Wang, & Anderson, 2000). As previously reported (Touahri et al., 2012), we found that, at E12.5 in wild‐type embryos, some Olig2+ pMN cells coexpress Sox10 (Figure 1c). Few Olig2+ cells that have already emigrated in the spinal parenchyma were also observed and all of them were found coexpressing Sox10 (Figure 1c). Similar pattern of Olig2 and Sox10 expression was observed in sulf2−/− littermates (Figure 1d) and cell counting indicated that the same numbers of Olig2+ pMN cells and Olig2+/Sox10+ parenchymal OPCs have been generated at E12.5 in sulf2−/− and wild‐type embryos (Figure 1e,f). Therefore, contrasting with previous study (Jiang et al., 2017), our data indicated that Sulf2 is dispensable for initiation of OPC production in the ventral spinal cord. To get further insight on this disparity of results, we next considered the possibility that Sulf2 might impair ongoing OPC production at later developmental stages. We then examined Olig2 and Sox10 expression 1 day later, at E13.5, and, again, found similar patterns of Olig2/Sox10 coexpression in sulf2−/− and wild‐type littermate embryos (Figure 1g–h″,k). We therefore concluded that Sulf2 activity is not required to sustain OPC generation. However, we made at that stage a quite unexpected observation. We found that, in wild‐type embryos, while coexpression of Olig2 and Sox10 was detected in most cells streaming away from the pMN domain, expression of Sox10 remained undetectable in a subset of parenchymal Olig2+ cells (Figure 1g–g‴, i–i″). These Olig2+/Sox10_ cells were predominantly found close to the progenitor zone, suggesting that they had been recently generated. In support of this, we never detected Olig2+/Sox10_ cells in the parenchyma at E12.5 (Figure 1c). Strikingly, only few Olig2+/Sox10_ parenchymal cells were detected in sulf2−/− littermates (Figure 1h–h‴, j–j″) and quantification of these cells indicated that their number was significantly reduced in sulf2−/− embryos compared to wild‐type littermates (Figure 1l). Deficient generation of Olig2+/Sox10_ cells but not of Olig2+/Sox10+ OPCs was still apparent at fetal stage E18.5, where Olig2+/Sox10_ cells were still found populating the ventral spinal cord (Figure 2a–f). These results therefore pointed out to a specific function of sulf2 in controlling generation of Olig2+ cells that do not activate Sox10 while populating the spinal parenchyma.
Figure 1.
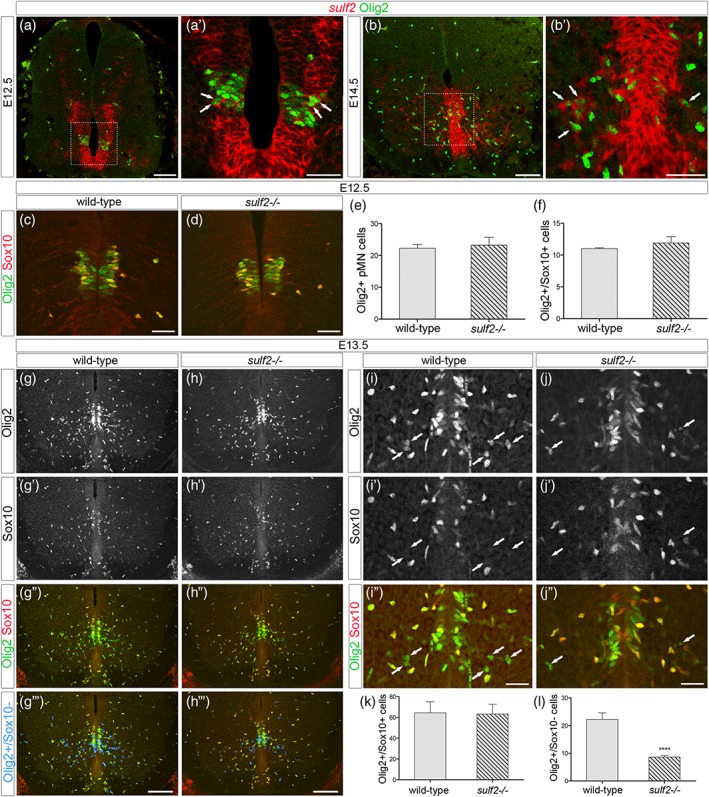
Sulf2 depletion impairs production of Olig2‐expressing precursor cells distinct from OPCs. All images show transverse spinal cord sections. (a–b′) Combined detection of sulf2 mRNA (red) and Olig2 (green) at E12.5 (a, a′) and E14.5 (b, b′). a′ and b′ show higher magnification of the areas framed in a and b, respectively. Arrows in a′ and b′ point to sulf2+/Olig2+ cells. (c, d, g–j″) Double detection of Olig2 (green) and Sox10 (red) at E12.5 (c, d) and E13.5 (g–j″) in wild‐type (c, g–g‴, i–i″) and sulf2−/− (d, h–h‴, j–j″) embryos. In g‴ and h‴, Olig2+/Sox10_ cells (green only) have been highlighted in blue using Colocalization Finder feature of ImageJ software. Arrows in i–j″ point to Olig2+/Sox10‐ cells. Note reduced number of Olig2+/Sox10_ cells in the spinal parenchyma of sulf2−/− (h″, h″’, j″) compared to wild‐type (g″, g‴, i″) E13.5 embryos. (e–f) Quantification of Olig2+ pMN cells (e, n = 4) and Olig2+/Sox10+ cells (f, n = 4) in E12.5 wild‐type and sulf2−/− embryos. (k, l) Quantification of Olig2+/Sox10+ cells (k) and Olig2+/Sox10_ cells (l) in the E13.5 spinal parenchyma of wild‐type (n = 6) and sulf2−/− (n = 8) embryos. Data are presented as mean ± SEM (****p < .0001). Scale bars = 100 μm in a, b, g–h‴ and 50 μm in a′, b′, c, d, i–j″ [Color figure can be viewed at wileyonlinelibrary.com]
Figure 2.
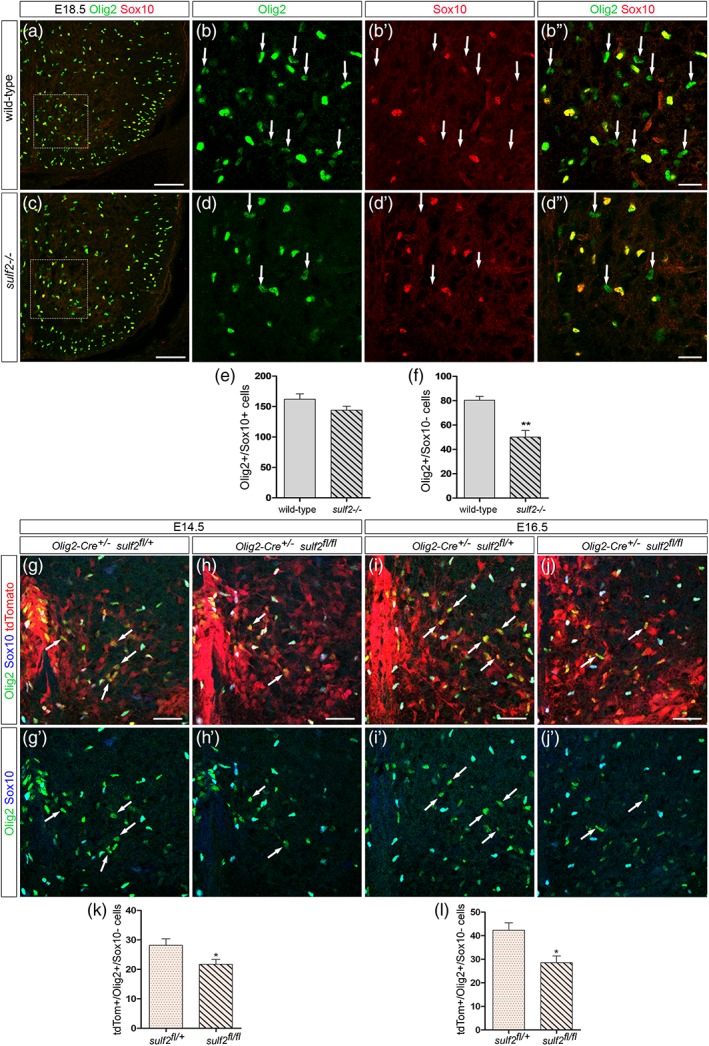
Deficient production of Olig2+/Sox10_ cells is detectable at late embryonic stage and in CreLox model deleting sulf2 specifically in Olig2+ neural progenitors All images show transverse sections of hemi‐ventral spinal cord. (a–d″) Double detection of Olig2 (green) and Sox10 (red) at E18.5 in wild‐type (a, b–b″) and sulf2−/− (c, d–d″) embryos. b–b″ and d–d″ show higher magnification of the areas framed in a and c, respectively. Arrows in b–b″ and d–d″ point to Olig2+/Sox10_ cells. (E–F) Quantification of Olig2+/Sox10+ cells (e) and Olig2+/Sox10_ cells (f) in the E18.5 spinal parenchyma of wild‐type (n = 5) and sulf2−/− (n = 5) embryos. (g–j′) Detection of Olig2 (green), Sox10 (blue) and tdTomato (red) at E14.5 (g, h) and E16.5 (i, j) in Olig2‐Cre +/− /sulf2 fl/+ (g, i) and Olig2‐cre +/− ;sulf2 fl/fl (h, j) embryos. g′–j′ show double staining of Olig2 and Sox10 corresponding to g–j. Note high level of tdTomato expression reflecting efficient recombinase activity of the Cre protein in the ventral spinal cord. (k, l) Quantification of Olig2+/Sox10_ cells in the spinal parenchyma of Olig2‐Cre +/− /sulf2 fl/+ and Olig2‐cre +/− ; sulf2 fl/fl embryos at E14.5 (k, n = 5 for each genotype) and at E16.5 (l, n = 5 for each genotype). Data are presented as mean ± SEM (**p < .01, *p < .05). Scale bars = 100 μm in a, c and g–j′ and 25 μm in b–b″, d–d″ [Color figure can be viewed at wileyonlinelibrary.com]
We next asked whether Sulf2 expression specifically in Olig2+ cells is required for proper generation of the Olig2+/Sox10_ cell subtype. To test this, we turned to a mouse line that carries a “floxed” allele of sulf2 (Tran et al., 2012). By breeding sulf2 “floxed” mice with olig2 promoter‐driven Cre (Olig2‐cre) knock‐in mice (Dessaud et al., 2007), we obtained sulf2 fl/+and sulf2 fl/fl littermate embryos on an Olig2‐Cre:Rosa‐tdTomato background. Counting of Olig2+/Sox10_ cells was performed in Olig2‐Cre +/− /sulf2 fl/+ and Olig2‐Cre +/− ; sulf2 fl/fl littermates at E14.5 and E16.5 (Figure 2g–l). We found that, at each stage, the number of Olig2+/Sox10_ cells was significantly reduced in Olig2‐Cre +/− ; sulf2 fl/fl embryos compared to Olig2‐Cre +/− /sulf2 fl/+ littermates (Figure 2k, l). These results therefore support the view that Sulf2 depletion specifically in Olig2+ cells is sufficient to impair generation of Olig2+/Sox10_ cells.
Together, our data point out two important issues: (a) two molecularly distinct Olig2+ precursor cell subtypes, namely the Olig2+/Sox10+ OPCs and a second cell population expressing Olig2 but not Sox10, are generated in the ventral spinal cord, (b) Sulf2 expression by Olig2+ cells specifically controls generation of the Olig2+/Sox10_ cell subtype.
3.2. Olig2+/Sox10_ cells express astroglial identity genes
The foregoing analysis opened the question of the Olig2+/Sox10_ cell identity. Supporting the view that these cells are distinct from OPCs, parenchymal Olig2+/Sox10_ cells neither expressed PDGFRα (Figure 3a–b″) nor Olig1 (Figure 3c–d″), both considered as early and specific hallmarks of OPCs in the spinal cord (Meijer et al., 2012; Pringle & Richardson, 1993). We then investigated the possibility that Olig2+/Sox10_ cells might be related to the astroglial lineage. For this, we combined immunodetection of Olig2 and in situ localization of fgfr3, aldh1L1, and tenascin‐C mRNAs, all reported to mark spinal APs as well as progenitors from which they originate (Cahoy et al., 2008; Karus, Denecke, ffrench‐Constant, Wiese, & Faissner, 2011; Pringle et al., 2003). At E13.5, fgfr3 mRNA was detected in the progenitor zone where its expression domain overlaps the Olig2+ pMN domain (Figure 3e). At this stage, few individual fgfr3+ cells were also detected in the spinal parenchyma and, noticeably, most, if not all, of them were positive for the Olig2 staining (Figure 3e), indicating that the first fgfr3+ cells to leave the progenitor zone express Olig2. By E15.5, a much greater number of fgfr3+/Olig2+ cells were found in the ventral parenchyma (Figure 3f). At this stage, fgfr3+/Olig2_ parenchymal cells were also observed emigrating from ventrally and dorsally located progenitor domains (Figure 3f). Similar results were obtained detecting aldh1L1 or tenascin‐C mRNAs and the Olig2 protein (Figure 3g,h). These data therefore revealed the existence of a ventral spinal cord AP subtype that distinguishes itself from others by Olig2 expression.
Figure 3.
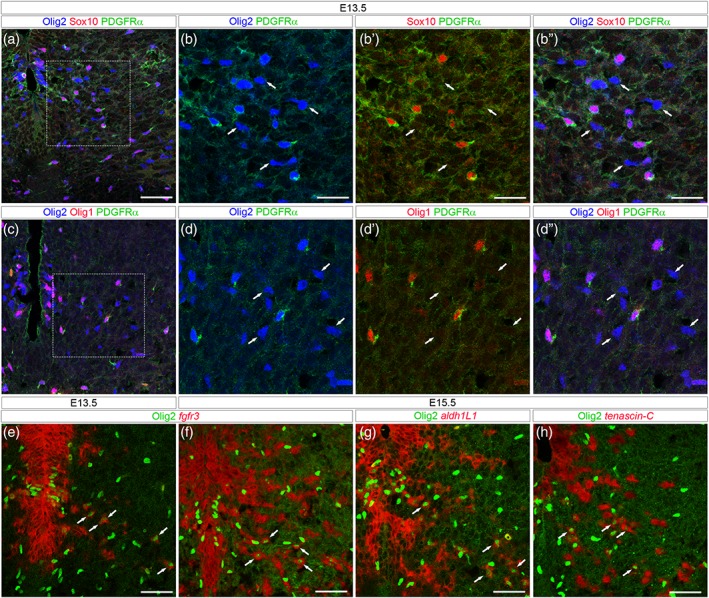
Olig2+/Sox10_ cells express AP but not OPC markers. All images show transverse spinal cord sections. (a–d″) Combined detection of Olig2 (blue), PDGFRα (green) and Sox10 (a, b–b″, red) or Olig1 (c, d–d″, red) viewed on E13.5 hemi‐ventral spinal cord. b–b″ and d–d″ show higher magnification of the area framed in a and c, respectively. Arrows point to Olig2+ cells that are not stained with the OPC markers. (e–h) Combined detection of Olig2 (green) and mRNAs (red) encoding for fgfr3 (e, f), aldh1L1 (g) and tenascin‐C (h) viewed on E13.5 (e) and E15.5 (f–h) hemi‐ventral spinal cords. Note, in all panels, the presence of Olig2+ cells stained with mRNA probes (arrows). Scale bars = 50 μm in a, c, e–h and 25 μm in b–b″, d–d″ [Color figure can be viewed at wileyonlinelibrary.com]
Due to the setback of combining Sox10 immunostaining with in situ hybridization, we were not able to directly define whether fgfr3+/Olig2+ APs match the Olig2+/Sox10_ cell population. We therefore turned to Aldh1L1‐GFP astrocyte‐specific reporter mice (Cahoy et al., 2008) and performed immunodetection of the reporter protein together with that of Olig2 and Sox10. We first focused on the progenitor zone of wild‐type E12.5 embryos and found the presence of Aldh1L1‐GFP+/Olig2+ included in the pMN domain (Figure 4a–a‴). We also observed at this stage some Aldh1L1‐GFP+/Olig2+ cells emigrating from the progenitor zone (Figure 4a′). Comparison with the Sox10 staining allowed detecting Olig2+/Sox10+ cells located both in the pMN domain and in the ventral parenchyma (Figure 4a‴). Confirming that OPCs do not activate expression of Aldh1L1‐GFP, Sox10 expression was never detected in Aldh1L1‐GFP+/Olig2+ cells (Figure 4a″,a‴). These data, by revealing a subtype of Olig2+ pMN cells expressing Aldh1L1 but not Sox10 at E12.5, bring direct evidence for generation of glial precursor cells displaying an AP identity in the pMN domain. We next asked whether generation of Aldh1L1‐GFP+/Olig2+/Sox10_ cells is impaired by sulf2 loss‐of‐function. At E13.5 in wild‐type embryos, Aldh1L1‐GFP+/Olig2+/Sox10_ cells were still detected in the ventral progenitor zone and were also found emigrating in the mantle layer (Figure 4b–b‴). Although Aldh1L1‐GFP+/Olig2+/Sox10_ cells were also detected in sulf2 mutant embryos (Figure 4c–c‴), cell counting indicated that they were reduced in number in these embryos compared to wild‐type littermates (Figure 4e) and no significant difference was found in the number of Aldh1L1‐GFP_/Olig2+/Sox10+ cells between wild‐type and sulf2−/− embryos (Figure 4d). Together, these data add further support to the view that Sulf2 plays a role in triggering generation of Olig2+ APs from progenitor cells of the pMN domain.
Figure 4.
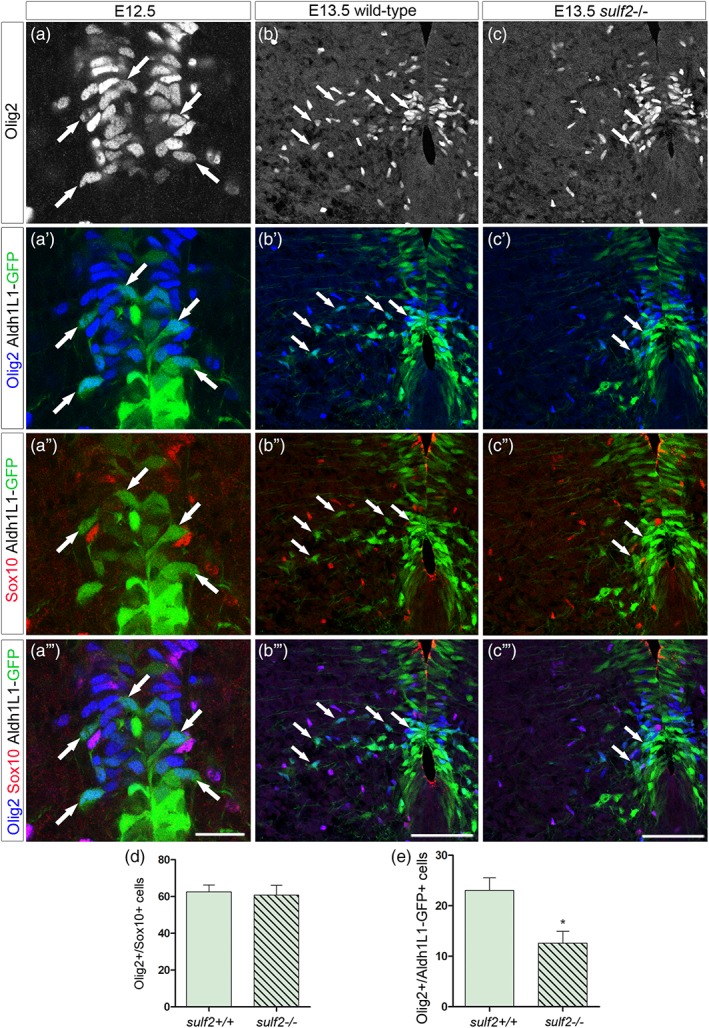
Sulf2 depletion impairs production of Aldh1L1‐GFP+/Olig2+ cells originating from the pMN domain. All images show transverse spinal cord sections. (a–c‴) Combined detection of Olig2 (blue), Sox10 (red), and GFP (green) on Aldh1L1‐GFP transgenic embryos on a sulf2 wild‐type background at E12.5 (a–a‴) and E13.5 (b–b″’) and on sulf2‐/‐ background at E13.5 (c–c‴). Vertical sets present successively Olig2 staining, Olig2 and Aldh1L1‐GFP staining, Sox10 and Aldh1L1‐GFP staining and the merged image. Arrows in all panels point to Olig2+/Sox10‐ cells. a–a‴ show high magnification of the ventral progenitor zone. Note the presence of GFP+/Olig2+/Sox10_ cells both in the progenitor zone (Olig2+ pMN domain) and in the adjacent spinal parenchyma. (d) Quantification of Olig2+/Sox10+ cells in E13.5 wild‐type and sulf2−/− embryos (n = 5 for each genotype). (e) Quantification of Aldh1L1‐GFP+/Olig2+/Sox10_ cells in E13.5 wild‐type and sulf2−/− embryos (n = 5 for each genotype). Data are presented as mean ± SEM (*p < .05). Scale bars = 25 μm in a–a‴ and 75 μm in b–c‴ [Color figure can be viewed at wileyonlinelibrary.com]
To learn more about the Olig2+ AP subtype, we next performed immunostaining using antibodies specific for Nkx6.1, expressed in APs originating from ventral progenitors, and for NFIA and Zeb1, two transcription factors broadly expressed in newly generated spinal APs (Deneen et al., 2006; Ohayon et al., 2016; Zhao et al., 2014). We found that essentially all Olig2+/Sox10_ cells coexpress these proteins at E14.5 (Figure 5), indicating that they share the same basic properties as previously identified ventral APs.
Figure 5.
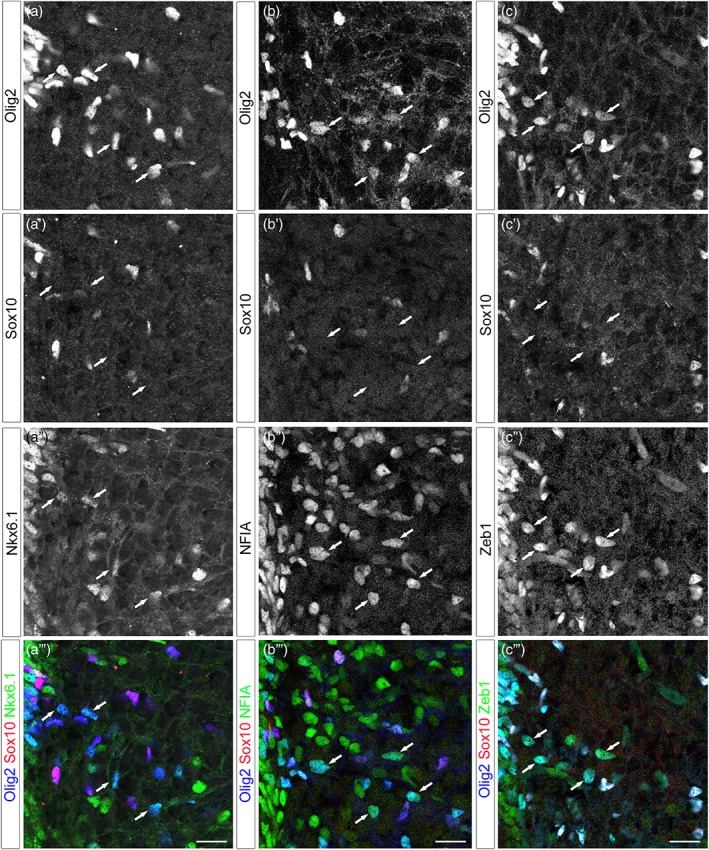
Olig2+/Sox10_ cells coexpress a set of AP nuclear proteins as they populate the ventral spinal parenchyma. All images show transverse sections of hemi‐ventral spinal cord, ventral progenitor zone is on the upper left. (a–c‴) Combined detection of Olig2 (blue), Sox10 (red), and Nkx6.1 (a″–a‴, green), NFIA (b″–b′″, green) or Zeb1 (c″–c‴, green) at E14.5. Vertical sets present successively Olig2 staining, Sox10 staining followed by astrocytic marker staining and the corresponding merged image. Arrows point to Olig2+/Sox10_ cells positive for the astrocytic markers. Scale bars = 50 μm [Color figure can be viewed at wileyonlinelibrary.com]
Together, these data reveal the existence of a spinal AP subtype that emerge from Olig2‐expressing progenitor cells and maintain Olig2 expression as they populate the ventral spinal parenchyma. For convenience, we refer to this sub‐population henceforth as Olig2+ APs.
3.3. Olig2+ APs maintain expression of Olig2 while activating astrocyte differentiation genes
We next examined whether, as previously reported for cortical white matter APs (Cai et al., 2007; Marshall, Novitch, & Goldman, 2005), expression of Olig2 in ventral spinal cord APs reflects an immature stage of astrocytogenesis. Specific markers for different stages of AP maturation are not available. However, acquisition of mature astrocyte identity in the spinal cord can be followed according to the previously reported stepwise process of their maturation (Tien et al., 2012). This process progresses through different stages: a proliferating intermediate AP stage, during which parenchymal APs remain proliferative (E14.5 to P3); a maturing postnatal astrocyte stage (from P5) in which APs no more proliferate and an adult astrocyte status. We therefore assessed Olig2 and Sox10 expression at early (P2) and late (P7) postnatal stages as well as in adult spinal cord, postulating that, if Olig2 expression in Olig2+ APs is downregulated as these cells undergo terminal differentiation, the population of Olig2+/Sox10_ cells should progressively disappear. Olig2+/Sox10_ cells were still detected in the ventral spinal cord at postnatal and adult stages (Figure 6A–C) and no significant difference was found at each stage in their rates relative to the Olig2+/Sox10+ oligodendroglial cell population (Figure 6D), indicating that the Olig2+/Sox10_ ventral spinal cell population persists at stages of astrocyte maturation. EdU injection over the period of P0 to P2 indicated that 20% of these cells incorporated EdU (Figure 6E–E″,F), corresponding to the expected proliferation profile for APs at early postnatal stages (Tien et al., 2012). We also found that, at postnatal and adult stages, Olig2 still showed colocalization with the AP marker fgfr3 (Figure 6G–J), indicating that Olig2 expression is retained in astroglial cells until adult stage.
Figure 6.
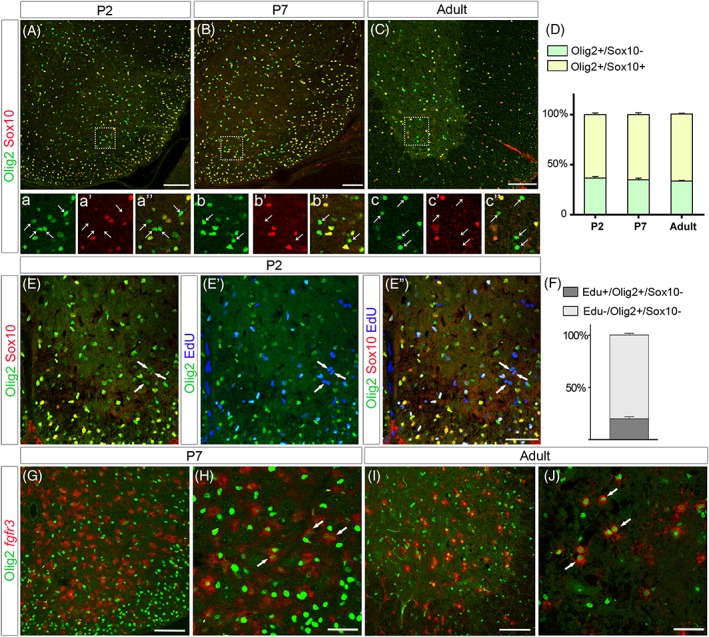
Olig2+/Sox10_ cells are maintained at postnatal and at adult stages
All images show transverse sections of hemi‐ventral spinal cord. (A–c″) Double detection of Olig2 (green) and Sox10 (red) at P2 (A, a–a″), P7 (B, b–b″), and adult (C, c–c″) stages. Images a–a″, b–b″, and c–c″ show higher magnification of the areas framed in A, B, and C, respectively and horizontal sets present successively Olig2 staining, Sox10 staining and the merged image. Note the presence of Olig2+/Sox10_ cells (arrows) at all stages. (D) Proportion of Sox10_ and Sox10+ cells on the Olig2+ cell population in the ventral spinal cord at P2 (n = 5), P7 (n = 5) and at adult stage (n = 5). (E–E″) Combined detection of Olig2 (green), Sox10 (red) and EdU (blue) at P2. Images E, E′, and E″ show successively Olig2/Sox10 staining, Olig2/EdU staining, and the merged image. Arrows point to Olig2+/Sox10_ cells positive for EdU. (F) Proportion of EdU+ cells on the Olig2+/Sox10‐ cell population at P2 (n = 4). (G–J) Double detection of Olig2 (green) and fgfr3 mRNA (red) viewed on ventral spinal cord hemi‐section at P7 (G, H) and at adult stage (I, J). H and J are high magnifications of the ventral spinal cord in which arrows point to Olig2+/fgfr3+ cells. Data are presented as mean ± SEM. Scale bars = 100 μm in A–E″, G, I and 50 μm in H and J [Color figure can be viewed at wileyonlinelibrary.com]
To further characterize the Olig2+ astroglial cell population, we analyzed expression of a set of glial cell markers in postnatal (P7) and adult Aldh1L1‐GFP spinal cords. We found extensive coexpression of Aldh1L1‐GFP and Olig2 but not Sox10 at P7 (Figure 7a–b′) and at adult stage (Figure 8a–b′). Supporting the view that they originate from ventral progenitors, Aldh1L1‐GFP+/Olig2+ cells were all found coexpressing Nkx6.1 (Figures 7c–d′ and 8c–d′). We next examined expression of the astrocytic marker AldoC and found that a fraction of ventral Aldh1L1‐GFP+/AldoC+ cells indeed express Olig2 at both stages (Figures 7e–f′ and 8e–f′). We then analyzed expression of the astrocyte maturation marker GFAP and also found Aldh1L1‐GFP+/Olig2+ cells expressing GFAP in P7 (Figure 7g–h′) and adult individuals (Figure 8g–h′). To confirm this, we analyzed expression Olig2 and Sox10 in GFAP‐GFP transgenic mice in which expression of the reporter allows complete staining of astrocyte cell bodies (Nolte et al., 2001). In these mice, GFAP‐GFP+ cells coexpressing Olig2 were also detected in the ventral spinal parenchyma at P7 (Figure 7i–j′). Finally, we examined expression of the connexin proteins C×43 and C×30, known to be differentially expressed in astrocytes of the adult rodent spinal cord (Orthmann‐Murphy, Abrams, & Scherer, 2008). Immunostainings of C×43 and C×30 together with that of Olig2 were performed on sections of adult Aldh1L1‐GFP spinal cords (Figure 8i–l′). Characteristic punctate immunoreactivity was observed for both C×43 and C×30 on Olig2+/Aldh1L1‐GFP+ cells whose soma and processes can be detected by the endogenous fluorescence of GFP (Figure 8j,j′ and l,l′).
Figure 7.
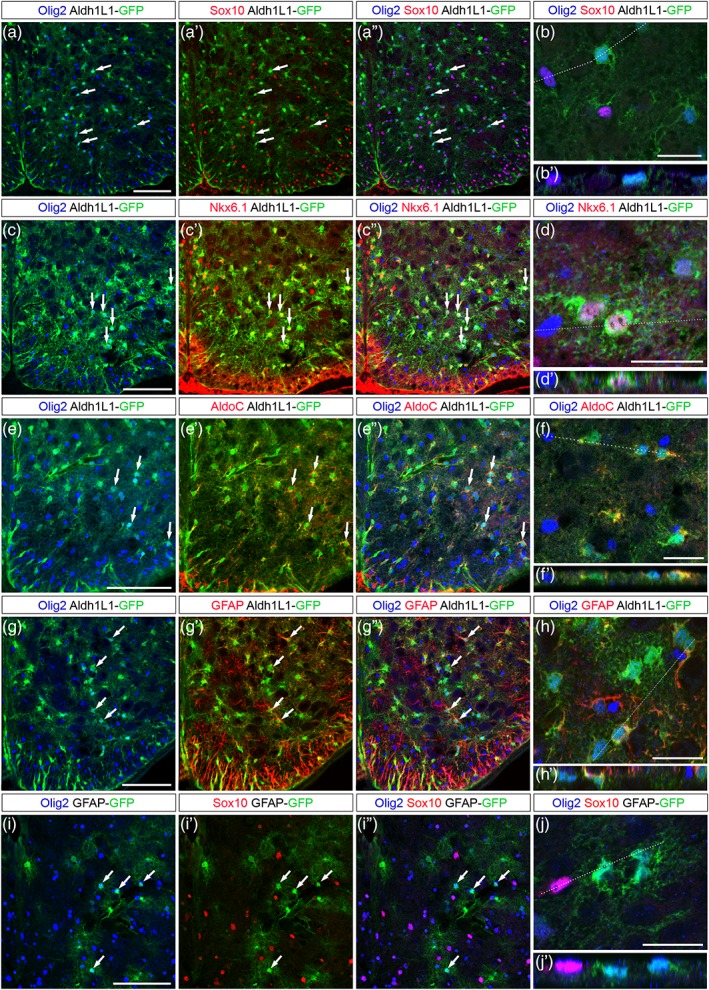
Olig2+ APs retain expression of Olig2 as they differentiate in the P7 ventral spinal cord. Images show transverse sections of P7 hemi‐ventral spinal cord. (a–j′) Combined detection of Olig2 (blue), Sox10 (a′–b′, i′–j′, red), Nkx6.1 (c′–d′, red), AldoC (e′–f′, red) or GFAP (g′–h′, red) in Aldh1L1‐GFP (a–h′, green) and GFAP‐GFP (i–j′, green) transgenic individuals. Horizontal sets present successively Olig2 and GFP staining, Sox10, Nkx6.1, AldoC or GFAP and GFP staining (prime) and the merged image (double prime). Arrows point to Olig2+ cells positive for the astrocytic markers. Panels b, d, f, h and j show higher magnification of the ventral grey matter and underneath prime panels show images in the Z‐plane, corresponding to the dashed line drawn in the X/Y plane above. Scale bars = 100 μm in a–a′, c–c′, e–e′, g–g′, and i–i′ and 25 μm in b, d, f, h, and j [Color figure can be viewed at wileyonlinelibrary.com]
Figure 8.
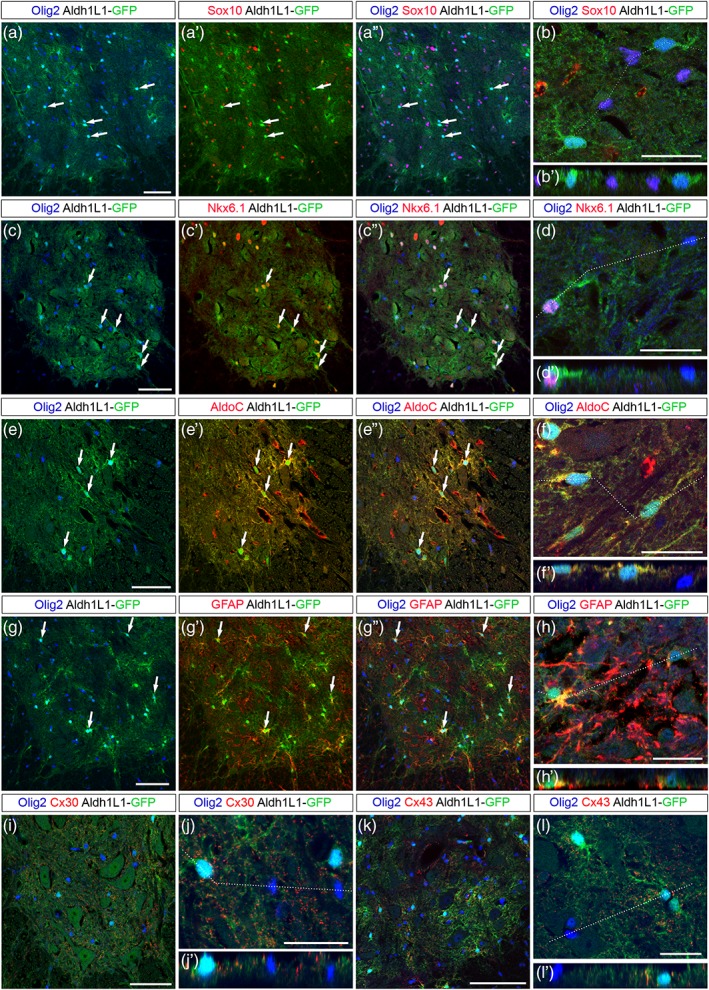
Olig2+ astrocytes are maintained in the grey matter of the adult spinal cord. Images show transverse sections of adult hemi‐ventral spinal cord. (a–h′) Combined detection of Olig2 (blue) and Sox10 (a′–b′, red), Nkx6.1 (c′–d′, red), AldoC (e′–f′, red) and GFAP (g′–h′, red) in Aldh1L1‐GFP (green) transgenic individuals. Horizontal sets present successively Olig2 and GFP staining, Sox10, Nkx6.1, AldoC or GFAP and GFP staining (prime) and the merged image (double prime). Arrows point to Olig2+ cells positive for the astrocytic markers. Panels b, d, f, and h show higher magnification of the ventral grey matter and underneath prime panels show images in the Z‐plane, corresponding to the dashed line drawn in the X/Y plane above. (i–l′) Combined detection of Olig2 (blue), GFP (green), and Cx30 (i–j′) or Cx43 (k–l′) on Aldh1L1‐GFP adult spinal cord. Panels j and l show higher magnification of the ventral grey matter and underneath prime panels show images in the Z‐plane, corresponding to the dashed line drawn in the X/Y plane above. Scale bars = 100 μm in a–a′ and K, 50 μm in c–c′, e–e′, g–g′ and i–i′ and 25 μm in b, d, f, h, j, and l [Color figure can be viewed at wileyonlinelibrary.com]
Together, these data, showing that Olig2 expression is retained at postnatal and adult stages in cells expressing the AP markers fgfr3, Aldlh1L1 and Nkx6.1 but also in cells that activate expression of functionally relevant astrocyte differentiation markers AldoC, GFAP, C×43, and C×30 support the view that Olig2+ APs maintain Olig2 expression as they maturate in the ventral spinal cord.
3.4. Sulf2 depletion while impairing the first wave of AP production does not cause major spinal cord AP deficit
Sulf2 expression is not restricted to the pMN domain, but extends ventrally and dorsally in progenitor domains known to generate V1 to V3 APs (see Figure 1a,b). Analysis of sulf2 expression over spinal cord development further revealed sulf2+ parenchymal cells streaming away at all levels of the ventral progenitor zone (Figure 9a–c), within a pattern very suggestive of sulf2 expression being maintained in most, if not all, ventral spinal cord APs. Confirming the AP identity of parenchymal sulf2+ cells, we found that these cells coexpress fgfr3 but not sox10 as they leave the progenitor zone (Figure 9d,e). We next logically asked whether Sulf2 might have a general influence on AP generation. To investigate this question, we compared the time course of fgfr3 expression in wild‐type and sulf2−/− embryos. At E13.5, when the first wave of AP production became discernible in wild‐type embryos (Figure 10a), only very few fgfr3+ cells were found emigrating from the progenitor zone in sulf2−/− embryos (Figure 10e) and cell quantification confirmed defective production of fgfr3+ cells in sulf2−/− embryos compared to wild‐type littermates (Figure 10i). Thus, as expected since the first APs to be generated correspond to the Olig2+ AP subtype (see Figure 3), Sulf2 depletion causes defective AP production at initiation of astrogenesis. At E14.5 and E15.5, although cell counting still indicated a decrease in the number of ventral fgfr3+ cells in mutant embryos (Figure 10j,k), numerous fgfr3+ individual cells were found populating the spinal parenchyma both in wild‐type (Figure 10b,c) and sulf2−/− embryos (Figure 10f,g). At E18.5, larger numbers of fgfr3+ cells were found populating the spinal cord parenchyma of wild‐type (Figure 10d) and sulf2−/− embryos (Figure 10h) and cell counting indicated a downward trend, although not significant, of their number in sulf2−/− compared to wild‐type individuals (Figure 10l), indicating no gross effect of Sulf2 depletion on astrocyte development. To distinguish Olig2+ APs among the overall population, we next combined Olig2 and GFAP detection in E18.5 wild‐type and sulf2−/− embryos. As expected, GFAP+/Olig2+ cells were detected in the ventral spinal parenchyma of wild‐type embryos (Figure 10m–m′). Only few GFAP+/Olig2+ cells were found in sulf2−/− embryos (Figure 10n–n′) and cell counting confirmed that their number was significantly reduced compared to wild‐type littermates (Figure 10o), thus confirming that proper generation of this astrocyte subtype depends on Sulf2 activity.
Figure 9.
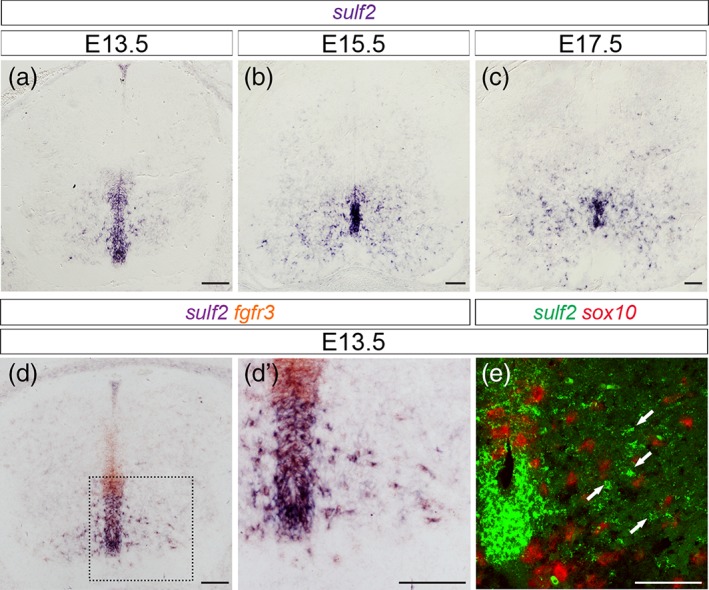
Sulf2 is broadly expressed in ventral APs as they populate the spinal parenchyma. All images show transverse spinal cord sections. (a–c) Temporal expression profile of sulf2 mRNA at E13.5 (a), E15.5 (b), and E17.5 (c). (d, d′) Combined detection of sulf2 (brown) and fgfr3 (purple) mRNAs. d′ shows higher magnification of the area framed in d. (e) Combined detection at confocal resolution of sulf2 (green) and sox10 (red) mRNAs in hemi‐ventral spinal cord at E13.5. Scale bars = 100 μm [Color figure can be viewed at wileyonlinelibrary.com]
Figure 10.
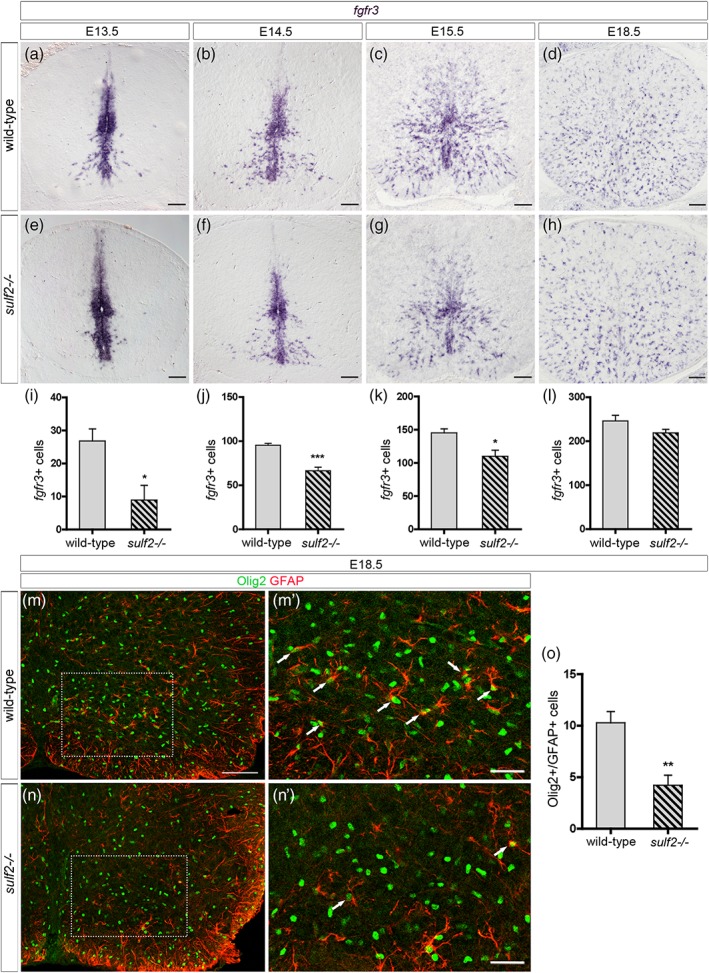
Sulf2 depletion impairs early wave of AP production but does not cause major defect of astrocytogenesis. All images show transverse spinal cord sections. (a–h) Detection of parenchymal cells expressing fgfr3 mRNA in wild‐type (a–d) and sulf2−/− (e–h) embryos at E13.5 (a, e), E14.5 (b, f), E15.5 (c, g), and E18.5 (d, h). (i–l) Quantification of fgfr3+ parenchymal cells in wild‐type and sulf2−/− embryos at E13.5 (i; wild‐type n = 7; sulf2−/− n = 5), E14.5 (j; wild‐type n = 6; sulf2−/− n = 4), E15.5 (k; n = 5 for each genotype) and E18.5 (l; n = 5 for each genotype). (m–n′) Double detection of Olig2 (green) and GFAP (red) in wild‐type (m, m′) and sulf2−/− (n, n′) spinal cords at E18.5. m′ and n′ show higher magnification of the area framed in m and n, respectively. Arrows in m′ and n′ point to Olig2+/GFAP+ cells. (o) Quantification of Olig2+/GFAP+ cells in the ventral spinal cord of E18.5 wild‐type and sulf2−/− embryos (n = 5 for each genotype). Data are presented as mean ± SEM (***p < .001, **p < .01, and *p < .05). Scale bars = 100 μm in a–h, m, n and 50 μm in m′, n′ [Color figure can be viewed at wileyonlinelibrary.com]
Therefore, Sulf2 depletion, while affecting the first wave of AP production, did not cause a massive defect in the generation of ventral spinal cord APs. These data not only reinforce the view that Olig2+ astrocytes depend on Sulf2 for their development but also highlight the role of Sulf2 for proper generation of this particular AP subtype.
4. DISCUSSION
Astrocytes are currently considered to be a remarkably heterogeneous population of cells exhibiting highly diverse functional properties that can impact their influence on neuronal transmission. However, although functional heterogeneity of astrocytes is a well‐established concept, how this diversity is generated still remains elusive. In particular, the question remains whether patterning transcription factors contribute to generate stable molecular and functional diversity in astrocytes, as they do in neurons. In the present work, by addressing the role of Sulf2 in the mouse embryonic spinal cord, we highlight a specific function for this enzyme in controlling production of a specific AP subtype, which, remarkably, expresses Olig2 at all stages of its development. We therefore characterized astrocytes displaying an “Olig2 identity” from initial stage of their generation to terminal differentiation in the ventral spinal cord and identified a specific player of their generation.
Our work reveals a previously undefined heterogeneity within the population of Olig2+ glial precursor cells generated in the early gliogenic spinal cord. So far, although pMN cells are recognized to generate some astroglial cells (Masahira et al., 2006; Tsai et al., 2012), the nature of their precursor cells in the spinal cord parenchyma was unknown. Thus, OPCs were the only pMN‐derived glial precursor cells to have been characterized. Typical OPC molecular identity relies on expression of Olig2 together with Sox10, Olig1, and PDGFRα, a set of genes activated as soon as pMN cells commit to the OPC fate (Bergles & Richardson, 2015). We identified a subtype of Olig2+ cells that do not activate this OPC program but, instead, adopt AP molecular features, activating expression of fgfr3, Aldh1L1, TenascinC, and Nkx6.1, adopting later on definitive astrocyte identity with expression of structural and functional proteins such as AldoC, GFAP, C×43 and C×30.
Noticeably, OPCs (Olig2+/Sox10+) and APs (Olig2+/Aldh1L1+) are already distinguishable within the pMN domain, indicating that they segregate early, before leaving the progenitor zone. We did not detect real distinct sub‐domains within the pMN domain, where the two Olig2+ cell subtypes appeared instead intermingled. This therefore questions the model of spatial segregation into distinct progenitor cell domains for glial‐cell subtype specification in the developing spinal cord. Several types of glial restricted precursor cells that can generate astrocytes and oligodendrocytes (O‐A precursor cells) have been identified in the past, including Glial‐Restricted Precursor cell (GRP) and O‐2A cells (Liu & Rao, 2004). However, their existence in vivo has been the subject of debate (Rowitch & Kriegstein, 2010). Our results showing that OPCs and APs develop in the same region of the neural tube are consistent with the O‐A precursor cell model, opening the possibility that Olig2+ progenitor cells of the pMN domain might share common features with these bipotent glial precursor cells at gliogenic stages. However, the possibility that Olig2+ cells are heterogeneous and that OPC and AP fate‐restricted Olig2+ progenitors are included in the pMN domain cannot be excluded.
A previous study by Jiang and collaborators (Jiang et al., 2017) reported that Sulf2 depletion caused deficient production of OPCs at the initial stage of gliogenesis, that is, E12.5. Here we show that lack of Sulf2 leads to defective Olig2+ AP generation but has no effect on OPC generation during all development, from E12.5 to E18.5. At first, differences in mouse models and/or markers used to identify OPCs could possibly account for these differences. However, both mouse lines from Nagamine and collaborators (Nagamine et al., 2012) in the study of Jiang and collaborators and from Ai and collaborators (Ai et al., 2007) in the present study, should be functionally similar inasmuch as they carry deletions in the region encoding the catalytic site of the enzyme. Also, cells expressing PDGFRα (as in the study of Jiang et al., 2017) or Olig2/Sox10 (as in our present work) are recognized as a single cell population in the embryonic ventral spinal cord (Finzsch, Stolt, Lommes, & Wegner, 2008; Zhou et al., 2000). Therefore, there is currently no evident explanation for this discrepancy. Nevertheless, the effect of the lack of Sulf2 on Olig2+ AP generation highlights the important role of Sulf2 in glial subtype development.
How Sulf2 participates in the control of Olig2+ AP generation remains to be established. Sulf2, together with Sulf1, are recognized as critical regulators of heparan sulfate (HS) activities through their ability to catalyze specific 6‐O‐desulfation of the polysaccharide (El Masri et al., 2017). HS, found at the cell surface and in the extracellular matrix, interact with many signaling cues and a major determinant of specificity and affinity of HS‐ligand interactions is 6‐O‐sulfation. Then, Sulf proteins by post‐synthetically modulating levels of 6‐O‐sulfation regulate HS binding properties toward numerous ligands important in development, including Shh, FGFs, Wnt, and BMPs. HS on heparan sulfate proteoglycans (HSPGs) are known to modulate signaling activities in many different ways: they can regulate ligand secretion, processing, and access to receptors but HSPGs can also act as receptors or co‐receptors (Rosen & Lemjabbar‐Alaoui, 2010). We know from our previous work that Sulf1, expressed by Shh secreting cells of the medial and lateral floor plate cells but not by the Olig2+ pMN responsive cells, contributes to induce OPC generation by controlling delivery of inductive signal provided by Shh (Al Oustah et al., 2014; Touahri et al., 2012). In contrast to Sulf1, Sulf2 is expressed by Olig2+ progenitor cells of the pMN domain, opening the possibility that Sulf2 might control gliogenesis acting cell autonomously on these cells. This is supported by our data showing that inactivation of Sulf2 specifically in Olig2‐expressing cells is sufficient to impair generation of Olig2+ APs. Noticeably, we found heterogeneity in levels of sulf2 expression among pMN cells. Therefore, an attractive hypothesis could be that high levels of Sulf2 expression in a subset of Olig2+ progenitor cells favor reception of astroglial inductive signal. A set of signaling factors, including members of the interleukin‐6 family of cytokines such as LIF and CNTF, which activate the JAK–STAT pathway as well as BMP–Smad and Notch signaling have been shown to promote astrocyte differentiation from neural stem/progenitor cells (Gallo & Deneen, 2014; Kanski, van Strien, van Tijn, & Hol, 2014). However, effects of these factors on astrocytogenesis have primarily been studied by their ability to induce GFAP expression and we now know that the specification of APs from neural progenitors occurs before GFAP induction during development. Thus, whether these factors contribute to actively instruct neural precursor cells to follow an astroglial fate and, thereby, whether they might correspond to signal regulated by Sulf2, remains an open question.
An important aspect of our study is the characterization of ventral spinal cord astrocytes that maintain expression of Olig2 until they reach their mature stage. Previous studies already noted the presence of GFAP+ astrocytes coexpressing Olig2 in the grey matter of the adult mouse spinal cord (Barnabé‐Heider et al., 2010; Guo et al., 2011). Even if some Olig2+ astrocytes were occasionally observed in the white matter, we also found that most Olig2+ astrocytes are located in the grey matter, supporting the view they are related to protoplasmic astrocytes. Similarly, subpopulation of glial cells co‐expressing astrocytic genes and Olig2 have been reported to populate the thalamus (Griemsmann et al., 2015), indicating that Olig2 expression is also maintained in at least subtypes of astrocytes of the adult brain. Therefore, our work, by uncovering production of Olig2+ APs at initial stages of gliogenesis, sheds light on the developmental origin of spinal cord Olig2+ astrocytes. The most striking aspect is that ventral spinal Olig2+ APs maintain expression of Olig2 as they acquire a mature astrocytic identity. It is therefore obvious that, at least for this particular astrocyte subtype, Olig2 does not behave as a repressor of astrocytogenesis. Nonetheless, this observation raises the question whether expression of Olig2 confers specific functional properties to these astrocytes. During the past decade, evidence has accumulated that astrocytes represent remarkably heterogeneous cells that differ in their morphology, developmental origin, gene expression profile, physiological properties, function, and response to injury and disease (Adams & Gallo, 2018; Ben Haim & Rowitch, 2017). Our study, by revealing the time of birth and molecular identity of pMN‐derived astrocytes as well as a key regulator of their production during development, therefore represents an important advance in clarifying astrocyte heterogeneity in the spinal cord.
ACKNOWLEDGMENTS
We especially thank X. Ai, N. Rouach, H. Kettenmann and B. Novitch for their generous sharing of Sulf2 mutant, Aldh1L1‐GFP, GFAP‐GFP, and Olig2‐Cre mice, respectively. We acknowledge B. Guiard for the gift anti‐Connexin antibodies and A. Pattyn for providing valuable reagents and for critical reading of the manuscript. We thank the ABC facility and ANEXPLO for housing mice and the Toulouse Regional Imaging Platform (TRI) for technical assistance in confocal microscopy. We acknowledge the Developmental Studies Hybridoma Bank, developed under the auspice of NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA, for supplying monoclonal antibodies. Work in C.S.’ lab was supported by grants from ANR, ARC, ARSEP, CNRS, and University of Toulouse. D. O. was supported by grants from ARSEP and ARC.
Ohayon D, Escalas N, Cochard P, Glise B, Danesin C, Soula C. Sulfatase 2 promotes generation of a spinal cord astrocyte subtype that stands out through the expression of Olig2. Glia. 2019;67:1478–1495. 10.1002/glia.23621
Funding information Agence Nationale de la Recherche, Grant/Award Number: ANR‐15‐CE16‐0014‐02; Centre National de la Recherche Scientifique; Fondation ARC pour la Recherche sur le Cancer; Fondation pour l'Aide à la Recherche sur la Sclérose en Plaques; Université de Toulouse
REFERENCES
- Adams, K. L. , & Gallo, V. (2018). The diversity and disparity of the glial scar. Nature Neuroscience, 21(1), 9–15. 10.1038/s41593-017-0033-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai, X. , Kitazawa, T. , Do, A. T. , Kusche‐Gullberg, M. , Labosky, P. A. , & Emerson, C. P. (2007). SULF1 and SULF2 regulate heparan sulfate‐mediated GDNF signaling for esophageal innervation. Development, 134(18), 3327–3338. doi:134/18/3327 [pii]. 10.1242/dev.007674 [DOI] [PubMed] [Google Scholar]
- Al Oustah, A. , Danesin, C. , Khouri‐Farah, N. , Farreny, M. A. , Escalas, N. , Cochard, P. , … Soula, C. (2014). Dynamics of sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring sulfatase 1. Development, 141(6), 1392–1403. 10.1242/dev.101717 [DOI] [PubMed] [Google Scholar]
- Barnabé‐Heider, F. , Göritz, C. , Sabelström, H. , Takebayashi, H. , Pfrieger, F. W. , Meletis, K. , & Frisén, J. (2010). Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell, 7(4), 470–482. 10.1016/j.stem.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Ben Haim, L. , & Rowitch, D. H. (2017). Functional diversity of astrocytes in neural circuit regulation. Nature Reviews. Neuroscience, 18(1), 31–41. 10.1038/nrn.2016.159 [DOI] [PubMed] [Google Scholar]
- Bergles, D. E. , & Richardson, W. D. (2015). Oligodendrocyte development and plasticity. Cold Spring Harbor Perspectives in Biology, 8(2), a020453 10.1101/cshperspect.a020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy, J. D. , Emery, B. , Kaushal, A. , Foo, L. C. , Zamanian, J. L. , Christopherson, K. S. , … Barres, B. A. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. The Journal of Neuroscience, 28(1), 264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, J. , Chen, Y. , Cai, W. H. , Hurlock, E. C. , Wu, H. , Kernie, S. G. , … Lu, Q. R. (2007). A crucial role for Olig2 in white matter astrocyte development. Development, 134(10), 1887–1899. 10.1242/dev.02847 [DOI] [PubMed] [Google Scholar]
- Danesin, C. , & Soula, C. (2017). Moving the Shh source over time: What impact on neural cell diversification in the developing spinal cord? Journal of Developmental Biology, 5(2), 4. 10.3390/jdb5020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen, B. , Ho, R. , Lukaszewicz, A. , Hochstim, C. J. , Gronostajski, R. M. , & Anderson, D. J. (2006). The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron, 52(6), 953–968. 10.1016/j.neuron.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Dessaud, E. , McMahon, A. P. , & Briscoe, J. (2008). Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen‐regulated transcriptional network. Development, 135(15), 2489–2503. 10.1242/dev.009324 [DOI] [PubMed] [Google Scholar]
- Dessaud, E. , Yang, L. L. , Hill, K. , Cox, B. , Ulloa, F. , Ribeiro, A. , … Briscoe, J. (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature, 450(7170), 717–720. doi:nature06347 [pii]. 10.1038/nature06347 [DOI] [PubMed] [Google Scholar]
- Dhoot, G. K. , Gustafsson, M. K. , Ai, X. , Sun, W. , Standiford, D. M. , & Emerson, C. P. (2001). Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science, 293(5535), 1663–1666. 10.1126/science.293.5535.1663 [DOI] [PubMed] [Google Scholar]
- El Masri, R. , Seffouh, A. , Lortat‐Jacob, H. , & Vivès, R. R. (2017). The "in and out" of glucosamine 6‐O‐sulfation: The 6th sense of heparan sulfate. Glycoconjugate Journal, 34(3), 285–298. 10.1007/s10719-016-9736-5 [DOI] [PubMed] [Google Scholar]
- Finzsch, M. , Stolt, C. C. , Lommes, P. , & Wegner, M. (2008). Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development, 135(4), 637–646. 10.1242/dev.010454 [DOI] [PubMed] [Google Scholar]
- Gallo, V. , & Deneen, B. (2014). Glial development: The crossroads of regeneration and repair in the CNS. Neuron, 83(2), 283–308. 10.1016/j.neuron.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, S. , Zheng, C. , Doughty, M. L. , Losos, K. , Didkovsky, N. , Schambra, U. B. , … Heintz, N. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature, 425(6961), 917–925. 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- Griemsmann, S. , Höft, S. P. , Bedner, P. , Zhang, J. , von Staden, E. , Beinhauer, A. , … Steinhäuser, C. (2015). Characterization of Panglial gap junction networks in the thalamus, neocortex, and hippocampus reveals a unique population of glial cells. Cerebral Cortex, 25(10), 3420–3433. 10.1093/cercor/bhu157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F. , Maeda, Y. , Ma, J. , Delgado, M. , Sohn, J. , Miers, L. , … Pleasure, D. (2011). Macroglial plasticity and the origins of reactive astroglia in experimental autoimmune encephalomyelitis. The Journal of Neuroscience, 31(33), 11914–11928. 10.1523/JNEUROSCI.1759-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz, N. (2004). Gene expression nervous system atlas (GENSAT). Nature Neuroscience, 7(5), 483 10.1038/nn0504-483 [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Ishino, Y. , Hashimoto, H. , Keino‐Masu, K. , Masu, M. , Uchimura, K. , … Ikenaka, K. (2017). Sulfatase 2 modulates fate change from motor neurons to Oligodendrocyte precursor cells through coordinated regulation of Shh signaling with Sulfatase 1. Developmental Neuroscience, 39, 361–374. 10.1159/000464284 [DOI] [PubMed] [Google Scholar]
- Kanski, R. , van Strien, M. E. , van Tijn, P. , & Hol, E. M. (2014). A star is born: New insights into the mechanism of astrogenesis. Cellular and Molecular Life Sciences, 71(3), 433–447. 10.1007/s00018-013-1435-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karus, M. , Denecke, B. , ffrench‐Constant, C. , Wiese, S. , & Faissner, A. (2011). The extracellular matrix molecule tenascin C modulates expression levels and territories of key patterning genes during spinal cord astrocyte specification. Development, 138(24), 5321–5331. 10.1242/dev.067413 [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt, K. , Herbarth, B. , Sock, E. , Hermans‐Borgmeyer, I. , & Wegner, M. (1998). Sox10, a novel transcriptional modulator in glial cells. The Journal of Neuroscience, 18(1), 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna, W. C. , Kalus, I. , Padva, M. , Baldwin, R. J. , Merry, C. L. , & Dierks, T. (2007). The heparanome‐‐The enigma of encoding and decoding heparan sulfate sulfation. Journal of Biotechnology, 129(2), 290–307. 10.1016/j.jbiotec.2007.01.022 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , & Rao, M. S. (2004). Glial progenitors in the CNS and possible lineage relationships among them. Biology of the Cell, 96(4), 279–290. 10.1016/j.biolcel.2004.02.001 [DOI] [PubMed] [Google Scholar]
- Lu, Q. R. , Sun, T. , Zhu, Z. , Ma, N. , Garcia, M. , Stiles, C. D. , & Rowitch, D. H. (2002). Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell, 109(1), 75–86. [DOI] [PubMed] [Google Scholar]
- Marshall, C. A. , Novitch, B. G. , & Goldman, J. E. (2005). Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. The Journal of Neuroscience, 25(32), 7289–7298. 10.1523/JNEUROSCI.1924-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahira, N. , Takebayashi, H. , Ono, K. , Watanabe, K. , Ding, L. , Furusho, M. , … Ikenaka, K. (2006). Olig2‐positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Developmental Biology, 293(2), 358–369. 10.1016/j.ydbio.2006.02.029 [DOI] [PubMed] [Google Scholar]
- Meijer, D. H. , Kane, M. F. , Mehta, S. , Liu, H. , Harrington, E. , Taylor, C. M. , … Rowitch, D. H. (2012). Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nature Reviews. Neuroscience, 13(12), 819–831. 10.1038/nrn3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine, S. , Tamba, M. , Ishimine, H. , Araki, K. , Shiomi, K. , Okada, T. , … Keino‐Masu, K. (2012). Organ‐specific sulfation patterns of heparan sulfate generated by extracellular sulfatases Sulf1 and Sulf2 in mice. The Journal of Biological Chemistry, 287(12), 9579–9590. 10.1074/jbc.M111.290262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte, C. , Matyash, M. , Pivneva, T. , Schipke, C. G. , Ohlemeyer, C. , Hanisch, U. K. , … Kettenmann, H. (2001). GFAP promoter‐controlled EGFP‐expressing transgenic mice: A tool to visualize astrocytes and astrogliosis in living brain tissue. Glia, 33(1), 72–86. [PubMed] [Google Scholar]
- Ohayon, D. , Garcès, A. , Joly, W. , Soukkarieh, C. , Takagi, T. , Sabourin, J. C. , … Pattyn, A. (2016). Onset of spinal cord astrocyte precursor emigration from the ventricular zone involves the Zeb1 transcription factor. Cell Reports, 17(6), 1473–1481. 10.1016/j.celrep.2016.10.016 [DOI] [PubMed] [Google Scholar]
- Orthmann‐Murphy, J. L. , Abrams, C. K. , & Scherer, S. S. (2008). Gap junctions couple astrocytes and oligodendrocytes. Journal of Molecular Neuroscience, 35(1), 101–116. 10.1007/s12031-007-9027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, N. P. , & Richardson, W. D. (1993). A singularity of PDGF alpha‐receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development, 117(2), 525–533. [DOI] [PubMed] [Google Scholar]
- Pringle, N. P. , Yu, W. P. , Howell, M. , Colvin, J. S. , Ornitz, D. M. , & Richardson, W. D. (2003). Fgfr3 expression by astrocytes and their precursors: Evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development, 130(1), 93–102. [DOI] [PubMed] [Google Scholar]
- Rosen, S. D. , & Lemjabbar‐Alaoui, H. (2010). Sulf‐2: An extracellular modulator of cell signaling and a cancer target candidate. Expert Opinion on Therapeutic Targets, 14, 935–949. 10.1517/14728222.2010.504718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch, D. H. , & Kriegstein, A. R. (2010). Developmental genetics of vertebrate glial‐cell specification. Nature, 468(7321), 214–222. 10.1038/nature09611 [DOI] [PubMed] [Google Scholar]
- Takebayashi, H. , Nabeshima, Y. , Yoshida, S. , Chisaka, O. , & Ikenaka, K. (2002). The basic helix‐loop‐helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Current Biology, 12(13), 1157–1163. [DOI] [PubMed] [Google Scholar]
- Tien, A. C. , Tsai, H. H. , Molofsky, A. V. , McMahon, M. , Foo, L. C. , Kaul, A. , … Rowitch, D. H. (2012). Regulated temporal‐spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development, 139(14), 2477–2487. 10.1242/dev.077214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touahri, Y. , Escalas, N. , Benazeraf, B. , Cochard, P. , Danesin, C. , & Soula, C. (2012). Sulfatase 1 promotes the motor neuron‐to‐oligodendrocyte fate switch by activating Shh signaling in Olig2 progenitors of the embryonic ventral spinal cord. The Journal of Neuroscience, 32(50), 18018–18034. 10.1523/JNEUROSCI.3553-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, T. H. , Shi, X. , Zaia, J. , & Ai, X. (2012). Heparan sulfate 6‐O‐endosulfatases (Sulfs) coordinate the Wnt signaling pathways to regulate myoblast fusion during skeletal muscle regeneration. The Journal of Biological Chemistry, 287(39), 32651–32664. 10.1074/jbc.M112.353243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, l. A. , McCutchen, M. D. , Bonner‐Fraser, M. , Fraser, S. E. , Bumm, L. A. , & McCauley, D. W. (2007). Fluorescent in situ hybridization employing the conventional NBT/BCIP chromogenic stain. BioTechniques, 42(6), 756–759. 10.2144/000112476 [DOI] [PubMed] [Google Scholar]
- Tsai, H. H. , Li, H. , Fuentealba, L. C. , Molofsky, A. V. , Taveira‐Marques, R. , Zhuang, H. , … Rowitch, D. H. (2012). Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science, 337(6092), 358–362. 10.1126/science.1222381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventéo, S. , Bourane, S. , Méchaly, I. , Sar, C. , Abdel Samad, O. , Puech, S. , … Carroll, P. (2012). Regulation of the Na,K‐ATPase gamma‐subunit FXYD2 by Runx1 and ret signaling in normal and injured non‐peptidergic nociceptive sensory neurons. PLoS ONE, 7(1), e29852. 10.1371/journal.pone.0029852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Chen, Y. , Zhu, Q. , Huang, H. , Teng, P. , Zheng, K. , … Qiu, M. (2014). Control of astrocyte progenitor specification, migration and maturation by Nkx6.1 homeodomain transcription factor. PLoS ONE, 9(10), e109171. 10.1371/journal.pone.0109171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , & Anderson, D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell, 109(1), 61–73. [DOI] [PubMed] [Google Scholar]
- Zhou, Q. , Wang, S. , & Anderson, D. J. (2000). Identification of a novel family of oligodendrocyte lineage‐specific basic helix‐loop‐helix transcription factors. Neuron, 25(2), 331–343. [DOI] [PubMed] [Google Scholar]


