Access to clinical data is critical for the advancement of translational research. However, the numerous regulations and policies that surround the use of clinical data, although critical to ensure patient privacy and protect against misuse, often present challenges to data access and sharing. In this article, we provide an overview of clinical data types and associated regulatory constraints and inferential limitations. We highlight several novel approaches that our team has developed for openly exposing clinical data.
Background
Recognizing the need to respect and protect patient privacy, numerous regulations have been established to govern the use of clinical data by researchers, including the federal Health Insurance Portability and Accountability Act of 1996 (HIPAA) and the European Union General Data Protection Regulation. Institution‐specific guidelines and governing bodies such as institutional review boards (IRBs) also address research involving patient data and other sensitive data available in electronic medical records (e.g., administrative data), in part as a result of concerns regarding the liability of healthcare providers and institutions.1, 2
The Biomedical Data Translator (Translator) program, funded by the National Center for Advancing Translational Sciences, aims to facilitate the transformation of basic science discoveries into clinically actionable knowledge and leverage clinical expertise to drive research innovations.3, 4 Access to clinical data is central to the vision of the program. Yet, the program's dedication to open science adds complexity to the regulatory, technical, and cultural challenges that already surround access to clinical data.
We review here the types of clinical data sets that can be derived from paper or electronic medical records, their applications and limitations, and their associated regulatory constraints, focusing primarily on compliance requirements mandated in the United States under HIPAA (Table 1 ). We briefly describe several clinical data types that are commonly employed in clinical and translational research, including fully identified clinical data, HIPAA‐limited clinical data, deidentified clinical data, and synthetic data. We highlight several novel approaches for openly exposing clinical data that we have developed as part of the Translator program, namely, HIPAA Safe Harbor Plus (HuSH+) clinical data, clinical profiles, Columbia Open Health Data (COHD), and the Integrated Clinical and Environmental Exposures Service (ICEES).
Table 1.
Clinical data types, regulatory access restrictions, and applications
| Clinical data type | Brief description | Regulatory access restrictions | Applications |
|---|---|---|---|
| Fully identified clinical data sets | Observational patient data derived from paper‐based or electronic medical records | IRB approval is required; an executed data use agreement is possibly requireda | Clinical interpretation and scientific inference and discovery |
| HIPAA‐limited clinical data sets | Observational patient data containing only a limited set of HIPAA‐defined PHI | IRB approval is required; an executed data use agreement is possibly requireda | Clinical interpretation and scientific inference and discovery, but with the understanding that certain data elements have been removed from the data and/or transformed |
| Deidentified clinical data sets | Observational patient data, but with all HIPAA‐defined PHI elements removed | IRB approval is not requiredb; IRB “Request for Determination of Human Subjects Research” is typically recommended; an executed data use agreement is possibly required | Clinical interpretation and scientific inference and discovery, but with the understanding that inferences regarding time and potentially other factors cannot be made |
| HuSH+ clinical data sets |
Observational patient data, fully compliant with HIPAA Safe Harbor, but unlike deidentified clinical data sets, HuSH+ clinical data sets have been altered such that (i) real patient identifiers (including geocodes) have been replaced with random patient identifiers and (ii) dates (including birth dates) have been shifted by a random number of days (maximum of ± 50 days), with all dates for a given patient shifted by the same number of days Data are derived from UNC Health Care System |
An executed data use agreement is requiredc | Clinical interpretation and scientific inference and discovery, but with the understanding that any inferences based on date/time and location (geocode) cannot be made with precision, and all other inferences must consider date/time and location as potentially hidden covariates |
| Clinical profiles |
Statistical profiles of disease and associated phenotypic presentation derived from observational patient data Data are derived from Johns Hopkins Medicine |
IRB approval is required to generate clinical profiles; no other restrictions apply | Clinical interpretation and scientific inference, but with the understanding that the data represent statistical profiles |
| Synthetic clinical data sets | Realistic, but not real, observational patient data generated statistically using population distributions of observational patient data | None | Feasibility assessments and algorithm validation; generation of clinical profiles |
| COHD |
Counts of observational clinical co‐occurrences (e.g., co‐occurrences of specific diagnoses and prescribed medications), as well as their relative frequency and observed–expected frequency ratio Data are derived from Columbia University Irving Medical Center |
None | Clinical interpretation and scientific inference, but with the understanding that the data are restricted to co‐occurrences |
| ICEES |
Patient‐level or visit‐level counts of observational patient data integrated at the patient and visit level with a variety of environmental exposures derived from multiple public data sources Data are derived from UNC Health Care System and a variety of public data sources on environmental exposures |
IRB approval is required to generate ICEES integrated feature tables; no other restrictions apply | Clinical interpretation and scientific inference, but with the understanding that the raw data have been transformed (e.g., binned or categorized) |
COHD, Columbia Open Health Data; HIPAA, Health Insurance Portability and Accountability Act; HuSH+, HIPAA Safe Harbor Plus; ICEES, Integrated Clinical and Environmental Exposures Service; IRB, institutional review board; PHI, protected health information; UNC, University of North Carolina.
aIndividual institutions may require a secure workspace for data access and use. bWhile HIPAA and IRB regulations do not apply, institutional approvals may be required. cHuSH+ clinical data sets were conceptualized and created by UNC as part of the National Center for Advancing Translational Sciences–funded Biomedical Data Translator program. The institution requires a fully executed data use agreement for access to the data.
Types of Clinical DATA SETS
Fully identified clinical data sets
Fully identified clinical data sets comprise observational patient data, including direct patient identifiers (i.e., protected health information (PHI)), as defined in the privacy rule issued under HIPAA. Access requires a specific research hypothesis, study approval by an IRB, a full or partial waiver of HIPAA‐informed consent, and typically a secure workspace. For investigators not affiliated with a specific institution, additional regulations and approvals may apply, including a data use agreement (DUA) with the provider institution. Fully identified clinical data sets may be used for clinical interpretation and scientific inference and discovery. However, as with all data sets but especially observational administrative data sets, issues of data quality and integrity must be taken into account when drawing conclusions.1
HIPAA‐limited clinical data sets
HIPAA‐limited clinical data sets comprise observational patient data with limited PHI: dates such as admission, discharge, service, and dates of birth and death; city, state, and five digits or more zip codes; and ages in years, months, days, or hours. HIPAA‐limited clinical data sets may be used or disclosed for purposes of research, public health, or healthcare operations without obtaining patient authorization or a waiver of HIPAA‐informed consent but with IRB approval and (in some cases) a fully executed DUA. HIPAA‐limited clinical data sets may be used for clinical interpretation and scientific inference and discovery but with the understanding that certain data elements have been removed from the data and/or transformed (e.g., age vs. birth date).
Deidentified clinical data sets
Deidentified clinical data sets comprise observational patient data from which all PHI elements have been removed. Access to deidentified clinical data sets does not require IRB approval, although an IRB Request for Determination of Human Subjects Research is advised. In addition, a fully executed DUA is sometimes required. Deidentified clinical data sets may be used for clinical interpretation and scientific inference and discovery but to a lesser extent than HIPAA‐limited clinical data sets because of the fact that key variables or covariates may have been removed from the data. For instance, dates are required to make inferences regarding seasonal patterns in clinical outcomes and correlations with natural disasters, system‐related issues such as protocol changes, and regulatory issues such as new black‐box warnings.
HuSH+ clinical data sets
HuSH+ clinical data sets were created by Translator team members as a hybrid deidentification approach that is completely compliant with HIPAA and provides restricted access to observational patient data from the UNC Health Care System. HuSH+ clinical data sets differ from deidentified clinical data sets in that (i) real patient identifiers (including geocodes) have been replaced with random patient identifiers and (ii) dates (including birth dates) have been shifted by a random number of days (maximum of ± 50 days), with all dates for a given patient shifted by the same number of days. Access to HuSH+ clinical data does not require IRB approval but does require a fully executed DUA per institutional mandate. HuSH+ clinical data sets may be used in a limited fashion for clinical interpretation and scientific inference and discovery. The main considerations are that any inferences based on date/time and location (geocode) cannot be made with precise accuracy or correlated with seasonal trends or specific events, and all other inferences must consider date/time and location as potentially hidden covariates.
Clinical profiles
Clinical profiles have been developed as part of the Translator program and represent statistical profiles of disease and associated phenotypic presentations derived from observational patient data from Johns Hopkins Medicine using the Health Level Seven International Fast Healthcare Interoperability Resources common data model. At present, clinical profiles include data on demographics, diagnoses, disease comorbidities, symptoms, medications, procedures, and laboratory measures. IRB approval is required to generate clinical profiles but once generated, clinical profiles can be openly shared. Institutional restrictions may apply, however. Clinical profiles can be used for clinical interpretation and scientific inference and discovery but with the understanding that they represent statistical summaries of patient populations and only indirectly represent patient‐level observations. Multiple computational tools and example output files are openly available for creating and using clinical profiles (see Supplemental Information on Clinical Profiles in Further Reading ).
Synthetic clinical data sets
Synthetic clinical data sets comprise realistic (but not real) data generated statistically by applying simulation techniques to population distributions of observational patient data. Synthetic clinical data sets can be openly shared. A publicly available example, the Synthetic Mass data set, was generated using the Synthea method5 to simulate patient‐level and population‐level data on patients who reside in the state of Massachusetts. A similar open effort is Simulacrum, which is based on observational patient data held by Public Health England's National Cancer Registration and Analysis Service. The data include realistic patient histories with clinically relevant patient encounters; as such, the data can be used for feasibility assessments and algorithm validation but not for clinical interpretation or scientific inference and discovery.
COHD
Translator team members have pioneered the use of clinical co‐occurrence tables as part of the COHD initiative.6 COHD provides open access to observational patient data from Columbia University Irving Medical Center in the form of co‐occurrence counts of pairs of concepts or clinical feature variables (e.g., medications and diagnoses), as well as their relative frequency and observed–expected frequency ratio. The data are publicly accessible via an open web interface or Application Programming Interface. Risks to patient privacy are mitigated by excluding rare features (counts ≤ 10) and perturbing the counts according to the Poisson distribution. The data can be used to derive insights into questions of clinical relevance and importance for translational research. For instance, an individual user may wish to know the frequency of asthma among African American patients (Figure 1 a). A search of the COHD service reveals that there are 11,716 African American patients with a diagnosis of asthma among 208,438 African American patients (5.62%). For comparison, a second search reveals that there are 29,913 white patients with a diagnosis of asthma among 601,167 white patients (4.98%).
Figure 1.
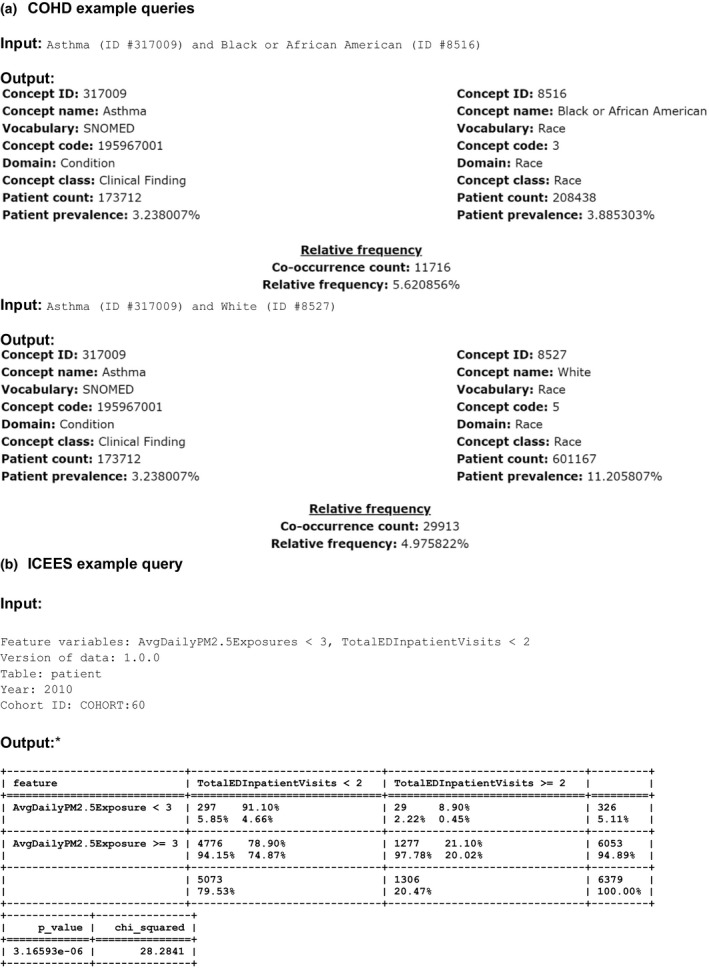
Example queries, including input parameters and output, for Columbia Open Health Data (COHD) (a) and the Integrated Clinical and Environmental Exposures Service (ICEES) (b). AvgDailyPM2.5Exposure = average daily patient exposure to PM2.5 (μg/m3) over a 1‐year study period; TotalEDInpatient Vists = total number of emergency department or inpatient visits for respiratory issues during a 1‐year study period. The study period shown here is for calendar year 2010. AvgDailyPM2.5Exposure <3 range: 1.58, 9.63 μg/m3; AvgDailyPM2.5Exposure ≥3 range: 9.63, 17.33 μg/m3. ID, identifier; PM2.5, airborne particulate matter ≤2.5 μm in diameter.
ICEES
ICEES was designed by Translator team members as a novel extension of COHD.7 Specifically, ICEES permits open access to observational patient data from the UNC Health Care System that have been integrated at the patient and visit level with environmental exposures data (e.g., airborne and roadway pollutants, socioeconomic factors) derived from multiple public sources. A complex data extraction and integration software pipeline has been developed to create ICEES integrated feature tables.8 The tables are generated using PHI (geocodes and dates), but the data are then binned or recoded and stripped of PHI. Thus, the ICEES pipeline must be executed under an approved IRB protocol, but subsequent steps are not subject to IRB regulation, and ICEES is publicly accessible via an Application Programming Interface. ICEES provides a number of functionalities for clinical interpretation and scientific inference and discovery. For example, Figure 1 b demonstrates that for COHORT:60 (African Americans with asthma‐like conditions in calendar year 2010), the percentage of patients with two or more annual emergency department or inpatient visits for respiratory issues is higher among patients with high average daily exposure to particulate matter ≤ 2.5 μm in diameter than among patients with low average daily exposure to particulate matter ≤ 2.5 μm in diameter (21.10% vs. 8.90%, P < 0.0001, N = 6,379), thus replicating published literature on the association between airborne pollutant exposures and asthma exacerbations.9 The data additionally suggest that African Americans with asthma‐like conditions have relatively high exposure to particulate matter, with ~ 95% of the cohort exposed to ≥ 9.63 μg/m3 average daily particulate matter ≤ 2.5 μm in diameter.
Clinical fingerprints
Although not a new clinical data type per se, Translator teams have been working to develop privacy‐preserving analytic approaches to visualize and compare patient data, including genomic data and clinical records in semistructured JavaScript Object Notation or eXtensible Markup Language formats. Genomic data typically consist of lists of variants relative to a reference allele sorted by position. Genome fingerprints capture the unique patterns generated by pairs of consecutive single‐nucleotide variants as patient‐level matrices or fingerprints.10 The correlation between two fingerprints reflects the degree of relatedness between two genomes. Clinical fingerprints similarly transform clinical records from the Fast Healthcare Interoperability Resources format into numerical vectors that greatly simplify their comparison. Translator team members are working to adapt this methodology for application to the ICEES integration pipeline and incorporation into the ICEES integrated feature tables.
Conclusion
In this article, we described various types of clinical data sets and associated inferential limitations and regulatory constraints, focusing primarily on compliance requirements mandated in the United States under HIPAA. We highlighted several novel approaches that we have developed as part of the Translator program to openly expose observational patient data, while respecting and protecting patient privacy. We recognize that each of these approaches retains a residual risk of patient reidentification; thus, we continue to work with experts in regulatory protections and computer security to ensure that those risks remain minimal. Although the Translator approaches are designed to be disease‐agnostic and generalizable, they were developed to comply with HIPAA and institutional guidelines; as such, our approaches may need to be modified prior to adoption elsewhere. Nonetheless, through these open services, we hope to accelerate clinical and translational science and foster biomedical discovery.
Funding
Support for this project was provided by the National Center for Advancing Translational Sciences, National Institutes of Health through the Biomedical Data Translator program (awards 1OT3TR002019, 1OT3TR002020, 1OT3TR002025, 1OT3TR002026, 1OT3TR002027, 1OT2TR002514, 1OT2TR002515, 1OT2TR002517, 1OT2TR002520, 1OT2TR002584) and the Clinical and Translational Sciences Award program (award UL1TR002489).
Conflict of Interest
All authors declared no competing interests for this work.
Supporting information
Clinical Data: Sources and Types, Regulatory Constraints, Applications.
Acknowledgments
The authors acknowledge and appreciate the contributions provided by the following individuals: Chris Bizon, Steve Cox, Ashok Krishnamurthy, Lisa Stillwell, and Hao Xu of the University of North Carolina Renaissance Computing Institute; James Champion of the North Carolina Translational and Clinical Sciences Institute; David B. Peden of the University of North Carolina School of Medicine; Sarav Arunachalam of the University of North Carolina Institute for the Environment; Max Robinson of the Institute for Systems Biology; and Stefano Rensi of Stanford University.
References
- 1. Harman, L.B. , Flite, C.A. & Bond, K. Electronic health records: privacy, confidentiality, and security. Virtual Mentor 14, 712–719 (2012). [DOI] [PubMed] [Google Scholar]
- 2. Na, L. , Yang, C. , Lo, C.C. , Zhao, F. , Fukuoka, Y. & Aswani, A. Feasibility of reidentifying individuals in large national physical activity data sets from which protected health information has been removed with use of machine learning. JAMA Network Open 1, e186040 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The Biomedical Data Translator Consortium. The Biomedical Data Translator program: conception, culture, and community. Clin. Transl. Sci. 12, 92–94 (2019). https://doi.org/(2018a10/1111/cts.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Biomedical Data Translator Consortium Toward a universal biomedical data translator. Clin. Transl. Sci. 12, 86–90 (2019). https://doi.org/(2018b10/1111/cts.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walonoski, J. et al Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J. Am. Med. Inform. Assoc. 25, 230–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ta, C. , Dumontier, M. , Hripcsak, G. , Tatonetti, N. & Weng, C. Columbia Open Health Data, clinical concept prevalence and co‐occurrence from electronic health records. Sci. Data 5, 180273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fecho, K. et al A novel approach for exposing and sharing clinical data: the Translator Integrated Clinical and Environmental Exposures Service. J. Am. Med. Inform. Assoc. (in press). 10.1093/jamia/ocz042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pfaff, E.R. et al All roads lead to FHIR: an extensible clinical data conversion pipeline. American Medical Informatics Association 2019 Informatics Summit, San Francisco, CA, March 25–28, 2019. Abstract.
- 9. Mirabelli, M.C. , Vaidyanathan, A. , Flanders, W.D. , Qin, X. & Garbe, P. Outdoor PM2.5, ambient air temperature, and asthma symptoms in the past 14 days among adults with active asthma. Environ. Health Perspect. 124, 1882–1890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glusman, G. , Mauldin, D.E. , Hood, L.E. & Robinson, M. Ultrafast comparison of personal genomes via precomputed genome fingerprints. Front. Genet. 8, 136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical Data: Sources and Types, Regulatory Constraints, Applications.


