Systems pharmacology models are having an increasing impact on pharmaceutical research and development from preclinical through postapproval phases, including use in regulatory interactions. Given the wide diversity among the models and the contexts of use, a common but flexible strategy for model assessment is needed to enable the appropriate interpretation of model‐based results. We present an approach to evaluate these models and discuss how it can be customized to available data and intended application.
Model Assessment Considerations
A wide range of modeling approaches, including empirical, mechanistic pharmacokinetic/pharmacodynamic, and quantitative systems pharmacology (QSP), can be applied toward pharmaceutical research and development. The evaluation of these models is critical to understanding their strengths/limitations and interpreting model results. Assessment typically involves evaluating fits to observed data and testing predictive capabilities where possible. Pharmacokinetic/pharmacodynamic models are routinely evaluated by goodness‐of‐fit plots, predictive checks, and external validations focused on capturing output data under the premise of parsimony.1 In contrast, QSP models focus on the representation of underlying biological systems and address questions that involve exploration of mechanism and extrapolation to novel scenarios. QSP models are thus frequently and by necessity complex and underconstrained, leading to confusion around how QSP models can be appropriately evaluated.2 Previous QSP tutorials have presented considerations in planning, developing, qualifying, and applying systems models.3, 4 Here, we focus on model assessment, defining four major assessment areas (biology, implementation, simulation, and robustness), and suggest activities that can be customized based on the context of the work, mapping these efforts to previously presented QSP workflow stages and qualification criteria (Table 1). We illustrate the tailored application of the assessment approach with two published models of cancer signaling.
Table 1.
Quantitative systems pharmacology model assessment in four key areas—biology, implementation, simulation, and robustness
| Assessment area | Workflow stage4 | MQM criterion3 | Assessment approach | ||
|---|---|---|---|---|---|
| Considerations | Specific assessments | Reporting | |||
| Biology | 1–2 | 1–2 | Biological relevance and plausibility |
|
|
| 1–2 | 1–2 | Main hypotheses and assumptions | |||
| 1–2 | 3–6 | Alternate hypotheses | |||
| Implementation | 3 | 2, 7–8 | Technical QA/QC |
|
|
| 3–4 | 2, 5–6 | Model structure and parameter ranges |
|
|
|
| 4 | 7 | Sensitivities and behaviors |
|
|
|
| Simulations | 4 | 7–8 | Reproduction of behaviors (calibration/training) |
|
|
| 4–5 | 8 | Prediction of behaviors (validation/testing) |
|
|
|
| Robustness | 5–6 | 3–8 | Evaluation of variability and uncertainty |
|
|
MQM, model qualification method; Assessment considerations in each key area are outlined and mapped to published guidances3, 4 alongside specific assessments of interest in quantitative systems pharmacology modeling and recommendations for reporting.
MQM, model qualification method; QA, quality assurance; QC, quality check; SA, sensitivity analysis; VPC, visual predictive check.
Biological Relevance
Assessment of the biological relevance is of critical importance in QSP, where utility requires that the biology included is appropriate to address the problem at hand and reflects relevant knowledge, data, and literature. Thus, literature support and input from biological and clinical experts are valuable in assessment. Mechanisms, hypotheses, behaviors, and phenotypes of interest should be articulated to ensure the adequacy of biological scope. QSP models typically include the representation of targets, drugs, biomarkers, and outcomes of interest. Although the scale, breadth, and depth of biological scope differ greatly among applications, a model should minimally include sufficient biological pathways to connect each target or drug to the relevant biomarkers and outcomes, potentially via intermediaries.
Model Implementation
Assessment of the model implementation involves evaluation of the mathematical formulation and quality checks on the accuracy and veracity of the model, its mathematical structure, the parameters, and their influence on model simulations. The choice of formalism must be consistent with the project goals. Ensuring that the implementation is technically accurate (e.g., correct coding, unit consistency) and appropriate for the mechanisms represented is also essential and may be required in regulatory submission. Once structure and implementation are confirmed, dynamical systems analyses can be used to explore inherent model dynamics and corresponding parameter ranges to assess their relevance. Although structural identifiability of model parameters can be difficult to assess or ensure, the impact of the parameters on the ability to reproduce critical behaviors can be determined via sensitivity analyses that determine how uncertainty in and variability around a given parameter set (local) or throughout parameter space (global) influence model outputs. Other approaches, such as Monte Carlo simulation, that explore model behavior under different parameterizations can also inform this question and ensure consistency with expectations. These methods are used to confirm the ability of the model to generate distinct qualitative features or phenotypes (e.g., ranges of treatment response, different dynamical signaling features)5 or to highlight needed revision of the model biology or mathematics.
Simulation Results
Assessment of simulation results gauges the qualitative and quantitative plausibility of model simulations with respect to data and biological understanding. Generally, during modeling, parameters are estimated such that model or subsystem outputs match a set of qualitative and/or quantitative criteria. Confirmation that the model satisfactorily recapitulates this training/calibration data is one critical step. However, confidence in model predictions further requires testing the model's ability to prospectively or retrospectively predict data or behaviors not used in model calibration. Ideally, these validation/test experiments should be orthogonal to the calibration data yet fall within the scope of the biology represented. When data are limited, alternative approaches such as leave‐one‐out cross‐validation or iterative calibration, validation, and updating can be considered. Sensitivity analysis that demonstrates appropriate responses to parameter modification can also provide confidence in simulated or predicted behaviors. Where validation against data or other biological knowledge is not demonstrated, prospective simulations should be considered explorations, hypotheses, or “potential outcomes” rather than predictions. In such scenarios, modeling can still provide value by increasing mechanistic understanding, highlighting potential outcomes, and identifying or reducing uncertainties and risks in pharmaceutical research and development.
Robustness of Results
Assessment of the robustness of results ties in many aspects of biology, implementation, and simulations to increase confidence in model‐based insight and predictions, specifically their robustness to biological variability and uncertainty (alternate hypotheses or quantitative differences). This assessment focuses on the extent to which the impact of variability or uncertainty in topology or parameters has been considered in predictions and the extent to which variability in data is captured. This can be done through explicit simulation of alternate parameterizations (“virtual subjects”)4 or collections thereof that cover input and/or output uncertainty and variability.4, 5 Note that exploration of parameter uncertainty helps address concerns related to parameter identifiabilty.
Context Dependence
The application of the model and the availability of data determine how and to what extent different assessment approaches are appropriate. Some applications (e.g., clinical trial design) require more robust assessment, whereas a more flexible approach may be sufficient for mechanistic exploration. Decisions with significant safety or financial implications also require more rigorous assessment, as do efforts where modeling is a primary driver for a decision, without parallel evidence. Abundant data enable separate calibration and validation data sets, whereas limited data may necessitate other approaches to testing the model and corresponding caution in interpretation of results. In addition, different mathematical formulations require different mathematical assessment techniques. Although context influences how and to what extent each major assessment area is addressed, all areas should be considered and discussed. Figure 1 shows how contextual considerations can influence the degree of rigor required in model assessment and indicates the different context surrounding example models of cancer signaling pathways.6, 7, 8, 9 Here, we discuss how context influences model assessment for two of these studies.6, 7
Figure 1.
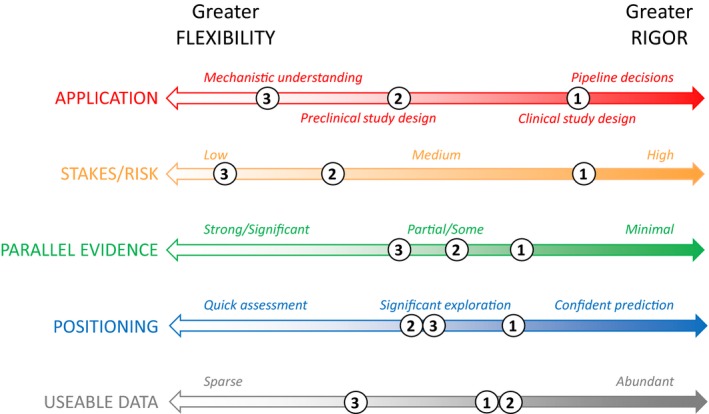
Context‐dependent considerations in assessment of Quantitative Systems Pharmacology models. The rigor required or flexibility acceptable in model assessment is influenced by context‐specific considerations, including: intended application of the model; financial, safety, or other risks involved; parallel evidence supporting model‐based recommendations; intended positioning of modeling work; and the nature and extent of the data available for the modeling effort. As illustration, we roughly indicate on the axes the different context of each of the following modeling efforts involving the mitogen‐activated protein kinase (MAPK) signaling pathway: (1) Kirouac et al.6 used preclinical and limited clinical data to support clinical strategy with potentially significant consequences and only preclinical parallel evidence on the drug/combo efficacy; (2) Eduati et al.7 used rich preclinical data to explore signaling diversity and resistance mechanisms and propose hypotheses for in vitro testing; (3) different dynamical models8, 9 have aimed more generally to understand the implications of mechanistic signaling topology and feedback on the pathway behavior.
Context
Many cancers display alterations in mitogen activated protein kinase (MAPK), PI3K, and other intracellular signaling pathways that promote tumor growth. Briefly, the canonical MAPK pathway proceeds from receptor engagement through RAS, RAF, MEK, and ERK phosphorylation to downstream effects on cell growth, survival, and protein translation. Kirouac et al.6 modeled the MAPK pathway based on rich preclinical and limited clinical data to explore the potential utility of a novel ERK inhibitor, especially in the treatment of RAF‐mutant BRAFV600E colorectal cancer, to support clinical strategy and ongoing phase I trials. Eduati et al.7 modeled multiple signaling pathways, including MAPK, using in vitro data from a broad set of colorectal cancer lines to investigate diversity in cellular signaling and mechanisms of resistance and to suggest sensitivities for possible therapeutic investigation.
Biology
In each case, the biology represented is appropriate for the goal. Kirouac et al.6 focus on the MAPK pathway, including targets and mutations of interest, hypothesized resistance mechanisms (receptor redundancy, bypass signaling, feedbacks), and a mechanistic link from signaling to cell/tumor growth. In contrast, Eduati et al.7 address a broader set of signaling pathways and interactions required to identify resistance mechanisms in different cell lines.
Implementation
Kirouac et al.6 use an ordinary diffferential equation formulation appropriate for signaling and growth dynamics, whereas the logic–ordinary diffferential equation implementation of signaling used by Eduati et al.7 facilitates exploration of differential signaling among cells lines while an elastic net model relates signaling model parameters to in vitro cell survival. Full model specifications are provided in each to enable thorough mathematical assessment. Although neither study presents formal structural or dynamical analysis, the Kirouac et al.6 model is shown to capture critical dynamic data, such as feedback‐driven ERK rebound. With respect to parameter sensitivity, both studies verify reasonable model sensitivities by demonstrating appropriate responses of diverse cells/tumors to different perturbations (drugs and stimuli), and Eduati et al.7 further analyzes which parameters are most correlated with survival.
Simulation
Both models are calibrated to rich data sets obtained using diverse preclinical models and treatments. Both studies include preclinical validation: Kirouac et al.6 by (retrospective) prediction of growth response for multiple drug combinations and preclinical models and Eduati et al.7 by (prospective) in vitro verification of a novel model‐predicted drug combination. To support clinical application, Kirouac et al.6 capture prior clinical trial data in a virtual population and validate quantitative predictions for ERK inhibitor efficacy against emerging clinical results. In contrast, clinical simulation is not the focus of the Eduati et al.7 effort, and thus clinical validation is neither required nor included; instead, they cite ongoing trials as evidence for the relevance of their predictions.
Robustness
Variability is explored in each study using different parameterizations for different cell lines/tumors. Eduati et al.7 emphasize variability in preclinical signaling pathway usage and graphically illustrate the inferred differences. Kirouac et al.6 focus on representing the diversity required to predict clinical response distributions and present both ranges of parameters sampled and the resulting virtual population output variability, although they do not report the final parameter ranges retained in the virtual population. Ultimately, the approaches taken in each study enabled the exploration of differential responsiveness and resistance in the corresponding contexts. Numerous other modeling studies (from Huang and Ferrell8 to Kochańczyk et al.9) have investigated MAPK pathway topology, signal propagation, feedback and crosstalk, and dynamical features. Such studies often perform detailed dynamical and parameter sensitivity analyses, but did not always include nor require extensive model calibration and validation given the exploratory intent of the studies, further illustrating how goals and context help influence assessment strategy.
Conclusion
We have proposed a customizable approach for QSP model assessment consistent with previous guidances and tutorials. A technical review of QSP efforts could include an assessment summary describing context of use and listing approaches in each of the four major areas, noting justification and limitations. This could accompany detailed reporting of assessment and modeling results as outlined in Table 1 and in a recent publication from the UK QSP Network.10 This uniform assessment approach, which allows for customization to context of use, could thus support communication and review, including regulatory interactions. Publication and transparent model sharing would further promote assessment and use by the community, as evidenced by the recently described reuse and utilization of a published QSP model by the US Food and Drug Administration.10 Moving forward, common language and libraries of visualizations, analysis scripts or tools, and metrics would facilitate the execution, communication, and review of QSP efforts as it has in the field of pharmacometrics.
Funding
No funding was received for this work.
Conflict of Interest
S.R. is an employee of Genentech Inc. J.R.C. is an employee of Eli Lilly and Co. C.M.F. is an employee of Rosa & Co LLC. C.J.T. is an employee of Bristol‐Myers Squibb Co.
References
- 1. Upton, R. & Mould, D. Basic concepts in population modeling, simulation, and model‐based drug development: part 3‐introduction to pharmacodynamics modeling methods. CPT Pharmacometrics Syst. Pharmacol. 3, e88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agoram, B. Evaluating systems pharmacology models is different from evaluating standard pharmacokinetic‐pharmacodynamic models. CPT Pharmacometrics Syst. Pharmacol. 3, e101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friedrich, C.M. A model qualification method for mechanistic physiological QSP models to support model‐informed drug development. CPT Pharmacometrics Syst. Pharmacol. 5, 43–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gadkar, K. et al A six‐stage workflow for robust application of systems pharmacology. CPT Pharmacometrics Syst. Pharmacol. 5, 235–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmidt, B.J. et al Alternate virtual populations elucidate the type I interferon signature predictive of the response to rituximab in rheumatoid arthritis. BMC Bioinformatics 14, 221–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirouac, D.C. et al Clinical responses to ERK inhibition in BRAFV600E‐mutant colorectal cancer predicted using a computational model. NPJ Syst. Biol. Appl. 3, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eduati, F. et al Drug resistance mechanisms in colorectal cancer dissected with cell type‐specific dynamic logic models. Cancer Res. 77, 3364–3375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang, C.Y. & Ferrell, J.E. Jr. Ultrasensitivity in the mitogen‐activated protein kinase cascade. Proc. Natl Acad. Sci. USA 93, 10078–10083 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kochańczyk, M. et al Relaxation oscillations and hierarchy of feedbacks in MAPK signaling. Sci. Rep. 7, 38244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cucurull‐Sanchez, L. et al Best practices to maximise the use and re‐use of QSP models Recommendations from the UK QSP Network. CPT Pharmacometrics Syst. Pharmacol. 10.1002/psp4.12381. [e‐pub ahead of print] . [DOI] [Google Scholar]


