Abstract
Background
The bHLH (basic helix-loop-helix) transcription factor is one of the largest families of transcription factors in plants, containing a large number of members with diverse functions. Chinese jujube (Ziziphus jujuba Mill.) is the species with the highest economic value in the family Rhamnaceae. However, the characteristics of the bHLH family in the jujube genome are still unclear. Hence, ZjbHLHs were first searched at a genome-wide level, their expression levels under various conditions were investigated systematically, and their protein-protein interaction networks were predicted.
Results
We identified 92 ZjbHLHs in the jujube genome, and these genes were classified into 16 classes according to bHLH domains. Ten ZjbHLHs with atypical bHLH domains were found. Seventy ZjbHLHs were mapped to but not evenly distributed on 12 pseudo- chromosomes. The domain sequences among ZjbHLHs were highly conserved, and their conserved residues were also identified. The tissue-specific expression of 37 ZjbHLH genes in jujube and wild jujube showed diverse patterns, revealing that these genes likely perform multiple functions. Many ZjbHLH genes were screened and found to be involved in flower and fruit development, especially in earlier developmental stages. A few genes responsive to phytoplasma invasion were also verified. Based on protein-protein interaction prediction and homology comparison, protein-protein interaction networks composed of 92 ZjbHLHs were also established.
Conclusions
This study provides a comprehensive bioinformatics analysis of 92 identified ZjbHLH genes. We explored their expression patterns in various tissues, the flowering process, and fruit ripening and under phytoplasma stress. The protein-protein interaction networks of ZjbHLHs provide valuable clues toward further studies of their biological functions.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5936-2) contains supplementary material, which is available to authorized users.
Keywords: ZjbHLHs, Chinese jujube, Tissue-specific expression, Flower and fruit development, Phytoplasma, Protein-protein interaction
Background
Transcription factors (TFs) are important regulatory factors in eukaryotes that interact with cis-elements to regulate the expression of specific genes in response to environmental stresses [1]. According to the sequence of arginine and lysine residues in the DNA binding region, the TFs of higher plants can be divided into four categories: zinc finger [2], helix-turn-helix (HTH) [3], basic leucine zipper (bZIP) [4], and helix-loop-helix (HLH) [5].
The basic helix-loop-helix (bHLH) family is one of the largest TF families in plants [6]. The bHLH domain is composed of approximately 50–60 conserved amino acid sequences and contains two functional regions: one is the basic amino acid region with a length of approximately 15 amino acids at the N-terminal, and the other is the HLH region at the C-terminal [7]. The basic region of approximately 15 amino acids is responsible for binding to the E-box (CANNTG) element. Studies have shown that two helices of the same transcription factor or different transcription factors interact to form homologous or heterologous dimers, which can combine with different parts of the gene promoter to regulate the target gene [8]. Moreover, some atypical bHLHs with a less basic region that is critical for DNA binding were further identified and characterized in Arabidopsis [9–11].
As an increasing number of genome sequences are being released, a variety of bHLH superfamily genes have been identified and analyzed in a wide range of plant species, such as Arabidopsis [12], pear [13], peach [1], apple [14], grape [15] and cotton [16]. Furthermore, the functions of many bHLH proteins in plants have been studied in detail. bHLHs play various roles in plant development [16–18], signal transduction [8], tolerance [19–21] and secondary metabolite production [22]. Identification of bHLHs at a genome-wide level is the first step toward further studies into their biological functions. However, the bHLH transcription factors in Chinese jujube (Ziziphus jujuba Mill.) have not been reported before and the related functions of ZjbHLHs are still unknown.
Chinese jujube is a species with high economic value in the family Rhamnaceae and is also one of the most representative national fruit trees in China. Wild jujube, the wild relative species of Chinese jujube, usually has smaller trees and fruits than the Chinese jujube. Both jujube and wild jujube trees have many agronomic advantages, such as early fruit production, long flowering season and high tolerance to various biotic and abiotic stresses. And the bHLH gene family has a variety of functions such as flower and fruit development and is necessary for the normal growth and development in many plants [8, 16–22]. Compared with other TFs, bHLHs are involved in more reaction pathways and acted as some co-regulators on gene expression together with many other proteins. Therefore, we want to figure out the functions of this gene family in jujube. The jujube genome database [23, 24] provided the possibilities and resources for searching the crucial gene families related to its biological characteristics at the genome level. For the wide and diverse biological roles of bHLHs in plant development, the gene number, classification, and gene structure of this family in the jujube genome and their expression under various conditions in jujube and wild jujube were systematically analyzed in this study, and the protein-protein interaction networks were also predicted. The results provide valuable clues for further revealing the functions of this family in jujube growth and development.
Results
Identification of ZjbHLHs
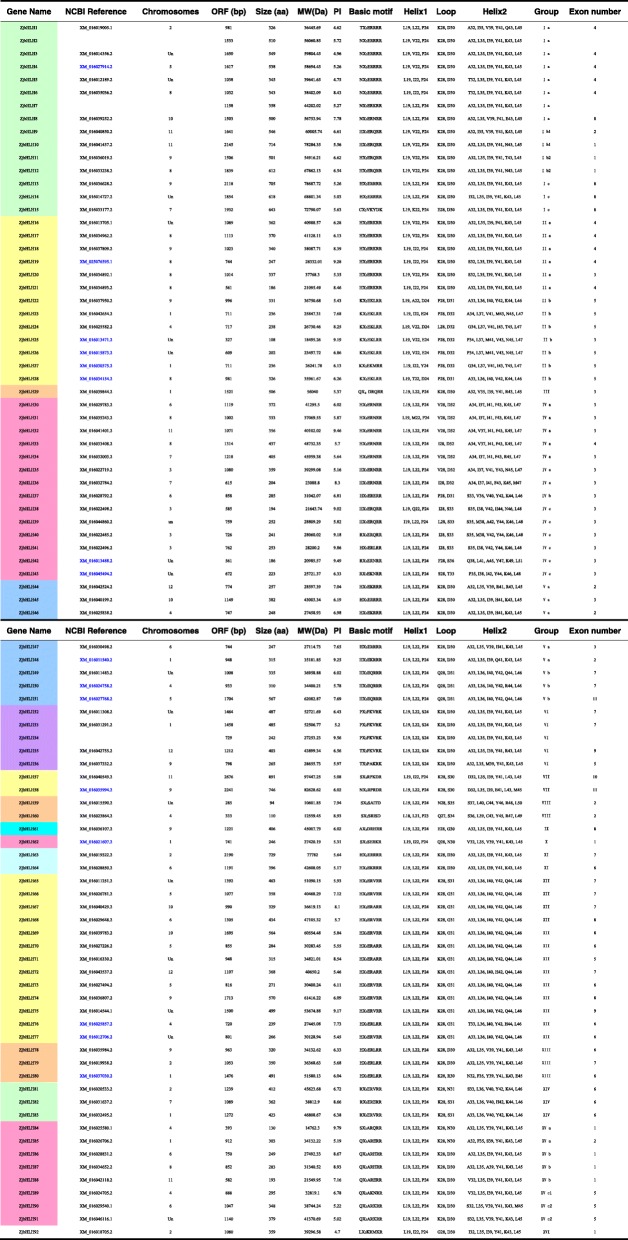
A total of 92 nonredundant putative bHLH transcripts (Table 1) were identified in the jujube genome sequence (https://www.ncbi.nlm.nih.gov/genome/?term=jujube) [23]. To verify the reliability of each sequence, 92 protein sequences were analyzed using the online CD-search and SMART tools, and 82 bHLH proteins were found to have a typical bHLH structure except for the 10 bHLH proteins with an atypical bHLH domain (Additional file 1: Figure S1). They were named from ZjbHLH1 to ZjbHLH92 according to their gene structure and motifs, and the ZjbHLH2, 7, and 54 sequences were new genes with no information in NCBI. The ORF length for ZjbHLH genes ranged from 285 bp (ZjbHLH59) to 2676 bp (ZjbHLH57), and the genes encoded proteins ranging from 94 (ZjbHLH59) to 891 (ZjbHLH57) amino acids (aa) in length, with predicted pIs ranging from 4.62 (ZjbHLH1) to 9.86 (ZjbHLH41). The proteins with an isoelectric point of less than 7 accounted for 66% of the total, which means that most of the ZjbHLH genes were weakly acidic. Based on their physical and chemical properties, the family members have different characteristics, indicating that they likely have multiple functions.
Table 1.
The information of bHLH gene family in Chinese jujube
Previous genome evolution studies showed that Chinese jujube is closely related to species of the Rosaceae family [23], so the number of bHLHs from three Rosaceae species (apple, pear and peach), grape, cotton and Arabidopsis were compared with ZjbHLHs (Additional file 2: Table S1). There were similar gene numbers in the bHLH family of jujube and peach, and this result was consistent with a previous study on the MADS-box family [25]. Compared with the number of bHLH genes found in other plant species, the number of bHLH genes found in jujube and peach was lower. The gene numbers may be related to evolutionary differences, genome replication, or the genome size of these plants [1].
Phylogenetic analysis of ZjbHLHs
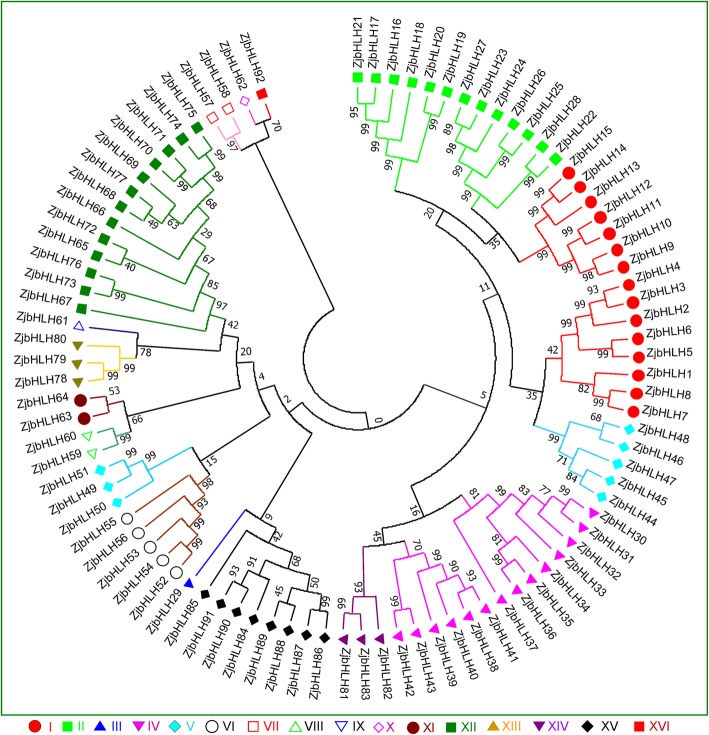
A phylogenetic tree of the ZjbHLH proteins was constructed by aligning multiple domain sequences (Fig. 1). A phylogenetic tree of bHLH genes of Arabidopsis and jujube was established, and ZjbHLHs were divided into 16 categories (Fig. 1). And the bHLH phylogenetic trees of peach [1] and jujube (Additional file 3: Figure S2) was constructed, which further verified the above classification result. The bHLH genes of the six species (jujube, apple, pear, peach, grape, cotton and Arabidopsis) showed a mixed pattern on the evolutionary tree (Additional file 4: Figure S3). These results indicated that the bHLH genes were present before the divergence of various plant species and then expanded in each species independently.
Fig. 1.
The phylogenetic analysis of ZjbHLHs. The NJ tree was constructed from the protein sequences of ZjbHLHs using MEGA7 with 1000 bootstrap copies
Multiple sequence alignment and conserved motifs in ZjbHLHs
Comparison of multiple sequences showed that most of the ZjbHLH genes had the same conserved domain structure except for the VI, VII and VIII ZjbHLH genes (Fig. 2). The domain sequences in the ZjbHLH gene family were highly conserved (Additional file 1: Figure S1). Five residues (His-1, Glu-5, Arg-6, Arg-8, and Arg-9), three residues (Leu-19, Leu-22, and Pro-24), two residues (Lys-28 and Asp-30) and six residues (Ala-32, Leu-35, Ile-39, Tyr-41, Lys-43, and Leu-45) made up the basic region, the first helix region, the loop region and the second helix region, respectively. There were 6 motifs among ZjbHLHs, and proteins in the same group had similar numbers and types of motifs (Additional file 5: Figure S4). The bHLH domains Motif 1 and Motif 2 were highly conserved among the 92 proteins (Additional file 6: Figure S5), and only ten (ZjbHLH15, ZjbHLH52~ZjbHLH60) of them contained variations. ZjbHLH59 and ZjbHLH60 were identified as atypical bHLH genes by the Conserved Domain Search Service (CD Search) [26].
Fig. 2.
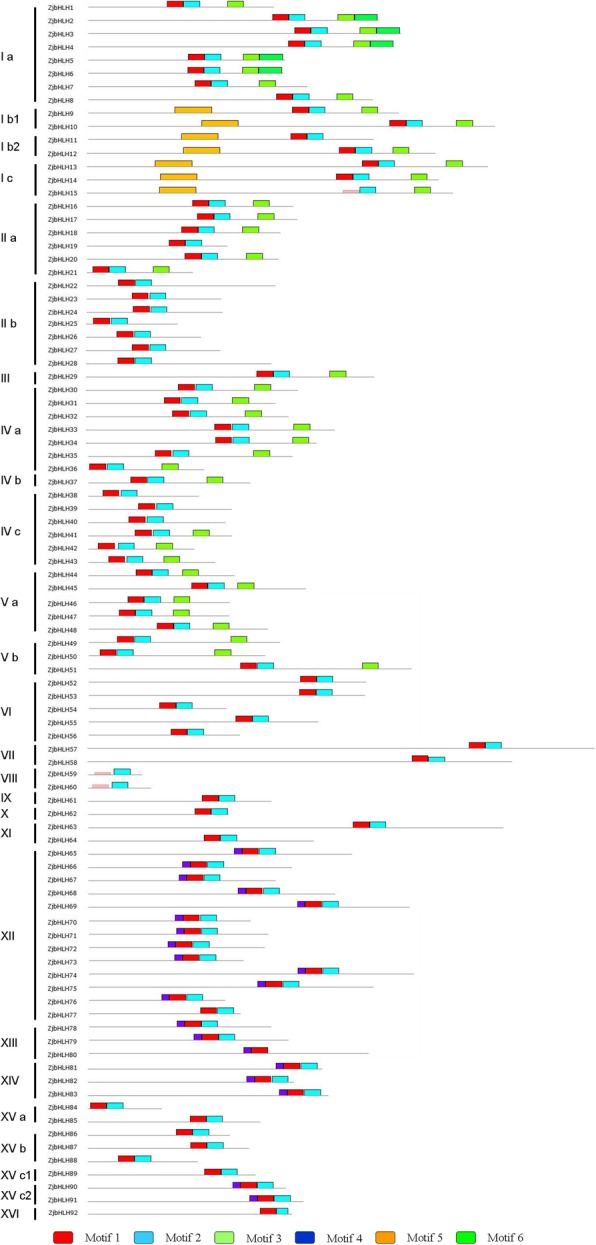
Conserved motifs of ZjbHLH proteins. The motifs in ZjbHLHs were identified by using Multiple Em for Motif Elicitation (MEME). In these ZjbHLH proteins, six conserved motifs were identified and displayed in different colors
The chromosomal location and gene structure of ZjbHLHs
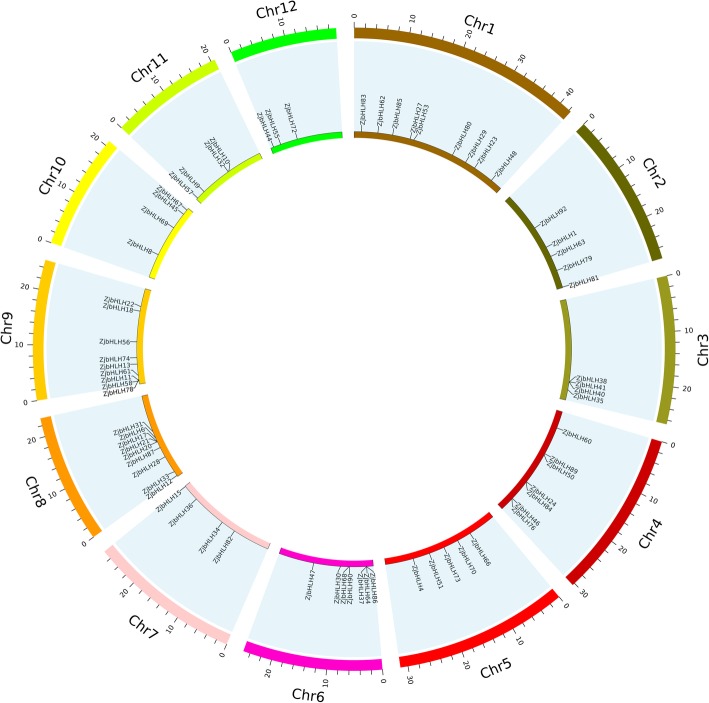
Of the 92 ZjbHLH genes, 70 were mapped to 12 pseudo-chromosomes in the jujube genome (Fig. 3), 17 genes were located on 12 scaffolds, and 5 genes were uncommented. ZjbHLHs were not evenly distributed across the 12 chromosomes. Nine ZjbHLH genes (9.8%) were on Chr. 1, 8, and 9, and 7 ZjbHLHs (7.6%) were located on Chr. 4 and 6. Furthermore, some ZjbHLHs concentrated on part of the chromosome, and some relatively high-density bHLH genes were observed in some chromosomal regions. Some genes were tightly packed into clusters to form tandem repeats (ZjbHLH27 and 53; ZjbHLH38, 40 and 41; ZjbHLH64 and 37; ZjbHLH17, 20 and 21). A previous study analyzed repeated events in rice and Arabidopsis [27], indicating that some bHLH subfamily members are most likely derived from repetitive events.
Fig. 3.
The chromosomal location of 70 ZjbHLH genes. Genes are mapped to jujube chromosomes by the Circos tool. The chromosomes of jujube are arranged in a circle
Additionally, the gene structure was highly conserved within each group (Fig. 4), except for the 3 uncommented ZjbHLHs (2, 7 and 54). We found that Group VI, VII, IX, XI, XII, XIII, and XIV genes contained more introns and were more complicated than genes in the other groups (Fig. 4).
Fig. 4.
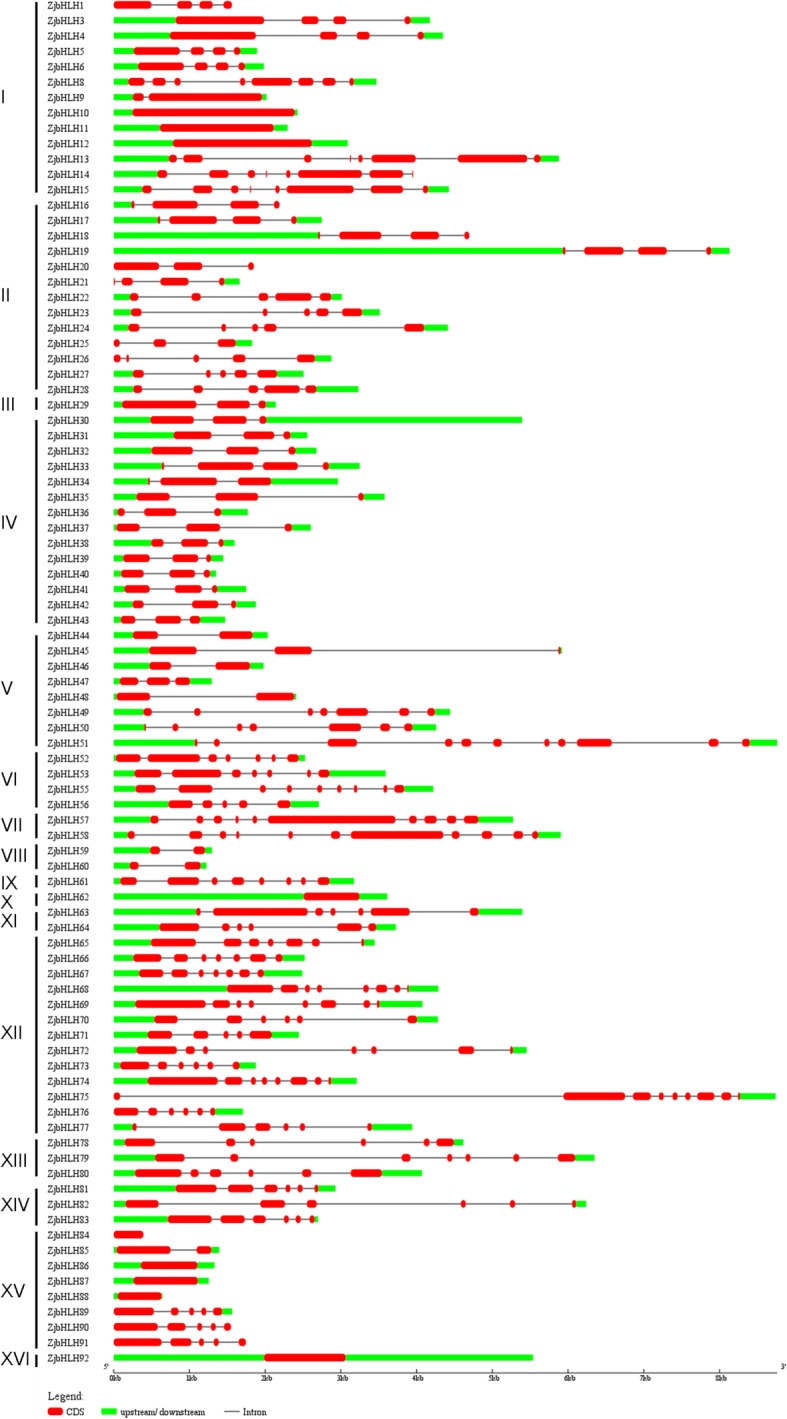
The gene structure of ZjbHLHs in Chinese jujube. Introns and exons are represented by black lines and red boxes respectively and upstream/downstream are represented by green boxes
Expression patterns of ZjbHLHs in various tissues/organs
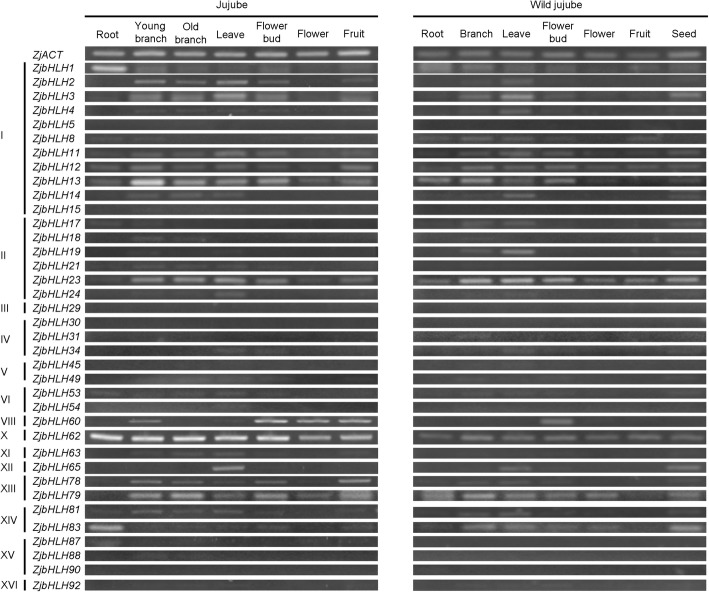
To explore the tissue-specific expression of ZjbHLHs, their expression patterns were determined in various tissues by semiquantitative PCR. The expression patterns of most examined ZjbHLHs were similar in jujube and wild jujube (Fig. 5), except for 8 ZjbHLHs (17, 21, 54, 60, 63, 79, 83, and 87). Some genes were mainly expressed in vegetative organs (ZjbHLH1, 2, 3, 4, 11, 63, 65, 81, 83, and 87) or reproductive organs (ZjbHLH60). In particular, ZibHLH62 was stably expressed in various organs of both jujube and wild jujube and can be used as a housekeeping gene. These results showed that most of the ZjbHLHs had diverse tissue-specific expression patterns, indicating that they play multiple roles in various organs.
Fig. 5.
Expression patterns of 37 ZjbHLH genes in seven tissues of jujube and wild type jujube by RT-PCR. ZjACT was used as an internal control. Left: jujube, from left to right: root, young branch, old branch, leaf, flower bud, flower, and fruit. Right: wild jujube, from left to right: root, branch, leaf, flower bud, flower, fruit and seed
In addition, the expression of ZjbHLH8 and 19 genes in the branches and leaves of jujube was significantly weaker than that in wild jujube. This differential expression indicated that some ZjbHLHs may have different functions between jujube and wild jujube.
ZjbHLHs involved in flower and fruit development
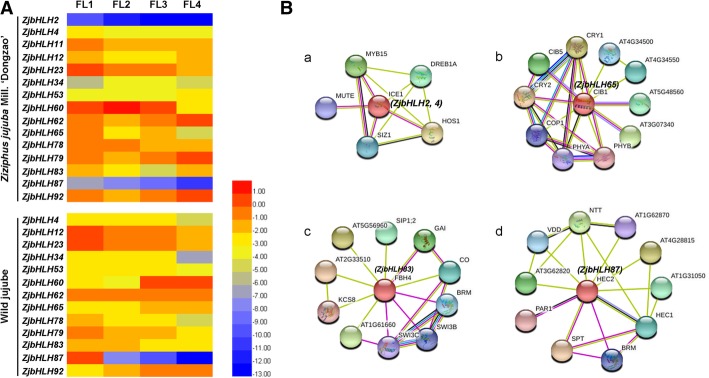
Based on the tissue-specific expression, the expression of ZjbHLHs was further detected at four floral developmental stages (Fig. 6a). Among them, ZjbHLH62 and ZjbHLH53 were expressed stably at the four stages in jujube and wild jujube. The expression levels of ZjbHLH4, 12, 23, 78 and 87 genes decreased gradually with flower development, and ZjbHLH92 had a high expression level at the later stages in both jujube and wild jujube. It is remarkable that the expressions of ZjbHLH4, 12, 34, 60, 62, 78, 79 and 83 genes in four stages showed opposite trends between jujube and wild jujube and four genes (ZjbHLH4, 12, 60 and 78) showing significantly different expression were screened out (Additional file 9: Figure S6), indicating that they may perform different functions during flower development in jujube and its wild-type species. Through protein-protein interaction prediction and homology comparison, it is predicted that ZjbHLH2, 4, 65, 83, and 87 genes have crucial functions during flower development (Fig. 6b).
Fig. 6.
a Heat maps of the relative expression of ZjbHLH genes during flower development. FL1, bud emergence stage; FL2, inflorescence emergence stage; FL3, yellow bud stage; FL4, petal spread stage. Scaled log2 expression values based on qRT-PCR data are shown from blue to red, indicating low to high expression. b The protein-protein interaction analysis of four ZjbHLHs by STRING database
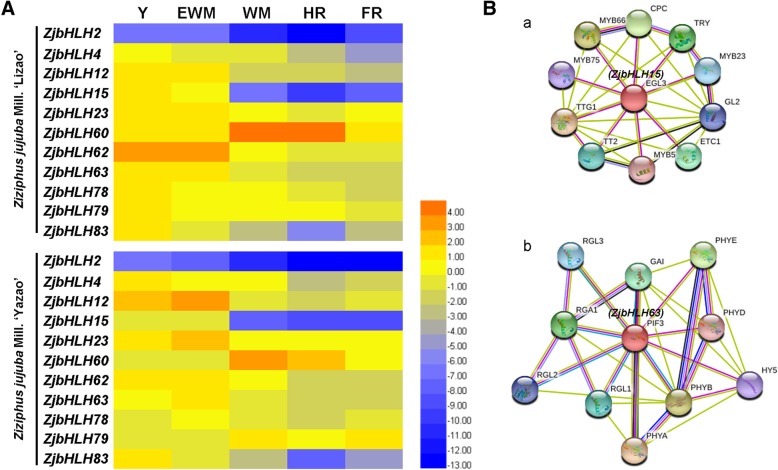
During jujube fruit development, some genes (ZjbHLH2, 4, 12, 15, 23, 62, 63, 78 and 83) were highly expressed at the first two stages and then significantly decreased at later stages (Fig. 7a). However, ZjbHLH60 was mainly expressed at the late stages. The protein interaction prediction and homology comparison also indicated that ZjbHLH15 and 63 might play important roles in fruit development (Fig. 7b). The above results indicated that some ZjbHLHs were truly involved in jujube flower and fruit development.
Fig. 7.
a Heat maps of the relative expression of ZjbHLH genes during fruit ripening in ‘Lizao’ and ‘Yazao’. Y, young fruit; EWM, early white mature fruit; WM, white mature fruit; HR, half-red fruit; FR, full red fruit. Scaled log2 expression values based on qRT-PCR data are shown from blue to red, indicating low to high expression. b The protein-protein interaction analysis of two ZjbHLHs by STRING database
ZjbHLHs participated in jujube-phytoplasma interactions
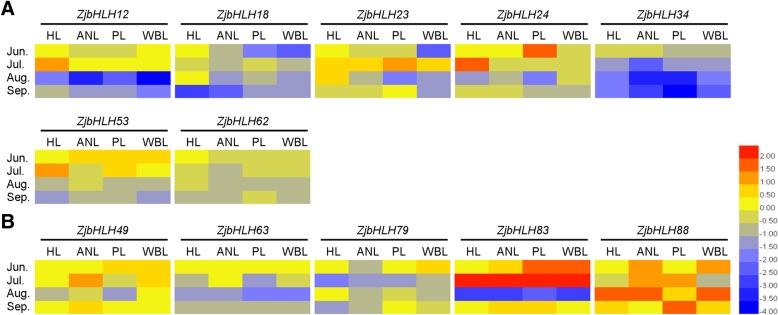
JWB caused by phytoplasma is a destructive disease in jujube production. Since bHLH genes have multiple functions in plants, whether they participate in jujube-phytoplasma interactions remains unclear. Hence, their expression changes were investigated in jujube under phytoplasma stress. Among the 23 ZjbHLH genes detected, the expression of ZjbHLH12, 18, 23, 24, 34, 53, and 62 genes in diseased leaves was significantly lower than the expression in healthy leaves (Fig. 8a). ZjbHLH49, 63, 79, 83, and 88 genes were highly expressed in diseased leaves (Fig. 8b). These results suggested that some ZjbHLHs participate in jujube-phytoplasma interactions.
Fig. 8.
Heat maps of the relative expression of ZjbHLH genes under phytoplasma stress. a The gene expression in diseased leaves was significantly lower than that in healthy leaves. b The gene expression in diseased leaves was significantly higher than that in healthy leaves. Scaled log2 expression values based on qRT-PCR data are shown from blue to red, indicating low to high expression, respectively
ZjbHLH protein-protein interaction network prediction
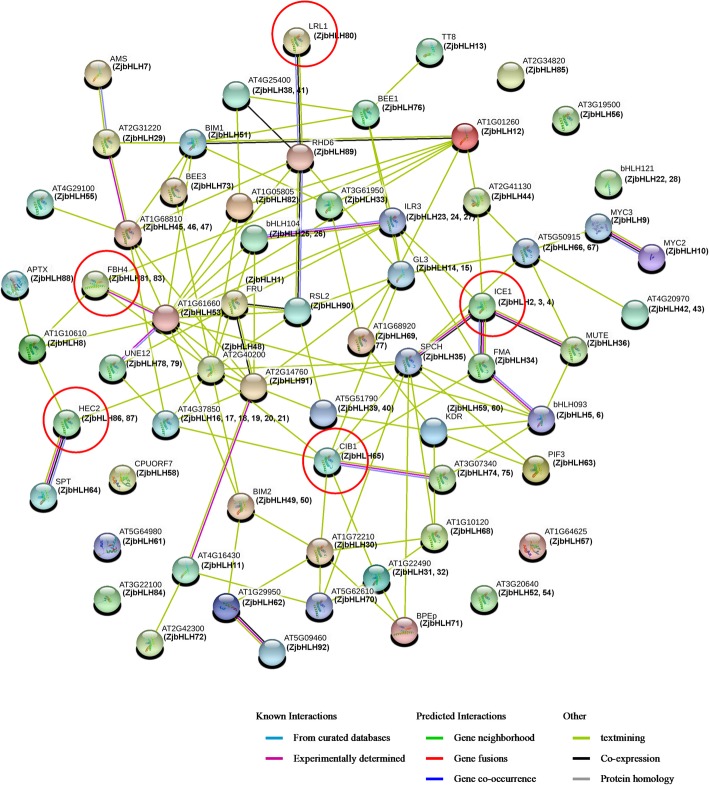
Based on the orthologs in Arabidopsis (Additional file 7: Table S2), it was predicted by STRING that many ZjbHLH proteins interacted with each other (Fig. 9), which is in accord with previous reports that the binding activity of bHLH proteins depends upon the formation of homodimers or heterodimers among bHLH proteins [28, 29]. Overall, several important interactions were predicted in Fig. 9. Both FBH4 (homolog of ZjbHLH81 and 83) and CIB1 (homolog of ZjbHLH65) were involved in the regulation of flowering time [30, 31], and HEC (homolog of ZjbHLH86 and 87) could interact with SPT (homolog of ZjbHLH64) to jointly regulate pistil development by regulating cytokinins and other hormones [32]. ICE1 (homolog of ZjbHLH2, 3, and 4) could interact with FMA (homolog of ZjbHLH34), SPCH (homolog of ZjbHLH35) and MUTE (homolog of ZjbHLH36) could regulate stomatal differentiation [33]. Moreover, ICE1 also regulated lateral bud growth and plant stress response [34, 35], and LRL1 (homolog of ZjbHLH80), RHD6 (homolog of ZjbHLH89) and RSL2 (homolog of ZjbHLH90) were involved in the regulation of root hair development [36, 37]. These results further proved the functional diversity of ZjbHLH genes. In addition, we also found that the functions of those genes contained more introns were mostly related to flower and root development (Additional file 7: Table S2). The predicted network provides some useful clues for functional studies, further experimental evidences should be needed.
Fig. 9.
A protein-protein interaction network for ZjbHLHs based on their orthologs in Arabidopsis. This network was predicted by online software STRING. ZjbHLH proteins were shown in brackets with Arabidopsis orthologs
Discussion
In this study, a total of 92 bHLH genes were identified in the jujube genome. Based on phylogenetic analysis, intron-exon gene structure, conserved protein motifs and amino acid physical and chemical property prediction, these ZjbHLHs were divided into 16 categories; most sequences have the same conservative sequence except for the VI, VII, and VIII groups. Among them, there are 10 atypical sequences, and this trait has also been demonstrated in other plants [25]. Furthermore, our BLAST results strongly supported our classifications of the ZjbHLHs, and detailed information about these orthologs was also summarized (Additional file 7: Table S2).
Many ZjbHLH proteins are involved in jujube flower development. ZjbHLH2 and ZjbHLH4 were expressed at higher levels at earlier flower development stages. These two genes belong to the ICE1 branch, which can regulate lateral bud growth [35]. A similar function was also confirmed by homologous protein interactions (Fig. 6B-a). ICE1 can interact with the HOS1 protein, which is an important regulator of flowering time [38]. ZjbHLH65, 83 and 87 proteins were homologous to CIB4, FBH4 and HEC2, respectively, and were three key regulatory factors in flower development (Fig. 6B-b, c, d). ZjbHLH83 is the homologous gene of FBH4 (At2g42280). Overexpression of FBH4 in Arabidopsis drastically elevated CO expression and caused early flowering regardless of the photoperiod [30]. Here, ZjbHLH83 showed high expression at earlier flower development stages (Fig. 6a) and was also predicted to interact with CO (Fig. 6B-c). In addition, a series of CIB genes (CIB1, 2, 3, 4, and 5) in Arabidopsis can activate FT transcription by interacting with CRY2 protein and mediate the regulation of flowering time [31]. There are also a series of CIB homologous genes in jujube, namely, ZjbHLH65 (homologs of CIB1), ZjbHLH74 and 75 (homologs of CIB3), ZjbHLH68 (homologs of CIB4), and ZjbHLH69 (homologs of CIL1). Since CIB genes have been proven to be conserved among plants [31], they are likely to have a similar function in flower development.
In fruit development, the expression patterns of most ZjbHLH genes detected in the two cultivars were in line with each other, which indicated that their functions in fruit development might be conserved among jujube varieties. ZjbHLH63 expression was significantly higher at the early stage of fruit development (Fig. 7a), which is the period of fruit enlargement. ZjbHLH63 is the homolog of AtPIF3 (Fig. 9), a key factor affecting light morphogenesis [39]. The homologous comparison and protein interaction prediction (Fig. 7B-b) also indicated that it might be involved in fruit enlargement. In addition, ZjbHLH15 homologous protein in Arabidopsis was predicted to be involved in fruit anthocyanin synthesis (Fig. 7B-a), but in this study, its expression decreased significantly at the fruit coloring stages (Fig. 7a). Therefore, we hypothesized that jujube fruit color changes might not correlate with the accumulation of anthocyanin.
In addition, the expression patterns of some bHLH genes in jujube and wild jujube were not the same, indicating that ZjbHLH genes participate in different regulation pathways between jujube and wild jujube, especially in flower development. Further studies are needed to elucidate the detailed interaction network of the growth and development of jujube and wild jujube.
Conclusions
This study described the bHLH gene family of Chinese jujube at the genome level. Their gene structure, chromosomal distribution, phylogenetic relationship, and tissue-specific expression patterns were presented. Ten ZjbHLHs with atypical bHLH domains were identified. Many ZjbHLH genes were confirmed to involve in flower and fruit development and responsive to phytoplasma stress. An integrated ZjbHLHs protein-protein interaction network was also predicted. These results are very meaningful to the future functional analysis of ZjbHLHs.
Methods
Plant materials
Chinese jujube and wild jujube trees used in this study are cultivated in the Experimental Station of Chinese Jujube, Hebei Agricultural University. No specific permits are required for the sample collection. They are not endangered or protected species.
Seven tissue types (roots, young branches, old branches, leaves, flower buds, flowers and young fruits) collected from three jujube trees and three wild jujube trees were used for organ-specific expression analysis. The flowers of jujube and wild jujube were used for qRT-PCR analysis. The four development stages sampled were the bud emergence stage (FL1), inflorescence emergence stage (FL2), yellow bud stage (FL3) and petal spread stage (FL4). The fruits of two jujube cultivars (‘Lizao’ and ‘Yazao’) were used to investigate the expression pattern of ZjbHLHs. Five developmental stages, including the young fruit stage (Y), early white mature fruit stage (EWM), white mature fruit stage (WM), half-red fruit stage (HR) and full-red fruit stage (FR), were sampled. Each treatment was collected from three biological replicates.
Three kinds of tissues representing disease symptoms of different severity of Jujube witches’ broom (JWB) disease (apparently normal leaves (ANL), phyllody leaves (PL), and witches’ broom leaves (WBL)) from diseased trees and healthy leaves (HL) from healthy trees were collected in four periods (June, July, August and September). All treatments were conducted with three biological replicates.
Identification and protein structure analysis of ZjbHLHs
The hidden Markov model (HMM) file of the bHLH domain (PF00010) was downloaded from the Pfam database (http://pfam.xfam.org/), and HMMER 3.1b2 software was used to find the ZjbHLH protein sequences in the jujube genome [23]. To further confirm our sequences, we used the online CD-search tool (NCBI database), the SMART tool (http://smart.embl-heidelberg.de/) and the website of PlantTFDB to screen sequences. Truncated and false genes were excluded from our analysis. The number of amino acids, molecular weight, and theoretical pI of ZjbHLH genes were predicted by NCBI and ProtParam (https://web.expasy.org/compute_pi/). The conserved motifs of ZjbHLH proteins were detected by MEME (http://meme-suite.org/) [40].
The chromosomal location and gene structure of ZjbHLHs
To determine the chromosomal location of the ZjbHLH genes, their gene sequences were used as query sequences in BLASTN searches against the jujube genome. Each ZjbHLH gene was mapped to the jujube genome according to its genome coordinates. Tandem duplications were identified as previously described [41].
The website GSDS (http://gsds.cbi.pku.edu.cn/) was used to predict the number of exons from the coding domain sequences (CDS) and DNA sequences of the ZjbHLH genes [42].
Multiple sequence alignment and phylogenetic tree construction
Multiple sequence alignment was analyzed by using ClustalX2 and edited by BioEdit. A phylogenetic tree of 92 ZjbHLHs was constructed based on their conserved domains. bHLH proteins of six other species (Arabidopsis thaliana, Prunus persica, Malus domestica, Pyrus bretschneideri, Vitis vinifera L. and Anemone vitifolia Buch.) were downloaded from NCBI. MEGA 7 software and the neighbor-joining statistical method were used to construct a rooted phylogenetic tree [43, 44]. The evolutionary distances were obtained using the p-distance method, and these distances were used to estimate the number of amino acid substitutions per site. The reliability of each phylogenetic tree was established by conducting 1000 bootstrap sampling iterations.
RNA isolation and expression analysis
Total RNA was extracted using an RNAprep Pure Plant Kit (TIANGEN) according to the manufacturer’s protocol. After genomic DNA was removed by RNase-free DNase I (TIANGEN), the RNA concentration and purity were checked on a NanoDrop2000 spectrophotometer. First-strand cDNA was synthesized by reverse transcribing 500 ng of total RNA with a FastQuant RT Super Mix Kit (TIANGEN). The cDNA was used as the template for gene expression analysis.
Gene expression was detected by semiquantitative PCR and qRT-PCR. The primers used in this study are listed in Additional file 8: Table S3. PCR products were amplified in triplicate using Bio-Rad iQ™5 with TransStart Top Green qPCR SuperMix AQ131 (TransGen Biotech, China) in 20 μL reactions. Each reaction contained 10 μL of 2 × TransStart® Top Green qPCR SuperMix, 0.4 μL each of 10 μM primers, 8.2 μL of ddH2O and 1 μL of cDNA. The thermal profile for RT-qPCR was as follows: preincubation for 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C, 10 s at 53–58 °C, and 10 s at 72 °C. Three biological replicates were performed for each treatment. Threshold cycle values were calculated using iCycler software, and ZjACT was used as an internal control [45]. Relative transcript levels were calculated according to the 2–ΔΔCT method [46].
Protein-protein interaction network prediction
Ninety-two ZjbHLH protein sequences were used as queries, and protein-protein interactions were predicted by the STRING website (https://string-db.org/). The orthologs of Arabidopsis thaliana were selected as references. After completing the BLAST step, the network was constructed using the highest score gene (bitscore). Finally, an interaction network among ZjbHLHs was constructed in this study.
Additional files
Figure S1. The multiple sequence alignment in ZjbHLH proteins. (DOC 3607 kb)
Table S1. Number of bHLH gene family from Chinese jujube and other six species. (DOC 50 kb)
Figure S2. The phylogenetic analysis of bHLH proteins of Ziziphus jujuba and Persica prunu. The NJ tree was constructed from the protein sequences of ZjbHLHs and PpbHLHs using MEGA7 with 1000 bootstrap copies. (DOC 483 kb)
Figure S3. The phylogenetic analysis of bHLH proteins of Ziziphus jujuba, Arabidopsis thaliana, Persica prunus, Malus domestica, Pyres bretschneideri, Vitis vinifera and Gossypium raimondii. There are 16 categories in total, and I, IV, XI and XV are selected for display. (DOC 1096 kb)
Figure S4. The amino acid sequences of 6 motifs among ZjbHLH proteins. (DOC 240 kb)
Figure S5. The major functional domain of ZjbHLH proteins. (DOC 384 kb)
Table S2. Summary information for 92 ZjbHLH proteins in STRING database. (XLS 95 kb)
Table S3. The primers of ZjbHLH genes used in this study. (DOC 142 kb)
Figure S6. Expression patterns of four ZjbHLH genes in flower development stage of jujube and wild jujube. FL1, bud emergence stage; FL2, inflorescence emergence stage; FL3, yellow bud stage; FL4, petal spread stage. The expression levels of eight treatments (four development stages in jujube and wild jujube, respectively) were compared and analyzed either between different stages of the same species or between different species of the same stage. All statistical analyses were performed with SPSS software 17.0. Duncan’s multiple range tests were used to assess differences between treatments. Different letters mean significant difference at 0.05 levels between the corresponding treatments. (DOC 319 kb)
Acknowledgements
Not applicable.
Abbreviations
- ANL
Apparently normal leaves
- bHLH
Basic helix-loop-helix
- CDS
Coding domain sequences
- EWM
Early white mature fruit stage
- FR
Full-red fruit stage
- HL
Healthy leaves
- HMM
Hidden Markov model
- HR
Half-red fruit stage
- JWB
Jujube witches’ broom disease
- PL
Phyllody leaves
- qRT-PCR
Quantitative real-time PCR
- TFs
Transcription factors
- WBL
Witches’-broom leaves
- white WM
mature fruit stage
- Y
Young fruit stage
Authors’ contributions
JZ designed the research; HL, WG, CX, YZ and JZ performed the experiments, analyzed the data and wrote the paper. ZL, ML and YZ participated in the data analysis. YZ and XM performed RT-PCR and RT-qPCR experiments. All authors read and approved the final the manuscript.
Funding
Supported by the National Key R&D Program Project Funding (2018YFD1000607), the National Natural Science Foundation of China (31772285), Foundation for 100 Innovative Talents of Hebei Province (SLRC2019031), Hebei Distinguished Young Scholar (C2016204145), and Hebei Special Funds for Intellectual Introduction and Talent Training (A2016002054). These funding bodies had no role in the design of the study, sample collection, analysis or interpretation of data and in writing the manuscript.
Availability of data and materials
All data and materials are presented in the main manuscript and additional supporting file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongtai Li and Weilin Gao contributed equally to this work.
Contributor Information
Hongtai Li, Email: 1061752104@qq.com.
Weilin Gao, Email: kissmiaomiao@outlook.com.
Chaoling Xue, Email: 18932666571@189.cn.
Yao Zhang, Email: 1104017540@qq.com.
Zhiguo Liu, Email: jujubeliu@163.com.
Yu Zhang, Email: 807583647@qq.com.
Xianwei Meng, Email: 1014654328@qq.com.
Mengjun Liu, Email: lmj1234567@aliyun.com.
Jin Zhao, Email: zhaojinbd@126.com.
References
- 1.Zhang C, Feng R, Ma R, Shen Z, Cai Z, Song Z, et al. Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS One. 2018;13(4):e0195974. doi: 10.1371/journal.pone.0195974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdullah M, Cheng X, Cao Y, Su X, Manzoor MA, Gao J, et al. Zinc finger-homeodomain transcriptional factors (ZHDs) in upland cotton (Gossypium hirsutum): genome-wide identification and expression analysis in fiber development. Front Genet. 2018;9:357. doi: 10.3389/fgene.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Anantharaman V, Balaji S, Mohan Babu M, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29(2):231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.An J, Qu F, Yao J, Wang X, You C, Wang X, Hao Y. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticult Res. 2017;4:17023. doi: 10.1038/hortres.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Zhao P, Kong N, Lu R, Pei Y, Huang C, et al. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes. 2018;9:54. doi: 10.3390/genes9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Hu Z, Zhao T, Yang Y, Chen T, Yang M, et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum) BMC Genomics. 2015;16(1):39. doi: 10.1186/s12864-015-1249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, et al. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015;83(6):976–990. doi: 10.1111/tpj.12942. [DOI] [PubMed] [Google Scholar]
- 9.Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome a signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- 10.Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyun Y, Lee IK. IDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61:283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- 12.Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin C, Huang X, Li K, Yin H, Li L, Yao Z, et al. Overexpression of a bHLH1 transcription factor of Pyrus ussuriensis confers enhanced cold tolerance and increases expression of stress-responsive genes. Front Plant Sci. 2016;7:441. doi: 10.3389/fpls.2016.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao K, Dong Q, Li C, Liu C, Ma F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci. 2017;8:480. doi: 10.3389/fpls.2017.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, Su L, Gao H, Jiang X, Wu X, Yi L, et al. Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front Plant Sci. 2018;9:64. doi: 10.3389/fpls.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Zhang J, Liu D, Wei Y, Wang Y, Li X. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum) BMC Plant Biol. 2018;18:304. doi: 10.1186/s12870-018-1523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang S, An H, Luo J, Wang X, Shi C, Xu F. Comparative analysis of transcriptomes to identify genes associated with fruit size in the early stage of fruit development in Pyrus pyrifolia. Int J Mol Sci. 2018;19(8):2342. doi: 10.3390/ijms19082342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing B, Yang D, Yu H, Zhang B, Yan K, Zhang X, et al. Overexpression of SmbHLH10 enhances tanshinones biosynthesis in Salvia miltiorrhiza hairy roots. Plant Sci. 2018;276:229–238. doi: 10.1016/j.plantsci.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress. Biochem Biophys Res Commun. 2018;503(2):397–401. doi: 10.1016/j.bbrc.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 20.Guan S, Xu Q, Ma D, Zhang W, Xu Z, Zhao M, Guo Z. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene. 2019;685:96–105. doi: 10.1016/j.gene.2018.10.066. [DOI] [PubMed] [Google Scholar]
- 21.Zhao K, Li S, Yao W, Zhou B, Li R, Jiang T. Characterization of the basic helix–loop–helix gene family and its tissue-differential expression in response to salt stress in poplar. Peer J. 2018;6:e4502. doi: 10.7717/peerj.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian M, Zhang X, Zhu Y, Xie G, Qin M. Global transcriptome analyses reveal differentially expressed genes of six organs and putative genes involved in (Iso) flavonoid biosynthesis in Belamcanda chinensis. Front Plant Sci. 2018;9:1160. doi: 10.3389/fpls.2018.01160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M, Zhao J, Cai Q, Liu G, Wang J, Zhao Z, et al. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 2014;5:5315. doi: 10.1038/ncomms6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Zhang C, Zhao X, Fei Z, Wan K, Zhang Z, et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016;12(12):e1006433. doi: 10.1371/journal.pgen.1006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang LM, Zhao J, Feng CF, Liu MJ, Wang JR, Hu YF. Genome-wide identification, characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Sci Rep. 2017;7(1):1025. doi: 10.1038/s41598-017-01159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21(12):3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141(4):1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27(4):862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J Cell Mol Biol. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 30.Ito S, Song Y, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. PNAS. 2012;109(9):3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013;9(10):e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuster C, Gaillochet C, Lohmann JU. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development. 2015;142(19):3343–3350. doi: 10.1242/dev.120444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi X, Torii KU. Hormonal and environmental signals guiding stomatal development. BMC Biol. 2018;16(1):21. doi: 10.1186/s12915-018-0488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y, Zhao Q, Zhao X, Hao Y, You C. Expression analysis of the MdCIbHLH1 gene in apple flower buds and seeds in the process of dormancy. Horticult Plant J. 2016;2(2):61–66. [Google Scholar]
- 35.Zhang T, Mo J, Zhou K, Chang Y, Liu Z. Overexpression of Brassica campestris BcICE1 gene increases abiotic stress tolerance in tobacco. Plant Physiol Biochem. 2018;132:515–523. doi: 10.1016/j.plaphy.2018.09.039. [DOI] [PubMed] [Google Scholar]
- 36.Breuninger H, Thamm A, Streubel S, Sakayama H, Nishiyama T, Dolan L. Diversification of a transcription factor family led to the evolution of antagonistically acting genetic regulators of root hair growth. Curr Biol. 2016;26(12):1622–1628. doi: 10.1016/j.cub.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Qi C, Luo J, Liu L, He Y, Chen L. Characterization of LRL5 as a key regulator of root hair growth in maize. Plant J. 2019;98(1):71–82. doi: 10.1111/tpj.14200. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Pan T, Liang W, Gao L, Wang X, Li H, Liang S. Overexpression of an orchid (Dendrobium nobile) SOC1/TM3-like ortholog, DnAGL19, in Arabidopsis regulates HOS1-FT expression. Front Plant Sci. 2016;7:99. doi: 10.3389/fpls.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Kekun, Zheng Ting, Zhu Xudong, Jiu Songtao, Liu Zhongjie, Guan Le, Jia Haifeng, Fang Jinggui. Genome-Wide Identification of PIFs in Grapes (Vitis vinifera L.) and Their Transcriptional Analysis under Lighting/Shading Conditions. Genes. 2018;9(9):451. doi: 10.3390/genes9090451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua Z, Doroodian P, Vu W. Contrasting duplication patterns reflect functional diversities of ubiquitin and ubiquitin-like protein modifiers in plants. Plant J. 2018;95(2):296–311. doi: 10.1111/tpj.13951. [DOI] [PubMed] [Google Scholar]
- 42.Guo A, Zhu Q, Chen X, Luo J. GSDS: a gene structure display server. Yi Chuan. 2007;29:1023–1026. [PubMed] [Google Scholar]
- 43.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 44.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Bu J, Zhao J, Liu M. Expression stabilities of candidate reference genes for RT-qPCR in Chinese jujube (Ziziphus jujuba mill.) under a variety of conditions. PLoS One. 2016;11(4):e0154212. doi: 10.1371/journal.pone.0154212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The multiple sequence alignment in ZjbHLH proteins. (DOC 3607 kb)
Table S1. Number of bHLH gene family from Chinese jujube and other six species. (DOC 50 kb)
Figure S2. The phylogenetic analysis of bHLH proteins of Ziziphus jujuba and Persica prunu. The NJ tree was constructed from the protein sequences of ZjbHLHs and PpbHLHs using MEGA7 with 1000 bootstrap copies. (DOC 483 kb)
Figure S3. The phylogenetic analysis of bHLH proteins of Ziziphus jujuba, Arabidopsis thaliana, Persica prunus, Malus domestica, Pyres bretschneideri, Vitis vinifera and Gossypium raimondii. There are 16 categories in total, and I, IV, XI and XV are selected for display. (DOC 1096 kb)
Figure S4. The amino acid sequences of 6 motifs among ZjbHLH proteins. (DOC 240 kb)
Figure S5. The major functional domain of ZjbHLH proteins. (DOC 384 kb)
Table S2. Summary information for 92 ZjbHLH proteins in STRING database. (XLS 95 kb)
Table S3. The primers of ZjbHLH genes used in this study. (DOC 142 kb)
Figure S6. Expression patterns of four ZjbHLH genes in flower development stage of jujube and wild jujube. FL1, bud emergence stage; FL2, inflorescence emergence stage; FL3, yellow bud stage; FL4, petal spread stage. The expression levels of eight treatments (four development stages in jujube and wild jujube, respectively) were compared and analyzed either between different stages of the same species or between different species of the same stage. All statistical analyses were performed with SPSS software 17.0. Duncan’s multiple range tests were used to assess differences between treatments. Different letters mean significant difference at 0.05 levels between the corresponding treatments. (DOC 319 kb)
Data Availability Statement
All data and materials are presented in the main manuscript and additional supporting file.