Summary
Overlapping runs of homozygosity (ROH islands) shared by the majority of a population are hypothesized to be the result of selection around a target locus. In this study we investigated the impact of selection for coat color within the Noriker horse on autozygosity and ROH patterns. We analyzed overlapping homozygous regions (ROH islands) for gene content in fragments shared by more than 50% of horses. Long‐term assortative mating of chestnut horses and the small effective population size of leopard spotted and tobiano horses resulted in higher mean genome‐wide ROH coverage (S ROH) within the range of 237.4–284.2 Mb, whereas for bay, black and roan horses, where rotation mating is commonly applied, lower autozygosity (S ROH from 176.4–180.0 Mb) was determined. We identified seven common ROH islands considering all Noriker horses from our dataset. Specific islands were documented for chestnut, leopard spotted, roan and bay horses. The ROH islands contained, among others, genes associated with body size (ZFAT, LASP1 and LCORL/NCAPG), coat color (MC1R in chestnut and the factor PATN1 in leopard spotted horses) and morphogenesis (HOXB cluster in all color strains except leopard spotted horses). This study demonstrates that within a closed population sharing the same founders and ancestors, selection on a single phenotypic trait, in this case coat color, can result in genetic fragmentation affecting levels of autozygosity and distribution of ROH islands and enclosed gene content.
Keywords: draught horse, LASP1, LCORL, MC1R, PATN1, ROH island, selection signature, ZFAT
Introduction
Runs of homozygosity (ROH) are long consecutive homozygous regions distributed across the genome that arise from identical‐by‐descendent haplotypes transmitted by common ancestors (Ceballos et al. 2018). Overlapping homozygous regions, shared by a higher percentage of individuals in a population (ROH islands), are commonly hypothesized to be the result of selection around a target locus or indicate a low recombination rate in specific genomic regions (Pemberton et al. 2012; Metzger et al. 2015; Peripolli et al. 2016, 2018). We chose the Noriker horse population to investigate the impact of selection on the size, frequency, distribution and localization of ROHs in the genome. This autochthonous Austrian draught horse breed is purebred since the closure of the studbook in the year 1903. The breeding program is based merely upon conformation traits, including a total of 11 traits (ratings of type, head, neck, forequarter, midquarter, hindquarter, forelegs, hindlegs, walk, trot and correctness of gaits). Within the breeding program pursuing a common breeding goal, the selection and maintenance of six different coat color strains (chestnut, bay, black, roan, leopard spotted and tobiano) represents a central objective and requires different mating strategies (assortative mating and rotation mating) (Druml et al. 2009).
Long‐term assortative mating is a common practice for breeding chestnut‐colored Noriker horses, which have been selected for the recessive variant of the melanocortin 1 receptor gene (MC1R) on ECA3 at position 36 259.552 (Marklund et al. 1996). Leopard spotted coat color is genetically determined by two loci—the leopard complex (LP) allele (ECA1:g.108 297.929–108 297.930ins1378) (Bellone et al. 2013) and the modifier pattern1 (PATN1) allele (SNP ECA3:g.23 658.447T>G in the 3′‐untranslated region of RFWD3) (Holl et al. 2016)—whereby only heterozygosis on the LP locus combined with the presence of the dominant modifier PATN1 results in the favored well‐marked leopard phenotype (Bellone et al. 2013; Holl et al. 2016; Druml et al. 2017a). Horses with non‐desired phenotypes, such as few spots (LP/LP PATN1/–) or snow cap blanket (LP/lp patn1/patn1), are not used for breeding purposes. This intensive selection scheme is responsible for a high prevalence of the genotype LP/lp PATN1/– (up to 96.9%) in leopard spotted Noriker horses (Druml et al. 2017a). Within the roan coat color strain, blue roan horses are strongly favored. The percentage of blue roans comprises 100% in breeding stallions and 95% in mares. Based upon the dominant mode of inheritance (Marklund et al. 1999), rotating matings between roan and black horses are commonly applied. Bay and black horses represent more than 60% of the Noriker horse population. In these strains, both breeding strategies—rotation and assortative matings—are usually applied (Fig. 1).
Figure 1.

In the Austrian Noriker horse breed six coat colors (bay, chestnut, black, blue roan, leopard spotted, tobiano) are bred systematically. Percentages of each color in the current population are indicated below the pictures (images by Thomas Druml).
Population structure analyses of the Noriker horse using pedigree data revealed that long‐term mating strategies has resulted in specific family structures (Druml et al. 2009). In particular, chestnut and leopard spotted horses differed in inbreeding levels, effective population size and different allele frequencies of the glycogen synthase 1 (GYS1) variant, whereas higher incidence of the GYS1 allele H variant, associated with polysaccharide myopathy type I, was documented for chestnut Noriker horses (Druml et al. 2017b). These pedigree‐based results were additionally confirmed by a genome‐wide population structure analysis (Druml et al. 2018). A morphological study of 497 Noriker horses using 31 different body measurements also revealed anatomical differences between the coat color strains (Druml et al. 2008). Black and roan horses are commonly characterized by a long format with a long neck, but black horses show significantly higher chest circumference than do roans. Chestnuts reach highest values for size and mass, represented by height at withers, circumference of chest, width of chest and a high caliber. The caliber index describes the relation of chest circumference and cannon bone circumference to height at withers. Leopard and tobianos characteristically are small‐framed, have shorter necks, a narrow chest and a lighter caliber, whereas tobianos typically show a long‐rectangular body format (Druml et al. 2008).
The aim of this study was to investigate the impact of coat color selection on autozygosity and ROH patterns. Furthermore, we analyzed overlapping homozygous regions (ROH islands), including annotated genes and their gene ontology in the different coat color strains. Besides coat color, body size represents one of the major selection criteria in Noriker horses during the last four decennia. Therefore, we examined several SNPs associated with body size that were located in ROH islands of the Noriker horse and compared their genotype frequencies with those of the smallest Middle European draught horse breed—the Posavina horse, an autochthonous draught horse originating from Croatia and Slovenia.
Material and methods
Sampling
The 174 Noriker horses included in this study were sampled in the years 2013 to 2016 in order to represent genealogical structures (sire lines and mare families), coat color strains and geographical distribution of the breed. Of these 174 horses, born between 1996 and 2014, 33 animals were black, 31 animals were bay and 36 animals were chestnut. Furthermore, the sample comprised 23 blue roan horses, 48 leopard spotted horses and three tobianos. In addition to the Noriker horses, we used SNP data of 28 Posavina horses from a previous study (Grilz‐Seger et al. 2018). The Posavina horse was sampled to represent family structure of the Slovenian Posavina population.
SNP genotyping
The SNP genotypes for the 202 horses were determined using the Affymetrix Axiom™ Equine genotyping array. The chromosomal positions of the SNPs were derived from the EquCab2 reference genome (Wade et al. 2009). We did not consider SNPs positioned on the sex chromosomes (X, 28 017 SNPs; Y, one SNP) and SNPs without known chromosomal positions (30 864 SNPs). SNPs with more than 10% missing genotypes were excluded. The remaining subset was further edited for minor allele frequency (>0.01). This resulted in a total of 533 268 SNPs that passed quality control and were used for the genetic analyses.
ROH analysis
ROH segments were determined with an overlapping window approach implemented in plink v1.7 (Purcell et al. 2007) based on the following settings: minimum SNP density was set to one SNP per 50 kb with a maximum gap length of 100 kb. The final segments were called runs of homozygosity (ROH) if the minimum length of the homozygous segment was greater than 500 kb and comprised more than 80 homozygous SNPs; one heterozygote and two missing genotypes were permitted within each segment.
The total number of ROH (N ROH), average length of ROH (L ROH) and sum of all ROH segments (S ROH) for each horse were summarized for the entire Noriker sample and for the different coat color strains. To analyze the ROH length distribution, ROH segments were divided into the following seven length classes: 0.5–1, >1–2, >2–4, >4–6, >6–8, >8–10 and >10 Mb. Genomic inbreeding (F ROH) was calculated following the method described by McQuillan et al. (2008):
where the length of the autosomal genome (L AUTO) was set to 2.243 GB.
The distribution of ROH segments across the genome was visualized using the R package detectrohs (www.r-project.org). Putative ROH islands were determined based upon overlapping homozygous regions within more than 50% of the horses. The map viewer of the equine Ensembl database EquCab2, available at www.ensembl.org, was used to identify genes located in ROH islands. For determination of Gene Ontology (GO) terms and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways of identified genes, we used the open source Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 package (Huang et al. 2009). For the GO analysis we chose the equine annotation file as background and a significance threshold of P < 0.05. Further statistical analyses and data preparation were performed using the sas v.9.1 software package (SAS 2009).
High‐resolution population network
To visualize the population structure of Noriker horses with respect to the different coat color strains, we performed a high‐resolution network analysis based upon an identical‐by‐state‐derived relationship matrix (G), following the description by Druml et al. (2018). Briefly, we computed genetic distances by subtracting pairwise relationships from 1 and applied the NetView approach in its default setting (number of k nearest neighbors k‐NN = 10) (Neuditschko et al. 2012; Steinig et al. 2015). Genetic relatedness between horses is marked by thickness of edges (connecting lines), thicker edges corresponding to lower genetic distances. The node size is in relation to the individual S ROH, thus depicting inbred or outbred horses within the population network, and the node color represents the respective coat color of the horses.
Results and discussion
ROH analysis and population structure
ROH analysis revealed, by the means of S ROH and N ROH, that coat color strains of the Noriker horse can be divided into two major groups. Chestnut, leopard spotted and tobiano horses were characterized by the highest S ROH (mean S ROH ranging from 237.4 Mb in chestnuts to 284.2 Mb in tobianos) and the highest N ROH (181.7 in chestnuts, followed by leopard spotted horses with 166.0 and tobianos with 158.7) (Table 1). Black, bay and roan horses, representing the second group, showed lower values, with S ROH ranging from 176.5 Mb in roan to 180.0 Mb in black horses. The lowest N ROH was detected within black horses with a value of 139.3, whereas the upper limit within this group was observed in roan horses with a N ROH of 151.5. On an individual level, maxima values for S ROH above 400 Mb were reached by a chestnut and a leopard spotted horse, indicating F ROH values greater than 18%.
Table 1.
Mean values, standard deviation (SD), minimum and maximum values for the variables S ROH (in Mb), N ROH, L ROH (in Mb) and F ROH for the entire sample of Noriker horses and the subpopulations defined by chestnut, bay, black, blue roan, leopard spotted and tobiano coat color.
| Variable | n | Mean | SD | Min. | Max. |
|---|---|---|---|---|---|
| Noriker total sample | |||||
| S ROH | 174 | 213.2 | 97.6 | 13.1 | 452.0 |
| N ROH | 159.2 | 52.3 | 19.0 | 320.0 | |
| L ROH | 1.3 | 0.4 | 0.6 | 2.4 | |
| F ROH | 0.10 | 0.04 | 0.01 | 0.20 | |
| Noriker chestnut | |||||
| S ROH | 36 | 237.4 | 102.0 | 20.7 | 432.0 |
| N ROH | 181.7 | 57.0 | 32.0 | 320.0 | |
| L ROH | 1.3 | 0.4 | 0.6 | 2.3 | |
| F ROH | 0.11 | 0.04 | 0.01 | 0.19 | |
| Noriker bay | |||||
| S ROH | 31 | 178.7 | 99.0 | 13.1 | 351.2 |
| N ROH | 149.6 | 61.6 | 20.0 | 245.0 | |
| L ROH | 1.1 | 0.3 | 0.6 | 1.8 | |
| F ROH | 0.08 | 0.04 | 0.01 | 0.16 | |
| Noriker black | |||||
| S ROH | 33 | 180.0 | 110.9 | 15.7 | 354.8 |
| N ROH | 139.3 | 65.4 | 19.0 | 227.0 | |
| L ROH | 1.2 | 0.4 | 0.7 | 1.9 | |
| F ROH | 0.08 | 0.05 | 0.01 | 0.16 | |
| Noriker blue roan | |||||
| S ROH | 23 | 176.5 | 72.8 | 16.4 | 306.2 |
| N ROH | 151.5 | 40.8 | 26.0 | 212.0 | |
| L ROH | 1.1 | 0.3 | 0.6 | 1.8 | |
| F ROH | 0.08 | 0.03 | 0.01 | 0.14 | |
| Noriker leopard spotted | |||||
| S ROH | 48 | 253.4 | 71.7 | 57.0 | 452.0 |
| N ROH | 166.0 | 27.3 | 77.0 | 224.0 | |
| L ROH | 1.5 | 0.3 | 0.7 | 2.4 | |
| F ROH | 0.11 | 0.03 | 0.02 | 0.20 | |
| Noriker tobiano | |||||
| S ROH | 3 | 284.2 | 100.6 | 24.6 | 397.2 |
| N ROH | 158.7 | 6.5 | 152.0 | 165.0 | |
| L ROH | 1.8 | 0.6 | 1.3 | 2.4 | |
| F ROH | 0.13 | 0.04 | 0.09 | 0.18 | |
The majority (60%–85%) of ROH segments in basic colored (bay, chestnut, black) and roan horses was shorter than 4 Mb. Leopard spotted horses and tobianos were characterized by the lowest proportion of ROH segments (19.9%) in the length class 0.5–1 Mb. The proportion of ROH segments greater than 10 Mb, indicating recent inbreeding, varied between 2.3% and 3.9% in basic‐colored and roan horses. In tobianos and leopards a two‐ to three‐times higher percentage of ROHs in this length class was observed (Fig. 2).
Figure 2.
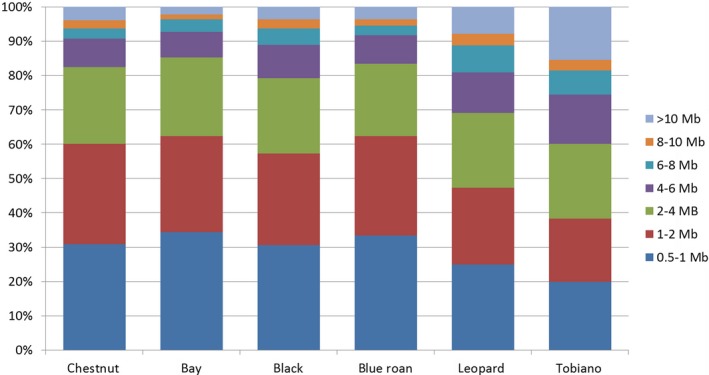
Distribution of runs of homozygosity (ROH) segments of different length classes (0.5–1, >1–2, >2–4, >4–6, >6–8, >8–10 and >10 Mb) per Noriker coat color strains (chestnut, bay, black, blue roan, leopard spotted, tobiano).
The high‐resolution population network, as illustrated in Fig. 3, clearly demonstrates that the Noriker breed is divided into genetic units that represent family structures that originated from systematical breeding for coat color over a long term. Leopard spotted horses are characterized by higher genetic distances to the main cluster (on the left), as they form a clearly distinct and separate group. Within the center of the main cluster, the higher degree of admixture between bay and black horses can be observed, a result of rotating mating strategies. The chestnut cluster (bottom left) illustrates the effects of assortative mating, as a closer relationship exists only within the chestnut group and it is linked to the center only at the top, simultaneously exhibiting a high genetic distance to the leopard spotted horses. Blue roan horses form a specific group (assortative mating), but they also generate a bridge between the core and the leopard cluster.
Figure 3.
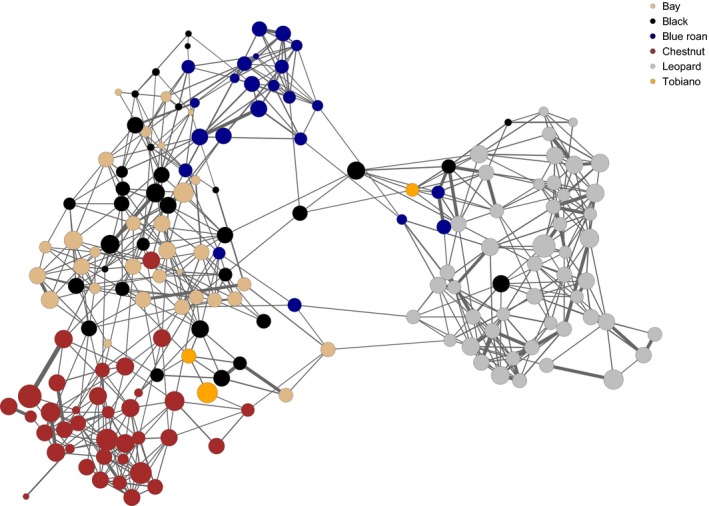
High‐resolution network visualization of Noriker horses. Each node represents an individual, node colors visualize the different coat colors of the horses (brown = chestnut, crème = bay, black = black, blue = blue roan, grey = leopard, orange = tobiano) and the size of node is in relation to S ROH. Links between nodes illustrate the genetic distance between individuals; thickness of links are proportional to genetic relationship.
The values of the parameters S ROH, N ROH and L ROH underline the influence of effective and actual population size and the effect of different mating strategies on the length and distribution of ROH (Table 1). In bay, black and blue roan horses, rotating matings are common. This exchange of breeding animals between the color strains has increased effective population size resulting in lower mean S ROH, N ROH, and F ROH values. Chestnut Noriker horses were characterized by relatively high S ROH, high N ROH, a high proportion of ROH segments shorter than 6 Mb (90.8%) and simultaneously a low proportion of long ROHs (>10 Mb) (3.9%). These findings can be interpreted as reflecting the result of a long‐term assortative mating strategy (true‐to‐color matings up to 12 generations back), increasing relatedness and the prevention of closer consanguinity matings. As a consequence, the relatively high F ROH of 0.11 (maxima up to 0.19) is composed of a higher number of shorter ROHs, indicating ‘older’ inbreeding. The ROH pattern in the leopard spotted sample reveals a profile typical for a bottlenecked and consanguineous population (Ceballos et al. 2018). Bottlenecks occurred in this coat color strain in the 1920s and 1960s, and the actual population size of leopard spotted Noriker horses comprises only 120 breeding animals. These findings are in concordance with the high‐resolution population network (Fig. 3), which illuminates that the application of systematical mating strategies (chestnut, assortative mating; blue roan, mostly assortative mating; leopard, assortative mating plus rotation mating; bay and black, rotation mating) results in well‐defined population structures (coat color strains) within one breed. Previous studies based upon pedigree and genome‐wide SNP data also illustrated genetic fragmentation of the Noriker population into different coat color strains (Druml et al. 2009, 2018).
ROH islands
Within the entire Noriker sample, we identified seven ROH islands on ECA3 (Fig. 4), ECA9 (Fig. S1) and ECA11 (Fig. S2) that were shared by more than 50% of horses, together covering 5.1 Mb of the whole genome. Four of these islands were located on ECA11. The percentage of animals sharing these overlapping homozygous regions ranged from 51.2% to 65.8% (Table 2). The most prominent common ROH island on ECA11 was present in all color strains, except leopard spotted horses, and harbored the homeobox‐B (HOXB) cluster among 29 other annotated genes.
Figure 4.
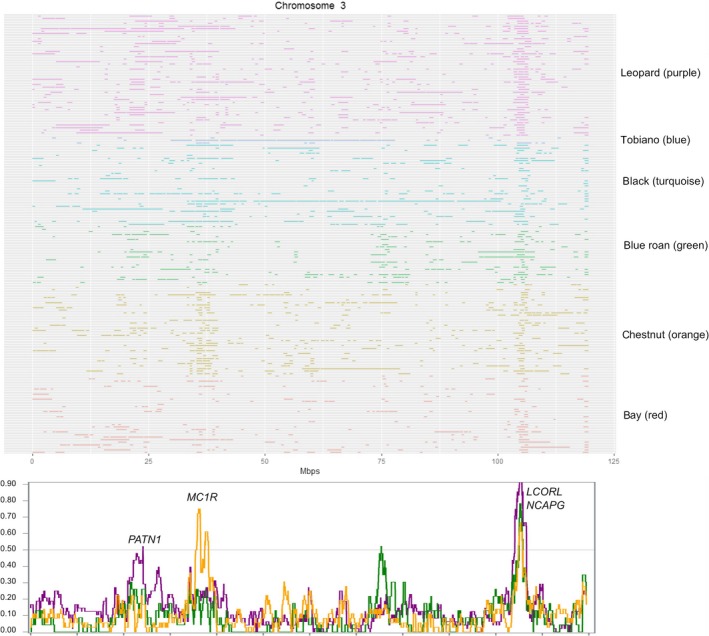
Distribution of runs of homozygosity (ROH) on ECA3 per individual. Line colors indicate coat color of horses (purple = leopard spotted, blue = tobiano, turquoise = black, green = roan, orange = chestnut, red = bay). In the line plot below, the frequencies of animals sharing a ROH are illustrated (purple = leopard spotted, green = roan, orange = chestnut).
Table 2.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the entire Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Annotated genes |
|---|---|---|---|---|---|
| 3 | 104 695.238 | 105 832.553 | 1137.315 | 61.1 | LCORL, NCAPG, DCAF16 |
| 3 | 106 272.331 | 106 310.866 | 38.535 | 50.6 | – |
| 9 | 74 971.396 | 75 661.910 | 690.514 | 61.1 | ZFAT |
| 11 | 23 203.615 | 23 575.186 | 371.571 | 52.0 | FBXO4, RPL23, LASP1, C11H17orf98, CWC25, PIP4K2B, PSMB3, PCGF2, CISD3, MLLT6 |
| 11 | 23 628.626 | 23 649.615 | 20.989 | 51.2 | SOCS7 |
| 11 | 23 712.507 | 25 010.061 | 1297.554 | 57.5 | NPEPPS, KPNB1, TBKBP1, OSBPL7, MRPL10, LRRC46, SCRN2, SP6, SP2, PNPO, PRR15L, CDK5RAP3, COPZ2, CBX1, SNX11, HOXB1, HOXB2, HOXB3, HOXB5, HOXB6, HOXB7, HOXB8, HOXB13, TTLL6, CALCOCO2, UBE2Z, ATP5MC1, SNF8, IGF2BP1 |
| 11 | 29 724.170 | 31 250.934 | 1526.764 | 65.8 | STXBP4, HLF, MMD, TMEM100, PCTP, ANKFN1, C11orfH17orf67 |
ROH islands in substructures
Investigating the coat color strains separately, we identified distinct ROH patterns. The highest number of ROH islands (n = 10) was detected in the chestnut sample and together covered a length of 7.7 Mb (Table 3). Four of these overlapping homozygous regions were private and were located on ECA2, ECA3 (two islands) and ECA10. According to our expectations, chestnut‐colored Noriker horses had an 851‐kb‐long homozygous region on ECA3, containing among 15 annotated genes melanocortin 1 receptor (MC1R), responsible for chestnut coat color (Fig. 4). Extended haplotypes around the MC1R locus have already been proven by McCue et al. (2012), Petersen et al. (2013) and Grilz‐Seger et al. (2018). A further private island on ECA3 was shared by 58.3% of chestnut animals and harbored the genes solute carrier family 39 member 8 (SLC39A8) and B‐cell scaffold protein with ankyrin repeats 1 (BANK1). The third overlapping homozygous region on ECA2 at position 28.66–28.86 Mb was present in 51% of chestnut horses, containing 10 annotated genes, and the fourth private island on ECA10 encompassed three members of the zink finger genes (ZNF304, ZNF772 and ZNF773) and the aurora kinase C (AURKC) gene (Table 3).
Table 3.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the chestnut Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Known genes |
|---|---|---|---|---|---|
| 2 | 28 516.118 | 28 526.437 | 10.32 | 53.6 | – |
| 2 | 28 660.340 | 28 868.113 | 267.77 | 54.0 | TENT5B, KDF1, NUDC, NROB2, GPATCH3, GPN2, SFN, ZDHHC18, PIGV, ARID1A |
| 3 | 35 704.817 | 36 555.935 | 851.12 | 68.6 | SPG7, RPL13, CPNE7, DPEP1, CDK10, VPS9D1, FANCA, SPATA2L, ZNF276, SPIRE2, TCF25, MCR1, DEF8, DBNDD1, GAS8 |
| 3 | 37 444.692 | 38 217.472 | 772.78 | 58.3 | SLC39A8, BANK1 |
| 3 | 104 849.802 | 105 720.637 | 807.84 | 61.9 | LCORL |
| 9 | 74 959.554 | 75 661.942 | 702.39 | 62.0 | ZFAT |
| 10 | 26 105.708 | 26 644.961 | 539.25 | 52.8 | AURKC, ZNF304, ZNF772, ZNF773 |
| 11 | 23 203.615 | 25 657.958 | 2257.60 | 62.3 | FBXO47, RPL23, LASP1, C11H17orf98, PCGF2, CISD3, MLLT6, CWC25, PIP4K2B, PSMB3, SRCIN1, SOCS7, GPR179, NPEPPS, KNPB1, TBKBP1, MRPL10, OSBPL7, LRRC46, SCRN2, SP6, SP2, PRR15L, PNPO, CDK5RAP3, COPZ2, SNX11, CBX1, HOXB1, HOXB2, HOXB3, HOXB5, HOXB6, HOXB7, HOXB8, HOXB13, TTLL6, CALCOCO2, ATP5MC1, UBE2Z, SNF8, IGF2BP1, BAGALNT2, GNGT2, ABI3, PHOSPHO1, ZNF652, NXPH3, PHB, NGFR, SPOP, SLC35B1, FAM117A, KAT7, TAC4 |
| 11 | 29 652.068 | 31 309.554 | 1657.49 | 81.4 | COX11, STXBP4, HLF, MMD, TMEM100, PCTP, ANKFN1, C11H17orf67, DGKE |
| 11 | 31 530.857 | 31 614.545 | 83.69 | 52.8 | AKAP1 |
Within the bay sample, seven ROH islands on ECA3, ECA9, ECA10 and ECA11 with a total length of 3.9 Mb were identified (Table 4). One island on ECA10 at position 24.68–24.80 Mb, shared by 50.8% of horses, was private for bay horses and contained six annotated genes. In the black sample, the lowest number of ROH islands (n = 4), together covering a total length 3.8 Mb, was found on ECA3, ECA9 and ECA11 (Table 5). These four islands overlap in general with those of the bay sample in position and length.
Table 4.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the bay Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Known genes |
|---|---|---|---|---|---|
| 3 | 105 152.868 | 105 210.249 | 57.38 | 51.6 | – |
| 3 | 105 473.521 | 106 323.816 | 850.30 | 51.6 | LCORL, NCAPG, DCAF16, FAM184B, MED28, LAP3 CLN2, QDPR |
| 9 | 74 981.480 | 75 661.942 | 680.46 | 62.0 | ZFAT |
| 10 | 24 684.675 | 24 808.945 | 124.27 | 50.8 | FIZ1, ZNF784, ZNF581, CCDC106, U2AF2, EPN1 |
| 11 | 24 123.810 | 25 002.125 | 878.32 | 58.1 | CDK5RAP3, COPZ2, CBX1, SNX11, HOXB1, HOXB2, HOXB3, HOXB5, HOXB6, HOXB7, HOXB8, HOXB13, TTLL6, CALCOCO2, UBE2Z, ATP5MC1, SNF8, IGF2BP1 |
| 11 | 25 612.643 | 25 657.958 | 5.32 | 51.6 | FAM117A, TAC4 |
| 11 | 29 877.648 | 31 220.694 | 1343.05 | 62.4 | HLF, MMD, TMEM100, PCTP, ANKFN1 |
Table 5.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the black Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Known genes |
|---|---|---|---|---|---|
| 3 | 105 046.433 | 105 630.163 | 583.73 | 53.3 | – |
| 9 | 75 089.448 | 75 661.942 | 572.49 | 55.4 | ZFAT |
| 11 | 23 732.591 | 25 010.061 | 1277.47 | 55.5 | NPEPPS, KPNB1, TBKBP1, OSBPL7, MRPL10, LRRC46, SCRN2, SP6, SP2, PNPO, PRR15L, CDK5RAP3, COPZ2, CBX1, SNX11, HOXB1, HOXB2, HOXB3, HOXB5, HOXB6, HOXB7, HOXB8, HOXB13, TTLL6, CALCOCO2, UBE2Z, ATP5MC1, SNF8, IGF2BP1 |
| 11 | 29 644.180 | 30 991.846 | 1347.67 | 64.0 | COX11, STXBP4, HLF, MMD, TMEM100, PCTP, ANKFN1 |
Within roan horses, eight islands on ECA2, ECA3, ECA9, ECA11 and ECA15 were identified, altogether covering a length of 5.8 Mb, and the island on ECA15 was also present in the leopard spotted sample. All other islands overlapped with those of the black sample and differed slightly in length and position (Table 6).
Table 6.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the blue roan Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Known genes |
|---|---|---|---|---|---|
| 2 | 28 521.277 | 28 524.547 | 3.27 | 52.2 | – |
| 3 | 75 358.079 | 75 400.511 | 42.43 | 52.2 | – |
| 3 | 104 943.947 | 105 767.304 | 823.36 | 68.2 | LCORL, NCAPG |
| 9 | 74 982.834 | 75 661.597 | 678.76 | 65.9 | ZFAT |
| 11 | 22 989.768 | 24 884.475 | 1894.71 | 61.9 | FBXL20, STAC2, CACNB1, ARL5C, PLXDC1, FBXO47, RPL23, LASP1, C11H17orf98, CWC25, CISD3, PIP4K2B, PSMB3, PCGF2, MLLT6, SRCIN1, SOCS7, GPR179, NPEPPS, KPNB1, TBKBP1, OSBPL7, MRPL10, LRRC46, SCRN2, SP6, SP2, PNPO, PRR15L, CDK5RAP3, COPZ2, CBX1, SNX11, HOXB1, HOXB2, HOXB3, HOXB5, HOXB6, HOXB7, HOXB8, HOXB13, TTLL6, CALCOCO2 |
| 11 | 24 901.862 | 25 022.125 | 120.26 | 52.7 | UBE2Z, SNF8, IGF2BP1, |
| 11 | 29 644.180 | 31 307.829 | 1663.65 | 66.0 | COX11, STXBP4, HLF, MMD, TMEM100, PCTP, ANKFN1, DGKE |
| 15 | 66 557.266 | 67 105.755 | 548.49 | 55.8 | – |
Within leopard spotted horses, nine islands on ECA2, ECA3, ECA6, ECA9, ECA11 and ECA15 with a total length of 7.2 Mb were identified (Table 7). Two of them, located on ECA6 and ECA11, were found only in this subpopulation. The overlapping homozygous region on ECA6 at position 28.80–30.11 Mb, containing the genes CACNA2D4, LRTM2, ADIPOR2, WNT5B, FBXL14, ERC1, RAD52, WNK1, NINJ2 and B4GALNT3, was shared by 76.3% of leopard spotted animals. Interestingly this ROH island was also found in a previous study in the Bosnian Mountain Horse, the oldest horse breed on the Balkan Peninsula (Grilz‐Seger et al. 2018). Leopard spotted coat color was detected in 25 000‐year‐old ancient DNA samples originating from wild, pre‐domestic horses of Western and Eastern Europe (Pruvost et al. 2011). In the Noriker breed this color variant was first mentioned in the year 1652, and its origin commonly has been traced to local horses from the central alpine region (Grilz‐Seger et al. 2017).
Table 7.
Chromosomal position, length and gene content of runs of homozygosity (ROH) islands shared by more than 50% (ROH frequency) of the leopard spotted Noriker sample.
| Chr. | Begin | End | Length (kb) | ROH frequency (%) | Known genes |
|---|---|---|---|---|---|
| 2 | 28 516.118 | 28 544.165 | 28.05 | 54.2 | – |
| 3 | 24 150.651 | 24 160.721 | 10.06 | 52.1 | – |
| 3 | 104 084.892 | 106 607.212 | 2522.32 | 78.0 | LCORL, NCAPG, DCAF16, FAM184b, MED28, LAP3, ODPR, CLRN2, LDB2 |
| 6 | 28 806.179 | 30 119.366 | 1313.19 | 76.3 | CACNA2D4, LRTM2, ADIPOR2, WNT5B, FBXL14, ERC1, RAD52, WNK1, NINJ2, B4GALNT3 |
| 9 | 74 959.554 | 75 442.848 | 483.29 | 62.9 | ZFAT |
| 11 | 23 203.615 | 23 213378 | 9.76 | 52.1 | FBXO47, RPL23 |
| 11 | 29 683.529 | 31 631.624 | 1948.10 | 62.6 | COX11, STXBP4, HLF, MMD, TMEM100, PCTP, ANKFN1, C11orfH17orf67, DGKE, COIL, SCPEP1, AKAP1 |
| 11 | 32 599.971 | 32 940.833 | 340.86 | 54.4 | SUPT4H1, RNF43, HSF5, MTMR4, SEPT4, C11H17orf47, TEX14, RAD51C |
| 15 | 66 695467 | 67 228.658 | 533.19 | 52.1 | – |
In the Noriker breed, a strong selection pressure toward a well‐marked Leopard spotted phenotype (genotype: LP/lp PATN1/‐) exists and non‐favored phenotypes, such as few spots (LP/LP PATN1/‐) and snow cap blankets (LP/lp patn1/patn1), are not used for breeding purposes. Due to this diversifying selection favoring heterozygous LP/lp animals (Bellone et al. 2013; Holl et al. 2016; Druml et al. 2017a), in our sample of leopard spotted horses no ROH islands were found, including the LP locus. However, selection for a well‐marked leopard spotted phenotype simultaneously increases the frequency of the PATN1 allele (Druml et al. 2017a; Grilz‐Seger et al. 2017). Thus, 42% of leopard spotted horses shared an overlapping homozygous region around the PATN1 locus (ECA3:22.08–24.19 Mb), harboring 28 annotated genes (Fig. 4). The homozygous state for PATN1 was also confirmed by genotype data analyses performed by Druml et al. (2017a) and Grilz‐Seger et al. (2017), supporting our findings on ECA3.
Gene ontology and enrichment analysis
Of 51 annotated genes located in ROH islands of the entire Noriker sample, six GO terms related to biological processes, three GO terms related to cellular components and two GO terms related to molecular functions were extracted (Table 8). The highest levels of significance (P < 0.0001) were observed for the terms GO:0048704—embryonic skeletal system morphogenesis and GO:0009952—anterior/posterior pattern specification, both based on the genes HOXB3, HOXB1, PCGF2, HOXB2, HOXB7, HOXB8, HOXB5 and HOXB6. Two terms (GO:0021570—rhombomere 4 development and GO:0021612—facial nerve structural organization), related to development of structural elements, and the term GO:0001525—angiogenesis were highlighted. A significant enrichment (P < 0.003) of several members of the HOXB cluster was found for sequence‐specific DNA binding and transcription factor activity, where also the gene ZFAT was pinpointed. GO analysis revealed, with the exception of the leopard sample and the skin‐related terms in chestnuts horses, highest prominence for the HOXB cluster in all investigated groups, including high significances for the terms GO:0048704—embryonic skeletal system morphogenesis and GO:0009952—anterior/posterior pattern specification ([Link], [Link], [Link], [Link], [Link]). The identical HOXB cluster and GO terms were also cited in a recent study for the Posavina horse breed (Grilz‐Seger et al. 2018). From the manifold functions HOX genes can have, the definition of axial identity on the anterior–posterior axis, morphological regulation and genetic control of body shape represent the most important ones (Pearson et al. 2005). According to recent studies in evolutionary genetics, HOX gene mutations are involved in morphological adaptation and diversification (Pearson et al. 2005). In an archaeogenetic study on genomic changes in early domestic horses, Librado et al. (2017) revealed significant functional enrichment for the development of the anterior–posterior axis in Scythian horse remains, simultaneously listing HOXD8 among other 120 candidate genes selected by Scythian breeders. Although Noriker and Posavina horses exhibit different conformations, selection in both breeds is based upon conformation traits in favor of breed‐typical appearance. Therefore, we can postulate that the highlighted HOXB cluster (including HOXB1–HOXB13) may be the result of strong selection intensity on a breed‐specific habitus. The absence of a homozygous HOXB cluster in leopard spotted horses supports this hypothesis, as within this subpopulation the focus relies more on a well‐marked spotting pattern than on a breed‐specific conformation.
Table 8.
Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within runs of homozygosity (ROH) islands for the entire sample of Noriker horses.
| Category | Term | P‐value | Genes | Fold enrichment | Bonferroni adjusted P‐value |
|---|---|---|---|---|---|
| Biological process | GO:0048704—embryonic skeletal system morphogenesis | <0.001 | HOXB3, HOXB1, PCGF2, HOXB2, HOXB7, HOXB8, HOXB5, HOXB6 | 91.26 | <0.001 |
| GO:0009952—anterior/posterior pattern specification | <0.001 | HOXB3, HOXB1, PCGF2, HOXB2, HOXB7, HOXB8, HOXB5, HOXB6 | 42.22 | <0.001 | |
| GO:0021570—rhombomere 4 development | 0.006 | HOXB1, HOXB2 | 353.62 | 0.508 | |
| GO:0021612—facial nerve structural organization | 0.022 | HOXB1, HOXB2 | 88.40 | 0.941 | |
| GO:0001525—angiogenesis | 0.029 | HOXB3, HOXB13, TMEM100 | 10.94 | 0.977 | |
| Cellular component | GO:0005776—autophagosome | 0.004 | OSBPL7, CALCOCO2, PIP4K2B | 31.45 | 0.162 |
| GO:0005654—nucleoplasm | 0.010 | CWC25, MRPL10, HOXB7, PSMB3, SNF8, PNPO, HOXB13, KPNB1, PIP4K2B | 2.83 | 0.358 | |
| Molecular function | GO:0043565—sequence‐specific DNA binding | 0.003 | HOXB1, HOXB2, HOXB7, HOXB6, HOXB13 | 7.64 | 0.113 |
| GO:0003700—transcription factor activity, sequence‐specific DNA binding | 0.012 | HOXB2, HOXB7, HOXB8, HOXB6, ZFAT | 5.31 | 0.349 |
For leopard spotted horses, only one GO term related to biological process and one KEGG pathway were extracted (Table S1). Both were based upon the genes RAD51C and RAD52. At a significance level of 0.009 the term GO:0000730—DNA recombinase assembly and the KEGG pathway ecb03440:homologous recombination were derived for this subpopulation. The genes RAD51C and RAD52 play an important role for DNA double‐strand break repair and homologous recombination (Chapman et al. 2012).
In the chestnut subpopulation (Table S2), additional GO terms related to skin development (GO:0010482—regulation of epidermal cell division, GO:0045606—positive regulation of epidermal cell differentiation and GO:0003334—keratinocyte development) and cell regulation (GO:0043410—positive regulation of MAPK cascade) were extracted. One term in chestnut horses was related to molecular function (GO:0043565—sequence‐specific DNA binding), which was also present in all samples except in the leopard spotted group. Additionally the chestnut‐specific KEGG pathway ecb03010:Ribosome was highlighted.
One term (GO:0002244—hematopoietic progenitor cell differentiation) related to biological processes was found to be specific for bay horses (Table S3).
Size‐associated SNPs within ROH islands
The Noriker horse, a medium‐sized draft horse breed, generally with an average height at withers ranging from 155 to 160 cm, was exposed to a selection pressure toward a higher height at withers and higher caliber within the last decades (from 155 cm average height in the year 1973 to 162 cm in the year 2004). Currently, individual maxima could reach a height at withers of up to 173 cm, and especially black and blue roans show a higher tendency toward a bigger size. Nevertheless, a variation in this trait exists, and tobiano and leopard spotted horses frequently exhibit a lower caliber (Druml et al. 2008). The caliber index describes the relation of chest circumference and cannon bone circumference to height at withers and is a well‐suited descriptor to differentiate between light and heavy horses (Druml et al. 2008). In Posavina, horse height at withers ranges from 138 to 148 cm, and it is therefore considered the smallest mid‐European draught horse breed. Although exhibiting a smaller frame, this breed is characterized by a high caliber.
From the entire Noriker sample, 60.3% of the horses shared a homozygous region on ECA9 harboring the single gene ZFAT (zinc finger and AT‐hook domain containing). Near this gene, Makvandi‐Nejad et al. (2012) identified a SNP at position ECA9:75 550 059, which was associated with body size in horses and explained 56.7% of phenotypic variation among 16 breeds. In addition to this SNP, the authors verified three further loci near the genes HMGA2 (high mobility group AT‐hook 2) and LCORL (ligand dependent nuclear receptor corepressor like) and the adjacent NCAPG (non‐SMC condensin I complex subunit G) and LASP1 (LIM and SH3 protein 1) genes. All four loci together explained 83% of size variation in horses. The association analysis of Makvandi‐Nejad et al. (2012) was based upon a previous study by Brooks et al. (2010), in which the majority (65.9%) of the variance in 33 linear and circular body measurements was explained by the first principal component describing overall body size. By use of F ST‐based statistics, Petersen et al. (2013) also identified putative targets of selection for size (mass and height) on ECA3 and ECA11, harboring the genes LCORL, NCAPG and LASP1, in draught and Miniature horses. All these aforementioned genes were identified within ROH islands of the entire Noriker sample. We extracted three out of four SNPs associated with variation in body size according to Makvandi‐Nejad et al. (2012) for the entire set of Noriker and Posavina samples (Table 9). At the SNP near the LCORL/NCAPG locus, 93.1% of Noriker horses were homozygous for the C/’Big’ allele. The other two SNPs, on ECA9 and ECA11, were homozygous for the ‘Big’ alleles in 68.9% and 70.1% of Noriker horses respectively.
Table 9.
Genotype frequencies of body size associated SNPs near the genes LCORL/NCAPG, ZFAT, LASP1 from the study of Makvandi‐Nejad et al. (2012) calculated for all Noriker horses (n = 174) and the Posavina sample (n = 28).
| Chr. | SNP | Gene | Position | Genotype frequency | HWE P‐value | Major allele |
|---|---|---|---|---|---|---|
| Posavina: mean height at withers = 138–148 cm; high caliber | ||||||
| 3 | AX‐103666681 | LCORL/NCAPG | 105 547.002 | 3.8% C/C; 39.3% C/T; 57.1% T/T | n.s. | T (small) |
| 9 | AX‐103185455 | ZFAT | 75 550.059 | 7.1% C/C; 25.0% C/T; 67.9% T/T | n.s. | T (big) |
| 11 | AX‐104097126 | LASP1 | 23 259.732 | 7.1% G/G; 35.7% A/G; 57.1% A/A | n.s. | A (big) |
| Noriker: mean height at withers = 158–165 cm; high caliber | ||||||
| 3 | AX‐103666681 | LCORL/NCAPG | 105 547.002 | 0% T/T; 6.9% T/C; 93.1% C/C | n.s. | C (big) |
| 9 | AX‐103185455 | ZFAT | 75 550.059 | 18.3% C/C; 12.8% T/C; 68.9% T/T | >0.001 | T (big) |
| 11 | AX‐104097126 | LASP1 | 23 259.732 | 1.8% G/G; 28.1% A/G; 70.1% A/A | n.s. | A (big) |
HWE, Hardy–Weinberg equilibrium.
Dividing the horses into the different color strains, the proportion of homozygous animals for the C allele (‘Big’) at the SNP near LCORL/NCAPG ranged from 81.1% in black to 97.9% in leopard spotted horses. None of the animals was homozygous for the T allele (‘Small’). The genotype frequencies within the subpopulations are shown in Fig. 5. The majority (63.6%–78.6%) of basic colored and blue roan horses were homozygous for the T allele (‘Big’) at the ZFAT locus. All tobianos and 33.3% of the leopard spotted horses were homozygous for the C allele, associated with small body size. For the third SNP at the LASP1 locus, the percentage of animals homozygous for the A allele (‘Big’) varied from 64.3% in roan to 86.1% in chestnut horses and 4.2% of leopard spotted horses and one out of three tobianos were homozygous for the G allele (‘Small’).
Figure 5.
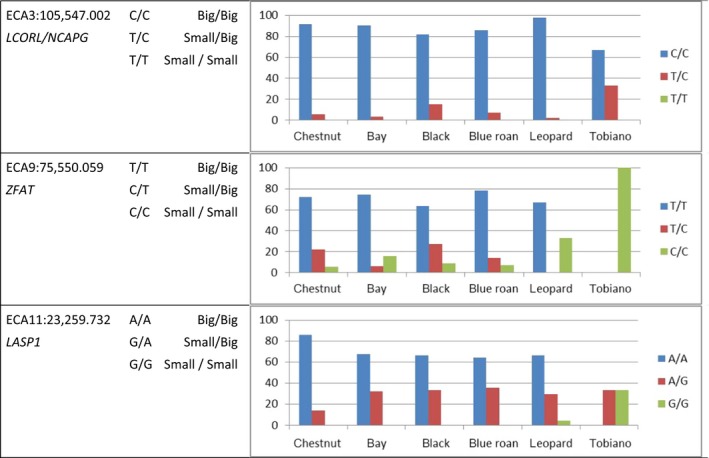
Distribution of three SNP genotypes associated with body size in horses from the study of Makvandi‐Nejad et al. (2012) for Noriker horses structured by color strains (36 chestnut, 31 bay, 33 black, 23 blue roan, 48 leopard three tobiano horses). The SNPs are located near the genes LCORL/NCAPG (AX‐103666681 at ECA3:105 547.002), ZFAT (AX‐103185455 at ECA9:75 550.059) and LASP1 (AX‐104097126 at ECA11:23 259.732).
The extracted size‐associated SNP genotypes for the Posavina horse revealed a different profile. At the LCORL/NCAPG locus, 57.1% of the animals were homozygous for the T allele (‘Small’). For the two other SNPs on ECA9 and ECA11, 67.9% and 57.1% of horses respectively were homozygous for the ‘Big’ alleles (Table 9). The inverse genotype relationship between Posavina horses and Noriker on the LCORL/NCAPG locus supports the assumption that those genes are associated with height at withers and that the other two loci, ZFAT and LASP1, are involved in size parameters (length, volume, caliber). Deviation from Hardy‐Weinberg equilibrium was observed for the ZFAT locus in the Noriker horse (P < 0.001) and revealed ongoing selection toward bigger body size.
Conclusion
The breeding objective of the Noriker horse is based mainly upon the evaluation of conformation traits. Coat color is an important additional selection trait, which is pursued in strict mating strategies applied by the breeders. Although various ROH islands highlighted the selection for morphological traits and size in Noriker horses, this study demonstrates that, within a closed population sharing the same founders and ancestors, selection that differs in only one single trait (coat color) can result in a genetic fragmentation affecting levels of autozygosity and ROH island patterns. Furthermore, we were able to show that specific selection for coat color might also affect other genomic regions besides the target locus, as pinpointed by deviating ROH islands and gene ontologies. We could confirm that genes enclosed in ROH islands represent targets of selection, exemplarily illustrated for SNPs associated with coat color and body size. Therefore, we suggest that, besides other methods, the analysis of ROH islands should be taken into account to scan the genome for selection signatures.
Conflict of interest
The authors declare that there are no conflicts of interest.
Availability of data
Genotypes are available for reproduction of the results upon signing of a material transfer agreement. ROH data are accessible via www.animalgenome.org/repository/pub/UVMV2019.0223/
Supporting information
Figure S1. Distribution of ROHs on ECA9 per individual.
Figure S2. Distribution of ROHs on ECA11 per individual.
Table S1. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the leopard spotted Noriker sample.
Table S2. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the chestnut Noriker sample.
Table S3. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the bay Noriker sample.
Table S4. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the black Noriker sample.
Table S5. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the blue roan Noriker sample.
Acknowledgements
This work was financially supported by the Austrian Research Promotion Agency (FFG, contract no. 843464), the Federal Ministry for Sustainability and Tourism (BMNT; contract no. 101332) and Slovenian Research Agency program P4‐0053 to M. Cotman.
References
- Bellone R.R., Holl H., Setulari V. et al (2013) Evidence for retroviral insertion in TRPM1 as the cause of congenital stationary night blindness and leopard complex spotting in the horse. PLoS ONE 8, e78280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S.A., Makvandi-Nejad S., Chu E., Allen J.J., Streeter C., Gu E., McCleery B., Murphy B.A., Bellone R. & Sutter N.B. (2010) Morphological variation in the horse: defining complex traits of body size and shape. Animal Genetics 41(Suppl. 2), 159–65. [DOI] [PubMed] [Google Scholar]
- Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M. & Wilson J. (2018) Runs of homozygosity: window into population history and trait architecture. Nature Reviews Genetics 19, 220–34. [DOI] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R.G. & Boulton S.J. (2012) Playing the end game: DNA double‐strand break repair pathway choice. Molecular Cell 47, 497–510. [DOI] [PubMed] [Google Scholar]
- Druml T., Baumung R. & Sölkner J. (2008) Morphological analysis and effect of selection for conformation in the Noriker draught horse population. Livestock Science 115, 118–28. [Google Scholar]
- Druml T., Baumung R. & Sölkner J. (2009) Pedigree analysis in the Austrian Noriker draught horse: genetic diversity and the impact of breeding for coat color on population structure. Journal of Animal Breeding and Genetics 126, 348–56. [DOI] [PubMed] [Google Scholar]
- Druml T., Neuditschko M., Grilz‐Seger G., Neuhauser B. & Brem G. (2017a) Phenotypic and genetic analysis of the leopard complex spotting in Noriker horses. Journal of Heredity 108, 505–14. [DOI] [PubMed] [Google Scholar]
- Druml T., Grilz‐Seger G., Neuditschko M. & Brem G. (2017b) Association between population structure and allele frequencies of the glycogen synthase 1 mutation in the Austrian Noriker draft horse. Animal Genetics 48, 108–12. [DOI] [PubMed] [Google Scholar]
- Druml T., Neuditschko M., Grilz‐Seger G., Horna M., Ricard A., Mesarič M., Cotman M., Pausch H. & Brem G. (2018) Population networks associated with runs of homozygosity reveal new insights into the breeding history of the Haflinger horse. Journal of Heredity 119, 384–92. [DOI] [PubMed] [Google Scholar]
- Grilz‐Seger G., Neuhauser B., Druml T. & Brem G. (2017) Classification and nomenclature of the leopard complex spotting in the Noriker horse breed and its relevance for the breeding for color. Züchtungskunde 89, 359–74. [Google Scholar]
- Grilz‐Seger G., Mesarič M., Cotman M., Neuditschko M., Druml T. & Brem G. (2018) Runs of homozygosity and population history of three horse breeds with limited population size. Journal of Equine Veterinary Science 71, 27–34. [Google Scholar]
- Holl H.M., Brooks S.A., Archer S., Brown K., Malvick J., Penedo M.C. & Bellone R.R. (2016) Variant in the RFWD3 gene associated with PATN1, a modifier of leopard complex spotting. Animal Genetics 47, 91–101. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T. & Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Librado P., Gamba C., Gaunitz C., Der Sarkissian C. & Pruvost M. (2017) Ancient genomic changes associated with domestication of the horse. Science 356, 442–5. [DOI] [PubMed] [Google Scholar]
- Makvandi‐Nejad S., Hoffman G.E., Allen J.J. et al (2012) Four loci explain 83% of size variation in the horse. PLoS ONE 7, e39929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund L., Moller M.J., Sandberg K. & Andersson L. (1996) A missense mutation in the gene for melanocyte‐stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mammalian Genome 7, 895–9. [DOI] [PubMed] [Google Scholar]
- Marklund L., Moller M.J., Sandberg K. & Andersson L. (1999) Close association between sequence polymorphism in the KIT gene and the roan coat color in horses. Mammalian Genome 10, 283–8. [DOI] [PubMed] [Google Scholar]
- McCue M.E., Bannasch D.L., Petersen J.L. et al (2012) A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genetics 8, e1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A., Abdel‐Rahman R. et al (2008) Runs of homozygosity in European populations. American Journal of Human Genetics 83, 359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger J., Karwath M., Tonda R., Beltran S., Águeda L., Gut M., Gut L.G. & Distl O. (2015) Runs of homozygostiy reveal signatures of positive selection for reproduction traits in breed and non‐breed horses. BMC Genomics 16, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuditschko M., Khatkar M.S. & Raadsma H.W. (2012) NetView: a high‐definition network‐visualization approach to detect fine scale population structures from genome‐wide patterns of variation. PLoS ONE 7, e48375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.C., Lemmons D. & McGinnis W. (2005) Modulating HOX gene functions during animal body patterning. Nature Reviews Genetics 6, 893–904. [DOI] [PubMed] [Google Scholar]
- Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A. & Li J.Z. (2012) Genomic patterns of homozygosity in worldwide human populations. American Journal of Human Genetics 91, 275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peripolli E., Munari D.P., Siva M.V.G.B., Lima A.L.F., Irgang R. & Baldi F. (2016) Runs of homozygosity: current knowledge and applications in livestock. Animal Genetics 48, 255–71. [DOI] [PubMed] [Google Scholar]
- Peripolli E., Stafuzza N.B., Munari D.P., Lima A.L.F., Irgang R., Machado M.A., Panetto J.C.C., Ventura R.V., Baldi F. & Silva M.V.G.B. (2018) Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics 19, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J.L., Mickelson J.R., Rendahl A.K., Valberg S.J., Andersson L.S., Axelson J., Bailey E‐ Bannesch D., Binns M.M. & Borges A.S. (2013) Genome‐wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genetics 9, e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruvost M., Bellone R., Benecke N. et al (2011) Genotypes of predomestic horses match phenotypes painted in Paleolithic works of cave art. Proceedings of the National Academy of Sciences of the United States of America 108, 18626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd‐Brown K. et al (2007) plink: a toolset for whole‐genome association and population‐based linkage analyses. American Journal of Human Genetics 81, 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (2009) sas version 9.1. SAS Institute, Inc., Cary, NC. [Google Scholar]
- Steinig E.J., Neuditschko M., Khatkar M.S., Raadsma H.W. & Zenger K.R. (2015) netview p: a network visualization tool to unravel complex population structure using genome‐wide SNPs. Molecular Ecology Resources 16, 216–27. [DOI] [PubMed] [Google Scholar]
- Wade C.M., Giulotto E., Sigurdsson S. et al (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Distribution of ROHs on ECA9 per individual.
Figure S2. Distribution of ROHs on ECA11 per individual.
Table S1. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the leopard spotted Noriker sample.
Table S2. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the chestnut Noriker sample.
Table S3. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the bay Noriker sample.
Table S4. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the black Noriker sample.
Table S5. Gene Ontology (GO) terms and KEGG pathways enriched (P < 0.05) based on annotated genes within ROH islands for the blue roan Noriker sample.
Data Availability Statement
Genotypes are available for reproduction of the results upon signing of a material transfer agreement. ROH data are accessible via www.animalgenome.org/repository/pub/UVMV2019.0223/


