Abstract
The c‐Myc protein is a transcription factor with oncogenic potential controlling fundamental cellular processes. Homologs of the human c‐myc protooncogene have been identified in the early diploblastic cnidarian Hydra (myc1, myc2). The ancestral Myc1 and Myc2 proteins display the principal design and biochemical properties of their vertebrate derivatives, suggesting that important Myc functions arose very early in metazoan evolution. c‐Myc is part of a transcription factor network regulated by several upstream pathways implicated in oncogenesis and development. One of these signaling cascades is the Wnt/β‐Catenin pathway driving cell differentiation and developmental patterning, but also tumorigenic processes including aberrant transcriptional activation of c‐myc in several human cancers. Here, we show that genetic or pharmacological stimulation of Wnt/β‐Catenin signaling in Hydra is accompanied by specific downregulation of myc1 at mRNA and protein levels. The myc1 and myc2 promoter regions contain consensus binding sites for the transcription factor Tcf, and Hydra Tcf binds to the regulatory regions of both promoters. The myc1 promoter is also specifically repressed in the presence of ectopic Hydra β‐Catenin/Tcf in avian cell culture. We propose that Hydra myc1 is a negative Wnt signaling target, in contrast to vertebrate c‐myc, which is one of the best studied genes activated by this pathway. On the contrary, myc2 is not suppressed by ectopic β‐Catenin in Hydra and presumably represents the structural and functional c‐myc ortholog. Our data implicate that the connection between β‐Catenin‐mediated signaling and myc1 and myc2 gene regulation is an ancestral metazoan feature. Its impact on decision making in Hydra interstitial stem cells is discussed.
Keywords: cnidarian, development, gene regulation, oncogene, signal transduction
Abbreviations
- Alp
alsterpaullone
- bHLHZip
basic region/helix‐loop‐helix/leucine zipper
- CEF
chicken embryo fibroblasts
- ChIP
chromatin immunoprecipitation
- ESI‐MS
electrospray ionization mass spectrometry
- GFP
green fluorescent protein
- GSK
glycogen synthase kinase
- HA
hemagglutinin
- myc
myelocytomatosis oncogene
- RACE
rapid amplification of cDNA ends
- Tcf/LEF
T‐cell factor/lymphoid enhancer‐binding factor
- Wnt
wingless/int
Introduction
The c‐myc gene has been originally identified in the form of the transforming determinant of avian acute leukemia virus MC29 1, 2. The highly oncogenic v‐myc allele is derived from the cellular chicken c‐myc protooncogene by retroviral transduction 1, 2, 3, 4, 5. The cellular c‐myc proto‐oncogene encodes the c‐Myc protein, a transcription factor with oncogenic potential representing the central hub of a network controlling global gene expression and regulating fundamental cellular processes like growth, proliferation, differentiation, metabolism, and apoptosis 2, 3, 5, 6, 7, 8. c‐Myc is a bHLHZip protein encompassing protein dimerization domains (helix‐loop‐helix, leucine zipper) and a DNA contact surface (basic region) that forms heterodimers with the Max (MAX) protein and binds typically to specific DNA sequence elements termed E‐boxes (5′‐CACGTG‐3′) 4, 5. Upstream acting signaling pathways regulate the c‐Myc transcription factor network like those triggered by mitogenic receptor tyrosine kinases, or by wingless/int (Wnt) 9, 10, 11, 12. Human T‐cell factor 4 (Tcf4), the effector of Wnt/β‐Catenin signaling has been identified as an oncogenic regulator of the c‐myc and cyclin D1 genes in colon cancer 13, 14, 15. The Wnt signaling pathway is highly conserved throughout animal evolution representing one of the key cascades to regulate development and stemness 14, 16, 17.
c‐Myc and Max homologs with conserved basic functions have been found in early diverging metazoans 18 and even in premetazoans 19, suggesting that principal functions of the c‐Myc master regulator arose very early in the evolution of multicellular animals. In the diploblastic cnidarian Hydra, two c‐myc homologs (myc1 and myc2) have been identified, which are transcriptionally activated in the interstitial stem cell system 18, 20, 21. Both Hydra Myc proteins show the same degree of overall sequence identity (32%) compared to human c‐Myc, but Myc2 is structurally more related to its human ortholog concerning protein size and the degree of Myc box conservation (MBI, MBIII) in the transactivation domain 18. Paradoxically, downregulation of myc1 by short interfering RNA or chemical inhibition promoted stem cell proliferation 22, suggesting a divergent role of Myc1 in the homeostasis of the interstitial stem cell lineage. In contrast to myc1, expression of the myc2 gene is not restricted to the interstitial stem cell system but also occurs in proliferating epithelial stem cells throughout the gastric region. Furthermore, myc2 is specifically activated in cycling precursor cells during early oogenesis and spermatogenesis, suggesting that the Myc2 protein has a possible nonredundant function in cell cycle progression 20.
Crucial components of the Wnt signaling pathway regulating c‐myc in vertebrates are conserved in Hydra such as Wnt3a, Frizzled, Dsh (disheveled), GSK (glycogen synthase kinase)‐3β, β‐Catenin, or Tcf (T‐cell factor)/Lef (lymphoid enhancer‐binding factor) 23, 24, 25, 26. Further studies have revealed a surprising complexity of cnidarian Wnt proteins that are implicated in fundamental morphogenetic processes emphasizing the important role of Wnt signaling in organismal patterning throughout the animal kingdom 16, 27, 28, 29, 30, 31. Here, we report that myc1 is downregulated in the gastric region upon ectopic activation of β‐Catenin signaling, whereas under the same conditions myc2 expression levels remain constant. Our results therefore imply that the Hydra myc1 gene represents a potential negative target of the Wnt/β‐Catenin/Tcf signaling pathway and that myc2 presumably represents the functional ortholog of human c‐myc.
Results
Activation of β‐Catenin signaling in Hydra leads to specific repression of myc1
Possible effects of nuclear β‐Catenin signaling on the expression of the myc1 and myc2 genes were analyzed by in situ hybridization. Polyps from transgenic Hydra (β‐cat‐Tg) express high levels of a β‐Catenin‐GFP fusion protein triggered by an actin promoter throughout their entire body columns. As a result of the enhanced nuclear signaling activity of β‐Catenin in all cells, β‐cat‐Tg animals form multiple ectopic head and foot structures 31, 32. These animals were compared with wild‐type polyps by testing for myc1 and myc2 expression. The in situ hybridization patterns showed that myc1 was significantly downregulated throughout the body column of β‐cat‐Tg animals, whereas myc2 expression remained unchanged or even appeared to be slightly upregulated in distinct cells (Fig. 1A). Expression of myc1 and myc2 was also analyzed after Alsterpaullone (Alp) treatment, and compared with nontreated wild‐type control animals. This compound specifically blocks the activity of GSK‐3β that normally contributes to β‐Catenin degradation by phosphorylation 28. Consequently, β‐Catenin levels in cell nuclei are elevated due to protein stabilization, and ectopic tentacles and heads form along the entire body column 28, 30. Equivalent to the results obtained in β‐cat‐Tg animals, expression of myc1 was downregulated after 48 h in Alp‐treated animals, whereas myc2 expression showed the same expression pattern as observed for the β‐cat‐Tg animals (Fig. 1B).
Figure 1.
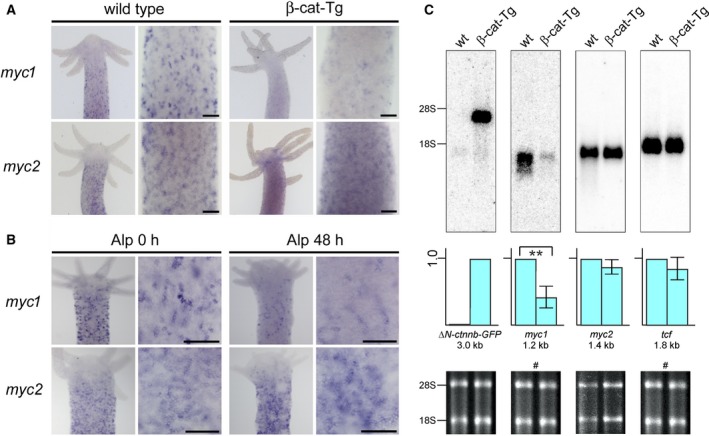
Specific repression of myc1 mRNA expression upon β‐Catenin activation. (A) Expression patterns of Hydra myc1 and myc2 in β‐Catenin transgenic animals (β‐cat‐Tg) compared to wild‐type polyps. Whole wild‐type and transgenic polyps have myc1 and myc2 expressed throughout their body columns, but not in head and foot structures. myc1 is downregulated in the transgenic polyps, whereas myc2 levels are not significantly changed. (B) Differential expression of Hydra myc1 and myc2 48 h after onset of treatment with the GSK‐3β inhibitor Alp. Inhibition of GSK‐3β results in activation of β‐Catenin in the canonical Wnt signaling pathway, and as a result in a reduction of the myc1 expression level. Upper halves of polyps and magnified views from gastric regions are shown. Bars in the magnified views represent 25 μm. (C) Northern analysis using aliquots (2.0 μg) of poly(A)+‐selected RNAs from whole wild‐type (wt) and transgenic Hydra animals, and Hydra myc1, myc2, or tcf‐specific cDNA probes derived from the relevant coding regions. Positions of residual ribosomal RNAs (28S, 18S) are given on the left site. Ethidium bromide‐stained RNAs used for blot analysis are shown below. The blot, which was hybridized to a second probe after filter washing is indicated by a hash sign (#). For transgenic mRNA detection, a GFP‐specific probe (ΔN‐ctnnb‐GFP) was applied (arbitrary expression level 1.0). Representative blots from three independent experiments are shown where myc1, myc2, and tcf levels from wild‐type animals were arbitrarily set to 1.0. Standard deviations (SD, n = 3) are shown by vertical bars. Statistical significance was assessed by using a paired Student t‐test (**P < 0.01).
To confirm the specific downregulation of myc1, poly(A)+‐selected RNAs isolated from whole β‐cat‐Tg animals and from whole wild‐type controls were analyzed by Northern hybridization using DNA probes derived from the coding regions of Hydra myc1, myc2, and tcf. As a control for detection of the transgenic ΔN‐ctnnb‐GFP mRNA, a probe derived from the GFP‐encoding portion was used. The Northern analysis confirmed that, compared to normal animals, the transgenic polyps contain distinctly lower amounts of myc1 mRNA, in contrast to myc2, or tcf whose overall expression levels were almost not affected (Fig. 1C). To test myc1 and myc2 expression also at the protein levels, immunoblot analysis was performed using cell extracts from whole animals and immunoglobulin G‐purified antisera directed against Myc1, or Myc2. Previous immunoprecipitation analyses using extracts from 35S‐methionine pulse‐labeled Hydra and a Myc1‐specific antibody have detected the endogenous Myc1 protein with an apparent molecular mass of 36 kDa and a minor 32‐kDa protein, whereas in vitro translated full‐length Myc1 had a size of 39 kDa 18, 20. The 36‐kDa Myc1 protein presumably results from the usage of an alternative translation start site 18. On the other hand, endogenous Myc2 was expressed as a 41‐kDa protein having the same size as the in vitro translated full‐length product 20. Here, immunoblot analysis using equal amounts of nonlabeled extracts from wild‐type and transgenic Hydra detected Myc1 and Myc2 proteins with similar apparent molecular masses as the previously detected proteins from pulse‐labeled cells, but in case of Myc1 the 32‐kDa protein band was the dominant one and the 36‐kDa isoform only weakly expressed (Fig. 2). Myc2, expressed at low levels as reported previously 20, displayed an apparent molecular mass of 41 kDa as expected. The 32‐kDa Myc1 isoform, which was downregulated in transgenic polyps, could result from proteolytic processing of the 36‐kDa protein. Although the nature of the smaller Myc1 protein needs yet to be determined, the results are in line with those obtained by in situ and Northern expression analyses (Fig. 1). The lower amounts of endogenous Myc2 versus Myc1 are probably due to the higher instability of the Myc2 protein as reported previously 20. Titer and specificities of the applied polyclonal antibodies were controlled using the carboxyl‐terminal Myc1 p16 and the full‐length Myc2 p41 protein (Fig. 2A,B). As expected, expression of the ΔN‐ctnnb‐GFP protein was exclusively detected in transgenic Hydra, whereas Tcf or Max were expressed at almost equal levels both in wild‐type and in transgenic animals (Fig. 2C).
Figure 2.
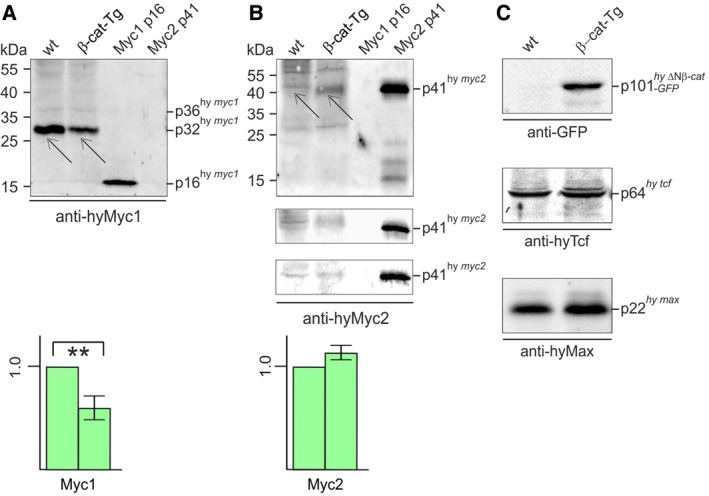
Specific repression of Myc1 protein expression upon β‐Catenin activation. (A, B) Expression of Hydra Myc1 (A) and Myc2 (B) proteins in β‐catenin transgenic animals (β‐cat‐Tg) compared to wild‐type (wt) polyps. (C) Expression control of the transgenic fusion protein ΔN‐Ctnnb‐GFP, and of Hydra Tcf or Hydra Max in β‐cat‐Tg compared to wt polyps. Protein expression was tested by immunoblot analysis using each 10 μg of cell extract. Proteins were resolved by SDS/PAGE (10%, wt/vol) and detected using antibodies directed against recombinant Myc1, Myc2, Max, or Tcf proteins, or against the green fluorescent protein (GFP). As specificity control, each 10 ng of recombinant Myc1 p16 or full‐length Myc2 (p41) were used in panels A and B. Representative blots from three independent experiments are shown, where Myc1 and Myc2 levels from wild‐type animals were arbitrarily set to 1.0. Thin arrows depict the endogenous Hydra Myc1 or Myc2 protein bands. Standard deviations (SD, n = 3) are shown by vertical bars. Statistical significance was assessed by using a paired Student t‐test (**P < 0.01). In case of Myc2, which is hardly detectable by immunoblotting, all three blots used for quantification are shown (P = 0.08).
Loss of myc1 expression could result from either transcriptional downregulation of this gene in interstitial cells including stem cells, which occur as single cells (1s) and cell pairs (2s), or from potential β‐Catenin‐induced disappearance of myc1‐expressing cells. We studied putative changes in the numbers of myc‐expressing interstitial stem cells in the body columns of β‐Catenin‐activated versus wild‐type polyps using the maceration technique (Fig. 3A). In a macerated cell suspension, cells maintain their original morphology, and thereby the different cell types can be clearly distinguished and precisely quantified (Fig. 3B) 33. The results in Fig. 3C show that in both, β‐cat‐Tg and Alp‐treated tissue, interstitial stem cells are embedded at higher densities in the ecto‐ and endodermal epithelial tissue layers. Thus, enhanced nuclear β‐Catenin signaling seems to stimulate interstitial stem cell maintenance. Furthermore, our results clearly support the view that loss of myc1 expression is most likely caused by β‐Catenin‐regulated transcriptional repression in these cells.
Figure 3.
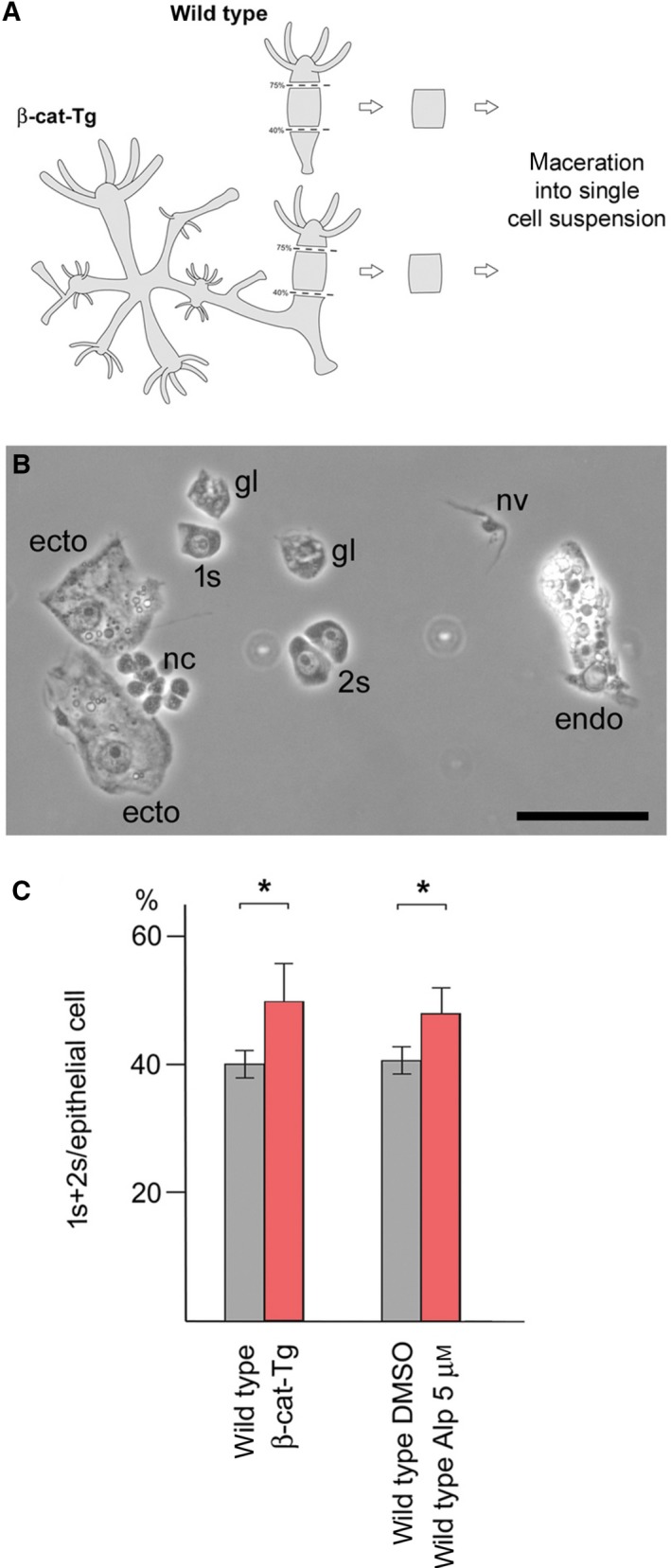
Interstitial stem cell densities in the body columns of β‐Catenin‐activated and wild‐type polyps. (A) Scheme of the position of macerated tissue pieces. Tissue samples representing roughly half of the tissue mass of a budless control polyp were excised from the mid body column. Equivalent tissue pieces were excised from the body columns of β‐cat‐Tg transgenes and polyps treated with Alp for 60 h. The tissue pieces were macerated, spread as single‐cell suspensions onto microscope slides, and analyzed using a phase contrast microscope. (B) Representative phase contrast image of macerated cells from a β‐cat‐Tg polyp. 1s: single interstitial stem cell; 2s: interstitial stem cell pair; ecto: ectodermal epithelial cell; endo: endodermal epithelial cell; nv: nerve cell; nc: nest of differentiating nematocytes; gl: gland cell. Bar: 25 μm. (C) Quantification of macerated tissue pieces reveals higher interstitial stem cell (1s+2s) densities in β‐Catenin‐activated tissues as compared with wild‐type controls. 1s+2s density is defined as the numbers of 1s+2s per epithelial cells. Bars represent the mean ± SD of three independent experiments. Statistical significance was assessed by using a paired Student t‐test (*P < 0.1).
Hydra Tcf binds to the promoters of Hydra myc1 and myc2
To investigate the mechanism of Hydra myc1 downregulation by Wnt/β‐Catenin signaling, the promoter region of myc1 was defined by transcription start site mapping. cDNA prepared from whole Hydra was subjected to 5′rapid amplification of cDNA ends (5′RACE). Two closely spaced transcription start sites were detected in the myc1 promoter region, whereas transcription of myc2 starts at one defined site (Fig. 4A) as reported recently 20. The sequence of the myc1 5′RACE product also allowed deduction of the relevant full‐length mRNA sequence. Alignment of this sequence with the published Hydra genome 34 led to definition of the myc1 gene topography. In contrast to myc2, which consists of three exons similar to the vertebrate c‐myc genes 20, Hydra myc1 has two exons only (Fig. 4B). Inspection of the 5′‐untranscribed regions of myc1 and myc2 revealed that both promoters contain potential binding elements termed TBE 13 for the Tcf transcription factor, closely matching the consensus site 5′‐CCTTTGWW‐3′ 35 (Fig. 4A). Whereas the myc2 promoter contains two TBE motifs located at positions −166 and −98 nucleotides (nt) upstream of the transcription start site, three canonical TBE sites are present in the myc1 promoter mapping to nt positions −128, −79, and +46 with reference to the proximal transcription start site (Fig. 4A). Interestingly, the myc2 promoter also contains a canonical Myc binding site (E‐box) at nt position −60 (Fig. 4A). Inspection of the promoters for further transcription factor binding sites revealed the presence of Oct‐1, C/EBPα, or NF‐1 motifs in the myc1, and of hunchback, C/EBPα, or NFκB motifs in the myc2 promoter, respectively (Fig. 5).
Figure 4.
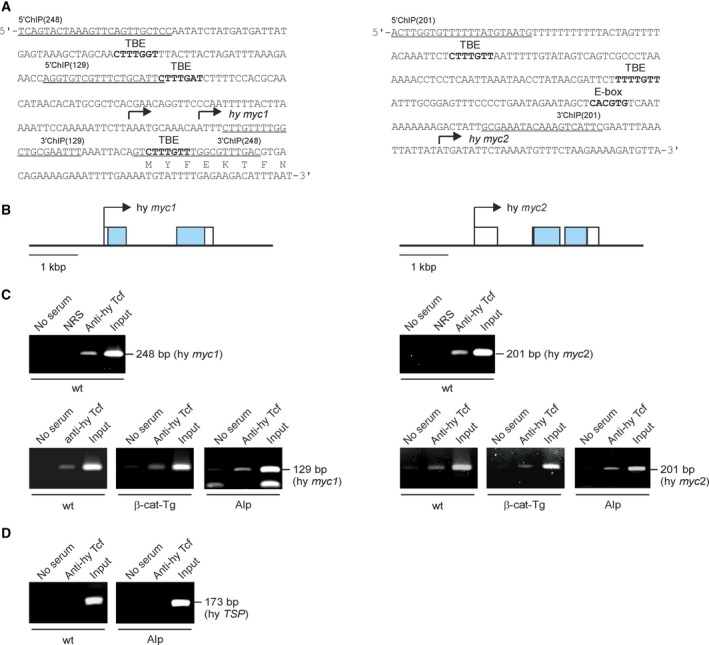
Structures of the Hydra myc1 and myc2 promoter regions and binding of Hydra Tcf. (A) Nucleotide sequences of the myc1 and myc2 regulatory regions. The transcription start sites mapped by 5′RACE (arrows), potential binding sites (in bold) for the transcription factors Tcf (TBE) or Myc (E‐box), and binding sites for 5′ and 3′ ChIP primers (underlined) are indicated. (B) Topographies of Hydra magnipapillata genomic loci (accession nos. NW_004167287, NW_004167363) containing the Hydra myc1 and myc2 genes. Exons are depicted by boxes with the coding regions shown in black. Arrows indicate the main transcription start sites. (C) ChIP of the Hydra myc1 and myc2 promoter regions using chromatin from whole Hydra wild type (wt) animals, from β‐catenin transgenic animals (β‐cat‐Tg), and from Alsterpaullone‐treated (Alp) Hydra. An antiserum directed against Hydra Tcf was used for precipitation, followed by PCR amplification of the indicated fragments from the myc1 or myc2 regulatory regions. Reactions with normal rabbit serum (NRS) or total chromatin (Input) were used as controls. (D) A fragment containing no Tcf binding site derived from the Hydra TSP regulatory region 36 was amplified as a control.
Figure 5.
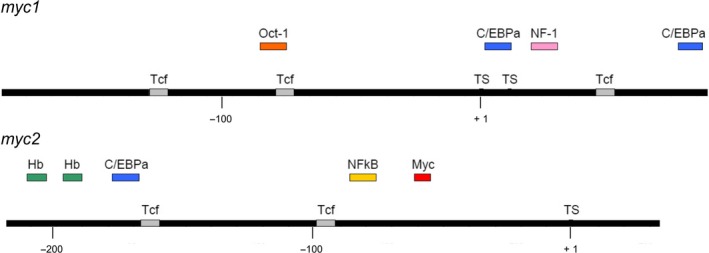
Promoter maps of Hydra myc1 and myc2. Bars represent the promoter regions. Positions of transcription factor binding sites were identified using the computer program AliBaba2 (gene‐regulation.com) and are depicted on or above the bars (Tcf, T‐cell‐specific transcription factor; C/EBPalpha, CCAAT enhancer‐binding protein alpha; NF‐1, nuclear factor 1; Hb, hunchback; NFkappaB, nuclear factor kappa B; Myc, myelocytomatosis viral oncogene protein product).
To test if promoter regions of Hydra myc1 and myc2 are bound by Tcf in vivo, ChIP analysis was performed using cross‐linked chromatin from whole wild‐type, transgenic, and Alp‐treated Hydra animals, and a Tcf‐specific antibody followed by PCR amplification of the specific DNA regions (Fig. 4C). To generate the antiserum directed against Hydra Tcf, a partial recombinant Hydra Tcf protein was expressed in Escherichia coli, purified, and then used as an immunogen (Fig. 6). Bound Hydra Tcf was detectable on both promoter segments (Fig. 4C) suggesting that a β‐Catenin/Tcf protein complex could indeed regulate the myc1 and myc2 genes. No significant changes in promoter occupancy were observed after β‐Catenin activation either by overexpression (β‐cat‐Tg) or GSK‐3β inhibition (Alp) (Fig. 4C). As a negative control for Tcf binding, a segment derived from the Hydra thrombospondin (TSP) promoter containing no Tcf binding site 36 was tested (Fig. 4D).
Figure 6.
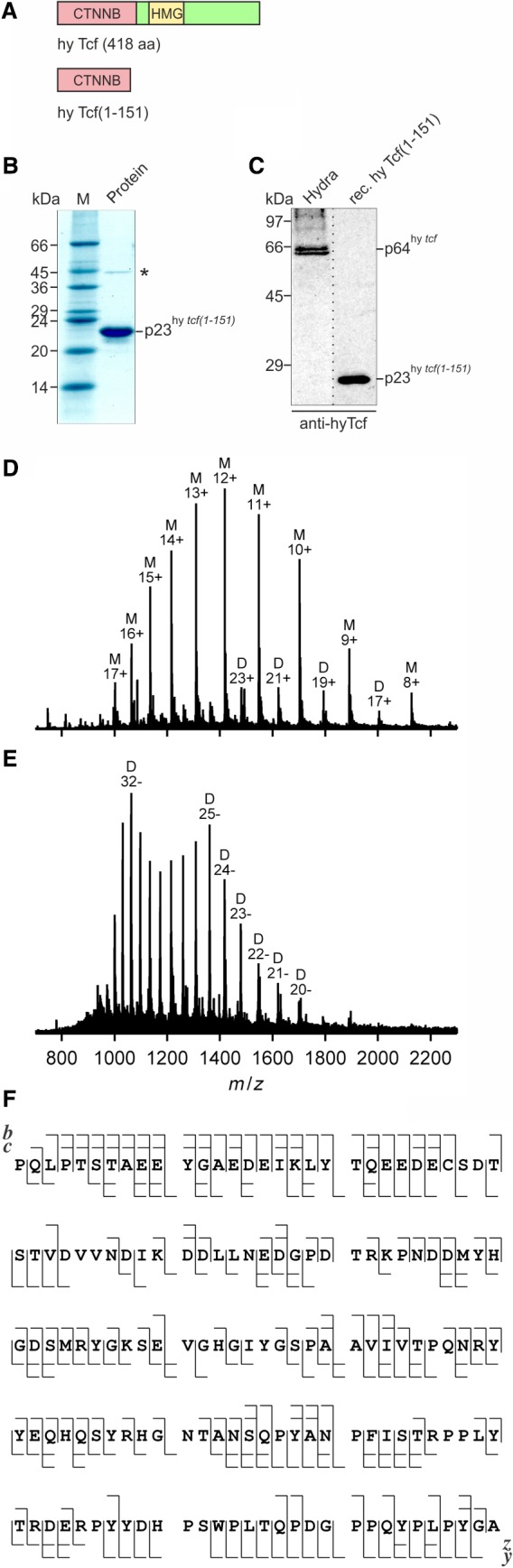
Analysis of the Hydra Tcf recombinant protein. (A) Schematic depiction of the Hydra (hy) Tcf protein product (GenBank accession no. XP_002159974). The positions of the high mobility group (HMG) representing the DNA binding domain, and the β‐Catenin binding site (CTNNB) are indicated. The amino‐terminal segment of Hydra Tcf [hy Tcf(1‐151)] was expressed in Escherichia coli and purified. (B) SDS/PAGE (12.5%, wt/vol) of 5 μg (Coomassie brilliant blue staining) purified recombinant Hydra Tcf(1‐151) p23. The upper faint band marked with an asterisk represents a cross‐linked dimer generated from p23hy tcf(1‐151) (see below). (C) Immunoblot analysis using 10 μg of cell extract from Hydra, and 5 ng of recombinant hy Tcf(1‐151). Proteins were resolved by SDS/PAGE (10%, wt/vol) and detected using an antiserum directed against recombinant hy Tcf(1‐151). The dotted line marks the splicing site in the blot image, from which one lane has been removed. (D–F) MS of recombinant hy Tcf(1‐151). The ESI‐MS of hy Tcf(1‐151) with a 7 Tesla Fourier transform ion cyclotron resonance (FT‐ICR) instrument (Bruker, Vienna, Austria) gives a mass value for the most abundant isotopic peak of 17 014.868 ± 0.006 Da (theoretical mass without initiating methionine: 17 014.874 Da; error 0.4 p.p.m.) using polyethylene glycol 1000 as calibrant. (D) ESI mass spectrum of hy Tcf‐NT in positive ion mode showing mostly (~ 95%) monomeric protein (M) but also protein dimers whose mass values (measured mass for the most abundant isotopic peak 34 027.727 Da) indicate covalent dimerization by formation of an intermolecular disulfide bond (theoretical mass for the most abundant isotopic peak 34 027.732 Da). (E) ESI of hy Tcf‐NT in negative ion mode gives predominantly (> 80%) dimer ions from intermolecular disulfide bond formation. (F) Fragment ion maps for the Hydra Tcf(1‐151) p23 protein showing 85% sequence coverage.
The myc1 promoter is selectively downregulated by ectopic β‐Catenin/Tcf
To test if the Hydra β‐Catenin and Tcf proteins are involved in transcriptional regulation of the myc1 and myc2 genes, luciferase reporter assays were performed using the chemically transformed quail cell line QT6 as test system. Cells were transiently co‐transfected with reporter plasmids containing the Hydra myc1 and myc2 promoters inserted into luciferase reporter vectors (pGL3‐hymyc1, pGL3‐hymyc2), and expression vectors (pRc) encoding Hydra Tcf or β‐Catenin proteins fused to a carboxyl‐terminal hemagglutinin (HA) tag (Fig. 7A). Ectopic protein expression was verified by immunoblotting. Overexpression of Tcf‐HA, β‐Catenin‐HA, or Tcf‐HA together with β‐Catenin‐HA led to partial repression of the myc1 promoter, whereas the myc2 promoter was not influenced. The results suggest that, in contrast to myc2, the myc1 promoter is repressed by β‐Catenin/Tcf although both promoters are bound by Tcf in vivo (Fig. 4C). Therefore, these biochemical analyses confirm the specific downregulation of myc1 upon activation of the β‐Catenin/Tcf signaling complex suggesting that myc2 but not myc1 represents the functional ortholog of vertebrate c‐myc.
Figure 7.
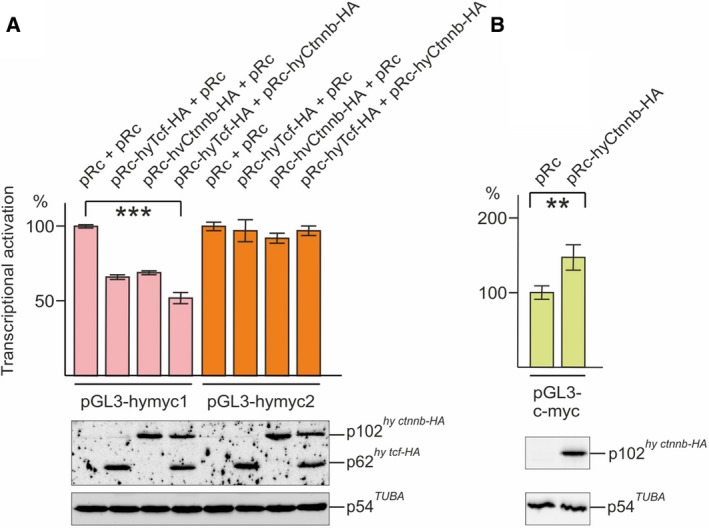
Transcriptional regulation of the Hydra myc1 and myc2 promoters. (A) Aliquots (0.5 μg) of the pGL3‐hymyc1 or pGL3‐hymyc2 reporter constructs were co‐transfected in triplicate with aliquots (0.5 μg) of pRc‐derived expression vectors encoding the Hydra Tcf‐HA, Ctnnb‐HA, or the empty expression vector (pRc) into the chemically transformed quail cell line QT6. (B) Aliquots (1.0 μg) of the pGL3‐c‐myc reporter construct were co‐transfected in triplicate with aliquots (1.0 μg) of pRc‐hyCtnnb‐HA, or the empty pRc vector into QT6 cells. Luciferase activities and protein concentrations were determined from cell extracts prepared 24 h after transfection. Relative luciferase activities and standard errors of the mean (SEM, n = 3) are visualized by bars and vertical lines, respectively. Statistical significance was assessed by using a paired Student t‐test (**P < 0.01, ***P < 0.001). For control of protein expression (lower panels), equal amounts of cell extracts (20 μL) were analyzed by SDS/PAGE (10% wt/vol). The ectopic HA‐tagged Hydra Tcf and Ctnnb proteins, and endogenous tubulin α were detected by immunoblotting.
To test, if Hydra β‐Catenin is capable at all to activate transcription in the applied cell system, a reporter construct containing the chicken c‐myc promoter containing multiple Tcf binding sites was co‐transfected with pRc‐hyCtnnb‐HA into QT6 cells. The result of this luciferase assay showed that, in contrast to the Hydra myc1 or myc2 promoters, Hydra β‐Catenin moderately activates this vertebrate c‐myc promoter (Fig. 7B). No luciferase activities were scored upon transfection of either the empty reporter plasmid, or the empty expression vector (data not shown). To investigate if overexpressed Hydra β‐Catenin also induces cell transformation in avian cells similar to its vertebrate counterparts 37, 38, the coding region of Hydra β‐catenin was inserted into the replication‐competent retroviral RCAS expression vector (Fig. 8A) and transfected into chicken embryo fibroblasts (CEF). As a positive control, an RCAS construct encoding a LEF1/β‐Catenin fusion protein (ΔLEF1ΔCTNNB) 37, 38 was employed. As expected, overexpression of ΔLEF1ΔCTNNB lead to efficient cell transformation manifested by colony formation in soft agar 37, 38 (Fig. 8B). Remarkably, also the cells overexpressing full‐length Hydra β‐Catenin were able to form colonies indicating a transformed phenotype (Fig. 8B). The ectopic proteins were efficiently expressed from their retroviral vectors, which was monitored by immunoblot analysis (Fig. 8C). This result shows that the principal oncogenic potential of β‐Catenin has been conserved through metazoan evolution.
Figure 8.
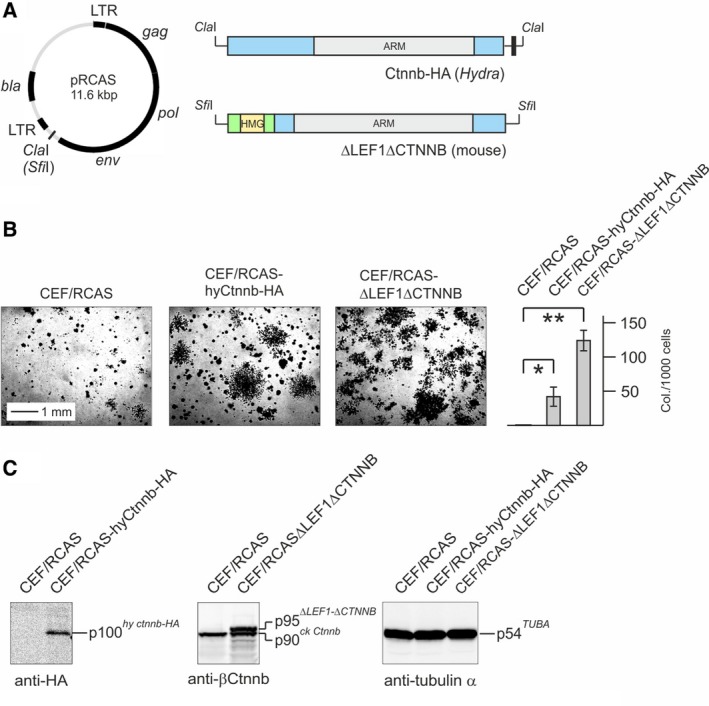
Oncogenic transforming activity of Hydra β‐Catenin. (A) Structures of the applied RCAS constructs (HA, hemagglutinin tag). (B) Agar colony formation of CEF transfected with the empty retroviral RCAS vector, or with RCAS constructs depicted under A. Each 1 × 104 cells were seeded in soft nutrient agar onto MP12 dishes and colonies were scored after 3 weeks. Untransformed control CEF infected with the empty RCAS vector produced very small background colonies, which has been also observed previously 37, 38. Colonies were counted from triplicate dishes. A representative experiment from two independent assays (n = 2) is shown. Vertical bars show standard deviations (SD). Statistical significance was assessed by using a paired Student t‐test (*P < 0.05, **P < 0.01). (C) Immunoblot analysis of ectopically expressed proteins using the indicated antibodies. The antibody directed against the mouse β‐Catenin portion present in the ΔLEF1ΔCTNNB fusion protein also detects the endogenous chicken (ck) Ctnnb (p90).
Discussion
Although substantial progress has been made in understanding the pleiotropic functions of the human c‐Myc protein in cellular proliferation, growth, energy metabolism, differentiation, and apoptosis 5, 6, 12, 39, many open questions remain regarding the underlying molecular mechanisms. The recent identification of c‐Myc to act as a possible general amplifier of gene expression controlling multiple transcriptional programs 8, 40, 41, 42, 43 even enhances the complexity of Myc biology. Furthermore, several upstream signaling pathways like the mitogenic Ras/Raf cascade, or the Wnt/β‐Catenin/Tcf4 axis regulate c‐Myc expression and activity in cell proliferation, but they are also relevant in malignant cell growth leading to aberrant c‐myc activation 2, 4, 5, 12.
A possibility to dissect the multiple Myc functions is the analysis of genetically defined invertebrate model organisms, like the simple eumetazoan Hydra, a classical diploblastic model system to study pattern formation, regeneration, and stem cell dynamics. About 600 million years ago, single‐cell premetazoans and simple metazoans were predominant, but during the Cambrian period, a rapid diversification of multicellular lifeforms began. Hydra represents one of the earliest multicellular animals to evolve out of single‐celled basal species, has a high regeneration potential, and is biologically immortal 21. This ancient organism could represent a suitable model system to study basal mechanisms in tumor biology because cancer is as old as multicellular life. Intriguingly, natural occurring tumors have been observed in Hydra, presumably resulting from differentiation‐arrested female gametes 44, a cell type in which myc2 is expressed at high levels 20. The occurrence of tumors in this early diverging organism is actually in line with the atavistic cancer model, in which the biological origin of malignant growth is traced back to the transition between unicellularity to multicellularity 45, 46. This theory was recently supported by molecular analyses of multiple sets of genes with common phylogenetic origin (phylostrata) in solid human tumors. The study showed that dormant genes conserved with unicellular organisms became strongly upregulated in tumors, whereas genes of metazoan origin were primarily inactivated 47.
Hydra is also one of the most basal metazoan organisms employed so far for analysis of the major cancer driver Myc and its signaling network 18, 20 revealing that biochemical and oncogenic properties of c‐Myc arose very early in metazoan evolution. In Hydra there are four myc‐related genes (myc1–4), which branched off from a basal position during cnidarian evolution 18, 20, 21. myc1 and myc2 show the closest homology to vertebrate c‐myc but significantly less homology to L‐myc or N‐myc 18, because the diversification of c‐, L‐, and N‐myc subfamilies occurred later within the vertebrate lineage 18. On the protein level, Hydra Myc1 shares 32%, 22%, or 20% amino acid sequence identity with the human c‐Myc, L‐Myc, or N‐Myc proteins, whereas Myc2 displays 32%, 22%, or 24% identity with c‐Myc, L‐Myc, or N‐Myc, respectively. As reported previously, the structures of Hydra Myc1 and Myc2 proteins display the same principal topography and similar evolutionary relationship with the human c‐Myc protein with equal sequence identities (32%), but share only 24% overall identities among themselves 18. Although Myc1 or Myc2 cannot be assigned directly as ancestor of any specific vertebrate homolog, comparison of the sizes and conserved Myc boxes in the transactivation domain between Hydra Myc1 or Myc2 and human c‐Myc suggests that Myc2 is most likely the closest ortholog of human c‐Myc 18. Furthermore, the myc2 gene is composed of three exons like human c‐myc, in contrast to myc1 containing two exons only (Fig. 4B). Concerning the differential regulation of the Hydra myc genes (Fig. 7), comparison of the promoter structures indicates, that besides Tcf/β‐Catenin additional transcription factors may be involved in mediating transmission of upstream acting signaling pathways (Fig. 5). In addition, the presence of a Myc binding site in the myc2 promoter suggests that Hydra Myc proteins themselves may participate in myc2‐specific gene regulation.
In mammalian Wnt/β‐Catenin signal transduction, the transcription factor Tcf4 represents a crucial nuclear effector 9, 14. Comparison of the Hydra Tcf protein sequence with homologs from chicken or human reveals the highest degree of sequence identity with the vertebrate Tcf4 (Fig. 9), regulating vertebrate c‐myc via TBE elements present in the relevant promoter regions 13. In Hydra, wnt and tcf genes are transcriptionally activated in early bud formation and head regeneration namely in the putative Hydra head organizer, the upper part of the hypostome 23, 28. Furthermore, elevated levels of β‐Catenin result in body columns that have high head organizer potency accounting for the formation of ectopic head structures in these animals 28 (Fig. 1A,B). Here, we present evidence that β‐Catenin has an additional function in interstitial cells of the gastric region. Using pharmacologically treated or genetically modified Hydra, we have shown that activation of β‐Catenin seems to enhance interstitial stem cell maintenance, and expression and promoter analyses demonstrate a specific repression of myc1, but not of myc2. According to our current working model (Fig. 10), this reduction in myc1 expression by nuclear β‐Catenin could in fact promote interstitial stem cell maintenance and self‐renewal based on an earlier observation that myc1 inhibition by myc1‐specific antisense RNAs has the same effect 22. The reason why in our biochemical analyses myc2 is not significantly induced upon ectopic β‐Catenin/Tcf activation is not yet clear. So far, our experiments were performed in intact polyps only. Therefore, it was not possible to resolve how myc2 expression responds to changes in nuclear β‐Catenin activity specifically in interstitial cells. A reason for the overall unchanged myc2 expression could be that myc2 is indeed activated in stem cells, but this is masked by a possible downregulation in epithelial cells. Since the two epithelial cell types form the major part of the Hydra body mass, a slight reduction in myc2 expression upon β‐Catenin activation could dilute a positive stimulatory effect in interstitial stem cells. Subtle shifts in Myc expression above or below distinct thresholds can trigger striking differences in biological output 48. In addition, myc2 expression may depend on possible Myc1‐triggered transcriptional activation mediated by the E‐box present in the myc2 promoter (Fig. 4A). Consequently, a decrease in Myc1 levels due to Wnt signal‐triggered myc1 downregulation could mask a possible stimulatory effect of β‐Catenin/Tcf on the myc2 promoter, a hypothesis, which has to be pursued by further investigations.
Figure 9.

Amino acid sequence alignments of Hydra magnipillata (hy) Tcf with the Homo sapiens (h) and Gallus gallus (ck) homologs. (A) Alignment with human TCF4 (TCF7L2) and chicken Tcf4 (Tcf7l2). (B) Alignment with human LEF‐1 and chicken Lef1. GenBank accession numbers are: hy Tcf, NP_001296662; h TCF‐4, CAG38811; ck Tcf‐4, NP_001193439; h LEF‐1, Q9UJU2; ck Lef‐1, NP990344. Identical residues are shaded in blue, and gaps are indicated by dashes. The β‐Catenin (CTNNB) and DNA binding (HMG) domains are boxed in pink or yellow, respectively. Sequence identities between hy Tcf and h LEF1 or ck Lef1 are 29%, and between hy Tcf and h TCF4 or ck Tcf4 42%. The alignment was generated by using the computer program (omega) clustalw with additional manual adjustments.
Figure 10.
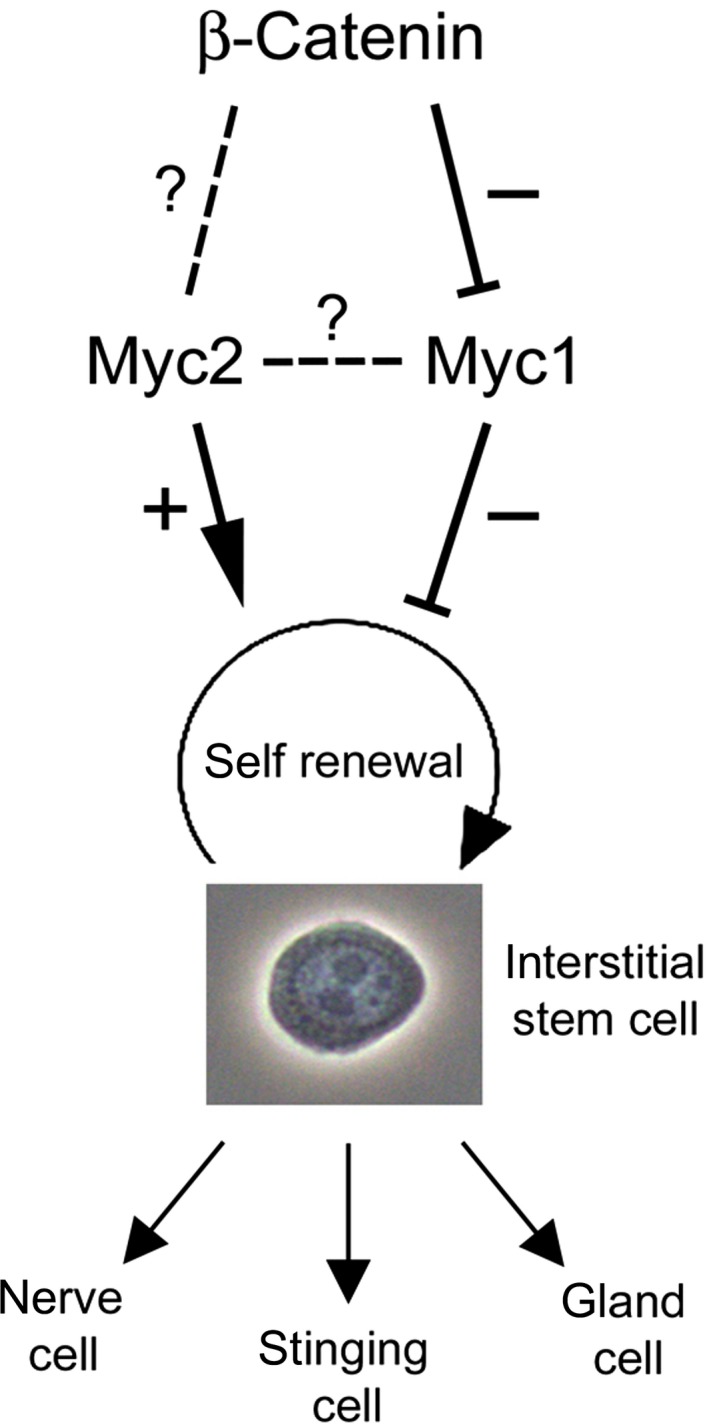
Hydra β‐Catenin/Myc interaction model. Interstitial stem cells permanently undergo self‐renewal and differentiation into three somatic products: nerve cells, stinging cells (nematocytes), and gland cells. We propose that Myc2 acts as maintenance factor in interstitial stem cell self‐renewal and that this is complemented by a double‐negative action of β‐Catenin and Myc1. The detailed actions of β‐Catenin on Myc2 and the cross‐talk between Myc1 and Myc2 are not yet known.
Our finding that Hydra myc1 is a negatively regulated target of β‐Catenin was unexpected, because c‐myc is one of the best studied targets activated by mammalian β‐Catenin. However, also in case of human N‐myc, a negative regulation of mRNA expression caused by activated Wnt signaling has been reported 49, 50. In murine embryonic stem cells, c‐Myc controls the balance between stem cell self‐renewal, pluripotency and differentiation 51, 52 thereby driving a regulatory network to maintain this stem cell identity 53. Furthermore, c‐Myc potentiates the Wnt/β‐Catenin signaling pathway by transcriptional repression of Wnt antagonists. This induces transcriptional activation of the endogenous myc family members, which in turn activate a Myc‐driven self‐reinforcing circuit 54. Depletion of c‐myc by gene knock‐out or by inactivation of a c‐myc‐driving superenhancer induces reversible dormancy in stem cells with differentiation‐arrested pluripotent progenitors 55, 56.
In colon cancer, mutation of the APC tumor suppressor activates β‐Catenin/Tcf signaling, leading to transcriptional induction of c‐myc and cyclin D1 in colon cancer 13, 15 illustrating the oncogenic potential of this signal transduction pathway. The ancestral Hydra β‐Catenin protein is highly conserved sharing 62% sequence identity to its human ortholog 23. Furthermore, Hydra β‐Catenin displays oncogenic potential similar to its vertebrate counterparts (Fig. 8). As reported previously, both Hydra Myc proteins have transforming activities 18, 20 demonstrating that structural elements with basic oncogenic properties must have been established very early in metazoan evolution. Further investigations involving appropriate transgenic and interfering RNA technologies will be performed to functionally characterize these transforming proteins in Hydra, and to fully elucidate upstream and downstream acting Myc signaling pathways.
Materials and methods
Animals
Hydra vulgaris strains Basel, Hydra magnipapillata wild‐type strain 105 and the Hydra β‐cat‐Tg strain containing a transgenic β‐catenin gene fused in frame to the coding sequence of the enhanced GFP 31 were used in this study. Mass cultures were kept as described 57. Experimental animals were collected 24 h after the last feeding. Treatment with the inhibitor Alp was done as described 28, and phenotypes of treated polyps were analyzed at 48 or 60 h after the onset of treatment.
Maceration of Hydra tissue
Maceration of excised tissue pieces from the gastric region was done as described using a maceration solution containing acetic acid (1 vol)/glycerol (1 vol)/water (7 vol) 33. Fixed macerated cell suspensions were then spread onto microscope slides, and analyzed under phase contrast optics. For a single measurement, 10 tissue pieces were macerated, and about 500 epithelial cells and a corresponding number of 1s+2s interstitial stem cells were counted.
Whole mount in situ hybridization
In situ hybridization with digoxigenin‐labeled RNA probes was done according to a protocol as described 58 using myc1‐ and myc2‐specific cDNA probes 18, 20.
DNA cloning and nucleic acid analysis
Molecular cloning, DNA sequencing, and Northern analysis have been described 18, 59. Total RNA isolation and poly(A)+‐RNA selection from ~ 600 Hydra animals was performed as described 18. The yield of total RNA per animal was ~ 2 μg. DNA fragments specific for Hydra myc1, and myc2 have been described 18, 20. To detect the Hydra tcf mRNA, a DNA fragment encompassing the tcf coding regions was applied. To obtain a DNA probe for detection of the transgenic ΔN‐ctnnb‐GFP mRNA, a 718‐nt EcoRI/SmaI fragment containing almost the entire GFP coding region was excised from the HoTG plasmid 60. To determine the myc1 transcription start site, 5′RACE was performed as described 61 using the primers 5′‐GGATCATCATTAGTTGGAAATGGCGAAG‐3′ and 5′‐GTCTCGCAGTACTTCTGAAGGAAAAACACTTG‐3′ for first strand cDNA synthesis and subsequent PCR, respectively. Mapping of the myc2 transcription start site using the primers 5′‐GTGTACACCAATTTGAACCAGTCATATCGA‐3′ and 5′‐AATTTATCCACAGCTATTATGTACACAATT‐3′ has been described previously 20.
Promoter analysis
ChIP analysis was carried out as described 20, 59, 61, 62 using sheared extracts from ~ 300 Hydra animals treated with formaldehyde for 30 min. Immunoprecipitations were performed with specific antibodies followed by PCR amplification of 248‐ and 201‐bp fragments from the Hydra myc1 or myc2 regulatory regions, respectively, using the specific primer pairs 5′‐TCAGTACTAAAGTTCAGTTGCTCC‐3′/5′‐GTCTTTGTTTGGCGTTTGAC‐3′ (myc1), and 5′‐ACTTGGTGTTTTTTATGTAATG‐3′/5′‐GAATGACTTTGTATTTCGC‐3′ (myc2). In addition, the primer pair 5′‐AGGTGTCGTTTCTGCATTC‐3′/5′‐AAATTCGCAGCCAAAACAAG‐3′ (myc1) was employed to amplify a 129‐bp fragment from the myc1 promoter region. The primers used to amplify a 173‐bp segment from the TSP promoter have been described 36.
Transcriptional transactivation analysis using the luciferase reporter system has been described 20, 59, 62. To generate the reporter constructs pGL3‐hymyc1 and pGL3‐hymyc2, a 416‐bp and a 319‐bp segment encompassing nucleotides −369 to +47 or nucleotides −216 to +103 of the Hydra myc1 or myc2 promoter regions, respectively, were amplified by PCR from genomic DNA using the primer pairs 5′‐GGATCCGGTACCCCCAATTATTAAAATTGTGGTGTG‐3′/5′‐GGATCCAAGCTTAGACTGTAATTTAAATTCGCAGCC‐3′, or 5′‐GGATCCGGTACCCTTGGTGTTTTTTATGTAATG‐3′/5′GGATCCAAGCTTTAGATCGGCTTTCATAGATA‐3′, and inserted into the KpnI and HindIII sites of the pGL3‐Basic vector (Promega, Vienna, Austria). To construct pGL3‐c‐myc, a 691‐bp fragment of the chicken c‐myc promoter (accession no. J00889) encompassing nucleotides −621 to +107 was amplified from CEF cDNA and inserted into pGL3‐Basic. This insert encompasses the transcription start site (+1) and four consensus Tcf binding sites (5′‐YCTTTGWW‐3′). To create pRc‐hyTcf‐HA and pRc‐hyCtnnb‐HA, the coding sequences of Hydra tcf and of β‐catenin (ctnnb) were amplified from Hydra cDNA as described 18 using the primer pairs 5′‐TCGGCGGCCGCATGCCCCAACTACCAACTTCAACAG‐3′/5′‐TCGTCTAGATTAAGCGTAATCTGGAACATCGTATGGGTATCTAGTTTCAATTGCCTGAAGCGT TGG‐3′ and 5′‐TCGGCGGCCGCATGATGGAGGATTCAACTGCTCAAATGAG‐3′/5′‐TCGTCTAGACTAAGCGTAATCTGGAACATCGTATGGGTACAAGTCAGGGTCAAACCAACCCTG‐3′. The 3′‐primers contain the coding sequence of the hemagglutinin epitope (HA) fused in frame to the 3′‐end of the tcf or ctnnb coding sequences to allow immunological detection of the ectopic proteins. The PCR products were digested with NotI/(blunt ended) and XbaI and inserted into the eukaryotic pRc/RSV expression vector (Thermo Fisher Scientific, Vienna, Austria) which had been opened with HindIII/(blunt ended)/XbaI. Calcium phosphate‐mediated DNA transfection, and luciferase assays were performed as described 20.
Cells and retroviruses
Cultivation of CEF and of the methylcholanthrene‐transformed cell line QT6 and cell transformation assays were performed as described 18, 59, 61. The construct pRCAS‐ΔLEF1ΔCTNNB (pRCAS‐ΔN‐bΔN) encoding a truncated mouse LEF1/CTNNB fusion protein has been described 38. To construct pRCAS‐hyCtnnb‐HA, the Hydra ctnnb insert from pRc‐hyCtnnb‐HA was released with XbaI/NotI/(blunt ended) and inserted into the ClaI/(blunt ended) pRCAS (BP) vector 63.
Protein expression, purification, and analysis
To construct pET11d‐hyTcf(1‐151) a DNA segment encoding amino acid residues 2‐151 from Hydra Tcf was amplified by PCR using the primers 5′‐CCCCAACTACCAACTTCA‐3′/5′‐AAGCTTGGATCCTTAAGCTCCATATGGCAAAGG‐3′ and the plasmid pGEX6p3‐hyTcf 28 as a template. The PCR product was digested with BamHI and inserted into pET11d vector yielding the construct pET11d‐hyTcf(1‐151) with the initiating methionine codon derived from the vector. The encoded Hydra Tcf(1‐151) protein (151 amino acids; M r = 17 146; pI = 4.13) represents a truncated version of the 418‐amino acid full‐length Hydra Tcf protein. DNA of the construct pET11d‐hyTcf(1‐151) was transformed into E. coli strain BL21 (DE3) CodonPlus‐RIL (Stratagene/Agilent, Vienna, Austria). To express recombinant hyTcf(1‐151) protein, bacteria from a single colony were grown overnight at 37 °C with shaking at 220 r.p.m. in 10 mL of LB medium containing 100 μg·mL−1 ampicillin and 25 μg·mL−1 chloramphenicol. The bacteria were transferred into 400 mL LB medium containing 100 μg·mL−1 ampicillin, and grown at 37 °C with shaking at 220 r.p.m. to an optical density of 0.5 (600 nm). To induce recombinant protein expression, isopropyl‐β‐d‐thiogalactopyranoside was added to a final concentration of 1 mm and bacteria were incubated as above for 3 h. The bacteria were pelleted and resuspended in 25 mL of buffer A (20 mm Tris HCl pH7.5, 80 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF), and then lysed at 1300 psi using a French Press. DNAse I was added to a final concentration of 5 μg·mL−1, and the lysate was incubated at 4 °C for 30 min. The sample was centrifuged at 18 000 g for 20 min at 4 °C. Ammonium sulfate was added to the clarified supernatant at 40% (w/v) saturation, and the solution was stirred on ice for 30 min. The precipitated proteins were pelleted by centrifugation at 11 000 g for 30 min at 4 °C. The pellet was dissolved in 10 mL of buffer A and dialyzed for 36 h at 4 °C against 2.5 L of buffer A, centrifuged at 18 000 g for 20 min, and then loaded onto a Mono Q anion exchange column using an automated liquid chromatography system (ÄKTA purifier; GE Healthcare). Chromatography was carried out with a linear gradient from 0 to 1 m NaCl in buffer A at a flow rate of 1 mL·min−1. Fractions containing hy Tcf(1‐151) protein were combined and applied onto a Superdex‐200 gel filtration column (GE Healthcare, Vienna, Austria) equilibrated with buffer A, and then eluted with the same buffer at a flow rate of 0.5 mL·min−1. hy Tcf(1‐151) containing fractions were pooled, dialyzed against PBS, and stored at −80 °C. The final yield of purified hy Tcf(1‐151) from the 400‐mL bacteria culture was approximately 500 μg. For electrospray ionization mass spectrometry (ESI‐MS), hy Tcf(1‐151) was desalted using Vivaspin 500 PES centrifugal concentrators (MWCO 5000) (Sartorius, Vienna, Austria). Centrifugal concentration and addition of 100 mm ammonium acetate in H2O (18 MΩ) to the supernatant was repeated six times, followed by six cycles of concentration and dilution with H2O (18 MΩ). The final protein concentration of the ESI solution (H2O : CH3OH vol/vol) was 1 μm. For ESI‐MS analysis in positive and negative ion mode, 1% vol/vol acetic acid and 0.1% 1,8‐diazabicyclo[5.4.0]undec‐7‐en (DBU) were added to aliquots of the ESI solution, respectively. SDS/PAGE and immunoblotting were done as described 18, 59. To prepare cell extracts from whole Hydra, ~ 10 animals were dissolved in 200 μL of RIPA buffer 59 and the extract clarified by centrifugation at 20 000 g for 1 h at 4 °C. The yield of total protein per animal was ~ 20 μg.
A polyclonal rabbit antiserum directed against the purified recombinant hyTcf(1‐151) protein was generated as described previously using 3 × 100 μg recombinant protein as immunogen 18, 20. The antiserum directed against Hydra Max has been described 18. The monoclonal mouse antibodies anti‐α‐tubulin, anti‐GFP, and anti‐HA were purchased from Sigma‐Aldrich, Vienna, Austria (T5168), Roche, Vienna, Austria (11 814 460 001), and Covance, Vienna, Austria (MMS‐101P), respectively. The polyclonal rabbit antibody directed against mouse anti‐β‐Catenin was purchased from Sigma‐Aldrich, Vienna, Austria (C2206).
Hydra Myc1‐ and Myc2‐specific antisera 18, 20 were subjected to immunoglobulin G (IgG) purification. To each 20 mL antiserum, an equal volume of saturated ammonium sulfate solution was added dropwise resulting in a final concentration of 50% (w/v), and stirred on ice for 3 h. After centrifugation at 3000 g for 20 min (4 °C), the pellets were washed twice in 50% (w/v) ammonium sulfate (20 mL), and centrifuged as above. Pellets were dissolved in 4 mL of PBS, and dialyzed against 20 mm sodium phosphate pH 7.0 for 48 h. Prior to protein G affinity chromatography, samples were clarified by centrifugation at 13 000 g for 30 min (4 °C), and then loaded onto a 5‐mL HiTrapG column (GE Healthcare, Vienna, Austria) equilibrated with 20 mm sodium phosphate pH 7.0. Bound IgG were eluted with 0.1 m glycine pH 2.7, and 1‐mL fractions immediately neutralized with 200 μL of 1 m Tris‐HCl pH 9.0. IgG fractions were pooled resulting in final volumes of 8 mL each, and dialyzed against 20 mm sodium phosphate pH 7.0 for 48 h. The IgG preparations were quantified (anti‐hy Myc1 540 μg·mL−1; anti‐hy Myc2 1080 μg·mL−1), and stored in aliquots at −80 °C. For immunoblotting, 1 : 50 dilutions of IgG‐purified antisera were applied.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
MH, KBi, and BH planned experiments, MH, SGl, SGu, AR, KP, and KBr performed experiments, MH, KBr, KBi, and BH analyzed data, MH and BH wrote the paper, with contributions from KBi.
Acknowledgements
We thank Hans Bode for providing β‐cat‐Tg polyps, and Masahiro Aoki and Peter K. Vogt for providing the construct RCAS‐ΔN‐bΔN. This work was supported by Austrian Science Fund (FWF) grants P23652 (to KBi), P20734 (to BH), and P27347 (to KBr). SG was a recipient of fellowships from the Austrian Academy of Sciences and from the University of Innsbruck.
Contributor Information
Markus Hartl, Email: markus.hartl@uibk.ac.at.
Bert Hobmayer, Email: bert.hobmayer@uibk.ac.at.
References
- 1. Bister K & Jansen HW (1986) Oncogenes in retroviruses and cells: biochemistry and molecular genetics. Adv Cancer Res 47, 99–188. [DOI] [PubMed] [Google Scholar]
- 2. Stefan E & Bister K (2017) MYC and RAF: key effectors in cellular signaling and major drivers in human cancer. Curr Top Microbiol Immunol 407, 117–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eisenman RN (2001) Deconstructing myc. Genes Dev 15, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 4. Adhikary S & Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6, 635–645. [DOI] [PubMed] [Google Scholar]
- 5. Conacci‐Sorrell M, McFerrin L & Eisenman RN (2014) An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4, a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eilers M & Eisenman RN (2008) Myc's broad reach. Genes Dev 22, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang CV (2012) MYC on the path to cancer. Cell 149, 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolf E, Lin CY, Eilers M & Levens DL (2015) Taming of the beast: shaping Myc‐dependent amplification. Trends Cell Biol 25, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dang CV (1999) c‐Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanahan D & Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 11. Reya T & Clevers H (2005) Wnt signalling in stem cells and cancer. Nature 434, 843–850. [DOI] [PubMed] [Google Scholar]
- 12. Dang CV (2010) Enigmatic MYC conducts an unfolding systems biology symphony. Genes Cancer 1, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B & Kinzler KW (1998) Identification of c‐MYC as a target of the APC pathway. Science 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- 14. Zhan T, Rindtorff N & Boutros M (2017) Wnt signaling in cancer. Oncogene 36, 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tetsu O & McCormick F (1999) Beta‐catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426. [DOI] [PubMed] [Google Scholar]
- 16. Clevers H & Nusse R (2012) Wnt/beta‐catenin signaling and disease. Cell 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- 17. Holland JD, Klaus A, Garratt AN & Birchmeier W (2013) Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol 25, 254–264. [DOI] [PubMed] [Google Scholar]
- 18. Hartl M, Mitterstiller AM, Valovka T, Breuker K, Hobmayer B & Bister K (2010) Stem cell‐specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan Hydra . Proc Natl Acad Sci USA 107, 4051–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young SL, Diolaiti D, Conacci‐Sorrell M, Ruiz‐Trillo I, Eisenman RN & King N (2011) Premetazoan ancestry of the Myc‐Max network. Mol Biol Evol 28, 2961–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hartl M, Glasauer S, Valovka T, Breuker K, Hobmayer B & Bister K (2014) Hydra myc2, a unique pre‐bilaterian member of the myc gene family, is activated in cell proliferation and gametogenesis. Biol Open 3, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hobmayer B, Jenewein M, Eder D, Eder MK, Glasauer S, Gufler S, Hartl M & Salvenmoser W (2012) Stemness in Hydra – a current perspective. Int J Dev Biol 56, 509–517. [DOI] [PubMed] [Google Scholar]
- 22. Ambrosone A, Marchesano V, Tino A, Hobmayer B & Tortiglione C (2012) Hymyc1 downregulation promotes stem cell proliferation in Hydra vulgaris . PLoS One 7, e30660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U & Holstein TW (2000) WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra . Nature 407, 186–189. [DOI] [PubMed] [Google Scholar]
- 24. Hobmayer E, Hatta M, Fischer R, Fujisawa T, Holstein TW & Sugiyama T (1996) Identification of a Hydra homologue of the beta‐catenin/plakoglobin/armadillo gene family. Gene 172, 155–159. [DOI] [PubMed] [Google Scholar]
- 25. Wenger Y & Galliot B (2013) RNAseq versus genome‐predicted transcriptomes: a large population of novel transcripts identified in an Illumina‐454 Hydra transcriptome. BMC Genom 14, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minobe S, Fei K, Yan L, Sarras M Jr & Werle M (2000) Identification and characterization of the epithelial polarity receptor “Frizzled” in Hydra vulgaris . Dev Genes Evol 210, 258–262. [DOI] [PubMed] [Google Scholar]
- 27. Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ et al (2005) Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160. [DOI] [PubMed] [Google Scholar]
- 28. Philipp I, Aufschnaiter R, Ozbek S, Pontasch S, Jenewein M, Watanabe H, Rentzsch F, Holstein TW & Hobmayer B (2009) Wnt/beta‐catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra . Proc Natl Acad Sci USA 106, 4290–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H & Holstein TW (2009) Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330, 186–199. [DOI] [PubMed] [Google Scholar]
- 30. Broun M, Gee L, Reinhardt B & Bode HR (2005) Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 132, 2907–2916. [DOI] [PubMed] [Google Scholar]
- 31. Gee L, Hartig J, Law L, Wittlieb J, Khalturin K, Bosch TC & Bode HR (2010) beta‐catenin plays a central role in setting up the head organizer in hydra. Dev Biol 340, 116–124. [DOI] [PubMed] [Google Scholar]
- 32. Gufler S, Artes B, Bielen H, Krainer I, Eder MK, Falschlunger J, Bollmann A, Ostermann T, Valovka T, Hartl M et al (2018) beta‐Catenin acts in a position‐independent regeneration response in the simple eumetazoan Hydra . Dev Biol 433, 310–323. [DOI] [PubMed] [Google Scholar]
- 33. David CN (1973) A quantitative method for maceration of hydra tissue. Wilhelm Roux Arch Entwickl Mech Org 171, 259–268. [DOI] [PubMed] [Google Scholar]
- 34. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D et al (2010) The dynamic genome of Hydra . Nature 464, 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arce L, Yokoyama NN & Waterman ML (2006) Diversity of LEF/TCF action in development and disease. Oncogene 25, 7492–7504. [DOI] [PubMed] [Google Scholar]
- 36. Lommel M, Strompen J, Hellewell AL, Balasubramanian GP, Christofidou ED, Thomson AR, Boyle AL, Woolfson DN, Puglisi K, Hartl M et al (2018) Hydra mesoglea proteome identifies thrombospondin as a conserved component active in head organizer restriction. Sci Rep 8, 11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aoki M, Hecht A, Kruse U, Kemler R & Vogt PK (1999) Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid‐enhancing factor 1. Proc Natl Acad Sci USA 96, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aoki M, Sobek V, Maslyar DJ, Hecht A & Vogt PK (2002) Oncogenic transformation by beta‐catenin: deletion analysis and characterization of selected target genes. Oncogene 21, 6983–6991. [DOI] [PubMed] [Google Scholar]
- 39. Herold S, Herkert B & Eilers M (2009) Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer 9, 441–444. [DOI] [PubMed] [Google Scholar]
- 40. Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R et al (2012) c‐Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dang CV (2014) Gene regulation: fine‐tuned amplification in cells. Nature 511, 417–418. [DOI] [PubMed] [Google Scholar]
- 42. Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI & Young RA (2012) Transcriptional amplification in tumor cells with elevated c‐Myc. Cell 151, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lewis LM, Edwards MC, Meyers ZR, Talbot CC Jr, Hao H, Blum D, Reproducibility Project: Cancer Biology , Iorns E, Tsui R, Denis A, Perfito N & Errington TM (2018) Replication study: transcriptional amplification in tumor cells with elevated c‐Myc. Elife 7, e30274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Domazet‐Loso T, Klimovich A, Anokhin B, Anton‐Erxleben F, Hamm MJ, Lange C & Bosch TC (2014) Naturally occurring tumours in the basal metazoan Hydra . Nat Commun 5, 4222. [DOI] [PubMed] [Google Scholar]
- 45. Zanni G & Opitz JM (2013) Annals of morphology. Atavisms: phylogenetic Lazarus? Am J Med Genet A 161A, 2822–2835. [DOI] [PubMed] [Google Scholar]
- 46. Davies PC & Lineweaver CH (2011) Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys Biol 8, 015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trigos AS, Pearson RB, Papenfuss AT & Goode DL (2017) Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc Natl Acad Sci USA 114, 6406–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown‐Swigart L, Johnson L & Evan GI (2008) Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 14, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Duffy DJ, Krstic A, Schwarzl T, Halasz M, Iljin K, Fey D, Haley B, Whilde J, Haapa‐Paananen S, Fey V et al (2016) Wnt signalling is a bi‐directional vulnerability of cancer cells. Oncotarget 7, 60310–60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duffy DJ, Krstic A, Schwarzl T, Higgins DG & Kolch W (2014) GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Mol Cancer Ther 13, 454–467. [DOI] [PubMed] [Google Scholar]
- 51. Laurenti E, Varnum‐Finney B, Wilson A, Ferrero I, Blanco‐Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN et al (2008) Hematopoietic stem cell function and survival depend on c‐Myc and N‐Myc activity. Cell Stem Cell 3, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR & Trumpp A (2004) c‐Myc controls the balance between hematopoietic stem cell self‐renewal and differentiation. Genes Dev 18, 2747–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Laurenti E, Wilson A & Trumpp A (2009) Myc's other life: stem cells and beyond. Curr Opin Cell Biol 21, 844–854. [DOI] [PubMed] [Google Scholar]
- 54. Fagnocchi L, Cherubini A, Hatsuda H, Fasciani A, Mazzoleni S, Poli V, Berno V, Rossi RL, Reinbold R, Endele M et al (2016) A Myc‐driven self‐reinforcing regulatory network maintains mouse embryonic stem cell identity. Nat Commun 7, 11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R et al (2018) A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520. [DOI] [PubMed] [Google Scholar]
- 56. Scognamiglio R, Cabezas‐Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgartner D, Carnevalli LS, Atzberger A, Haas S et al (2016) Myc depletion induces a pluripotent dormant state mimicking diapause. Cell 164, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hobmayer B, Holstein TW & David CN (1997) Stimulation of tentacle and bud formation by the neuropeptide head activator in Hydra magnipapillata . Dev Biol 183, 1–8. [DOI] [PubMed] [Google Scholar]
- 58. Philipp I, Holstein TW & Hobmayer B (2005) HvJNK, a Hydra member of the c‐Jun NH2‐terminal kinase gene family, is expressed during nematocyte differentiation. Gene Expr Patterns 5, 397–402. [DOI] [PubMed] [Google Scholar]
- 59. Hartl M, Nist A, Khan MI, Valovka T & Bister K (2009) Inhibition of Myc‐induced cell transformation by brain acid‐soluble protein 1 (BASP1). Proc Natl Acad Sci USA 106, 5604–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Khalturin K, Anton‐Erxleben F, Sassmann S, Wittlieb J, Hemmrich G & Bosch TC (2008) A novel gene family controls species‐specific morphological traits in Hydra . PLoS Biol 6, e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reiter F, Hartl M, Karagiannidis AI & Bister K (2007) WS5, a direct target of oncogenic transcription factor Myc, is related to human melanoma glycoprotein genes and has oncogenic potential. Oncogene 26, 1769–1779. [DOI] [PubMed] [Google Scholar]
- 62. Valovka T, Schonfeld M, Raffeiner P, Breuker K, Dunzendorfer‐Matt T, Hartl M & Bister K (2013) Transcriptional control of DNA replication licensing by Myc. Sci Rep 3, 3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Petropoulos CJ & Hughes SH (1991) Replication‐competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol 65, 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]


