Abstract
Background
Over 1 million men are diagnosed with prostate cancer each year worldwide, with a wide range of research programs requiring access to patient tissue samples for development of improved diagnoses and treatments. A random sampling of prostate tissue is sufficient for certain research studies; however, there is growing research need to target areas of the aggressive tumor as fresh tissue. Here we set out to develop a new pathway “PEOPLE: PatiEnt prOstate samPLes for rEsearch” to collect high‐quality fresh tissue for research use, using magnetic resonance imaging (MRI) to target areas of tumor and benign tissue.
Methods
Prostate tissue was sampled following robotic radical prostatectomy, using MRI data to target areas of benign and tumor tissue. Initially, 25 cases were sampled using MRI information from clinical notes. A further 59 cases were sampled using an optimized method that included specific MRI measurements of tumor location along with additional exclusion criteria. All cases were reviewed in batches with detailed clinical and histopathological data recorded. For one subset of samples, DNA was extracted and underwent quality control. Ex vivo culture was carried out using the gelatin sponge method for an additional subset.
Results
Tumor was successfully fully or partially targeted in 64% of the initial cohort and 70% of the optimized cohort. DNA of high quality and concentration was isolated from 39 tumor samples, and ex vivo culture was successfully carried out in three cases with tissue morphology, proliferation, and apoptosis remaining comparable before and after 72 hours culture.
Conclusion
Here we report initial data from the PEOPLE pathway; using a method for targeting areas of tumor within prostate samples using MRI. This method operates alongside the standard clinical pathway and minimizes additional input from surgical, radiological, and pathological teams, while preserving surgical margins and diagnostic tissue.
Keywords: biobank, cancer, magnetic resonance imaging, prostate, sampling, specimen, tissue
1. INTRODUCTION
Prostate cancer is the second most common cancer in men worldwide, with an estimated 1.1 million men having been diagnosed with the disease in 2012.1 Current prostate cancer research programmes are often focused on better ways to diagnose and treat aggressive prostate cancer, while sparing men with more indolent cancers from undergoing unnecessary procedures and treatments. To best address these priorities, there is an increasing research need for high‐quality prostate tissue, which represents the index lesion. With a wide range of emerging technologies being adopted in prostate cancer research, there is also a growing need for fresh tissue that can either be used immediately or banked for larger studies, for example, in ex vivo culture, ex vivo magnetic resonance imaging (MRI) for assessment of new treatments and/or biomarkers or for large cohort genomic studies such as the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA). For studies requiring live cells, researchers must be able to identify areas of tumor and benign tissue for experimental use immediately upon tissue collection, rather than fix or freeze tissue and wait for results from the histological analysis.
Numerous methods have been published for sampling radical prostatectomy specimens for research without impeding on the diagnostic pathway. Key considerations across each published method include the minimization of ischemia time, control of temperature during transport and processing, preservation of surgical margins for pathological examination and careful recording of positions of samples within the prostate. One early method involved slicing the prostate in half immediately in theater, taking research biopsies for freezing and then suturing the prostate back together for fixation.2 Another method involved slicing the whole prostate fresh in 4‐mm slices, and storing one whole slice for research in RNAlater, while the rest were fixed and stored as formalin‐fixed paraffin‐embedded (FFPE) blocks for pathological assessment.3 Sooriakumaran et al4 published a method in 2011 which involved slicing and taking mapped punch biopsies for immediate storage in RNAlater in theater, followed by fixation of remaining sections in histology cassettes. The team later confirmed consistently high RNA integrity independent of ischemia time.5 To generate larger frozen specimens suitable for inclusion in the ICGC, another group quartered and flash froze alternate slices of the prostate for biobanking. This resulted in the generation of large amounts research specimens, but also large proportions of surgical margins not being available for routine histological processing, although the banked samples could be accessed later by pathology if required for further diagnostic tests.6
Our previous prostate sampling publication7 built upon these published methods to generate large quantities of high‐quality biopsy punches for research from radical prostatectomy specimens without affecting surgical margins. The method included pinning the prostate capsule to a cork board following sampling to ensure surgical margins did not become warped during fixation. Samples were taken randomly across a full transverse slice and stored without knowledge of tumor content. The method was highly successful for the ICGC study, where tumor content could be assessed at a later date. However, the ability to be reasonably confident of tumor content in real time is essential for many research applications requiring fresh tissue. The ability to use tissue immediately with high confidence of tumor content opens up a wide range of new technologies for prostate cancer research, in particular, new imaging techniques and ex vivo culture.
With numerous MRI clinical trials and studies reporting the efficacy of MRI in diagnosing clinically significant prostate cancer, there is an increasing trend towards using MRI before transperineal template biopsy to diagnose patients with suspected prostate cancer.8, 9, 10, 11 This represents an ideal opportunity to better target research samples, by incorporating a review of MRI data into the sampling procedure.
Here we set out to use MRI to build upon previously published prostate sampling methods, enabling researchers to target specific areas of tumor and benign tissue within radical prostatectomy specimens, and use tissue either immediately fresh, or frozen or fixed as applicable to the specific study requirements.
2. MATERIALS AND METHODS
2.1. Tissue samples
Patients were recruited and consented under Genomics England's 100,000 Genome Project Ethics at University College Hospital between March 2016 and July 2017, with a subset of patients also consented under UCL/UCLH Biobank Ethics (REC 15/YH/0311). A summary of the clinical characteristics of these patients are included in Table 1.
Table 1.
Patient characteristics
| Patient characteristics | N = 84 |
|---|---|
| Age at surgery, y | 62.1 ± 6.77 |
| Tumor volume, mL | 3.57 ± 2.14 |
| Prostate volume, mL | 39.18 ± 14.59 |
| PSA, ng/dL | 10.86 ± 8.07 |
| Gleason grade (%) | 4 (4.76) |
| 3 + 3 | 50 (59.52) |
| 3 + 4 | 22 (26.19) |
| 4 + 3 | 7 (8.33) |
| 4 + 5 | 1 (1.19) |
| MRI grade (Likert) (%) | |
| 1 | 5 (5.95) |
| 2 | 19 (22.62) |
| 3 | 19 (22.62) |
| 4 | 27 (32.14) |
| 5 | 6 (7.14) |
| Unknown | 8 (9.52) |
| Pathological staging (%) | |
| pT1 | 1 (0.01) |
| pT2 | 36 (42.86) |
| pT3 | 39 (46.42) |
| Unknown | 8 (9.52) |
Research specimens were collected from 84 prostate cancer patients who underwent robot assisted radical prostatectomy at UCLH between March 2016 and July 2017. Key clinical characteristics are noted here with additional information in Supporting Information Data File 1.
Abbreviation: MRI, magnetic resonance imaging.
2.2. Tissue sampling from radical prostatectomy specimens
Prostates were collected from theaters immediately upon removal from the patient during robotic radical prostatectomy at UCLH and transported under UN3373 guidelines to the UCLH Pathology Department. If suitable staff were not available to sample the prostate immediately, the specimen was stored at 4°C for up to 24 hours in accordance with Genomics England guidelines. The prostate was weighed and inked, and a 5‐mm transverse slice was removed using the parallel blade device as previously published.7 Tumor and benign areas were identified and one to two samples of each removed as per the requirements of each study, using 3‐ or 6‐mm biopsy punches. Tumor regions typically felt slightly denser and occasionally looked paler and this was taken into account when selecting regions to sample.
The remaining tissue was pinned to cork, fixed in 10% neutral buffered formalin and processed as per local protocols. This process was initially carried out by pathologists, and later two trained postdoctoral researchers signed off as competent to carry out the procedure, under the supervision of a pathologist.
2.3. Tumor targeting using MRI
All patients received a multiparametric prostate MRI before their robotic radical prostatectomy, either at UCLH or at their referring hospital (Table 2). Two methods were used to target tumor samples using MRI data (Table 3).
Table 2.
MRI location
| MRI location | Patients, n (%) |
|---|---|
| Royal Free Hospital (North West London) | 14 (16.66) |
| St. Bartholomew's Hospital (Central London) | 12 (14.29) |
| Barnet General Hospital (North London) | 11 (13.1) |
| Princess Alexandra Hospital (Harlow, Essex) | 10 (11.9) |
| University College London Hospitals (Central London) | 10 (11.9) |
| King George Hospital (East London) | 9 (10.7) |
| North Middlesex University Hospital (North London) | 5 (5.95) |
| Whipps Cross University Hospital (East London) | 3 (3.57) |
| Other | 7 (8.33) |
Although all patients underwent surgery at UCLH, MRI was carried out across 13 hospitals with the majority of patients referred to UCLH post‐MRI. The number of patients from each hospital is given with the percentage of the overall cohort in brackets. Hospitals where less than three patients underwent MRI were grouped to avoid potential identification of patients, and include Homerton University Hospital, Whittington Hospital, Enfield Alliance Hospital, Queen's Hospital, and St. Margaret's Hospital.
Abbreviation: MRI, magnetic resonance imaging.
Table 3.
Tumor targeting methods
| Methods | Description | Exclusion criteria |
|---|---|---|
| Initial method | Surgical planning sheet reviewed (biopsy and MRI notes and sketches from radiologists, pathologists, and surgeons) | … |
| Transverse slice taken at base, mid or apex of prostate, and biopsy punches taken based on surgical planning sheet sketches | … | |
| Optimized method | Surgical planning sheet reviewed (biopsy and MRI notes and sketches from radiologists, pathologists and surgeons) | Prior therapy |
| MRI review: Identification of index lesion based on surgical planning sheet, measurement of prostate length (base–apex, mm) and distance from base to desired transverse slice (mm) | No observable lesion, or only diffuse changes by MRI | |
| Prostate measured, correction factor applied if shrinkage had occurred and transverse slice taken at measured position from base, then biopsy punches taken based on surgical planning sheet sketches and visibility or palpability of tumor where possible | Lesion smaller than 5 mm |
The first 25 patients recruited under PEOPLE were part of an initial cohort, where a basic tumor targeting approach was used and outlined here. The following 59 specimens were sampled using an optimized approach with more specific MRI targeting, also outlined here.
Abbreviation: MRI, magnetic resonance imaging; PEOPLE, PatiEnt prOstate samPLes for rEsearch.
2.3.1. Initial method
All patients undergoing robotic radical prostatectomy at UCLH are discussed at a surgical planning meeting, where a surgical planning sheet was filled out with details of tumor location according to both biopsy and MRI data (Figure 1A). This sheet was uploaded centrally and accessed when planning the tissue sampling to take a transverse slice towards the apex, mid‐gland or base as indicated, and then take a 3‐ or 6‐mm tissue punch from the region identified as containing a suspicious lesion.
Figure 1.
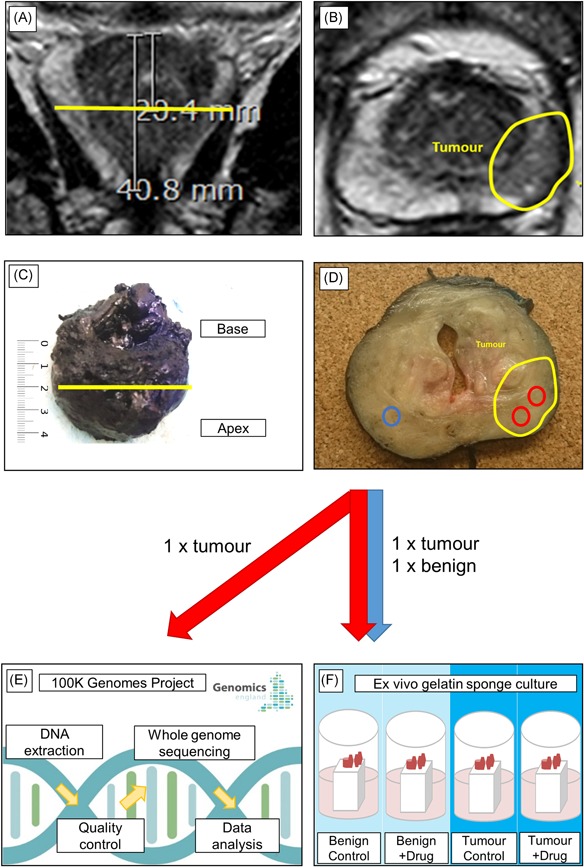
Tumor targeting methods. Following review of the surgical planning sheet, the index lesion is identified in MRI images. The position of the optimal transverse slice is identified and a measurement is taken from the base to this position and from the base to apex using a coronal T2‐weighted image (A). The position of the tumor is noted, here in an axial T2 weighted image (B). Following radical prostatectomy, the full surgical margins are inked right side blue, left side black according to local protocol, then the prostate is measured from base to apex and a correction factor is applied if the prostate has shrunk in comparison with the MRI image. The position of the desired 5‐mm transverse slice is identified (C). Following slicing, the area of expected tumor is confirmed if possible visually and/or palpably, with some tumors appearing paler and denser than benign tissue. Six millimeter biopsy punches are used to remove samples from indicated tumor and benign areas (D). One tumor sample is submitted to Genomics England for DNA extraction, quality control, whole genome sequencing and data analysis as part of the 100,000 Genomes Project (E). Matched tumor and benign samples are submitted for use in ex vivo gelatin sponge culture, or other ethically approved research projects in a subset of cases (F) [Color figure can be viewed at wileyonlinelibrary.com]
2.3.2. Optimized method
Following review of the surgical planning sheet, MRI images were visualized using IMPAX software (version 6.5; AGFA‐Gevaert, Mortsel, Belgium) and T2‐weighted axial, T2‐weighted coronal, diffusion‐weighted imaging, and apparent diffusion coefficient (ADC) images used to identify the index lesion, defined as the largest tumor focus within the prostate. A measurement was then taken from the base of the prostate to the target lesion in mm within the software, using the image where the tumor was most visible—typically the T2‐weighted axial image (Figure 1B). Once trained by a board‐certified radiologist (EJ), a postdoctoral researcher could carry out this procedure in less than 5 min/patient. During the sampling procedure, a transverse slice was removed at the position identified previously using the MRI image, measured from the base of the prostate by a ruler in mm (Figure 1C). After the first six cases were sampled, an additional step was carried out to identify any shrinkage that could affect the accuracy of this method. Here, length of the prostate was measured from base to apex on the coronal T2 MRI image, and compared with the length of the prostate measured by ruler immediately before slicing. Based upon the amount of shrinkage observed an ad hoc correction was applied before identifying the position to slice. Exclusion criteria were applied to omit patients who had undergone prior therapy, had no distinct lesion by MRI (ie, only diffuse changes) and patients who had lesions smaller than 5 mm visible on MRI.
2.4. Tissue storage
Tissue cores were stored depending on the individual study. At least one core from each patient was embedded in optimized cutting temperature compound (OCT), snap frozen in liquid nitrogen immediately and then stored at −80°C. For subsets of cases, additional benign or tumor tissue was either snap frozen dry in cryovials in liquid nitrogen, fixed immediately in 10% neutral buffered formalin and stored as FFPE blocks, or transported in warm media for ex vivo culture (ex vivo culture was not carried out where prostates had been stored overnight before sampling).
2.5. Assessment of tumor content
All 84 samples assessed for tumor content had been stored frozen in OCT. Samples were sectioned at 5 μm thickness using the sterile technique on the cryostat, then stained immediately with haematoxylin and eosin (H&E) as per 100,000 Genomes Project standard operating procedures. Slides were assessed by an experienced consultant uropathologist and tumor content was reported as 0%, 5%, 10%, 15%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, or 100%.
2.6. Tissue data review
All specimens were reviewed in batches, by a multidisciplinary team including at least one consultant uropathologist along with a pathology registrar, postdoctoral researchers and technicians. All anonymised data is included in Supporting Information Data File 1 and includes whether the specimen was refrigerated before sampling, number of samples taken, whether the initial or optimized tumor targeting method was used, which member of the pathology team supervised cut up, which area(s) were sampled, which level the sample was taken, distance to tumor on the same level if tumor was missed, whether the tumor could have been hit if samples were taken on the next/previous level in the same position, the total number of levels, the number of levels the tumor was present in, the level where tumor(s) reached maximum dimension, the maximum tumor dimension, the number of tumors in the specimen, the most affected area of the prostate by MRI versus biopsy, prostate volume and weight, tumor volume, Gleason grade, pathological tumor staging, MRI (Likert), the hospital the MRI was carried out in, tumor volume, patient age, PSA, base‐apex length (MRI), base‐apex length (ruler), base‐apex shrinkage (%), cellularity (very low [<700 total cells], low [4000 cells], medium [4000‐10 000 total cells], high [>10 000 total cells], very high [>50 000 total cells]), number of 10 µm sections submitted for DNA extraction, which sample used for frozen sections (if more than one was available), DNA concentration and 260:280 ratio.
2.7. DNA isolation
DNA was isolated from 20 to 40 frozen tissue sections per sample (10 μm) from 39 cases, taken adjacent to sections used for tumor content assessment, as per 100 000 Genomes Project standard operating procedures. The Maxwell 16 LEV Blood DNA Kit (AS1290; Promega) was used to extract DNA. Before extraction samples were homogenized by the addition of 300 μL lysis buffer and 30 μL Proteinase K (Promega) to each sample, followed by incubation at 56°C for at 30 minutes. Samples were then transferred to Maxwell 16 LEV Cartridges for extraction, according to the manufacturer's protocol. DNA was eluted in 70 μL Elution Buffer (Promega). Concentration was assessed using the Qubit dsDNA HS Assay Kit with the Qubit Fluorometer (Thermo Fisher Scientific) and 260:280 ratio assessed using the Nanodrop Fluorometer (Thermo Fisher Scientific) according to manufacturer's instructions.
2.8. Ex vivo culture
Ex vivo culture was carried out using the gelatin sponge method.12 Sponges were placed in a 24‐well plate with 200 μL/well Roswell Park Memorial Institute media supplemented with 10% fetal bovine serum 2 to 3 hours before tissue collection to allow the sponges to draw up the media in a 37°C, 5% CO2 incubator. Fresh 3 or 6 mm cores were divided as appropriate using a sterile scalpel and placed on the damp sponges, then incubated for 72 hours. Both uncultured control samples and samples that were cultured for 72 hours were fixed in 10% neutral buffered formalin and stored as FFPE blocks. Four micrometer sections from the FFPE blocks were stained with H&E, or assessed for Ki67 (m7240, 1:100 dilution, F. Dabe 1 minute haematoxylin, ER2 30 minutes; Dako) by immunohistochemistry performed on the BondMax autostainer (Leica) and cleaved caspase 3 (9664; 1:200 dilution, F. Dabe 1 min haematoxylin, ER2 20 minutes; Cell Signalling Technology).
3. RESULTS
Eighty‐four prostates were sliced and sampled for various research projects under the PEOPLE pathway, with tumors targeted using the initial method for 25 prostates and optimized method for 59 prostates (Figure 1). Patient characteristics are reported in Table 1. All surgery was carried out in UCLH and MRIs were carried out in 13 different hospitals across Greater London and Essex (Table 2).
After the first 25 cases a detailed case review was carried out and seven cases were identified where the tumor was missed. All could potentially have been accurately targeted had the transverse slice been taken more towards the base or apex of the prostate (Supporting Information Data File 1). This resulted in a change to the optimized method where measurements from the MRI image were used to target the transverse slice and tumor.
To account for shrinkage that can occur following surgery, 46 prostates were measured using a ruler from base to apex, and compared with MRI measurements of the same distance. The majority of prostates did shrink, with a mean shrinkage of 5.71% ± 10.57% and a trend towards larger prostates shrinking more and smaller prostates shrinking less (P = 0.0002) (Figure 2A). Some prostates increased in size, possibly due to growth between the time the MRI image was taken and surgery.
Figure 2.
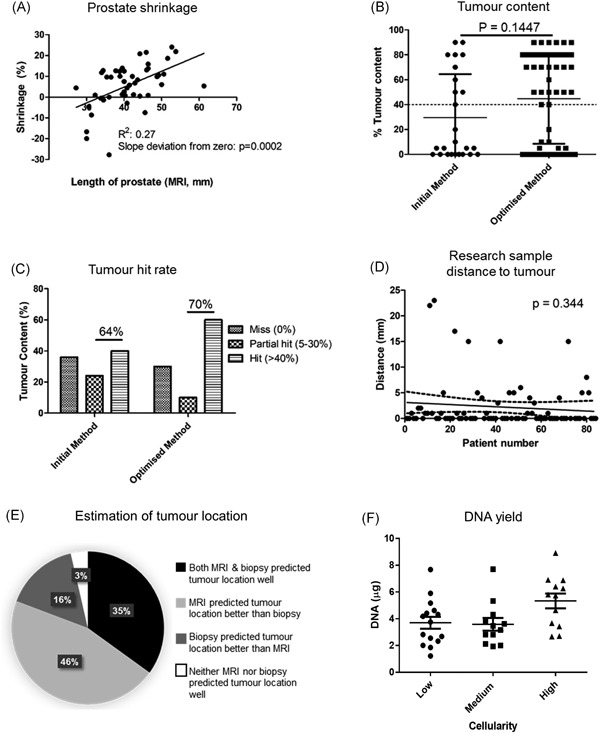
Key data from case review. Prostate length was measured in mm in MRI images and using a ruler postsurgery and an ad hoc correction for shrinkage applied each time before slicing (A). Tumor content was assessed by a uropathologist based on H&E staining of frozen sections of tumor targeted samples. Mean tumor content was 29.6% for the initial targeting method and 44.8% for the optimized targeting method. All samples above the threshold of 40% tumor content were deemed eligible for next generation sequencing as per 100 000 Genome Project Guidelines. A two‐tailed Mann‐Whitney was performed and the difference between the two cohorts was deemed nonsignificant (B). Samples where 5% to 40% tumor was identified were noted as “partial hits” and could be macrodissected to be submitted for next generation sequencing. (C). Distance from punch taken to nearest tumor on the same slice was measured and plotted sequentially in the order of patients sampled, with linear regression not identifying a significantly nonzero slope (D). The location of most significant tumor was recorded based on MRI, biopsy and radical prostatectomy data. It was recorded whether MRI, biopsy, both or neither best indicated the location of most significant disease postradical prostatectomy (E). DNA was isolated from 39 cases for next generation sequencing under the 100 000 Genomes Project. Almost all cases yielded concentrations over 20 ng/μL and those that did not were repeated with more sections and submitted for sequencing. Data is noted for the first DNA isolation of each case, with nonsignificant q values from a one way ANOVA and Tukey's test (F). ANOVA, analysis of variance; H&E: haematoxylin and eosin; MRI, magnetic resonance imaging
All tumor samples were assessed for tumor content by a consultant uropathologist, using frozen H&E sections. Forty percent of samples taken using the initial method and 60% of samples taken using the optimized method had tumor content of at least 40%, and therefore could be submitted without macrodissection for sequencing under the 100,000 Genomes Project (Figure 2B). Samples that had between 5% and 30% tumor content were considered “partial hits” and could potentially be used for sequencing following macrodissection. The sum of hits and partial hits for the initial method was 64% and for the optimized method was 70% (Figure 2C).
As well as the risk of missing the tumor when taking the transverse slice, there was also a risk of missing the tumor within the slice when taking punch biopsies. It appeared that accuracy in hitting tumor within the slice improved slightly over time, although this trend was not statistically significant (P = 0.344), based on measurements taken from the sampled area to the nearest tumor within that slice (Figure 2D).
The crucial aspect of the PEOPLE method is tumor targeting, and to maximize the efficacy of this we used both MRI and biopsy data to assess tumor location within the prostate for sampling. Where biopsy and MRI data did not agree, both indicated areas were targeted if possible, and if not, MRI data were used alone. During retrospective histopathological assessment, the consultant pathologist noted which area of the radical prostatectomy slides was most affected, for example, left posterior. This was subsequently compared with the planning protocol. In cases where the surgical planning sheet estimated by both biopsy and MRI that the tumor was in this position (left posterior), it was considered that both biopsy and MRI estimated tumor position well. If only one or neither of these indicated accurately the most affected area of the prostate, this was noted (Supporting Information Data File 1 and Table 2). Following assessment of all samples it was found that both MRI and biopsy data only correctly estimated the same location of highest Gleason tumor in 35% of cases, with MRI outperforming biopsy in 46% of cases and biopsy outperforming MRI in 16% of cases (Figure 2E). There were three cases where neither the MRI nor the biopsy correctly estimated the location of the tumor. In all three cases the tumor was present in the transverse slice, but the tissue sample was not taken from the area of the slice where the tumor was present. Two of these cases were reported Likert 2 by MRI (low likelihood of tumor presence) and one was reported Likert 3 (equivocal likelihood of tumor presence). These three prostates were also low in weight (mean, 36 vs 48 g cohort mean) and tumor volume (mean, 2.6 vs 4.4 mL cohort mean) (highlighted orange in Supporting Information Data File 1).
Hit rate was found not to significantly differ by tumor volume (Figure S1A), prostate volume (Figure S1B) or ISUP grade (Figure S1C). MRI (Likert) did not correlate well with Gleason grade (Figure S1D) or tumor volume (Figure S1E), although this cohort is underpowered to consider the accuracy of MRI in this regard. The initial and optimized method did not differ by ISUP grade (Figure S1F).
Thirty‐nine samples were submitted for sequencing under the 100,000 Genomes Project. DNA was eluted in a final volume of 70 μL, and was analyzed using spectrophotometry. All had 260:280 ratios of 1.8 to 2.0, with an average concentration of 41.7 ± 18.92 ng/μL. Cellularity of each sample was noted by a pathologist as very low (<700 total cells), low (4000 cells), medium (4000‐10 000 total cells), high (>10 000 total cells), very high (>50 000 total cells), and there was no significant difference in DNA concentration between each cellularity grouping (Figure 2F).
Three samples were cultured ex vivo using the gelatin sponge method.12 Control samples were stored as FFPE blocks at the time of sampling and after 72 hours culture at 37°C, and 4‐μm sections were stained with H&E, the proliferation marker Ki67 and apoptosis marker cleaved caspase 3. Minimal‐no differences in morphology, proliferation or apoptosis were noted between cultured and uncultured samples in cases 1 and 2, while some loss of tissue integrity was noted for patient 3, which could be optimized in future using different media or shorter culture (Figure 3).
Figure 3.
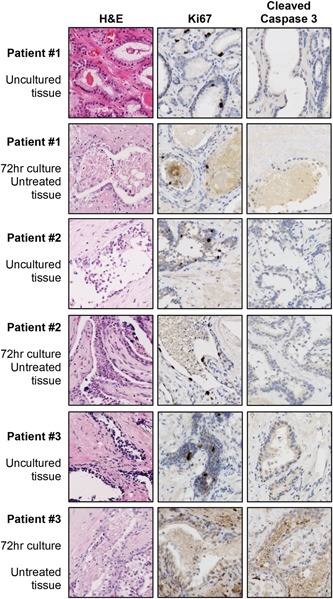
Ex vivo culture. Tissue from three cases was cultured ex vivo for 72 hours using the gelatin sponge method. Samples were stored as FFPE blocks after 72 hours (untreated tissue), with matched uncultured samples also stored FFPE (uncultured tissue). Sections were then stained with H&E (morphology), Ki67 (proliferation) and cleaved caspase 3 (apoptosis) to assess whether tissue from PEOPLE is of sufficient quality for culture. FFPE, formalin‐fixed paraffin‐embedded; PEOPLE, PatiEnt prOstate samPLes for rEsearch [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
A number of methods have been described for the collection of prostate cancer tissue for research following radical prostatectomy. A 2017 review on these existing methods highlighted the importance of reducing numbers of samples taken from surgical specimens, and suggested the use of imaging and biopsy data to better target lesions for sampling.13 Here, we address this by using MRI to target specific areas of tissue that can then be used immediately for research, vastly increasing the range of experimental techniques that can be exploited, while still integrating well within the clinical pathway.
Previous prostate tissue collection methods have sampled areas of the gland randomly, or by palpating or viewing the tumor as slightly more dense or pale. These methods are sufficient for certain downstream techniques, but are unlikely to be reliable for routine collection of high tumor content samples based upon our data that demonstrates that the average tumor only occupies 11% of the overall prostate volume, and is often not discernible by eye or touch from the surrounding benign tissue (Figure 1D). MRI has been heralded as a major step forward for prostate cancer diagnosis, with findings from multiple clinical trials advocating its routine use.8, 9, 10, 11 With this gain in popularity comes an opportunity for researchers to exploit MRI data to improve fresh tissue collection, here allowing us to successfully target tumor in 70% of samples (Figure 2C). We have incorporated MRI into the sampling method To improve tumor targeting and whilst UCLH is a world leader in MRI, most patients in our cohort had imaging at one of 12 other hospitals across the Greater London/Essex area before coming for surgery at UCLH. The varying quality of the MRI data did not impact on our ability to target tumors and we predict that this method can be successful wherever MRI is carried out before surgery (Table 2).
The major disadvantage of previous methods was the lack of confidence in the pathology of the sampled tissue that limited its utility and often required extensive sampling and/or pathologist time to identify samples with high tumor content.2, 3 The key benefit of incorporating MRI‐guided tumor targeting into the sample collection pathway is the increased freedom this provides to carry out a much wider range of downstream experiments. As with previous methodologies, samples can be stored in RNAlater for expression analysis, fixed and stored as FFPE for immunohistochemistry, or frozen for genomics studies, all with greater confidence in the content of each sample before histological assessment.4, 5 Here, tumor samples were frozen and DNA of consistently high purity and quantity for sequencing was isolated for the 100,000 Genomes Project (Figure 2F). Crucially, this targeting method also allows for the development of new methodologies which require fresh matched tumor and benign tissue. For example, ex vivo MRI or other imaging techniques can be carried out on fresh tissue quickly following surgery, allowing researchers to more effectively study density and other physical properties than with fixed or frozen tissue.14 The method we describe here allows techniques such as ex vivo culture, more widely used in breast cancer research, to be carried out to test new drugs or biomarkers.12, 15, 16, 17 We cultured tumor and benign tissue for 72 hours using the gelatin sponge method and were able to show that untreated tissue postculture had minimal degradation in morphology, proliferation, and apoptosis as uncultured tissue (Figure 3). As prostate cancer research progresses, we predict more methods will emerge, which require high quality fresh tumor tissue. To do these experiments using previously described tissue collection methods, researchers would need to overcollect samples and use a substantial excess to ensure presence of tumor. This is unfeasible in terms of both cost, time and an unethical use of human tissue that could be better utilized in other studies.
Although this method contains additional steps compared with previously published methods, including our own, we succeeded in reducing the time burden on clinical staff using a protocol that dovetailed with the standard clinical pathway and could be performed by trained postdoctoral researchers and technicians.7 This included measurement of MRI data, meaning the only radiology department input required was for training at the beginning of the study. This reduced the burden on the overall clinical system and allowed for flexible sample collection for research projects when required by the researchers. Additionally, surgical margins and diagnostic tissue are preserved as before.7
Although this new method does allow for a wider range of downstream applications and minimizes any additional burden on the pathologist, it does not significantly improve upon previously published tumor hit rates overall. This is due to the reduced tissue sampling carried out here, where one to two indicated tumor and matched benign punch biopsies are taken for immediate experimentation, leading to a tumor hit rate of 70%, rather than the previous methods of taking many samples, often from several prostate slices and assessing tumor content of each sample later, which has led to tumor hit rates from 69% to 100%.13 It remains useful to discuss the 30% of cases where tumor was missed here, with a view to further refinement of the pathway for implementation in new centers. Interestingly, in the cases where tumor was missed, there was no discernible difference in ISUP grade, MRI (Likert), tumor volume, tumor location or prostate volume, and as such we have not added further restrictions to our recruitment criteria based on these parameters. There were three cases where neither the biopsy nor the MRI indicated the location of the tumor well. These cases had lower tumor volume, prostate weight and MRI Likert score, none of which were out of the range of this cohort, but perhaps these criteria together, and taken in the context of differing biopsy and MRI results could be used to identify potential patients to avoid recruiting in future iterations of this method.
High quality prostate cancer research relies on access to high quality human tissue for experimentation. Here we have built upon existing tissue sampling methodologies to introduce the PEOPLE pathway, providing high quality MRI‐targeted fresh tissue for a wide range of research.
5. CONCLUSIONS
We report our initial data from the PEOPLE pathway, an improvement to our previously published prostate slicing method, where we utilize MRI data to target and collect high‐quality tumor and benign prostate tissue that can be used for a wide variety of research applications such as next‐generation sequencing or ex vivo culture, with minimal impact on the clinical pathway.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
The authors would like to thank the Genomics England 100,000 genomes project team for their support in this study, in particular Lyn Chitty, Manuel Rodriguez‐Justo and Adrienne Flanagan. The authors wish to acknowledge Prostate Cancer UK for funding SH under the Prostate Cancer UK (Traveling Prize Fellowship TLD‐PF16‐004) and HP and EWJ under INNOVATE (PG14‐018‐TR2). This workstudy was supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Center.
Heavey S, Costa H, Pye H, et al. PEOPLE: PatiEnt prOstate samPLes for rEsearch, a tissue collection pathway utilizing magnetic resonance imaging data to target tumor and benign tissue in fresh radical prostatectomy specimens. The Prostate. 2019;79:768‐777. 10.1002/pros.23782
The copyright line for this article was changed on 12 June 2019 after original online publication.
References
REFERENCES
- 1. http://globocan.iarc.fr/old/FactSheets/cancers/prostate‐new.asp.
- 2. Wheeler TM, Lebovitz RM. Fresh tissue harvest for research from prostatectomy specimens. Prostate. 1994;25(5):274‐279. [DOI] [PubMed] [Google Scholar]
- 3. Jhavar SG, Fisher C, Jackson A, et al. Processing of radical prostatectomy specimens for correlation of data from histopathological, molecular biological, and radiological studies: a new whole organ technique. J Clin Pathol. 2005;58(5):504‐508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sooriakumaran P, Henderson A, Denham P, Langley SE. A novel method of obtaining prostate tissue for gene expression profiling. Int J Surg Pathol. 2009;17(3):238‐243. [DOI] [PubMed] [Google Scholar]
- 5. Dev H, Rickman D, Sooriakumaran P, et al. Biobanking after robotic‐assisted radical prostatectomy: a quality assessment of providing prostate tissue for RNA studies. J Transl Med. 2011;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esgueva R, Park K, Kim R, et al. Next‐generation prostate cancer biobanking: toward a processing protocol amenable for the International Cancer Genome Consortium. Diagn Mol Pathol. 2012;21(2):61‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warren AY, Whitaker HC, Haynes B, et al. Method for sampling tissue for research which preserves pathological data in radical prostatectomy. Prostate. 2013;73(2):194‐202. [DOI] [PubMed] [Google Scholar]
- 8. Ahmed HU, El‐Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi‐parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389(10071):815‐822. [DOI] [PubMed] [Google Scholar]
- 9. Simmons LAM, Kanthabalan A, Arya M, et al. The PICTURE study: diagnostic accuracy of multiparametric MRI in men requiring a repeat prostate biopsy. Br J Cancer. 2017;116(9):1159‐1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Porpiglia F, Manfredi M, Mele F, et al. Diagnostic pathway with multiparametric magnetic resonance imaging versus standard pathway: results from a randomized prospective study in biopsy‐naive patients with suspected prostate cancer. Eur Urol. 2017;72(2):282‐288. [DOI] [PubMed] [Google Scholar]
- 11. Shah TT, To WKL, Ahmed HU. Magnetic resonance imaging in the early detection of prostate cancer and review of the literature on magnetic resonance imaging‐stratified clinical pathways. Expert Rev Anticancer Ther. 2017;17(12):1159‐1168. [DOI] [PubMed] [Google Scholar]
- 12. Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10(8):483‐487. [DOI] [PubMed] [Google Scholar]
- 13. Tolkach Y, Eminaga O, Wötzel F, et al. Blind biobanking of the prostatectomy specimen: critical evaluation of the existing techniques and development of the New 4‐Level tissue extraction model with high sampling efficacy. Prostate. 2017;77(4):396‐405. [DOI] [PubMed] [Google Scholar]
- 14. Bourne RM, Bailey C, Johnston EW, et al. Apparatus for histological validation of in vivo and ex vivo magnetic resonance imaging of the human prostate. Front Oncol. 2017;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van de Merbel AF, van der Horst G, van der Mark MH, et al. An ex vivo tissue culture model for the assessment of individualized drug responses in prostate and bladder cancer. Front Oncol. 2018;8:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miserocchi G, Mercatali L, Liverani C, et al. Management and potentialities of primary cancer cultures in preclinical and translational studies. J Transl Med. 2017;15(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnston E, Pye H, Bonet‐Carne E, et al. INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion‐weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer. 2016;16(1):816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information


