Abstract
Anti‐nuclear antibodies to speckled 100 kDa (sp100) and glycoprotein 210 (gp210) are specific serologic markers of primary biliary cholangitis (PBC) of uncertain/controversial clinical or prognostic significance. To study the genetic determinants associated with sp100 and gp210 autoantibody subphenotypes, we performed a genome‐wide association analysis of 930 PBC cases based on their autoantibody status, followed by a replication study in 1,252 PBC cases. We confirmed single‐nucleotide polymorphisms rs492899 (P = 3.27 × 10−22; odds ratio [OR], 2.90; 95% confidence interval [CI], 2.34‐3.66) and rs1794280 (P = 5.78 × 10−28; OR, 3.89; 95% CI, 3.05‐4.96) in the human major histocompatibility complex (MHC) region associated with the sp100 autoantibody. However, no genetic variant was identified as being associated with the gp210 autoantibody. To further define specific classical human leukocyte antigen (HLA) alleles or amino acids associated with the sp100 autoantibody, we imputed 922 PBC cases (211 anti‐sp100‐positive versus 711 negative cases) using a Han Chinese MHC reference database. Conditional analysis identified that HLA‐DRβ1‐Asn77/Arg74, DRβ1‐Ser37, and DPβ1‐Lys65 were major determinants for sp100 production. For the classical HLA alleles, the strongest association was with DRB1*03:01 (P = 1.51 × 10−9; OR, 2.97; 95% CI, 2.06‐4.29). Regression analysis with classical HLA alleles identified DRB1*03:01, DRB1*15:01, DRB1*01, and DPB1*03:01 alleles can explain most of the HLA association with sp100 autoantibody. Conclusion: This study indicated significant genetic predisposition to the sp100 autoantibody, but not the gp210 autoantibody, subphenotype in PBC patients. Additional studies will be necessary to determine if these findings have clinical significance to PBC pathogenesis and/or therapeutics.
Abbreviations
- AA
amino acid
- AMA
anti‐mitochondrial antibody
- ANA
anti‐nuclear antibody
- CI
confidence interval
- gp210
glycoprotein 210
- GWA
genome‐wide association
- HLA
human leukocyte antigen
- IL12
interleukin 12
- IL12R
IL12 receptor
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MHC
major histocompatibility complex
- OD
optical density
- OR
odds ratio
- PBC
primary biliary cholangitis
- PBS
phosphate‐buffered saline
- SNP
single‐nucleotide polymorphism
- sp100
speckled 100 kDa
Primary biliary cholangitis (PBC) is an autoimmune disease characterized by chronic inflammation and destruction of intrahepatic bile ducts. PBC is mainly observed in middle‐aged women. Patients with PBC generally present with abnormal liver tests including elevation of alkaline phosphatase (ALP), gamma glutamyltranspeptidase (GGT), and combinations of symptoms, including pruritus, fatigue, and portal hypertension.1 Anti‐mitochondrial antibodies (AMA) are the serological hallmark for PBC and are found in nearly 95% of patients.2 Several large‐scale genome‐wide association (GWA) studies in European, Japanese, and Han Chinese PBC cohorts have pointed to a strong genetic predisposition to PBC.3, 4, 5, 6, 7 Specific human leukocyte antigen (HLA) alleles and common polymorphisms in interleukin 12 (IL12), IL12 receptor (IL12R), tumor necrosis factor superfamily member 15 (TNFSF15), IL21, IL21R, and other immune‐associated genes have been implicated in PBC pathogenesis.3, 5, 6 Although these studies provided many insights in our understanding of PBC pathogenesis, no further studies have examined the genetics of subphenotypes in PBC.
Other than AMA, autoantibodies to glycoprotein 210 (gp210) and speckled 100 kDa (sp100), the two subtypes of anti‐nuclear antibodies (ANA), are also highly specific to PBC and can be detected in 10%‐40% of PBC patients.8, 9, 10 Anti‐gp210 and sp100‐positive serum samples present typical rim and multinuclear dot staining patterns, respectively. Anti‐gp210 antibody recognizes a single epitope containing 25 amino acids (AAs) in the C‐terminal tail of nuclear pore gp210 protein.11 Several studies have indicated that autoantibody to gp210 can serve as an indicator of poor prognosis. Muratori et al. found that anti‐gp210 positivity was associated with higher serum cholestatic parameters including ALP, GGT, and bilirubin.8 Unfavorable clinical outcome, more rapid disease progression, and significantly higher hepatic failure were found in anti‐gp210‐positive patients.11, 12, 13 Recently, Yang et al. found that anti‐gp210 positivity is associated with more severe forms of PBC in Chinese patients with prognostic value.14, 15 Sp100 is a component of promyelocytic leukemia protein (PML) and sp100‐containing nuclear bodies. Anti‐sp100 antibodies in PBC patients recognize multiple epitopes in sp100.16 Studies on the clinical significance of anti‐sp100 antibody have thus far generated mixed results.17, 18, 19, 20 A few early studies indicated that the anti‐sp100‐positive patient group tended to have earlier disease onset and more frequently had cirrhosis compared to anti‐sp100‐negative patients. Studies by Nakamura et al.12 and Yang et al.14 found no correlation of anti‐sp100 positivity with disease prognosis, but neither study investigated the subtypes of autoantibodies. Two semiquantitative analyses of anti‐sp100 antibody titers during the course of the treatment found a correlation of anti‐sp100 titers with Mayo risk score, an indicator of disease severity.17, 19 Because sp100 autoantibodies recognized multiple epitopes on sp100, an earlier study found a change of some anti‐sp100 epitopes during the course of disease progression and ursodesoxycholic acid (UDCA) treatment in some PBC patients, but the overall anti‐sp100 status remained unchanged.21 Unfortunately, no subsequent follow‐up or replication studies were reported by other researchers. Therefore, it remains undetermined whether any anti‐sp100 immunoglobulin subtypes or titers can serve as indicators of disease progression.
We recently performed a large‐scale GWA analysis in a Han Chinese PBC cohort and identified multiple common variants in major histocompatibility complex (MHC) region, TNFSF15, IL21, IL21R, chemokine (C‐X‐C motif) receptor 5, cluster of differentiation 58, signal transducer and activator of transcription 4, and other immune‐related genes associated with PBC.6 To further define the role of genetic factors associated with different PBC subphenotypes (anti‐gp210/anti‐sp100) at the whole‐genome level, we undertook serological analysis of anti‐gp210/anti‐sp100 status in our PBC cases. Combining the serological results and genome‐wide single‐nucleotide polymorphism (SNP) data, we systematically performed the association analysis to pinpoint the genetic variants associated with gp210 and sp100 autoantibody subphenotypes in PBC. The results presented in this report indicate a strong genetic predisposition to the sp100 autoantibody subphenotype in PBC patients.
Patients and Methods
Patient Information and Genome‐Wide Genotyping
The PBC patients recruited in this study have been reported previously.6 The PBC patients were recruited with the approval of the research ethics boards of Southeast University and Jiaotong University and in accordance with the guidelines of the Declaration of Helsinki (2008). Informed consent was obtained from all subjects recruited. In brief, all patients self‐reported Han Chinese origin and were diagnosed based on the criteria recommended by the American Association for the Study of Liver Diseases.22 SNP genotyping was performed using Han Chinese population–specific HumanOmniZhongHua‐8 BeadChip (Illumina, San Diego, CA). The quality control of genotyping results was detailed in our GWA report.6 We removed SNPs with a call rate of <90%, a minor allele frequency (MAF) < 1%, and a Hardy‐Weinberg equilibrium P value < 1 × 10−4. Potential genetic relatedness based on pairwise identity by descent for all of the successfully genotyped individuals was examined using PLINK 1.07 software.23 If first‐degree or second‐degree relative pairs were identified (PI‐HAT value > 0.25), we removed the samples with the lower genotyping call rates of the related individuals. SNPs located on the X, Y, or mitochondrial chromosomes and all copy number variation probes were excluded.
Enzyme‐Linked Immunosorbent Assay for ANTI‐sp100 and ANTI‐gp210 Antibodies
Serum samples were stored at −80°C in aliquot before use. Enzyme‐linked immunosorbent assay (ELISA) plates (96‐well, 100 ng/well) were coated overnight at 4°C with purified sp100 antigen (AA position 296‐386) or gp210 antigen (AA position 1863‐1887). The plate was further incubated with 1% bovine serum albumin (BSA) for 2 hours at room temperature and washed 3 times with phosphate‐buffered saline (PBS). Serum samples were diluted (1:200) with PBS buffer containing 1% BSA. After incubation for 1 hour at 37°C, the plate was washed 4 times with PBS and incubated with 1:40,000 diluted peroxidase‐conjugated antihuman immunoglobulin G antibody (Genscript, China) at room temperature for 1 hour. Next, after washing 4 times with PBS, the plate was incubated with 100 μL of chromogenic substrate (3,3,5,5‐tetramethylbenzidine) for 20 minutes, with addition of 2 M sulfuric acid (50 μL/well) as the stop solution. Optical density (OD) was read by an ELISA plate reader with absorbance at 450 nm. The results with an OD ratio higher than 3 times over negative controls were defined as antibody‐positive. Samples with an OD ratio between 2 and 3 times were retested and excluded if ambiguous on multiple testing with different serum dilutions.
SNP Genotyping
SNP genotyping in the replication study was performed by TaqMan assay for rs492899 and rs9380343 and by Sanger DNA sequencing for rs1794280. The primer and probe sequences are listed in Supporting Table S1. The TaqMan assay was performed using a Bio‐Rad CFX Manager Real‐Time PCR system in 10 μL of reaction mixtures containing 1 × PCR buffer, 2 mM deoxyribonucleotide triphosphate mix, 2 μM of each primer, 3 μM of labeled probe, 0.25 U Taq HS polymerase (Takara, Dalian), 5 ng of genomic DNA, and 6.15 μL deionized water. PCR conditions were as follows: 95°C for 10 minutes, followed by 40 cycles of 95°C for 30 seconds and 60ºC or 62°C for 30 seconds.
Imputation of HLA Alleles, AAs, and SNPs in MHC Region and Association Analysis
The genotyped 7,889 SNPs in the MHC region (chromosome 6 region at 29‐34 Mb, University of California Santa Cruz hg19 assembly) included on the HumanOmniZhongHua‐8 BeadChip were extracted for analysis. Imputation was performed using a Han MHC reference panel built by deep sequencing that contains 29,948 variants for 10,689 healthy Han Chinese subjects.24 SNP2HLA and Beagle software was used to impute SNPs, two‐digit and four‐digit HLA alleles, and HLA AAs.25, 26 We applied postimputation quality control criteria of MAF > 0.01, imputation quality (r 2 > 0.3). Finally, we excluded subjects and SNPs with < 90% genotyping completion and/or Hardy‐Weinberg equilibrium P < 1 × 10−4 or MAF < 0.01. A total of 23,137 variants were imputed. Among them, there were 22,317 SNP or insertion/deletions (including 7,889 input SNPs from GWA), 657 AA variants from HLA class I and II antigens, and 163 classical HLA alleles (66 two‐digit alleles and 97 four‐digit alleles). Linkage disequilibrium (LD) analysis for selected SNPs was performed using the Haploview program.27
Statistical Analysis
Variant associations were tested using logistic regression models with autoantibody status (positive/negative) as the outcome variable. The statistical results were presented without genomic control correction as there is negligible evidence of population stratification in our previous study.6 Stepwise conditional analysis were used to identify independent variants of the HLA allele or AA polymorphisms for two subphenotypes of PBC by selecting the most significant associated variant as a covariate until no variant satisfied the threshold (P < 2.11 × 10−6). Meta‐analysis based on a fixed‐effect model for the two independent PBC cohorts was performed using Stata12.0 software (Stata Corporation, College Station, TX).
Results
Determination of ANTI‐sp100 and ANTI‐gp210 Status in PBC GWA and Replication Cohorts
Anti‐gp210 was detected in 350/912 (38.38%) of the cases, while anti‐sp100 was found in 211/922 (22.89%) subjects. Of the 904 subjects successfully tested for both gp210 and sp100 antibodies, 87 (9.62%) were positive for both autoantibodies and 432 (47.79%) were negative for both antibodies. In the replication cohort, 1,252 subjects were analyzed. Anti‐gp210 was detected in 444 (35.46%) of the cases, while anti‐sp100 was found in 234 (18.69%) subjects. Among them, 122 (9.74%) were positive for both autoantibodies, and 696 (55.59%) were negative for both antibodies (Table 1).
Table 1.
Sample Information for the Discovery and Replication Study for Anti‐sp100 and Anti‐gp210 Among PBC Patients
| Number | Anti‐gp210 | Anti‐sp100 | Anti‐gp210/Anti‐sp100 | Anti‐gp210/Anti‐sp100 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Missing | Positive | Negative | Missing | Both Positive | Both Negative | ||
| Total cases | 2,182 | 794 | 1,370 | 18 | 445 | 1,729 | 8 | 209 | 1,128 |
| GWAS cases | 930 | 350 | 562 | 18 | 211 | 711 | 8 | 87 | 432 |
| Male | 3 | 1 | 2 | 0 | 3 | 0 | 2 | ||
| Female | 927 | 349 | 562 | 211 | 708 | 87 | 430 | ||
| Age (mean ± SD) | 54.20 ± 6.36 | 54.04 ± 11.31 | 54.47 ± 6.36 | 54.91 ± 2.83 | 53.97 ± 0.71 | 54.66 ± 7.79 | 56.51 ± 2.12 | ||
| Replication cases | 1,252 | 444 | 808 | 0 | 234 | 1,018 | 0 | 122 | 696 |
| Male | 185 | 82 | 103 | 43 | 142 | 29 | 89 | ||
| Female | 1,067 | 362 | 705 | 191 | 876 | 93 | 607 | ||
| Age (mean ± SD) | 55.93 ± 2.83 | 56.75 ± 6.36 | 55.48 ± 7.79 | 57.16 ± 13.44 | 55.65 ± 2.83 | 57.85 ± 16.26 | 55.31 ± 7.78 | ||
GWA Analysis and Replication Study for ANTI‐gp210 Subphenotype
In total 794,694 autosomal SNPs were analyzed between 350 anti‐gp210‐positive and 562 negative subjects. No SNP achieved genome‐wide significance (P < 5 × 10−8) (Fig. 1 and Table 2), and the strongest association was the HLA class II SNP rs1794280 (P = 1.86 × 10−6; odds ratio [OR], 0.40; 95% confidence interval [CI], 0.27‐0.59). There were 15 SNPs in the MHC region with P < 1.0 × 10−5. We performed LD analysis using these SNPs and identified two separate LD blocks (Supporting Fig. S1). To further test for possible HLA associations, we selected one SNP from each of two separate LD blocks (rs1794280 and rs9380343) for replication in the second PBC cohort with 444 anti‐gp210‐positive and 808 anti‐gp210‐negative subjects. We found no significant association with rs1794289 (P = 0.11; OR, 0.75; 95% CI, 0.53‐1.07) and rs9280343 (P = 0.08; OR, 1.16; 95% CI, 0.98‐1.37) in the replication cohort. Meta‐analysis, including 794 anti‐gp210‐positive and 1,370 negative subjects showed decreased association compared to the GWA result (Table 3). Our results did not confirm the association of rs1794280 and rs9380343 with anti‐gp210 phenotype.
Figure 1.
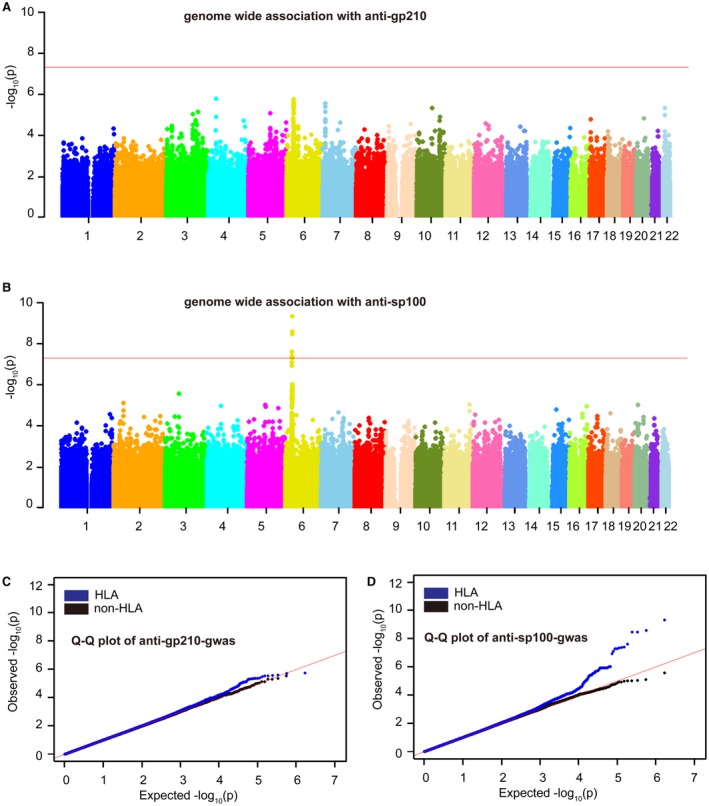
Manhattan and Q‐Q plots of anti‐gp210 and anti‐sp100 GWA results. (A) Manhattan plot of anti‐gp210‐GWA results; (B) Manhattan plot of anti‐sp100 GWA results; (C) Q‐Q plot of anti‐gp210‐GWA results; (D) Q‐Q plot of anti‐sp100 GWA results. (A,B) The horizontal axis shows genomic position, and the vertical axis shows the −log P values for association. The horizontal red line (P = 5 × 10−8) was set as the GWA significance threshold. (C,D) The red line is for null expectation, and the plot in blue is for P values including HLA SNPs, whereas the plot in black is for the P value after excluding SNPs in the MHC region.
Table 2.
Genetic Association Analysis of Anti‐gp210 or Anti‐sp100 With GWAS Data in Han Chinese PBC
| Association | SNP | Position (hg19) | A1/A2 | Antibody‐Positive | Antibody‐Negative | P | OR (95% CI) | Location |
|---|---|---|---|---|---|---|---|---|
| Anti‐gp210* | rs1794280 | 32667171 | T/A | 0.041 | 0.098 | 1.86 × 10−6 | 0.40 (0.27‐0.59) | Intergenic between HLA‐DQB1 and HLA‐DQA2 |
| kgp1505269 | 33053609 | A/G | 0.492 | 0.387 | 2.50 × 10−6 | 1.53 (1.28–1.83) | Missense variant of HLA‐DPB1 | |
| rs9380343 | 33079166 | A/G | 0.498 | 0.394 | 2.64 × 10−6 | 1.53 (1.28–1.82) | Upstream of HLA‐DPB2 | |
| rs9348906 | 33080360 | C/A | 0.496 | 0.393 | 2.92 × 10−6 | 1.53 (1.28–1.82) | Missense variant of HLA‐DPB2 | |
| rs10484569 | 33058952 | A/G | 0.485 | 0.383 | 3.29 × 10−6 | 1.52 (1.27–1.81) | Downstream of HLA‐DPB1 | |
| rs2068204 | 33058718 | A/G | 0.485 | 0.384 | 3.91 × 10−6 | 1.52 (1.27–1.81) | Downstream of HLA‐DPB1 | |
| rs4282438 | 33072172 | C/A | 0.484 | 0.383 | 4.27 × 10−6 | 1.51 (1.27–1.81) | Upstream of HLA‐DPB2 | |
| rs3097649 | 33056962 | A/G | 0.484 | 0.383 | 4.38 × 10−6 | 1.51 (1.27–1.81) | 3UTR of HLA‐DPB1 | |
| rs9296081 | 33076661 | C/G | 0.485 | 0.385 | 4.64 × 10−6 | 1.51 (1.27–1.80) | Upstream of HLA‐DPB2 | |
| rs6457713 | 33077776 | A/G | 0.485 | 0.385 | 4.64 × 10−6 | 1.51 (1.27–1.80) | Upstream of HLA‐DPB2 | |
| rs9296080 | 33076539 | A/G | 0.485 | 0.385 | 5.51 × 10−6 | 1.51 (1.27–1.80) | Upstream of HLA‐DPB2 | |
| rs3134977 | 32651641 | A/C | 0.041 | 0.093 | 7.97 × 10−6 | 0.42 (0.28‐0.62) | Intergenic between HLA‐DQB1 and HLA‐DQB2 | |
| rs3129716 | 32657436 | G/A | 0.041 | 0.093 | 7.97 × 10−6 | 0.42 (0.28‐0.62) | Intergenic between HLA‐DQB1 and HLA‐DQB2 | |
| rs7770370 | 33048921 | A/G | 0.368 | 0.466 | 8.60 × 10−6 | 0.67 (0.56‐0.80) | Intron variant of HLA‐DPB1 | |
| rs9357156 | 33041073 | A/C | 0.275 | 0.368 | 9.06 × 10−6 | 0.65 (0.54‐0.79) | Intron variant of HLA‐DPA1 | |
| Anti‐sp100† | rs1794280 | 32667171 | T/A | 0.140 | 0.055 | 4.83 × 10−10 | 2.79 (2.00‐3.90) | Intergenic between HLA‐DQB1 and HLA‐DQA2 |
| rs3134977 | 32651641 | A/C | 0.132 | 0.053 | 3.52 × 10−9 | 2.69 (1.92‐3.78) | Intergenic between HLA‐DQB1 and HLA‐DQB2 | |
| rs3129716 | 32657436 | G/A | 0.132 | 0.053 | 3.52 × 10−9 | 2.69 (1.92‐3.78) | Intergenic between HLA‐DQB1 and HLA‐DQB2 | |
| rs492899 | 31933518 | G/A | 0.169 | 0.082 | 2.49 × 10−8 | 2.28 (1.70‐3.06) | Intron variant of SKIV2L | |
| rs9268577 | 32395942 | A/G | 0.126 | 0.053 | 4.01 × 10−8 | 2.55 (1.81‐3.59) | Between BTNL2 and HLA‐DRA | |
| rs9986640 | 32764970 | A/G | 0.101 | 0.038 | 4.50 × 10−8 | 2.84 (1.93‐4.18) | Intergenic between HLA‐DQB2 and HLA‐DOB | |
| rs9501212 | 32760006 | G/A | 0.101 | 0.038 | 5.18 × 10−8 | 2.83 (1.92‐4.16) | Intergenic between HLA‐DQB2 and HLA‐DOB | |
| rs6925976 | 32755092 | C/A | 0.101 | 0.038 | 5.18 × 10−8 | 2.83 (1.92‐4.16) | Intergenic between HLA‐DQB2 and HLA‐DOB | |
| rs474534 | 31938107 | G/A | 0.185 | 0.096 | 8.19 × 10−8 | 2.13 (1.61–2.82) | Intron variant of DOMSZ |
P values are calculated based on a data set of 350 anti‐gp210‐positive and 562 anti‐gp210‐negative PBC cases. All SNPs in the chromosome 6 MHC region with P values < 1 × 10−5 are shown.
P values are calculated based on a data set of 211 anti‐sp100‐positive and 711 anti‐sp100‐negative PBC cases. All SNPs in the chromosome 6 MHC region with P values < 1 × 10−7 are shown.
Abbreviations: A1, minor allele; A2, major allele; UTR, untranslated region.
Table 3.
Summary of ANA (Anti‐sp100 and Anti‐gp210) Association for Genotyped Variants in PBC Cohorts at Two‐Stage Study
| Anti‐sp100 | Anti‐gp210 | |||||||
|---|---|---|---|---|---|---|---|---|
| rs1794280 | rs492899 | rs1794280 | rs9380343 | |||||
| Position (hg19) | 32667171 | 31933518 | 32667171 | 33079166 | ||||
| A1/A2 | T/A | G/A | T/A | T/C | ||||
| Subgroups | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative |
| GWAS | ||||||||
| Number | 211 | 711 | 211 | 711 | 350 | 562 | 350 | 562 |
| Genotypes (AA/AB/BB/missing) | 3/53/152/3 | 4/65/638/4 | 5/62/144/0 | 5/98/608/0 | 0/26/322/3 | 7/94/456/4 | 92/159/98/1 | 102/233/226/1 |
| MAF | 0.140 | 0.055 | 0.169 | 0.081 | 0.041 | 0.098 | 0.498 | 0.394 |
| OR (95% CI) | 2.79 (2.00‐3.90) | 2.28 (1.70‐3.06) | 0.40 (0.27‐0.59) | 1.53 (1.28–1.82) | ||||
| P | 4.83 × 10−10 | 2.49 × 10−8 | 1.86 × 10−6 | 2.64 × 10−6 | ||||
| Replication | ||||||||
| Number | 218 | 951 | 230 | 996 | 414 | 755 | 436 | 782 |
| Genotypes (AA/AB/BB) | 5/69/144 | 5/73/873 | 7/75/148 | 13/130/853 | 5/38/371 | 5/104/646 | 91/216/129 | 154/354/274 |
| MAF | 0.181 | 0.044 | 0.193 | 0.078 | 0.058 | 0.075 | 0.456 | 0.423 |
| OR (95% CI) | 4.85 (3.49‐6.73) | 2.82 (2.13‐3.75) | 0.75 (0.53‐1.07) | 1.16 (0.98‐1.37) | ||||
| P | 4.25 × 10−21 | 1.14 × 10−13 | 0.11 | 0.08 | ||||
| Meta‐analysis | ||||||||
| OR (95% CI) | 3.89 (3.05‐4.96) | 2.90 (2.34‐3.66) | 0.55 (0.42‐0.72) | 1.29 (1.15‐1.47) | ||||
| P | 5.78 × 10−28 | 3.27 × 10−22 | 1.71 × 10−5 | 3.96 × 10−5 | ||||
Abbreviations: A1, minor allele; A2, major allele.
GWA Analysis and Replication Study for ANTI‐sp100 Subphenotype
In total, 794,531 autosomal SNPs were analyzed between 211 anti‐sp100‐positive and 711 negative subjects. We observed a strong association in the HLA class II gene region, with six SNPs reaching genome‐wide significance (Fig. 1). The strongest association was also with rs1794280 (P = 4.83 × 10−10; OR, 2.79; 95% CI, 2.00‐3.90) but with the minor allele as the predisposing variant (Table 2). For the top nine SNPs with P < 1 × 10−7, we performed LD analysis and identified two independent LD groups (Supporting Fig. S2). The top SNPs from each group (rs1794280 and rs492899) were selected for replication using the additional 234 anti‐sp100‐positive and 1,018 negative subjects. In the replication study, both rs1794280 and rs492899 also showed significant associations with P = 4.25 × 10−21 (OR, 4.85; 95% CI, 3.49‐6.73) and 1.14 × 10−13 (OR, 2.82; 95% CI, 2.13‐3.75), respectively. Meta‐analysis using both data sets (445 anti‐sp100‐positive and 1,729 negative subjects in total) for rs1794280 and rs492899 showed P = 5.78 × 10−28 (OR, 3.89; 95% CI, 3.05‐4.96) and 3.27 × 10−22 (OR, 2.90; 95% CI, 2.34‐3.66), respectively (Table 3). These results confirmed a strong association of HLA class II gene polymorphisms with the generation of sp100 autoantibody in PBC patients.
Imputation Analysis of HLA SNP, HLA Antigen Variants, and Classical HLA Alleles With sp100 Autoantibody
To further define the genetic basis of sp100 autoantibody production in the MHC region, we conducted imputation analysis using a Han‐MHC reference panel built by sequencing 10,689 healthy control Han Chinese subjects.24 Analysis using 211 anti‐sp100‐positive versus 711 negative subjects identified 46 SNPs, five AA variants, and four HLA alleles (one of two digits and three of four digits), all in the HLA class II gene region, reaching genome‐wide significance (Table 4; Supporting Table S3). Among the most significant SNPs, three (rs77689370, rs9269941, and rs67476479) were coding variants for the DRB1 gene.
Table 4.
Summary of the Classic HLA Alleles and AA Polymorphisms Associated With Anti‐sp100
| Variants | Position (hg19) | A1/A2 | Reference of A1 | P | OR (95% CI) | Region | |
|---|---|---|---|---|---|---|---|
| Anti‐sp100+ | Anti‐sp100− | ||||||
| SNP | |||||||
| rs1794280 | 32667171 | T/A | 0.140 | 0.051 | 5.98 × 10−10 | 3.00 (2.09‐4.31) | Between DQB1 and DQA2 |
| 6‐32723737 | 32723737 | −CC/A | 0.111 | 0.036 | 1.26 × 10−9 | 3.37 (2.23‐5.09) | DQB2 |
| rs113547322 | 32206619 | T/G | 0.313 | 0.177 | 1.36 × 10−9 | 2.12 (1.66–2.72) | Close to NOTCH4 |
| rs9269173 | 32447188 | A/T | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | Between DRA and DRB5 |
| rs77689370 | 32549563 | A/G | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| rs9269941 | 32551939 | T/G | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| rs67476479 | 32551948 | P/A | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| HLA alleles | |||||||
| HLA_DRB1_03 | 32552064 | P/A | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| HLA_DRB1_0301 | 32552064 | P/A | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| HLA_DQA1_0102 | 32608306 | P/A | 0.313 | 0.182 | 8.10 × 10−9 | 2.04 (1.60–2.61) | DQA1 noncoding |
| HLA_DQB1_0201 | 32631061 | P/A | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DQB1 noncoding |
| HLA_DPB1_03 | 33049368 | P/A | 0.100 | 0.039 | 1.32 × 10−6 | 2.70 (1.78‐4.09) | DPB1 noncoding |
| HLA_DPB1_0301 | 33049368 | P/A | 0.100 | 0.039 | 1.32 × 10−6 | 2.70 (1.78‐4.09) | DPB1 noncoding |
| AA polymorphisms | |||||||
| AA_DRβ1_77_32551939 | 32551939 | N/T | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| AA_DRβ1_74_32551948_R | 32551948 | P/A | 0.135 | 0.050 | 1.51 × 10−9 | 2.97 (2.06‐4.29) | DRB1 coding |
| AA_DQα1_207_32610462_M | 32610462 | P/A | 0.313 | 0.182 | 8.10 × 10−9 | 2.04 (1.60–2.61) | DQA1 coding |
| AA_DQα1_207_32610462_V | 32610462 | A/P | 0.313 | 0.182 | 8.10 × 10−9 | 2.04 (1.60–2.61) | DQA1 coding |
| AA_DRβ1_71_32551957_K | 32551957 | P/A | 0.135 | 0.054 | 1.87 × 10−8 | 2.73 (1.90‐3.92) | DRB1 coding |
| AA_DRβ1_70_32551960_Q | 32551960 | P/A | 0.410 | 0.276 | 1.75 × 10−7 | 1.82 (1.45–2.28) | DRB1 coding |
Abbreviations: A1, effective allele; A2, alternative allele; A, absent; P, present.
Strong associations with specific AAs corresponding to the key residues that distinguish the specific classical alleles were observed, including DRβ1‐AAs 77, 74, 71, and 70, located within the third hypervariable region (AAs 67‐74) on the alpha helix of the DRβ1 chain, where they directly interact with antigens presented to helper T cells (Fig. 2). The most associated AA was in DRβ1 at position 74 with the AA arginine and at position 77 with asparagine (N), having predisposing effects (P = 1.51 × 10−9; OR, 2.97; 95% CI, 2.06‐4.29). The strong association in DRβ1 was also found at position 71 with a lysine residue (P = 1.87 × 10−8; OR, 2.73; 95% CI, 1.90‐3.92) and at position 70, with a glutamine residue (P = 1.75 × 10−7; OR, 1.82; 95% CI, 1.45‐2.88). The strong predisposing effect was also observed in DQα1 at position 207 with the presence of the AA methionine (M) or absence of valine (V).
Figure 2.
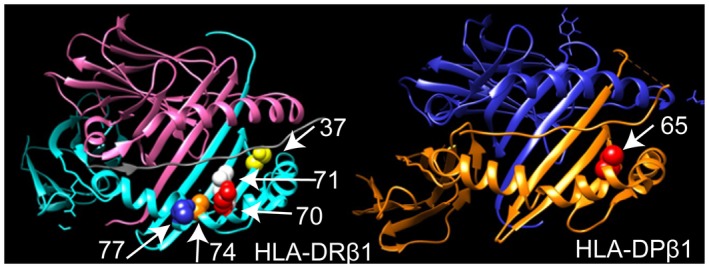
Three‐dimensional ribbon models for the HLA‐DR and HLA‐DP proteins associated with the production of anti‐sp100 autoantibodies. HLA‐DR and HLA‐DP protein structures were based on Protein Data Bank entries 3pdo and 3lqz with a direct view of the peptide‐binding groove, respectively, prepared using the program UCSF Chimera. AA positions identified by the association analysis are shown as spheres.
Among 163 classical HLA alleles imputed, the strongest association was with DRB1*0301 (P = 1.51 × 10−9; OR, 2.97; 95% CI, 2.06‐4.29). DQB1*0201 is in complete LD with DRB1*0301 and DRβ1‐Arg74 and showed the identical signal. A strong association was also observed for DQA1*0102 (P = 8.10 × 10−9; OR, 2.04; 95% CI, 1.60‐2.61), which is in complete LD with DQα1‐Met207. A smaller signal (not in LD with DRB1*0301) was observed with DPB1*0301 (P = 1.32 × 10−6). A weak association was observed in the HLA‐B and HLA‐C alleles (P > 1 × 10−4). There was no association (P < 1 × 10−2) observed for the HLA‐A and HLA‐DPA1 alleles.
DRβ1‐Asn77/Arg74, DRβ1‐Ser37, and DPβ1‐Lys65 Were Major Determinants for sp100 Autoantibody
To further define the association of specific AAs within different HLA antigens and identify putative functional/causal variants, we first tested each AA within HLA‐DRβ1 using logistic regression analysis. Due to complete LD between DRβ1 Arg74 and Asn77 (r 2 = 1), we selected Asn77 for conditional analysis because the only polymorphic AA residues were Asn and Thr in the Han Chinese cohort. Conditioning on DRβ1‐AA77 eliminated the effects of DRβ1‐AAs 74, 71, and 70. The remaining association within DRβ1 was observed at AA position 37 with a serine residue (P = 6.87 × 10−4), located in the P9 peptide‐binding pockets. Further conditional analysis with both DRβ1‐Asn77 and Ser37 eliminated the effect within DRβ1 (Fig. 3A‐C; Supporting Figs. S3 and S4). Thus, DRβ1‐Asn77 (and/or Arg74) and Ser37 account for all of the observed associations within HLA‐DRβ1.
Figure 3.
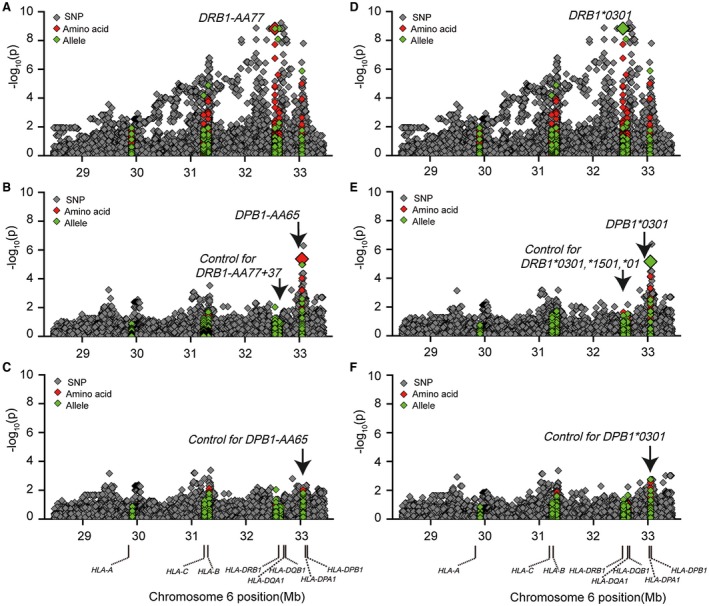
Conditional analysis of genotyped and imputed HLA SNPs, AAs, and classical alleles with anti‐sp100 subphenotype in the PBC GWA cohort. (A) Association results before conditional analysis, marked with AA77 in HLA‐DRβ1. (B) Conditional analyses controlling for HLA‐DRβ1 AA77 and AA37. (C) Conditional analysis by further controlling for HLA‐DPβ1 AA65. (D) Association results before conditional analysis, marked with DRB1*0301 in the DRB1 locus. (E) Conditional analysis by controlling for DRB1*0301, DRB1*1501, and DRB1*01. (F) Conditional analysis by further controlling for DPB1*0301.
After conditioning on DRβ1‐Asn77 and Ser37 in the MHC region, we only observed an association (P < 1 × 10−4) within the HLA‐DPB1 region, with the strongest association coming from rs3128928 (P = 5.04 × 10−7) and DPβ1‐Lys65 (P = 4.21 × 10−6). Further conditioning on DPβ1‐Lys65 or rs3128928 eliminated all of the effects within the HLA‐DPβ1 region (Fig. 3A‐C; Supporting Fig. S3). These results indicate that DRβ1‐Asn77/Arg74, DRβ1‐Ser37, and DPβ1‐Lys65 were likely the major determinants for sp100 production. We note that there was a small residual signal at rs2442753 (P = 4.1 × 10−4) (Supporting Table S3) approximately 20 kb proximal to MHC class I–related chain A and within the intron of a putative noncoding RNA (LOC105379664).
Classical HLA‐DRB1*03:01, HLA‐DRB1*15:01, and HLA‐DRB1*01 Alleles and DPB1*03:01 Allele Can Explain the Differential Production of ANTI‐sp100 Antibody in PBC Cohorts
We also tried to further characterize the association of sp100 autoantibody with classical HLA alleles. Because the most significant associations were with HLA‐DRβ1 AAs (77, 74, 71, and 70) and alleles (DRB1*0301), we first performed stepwise regression conditioning on different DRB1 alleles as covariates within the DRB1 region. When conditioning on DRB1*03:01 (P = 1.51 × 10−9), the next most significant allele is DRB1*15:01 (P = 3.41 × 10−3; OR = 1.72; 95% CI, 1.20‐2.47). When conditioning on DRB1*03:01 and DRB1*15:01, the remaining most associated allele is DRB1*01 (P = 3.38 × 10−3, OR = 3.52, 95% CI, 1.52‐8.16). Afterward, there is no significant association observed within the DRB1 region (Fig. 3D‐F; Supporting Table S3). These results indicated that there are three independent alleles for the sp100 autoantibody within the DRB1 region. After conditioning on these three alleles (DRB1*03:01, DRB1*15:01, and DRB1*01), the remaining signals (P < 1 × 10−4) within the MHC region only resided in the HLA‐DPB1 region, peaked with SNP rs3128928 (P = 4.30 × 10−7; OR, 2.60; 95% CI, 1.80‐3.77) or the classical allele DPB1*03:01 (P = 7.01 × 10−6; OR, 2.79; 95% CI, 1.78‐4.36). Conditioning on a combination of DRB1 (*03:01, *15:01 and *01) and DPB1*03:01 alleles, we observed no significant residue signal remaining in the HLA‐DPB1 region (Fig. 3F; Supporting Table S3). Similar to the AA results presented above, there was a small residual signal (P = 4.1 × 10−4) with rs2442753. These results clearly indicate that production of sp100 autoantibody can be mostly explained by a combination of risk alleles of DRB1 (*03:01, *15:01, and *01) and DPB1*03:01, with DRB1*03:01 possessing the major effect.
Discussion
A large‐scale GWA analysis conducted by our group indicated a strong genetic predisposition to PBC in Han Chinese.6 Here, we report a genetic study of PBC subphenotypes based on PBC‐specific ANAs against gp210 and sp100. A common feature in autoimmune diseases is the generation of high‐titer ANAs. Among the many ANAs identified so far, some are not disease‐specific, existing in many different autoimmune diseases and even some cancers, lung diseases, and infectious diseases; but there are also disease‐specific ANAs. ANAs to gp210 and sp100 are two autoantibodies highly specific to PBC.10 Several GWA studies in different autoimmune diseases have identified significant associations of different ANAs with specific polymorphisms and classical HLA alleles.28, 29, 30 Previously, two studies investigated the association of specific HLA class I and II alleles with ANAs in PBC, but these studies have been confined to small sample sets and limited HLA alleles.31, 32 The results presented in this report also have merits over previous reported imputations in other HLA fine‐mapping studies. The first lies in the population‐specific gene chips used in GWA with 7,889 markers in the MHC region included on the Illumina HumanOmniZhongHua‐8 BeadChip. The second is the Han Chinese–specific MHC panel generated by deep sequencing of 10,689 Han Chinese individuals.24 Therefore, we were able to perform the association analysis during the GWA stage using a large number of SNPs (including population‐specific SNPs) in the MHC region and apply these SNPs in accurate imputation of the remaining SNPs, AA variation in HLA antigens, and classical HLA alleles.
In this study, we observed striking differences in the association of genetic predisposition of two PBC‐specific ANAs to gp210 and sp100. A highly significant association was only observed with sp100 autoantibody but not with gp210. Anti‐gp210 antibody in PBC patients recognizes a single epitope within a 15‐AA stretch in the carboxyl terminus of gp210, a member of the nuclear pore complex that regulates the flow of macromolecules between the nucleus and the cytoplasm.33, 34, 35 The mechanism of generation of anti‐gp210 antibody is still a matter of debate and could arise by molecular mimicry of the Escherichia coli mutY gene product containing a similar antigenic determinant or by abnormal expression of gp210 in patients with PBC.33 A lack of association in HLA polymorphism with gp210 autoantibody may not support the notion of a molecular mimic of external bacterial protein epitopes in the generation of the gp210 autoantibody because this requires extended processing of bacterial protein fragments and presentation by antigen‐presenting cells and T cells. The autoimmune target of anti‐sp100 is the sp100 nuclear antigen, a major component of PML‐sp100 nuclear bodies. Different from anti‐gp210, sp100 autoantibody in PBC patients recognized multiple epitopes, including linear and conformational epitopes.36 Two major linear epitope sequences recognized by anti‐sp100 share sequence similarities with several viral proteins found in the Epstein‐Barr virus protein sequences.37 In our analysis, we found HLA‐DRβ1‐Asn77/Arg74, DRβ1‐Ser37, and HLA‐DPβ1‐Lys65 associated with the sp100 autoantibody. All of these AAs locate within the peptide‐binding grooves, which imply a functional impact of sp100 epitope presentation in autoantibody production. Therefore, our results may suggest significantly different mechanisms in the pathogenesis of two subphenotypes associated with ANAs to sp100 and gp210. The HLA association for anti‐sp100 autoantibody indicates that anti‐sp100 production is an adoptive immune response and T cell–dependent. The lack of specific HLA association for anti‐gp210 may suggest a T cell–independent response.
Our data show that although the sp100 autoantibody is highly specific to PBC, genotypes in HLA genes that are most strongly associated with sp100 autoantibody are not the same as those associated with PBC disease per se (Supporting Table S2). Indeed, this is also true for HLA antigens and AAs; HLA‐DRB1*08:03, HLA‐DRB1*07:01, and DPB1*17:01 are the major predisposing alleles for PBC (manuscript in preparation). One speculative explanation for these surprising discrepancies is that HLA‐DRB1*08:03, HLA‐DRB1*07:01, and DPB1*17:01 may present specific mitochondrial antigens that are critical for disease susceptibility, whereas these HLA antigens do not preferentially present either sp100 or gp210. AMA‐M2 autoantibodies (pathognomonic for PBC) recognize multiple different proteins, mainly the E2 subunits of the 2‐oxo‐acid dehydrogenase complex (ODGC) including pyruvate dehydrogenase complex, 2‐oxoglutarate dehydrogenase complex, and branched‐chain 2‐oxoacid dehydrogenase complex, with, to a less extent, the E1 and E3 subunits of ODGC. Therefore, HLA‐DRB1*08:03, HLA‐DRB1*07:01, and DPB1*17:01 may highly predispose for PBC but have very little effect on positivity for gp210 and sp100.
A potential limitation in our study lies in the measurement of both antibodies only at a single time point at or after diagnosis for more than half of our PBC cases. There were reports that change of anti‐sp100 titers has been found during the course of the treatment, but conversion of anti‐sp100 positivity was rarely found.19 But conversion of anti‐gp210 positivity was reported in some of the patients treated with UDCA in Nakamura et al.'s study of Japanese PBC cohort, although this report was not confirmed by other studies.38 This conversion during the treatment may increase the false‐negative rate in the study cohort, which may reduce the power of our study to detect a genetic association for anti‐gp210. However, this may not be a critical factor in the lack of association observed in our gp210 groups because both of our GWA and replication cohorts exhibit significantly higher anti‐gp210‐positive rates compared with the study by Nakamura et al. (38.4% and 35.5% versus 26.1%). At the same time, the sample size (930 GWA and 1,252 replication samples) in the current study should provide sufficient power to detect genetic associations. We also note that the current study is confined to PBC patients in the Han Chinese population. Additional studies will be necessary to ascertain whether the association of sp100 to particular HLA antigens and amino acids found in our study is also present in other populations.
As reviewed at the beginning of this article, the clinical significance of anti‐sp100 and anti‐gp210 is uncertain.8, 11, 12, 17, 18, 19, 20 Further studies will be needed to determine if PBC disease progression, endophenotypes, and, importantly, responses to different therapeutic regimens are associated with the presence or titers of these autoantibodies. We also note that in future PBC studies it may be useful not only to stratify patient groups based on variants that predispose to PBC itself but also to examine whether stratification based on the HLA associations of sp100 are associated with particular endophenotypes or therapeutic responses.
In summary, the current study addressed genetic determinants associated with sp100 and gp210 autoantibody subphenotypes in PBC and identified specific SNPs, HLA alleles, and AAs associated with the sp100 autoantibody but not with the gp210 autoantibody. The striking difference in genetic predisposition observed between the sp100 and gp210 subphenotypes may indicate distinct roles of these two autoantibodies in the pathogenesis of PBC.
Supporting information
Acknowledgment
We thank the patients for their participation in this study. Patient samples and clinical information were provided by the Jiangsu Provincial PBC Collaboration Group, Shanghai Renji Hospital, and Shuguang Hospital.
Supported in part by grants from the National Natural Science Foundation of China (81870397 to X.D.L; 81620108002, 81771732, 81830016, to X.M.; 81570469 to R.Q.T.) and from the Jiangsu Provincial Research Fund (BE2017713, to X.D.L.; BL2018657, to Y.T.).
Potential conflict of interest: Nothing to report.
Contributor Information
Xiangdong Liu, Email: xiangdongliu@seu.edu.cn.
Liangdan Sun, Email: ahmusld@163.com.
Xiong Ma, Email: maxiongmd@163.com.
References
Author names in bold designate shared co‐first authorship.
- 1. Lleo A, Marzorati S, Anaya JM, Gershwin ME. Primary biliary cholangitis: a comprehensive overview. Hepatol Int 2017;11:485‐499. [DOI] [PubMed] [Google Scholar]
- 2. Carey EJ, Ali AH, Lindor KD. Primary biliary cirrhosis. Lancet 2015;386:1565‐1575. [DOI] [PubMed] [Google Scholar]
- 3. Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med 2009;360:2544‐2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C, et al. Variants at IRF5‐TNPO3, 17q12‐21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet 2010;42:655‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, et al. Genome‐wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet 2012;91:721‐728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qiu F, Tang R, Zuo X, Shi X, Wei Y, Zheng X, et al. A genome‐wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun 2017;8:14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawashima M, Hitomi Y, Aiba Y, Nishida N, Kojima K, Kawai Y, et al. Genome‐wide association studies identify PRKCB as a novel genetic susceptibility locus for primary biliary cholangitis in the Japanese population. Hum Mol Genet 2017;26:650‐659. [DOI] [PubMed] [Google Scholar]
- 8. Muratori P, Muratori L, Ferrari R, Cassani F, Bianchi G, Lenzi M, et al. Characterization and clinical impact of antinuclear antibodies in primary biliary cirrhosis. Am J Gastroenterol 2003;98:431‐437. [DOI] [PubMed] [Google Scholar]
- 9. Bogdanos DP, Komorowski L. Disease‐specific autoantibodies in primary biliary cirrhosis. Clin Chim Acta 2011;412:502‐512. [DOI] [PubMed] [Google Scholar]
- 10. Worman HJ, Courvalin JC. Antinuclear antibodies specific for primary biliary cirrhosis. Autoimmun Rev 2003;2:211‐217. [DOI] [PubMed] [Google Scholar]
- 11. Nakamura M, Shimizu‐Yoshida Y, Takii Y, Komori A, Yokoyama T, Ueki T, et al. Antibody titer to gp210‐C terminal peptide as a clinical parameter for monitoring primary biliary cirrhosis. J Hepatol 2005;42:386‐392. [DOI] [PubMed] [Google Scholar]
- 12. Nakamura M, Kondo H, Mori T, Komori A, Matsuyama M, Ito M, et al. Anti‐gp210 and anti‐centromere antibodies are different risk factors for the progression of primary biliary cirrhosis. Hepatology 2007;45:118‐127. [DOI] [PubMed] [Google Scholar]
- 13. Wesierska‐Gadek J, Penner E, Battezzati PM, Selmi C, Zuin M, Hitchman E, et al. Correlation of initial autoantibody profile and clinical outcome in primary biliary cirrhosis. Hepatology 2006;43:1135‐1144. [DOI] [PubMed] [Google Scholar]
- 14. Yang F, Yang Y, Wang Q, Wang Z, Miao Q, Xiao X, et al. The risk predictive values of UK‐PBC and GLOBE scoring system in Chinese patients with primary biliary cholangitis: the additional effect of anti‐gp210. Aliment Pharmacol Ther 2017;45:733‐743. [DOI] [PubMed] [Google Scholar]
- 15. European Association for the Study of the Liver . EASL clinical practice guidelines: the diagnosis and management of patients with primary biliary cholangitis. J Hepatol 2017;67:145‐172. [DOI] [PubMed] [Google Scholar]
- 16. Szostecki C, Will H, Netter HJ, Guldner HH. Autoantibodies to the nuclear sp100 protein in primary biliary cirrhosis and associated diseases: epitope specificity and immunoglobulin class distribution. Scand J Immunol 1992;36:555‐564. [DOI] [PubMed] [Google Scholar]
- 17. Mytilinaiou MG, Meyer W, Scheper T, Rigopoulou EI, Probst C, Koutsoumpas AL, et al. Diagnostic and clinical utility of antibodies against the nuclear body promyelocytic leukaemia and sp100 antigens in patients with primary biliary cirrhosis. Clin Chim Acta 2012;413:1211‐1216. [DOI] [PubMed] [Google Scholar]
- 18. Tana MM, Shums Z, Milo J, Norman GL, Leung PS, Gershwin ME, et al. The significance of autoantibody changes over time in primary biliary cirrhosis. Am J Clin Pathol 2015;144:601‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gatselis NK, Zachou K, Norman GL, Gabeta S, Papamichalis P, Koukoulis GK, et al. Clinical significance of the fluctuation of primary biliary cirrhosis–related autoantibodies during the course of the disease. Autoimmunity 2013;46:471‐479. [DOI] [PubMed] [Google Scholar]
- 20. Hu CJ, Deng CW, Song G, Zhang W, Zhang SL, Li X, et al. Prevalence of autoimmune liver disease related autoantibodies in Chinese patients with primary biliary cirrhosis. Dig Dis Sci 2011;56:3357‐3363. [DOI] [PubMed] [Google Scholar]
- 21. Zuchner D, Sternsdorf T, Szostecki C, Heathcote EJ, Cauch‐Dudek K, Will H. Prevalence, kinetics, and therapeutic modulation of autoantibodies against sp100 and promyelocytic leukemia protein in a large cohort of patients with primary biliary cirrhosis. Hepatology 1997;26:1123‐1130. [DOI] [PubMed] [Google Scholar]
- 22. Lindor KD, Gershwin ME, Poupon R, Kaplan M, Bergasa NV, Heathcote EJ, et al. Primary biliary cirrhosis. Hepatology 2009;50:291‐308. [DOI] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet 2007;81:559‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou F, Cao H, Zuo X, Zhang T, Zhang X, Liu X, et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet 2016;48:740‐746. [DOI] [PubMed] [Google Scholar]
- 25. Jia X, Han B, Onengut‐Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Browning BL, Browning SR. A unified approach to genotype imputation and haplotype‐phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009;84:210‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263‐265. [DOI] [PubMed] [Google Scholar]
- 28. Gorlova O, Martin JE, Rueda B, Koeleman BPC, Ying J, Teruel M, et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome‐wide association strategy. PLoS Genet 2011;7:e1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morris DL, Fernando MM, Taylor KE, Chung SA, Nititham J, Alarcon‐Riquelme ME, et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun 2014;15:210‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu X, Yang M, Song ZJ, Dong Y, Li C, Shen M, et al. Fine mapping MHC associations in Graves' disease and its clinical subtypes in Han Chinese. J Med Genet 2018;55:685‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakamura M, Yasunami M, Kondo H, Horie H, Aiba Y, Komori A, et al. Analysis of HLA‐DRB1 polymorphisms in Japanese patients with primary biliary cirrhosis (PBC): the HLA‐DRB1 polymorphism determines the relative risk of antinuclear antibodies for disease progression in PBC. Hepatol Res 2010;40:494‐504. [DOI] [PubMed] [Google Scholar]
- 32. Zhao DT, Liao HY, Zhang X, Liu YM, Zhao Y, Zhang HP, et al. Human leucocyte antigen alleles and haplotypes and their associations with antinuclear antibodies features in Chinese patients with primary biliary cirrhosis. Liver Int 2014;34:220‐226. [DOI] [PubMed] [Google Scholar]
- 33. Nickowitz RE, Worman HJ. Autoantibodies from patients with primary biliary‐cirrhosis recognize a restricted region within the cytoplasmic tail of nuclear‐pore membrane glycoprotein gp210. J Exp Med 1993;178:2237‐2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wozniak RW, Bartnik E, Blobel G. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J Cell Biol 1989;108:2083‐2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear‐pore complex is a membrane‐spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J 1990;9:1495‐1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin‐like modifiers. J Biol Chem 1999;274:12555‐12566. [DOI] [PubMed] [Google Scholar]
- 37. Xie K, Snyder M. Two short autoepitopes on the nuclear dot antigen are similar to epitopes encoded by the Epstein‐Barr virus. Proc Natl Acad Sci USA 1995;92:1639‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakamura M, Kondo H, Tanaka A, Komori A, Ito M, Yamamoto K, et al. Autoantibody status and histological variables influence biochemical response to treatment and long‐term outcomes in Japanese patients with primary biliary cirrhosis. Hepatol Res 2015;45:846‐855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


