Abstract
Four‐way junctions (4WJs) are supramolecular DNA assemblies comprising four interacting DNA strands that in biology are involved in DNA‐damage repair. In this study, a new mononuclear platinum(II) complex 1 was prepared that is capable of driving the crystallization of the DNA oligomer 5′‐d(CGTACG)‐3′ specifically into a 4WJ‐like motif. In the crystal structure of the 1–CGTACG adduct, the distorted‐square‐planar platinum complex binds to the core of the 4WJ‐like motif through π–π stacking and hydrogen bonding, without forming any platinum–nitrogen coordination bonds. Our observations suggest that the specific molecular properties of the metal complex are crucially responsible for triggering the selective assembly of this peculiar DNA superstructure.
Keywords: hydrogen bonding, metallodrugs, nucleic acids, platinum, supramolecular chemistry
Interactions between platinum‐based compounds and nucleic acids have been extensively studied because the widely used anticancer drugs cisplatin, oxaliplatin, and carboplatin bind tightly to nuclear DNA, which is also reputed to be their primary target.1 Indeed, following the hydrolysis of two cis Pt−Cl bonds, cisplatin typically forms two coordination bonds with nitrogen atoms of adjacent DNA base pairs, thus producing local DNA damage, which initiates cancer‐cell death.2 Many cytotoxic aromatic compounds, whether they contain platinum or not, interact with DNA by intercalation of their flat, aromatic rings into the π–π stacked DNA base pairs of double‐stranded DNA. Overall, the development of molecules of different shapes, coordination and supramolecular properties, that recognize or induce the formation of different types of DNA motifs is an active field of research: trans platinum compounds,3 compounds that interact with the sugar phosphate backbone of DNA,4 and those that recognize nucleic acid superstructures that are different from double‐stranded DNA, such as DNA mismatches,5 G‐quadruplexes,6 DNA bulges,7 viral RNAs,8 and three‐way junctions, have been extensively studied.9 In particular, binuclear metal complexes developed by the Hannon group,10 which recognize three‐way junctions, have attracted considerable attention in the last decade. This series of supramolecular cylinders interact with the DNA palindromic hexanucleotide 5′‐d(CGTACG)‐3′ to form three‐way junctions with the metal complex in the central hydrophobic cavity of the DNA assembly.11 A later study demonstrated the ability of RNA three‐way junctions, such as 5′‐CGUACG‐3′, to also interact specifically with these metal complexes.12
Four‐way junctions (4WJs) are another form of noncanonical DNA superstructures characterized by four interconnected DNA fragments. Many supramolecular isomers of these highly flexible assemblies have been described. Some of these isomers, called Holliday junctions (HJs), are biologically important as they are involved in DNA repair.13 Processes such as double HJ dissolution are crucial for suppressing specific DNA damage that otherwise causes cancer predisposition and/or premature ageing, for example, in Bloom syndrome.14 HJs have even been proposed as a target for the design of selective inhibitors of HJ resolution, which would benefit from compounds capable of stabilizing 4WJs. The first structural characterization of a 4WJ was reported in 1999 with the sequence 5′‐d(CCGGGACCGG)‐3′.15 Since then, several protein–4WJ interactions have been characterized,16 and two crystal structures of 4WJs interacting with small organic molecules have been published.17 We now report the induction of a 4WJ‐like DNA structure by a new mononuclear platinum compound, [Pt(H2bapbpy)](PF6)2 (1, in which H2bapbpy is N‐(6‐(6‐(pyridin‐2‐ylamino)pyridin‐2‐yl)pyridin‐2‐yl)pyridin‐2‐amine).
Complex 1 was synthesized in 75 % yield by heating the tetrapyridyl ligand H2bapbpy18 with K2PtCl4 in an ethanol–water mixture. Single crystals suitable for X‐ray crystal‐structure determination were obtained from acetonitrile. In the crystal structure of [Pt(Hbapbpy)]PF6 (Figure 1; see also Table S1 in the Supporting Information), the d8 metal complex shows a strongly distorted, helical square‐planar coordination, similar to that observed in octahedral iron(II) complexes of the same ligand.18 In contrast to previously described octahedral metal complexes of this ligand, however, in 1 the H2bapbpy ligand was clearly deprotonated, thus indicating that the protons borne by the noncoordinating NH bridges of the ligand become quite acidic upon platinum(II) coordination. In solution, deprotonation of these NH bridges (pKa1=8.3; see Figure S1 in the Supporting Information) can interconvert the metal complex between its monocationic ([Pt(Hbapbpy)]+) and dicationic ([Pt(H2bapbpy)]2+) forms, and under neutral conditions the complex is mostly in the dicationic state. Owing to the tetradentate nature of the H2bapbpy ligand and the square‐planar preferred geometry of d8 metal centers, the coordination sphere of platinum in 1 is saturated and very stable. Stability studies in water, phosphate buffer, and cell growth medium, showed that over 48 h only deprotonation occurred, that is, no hydrolysis took place (see Figure S2).
Figure 1.
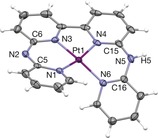
Displacement ellipsoid plot (50 % probability level) of the cationic moiety [Pt(Hbapbpy)]+ as observed in the crystal structure of [Pt(Hbapbpy)]PF6. Counteranions have been omitted for clarity.21
When 1 was added to a solution of the palindromic hexanucleotide 5′‐d(CGTACG)‐3′, single crystals of an adduct 1–CGTACG were grown that could be subjected to crystal‐structure determination. The 1–CGTACG adduct crystallized in the P3221 space group (see Table S2). The asymmetric unit shows one molecule of 1 per four DNA strands. The four crystallographically independent DNA strands form pairs of B‐type double‐helix units (Figure 2 a) that stack on each other to form columns growing in the [110], [010], and [100] crystal directions. The 5′‐terminal cytosines C1A and C1C were not localized, whereas C7B and C7D protrude out from the helix and form a connection with an adjacent symmetry‐related column in the packing structure. No coordination was found between the DNA base pairs and the platinum atom, which remained coordinated to the four nitrogen atoms of the tetrapyridyl ligand. The DNA‐binding site of 1 is located at the interface between couples of B‐type helices in each DNA column, where the C7 cytosine residues are twisted away. Although the “almost flat”, aromatic structure of 1 made us initially expect simple intercalation between the DNA base pairs, the binding site of 1 unexpectedly led to a remarkable distortion of the double‐helix structures (the rotational parameters of the base pairs are shown in Table S3).19 The resulting noncanonical binding site is formed by two tetrads (the two gray rectangles shown in Figure 3 a in each column), which sandwich the metal complex through π–π stacking. Each tetrad is built up of two C–G pairs formed with the contribution of a cytosine residue from the symmetry‐related column (C5A–G8B and G6C–C7B; G6A–C7D and C5C–G8D). The two C–G pairs are then connected by a double N2−H⋅⋅⋅N3 hydrogen bond established by the G6 and G8 guanine residues. Notably, the connections provided by C7B and C7D result overall in a 4WJ‐like structure (Figure 3 a,b).
Figure 2.
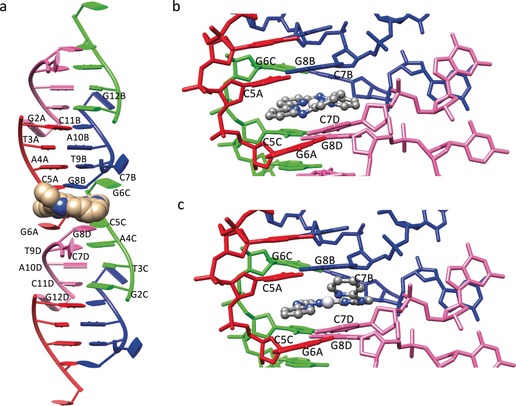
Crystal structure of the adduct 1–CGTACG. a) B‐Type double‐helix units arranged in a column in which each B‐type double‐helix unit is formed by two contiguous ds‐CGTACG moieties. Complex 1 is shown in CPK style. b, c) Binding site of the platinum complex 1 in the 1‐F and 1‐G conformation, respectively (see text). The four colours red, blue, green, and pink of the DNA strands correspond to the four crystallographically independent DNA oligomers (denoted A–D).
Figure 3.
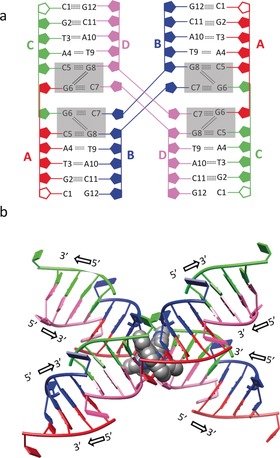
Different views of the four‐way‐junction DNA arrangement in the crystal structure of 1–CGTACG. a) Base‐pair arrangement and hydrogen‐bonding interactions. Complex 1 is not shown; the binding site in each column is shown as two gray rectangles. The C1A and C1C residues, not localized in the electron‐density map, are represented by empty (not colored) ribose rings. b) “X” conformation and 5′ and 3′ DNA growth directions; the two molecules of 1 are shown as a CPK model (only conformer F is shown for clarity).
Despite the significant differences characterizing the 6‐mer DNA sequence used in the study with respect to longer sequences found to generate real HJs, in the structure of 1–CGTACG the binding site of the platinum complex was found precisely at this 4WJ connection. The nonclassical intercalation site hosts one platinum complex, which was found to be at two equally occupied positions, with the platinum centers about 2.5 Å from each other. The best results in terms of B factors and residual densities in the Fo–Fc maps, which were affected by low resolution of diffraction and twinning of the crystals, were obtained by introducing two different conformations of the complex in the model, indicated below as 1‐F and 1‐G (Figure 2 b,c). At the end of refinement, the two molecules of 1 were found to be helical enantiomers. The two molecules were in an almost coplanar arrangement and approximately rotated by about 90° with respect to each other about the helical axis (see Figure S3). The presence of two equally probable positions for 1 in the 4WJ is most likely possible because the main interaction of the complex with the DNA bases is π–π stacking, with mean interplanar distances of about 3.4 Å for both conformations, so that the different chirality does not introduce any destabilization. Besides π–π stacking, coulombic interactions between the positively charged platinum complexes and the negative electron density of DNA must play a role in the stabilization of the adduct as well, whereas the protonation state of the amine bridges of the tetrapyridyl ligands could not be determined crystallographically.
To obtain a more detailed view of the interactions taking place between the two twisted molecules of 1 and the DNA 4WJ‐like structure, we performed quantum mechanics/molecular mechanics (QM/MM) calculations starting from the crystallographic coordinates of both conformations 1‐F and 1‐G. Overall, in silico relaxation did not lead to significant variations of the conformation of the model (see Figures S4 and S5) as compared to that of the crystal structure (Figure 4 a,b). Interestingly, the model clearly suggests that one of the two noncoordinating amines of the H2bapbpy ligand of complex 1 may be engaged in H‐bonding with DNA; however, the corresponding N⋅⋅⋅O contacts from the XRD analysis were too long (2.6–4.1 Å) to conclude that a strong H‐bond interaction was present considering the insufficient quality of the electron‐density map for that level of description.
Figure 4.
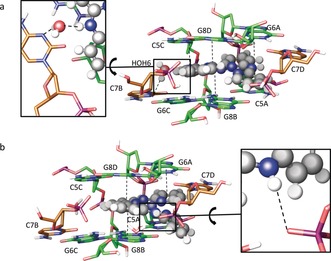
Binding site and selected interactions from QM/MM calculations for a) complex 1‐F and b) complex 1‐G. Carbon atoms of residues in the main column are shown in green, and carbon atoms of residues of symmetry‐related columns in brown.
The unique structure of the 1–CGTACG adduct prompted us to test the antiproliferative activity of 1 in cancer cell lines, and its ability to reach the cell nucleus. The cancer cell growth inhibition concentration (IC50) of 1 was investigated in two cisplatin‐sensitive cancer cell lines (A549, MCF‐7), one cisplatin‐resistant cancer cell line (MDA‐MB231), and one nonmalignant lung cell line (MRC‐5) by using a previously reported protocol.20 IC50 values as low as or much lower than that of cisplatin were found, also in noncancerous MRC‐5 cells (Table 1). The high toxicity of 1 in cisplatin‐resistant MDA‐MB231 cancer cells suggests a different mode of action (MoA) of 1 as compared to cisplatin. The uptake and localization of 1 in the cytosol, membranes, nuclei, and cytoskeleton of A549 cells were determined 24 h after treatment at 1.0 μm by cell fractionation followed by ICP‐MS analysis (see Table S4). Of the total uptake of 1586 pmol Pt per million cells, 15 % of the metal was found in the nucleus, similarly to cisplatin (14 %). When 1 was incubated for 24 h with a 2686 bp pUC19 plasmid, very weak interactions were observed at a 20:1 base pair/metal complex ratio, but at a 7.5:1 or lower ratio, migration of the supercoiled plasmid was severely retarded (see Figure S6). Such retardation implies that 1 does indeed interact with pUC19 plasmid DNA, but we cannot discriminate, at this stage, between specific interaction of 1 with the 4WJ in solution, and nonspecific interaction with dsDNA.
Table 1.
Cell growth inhibition concentrations (IC50 with 95 % confidence intervals, in μm) of 1 and cisplatin for cancer (A549, MCF‐7, MDA‐MB231), and noncancer (MRC‐5) cell lines.
| Cell line | 1 | Cisplatin | ||||
|---|---|---|---|---|---|---|
| A549 | 0.91 | −0.11 +0.12 |
3.1 | −0.98 +1.4 |
||
| MCF‐7 | 0.80 | −0.12 +0.14 |
4.0 | −1.5 +2.4 |
||
| MDA‐MB231 | 1.3 | −0.48 +0.75 |
>25 | – | ||
| MRC‐5 | 1.4 | −0.43 +0.63 |
12 | −3.9 +5.9 |
||
In conclusion, 1 is the first example of a metal complex capable of inducing a noncanonical DNA structure resembling a 4WJ in the DNA palindromic hexanucleotide 5′‐d(CGTACG). Details of the interaction have been elucidated at the atomic level. At present, the exact role of the distorted conformation of 1 and its noncoordinated NH bridges in the stabilization of the 4WJ‐like structure still needs to be assessed. Our preliminary biological results show that 1 has high biological activity and, because it goes to the nuclei, that it can interact with nuclear DNA. However, at this stage we can by no means assess whether the 4WJ‐like DNA structure observed in the solid state also forms in solution (see Figure S7), and whether it is responsible for the toxicity of 1. Meanwhile, the fact that the DNA 6‐mer used in our study is identical to that used by Hannon and co‐workers to demonstrate the specific interaction of DNA three‐way junctions with dinuclear compounds11a opens fascinating perspectives, as the same DNA sequence is triggered to adopt different three‐dimensional conformations depending on the nature of the interacting metal complex. Chemically speaking, the results reported herein suggest that it may be possible to design compounds with specific molecular shapes that develop selective supramolecular interactions with DNA, thus leading to the self‐assembly of different DNA superstructures. Overall, understanding the link between the specific shape and supramolecular properties of 1, and its unique ability to drive the crystallization of a DNA 4WJ‐like structure, may lead to unforeseen applications in supramolecular chemistry, biosensing, and/or medicinal chemistry.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This research was supported by the Dutch Organisation for Scientific Research (NWO‐CW) through a VIDI grant to S.B. The European Research Council is also kindly acknowledged for a Starting Grant to S.B. Prof. E. Bouwman is kindly acknowledged for her support and input. The COST action CM1105 “Functional metal complexes that bind to biomolecules” is gratefully acknowledged for stimulating scientific discussion and for a Short‐Term Scientific Mission for V.H.S.v.R. to work in the laboratory of L.M. L.M. and T.M. gratefully acknowledge Beneficentia Stiftung (Vaduz). T.M. thanks the University of Pisa (Rating di Ateneo 2018) and Fondazione AIRC per la Ricerca sul Cancro (Project Code: 18044) for financial support. Ente Cassa di Risparmio di Firenze, Italy, is gratefully acknowledged for a grant to F.P. (2014.0309). We thank Dr. Damiano Cirri (Dept. of Chemistry, University of Florence) for help with UV/Vis experiments.
V. H. S. van Rixel, A. Busemann, M. F. Wissingh, S. L. Hopkins, B. Siewert, C. van de Griend, M. A. Siegler, T. Marzo, F. Papi, M. Ferraroni, P. Gratteri, C. Bazzicalupi, L. Messori, S. Bonnet, Angew. Chem. Int. Ed. 2019, 58, 9378.
References
- 1.
- 1a. Kelland L., Nat. Rev. Cancer 2007, 7, 573–584; [DOI] [PubMed] [Google Scholar]
- 1b. Einhorn E. H., Clin. Cancer Res. 1997, 3, 2630–2632; [PubMed] [Google Scholar]
- 1c. Wheate N. J., Walker S., Craig G. E., Oun R., Dalton Trans. 2010, 39, 8113–8127; [DOI] [PubMed] [Google Scholar]
- 1d. Lippert B., Coord. Chem. Rev. 1999, 182, 263–295. [Google Scholar]
- 2.
- 2a. Reedijk J., Proc. Natl. Acad. Sci. USA 2003, 100, 3611–3616; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Sherman S. E., Lippard S. J., Chem. Rev. 1987, 87, 1153–1181; [Google Scholar]
- 2c. Wang F., Habtemariam A., van der Geer E. P. L., Fernández R., Melchart M., Deeth R. J., Aird R., Guichard S., Fabbiani F. P. A., Lozano-Casal P., Oswald I. D. H., Jodrell D. I., Parsons S., Sadler P. J., Proc. Natl. Acad. Sci. USA 2005, 102, 18269–18274; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2d. Jung Y., Lippard S. J., Chem. Rev. 2007, 107, 1387–1407; [DOI] [PubMed] [Google Scholar]
- 2e. Lippert B., BioMetals 1992, 5, 195–208; [DOI] [PubMed] [Google Scholar]
- 2f. Lippert B., Miguel P. J. S., Coord. Chem. Rev. 2016, 327–328, 333–348. [Google Scholar]
- 3.
- 3a. Radulovic S., Tesic Z., Manic S., Curr. Med. Chem. 2002, 9, 1611–1618; [DOI] [PubMed] [Google Scholar]
- 3b. Quiroga A. G., J. Inorg. Biochem. 2012, 114, 106–112. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Takahara P. M., Rosenzweig A. C., Frederick C. A., Lippard S. J., Nature 1995, 377, 649–652; [DOI] [PubMed] [Google Scholar]
- 4b. Komeda S., Qu Y., Mangrum J. B., Hegmans A., Williams L. D., Farrell N. P., Inorg. Chim. Acta 2016, 452, 25–33. [Google Scholar]
- 5. Song H., Kaiser J. T., Barton J. K., Nat. Chem. 2012, 4, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Han H., Hurley L. H., Trends Pharmacol. Sci. 2000, 21, 136–142; [DOI] [PubMed] [Google Scholar]
- 6b. Bazzicalupi C., Ferraroni M., Papi F., Massai L., Bertrand B., Messori L., Gratteri P., Casini A., Angew. Chem. Int. Ed. 2016, 55, 4256–4259; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 4328–4331; [Google Scholar]
- 6c. Neidle S., J. Med. Chem. 2016, 59, 5987–6011; [DOI] [PubMed] [Google Scholar]
- 6d. Cao Q., Li Y., Freisinger E., Qin P. Z., Sigel R. K. O., Mao Z.-W., Inorg. Chem. Front. 2017, 4, 10–32; [Google Scholar]
- 6e. Yu H., Wang X., Fu M., Ren J., Qu X., Nucleic Acids Res. 2008, 36, 5695–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Malina J., Hannon M. J., Brabec V., FEBS J. 2014, 281, 987–997; [DOI] [PubMed] [Google Scholar]
- 7b. Malina J., Scott P., Brabec V., Dalton Trans. 2015, 44, 14656–14665. [DOI] [PubMed] [Google Scholar]
- 8. Malina J., Hannon M. J., Brabec V., Sci. Rep. 2016, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Komor A. C., Barton J. K., Chem. Commun. 2013, 49, 3617–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Hotze A. C. G., Kariuki B. M., Hannon M. J., Angew. Chem. Int. Ed. 2006, 45, 4839–4842; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2006, 118, 4957–4960; [Google Scholar]
- 10b. Cerasino L., Hannon M. J., Sletten E., Inorg. Chem. 2007, 46, 6245–6251; [DOI] [PubMed] [Google Scholar]
- 10c. Malina J., Hannon M. J., Brabec V., Chem. Eur. J. 2007, 13, 3871–3877; [DOI] [PubMed] [Google Scholar]
- 10d. Boer D. R., Kerckhoffs J. M. C. A., Parajo Y., Pascu M., Usón I., Lincoln P., Hannon M. J., Coll M., Angew. Chem. Int. Ed. 2010, 49, 2336–2339; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 2386–2389. [Google Scholar]
- 11.
- 11a. Oleksi A., Blanco A. G., Boer R., Usón I., Aymamí J., Rodger A., Hannon M. J., Coll M., Angew. Chem. Int. Ed. 2006, 45, 1227–1231; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2006, 118, 1249–1253; [Google Scholar]
- 11b. Ducani C., Leczkowska A., Hodges N. J., Hannon M. J., Angew. Chem. Int. Ed. 2010, 49, 8942–8945; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 9126–9129. [Google Scholar]
- 12. Phongtongpasuk S., Paulus S., Schnabl J., Sigel R. K. O., Spingler B., Hannon M. J., Freisinger E., Angew. Chem. Int. Ed. 2013, 52, 11513–11516; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 11727–11730. [Google Scholar]
- 13. Holliday R., Gen. Res. 1964, 5, 282–304. [Google Scholar]
- 14. Larsen N. B., Hickson I. D., DNA Helicases and DNA Motor Proteins, Adv. Exp. Med. Biol., Vol. 973 (Ed.: M. Spies), Springer, New York, 2013, pp. 161–184. [Google Scholar]
- 15. Ortiz-Lombardía M., González A., Eritja R., Aymamí J., Azorín F., Coll M., Nat. Struct. Mol. Biol. 1999, 6, 913–917. [DOI] [PubMed] [Google Scholar]
- 16. Kitano K., Front. Genet. 2014, 5, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a. Liu Y., West S. C., Nat. Rev. Mol. Cell Biol. 2004, 5, 937–944; [DOI] [PubMed] [Google Scholar]
- 17b. Hays F. A., Watson J., Ho P. S., J. Biol. Chem. 2003, 278, 49663–49666; [DOI] [PubMed] [Google Scholar]
- 17c. Canals A., Arribas-Bosacoma R., Albericio F., Álvarez M., Aymamí J., Coll M., Sci. Rep. 2016, 7, 39680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonnet S., Siegler M. A., Costa J. S., Molnar G., Bousseksou A., Spek A. L., Gamez P., Reedijk J., Chem. Commun. 2008, 5619–5621. [DOI] [PubMed] [Google Scholar]
- 19.
- 19a. Lu X. J., Olson W. K., Nucleic Acids Res. 2003, 31, 5108–5121; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19b. Lu X. J., Olson W. K., Nat. Protoc. 2008, 3, 1213–1227; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19c. Zheng G. H., Lu X. J., Olson W. K., Nucleic Acids Res. 2009, 37, W240–W246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopkins S. L., Siewert B., Askes S. H. C., Veldhuizen P., Zwier R., Heger M., Bonnet S., Photochem. Photobiol. Sci. 2016, 15, 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CCDC 1844330 contains the supplementary crystallographic data for this paper. These data are provided free of charge by The Cambridge Crystallographic Data Centre.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


