Abstract
Viral infections can cause serious diseases for humans and animals. Accurate and early detection of viruses is often crucial for clinical diagnosis and therapy. Aptamers are mostly single-stranded nucleotide sequences that are artificially synthesized by an in vitro technology known as the Systematic Evolution of Ligands by Exponential Enrichment (SELEX). Similar to antibodies, aptamers bind specifically to their targets. However, compared with antibody, aptamers are easy to synthesize and modify and can bind to a broad range of targets. Thus, aptamers are promising for detecting viruses and treating viral infections. In this review, we briefly introduce aptamer-based biosensors (aptasensors) and describe their applications in rapid detection of viruses and as antiviral agents in treating infections. We summarize available data about the use of aptamers to detect and inhibit viruses. Furthermore, for the first time, we list aptamers specific to different viruses that have been screened out but have not yet been used for detecting viruses or treating viral infections. Finally, we analyze barriers and developing perspectives in the application of aptamer-based virus detection and therapeutics.
Keywords: aptamers, SELEX, aptasensors, virus detection, antiviral therapy
Introduction
Aptamers are small single-stranded artificial nucleotides (DNA or RNA), in the range of 10–100 nucleotides (nt), that have a remarkable ability to bind to their targets. Aptamer targets include a variety of small molecules such as amino acids, nucleotides, and antibiotics (Ellington and Szostak, 1992), but can also be larger, including proteins (Schneider et al., 1992), viruses and bacteria (Torres-Chavolla and Alocilja, 2009) as well as other cells (Ku et al., 2015). The secondary and tertiary structures of aptamers ensure the binding specificity to their targets via aptamer-target recognition, and may involve aromatic rings, π-π system stacking, van der Waals forces, electrostatic interactions or hydrogen bonding (Szpechcinski and Grzanka, 2006; Ku et al., 2015). Because of their binding specificity to their targets, aptamers are often compared to antibodies and are also known as chemical antibodies or artificial antibodies (Banerjee, 2010; Wang et al., 2016).
The selection method of aptamers, Systematic Evolution of Ligands by Exponential Enrichment (SELEX), is an in vitro process. Briefly, SELEX is based on iterative cycles of binding, separating and amplification of nucleotides. The basic mechanism of SELEX is shown in Figure 1. The first step of conventional SELEX is to incubate the sequence pool with the target (protein, nucleic acid, etc.). The sequence pool is a nucleic acid library containing 1014-1015 variants of random 30–100 nucleotides flanked by constant sequences at both ends. The random region contains the sequences that will be tested for high specificity and affinity to the target. Second, sequences that bind the target is kept, while unbound nucleotides are removed. The third step is to purify and amplify the bound sequences to form a new sequence pool for the next cycle. This cyclic process is typically repeated 8–15 times before achieving the desired aptamer sequence pool (Torres-Chavolla and Alocilja, 2009; Davydova et al., 2016). A negative selection, or counter selection, step involves incubating the sequence pool with target analogs, undesired subtypes, or the unbound sequences. This step can take place before or after target incubation to improve the specificity of candidates (Haller and Sarnow, 1997; Iwagawa et al., 2012). Biotechnology companies provide aptamer-related services, including the construction of sequence pools, aptamer selection, aptamer synthesis, and aptamer modification.
Figure 1.
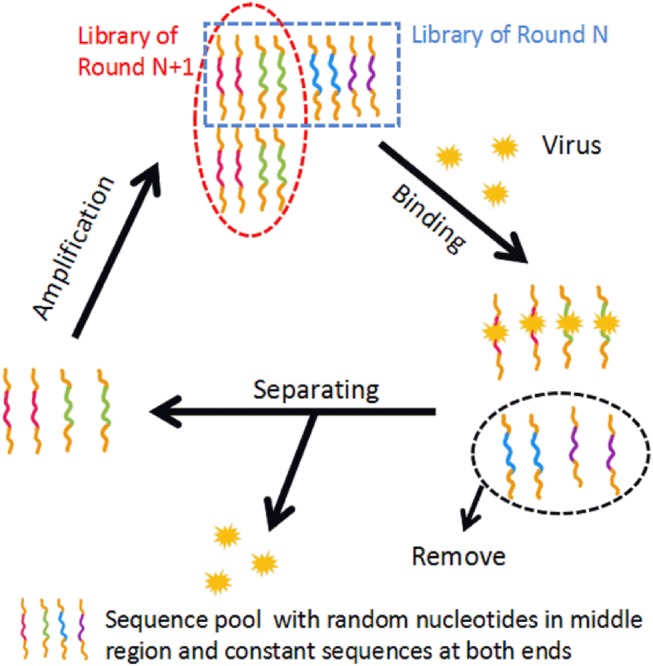
The basic mechanism of conventional SELEX. In the binding step, the sequence pool is incubated with the target. In the separating step, unbound sequences are removed and bound sequences are separated from the target. In the amplification step, the separated sequences are amplified, building a new sequence pool for the next iteration of SELEX.
Currently, viral infection is a serious threat for human beings. Although antibody-based detection methods and drugs are widely used in clinics, their popularity is hindered by high cost, antibody instability and the limitation of target types (Resch, 2017; Seo and Gu, 2017). A comparison between aptamers and antibodies is shown in Table 1. Aptamers have great potential as a feasible tool in virus detection and therapeutics.
Table 1.
Comparative properties of aptamers and antibodies.
| Property | Aptamer | Antibody | References |
|---|---|---|---|
| Time needed for selection | Several weeks | Several months | - |
| Cost of the selection | ~$4000 for individual aptamer sequences | ~$8,000 for mouse monoclonal antibody ~$20,000 for rabbit monoclonal antibody |
- |
| Synthesis and manufacture | Chemically synthesis in vitro | Produced in animal and then by recombinant methods | - |
| Modification | Easy and controllable | Limited and uncontrollable | Binning et al., 2012; Wandtke et al., 2015 |
| Stability | Stable in different environmental conditions | Requiring special conditions for the storage and handling | Davydova et al., 2016; Wang et al., 2016 |
| Batch-to-batch variation | Little or no | Difficult to avoid | Torres-Chavolla and Alocilja, 2009; Davydova et al., 2016 |
| Size | 5–25 kDa | Usually more than 125 kDa | Banerjee, 2010 |
| Chemical property | Mainly nucleic acids | Protein | - |
| Target range | Wide to almost anything | Limited to antigenic targets | O'Sullivan, 2002; Proske et al., 2005; Torres-Chavolla and Alocilja, 2009; Wandtke et al., 2015; Davydova et al., 2016 |
| In vivo complications | No intrinsic immune response | May lead immune response | Szpechcinski and Grzanka, 2006 |
| Specificity and affinity | High | High | - |
| Clinical application | Immature | Mature | - |
Applications of Aptamers In Virus Detection
Current techniques to diagnose viral infections include virus isolation in tissue cultures, immunological and molecular methods. However, these methods have a variety of limitations; for example, they are technically demanding, costly and can produce false positive or false negative results, whereas aptamer-based assay for virus detection may improve these drawbacks to some extent (Li et al., 2016; Vidic et al., 2017).
A biosensor is an analytical device that combines a bioreceptor and a transducer. The bioreceptor recognizes and binds the target with high sensitivity and selectivity, and averts interference from other microorganisms or molecules (Hong et al., 2012). The transducer then translates and outputs biological signals from the interaction between the analyte and the bioreceptor (Han et al., 2010). Aptamer-based biosensors, also called aptasensors, use aptamer as bioreceptors (also named capturing aptamer/probe) or transducers (also named signal aptamer/probe) (Cheng et al., 2009; Hianik et al., 2009). Aptasensors are mainly classified into optical and electronic aptasensors based on the type of transducer.
Optical Aptasensors
Optical aptasensors for virus detection can be classified into six categories based on the optical principles used for material detection. These categories are surface plasmon resonance (SPR) aptasensors, colorimetric aptasensors, chemiluminescence (CL) aptasensors, fluorescence aptasensors, surface-enhanced Raman scattering (SERS) aptasensors, and interferometry aptasensors.
SPR-Based Aptasensors
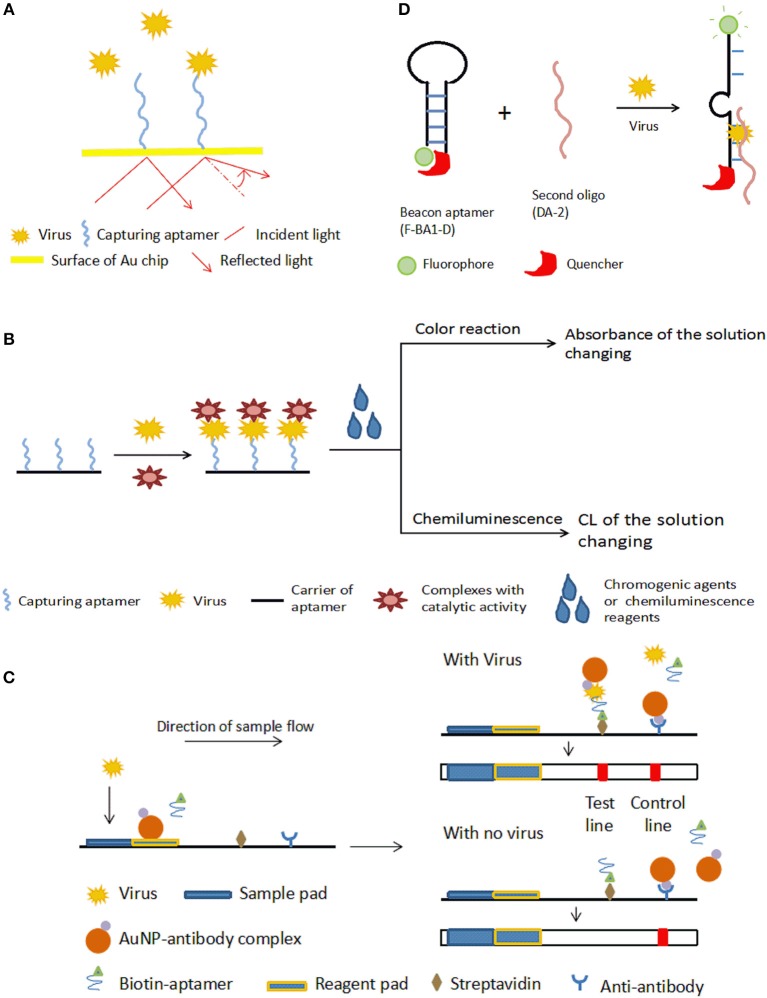
SPR measures the resonance of free electrons in some metal films by measuring the change of refractivity of the material bound on a surface (Adamczyk et al., 1999). For a typical SPR aptasensor, the capturing aptamer is immobilized on a metal surface, most often gold. The binding between viruses and aptamers changes the thickness of the gold surface, and as a result, the refractive index varies. The bound target on the surface can be quantified by monitoring the angles or intensity of the polarized light (Nguyen et al., 2015). The principle of SPR aptasensors is shown in Figure 2A. SPR sensors have certain advantages, including that no marking is required, miniaturization and automation (Skottrup et al., 2008).
Figure 2.
Schematic illustration of optical aptasensors. (A) Mechanism of SPR aptasensors. The aptamer-virus interaction changes the angle of reflected light, which indicates the amount of virus captured by the aptamers. (B) Mechanism of typical colorimetric-based aptasensor and CL aptamers. The aptamer is incubated with the virus, then catalytic-active complexes that bind the captured virus are added. Appropriate chromogenic or CL reagents are added to affect the color or luminous intensity of the sample. The change in color or luminous intensity is correlated to the amount of virus in the sample. (C) Mechanism of LFA-based aptasensor. In the presence of the target virus, both the aptamer and the AuNP-antibody complex bind to the virus, and the biotin on the aptamer enables the complex to be bound onto the streptavidin on the test line, allowing detection by the color of GNPs. The color of the test line does not change if there is no target in the sample. With or without virus, the AuNP-antibody complex is caught by the anti-antibody on the control line to cause a color change as a control. (D) Mechanism of the fluorescent quench method. Target virus can bind with F-BA1-D and DA-2 and change the structure of F-BA1-D, separating the fluorophore and the quencher and releasing fluorescence.
Bai et al. (2012) developed an SPR aptasensor for quickly detecting avian influenza virus (AIV) H5N1 within 1.5 h, with a detection range from 0.128 to 1.28 hemagglutination units (HAUs). Compared with other detection methods, this aptasensor was fast and portable, but the sensitivity was inferior to virus isolation and PCR methods. Similarly, Tombelli et al. (2005) proposed an SPR aptasensor for detecting the HIV-1 Tat protein. In another study by Nguyen et al. (2016), a pair of aptamers, IF10 and IF22, bound different sites of the same H5N1 virus, acting as the capturing probe and signal probe, respectively. This built a sandwich-type SPR biosensor platform for the sensitive detection of H5N1 viruses. In this aptasensor, the H5N1 virus was first bound by biotin-labeled aptamer IF10, which was fixed on the surface of the streptavidin-coated SPR gold chips. Then, the report aptamer IF22 linked with gold nanoparticles (AuNPs) combined with the virus captured on the SPR chips, and the AuNPs on IF22 enhanced the angle shift. By amplifying the signal with the sandwich system, the detection sensitivity of this biosensor was found to be 200 EID50/ml (50% embryo infective dose/ml) for H5N1 virus in feces samples, comparable with the sensitivity of ELISA.
Colorimetric-Based Aptasensors
In colorimetric detection, a shift of color is measured, which is either directly observed by eye or using a spectrophotometer. Colorimetric methods have merits, such as their low cost, simplicity, and portability, and thus have been widely applied in aptasensors (Feng et al., 2014; Ng et al., 2016). The principle of typical colorimetric-based aptasensors is shown in Figure 2B.
Nanomaterial-assisted colorimetric aptasensors
For this type of aptasensor, nanomaterials support the capturing aptamer, and some also take part in the signal conversion. To fabricate an aptasensor for detecting the influenza A virus, Chen et al. (2016) used an H3N2-specific aptamer modified with magnetic beads to capture the virus. AuNPs were modified with glucose oxidase (GOx) and concanavalin A (Con A), and these Con A-GOx-AuNP complexes were used for the output signal. The complexes bound the virus through a Con A-glycan interaction, and the GOx transformed the chemical signal into a color signal. This aptasensor detected the H3N2 virus at levels as low as 11.16 μg/ml with the help of a UV-vis instrument.
The hydrothermal reaction of HAuCl4 and graphene oxide produces graphene/AuNPs (Wang et al., 2010). In addition, the graphene/AuNPs have a peroxidase-like activity, mediating a catalytic reaction that is accompanied by a color change (He et al., 2011; Liang et al., 2011). Based on the graphene/AuNP hybrids, Liu et al. (2012) proposed a label-free aptasensor for detecting hepatitis C virus (HCV). In this system, the ssDNA aptamer prevents the peroxidase substrates from contacting the active interface and depresses the catalytic ability of the graphene/AuNPs. However, catalytic activity is recovered when viruses are present because the combination of the aptamer and virus reduces this catalytic hindrance. Finally, the substrate 3,3′,5,5′-tetramethylbenzidine is added to the system to visualize the result. The resulting color changes are highly correlated to the amount of virus.
Enzyme-linked aptamer assays (ELAA)
An ELISA is a basic diagnostic method for detecting complex target molecules. In ELAA, aptamers are used as a substitution for antibodies as the bio-receptor or the transducer (Nie et al., 2013). ELAA is also known as an enzyme-linked oligonucleotide adsorption test (ELOSA or ELONA) or an enzyme-linked aptasorbent assay (ELASA) (Rasoulinejad et al., 2016; Stoltenburg et al., 2016).
An ELAA for detecting the influenza A virus H5N1 used the aptamer RHA0006, which targets the hemagglutinin (HA) protein (Shiratori et al., 2014). In this aptasensor, the aptamer was immobilized on wells to capture the HA protein, and another 3′-biotinylated aptamer induced a color reaction in cooperation with streptavidin (SA)-horseradish peroxidase and the chromogen reagent 3,3′,5,5′-tetramethylbenzidine. This sandwich enzyme-linked aptasensor also recognized the H1N1 and H3N2 subtypes. The lower limit of detection reached 0.1 μg/well. Analogous ELAAs have been used to detect human norovirus (Escudero-Abarca et al., 2014), Zika virus (Lee and Zeng, 2017), and HCV (Park et al., 2013). In developing the Zika ELAA, researchers tested different pairs of capturing agent and detection agent. The aptamer/antibody pair exhibited the best detection, comparable to capacitive or impedimetric immunoassays and antibody-based ELISA kits. The detection limit of the aptamer1/aptamer2 pair was worse than the aptamer/antibody pair, but the author postulated that further research to optimize the aptamer/antibody pair may improve the detection effect. The aptamer has lower production cost and displayed a high degree of batch-to-batch consistency (Lee and Zeng, 2017).
Aptasensors based on lateral flow assay (LFA)
A lateral flow immunochromatographic assay (LFA) takes advantage of a series of capillary beds that transport fluid. LFA is widely used in clinical point-of-care detection, such as detecting levels of human chorionic gonadotrophin, HIV, HBV, and so on. Based on LFA, Le et al., 2017 put forward a method for detecting a multiplex strain-specific influenza virus. In this aptasensor, the virus was added into the sample pad and was conjugated with a biotinylated aptamer and an AuNP-labeled monoclonal antibody to form a complex at the conjugate pad. When the fluid reached the text line where SA was located, the conjugate was bound by biotin-SA, leading to a visible color change in the text line. The detection limit was about 2 × 106 virus particles. The working mechanism of aptamer-based LFA is shown in Figure 2C.
CL Aptasensors
CL is defined as material molecules generating optical radiation after absorbing chemical energy. In CL methods, the intensity of the luminous radiation reflects the concentration of the analytes. CL analysis has high sensitivity (detection limit of 10−12 to 10−21 mol) due to the ability to carry out photon metering without interference from scattered light background when an external excitation source exists, as well as a wide linear range (3–6 orders of magnitude). CL assays are another technology extensively applied in clinical diagnosis. The detecting principle of a typical CL aptasensor is similar to the principle of colorimetric-based aptasensors, shown in Figure 2B.
Based on a CL immunosorbent assay, Ahn et al. (2009) developed an aptasensor to detect severe acute respiratory syndrome coronavirus (SARS-CoV), with an aptamer capturing the SARS-CoV N protein. An enzyme-labeled secondary antibody to the N protein was employed to transduce the signal. This aptasensor detected SARS-CoV N protein at levels as low as 2 pg/ml. According to an analogous principle, Xi et al. (2015) constructed an aptasensor for detecting hepatitis B surface antigen (HBsAg). In this aptasensor, Fe3O4-SiO2 magnetic NPs were connected with the aptamer to help separate the targets from the sample. The linear range of this aptasensor was 1–200 ng/ml. This aptasensor had a lower detection limit than the limit of the ELISA used in clinical applications.
Fluorescence Aptasensors
Fluorescent aptamer biosensors use fluorophores as the signal output element. The outcome is reflected by changes in the fluorescence intensity or by the production of fluorescence polarization (Dwivedi et al., 2010; Ohk et al., 2010).
Aptasensor response with fluorescence intensity
Wang et al. (2016) applied a fluorescent-labeled universal aptamer to build an integrated microfluidic detection device for multivirus diagnosis. In this aptasensor, aptamers distinguished influenza A H1N1, H3N2, and influenza B viruses. For this aptasensor, an aptamer was modified on magnetic beads to catch the virus, and another fluorescence-labeled universal aptamer marked the captured analyte. Detection could be finished in 20 min, enabling point-of-care identification of influenza infection. In another study, a sol-gel protein chip was generated for detecting HCV core antigen in patient serum. In this chip, the aptamer was used to capture the virus, and anti-HCV and Cy3-labeled goat secondary antibodies were applied as signal probes (Lee et al., 2007).
Hmila described an aptamer-real-time-PCR method to detect the H9N2 influenza virus (Hmila et al., 2017). The capturing aptamer, specific to H9N2, was attached onto a particular strip. After virus binding by the capturing aptamer, a reporter aptamer was added into the system to bind the virus. The content of virus was calculated by measuring the bound reporter aptamer using the TaqMan RT-PCR reaction. This PCR method directly used swab samples without extracting nucleic acids, yielding a limit 1000-fold lower than a clinical ELISA. Liu et al. (2019) designed an aptamer selection strategy and identified two candidates for human noroviruses. These aptamers successfully detected human noroviruses from clinical samples as part of an in situ capture RT-qPCR assay.
In 2000, Yamamoto and his team reported a detection method to analyze the Tat protein of HIV-1 using aptamer-derived oligomers (Yamamoto et al., 2000a). They selected an aptamer RNATat specific to the HIV Tat protein (Yamamoto et al., 2000b). To build a molecular beacon aptamer, the aptamer RNATat was split into two oligomers. The beacon aptamer, named F-BA1-D, had a hairpin structure in its body region, a fluorophore at the 5′-end, and a quencher at the 3′-end. The hairpin structure placed the quencher and fluorophore near each other, inhibiting fluorophore emission. The other oligomer, DA-2, was a non-structured oligomer. As shown in Figure 2D, when the HIV-1 Tat protein was present, a stabilized ternary complex (Tat/F-BA1-D/DA-2) formed, in which the fluorophore and the quencher were separated, and fluorescent light was released (Yamamoto et al., 2000a). In Xiao's research, an aptamer specific to the prion protein PrPC similar to the aptamer RNATat mentioned above was designed. The detection range of this fluorescence aptamer sensor was 1.1-44.7 g/l, and the minimum limit of detection was 0.3 g/l (Xiao et al., 2009). This fluorescence quench assay was used to detect Influenza A virus DNA and the dengue virus genome (Fletcher et al., 2010; Liu et al., 2017).
Metal-enhanced fluorescence occurs when the emission of the fluorophore is enhanced around specific metal materials, modifying spectral characteristics and reducing photophysical constraints. Pang et al. (2015) applied metal-enhanced fluorescence to design an aptasensor for detecting H5N1. The main reagents included a core–shell of Ag-SiO2 NPs, aptamers and thiazole orange. When the H5N1 or HA protein was captured by the aptamers, the conformation of the aptamers changed into a G-quadruplex structure, causing thiazole orange fluorescence. This aptasensor detected H5N1 in both aqueous solution and patient serum. The detection process could be completed in under 30 min.
Utilizing the chemiluminescent resonance transfer strategy, Kim et al. (2018) designed an aptasensor for detecting norovirus GII. In this aptasensor, guanine-modified DNA aptamers were used to capture the target. In the presence of tetra-n-propylammonium hydroxide and dimethylformamide, the guanine of single-stranded DNA reacted with 3,4,5-trimethoxylphenylglyoxal, producing a high-energy intermediate. This intermediate then delivered energy to fluorescent dye (e.g., fluorescein, 6-FAM), which in turn emitted detectable light. The detection limit was 80 ng/ml in tap water.
Aptasensor response with fluorescence polarization
Szakács et al. (2018) proposed an aptamer-based fluorescent NP tracking analysis of viruses. In this study, human respiratory syncytial virus (RSV) was the analyte. The fluorescent aptamer bound viral glycoproteins to mark RSV. RSV could then be identified and counted using fluorescent nanoparticle tracking analysis. This analysis method was able to detect viruses larger than ~80–100 nm.
Quantum dots (QDs), also referred to as artificial atoms, are spherical-like inorganic semiconductor fluorescent nanocrystals. Compared with traditional organic dyes, QDs as fluorophores have good stability, perform well in multi-signal detection, and have other advantages as well (Michalet et al., 2005; Ikanovic et al., 2007). QDs are extensively used in fluorescent detection. Based on fluorescence polarization technology, Zhang et al. (2013) utilized a bifunctional DNA aptamer and QDs to develop an aptasensor for detecting H1N1. Briefly, a DNA sequence specific to H1N1 was modified on QDs to build a capturing probe. Another aptamer, specific to both the H1N1 protein and SA, amplified the fluorescence polarization value. This aptasensor detected H1N1 at levels as low as 3.45 nM.
SERS-Based Aptasensors
Raman spectroscopy is a type of scattered spectrum that provides “the unique chemical dactylogram” of molecules. When a laser light penetrates the medium, photons collide with the molecule, allowing the interaction of photons and the molecular vibrational energy or rotational energy. The energy of the photons can be adjusted, and the resulting energy changes reveal characteristics about the medium (Sassolas et al., 2011). However, its low sensitivity limits the application of Raman scattering. SERS overcomes this weak point by adsorbing molecules on rough metal surfaces or nanostructures (Otto, 1991; Kneipp et al., 1999; Sassolas et al., 2011; Xu et al., 2013).
Negri et al. (2012) developed a label-free SERS-based aptasensor to detect the viral nucleoprotein of influenza. In the system, Ag nanorods acted as the active substrates, and polyvalent anti-influenza aptamers were immobilized on the surface. The binding of the target and aptamer changed the nucleotide secondary structure, which was sensed by SERS.
Interferometry Aptasensors and Other Optical Aptasensors
Interferometry is a label-free technique that measures light intensity generated by the interference of different light beams. The information includes the index of refraction or physical properties, for example, the thickness of a film (Roh et al., 2011; Shah and Duncan, 2014). Roh et al. (2011) detected HCV with an Octet optical platform, where the HCV-specific RNA aptamer was coated on the optical organic film layer of a tip by biotin-SA binding. When the virus attached to the aptamer, the thickness of the organic film changed, and as a result, the signal spectrum changed. This platform had a detection limit of 700 pg/ml.
Electrical Aptasensors
Electrical aptasensors detect targets as the binding between the aptamer and target causes or changes an electrical signal. These aptasensors are classified as electrochemical aptasensors or piezoelectric transducers based on their detection mechanism.
Electrochemical Aptasensors
Typical electrochemical aptasensors immobilize the capturing aptamer on the electrode. Electrochemical aptasensors are categorized based on their method of producing electrical signals. First, in aptasensors without enzymes, the binding of aptamers to targets directly leads to an electrical signal change. Second, in aptasensors with enzymes, the electrical signal change is aided by enzyme catalysis. The third is based on a field-effect transistor (FET).
Aptasensors without enzymes
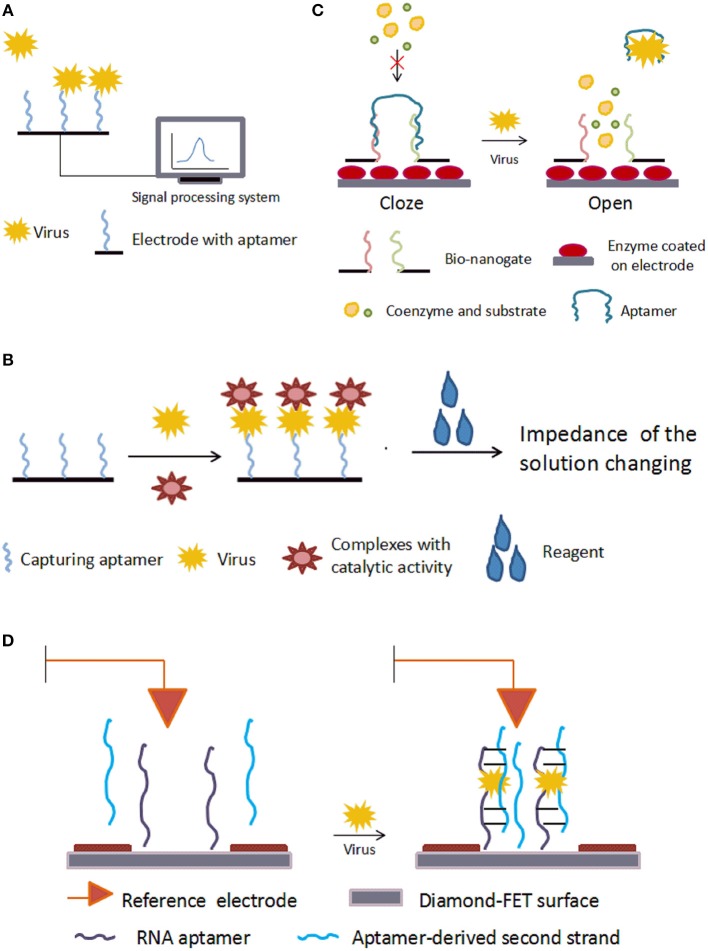
In aptasensors without enzymes, aptamers are immobilized on the electrode and the binding of aptamer to its target changes the impedance directly. The basic mechanism is shown in Figure 3A. Using microfluidic chips, Lum and his team built an impedance aptasensor to detect AIV H5N1 (Lum et al., 2015). In this biosensor, an SA-covered gold microelectrode was embedded on a microfluidic biochip. The chip was connected to the test-sense and the counter-reference probes of the impedance analyzer. Biotin-modified DNA aptamers were immobilized on the electrode. When the virus sample flowed through the microfluidic module, the aptamer captured the target, increasing the impedance. Detection took under 30 min, and the minimum detection limitation was 0.0128 HAU. Following the same principle, aptasensors were developed to detect influenza A virus (Kirkegaard and Rozlosnik, 2017) and vaccinia virus (Labib et al., 2012). Karash et al. (2016) designed an analogous aptasensor to detect H5N1 in chicken tracheal samples. Unlike previous methods, they used network-like thiocyanic acid/AuNPs to amplify the signal. This aptasensor finished detection within 1 h with a detection limit of 1 HAU for the H5N1(+) tracheal chicken swab samples, comparable to the conventional RT-PCR method.
Figure 3.
Schematic illustration of electrical aptasensors. (A) Mechanism of typical electrochemical aptasensor without enzymes. The combination of aptamer and target virus changes the electrical signal on the electrode. (B) Mechanism of electrochemical aptasensor with enzymes. (C) Mechanism of a nanogate. With no target virus, the aptamer binds the bio-nanogate to close the “door” and keep the enzyme away from the coenzyme and substrate. The target virus can grab the aptamer from the nanogate, and the enzyme on the electrode reacts with the coenzyme and substrate, leading to a change in the electrical signal on the electrode. (D) Mechanism of a diamond-aptamer FET sensor. The aptamer probe is on the diamond-FET surface. In the presence of virus, the aptamer captures the virus and forms a complex with a second aptamer strand. This causes changes in the electric charges on the surface, which is sensed by the electrode.
Giamberardino et al. (2013) fabricated an electrochemical impedance aptasensor for rapidly detecting noroviruses from clinical samples. This aptasensor employs an AuNP-modified screen-printed carbon electrode modified with an aptamer. The working mechanism was similar to the sensors introduced above. The detecting signal was a decreased redox current, measured by square wave voltammetry. The detection limit of this aptasensor was 180 virus particles. Although the aptasensor showed promise in on-site application, it needs further development before clinical applications would be feasible.
Aptasensors with enzymes
The basic mechanism of aptasensors with enzymes is shown in Figure 3B. Electrochemical impedance spectroscopy (EIS) is an electrochemical measuring method that applies a small amplitude sinusoidal potential or current as a disturbance signal. In EIS bio-sensors, the incident sinusoidal wave changes when it passes through the electrode, and these changes reflect the characteristics of the electrode. Bai et al. (2018) modified the gold working electrode with aptamers to build an EIS biosensor for detecting H1N1 virus. The detection limit was 0.9 pg/μL. Based on an increased ion strength, Fu et al. (2014) proposed an aptasensor for detecting H5N1. In this aptasensor, the capturing aptamer was fixed on magnetic beads, and AuNPs-GOx-ConA complexes were used to trigger an enzyme catalysis reaction, which increased the ionic strength and decreased the impedance. The change in impedance was detected by EIS. This aptasensor detected H5N1 as low as 8 × 10−4 HAU in 200μL samples. The EIS strategy was also adopted in aptasensors for detecting HCV (Ghanbari et al., 2017), in which a glassy carbon electrode was modified with graphene quantum dots. The capturing aptamer, specific to the HCV core antigen, was immobilized onto the glassy carbon electrodes by the noncovalent electrostatic interactions, hydrogen-bonding and π-π stacking. The ferricyanide/ferrocyanide was employed as the signal reporter. This compound slightly inhibited electron transfer caused by redox. Once the HCV core antigen combined with the aptamer, the complex strengthened the inhibition. These changes were measured by EIS. This aptasensor detected 3.3 pg/ml, with two different linearity ranges, 10–70 pg/ml and 70–400 pg/ml, much more effective than other reported PCR or EIS methods.
Nanogates are nanodevices that control chemical or biological reactions. The basic mechanism is shown in Figure 3C. Utilizing this technology, Wang et al. (2015) developed a label-free aptasensor for AIV H5N1. To fabricate the bio-nanogate, two thiolated ssDNA probes hybridized with the aptamer and were fixed on a nanoporous gold film. Then, this “gate” covered the surface of a lactate dehydrogenase-coated glassy carbon electrode. The hybridization of ssDNA and aptamer restricted the enzymatic reaction by keeping lactate dehydrogenase from its coenzymes and substrates in the testing solution. The binding of the virus to aptamers caused the aptamers to dissociate from the ssDNAs, “opening” the gate and allowing lactate dehydrogenase to contact the testing solution and react with its coenzymes and substrates. The enzymatic reaction produced a current signal on the electrode. This biosensor detected H5N1 as low as 2−9 HAU in about 1 h and had a linear range of 2−10-22 HAU.
Another electrochemical aptasensor, based on an enzymatic reaction for detecting H5N1, involves the AuNP-modified electrode functionalized with 3-mercaptopropionic acid and coated with a DNA aptamer to recognize the targets (Diba et al., 2015). An anti-H5N1 antibody, modified with alkaline phosphatase, generated an electrocatalytic reaction with the substrates. The lowest detectable concentration of this biosensor was 100 fM, and the linear range was 100 fM to 10 pM.
FET aptasensors
FET is a type of voltage-controlled semiconductor device that regulates electrical behaviors with an electric field. FET aptasensors immobilize aptamers on FET to detect changes in the charge distribution as a result of the binding of aptamers and target molecules. A diamond-aptamer FET sensor was investigated for detecting HIV-1 (Rahim Ruslinda et al., 2013). The mechanism of this diamond-FET aptasensor is shown in Figure 3D. In this sensor, the aptamer RNATat, against the Tat protein of HIV-1, was linked to an aminated diamond surface by terephthalic acid. When virus was captured by the RNATat aptamer, a second strand, which could also bind the Tat protein, was added to change the potential gate voltage by transforming the duplex structure of itself and of the initial aptamer. The change in the gate potential reflected the binding of aptamer and analyte. This aptasensor detected 1 nM HIV-1 Tat protein in samples.
Piezoelectric Transducers
Piezoelectric effect is the ability of certain materials to generate an electric charge in response to applied mechanical stress. Quartz crystal microbalance (QCM), a type of piezoelectric transducer, uses the piezoelectric properties of quartz crystals to translate changes on the quartz crystal electrode surface into changes in the output signal frequency. In QCM aptasensors, the aptamer is fixed on the quartz crystal electrode to capture the target, and the combination of the aptamer and target changes the quality of the pole, which is then transduced into detectable frequency changes. Minunni et al. (2004) used an RNA aptamer to develop a biosensor for detecting the Tat protein of HIV-1. The lowest detectable concentration was 0.25 mg/L, and the sensor was regenerated with NaOH and alcohol. Comparing an antibody-based sensor with this aptasensor, the antibody-based sensor had a wider linear range but a lower sensitivity. This method was reproducible. Wang and Li (2013) employed an ssDNA crosslinked polymeric hydrogel in a QCM aptasensor for rapid and accurate detection of AIV H5N1. The aptamer, specific to the surface protein of H5N1, was hybridized with an ssDNA and crosslinked with the polymer hydrogel, a network of water-insoluble polymer chains. The aptamer-ssDNA gel was fixed on a gold surface using a self-assembled monolayer method. In the absence of target virus, the gel retained a shrunken state. The combination of aptamer and the virus disrupted the connection between the aptamer and the ssDNA, causing the gel to swell. These changes were transduced to a detectable decreased frequency. The detection process took 30 min, with a detection limit of 0.0128 HAU. Compared with the antibody-based QCM sensor, the aptasensor had an improved detection time and detection limit.
Atomic force microscopy is a type of scanning probe microscopy with excellent resolution. This technology works by controlling and detecting the interactions between the sample and the mechanical probe. In a study by Pleshakova et al. (2018), an aptamer specific to the HCV core antigen was immobilized on an atomic force microscopy chip, and after incubation with the antigen, the chip underwent atomic force microscopy scanning for mass spectral analysis. The detection limit was as low as 0.1 pM.
Other Electrical Aptasensors
A single-molecule real-time aptasensor for detecting HIV-1 was introduced by Niedzwiecki et al. (2013). This study used nanopores, the resistive-pulse technique, and an RNA aptamer with specificity to the HIV-1 nucleocapsid protein 7 called SL3. A voltage was applied across a silicon nitride membrane, and the ionic current passing through the nanopores on the membrane was tested. When the aptamer-protein complex passed through the membrane, the current was interrupted and was replaced by a translocation event signal.
Other
A direct virus detection method was introduced by Le et al. (2014). According to this study, RNA aptamer-modified AuNPs coated a viral envelope to form a gold nanoshell, which was visualized using transmission electron microscopy. This aptamer-based method successfully detected influenza H3N2 viral particles. Aptasensors applied in virus detection are summarized in Table 2.
Table 2.
Summary of aptasensors applied in virus detection.
| Virus | Target site | Detection technique | Detection limit | Aptamer sequence | References |
|---|---|---|---|---|---|
| H1N1 | HA | Impedance aptasensor | - | - | Kirkegaard and Rozlosnik, 2017 |
| - | Fluorescence method | 3.45 nM | 5′-ACACAAATCCTATTGACCGCTGTGTGACGCAACACTCAAT-3′ | Zhang et al., 2013 | |
| Influenza A virus genes | Fluorescence method | - | 5′-CCCTTTAACCCCTTCTTCATCGAGAGTGTAGTCGGAAGAA-3′ | Liu et al., 2017 | |
| HA | Electrochemical method | 103 pfu/ml | 5′-AATTAACCCTCACTAAAGGGCTGAGTCTCAAAACCGCAATACACTGGTTGTATGGTCGAATAAGTTAA-3′ | Kiilerich-Pedersen et al., 2013 | |
| - | EIS method | 0.9pg/μL | - | Bai et al., 2018 | |
| H3N2 | Surface protein | Colorimetry | 11.16 μg/ml | 5′-AATTAACCCATCACTAAAGGGCTGAGTCTCAAAACCGCAATAACTGGTTGTATGGTCGAATAAGTTAA-3′ | Chen et al., 2016 |
| H5N1 | HA | SPR | 0.128 HAU | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGATACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCCTGTTG-3′ | Bai et al., 2012 |
| HA | Impedance method | 0.0128 HAU | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGATACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCCTGTTG-3′ | Lum et al., 2015 | |
| HA | Impedance method | 0.25 HAU for pure virus solution | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGATACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCCTGTTG-3′ | Karash et al., 2016 | |
| HA | Electrochemical method | 2−9 HAU | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGATACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCCTGTTG-3′ | Wang et al., 2015 | |
| HA | Electrochemical method | 100 fM | 5′-TTGGGGTTATTTTGGGAGGGCGGGGGTT-3 | Diba et al., 2015 | |
| HA | QCM | 0.0128 HAU | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGTAACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCGTGTTG-3′ | Wang and Li, 2013 | |
| HA | ELASA | 0.1 μg/well | 5′-GGGTTTGGGTTGGGTTGGGTTTTTGGGTTTGGGTTGGGTTGGGAAAAA-3′ | Shiratori et al., 2014 | |
| - | SPR | 200 EID50/ml | 5′-CGTACGGTCGACGCTAGCCGAAGGTTGGAGTAGGCTAAATTGGGTGTGCACGTGGAGCTCGGATCC-3′ | Nguyen et al., 2016 | |
| HA | MEF | 2 ng/ml in aqueous solution; 3 ng/ml in human serum |
5′-TTGGGGGCGGGAGGGTTTATTGGGGTT-3′ | Pang et al., 2015 | |
| HA | Impedance aptasensor | 0.0008 HAU in 200 μL sample | 5′-GTGTGCATGGATAGCACGTAACGGTGTAGTAGTAACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCGTGTTG-3′ | Fu et al., 2014 | |
| H9N2 | - | PCR | 102 TCID50/ml | 5′-CCTTGTTCTATTGAACCTCTTAGTCTGGTCCTCAGTTGGG-3′ | Hmila et al., 2017 |
| Influenza A viruses and influenza B viruses | Viral particles | Microfluidic system | - | 5′-ACAGCACCACAGACCACCCGCGGATGCCGGTCCCTACGCGTCGCTGTCACGCTGGCTGTTTGTCTTCCTGCC-3′ | Wang et al., 2016 |
| Influenza virus | HA | TEM | 3 × 108 viral particles | - | Le et al., 2014 |
| Nucleoprotein | SERS | - | 5′-TACgACTCACTATAgggATCCTgTATATATTTTgCAACTAATTgAATTCCCTTTAgTgAgggTT-3′ | Nitsche et al., 2007; Negri et al., 2012 | |
| HIV-1 | Tat protein | QCM | 0.25 ppm | 5′-ACGAAGCUUGAUCCCGUUUGCCGGUCGAUCGCUUCGA-3′ | Tombelli et al., 2005 |
| Tat protein | SPR | 0.12 ppm | 5′-ACGAAGCUUGAUCCCGUUUGCCGGUCGAUCGCUUCGA-3′ | Tombelli et al., 2005 | |
| Reverse transcriptase (RT) | CE/LIF assay | 50nM | 5′-ATCCGCCTGATTAGCGATACTTACGTGAGCGTGCTGTCCCCTAAAGGTGATACGTCACTTGAGCAAAATCACCTGCAGGGG-3′ | Pavski and Le, 2001 | |
| RT | Radioactivity-based RT nucleotide incorporation assays | - | - | DeStefano and Alves Ferreira-Bravo, 2018 | |
| Tat protein | Fluorescence method | - | 5′-ACGAAGCUUGAUCCCGUUUGCCGGUCGAUCGCUUCGA-3′ | Yamamoto et al., 2000a | |
| Tat protein | FET | 1nM | 5′-UCGGUCGAUCGCUUCAUAA-3′ | Rahim Ruslinda et al., 2013 | |
| Tat protein | QCM | 0.25 ppm | 5′-ACGAAGCUUGAUCCCGUUUGCCGGUCGAUCGCUUCGA-3′ | Minunni et al., 2004 | |
| NLC protein 7 (NCp7) | Nanopore and resistive-pulse technique | - | 5′-GGACUAGCGGAGGCUAGUCC-3′ | Niedzwiecki et al., 2013 | |
| HBV | HbsAg | Chemiluminescence | 0.1 ng/ml | 5′-GGGAATTCGAGCTCGGTACCCACAGCGAACAGCGGCGGACATAATAGTGCTTACTACGACCTGCAGGCATGCAAGCTTGG-3′ | Xi et al., 2015 |
| HCV | Envelope protein E2 | ELASA | 3.8–7.8 × 102 FFU/ml | - | Park et al., 2013 |
| DNA | Colorimetry | 11 nM | - | Liu et al., 2012 | |
| Core antigen | Fluorescence assay | - | 5′-GGGCCGTTCGAACCGAGCATGGATCGAGGATGGGAACACCCAGTAGGAGGATGGGCATGGCCGGACCCAAA-3′ATTAGCAGTGGGACAGTACTCAGGTCATCCTAGG-3′ | Lee et al., 2007 | |
| Core protein | ELASA | - | 5′-ACTATACACAAAAATAACACGACCGACGAAAAAACACAACC-3′ | Shi et al., 2014 | |
| Core antigen | LFA | 10 pg/ml with reader; 100 pg/ml with unaided eye |
5′-GATCGAGGATGGGAACACCCAGTAGGAGGATGGGCATGGCCGGACCCAAAATTAGCAGTAAAAAAAAAAAAAAAAAA-3′ | Wang et al., 2013 | |
| NS5B protein | Octet aptasensor | 700 pg/ml | 5′-GGCCACAUUGUGAGGGGCUC-3′ | Roh et al., 2011 | |
| Core antigen | Electrochemical method | 3.3 pg/ml | 5′-ACTATACACAAAAATAACACGACCGACGAAAAAACACAACC-3′ | Ghanbari et al., 2017 | |
| Helicase | Fluorescence method | - | 5′-GGGAGAGCGGAAGCGUGCUGGGCCACAUUGUGAGGGGCUCAGGUGGAUCGCAUGGCCGUGUCCAU-3′ | Cho et al., 2004; Jun et al., 2010 | |
| Core antigen | AFM-scanning | 10−14 M | 5′-ACGCTCGGATGCCACTACAGTAACACACACAACTTAAAATCATACAAAAAAGAGTAAATGCCTCATGGACGTGCTGGTGA-3′ | Pleshakova et al., 2018 | |
| Norovirus GII | - | Chemiluminescence | 80 ng/ml | 5′-GGGGGTTTTCATCTGTGTGAAGACTATATGGCGCTCACATATTTCTTTC-3′ | Kim et al., 2018 |
| Norovirus GII.3 | Capsids protein | Electrochemical method | 180 virus particles | 5′-GCTAGCGAATTCCGTACGAAGGGCGAATTCCACATTGGGCTGCAGCCCGGGGGATCC-3′ | Giamberardino et al., 2013 |
| Norovirus GII.4 | P particles | In situ capture RT-qPCR assay | - | 5′-CGATCAAACGTTCAAGCGGGGCCCGGAGGCGTGACTTGGACAGGCAGGCGTTACGATGCATCCCGCAAATGACGCATGA-3′ | Liu et al., 2019 |
| Dengue virus | EcoRI | Fluorescence method | - | 5′-CCGACGAGCAAGTAGCTCCAAGACGAGTTCAACCCCAGAATCAGGTCGG-3′ | Fletcher et al., 2010 |
| SARS coronavirus | N protein | Chemiluminescence | 2 pg/ml | 5′-GGGAGAGCGGAAGCGUGCUGGGCCUGUCGUUCGCUGUCUUGCUACGUUACGUUACACGGUUGGCAUAACCCAGAGGUCGAUGG-3′ | Ahn et al., 2009 |
| Bovine viral diarrhea virus type 1 | - | SPR | 500 TCID50/ml | 5′-CGTACGGAATTCGCTAGCTGCGCATCCACAAATGTATTGTCGGGGGATGGATCCGAGCTCCACGTG-3′ | Park et al., 2014 |
| Vaccinia | Intact virus particles | Impedimetric method | 60 virions/ml | 5′-CTCCTCTGACTGTAACCACGCGCGCCCCCGCTGTTCGAGCCGATAGAGGGCTAGTGTCATGCATAGGTAGTCCAGAAGCC-3′ | Labib et al., 2012 |
| HA | Fluorescence method | - | 5′-ATCCAGAGTGACGCAGCACGAGCCAGACATCTCACACCTGTTGCATATACATTTTGCATGGACACGGTGGCTTAGT-3′ | Parekh et al., 2010 | |
| Prion | Cellular prion protein | Fluorescence method | 0.3 mg/ml | - | Xiao et al., 2009 |
| Zika virus | NS1 protein | ELISA | ng/ml in solution; 10 ng/ml in serum | 5′-GATAGAATTCGAGCTCGGGCACTAGGTTGCAGGGGACTGCTCGGGATTGCGGATCAACCTAGTTGCTTCTCTCGTATGATGCGGGTCGACAAGCTTTAAT-3′ | Lee and Zeng, 2017 |
| RSV | Glycoprotein | Fluorescence polarization measurement | - | 5′-TAGGGAAGAGAAGGACATATGATAGTGCGGTGAGCCGTCGGACATACAAATACTTGACTAGTACATGACCACTTGA-3′ | Szakács et al., 2018 |
-, no data.
Aptamers in Antiviral Therapy
Viral infection is an intractable problem for human health, which has been highlighted in recent years. Efficient and early treatment improves the prognosis, but current treatment of viral infections is not satisfactory. Many antiviral drugs and vaccines are inefficient due to frequent virus mutations and viruses escaping the host immune system (Dunning et al., 2014; Marascio et al., 2014; Sahu, 2015). Moreover, many antiviral drugs have strong side effects, such as rashes, central nervous system disorders, influenza-like symptoms, hematologic abnormalities, or organ damage (Vcev, 2009; Frasca et al., 2012). At the same time, antiviral drugs may interact with other drugs, leading to even lower efficacy (Soriano et al., 2015).
Viral infection involves adsorption, penetration, uncoating, synthesis of macromolecule, assembly, and release. These processes may be inhibited by using specific molecules that target virus-infected cells or virus components. As a novel targeted molecule, aptamers could be applied to antiviral therapy. Several mechanisms such as clathrin- and caveolae-mediated endocytosis, macropinocytosis and phagocytosis could aid in aptamer uptake. Aptamers are distributed to subcellular compartments by endocytic vesicles according to the physiology of the host cells (Yoon and Rossi, 2018). In the following sections, we introduce antiviral aptamers that employ various mechanisms.
Suppressing Virus Attachment to Host Cells
Aptamers can impede virus entry into cells by affecting the virus or cell-surface receptors. The cellular protein nucleolin is thought to be involved in the attachment or entry of different viruses (Hovanessian, 2006; Xiao et al., 2011; Thongtan et al., 2012). Nucleolin interacts with the dengue virus capsid protein, taking part in the formation of infectious virus particles. This interaction was disturbed by the RNA aptamer AS1411, which bound to nucleolin (Balinsky et al., 2013). The influenza virus surface glycoprotein HA attaches to the sialic acid receptor of the host cell, playing a significant part in an early step of influenza infection (Skehel and Wiley, 2000; Eckert and Kim, 2001). An RNA aptamer, HA12-16, obstructed influenza virus infection in vulnerable cells by disabling the receptor-binding domain of the HA protein and enhancing cell viability (Kwon et al., 2014). A modified DNA aptamer, C7-35M, directly targeted the globular region of the AIV H9-type HA protein, suppressing virus attaching to host cells (Choi et al., 2011). In the penetration process of herpes simplex virus, the gD protein plays a key role by recognizing two protein receptors on target cells, herpes virus entry mediator and nectin-1 (Carfí et al., 2001). Based on this theory, two anti-herpes simplex virus-1 RNA aptamers were selected, which disturbed the interaction of the gD protein and the herpes virus entry mediator. This interference was dose-dependent (Gopinath et al., 2012). Similarly, another DNA aptamer targeting gD was selected for curbing herpes simplex virus-1 infection (Yadavalli et al., 2017).
Inhibiting Replication of Viruses
Various enzymes play different and significant roles in the virus replication cycle. Enzymes or their corresponding substrates could be targeted by antiviral aptamers. Inhibiting the replication of viral nucleic acid is another method in antiviral therapy.
In Japanese encephalitis virus, a single methyltransferase domain catalyzes the methylation of the RNA cap in the cytoplasm. This domain is on the N-terminal region of the viral non-structural protein NS5. A 24-mer truncated RNA aptamer modified with 2'-O-methyl pyrimidines against the Japanese encephalitis virus methyltransferase restrained viral production in host cells (Han and Lee, 2017). Jung et al. (2018) reported an analogous study in dengue virus.
The HCV non-structural 5B polymerase is an important RNA-dependent RNA polymerase that catalyzes HCV RNA replication (Luo et al., 2000; Cheney et al., 2002). Biroccio et al. (2002) selected an RNA aptamer B.2., characterized by a stem-loop structure, that potently inhibited the non-structural 5B polymerase. The B.2. aptamer and the template RNA have different binding domains on the RNA-dependent RNA polymerase, and B.2. could noncompetitively bind the RNA polymerase, weakening its activity. Two RNA aptamers, 27v and 127v, specific to the non-structural 5B polymerase, inhibited HCV polymerase activity in vitro (Bellecave et al., 2008). By competing for the binding sites of the polymerase with viral RNA template, the aptamer 27v blocked both the initiation and the elongation of viral RNA synthesis, while the aptamer 127v inhibited the initiation and postinitiation events.
The multifunctional regulatory protein pUL84 is fundamental in the early phase of human cytomegalovirus replication. By mediating the cellular importin-α/β pathway, nuclear localization signal is involved in the nuclear trafficking of pUL84. In Kaiser's research, peptide aptamers aimed at the nuclear localization signal domain of pUL84 abrogated the nuclear translocation of this viral replication factor by restraining the interaction between importin-α proteins and pUL84 (Kaiser et al., 2009).
The HBV core protein is significant in the production of the HBV nucleocapsid and affects viral envelopment (Deres et al., 2003; Roseman et al., 2005). Aptamer No.28 efficiently impeded HBV nucleocapsid assembly and suppressed viral replication (Zhang et al., 2014). Similarly, under intracellular conditions, a peptide aptamer against the HBV core protein prevented viral replication by disturbing capsid formation (Butz et al., 2001).
The HIV-1 nucleocapsid protein is crucial in the encapsidation of virus nucleic acids and the installment of virus particles (Kim et al., 2002). The retroviral psi packaging element is a cis-acting RNA element in the genome of HIV and is involved with regulating the packaging process of the viral genome in replication (Lever et al., 1989; McBride and Panganiban, 1997; McBride et al., 1997; Lever, 2007). Based on this, RNA aptamers specific to the HIV nucleocapsid protein were selected for disturbing viral packaging. The aptamers worked by competing for the psi RNA (Kim et al., 2002).
As a Delivery Tool
Small interfering RNA (siRNA) is a category of dsRNA 20-25 base pairs in size. By inducing degradation of mRNA after transcription, siRNA can inhibit the expression and translation of corresponding genes (Agrawal et al., 2003). SiRNA can also interfere with the formation of the chromatin structure of a genome (Hamilton and Baulcombe, 1999; Elbashir et al., 2001). SiRNA has shown great value in biomedical research and drug development since its discovery. However, off-targeting restricts siRNA applications in therapies (Shen et al., 2012). Aptamers are a desirable siRNA delivery tool of siRNA due to their high specificity, affinity to targets, and low toxicity.
The application of aptamer-siRNA in HIV-1 therapy has been a hot topic in recent years. Envelope glycoprotein GP120 (gp120) is a glycoprotein expressed on the HIV envelope. By attaching to the specific cell surface receptors, gp120 participates in the process of virus entry into cells (Dalgleish et al., 1984; Curtis et al., 1992). Zhou and coworkers employed the anti-gp120 aptamer-siRNA chimera for HIV-1 treatment. The aptamer carried the siRNA to cells infected with HIV-1, then the siRNA inhibited HIV replication (Zhou et al., 2008). In later studies, Zhou found that the aptamer could neutralize virus infection and transfer functional siRNAs to HIV-1 infected cells (Zhou et al., 2011). To improve the transport capacity of aptamers as siRNA carriers, researchers modified gp120-specific aptamers with a 3′ 7-carbon linker, which was bound with a 16-nucleotide 2′ OMe/2′ Fl GC-rich bridge sequence. The sequence promoted the non-covalent combination and interaction of various siRNAs with the aptamers (Zhou et al., 2013). The aptamer-siRNA system has also been studied by other researchers (Catuogno et al., 2015).
Others
To mitigate HIV-associated cardiomyopathy, Lopes de Campos et al. (2014) employed an anti-gp120 aptamer UCLA1. By directly binding to HIV-1 and neutralizing the virus, the aptamer protected cardiomyocytes from apoptosis and indirectly prevented infection of monocyte-derived macrophages.
Aptamers applied in antiviral therapy are summarized in Table 3.
Table 3.
Summary of aptamers and aptamer-based experiments in antiviral therapy.
| Virus | Aptamer type | Application | Aptamer sequence | References |
|---|---|---|---|---|
| DENV | RNA | Blocking the interaction between NCL and DENV capsid protein | 5′-GGTGGTGGTGGTTGTGGTGGTGGTGG-3′ | Balinsky et al., 2013 |
| RNA | Binding and inhibiting the methylation activity of MTase | 5′-GGGAGAGCGGAAGCGUGCUGGGCCCAGUGGUUGGGCACAUAUAGACUGUGUAAUUCGUAUAGUGUGCAUAACCCAGAGGUCGAUGGAUCCCC-3′ | Jung et al., 2018 | |
| Influenza virus | ssDNA | Targeting the HA | 5′-AACGCTCACTCCCCCAAGAAGAACCCCCCCCCCCCCCCCCCCCCCAGTGAGCGTT-3′ | Musafia et al., 2014 |
| Influenza virus (H5N1) | DNA | Binding to the HA1 protein to disrupt virus entry | 5′-GAATTCAGTCGGACAGCGGGGTTCCCATGCGGATGTTATAAAGCAGTCGCTTATAAGGGATGGACGAATATCGTCTCCC-3′ | Cheng et al., 2008 |
| Influenza A virus | DNA | Binding and inhibiting the endonuclease | 5′-CCGTAATACGACTCACTATAGGGGAGCTCGGTACCGAATTCGCAAGCGTCTGCATCCCGGTGGGACCATTAAAGCTTTGCAGAGAGGATCCTT-3′ | Yuan et al., 2015 |
| AIV | DNA | Hindering viral absorption or inhibiting HA-mediated membrane fusion by binding to HA | 5′-GCTGCAATACTCATGGACAGCCTCCTGGGGTCAGGCTCAGACATTGATAAAGCGACATCGGTCTGGAGTACGACCCTGAA-3′ or 5′-GCTGCAATACTCATGGACAGGGGCCGCGCCTGGTCGGTTGGGTGGGTGGCGCCCGGGACGGTCTGGAGTACGACCCTGAA-3′ | Zhang et al., 2015 |
| DNA | Recognize the HA protein and inhibit the binding of the virus | 5′-ATTAACCCTCACTAAAGGGAGGTAGTTATAGTATATGGAAGGGGGTGTTATGGTCGAATAAGTTAACG-3′ | Jeon et al., 2004; Choi et al., 2011 | |
| RNA | Neutralizing the receptor-binding domain of HA | 5′-GCUUGACGGAGAUCAAGGGCGAGUCUCAUACCAAGUUGAUGGGG-3′ | Kwon et al., 2014 | |
| HSV-1 | RNA | Binding to the gD protein to interfere with the binding of gD and the host receptors | 5′-GGGAGCUCAGCCUUCACUGCACGAGAGAGGUCGUCCCCAGGGGAGAACUCGUGCUCCUGGAGGCAAGUUGACUGCUCGCUCUCAGCUGGUCAAGGGCACCACGGUCGGAUCCUG-3′ | Gopinath et al., 2012 |
| DNA | Binding to the gD protein to interfere with the binding of gD and the host receptors | 5′-GGGCACGAGAGAGGTCGTCCCCAGGGGAGAACTCGTGCTCCTGG-3′ | Yadavalli et al., 2017 | |
| JEV | RNA | Suppressing JEV MTase to inhibit viral cap methylation | 5′-CCACGACAGCAUGCCAAUAGAUGCGCAUGGAGACGACAGCAU-3′ | Han and Lee, 2017 |
| HBV | DNA | Targeting the HBV core protein to reduce the synthesis of extracellular HBV DNA | 5′-ACGCTCGGATGCCACTACAGCTTCCCCTAATCTGGCGCTCTCATCTAATTTCCCTTCCTGCTCATGGACGTGCTGGTGAC-3′ | Zhang et al., 2014 |
| RNA | Interfering with viral P-ε complex formation | 5′-UGUUCAUGUCCUACUGUUCAAACAAAAAAACUGUGCACAAAAAUAAAUUGGGGCAUGGACA-3′ | Feng et al., 2011 | |
| DNA | Impairing virion formation by inhibiting the matrix domain- matrix binding domain interaction | 5′-gcgggtcgacgtttgCACACGCGAGCCGCCATGTCTGGGCcacatccatgggcgg-3′ | Orabi et al., 2015 | |
| Recombinant proteins | Working on the core protein to disturb viral capsid formation and DNA replication | - | Butz et al., 2001; Zhang et al., 2009 | |
| Recombinant proteins | Redistributing intracellular target protein into perinuclear inclusion bodies to inhibit viral capsid formation | - | Tomai et al., 2006 | |
| HCV | RNA | Suppressing HCV NS5B replicase | 5′-UUGAACGAUUGGUAGUAGAAUAUCGUCAG-3′ | Lee et al., 2013 |
| RNA | Recognizing the GTP binding site of NS5B to suppress the activity of polymerase | 5′-CGAAGCCGCUAUGGACCAGUGGCGCGGCUUCGGCCCGACGGAGUG-3′ | Biroccio et al., 2002 | |
| DNA | Binding to NS5B and inhibiting its polymerase activity | - | Bellecave et al., 2008 | |
| DNA | Inhibiting E2 protein binding to CD81 | 5′-GCGGAATTCTAATACGACTCACTATAGGGAACAGTCCGAGCCGAATGAGGAATAATCTAGCT CCTTCGCTGAGGGTCAATGCGTCATAGGATCCCCC-3′ |
Chen et al., 2009 | |
| Cytomegalovirus | Recombinant proteins | Neutralizing the NLS sites of pUL84 to interfere viral replication and production | - | Kaiser et al., 2009 |
| Rabies virus | ssDNA | Recognizing the RABV glycoprotein on infected cells to inhibit the earliest stages of infection | 5′-TATTTTTATATTTGTTTGACAGTCGCTTGCTTGTGTAGGCGTT-3′ | Liang et al., 2014 |
| HIV-1 | RNA | Working on the nucleocapsid protein | - | Kim et al., 2002 |
| RNA | Preventing the gp120 from interacting with the chemokine receptor | - | Lopes de Campos et al., 2014 | |
| RNA | - | 5′-UAAUACGACUCACUAUAGGGAGACAAGACUAGACGCUCAACAGGACCGAGAGAUGCAAC UAGUGAUUUCCCUCAUAAUCAUUCUAAGAGCUUCGACAUGAGA CUCACAACAGUUCCCUUUAGUGAGGGUUAAUU-3′ |
London et al., 2015 | |
| DNA | Inhibiting the RT | 5′-cgcctgattagcgatactCAGGCGTTGGGGGGGGGGGG-3′ or 5′-atccgcctgattagcgatatCAGAAGGATAAACTGTCCAGAAC-3′ |
Ditzler et al., 2011 | |
| RNA | Inhibiting the RT | 5′-GACAGGGCCCGTTTTCCAGTGTTTTCCCCTTTATCTCCTGGGTTCGTAGGGAATTCAG-3′ | Lange and Burke, 2014 | |
| DNA | Inhibiting the RT | 5′-GGGGGTGGGAGGAGGGTAGGCCTTAGGTTTCTGA-3′ | Shiang et al., 2013 | |
| DNA | Inhibiting both HIV infection and HIV-1 integrase | - | Magbanua et al., 2013 | |
| RNA | Inhibiting RT activity by competing with the primer/template for access to RT | 5′-GGGCAACCGGUGUCUACCGGGCUUCGGCCCGGUUCAAGGACACCGCCACUGC-3′ | Whatley et al., 2013 | |
| HIV | DNA | Targeting delivery of siRNAs | 5′-GTGACGTCCTGATCGATTGTCGCATTCGGTGTGACGATCTGCUCUAUUAGAUACAGGAGtt-3′ | Zhu et al., 2012 |
| DNA | Inhibiting both HIV infection and HIV-1 integrase | - | Magbanua et al., 2013 | |
| RNA | Binding to CycT1 to restrict the production of transcription elongation factor B | 5′-GGTAATACGACTCACTATAGGGAGATACCAGCTTATTCAATTCCUACCAA AUACGAGCCCAUCGUCACGUUCUCUUAUCUACAGATAGTAAGTGCAATCT-3′ |
Um et al., 2012 | |
| Ebola virus | RNA | Depressing the activity of the polymerase by interfering with the binding of gD and the host receptors | 5′-GGGAGACAAGAAUAAACGCUCAAGGCAUUUCUGCUAGUCUGGUUGUAA GAUAUUCAACACGUGAGUUUCGACAGGAGGCUCACAACAGGC-3′ or 5′-GGGAGACAAGAAUAAACGCUCAACGUUCAGUAUAACAGUCCGAGUCUA ACACACAAUGGGACACUGAAUUCGACAGGAGGCUCACAACAGGC-3′ |
Binning et al., 2013 |
| HPV | RNA | Binding to virus capsid and interrupting the binding of the virus capsid to heparan sulfate (HS) receptors | 5′-GGGAACAAAAGCUGCACAGGUUACCCCCGCUUGGGUCUCCCUAUAGUGAGUCGUAUUA-3′ or 5′-GGGAACAAAAGCUGCACAGGUUACCCCCGCUUGGGUCUCC-3′ |
Valencia-Reséndiz et al., 2018 |
Virus-Targeting Aptamers
In addition to the aptamers mentioned above, there are many aptamers for detecting different viruses that have not been used in virus detection or antiviral therapy. Table 4 summarizes aptamers that target different viruses.
Table 4.
Summary of aptamers that have not been used in virus detection or antiviral therapy.
| Virus | Binding site | Sequence | References |
|---|---|---|---|
| HPV-16 | L1 protein | 5′-GGGAACAAAAGCUGCACAGGUUACCCCCGCUUGGGUCUCCCUAUAGU GAGUCGUAUUA-3′ |
Leija-Montoya et al., 2014 |
| HBV | HBsAg | 5′-GTTGATTGCGTGTCAATCATGGCCGTCTATAATGATCG TAAACGACGGGTCATGTGTATGTTGGGGATTGGGACCTGATTGAGTTCAGCCCACATAC-3′ |
Liu et al., 2010 |
| HCV | Envelope glycoprotein E2 | 5′-GAATGAGGAATAATCTAGCTCCTTCGCTGA-3′ | Chen et al., 2009 |
| Human noroviruses (GII.2 and GII.4) | Capsid protein | 5′-GTCTGTAGTAGGGAGGATGGTCCGGGGCCCCGAGACGACGTTATCAGGC-3′ | Beier et al., 2014 |
| Dengue virus | Envelop protein domain III | 5′-GCACCGGGCAGGACGTCCGGGGTCCTCGGGGGGC-3′ | Chen et al., 2015 |
| Envelop protein domain III | 5′-CGGCATTCTCCTGCTACGAGG-CGCTGCGGTACACCCCGACTCCAC – GAGCCACTGTCTACGGACATCTG-3′ |
Gandham et al., 2014 | |
| SARS CoV | Nucleocapsid protein | 5′-GCAATGGTACGGTACTTCCGGATGCGGAAACTGGCTAATTGGTGAGGC TGGGGCGGTCGTGCAGCAAAAGTGCACGCTACTTTGCTAA-3 |
Cho et al., 2011 |
| Vaccinia | Surface protein | 5′-ATCGTCTGCTCCGTCCAATAGTGCATTGAAACTTCTGCATCCTCGTTTGGT GTGAGGTCGTGC-3′ |
Tang et al., 2009 |
| Ebolavirus | Soluble glycoprotein | 5′-GGGCGCUCAAUUUUUUAUUGCAUUUUUCUUUGAGCGCCC-3′ | Shubham et al., 2018 |
Conclusions and Future Perspectives
Aptamer technologies are being increasingly applied in research, for diagnosis and therapy, because of the high binding specificity and affinity, and other advantages of aptamers (Table 1). Although many studies have been published on virus detection and treatment, few aptamer-based products are commercially available for clinical diagnosis and therapy (González et al., 2016). In addition, several of these studies have compared the aptasensor with other detection methods in detail (Minunni et al., 2004; Wang and Li, 2013; Ghanbari et al., 2017). Nevertheless, aptamer technologies still face many impediments; for instance, the aptamer screening process is difficult. Even though the principles of SELEX are the same for diverse targets, the experimental details are often quite different, requiring significant time and effort to establish suitable reaction conditions. Also, selection failure is common due to significant uncertainties in PCR bias, PCR artifacts, and background binders (Rozenblum et al., 2016). Another difficulty in developing aptamer technologies is that aptamers are screened under certain conditions which do not always exactly replicate the conditions of complex clinical samples, so the structure, function, the binding affinity and specificity of aptamer could possibly be changed in clinical samples. Another hurdle for aptamer technologies is that special bases are used to construct aptamers to optimize their affinity and specificity, causing increased synthesis costs. To optimize the selection process and aptamer properties, researchers have proposed improved strategies, such as SOMAmer, bead-based selection, Cell-SELEX and microfluidics technology, and have achieved remarkable results (Sun and Zu, 2015). In addition, diverse chemical modifications to the nucleotide composition of aptamers, including pegylation, have improved the metabolic stability of aptamers. Aptamer applications in virus detection and therapies can be improved by (1) improving aptamer screening technologies; (2) further understanding the 3D models and the factors influencing the binding of aptamers and their targets; and (3) further verifying aptamers as diagnostic and therapeutic agents both in vitro and in vivo. In conclusion, while there are still some gaps in developing aptamers for clinical applications, aptamers will be widely used in virus detection and therapy with the improvement of the relevant technologies.
Author Contributions
XZ, JW, JG, and LS conceived the work and discussed the content. XZ drafted the manuscript. JW, JG, and LS were responsible for revising it. LM critically reviewed, edited, and finalized the manuscript for submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work has been supported by National Natural Science Foundation of China (Grant No. 81601751), Jiangsu Province Medical Talents Program (Grant No. QNRC2016453), and Jiangsu Province 333 Project (Grant No. BRA2017144).
References
- Adamczyk M., Mattingly P. G., Shreder K., Yu Z. (1999). Surface plasmon resonance (SPR) as a tool for antibody conjugate analysis. Bioconjug. Chem. 10, 1032–1037. 10.1021/bc990057e [DOI] [PubMed] [Google Scholar]
- Agrawal N., Dasaradhi P. V., Mohmmed A., Malhotra P., Bhatnagar R. K., Mukherjee S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. 10.1128/MMBR.67.4.657-685.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D. G., Jeon I. J., Kim J. D., Song M. S., Han S. R., Lee S. W., et al. (2009). RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 134, 1896–1901. 10.1039/b906788d [DOI] [PubMed] [Google Scholar]
- Bai C., Lu Z., Jiang H., Yang Z., Liu X., Ding H., et al. (2018). Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 110, 162–167. 10.1016/j.bios.2018.03.047 [DOI] [PubMed] [Google Scholar]
- Bai H., Wang R., Hargis B., Lu H., Li Y. (2012). A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 12, 12506–12518. 10.3390/s120912506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky C. A., Schmeisser H., Ganesan S., Singh K., Pierson T. C., Zoon K. C. (2013). Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 87, 13094–13106. 10.1128/JVI.00704-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee J. (2010). Antibodies are challenged. Indian J. Med. Sci. 64, 144–147. [PubMed] [Google Scholar]
- Beier R., Pahlke C., Quenzel P., Henseleit A., Boschke E., Cuniberti G., et al. (2014). Selection of a DNA aptamer against norovirus capsid protein VP1. FEMS Microbiol. Lett. 351, 162–169. 10.1111/1574-6968.12366 [DOI] [PubMed] [Google Scholar]
- Bellecave P., Cazenave C., Rumi J., Staedel C., Cosnefroy O., Andreola M. L., et al. (2008). Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother. 52, 2097–2110. 10.1128/AAC.01227-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binning J. M., Leung D. W., Amarasinghe G. K. (2012). Aptamers in virology: recent advances and challenges. Front. Microbiol. 3:29. 10.3389/fmicb.2012.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binning J. M., Wang T., Luthra P., Shabman R. S., Borek D. M., Liu G., et al. (2013). Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 52, 8406–8419. 10.1021/bi400704d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biroccio A., Hamm J., Incitti I., De Francesco R., Tomei L. (2002). Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76, 3688–3696. 10.1128/JVI.76.8.3688-3696.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz K., Denk C., Fitscher B., Crnkovic-Mertens I., Ullmann A., Schröder C. H., et al. (2001). Peptide aptamers targeting the hepatitis B virus core protein: a new class of molecules with antiviral activity. Oncogene 20, 6579–6586. 10.1038/sj.onc.1204805 [DOI] [PubMed] [Google Scholar]
- Carfí A., Willis S. H., Whitbeck J. C., Krummenacher C., Cohen G. H., Eisenberg R. J., et al. (2001). Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8, 169–179. 10.1016/S1097-2765(01)00298-2 [DOI] [PubMed] [Google Scholar]
- Catuogno S., Esposito C. L., de Franciscis V. (2015). A trojan horse for human immunodeficiency virus. Chem. Biol. 22, 313–314. 10.1016/j.chembiol.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Chen C., Zou Z., Chen L., Ji X., He Z. (2016). Functionalized magnetic microparticle-based colorimetric platform for influenza A virus detection. Nanotechnology 27, 435102. 10.1088/0957-4484/27/43/435102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Hu Y., Li D., Chen H., Zhang X. L. (2009). CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2. PLoS ONE 4:e8142. 10.1371/journal.pone.0008142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. L., Hsiao W. H., Lee H. C., Wu S. C., Cheng J. W. (2015). Selection and characterization of DNA aptamers targeting all four serotypes of dengue viruses. PLoS ONE 10:e0131240. 10.1371/journal.pone.0131240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney I. W., Naim S., Lai V. C., Dempsey S., Bellows D., Walker M. P., et al. (2002). Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297, 298–306. 10.1006/viro.2002.1461 [DOI] [PubMed] [Google Scholar]
- Cheng A. K., Sen D., Yu H. Z. (2009). Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 77, 1–12. 10.1016/j.bioelechem.2009.04.007 [DOI] [PubMed] [Google Scholar]
- Cheng C., Dong J., Yao L., Chen A., Jia R., Huan L., et al. (2008). Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX Biochem. Biophys. Res. Commun. 366, 670–674. 10.1016/j.bbrc.2007.11.183 [DOI] [PubMed] [Google Scholar]
- Cho S., Lee S. H., Chung W. J., Kim Y. K., Lee Y. S., Kim B. G. (2004). Microbead-based affinity chromatography chip using RNA aptamer modified with photocleavable linker. Electrophoresis 25, 3730–3739. 10.1002/elps.200406103 [DOI] [PubMed] [Google Scholar]
- Cho S. J., Woo H. M., Kim K. S., Oh J. W., Jeong Y. J. (2011). Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 112, 535–540. 10.1016/j.jbiosc.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. K., Lee C., Lee K. S., Choe S. Y., Mo I. P., Seong R. H., et al. (2011). DNA aptamers against the receptor binding region of hemagglutinin prevent avian influenza viral infection. Mol. Cells 32, 527–533. 10.1007/s10059-011-0156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis B. M., Scharnowske S., Watson A. J. (1992). Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U.S.A. 89, 8356–8360. 10.1073/pnas.89.17.8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish A. G., Beverley P. C., Clapham P. R., Crawford D. H., Greaves M. F., Weiss R. A. (1984). The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767. 10.1038/312763a0 [DOI] [PubMed] [Google Scholar]
- Davydova A., Vorobjeva M., Pyshnyi D., Altman S., Vlassov V., Venyaminova A. (2016). Aptamers against pathogenic microorganisms. Crit. Rev. Microbiol. 42, 847–865. 10.3109/1040841X.2015.1070115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deres K., Schröder C. H., Paessens A., Goldmann S., Hacker H. J., Weber O., et al. (2003). Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299, 893–896. 10.1126/science.1077215 [DOI] [PubMed] [Google Scholar]
- DeStefano J. J., Alves Ferreira-Bravo I. (2018). A highly sensitive aptamer-based HIV reverse transcriptase detection assay. J. Virol. Methods 257, 22–28. 10.1016/j.jviromet.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diba F. S., Kim S., Lee H. J. (2015). Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 72, 355–361. 10.1016/j.bios.2015.05.020 [DOI] [PubMed] [Google Scholar]
- Ditzler M. A., Bose D., Shkriabai N., Marchand B., Sarafianos S. G., Kvaratskhelia M., et al. (2011). Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res. 39, 8237–8247. 10.1093/nar/gkr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning J., Baillie J. K., Cao B., Hayden F. G. (2014). Antiviral combinations for severe influenza. Lancet Infect. Dis. 14, 1259–1270. 10.1016/S1473-3099(14)70821-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi H. P., Smiley R. D., Jaykus L. A. (2010). Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 87, 2323–2334. 10.1007/s00253-010-2728-7 [DOI] [PubMed] [Google Scholar]
- Eckert D. M., Kim P. S. (2001). Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810. 10.1146/annurev.biochem.70.1.777 [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. (1992). Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852. 10.1038/355850a0 [DOI] [PubMed] [Google Scholar]
- Escudero-Abarca B. I., Suh S. H., Moore M. D., Dwivedi H. P., Jaykus L. A. (2014). Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PLoS ONE 9:e106805. 10.1371/journal.pone.0106805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Dai S., Wang L. (2014). Optical aptasensors for quantitative detection of small biomolecules: a review. Biosens. Bioelectron. 59, 64–74. 10.1016/j.bios.2014.03.014 [DOI] [PubMed] [Google Scholar]
- Feng H., Beck J., Nassal M., Hu K. H. (2011). A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS ONE 6:e27862. 10.1371/journal.pone.0027862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S. J., Phillips L. W., Milligan A. S., Rodda S. J. (2010). Toward specific detection of Dengue virus serotypes using a novel modular biosensor. Biosens. Bioelectron. 26, 1696–1700. 10.1016/j.bios.2010.07.046 [DOI] [PubMed] [Google Scholar]
- Frasca G. M., Balestra E., Tavio M., Morroni M., Manarini G., Brigante F. (2012). [Renal toxicity of antiviral drugs]. G. Ital. Nefrol. 29 (Suppl. 56), S109–S114. [PubMed] [Google Scholar]
- Fu Y., Callaway Z., Lum J., Wang R., Lin J., Li Y. (2014). Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem. 86, 1965–1971. 10.1021/ac402550f [DOI] [PubMed] [Google Scholar]
- Gandham S. H., Volk D. E., Lokesh G. L., Neerathilingam M., Gorenstein D. G. (2014). Thioaptamers targeting dengue virus type-2 envelope protein domain III. Biochem. Biophys. Res. Commun. 453, 309–315. 10.1016/j.bbrc.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari K., Roushani M., Azadbakht A. (2017). Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 534, 64–69. 10.1016/j.ab.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Giamberardino A., Labib M., Hassan E. M., Tetro J. A., Springthorpe S., Sattar S. A., et al. (2013). Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE 8:e79087. 10.1371/journal.pone.0079087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González V. M., Martín M. E., Fernández G., García-Sacristán A. (2016). Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals 9:78. 10.3390/ph9040078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S. C., Hayashi K., Kumar P. K. (2012). Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 86, 6732–6744. 10.1128/JVI.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A. A., Sarnow P. (1997). In vitro selection of a 7-methyl-guanosine binding RNA that inhibits translation of capped mRNA molecules. Proc. Natl. Acad. Sci. U.S.A. 94, 8521–8526. 10.1073/pnas.94.16.8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Han K., Liang Z., Zhou N. (2010). Design strategies for aptamer-based biosensors. Sensors 10, 4541–4557. 10.3390/s100504541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. R., Lee S. W. (2017). Inhibition of Japanese encephalitis virus (JEV) replication by specific RNA aptamer against JEV methyltransferase. Biochem. Biophys. Res. Commun. 483, 687–693. 10.1016/j.bbrc.2016.12.081 [DOI] [PubMed] [Google Scholar]
- He W., Liu Y., Yuan J., Yin J. J., Wu X., Hu X., et al. (2011). Au@Pt nanostructures as oxidase and peroxidase mimetics for use in immunoassays. Biomaterials 32, 1139–1147. 10.1016/j.biomaterials.2010.09.040 [DOI] [PubMed] [Google Scholar]
- Hianik T., Porfireva A., Grman I., Evtugyn G. (2009). EQCM biosensors based on DNA aptamers and antibodies for rapid detection of prions. Protein Pept. Lett. 16, 363–367. 10.2174/092986609787848090 [DOI] [PubMed] [Google Scholar]
- Hmila I., Wongphatcharachai M., Laamiri N., Aouini R., Marnissi B., Arbi M., et al. (2017). A novel method for detection of H9N2 influenza viruses by an aptamer-real time-PCR. J. Virol. Methods 243, 83–91. 10.1016/j.jviromet.2017.01.024 [DOI] [PubMed] [Google Scholar]
- Hong P., Li W., Li J. (2012). Applications of aptasensors in clinical diagnostics. Sensors 12, 1181–1193. 10.3390/s120201181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian A. G. (2006). Midkine, a cytokine that inhibits HIV infection by binding to the cell surface expressed nucleolin. Cell Res. 16, 174–181. 10.1038/sj.cr.7310024 [DOI] [PubMed] [Google Scholar]
- Ikanovic M., Rudzinski W. E., Bruno J. G., Allman A., Carrillo M. P., Dwarakanath S., et al. (2007). Fluorescence assay based on aptamer-quantum dot binding to Bacillus thuringiensis spores. J. Fluoresc. 17, 193–199. 10.1007/s10895-007-0158-4 [DOI] [PubMed] [Google Scholar]
- Iwagawa T., Ohuchi S. P., Watanabe S., Nakamura Y. (2012). Selection of RNA aptamers against mouse embryonic stem cells. Biochimie 94, 250–257. 10.1016/j.biochi.2011.10.017 [DOI] [PubMed] [Google Scholar]
- Jeon S. H., Kayhan B., Ben-Yedidia T., Arnon R. (2004). A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 279, 48410–48419. 10.1074/jbc.M409059200 [DOI] [PubMed] [Google Scholar]
- Jun B. H., Rho C., Byun J. W., Kim J. H., Chung W. J., Kang H., et al. (2010). Multilayer fluorescence optically encoded beads for protein detection. Anal. Biochem. 396, 313–315. 10.1016/j.ab.2009.05.052 [DOI] [PubMed] [Google Scholar]
- Jung J. I., Han S. R., Lee S. W. (2018). Development of RNA aptamer that inhibits methyltransferase activity of dengue virus. Biotechnol. Lett. 40, 315–324. 10.1007/s10529-017-2462-7 [DOI] [PubMed] [Google Scholar]
- Kaiser N., Lischka P., Wagenknecht N., Stamminger T. (2009). Inhibition of human cytomegalovirus replication via peptide aptamers directed against the nonconventional nuclear localization signal of the essential viral replication factor pUL84. J. Virol. 83, 11902–11913. 10.1128/JVI.01378-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karash S., Wang R., Kelso L., Lu H., Huang T. J., Li Y. (2016). Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J. Virol. Methods 236, 147–156. 10.1016/j.jviromet.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Kiilerich-Pedersen K., Daprà J., Cherré S., Rozlosnik N. (2013). High sensitivity point-of-care device for direct virus diagnostics. Biosens. Bioelectron. 49, 374–379. 10.1016/j.bios.2013.05.046 [DOI] [PubMed] [Google Scholar]
- Kim B., Chung K. W., Lee J. H. (2018). Non-stop aptasensor capable of rapidly monitoring norovirus in a sample. J. Pharm. Biomed. Anal. 152, 315–321. 10.1016/j.jpba.2018.02.022 [DOI] [PubMed] [Google Scholar]
- Kim S. J., Kim M. Y., Lee J. H., You J. C., Jeong S. (2002). Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 291, 925–931. 10.1006/bbrc.2002.6521 [DOI] [PubMed] [Google Scholar]
- Kirkegaard J., Rozlosnik N. (2017). Screen-printed all-polymer aptasensor for impedance based detection of influenza A virus. Methods Mol. Biol. 1572, 55–70. 10.1007/978-1-4939-6911-1_5 [DOI] [PubMed] [Google Scholar]
- Kneipp K., Kneipp H., Itzkan I., Ramachandra R., Dasari S, Feld M. S. (1999). Ultrasensitive chemical analysis by raman spectroscopy. Chem. Rev. 99, 2957–2976. 10.1021/cr980133r [DOI] [PubMed] [Google Scholar]
- Ku T. H., Zhang T., Luo H., Yen T. M., Chen P. W., Han Y., et al. (2015). Nucleic acid aptamers: an emerging tool for biotechnology and biomedical sensing. Sensors 15, 16281–16313. 10.3390/s150716281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H. M., Lee K. H., Han B. W., Han M. R., Kim D. H., Kim D. E. (2014). An RNA aptamer that specifically binds to the glycosylated hemagglutinin of avian influenza virus and suppresses viral infection in cells. PLoS ONE 9:e97574. 10.1371/journal.pone.0097574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib M., Zamay A. S., Muharemagic D., Chechik A. V., Bell J. C., Berezovski M. V. (2012). Aptamer-based viability impedimetric sensor for viruses. Anal. Chem. 84, 1813–1816. 10.1021/ac203412m [DOI] [PubMed] [Google Scholar]
- Lange M. J., Burke D. H. (2014). Screening inhibitory potential of anti-HIV RT RNA aptamers. Methods Mol. Biol. 1103, 11–29. 10.1007/978-1-62703-730-3_2 [DOI] [PubMed] [Google Scholar]
- Le T. T., Adamiak B., Benton D. J., Johnson C. J., Sharma S., Fenton R., et al. (2014). Aptamer-based biosensors for the rapid visual detection of flu viruses. Chem. Commun. 50, 15533–15536. 10.1039/C4CC07888H [DOI] [PubMed] [Google Scholar]
- Le T. T., Chang P., Benton D. J., McCauley J. W., Iqbal M., Cass A. E. G., et al. (2017). Dual Recognition Element Lateral Flow Assay Toward Multiplex Strain Specific Influenza Virus Detection. Anal. Chem. 89, 6781–6786. 10.1021/acs.analchem.7b01149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Lee Y. J., Kim J. H., Lim J. H., Han W., Lee S. H., et al. (2013). Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J. Virol. 87, 7064–7074. 10.1128/JVI.00405-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Zeng H. (2017). Aptamer-based ELISA assay for highly specific and sensitive detection of Zika NS1 Protein. Anal. Chem. 89, 12743–12748. 10.1021/acs.analchem.7b02862 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim Y. S., Jo M., Jin M., Lee D. K., Kim S. (2007). Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 358, 47–52. 10.1016/j.bbrc.2007.04.057 [DOI] [PubMed] [Google Scholar]
- Leija-Montoya A. G., Benítez-Hess M. L., Toscano-Garibay J. D., Alvarez-Salas L. M. (2014). Characterization of an RNA aptamer against HPV-16 L1 virus-like particles. Nucleic Acid Ther. 24, 344–355. 10.1089/nat.2013.0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. (1989). Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63, 4085–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A. M. (2007). HIV-1 RNA packaging. Adv. Pharmacol. 55, 1–32. 10.1016/S1054-3589(07)55001-5 [DOI] [PubMed] [Google Scholar]
- Li P., Zhou L., Wei J., Yu Y., Yang M., Wei S., et al. (2016). Development and characterization of aptamer-based enzyme-linked apta-sorbent assay for the detection of Singapore grouper iridovirus infection. J. Appl. Microbiol. 121, 634–643. 10.1111/jam.13161 [DOI] [PubMed] [Google Scholar]
- Liang H. R., Hu G. Q., Xue X. H., Li L., Zheng X. X., Gao Y. W., et al. (2014). Selection of an aptamer against rabies virus: a new class of molecules with antiviral activity. Virus Res. 184, 7–13. 10.1016/j.virusres.2014.01.021 [DOI] [PubMed] [Google Scholar]
- Liang Y. Y., Li Y. G., Wang H. L., Zhou J. G., Wang J., Regier T., et al. (2011). Co3O4 nanocrystals on graphene as a synergistic catalyst for oxygen reduction reaction. Nat. Mater. 10, 780–786. 10.1038/nmat3087 [DOI] [PubMed] [Google Scholar]
- Liu D., Zhang Z., Yin Y., Jia F., Wu Q., Tian P., et al. (2019). Development and evaluation of a novel in situ target-capture approach for aptamer selection of human noroviruses. Talanta 193, 199–205. 10.1016/j.talanta.2018.09.084 [DOI] [PubMed] [Google Scholar]
- Liu G., Li J., Feng D. Q., Zhu J. J., Wang W. (2017). Silver nanoclusters beacon as stimuli-responsive versatile platform for multiplex dnas detection and aptamer-substrate complexes sensing. Anal. Chem. 89, 1002–1008. 10.1021/acs.analchem.6b04362 [DOI] [PubMed] [Google Scholar]
- Liu J., Yang Y., Hu B., Ma Z. Y., Huang H. P., Yu Y., et al. (2010). Development of HBsAg-binding aptamers that bind HepG2.2.15 cells via HBV surface antigen. Virol. Sin. 25, 27–35. 10.1007/s12250-010-3091-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhao H., Chen S., Yu H., Quan X. (2012). Interface engineering catalytic graphene for smart colorimetric biosensing. ACS Nano 6, 3142–3151. 10.1021/nn3010922 [DOI] [PubMed] [Google Scholar]
- London G. M., Mayosi B. M., Khati M. (2015). Isolation and characterization of 2′-F-RNA aptamers against whole HIV-1 subtype C envelope pseudovirus. Biochem. Biophys. Res. Commun. 456, 428–433. 10.1016/j.bbrc.2014.11.101 [DOI] [PubMed] [Google Scholar]
- Lopes de Campos W. R., Chirwa N., London G., Rotherham L. S., Morris L., Mayosi B. M., et al. (2014). HIV-1 subtype C unproductively infects human cardiomyocytes in vitro and induces apoptosis mitigated by an anti-Gp120 aptamer. PLoS ONE 9:e110930. 10.1371/journal.pone.0110930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J., Wang R., Hargis B., Tung S., Bottje W., Lu H., et al. (2015). An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 15, 18565–18578. 10.3390/s150818565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Hamatake R. K., Mathis D. M., Racela J., Rigat K. L., Lemm J., et al. (2000). De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74, 851–863. 10.1128/JVI.74.2.851-863.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua E., Zivkovic T., Hansen B., Beschorner N., Meyer C., Lorenzen I., et al. (2013). d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 10, 216–227. 10.4161/rna.22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marascio N., Torti C., Liberto M., Focà A. (2014). Update on different aspects of HCV variability: focus on NS5B polymerase. BMC Infect Dis. 14 (Suppl. 5):S1. 10.1186/1471-2334-14-S5-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. S., Panganiban A. T. (1997). Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71, 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. S., Schwartz M. D., Panganiban A. T. (1997). Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71, 4544–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., et al. (2005). Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544. 10.1126/science.1104274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minunni M., Tombelli S., Gullotto A., Luzi E., Mascini M. (2004). Development of biosensors with aptamers as bio-recognition element: the case of HIV-1 Tat protein. Biosens. Bioelectron. 20, 1149–1156. 10.1016/j.bios.2004.03.037 [DOI] [PubMed] [Google Scholar]
- Musafia B., Oren-Banaroya R., Noiman S. (2014). Designing anti-influenza aptamers: novel quantitative structure activity relationship approach gives insights into aptamer-virus interaction. PLoS ONE 9:e97696. 10.1371/journal.pone.0097696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri P., Chen G., Kage A., Nitsche A., Naumann D., Xu B., et al. (2012). Direct optical detection of viral nucleoprotein binding to an anti-influenza aptamer. Anal. Chem. 84, 5501–5508. 10.1021/ac202427e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Lim H. S., Ma Q., Gao Z. (2016). Optical aptasensors for adenosine triphosphate. Theranostics 6, 1683–1702. 10.7150/thno.15850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. H., Park J., Kang S., Kim M. (2015). Surface plasmon resonance: a versatile technique for biosensor applications. Sensors 15, 10481–10510. 10.3390/s150510481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. T., Seo H. B., Kim B. C., Kim S. K., Song C. S., Gu M. B. (2016). Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 86, 293–300. 10.1016/j.bios.2016.06.064 [DOI] [PubMed] [Google Scholar]
- Nie J., Deng Y., Deng Q. P., Zhang D. W., Zhou Y. L., Zhang X. X. (2013). A self-assemble aptamer fragment/target complex based high-throughput colorimetric aptasensor using enzyme linked aptamer assay. Talanta 106, 309–314. 10.1016/j.talanta.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Niedzwiecki D. J., Iyer R., Borer P. N., Movileanu L. (2013). Sampling a biomarker of the human immunodeficiency virus across a synthetic nanopore. ACS Nano 7, 3341–3350. 10.1021/nn400125c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche A., Kurth A., Dunkhorst A., Pänke O., Sielaff H., Junge W., et al. (2007). One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 7:48. 10.1186/1472-6750-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohk S. H., Koo O. K., Sen T., Yamamoto C. M., Bhunia A. K. (2010). Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 109, 808–817. 10.1111/j.1365-2672.2010.04709.x [DOI] [PubMed] [Google Scholar]
- Orabi A., Bieringer M., Geerlof A., Bruss V. (2015). An aptamer against the matrix binding domain on the hepatitis B virus capsid impairs virion formation. J. Virol. 89, 9281–9287. 10.1128/JVI.00466-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan C. K. (2002). Aptasensors–the future of biosensing? Anal. Bioanal. Chem. 372, 44–48. 10.1007/s00216-001-1189-3 [DOI] [PubMed] [Google Scholar]
- Otto A. (1991). Surface-enhanced Raman scattering of adsorbates. J. Raman Spectrosc. 22, 743–752. 10.1002/jrs.1250221204 [DOI] [Google Scholar]
- Pang Y., Rong Z., Wang J., Xiao R., Wang S. (2015). A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron. 66, 527–532. 10.1016/j.bios.2014.10.052 [DOI] [PubMed] [Google Scholar]
- Parekh P., Tang Z., Turner P. C., Moyer R. W., Tan W. (2010). Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 82, 8642–8649. 10.1021/ac101801j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Jee M. H., Kwon O. S., Keum S. J., Jang S. K. (2013). Infectivity of hepatitis C virus correlates with the amount of envelope protein E2: development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology 439, 13–22. 10.1016/j.virol.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Park J. W., Jin Lee S., Choi E. J., Kim J., Song J. Y., Bock Gu M. (2014). An ultra-sensitive detection of a whole virus using dual aptamers developed by immobilization-free screening. Biosens. Bioelectron. 51, 324–329. 10.1016/j.bios.2013.07.052 [DOI] [PubMed] [Google Scholar]
- Pavski V., Le X. C. (2001). Detection of human immunodeficiency virus type 1 reverse transcriptase using aptamers as probes in affinity capillary electrophoresis. Anal. Chem. 73, 6070–6076. 10.1021/ac0107305 [DOI] [PubMed] [Google Scholar]
- Pleshakova T. O., Kaysheva A. L. C, Bayzyanova Jcapital Em C., Anashkina capital A, Uchaikin V. F., Ziborov V. S., et al. (2018). The detection of hepatitis c virus core antigen using afm chips with immobolized aptamers. J. Virol. Methods 251, 99–105. 10.1016/j.jviromet.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Proske D., Blank M., Buhmann R., Resch A. (2005). Aptamers–basic research, drug development, and clinical applications. Appl. Microbiol. Biotechnol. 69, 367–374. 10.1007/s00253-005-0193-5 [DOI] [PubMed] [Google Scholar]
- Rahim Ruslinda A., Tanabe K., Ibori S., Wang X., Kawarada H. (2013). Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 Tat protein. Biosens. Bioelectron. 40, 277–282. 10.1016/j.bios.2012.07.048 [DOI] [PubMed] [Google Scholar]
- Rasoulinejad S. S., Gargari S. L. M. (2016). Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol. 231, 46–54. 10.1016/j.jbiotec.2016.05.024 [DOI] [PubMed] [Google Scholar]
- Resch B. (2017). Product review on the monoclonal antibody palivizumab for prevention of respiratory syncytial virus infection. Hum Vaccines Immunother. 13, 2138–2149. 10.1080/21645515.2017.1337614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Kim S. E., Jo S. K. (2011). Label free inhibitor screening of hepatitis C virus (HCV) NS5B viral protein using RNA oligonucleotide. Sensors 11, 6685–6696. 10.3390/s110706685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman A. M., Berriman J. A., Wynne S. A., Butler P. J., Crowther R. A. (2005). A structural model for maturation of the hepatitis B virus core. Proc. Natl. Acad. Sci. U.S.A. 102, 15821–15826. 10.1073/pnas.0504874102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum G. T., Lopez V. G., Vitullo A. D., Radrizzani M. (2016). Aptamers: current challenges and future prospects. Expert Opin. Drug Discov. 11, 127–135. 10.1517/17460441.2016.1126244 [DOI] [PubMed] [Google Scholar]
- Sahu G. K. (2015). Potential implication of residual viremia in patients on effective antiretroviral therapy. AIDS Res. Hum. Retroviruses 31, 25–35. 10.1089/aid.2014.0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassolas A., Blum L. J., Leca-Bouvier B. D. (2011). Optical detection systems using immobilized aptamers. Biosens. Bioelectron. 26, 3725–3736. 10.1016/j.bios.2011.02.031 [DOI] [PubMed] [Google Scholar]
- Schneider D., Tuerk C., Gold L. (1992). Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol. 228, 862–869. 10.1016/0022-2836(92)90870-P [DOI] [PubMed] [Google Scholar]
- Seo H. B., Gu M. B. (2017). Aptamer-based sandwich-type biosensors. J. Biol. Eng. 11:11. 10.1186/s13036-017-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. B., Duncan T. M. (2014). Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis. Exp. e51383. 10.3791/51383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Sun T., Ferrari M. (2012). Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 19, 367–373. 10.1038/cgt.2012.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Yu X., Gao Y., Xue B., Wu X., Wang X., et al. (2014). Inhibition of hepatitis C virus production by aptamers against the core protein. J. Virol. 88, 1990–1999. 10.1128/JVI.03312-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiang Y. C., Ou C. M., Chen S. J., Ou T. Y., Lin H. J., Huang C. C., et al. (2013). Highly efficient inhibition of human immunodeficiency virus type 1 reverse transcriptase by aptamers functionalized gold nanoparticles. Nanoscale 5, 2756–2764. 10.1039/c3nr33403a [DOI] [PubMed] [Google Scholar]
- Shiratori I., Akitomi J., Boltz D. A., Horii K., Furuichi M., Waga I. (2014). Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 443, 37–41. 10.1016/j.bbrc.2013.11.041 [DOI] [PubMed] [Google Scholar]
- Shubham S., Hoinka J., Banerjee S., Swanson E., Dillard J. A., Lennemann N. J., et al. (2018). A 2'FY-RNA motif defines an aptamer for ebolavirus secreted protein. Sci. Rep. 8:12373. 10.1038/s41598-018-30590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. J., Wiley D. C. (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569. 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- Skottrup P. D., Nicolaisen M., Justesen A. F. (2008). Towards on-site pathogen detection using antibody-based sensors. Biosensors Bioelectr. 24, 339–348. 10.1016/j.bios.2008.06.045 [DOI] [PubMed] [Google Scholar]
- Soriano V., Labarga P., Barreiro P., Fernandez-Montero J. V., de Mendoza C., Esposito I., et al. (2015). Drug interactions with new hepatitis C oral drugs. Expert Opin. Drug Metab. Toxicol. 11, 333–341. 10.1517/17425255.2015.998997 [DOI] [PubMed] [Google Scholar]
- Stoltenburg R., Krafciková P., Víglaský V., Strehlitz B. (2016). G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 6:33812. 10.1038/srep33812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Zu Y. (2015). A highlight of recent advances in aptamer technology and its application. Molecules 20, 11959–11980. 10.3390/molecules200711959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakács Z., Mészáros T., de Jonge M. I., Gyurcsányi R. E. (2018). Selective counting and sizing of single virus particles using fluorescent aptamer-based nanoparticle tracking analysis. Nanoscale 10, 13942–13948. 10.1039/C8NR01310A [DOI] [PubMed] [Google Scholar]
- Szpechcinski A., Grzanka A. (2006). [Aptamers in clinical diagnostics]. Postepy Biochem. 52, 260–270. [PubMed] [Google Scholar]
- Tang Z., Parekh P., Turner P., Moyer R. W., Tan W. (2009). Generating aptamers for recognition of virus-infected cells. Clin. Chem. 55, 813–822. 10.1373/clinchem.2008.113514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongtan T., Wikan N., Wintachai P., Rattanarungsan C., Srisomsap C., Cheepsunthorn P., et al. (2012). Characterization of putative Japanese encephalitis virus receptor molecules on microglial cells. J. Med. Virol. 84, 615–623. 10.1002/jmv.23248 [DOI] [PubMed] [Google Scholar]
- Tomai E., Butz K., Lohrey C., von Weizsäcker F., Zentgraf H., Hoppe-Seyler F. (2006). Peptide aptamer-mediated inhibition of target proteins by sequestration into aggresomes. J. Biol. Chem. 281, 21345–21352. 10.1074/jbc.M604258200 [DOI] [PubMed] [Google Scholar]
- Tombelli S., Minunni M., Luzi E., Mascini M. (2005). Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 67, 135–141. 10.1016/j.bioelechem.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Torres-Chavolla E., Alocilja E. C. (2009). Aptasensors for detection of microbial and viral pathogens. Biosensors Bioelectr. 24, 3175–3182. 10.1016/j.bios.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um H. J., Kim M., Lee S. H., Kim Y. H. (2012). Preventing the formation of positive transcription elongation factor b by human cyclin T1-binding RNA aptamer for anti-HIV transcription. AIDS 26, 1599–1605. 10.1097/QAD.0b013e3283554f7d [DOI] [PubMed] [Google Scholar]
- Valencia-Reséndiz D. G., Palomino-Vizcaino G., Tapia-Vieyra J. V., Benítez-Hess M. L., Leija-Montoya A. G., Alvarez-Salas L. M. (2018). Inhibition of human papillomavirus type 16 infection using an RNA aptamer. Nucleic Acid Ther. 28, 97–105. 10.1089/nat.2017.0687 [DOI] [PubMed] [Google Scholar]
- Vcev A. (2009). [Management of side effects during antiviral therapy]. Acta Med. Croatica 63, 463–467. [PubMed] [Google Scholar]
- Vidic J., Manzano M., Chang C. M., Jaffrezic-Renault N. (2017). Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 48:11. 10.1186/s13567-017-0418-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandtke T., Wozniak J., Kopinski P. (2015). Aptamers in diagnostics and treatment of viral infections. Viruses 7, 751–780. 10.3390/v7020751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhang L., Shen X. (2013). Development of a nucleic acid lateral flow strip for detection of hepatitis C virus (HCV) core antigen. Nucleosides Nucleotides Nucleic Acids 32, 59–68. 10.1080/15257770.2013.763976 [DOI] [PubMed] [Google Scholar]
- Wang C. H., Chang C. P., Lee G. B. (2016). Integrated microfluidic device using a single universal aptamer to detect multiple types of influenza viruses. Biosensors Bioelectr. 86, 247–254. 10.1016/j.bios.2016.06.071 [DOI] [PubMed] [Google Scholar]
- Wang H., Cui L. F., Yang Y., Sanchez Casalongue H., Robinson J. T., Liang Y., et al. (2010). Mn3O4-graphene hybrid as a high-capacity anode material for lithium ion batteries. J. Am. Chem. Soc. 132, 13978–13980. 10.1021/ja105296a [DOI] [PubMed] [Google Scholar]
- Wang R., Li Y. (2013). Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 42, 148–155. 10.1016/j.bios.2012.10.038 [DOI] [PubMed] [Google Scholar]
- Wang R., Xu L., Li Y. (2015). Bio-nanogate controlled enzymatic reaction for virus sensing. Biosens. Bioelectron. 67, 400–407. 10.1016/j.bios.2014.08.071 [DOI] [PubMed] [Google Scholar]
- Whatley A. S., Ditzler M. A., Lange M. J., Biondi E., Sawyer A. W., Chang J. L., et al. (2013). Potent Inhibition of HIV-1 reverse transcriptase and replication by nonpseudoknot, “UCAA-motif” RNA Aptamers. Mol. Ther. Nucleic Acids 2:e71. 10.1038/mtna.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Huang R., Li Z., He N., Wang T., Su E., et al. (2015). Selection of HBsAg-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis B Virus Infection. ACS Appl. Mater. Interfaces 7, 11215–11223. 10.1021/acsami.5b01180 [DOI] [PubMed] [Google Scholar]
- Xiao S. J., Hu P. P., Li Y. F., Huang C. Z., Huang T., Xiao G. F. (2009). Aptamer-mediated turn-on fluorescence assay for prion protein based on guanine quenched fluophor. Talanta 79, 1283–1286. 10.1016/j.talanta.2009.05.040 [DOI] [PubMed] [Google Scholar]
- Xiao X., Feng Y., Zhu Z., Dimitrov D. S. (2011). Identification of a putative Crimean-Congo hemorrhagic fever virus entry factor. Biochem. Biophys. Res. Commun. 411, 253–258. 10.1016/j.bbrc.2011.06.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Li H., Dihan H., Ruoff R. S., Wang A. X., Fan D. L. (2013). Plasmonics: near-field enhanced plasmonic-magnetic bifunctional nanotubes for single cell bioanalysis (Adv. Funct. Mater. 35/2013). Adv. Funct. Mater. 23, 4332–4338. 10.1002/adfm.201203822 [DOI] [Google Scholar]
- Yadavalli T., Agelidis A., Jaishankar D., Mangano K., Thakkar N., Penmetcha K., et al. (2017). Targeting Herpes Simplex Virus-1 gD by a DNA Aptamer Can Be an effective new strategy to curb viral infection. Mol. Ther. Nucleic Acids 9, 365–378. 10.1016/j.omtn.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R., Baba T., Kumar P. K. (2000a). Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells 5, 389–396. 10.1046/j.1365-2443.2000.00331.x [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Katahira M., Nishikawa S., Baba T., Taira K., Kumar P. K. (2000b). A novel RNA motif that binds efficiently and specifically to the Ttat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells 5, 371–388. 10.1046/j.1365-2443.2000.00330.x [DOI] [PubMed] [Google Scholar]
- Yoon S., Rossi J. J. (2018). Aptamers: Uptake mechanisms and intracellular applications. Adv. Drug Deliv. Rev. 134, 22–35. 10.1016/j.addr.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Zhang N., Singh K., Shuai H., Chu H., Zhou J., et al. (2015). Cross-protection of influenza A virus infection by a DNA aptamer targeting the PA endonuclease domain. Antimicrob. Agents Chemother. 59, 4082–4093. 10.1128/AAC.00306-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tian J., He Y., Chen S., Jiang Y., Zhao Y., et al. (2013). Protein-binding aptamer assisted signal amplification for the detection of influenza A (H1N1) DNA sequences based on quantum dot fluorescence polarization analysis. Analyst 138, 4722–4727. 10.1039/c3an00830d [DOI] [PubMed] [Google Scholar]
- Zhang W., Ke W., Wu S. S., Gan L., Zhou R., Sun C. Y., et al. (2009). An adenovirus-delivered peptide aptamer C1-1 targeting the core protein of hepatitis B virus inhibits viral DNA replication and production in vitro and in vivo. Peptides 30, 1816–1821. 10.1016/j.peptides.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu Z., Jiang F., Fu P., Shen J., Wu W., et al. (2015). Two DNA aptamers against avian influenza H9N2 virus prevent viral infection in cells. PLoS ONE 10:e0123060. 10.1371/journal.pone.0123060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang J., Pei X., Zhang Q., Lu B., Zhang X., et al. (2014). An aptamer targets HBV core protein and suppresses HBV replication in HepG2.2.15 cells. Int. J. Mol. Med. 34, 1423–1429. 10.3892/ijmm.2014.1908 [DOI] [PubMed] [Google Scholar]
- Zhou J., Li H., Li S., Zaia J., Rossi J. J. (2008). Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 16, 1481–1489. 10.1038/mt.2008.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li H., Zhang J., Piotr S., Rossi J. (2011). Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J. Vis. Exp. 2954. 10.3791/2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Neff C. P., Swiderski P., Li H., Smith D. D., Aboellail T., et al. (2013). Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 21, 192–200. 10.1038/mt.2012.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Shibata T., Kabashima T., Kai M. (2012). Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem. 56, 396–399. 10.1016/j.ejmech.2012.07.045 [DOI] [PubMed] [Google Scholar]