Abstract
The human brain shows several characteristics of an efficient communication network architecture, including short communication paths and the existence of modules interlinked by a small set of highly connected regions. Studies of structural networks comprising macroscopic white matter projections have shown that these putative hubs are densely interconnected, giving rise to a spatially distributed and topologically central collective called the “rich club.” In parallel, studies of intrinsic brain activity have consistently revealed distinct functional communities or resting-state networks (RSNs), indicative of specialized processing and segregation of neuronal information. However, the pattern of structural connectivity interconnecting these functional RSNs and how such inter-RSN structural connections might bring about functional integration between RSNs remain largely unknown. Combining high-resolution diffusion weighted imaging with resting-state fMRI, we present novel evidence suggesting that the rich club structure plays a central role in cross-linking macroscopic RSNs of the human brain. Rich club hub nodes were present in all functional networks, accounted for a large proportion of “connector nodes,” and were found to coincide with regions in which multiple networks overlap. In addition, a large proportion of all inter-RSN connections were found to involve rich club nodes, and these connections participated in a disproportionate number of communication paths linking nodes in different RSNs. Our findings suggest that the brain's rich club serves as a macroscopic anatomical substrate to cross-link functional networks and thus plays an important role in the integration of information between segregated functional domains of the human cortex.
Introduction
Brain function depends on efficient neural communication and integration of information within a complex network of connected brain regions, the human connectome (Sporns et al., 2005). On a macroscopic scale, the human connectome consists of a large number of anatomically distinct regions, interlinked by a complex web of structural white matter pathways. Studies examining the topology of this interregional network have revealed several characteristics of an efficient architecture, including economical wiring, short global communication paths, a modular architecture of distributed and functionally coupled communities, as well as a small number of highly connected and highly central hubs (Chklovskii et al., 2002; Kaiser and Hilgetag, 2006; Hagmann et al., 2008; Iturria-Medina et al., 2008; van den Heuvel and Hulshoff Pol, 2010; Bullmore and Sporns, 2012). In parallel, recordings of endogenously driven brain activity consistently show distinct functional communities, called resting-state networks (RSNs). RSNs exhibit high levels of within-community functional coupling while maintaining low or intermittent coupling between communities, indicative of specialized processing and segregation of neural information. Several large-scale RSNs have been identified (e.g., Damoiseaux et al., 2006; Dosenbach et al., 2010), all overlapping with distinct specialized functional subsystems of the human brain (Smith et al., 2009) and exhibiting internally dense white matter connectivity (Greicius et al., 2008; Honey et al., 2009; van den Heuvel et al., 2009). However, the pattern of structural connectivity enabling the integration of information between these different functional RSN communities, a crucial aspect of healthy brain function, remains unknown.
From a perspective of information integration, the existence of highly connected hubs in the mammalian brain is of particular interest (Bullmore and Sporns, 2009; Tomasi and Volkow, 2010). Recent studies on macroscopic anatomical brain connectivity have shown that brain hubs are not only individually rich in connections but are also densely interconnected, giving rise to the formation of a central and anatomically coherent “rich club” (Zamora-Lopez et al., 2009; Kaiser and Varier, 2011; van den Heuvel and Sporns, 2011; Harriger et al., 2012; Collin et al., 2013). In human cerebral cortex, the rich club comprises a collection of widely distributed and highly connected regions, with connections linking rich club members to one another forming a high-capacity backbone for brain communication (van den Heuvel et al., 2012). The relationship between the brain's rich club and its functional RSNs remains unclear. One possible hypothesis suggests that the structural rich club overlaps with a single or a small set of functional RSNs. Possible candidates include the default mode network or a small number of networks associated with attention and cognitive control. An alternative hypothesis suggests that the structural rich club intersects with the majority of functional RSNs (or all of them), thus forming an anatomical infrastructure for communication between RSNs, including those engaged in sensorimotor and higher cognitive processing.
Here, combining high resolution structural diffusion weighted imaging/tractography and resting-state functional MRI, we show that the brain's rich club cross-links all functional RSNs in the human brain, supporting the hypothesis of the rich club to form an anatomical substrate for integration of information between segregated functional subsystems.
Materials and Methods
Participants
Seventy-five healthy volunteers (male/female: 43/32; age mean ± SD, 28.2 ± 8.0 years) participated in this study after written consent as approved by the medical ethics committee for research in humans of the University Medical Center Utrecht, Utrecht, The Netherlands. Participants underwent a 45 min scanning session on a 3 tesla medical scanner (van den Heuvel et al., 2012). To assess the structural properties (i.e., rich club organization) as well as functional dynamics (RSNs) of the brain's connectome, an anatomical scan (T1), two structural diffusion weighted imaging (DWI) scans and a resting-state fMRI scan were acquired.
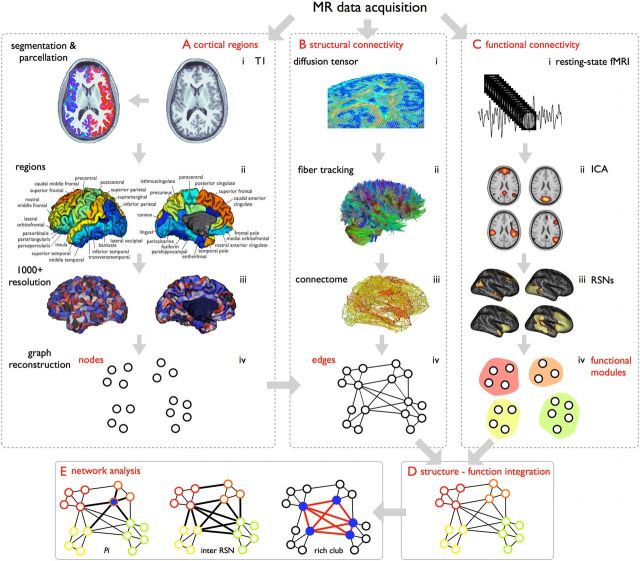
Figure 1 presents a schematic summary of the workflow of the cross-modal structural–functional analysis. The acquisition and analysis procedures of both the structural and functional scans are described in detail below (van den Heuvel et al., 2009; van den Heuvel and Sporns, 2011).
Figure 1.
Cross-modal structural–functional analysis. To assess the structural properties (i.e., rich club organization) as well as functional dynamics (i.e., RSNs) of the brain's connectome, a T1 anatomical scan (A), structural DWI (B), and resting-state fMRI were acquired (C). A, Cortical nodes. The cortical mantle was automatically segmented (i) into small parcels (1170) of approximately equal size (Fischl et al., 2004), overlapping with macroscopic cortical regions (ii,iii). These cortical parcels formed the nodes of the structural network (iv). B, Structural connectivity. Structural connectivity between brain regions was assessed using diffusion imaging (i), including the reconstruction of white matter tracts (ii), forming the connections of the network (iii,iv). C, Functional connectivity. Resting-state time-series were preprocessed (i), and 11 RSNs were selected based on ICA decomposition (ii,iii), forming a functional partitioning of the nodes of the network (iv). D, Cross-modal structural–functional integration. Functional RSNs defined a modular partition of the brain. Structural relations between these functional communities were analyzed by overlaying the structural network of reconstructed white matter pathways onto the functional partition. E, Network analysis. Network analysis was used to examine the structure–function interaction, including the examination of several cross-modal metrics of this interaction.
Structural connectivity
Anatomical T1-weighed scan.
A T1 weighed image (3D FFE using parallel imaging; TR/TE 10 ms/4.6 ms; FOV 240 × 240 mm, 200 slices, 0.75 mm isotropic voxelsize) was acquired. Tissue classification (including gray/white matter segmentation) and automatic cortical segmentation were performed using the Freesurfer suite (V5, http://surfer.nmr.mgh.harvard.edu/) (Fischl et al., 2004), parcellating the neocortex into 1170 distinct parcels of equal size, forming the nodes of the structural brain network (van den Heuvel and Sporns, 2011; van den Heuvel et al., 2009).
Structural connectivity: data acquisition and preprocessing.
Two sets of each 30 weighted diffusion scans (b = 1000 s/mm2) with each set consisting of 5 unweighted B0 scans (b = 0 s/mm2) and 30 weighted scans were acquired for each participant (SENSE, p-reduction 3; gradient set of 30 weighting directions; TR = 7035 ms, TE = 68 ms, EPI factor 35; FOV 240 × 240 mm, 2 mm isotropic, 75 slices, second diffusion set acquired with a reversed k-space readout).
Preprocessing.
Diffusion image preprocessing included the following steps: (1) two mean b = 0 images (one per set) were computed as the mean of the five b = 0 images per set, to improve signal-to-noise. (2) DWI images were corrected for susceptibility distortions by computing and applying a field distortion map determined on the two in opposite k-space readout acquired (averaged) b = 0 images (Andersson et al., 2003), (3) corrected for eddy-current distortions (Andersson and Skare, 2002), and (4) realigned to the b = 0 image, resulting in a single corrected set of corrected DWI scans (30 weighting directions and one b = 0 image). (5) The diffusion profile within each voxel was fitted a tensor using a robust tensor fitting method (RESTORE) (Chang et al., 2005), and the main diffusion direction was determined as the first principal eigenvector of the eigenvalue decomposition of the tensor. (6) White matter pathways were reconstructed using deterministic streamline tractography, based on the Fiber Assignment by Continuous Tracking algorithm (Mori and van Zijl, 2002). Within each voxel in the brain mask (i.e., the total of the segmented gray and white matter tissue), eight seeds were started following the main diffusion direction from voxel to voxel. The fiber reconstruction continued until one of three stopping criteria were met: (1) the streamline trajectory left the brain mask; (2) the streamline reached a voxel with a low fractional anisotropy value (FA < 0.1); or (3) the streamline made a turn larger than 45 degrees. Starting from eight seeds within each voxel in the brain mask, all reconstructable streamlines were collected, the total set reflecting the macroscopic white matter connectivity of the human brain.
Structural connectome reconstruction.
For each individual dataset, a subject-specific structural brain network was reconstructed. To this end, for each pair of two parcellated regions i and j, it was determined whether and, if so, how many streamlines of the subject's specific total collection of reconstructed streamlines touched both nodes i and j. If so, nodes i and j were determined to be structurally connected, and their connectivity was represented by a one in the N × N adjacency matrix A. The number of interconnecting streamlines was taken as the weight of the interconnecting edge between node i and j in the network. If no interconnecting streamline(s) were found, region i and j were taken to be nonconnected and the corresponding entry in a weighted connectivity matrix M was set to zero.
Rich club organization.
Structural rich club organization of the human connectome was described previously (van den Heuvel and Sporns, 2011). A rich club reflects a set of nodes in the network of whose level of interconnectivity exceeds the level of connectivity that can be expected on basis of chance alone. In short, the weighted rich club coefficient Φ(k) is computed as the ratio between the weights of connections present within the subnetwork S of nodes with a degree > k and the total sum of weights present within a subset of same size of the top ranking connections in the network. A normalized rich club coefficient Φnorm(k) is computed by dividing Φ(k) by Φrandom(k), with Φrandom (k) computed as the average rich club coefficient for each k of a set of 1000 randomized graphs (acquired by randomizing the adjacency matrix A preserving the degree sequence) (Rubinov and Sporns, 2010). A network is said to display rich club organization when Φnorm (k) exceeds the level of 1, for a range of increasing k.
Classification of nodes.
Group rich club members were selected based on a group-averaged network, averaging the individual adjacency matrices across the group of participants, and selecting rich club members as the nodes at the rich club level of k > 10 (van den Heuvel et al., 2012). As is the case for small-world and scale-free organization, the presence of rich club organization should be regarded as a topological property of the network as a whole, with rich club organization associated with a range of degrees k for which Φnorm(k) exceeds 1 (Colizza et al., 2006; van den Heuvel and Sporns, 2011). Previous studies have shown consistent results when examining rich clubs across different selection thresholds (Harriger et al., 2012; van den Heuvel et al., 2013). In this study, the rich club was selected at k > 10, but highly consistent and qualitatively similar results were found at both lower (e.g., k > 8, k > 9) and higher levels (e.g., k > 11, k > 12) of rich club selection.
Classification of connections.
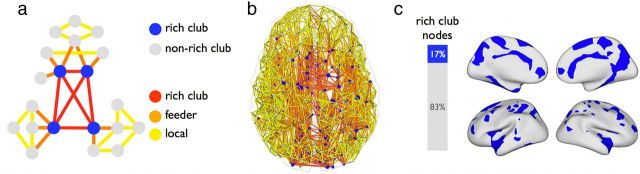
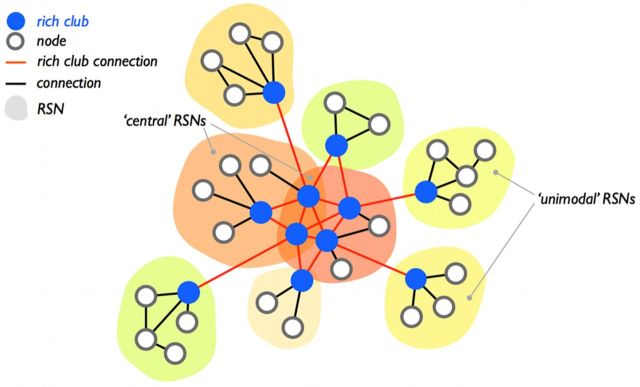
Rich club selection allows for the classification of the edges of the network into three connection classes, being “rich club connections,” linking members of the rich club, “feeder connections,” linking rich club nodes to nonrich club nodes, and “local connections,” describing the connections of the network that linked nonrich club nodes (van den Heuvel et al., 2012). Figure 2a provides a schematic illustration of the two classes of nodes and the three classes of connections.
Figure 2.
Structural rich club formation. a, Schematic illustration of the rich club and nonrich club nodes of the network and the three classes of connections (i.e., rich club, feeder, and local connections). b, Network representation of the group-averaged structural brain network, with the “nodes” of the network expressing center-of-mass of the cortical regions and the “connections” representing the reconstructed corticocortical white matter projections between these regions. Nodes are colored according to whether they participate in the rich club (blue nodes) or not (gray nodes), similar to the color scheme used in a. Connections between the nodes are color coded according to their connection class, with rich club (red), feeder connections (orange), and local connections (yellow). c, Projection of the structural rich club nodes on the cortical surface (map spatially smoothed for visualization).
Functional connectivity: data acquisition and preprocessing.
An 8 min time-series of resting-state BOLD signals was recorded using a fast fMRI sequence (3D PRESTO-SENSE p/s-reduction 2/2; TR/TE 22 ms/32 ms using shifted echo, flip-angle 9 degrees; dynamic scan time 0.5 s, 1000 timeframes; FOV 256 × 256 mm, 4 mm isotropic voxelsize, 32 slices). Image preprocessing of the functional resting-state time-series included the following steps (Fig. 1). Functional volumes were (1) realigned to correct for small head movements during acquisition using SPM8 (http://www.fil.ion.ucl.ac.uk) and (2) coregistered with the T1 scan for anatomical overlap. (3) To correct for influences of global signals and head movement, the global mean signal, the average signal of the ventricles, the average white matter signal, and six motion parameters were regressed out of the voxelwise time-series. (4) Resting-state time-series were bandpass filtered, selecting the resting-state frequencies of interest (0.01–0.1 Hz) and filtering out high-frequency factors of noninterest (e.g., respiratory frequencies of ∼0.3 Hz and cardiac frequencies up to 1 Hz). (5) For each individual dataset per segmented parcel, an averaged time-series was computed by averaging the time-series of the voxels included in each of the segments. Functional connectivity between node pairs in the network was computed as the Pearson correlation coefficient between the two regional time-series.
Functional analysis: RSNs.
Group-wise RSNs were determined using independent component analysis (ICA), using the Multivariate Exploratory Linear Optimized Decomposition into Independent Components (MELODIC) software as implemented in the FMRIB Software Library (FSL) (Beckmann et al., 2005). A similar approach as described previously (Damoiseaux et al., 2006) was used. The fMRI time-series were high (>0.01 Hz) and low pass (<0.1 Hz) filtered and concatenated in the temporal domain, followed by a multisession temporal concatenation ICA decomposition (Beckmann et al., 2005). The resulting ICA components were sorted into those that represented imaging artifacts (e.g., motion) and those that were likely to reflect a biological meaningful component. The latter were selected on the basis of their spatial overlap with RSNs identified in previous studies (Beckmann et al., 2005; Salvador et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; van den Heuvel et al., 2008; Smith et al., 2009). In total, ICA detected 54 components, of which in total 13 components were found to show strong agreement with commonly reported functional networks. These 13 components together represented 11 unique RSNs (two sets of components overlapped the same RSN and were therefore merged; see Results).
RSN node assignment and RSN overlap
Based on the selected RSNs, each structural node in the network (i.e., cortex parcel) was assigned an RSN following a majority-vote “winner-take-all” rule, by assigning each node to a single RSN based on the majority of the voxels comprising its corresponding parcel. In addition to this “winner-take-all” assignment, for each node a proportional assignment was determined, computed as the number of suprathreshold voxels belonging to individual RSNs within a node's parcel. Group ICA-based RSN selection allows for the examination of the level of participation of each cortical parcel (i.e., brain node) in multiple RSNs on a continuous scale, with each structural cortical parcel comprising a set of fMRI voxels. As such, for each parcel i, the level of overlap with each of the 11 RSNs was determined by the number voxels in parcel i that were assigned to each of the RSNs. This resulted in an “overlap score” expressing the level of overlap of each parcel with each of the 11 RSNs, ranging from 0, indicating no participation of parcel i in RSN s, to 1, indicating that all voxels of parcel i participated in RSNs. Participation levels of 10% and higher were taken into account for analysis.
Intra-RSN and inter-RSN connection assignment
Based on the winner-take-all RSN assignment, structural connections of the network were categorized into “inter-RSN” connections, being those structural connections in the network that linked nodes in two different RSNs; and “intra-RSN” connections, being those connections that linked nodes belonging to the same RSN.
Cross-modal structural–functional graph analysis
After the selection of the structural rich club and the functional communities of the connectome, the overlap between the two phenomena was examined, in particular the hypothesized integrative role of the rich club in binding distinct functional RSNs. With each node of the network assigned to a functional RSN, a cross-modal structural functional graph analysis was carried out, in which the functional RSNs described a modular partitioning of the nodes of the brain network (from here on referred to as “modules”), and the edges of the network described the structural connections (i.e., reconstructed white matter pathways) of the brain's network (from here on referred to as “connections”). Two analysis pathways were followed: (1) examining rich club participation in RSNs in terms of the “nodes” of the network; and (2) examining rich club involvement in terms of “connections” between the nodes and between the RSNs of the brain network.
Nodes: participation of structural rich club nodes in functional RSNs
Within-module degree.
For each node in the network, the within-module degree z-score (Guimera and Nunes Amaral, 2005) was computed. The within-module degree z-score expresses the level at which node i is structurally connected to the other nodes in its module (i.e., RSN), compared with the within-module degree of the other nodes in the module, formally described as follows:
 |
with κiS the number of structural connections of node i to the other nodes in module S (i.e., RSN) and κS and σSi denoting the mean and SD of the within-module degree over all nodes in S. High within-module degree z-scores (zi > 1.5) indicate a high level intramodular connectivity of a node.
Participation index.
The participation index Pi provides insight in the level of connectivity diversity of a node, quantifying to what extent a node connects across all modules (Guimera and Nunes Amaral, 2005) and is given by the following:
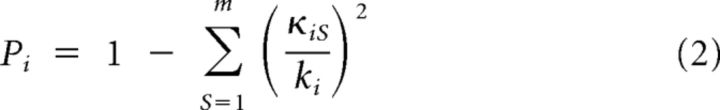 |
with κiS the number of structural links from node i to nodes in functional module S, m the number of modules (here 11 RSNs), and ki the total degree of node i. Low values of the participation index (Pi < 0.3) indicate that most of the links of i connect to nodes across a single or a small number of functional modules, and high values of the participation index (Pi > 0.3) indicate that the structural connections of node i link to a large proportion of modules in the network.
Node classification.
Network nodes were classified into four groups based on their zi and Pi scores (Guimera and Nunes Amaral, 2005): “low zi /low Pi” (low intra-RSN and low inter-RSN connectivity); “low zi /high Pi” (low intra-RSN but high inter-RSN connectivity); “high zi/low Pi” (“functional provincial hubs” with high intra-RSN but low inter-RSN connectivity); and “high zi/high Pi” (“functional connector hubs” with high intra-RSN and high inter-RSN connectivity), with zi > 1.5 corresponding to “high zi” and Pi > 0.3 corresponding to “high Pi” (Pan et al., 2010).
Functional neighborhood.
The functional neighborhood of a node was computed as the number of RSNs it is directly structurally connected to.
Rich club connections: relationship between rich club, feeder, and local connections, and RSNs
Intra-RSN and inter-RSN connectivity.
The proportion of inter-RSN and intra-RSN connections across each connection class (i.e., rich club, feeder, and local) was computed as the percentage of connections within each connection class that linked nodes in different RSNs and the percentage of connections linking two nodes in the same RSN, respectively.
Edge usage.
The level of edge usage of each connection and each connection class was computed as the number of times it was crossed in all shortest communication paths between node pairs in the network.
Structural–functional modularity.
The modularity index Q (Newman, 2006) describes the fraction of the number of edges in a network that connect nodes within the same community minus the expected value if the connections were distributed at random across the network. Based on the modularity score Q, a structural–functional modularity score Qsc_fc was computed, with the functional RSNs defining a module partition of the brain and the edges of the network represented by its structural connections, formally given as follows:
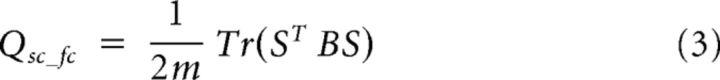 |
with S the matrix with the element Sis describing whether node i is in module s (i.e., here RSN s), Tr the trace of the matrix, and m the number of modules (i.e., RSNs), and B the modularity matrix given as follows:
 |
As such, Qsc_fc describes a metric of overlap between functionally defined communities of the network and the level of structural connectivity between and within these communities.
Vulnerability analysis.
To further examine the role of rich club connections on Qsc_fc, the weights of the connections in the rich club connection class were decreased by 10%, 25%, 50%, 75%, and 100% of the total weight, and the effect of this decrease on Qsc_fc was computed. A 100% decrease corresponded to the complete removal of the connections in the connection class. Concordantly, the effect of reducing the weights of feeder and local connections on Qsc_fc was also examined. The weights of connections within the class of feeder and local connections, respectively, were decreased by equal proportions (randomly selecting a set of connections with a total sum of weights equal to the set of rich club connections, 1000 random runs were performed), and the effect on Qsc_fc was examined. For comparison, the effect of reducing the weights by corresponding proportions across a set of 1000 randomly selected sets of connections was computed.
Statistical evaluation
Statistical evaluation was performed using permutation testing. For each examined effect (e.g., rich club vs nonrich club nodes or rich club connections vs feeder connections), the difference between the two group means was computed. Next, for 10,000 permutations, two random groups were formed by randomizing the class assignment (i.e., rich club/nonrich club or rich club/feeder/local), keeping group size intact, and the difference between the group means was computed. Over a set of permutations (10,000 permutations performed per test), this resulted in a distribution of class differences that could occur by chance (i.e., when no true difference would be present between the two original classes). This yielded an empirical null distribution of effects under the null hypothesis. A p value was then assigned to the original class difference by determining the percentage of the computed null distribution that exceeded the empirically obtained value.
Results
The integrative role of the brain's rich club in cross-linking different functional subsystems was examined by testing the relation between the brain's rich club, as defined in structural space on the basis of diffusion imaging data, and the set of functional networks (i.e., RSNs), defined in functional space by means of RSN decomposition of recordings of intrinsic brain activity.
Structural analysis: rich club
Consistent with previous reports (van den Heuvel and Sporns, 2011; van den Heuvel et al., 2012), the structural network of the human cerebral cortex showed a rich club organization, for a range of node degrees from k = 7 to k = 15, as reflected by a Φnorm(k) >1 (p < 0.001, 10,000 permutations). For all further analysis, nodes were selected to be members of the rich club at a level of k > 10 (van den Heuvel et al., 2012), comprising 17% of the network nodes. Nonrich club nodes comprised the remaining 83%. Rich club nodes were found to be widely distributed across the cortical surface (Fig. 2), including high degree regions in the parietal, frontal, temporal, and occipital lobes. Of all network connections, 13% linked pairs of rich club nodes (“rich club connections”), 26% linked a rich club node to a nonrich club node (“feeder connections”), and 61% linked nonrich club nodes to each other (“local connections”). Rich club connections accounted for 17% of all tractography streamlines (“network density”) and 27% of the network's total connection cost (the product of network density and the metric length of streamline trajectories).
Functional analysis: RSNs
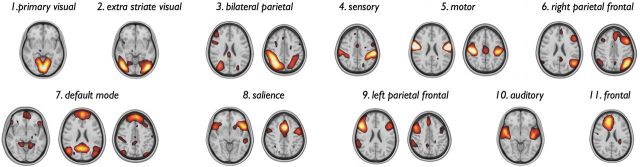
ICA analysis revealed 54 components, 13 of which were selected as biological meaningful on the basis of their spatial overlap with RSNs as reported in previous studies (Calhoun et al., 2001; Damoiseaux et al., 2006; Smith et al., 2009). Of the 13 components, two both overlapped the motor network, with one component including bilateral inferior regions of the precentral gyrus and one component predominantly including bilateral superior regions of the precentral gyrus. In addition, ICA revealed two overlapping components that both described the DMN (both ICA maps included anterior and posterior parts of the DMN). For further analysis, the superior and inferior motor components were merged into one single RSN, and the two overlapping components both describing the DMN were merged into one RSN. This resulted in a total of 11 examined RSNs (Fig. 3), including a primary visual, extrastriate visual, motor, somatosensory, and auditory network, the DMN (associated with spontaneous cognition) (Fox et al., 2005), two unilateral parietal–frontal networks (often referred to as “executive-control” or “task-activation” networks and suggested to be engaged in attentional processing and cognitive demanding tasks), the salience processing network (a network suggested to play an important role in the processing of salient information), and a frontal network. Together, these 11 RSNs covered 98.6% of the cortical surface.
Figure 3.
RSNs. ICA decomposition of the voxelwise resting-state fMRI time series resulted in the extraction of 11 RSNs. Consistent with other reports, the functional networks comprised the primary visual network (top row, from left to right), extrastriate visual, bilateral parietal, dorsal attention network, primary sensory, primary motor network, the right frontal parietal network, the default mode network (bottom row, from left to right), the salience processing network, the left frontal parietal network, auditory network, and a frontal network.
Cross-modal structural–functional graph analysis
In what follows, we (1) describe the cross-modal results pertaining to the interaction between rich club nodes and RSNs, and (2) followed by results pertaining to the interaction of rich club connections and functional RSNs.
Rich club nodes: involvement of rich club and nonrich club members in functional RSNs
Figure 4 shows the spatial layout of the RSNs (red) together with their respective structural rich club nodes (blue). Rich club nodes were encountered in all functional RSNs. RSN involvement in the rich club varied from a minimum of 3–9% (extrastriate visual, motor, sensory, auditory network) to a maximum of 22% (DMN) (Fig. 5a). The posterior part of the DMN (including portions of the cuneus/precuneus and posterior cingulate cortex) accounted for 75% (16% of total) of the DMN′s rich club nodes, whereas the anterior part (including portions of the frontal lobe) accounted for the remaining 25% (6% of total) (Fig. 5a). The number of rich club nodes per RSN varied between 6 and 44 (Fig. 5b), and on average, the proportion of rich club nodes across all RSNs was 19%, ranging from a minimum of 6–8% (extra-striate visual network and motor network) to a maximum of 23–36% (visual, DMN, salience, and frontal network) (Fig. 5b). Proportional to RSN size, the number of nodes belonging to the rich club was significantly below chance level for motor and extrastriate visual networks (p = 0.020 and p = 0.002, respectively; 10,000 permutations) and left and right parietal–frontal attention networks (p = 0.041 and p = 0.007, respectively) and significantly above chance level for the DMN, the salience network, and the frontal network (p = 0.020, p < 0.001, and p = 0.003, respectively).
Figure 4.
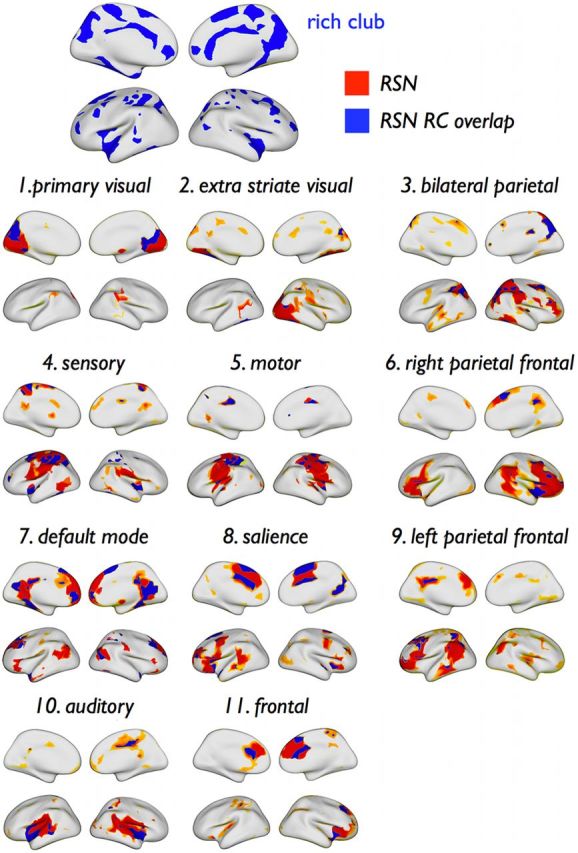
Spatial overlap of rich club and RSNs. Spatial overlap between members of the rich club and all RSNs, projected on the cortical surface. Parts of the surface assigned to a RSN are colored in red and blue, with blue regions indicating overlap between the RSN and the rich club.
Figure 5.
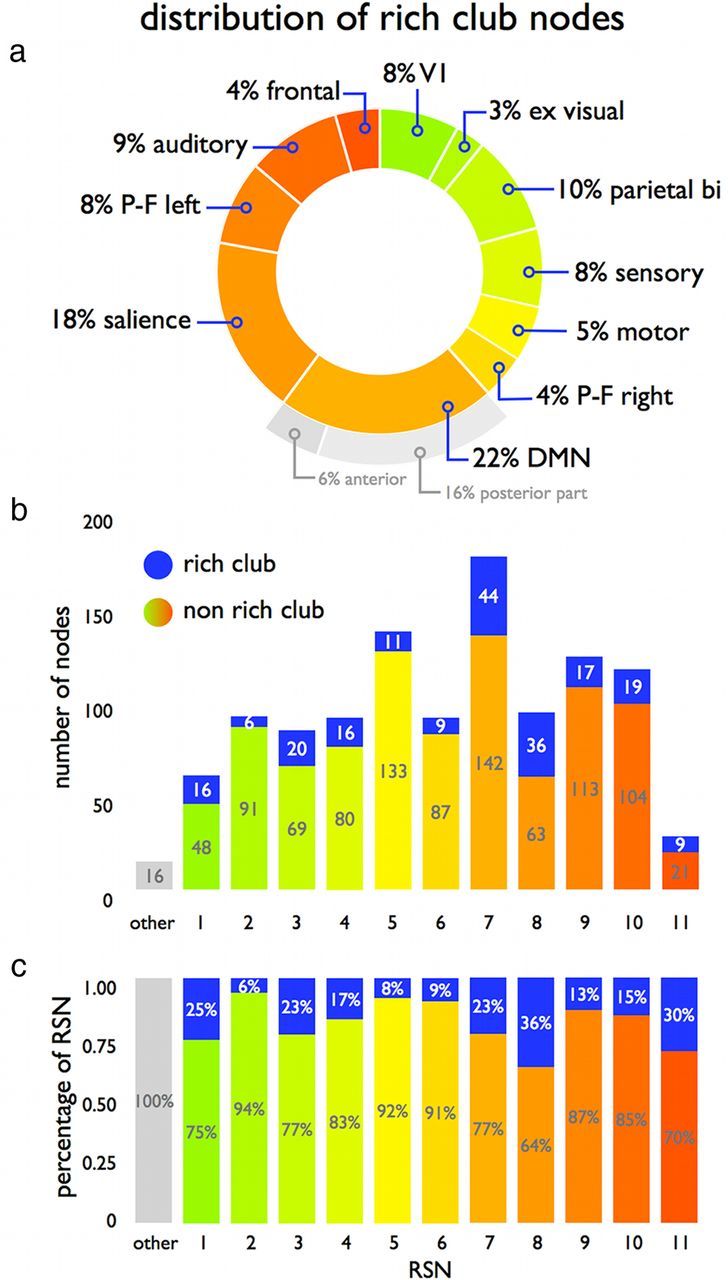
Rich club involvement in RSNs. a, Proportional involvement of RSNs in the rich club. b, Absolute number of rich club (blue) and nonrich club nodes (color coded according to RSN) involved in each of the 11 RSNs. Sixteen structural nodes were found not to overlap with any of the RSNs (i.e., RSNs overlapped 98% of the nodes of the cortex). c, Involvement of rich club nodes within each RSN, relative to RSN size.
Within-module degree z-score, community participation index, and functional neighborhood
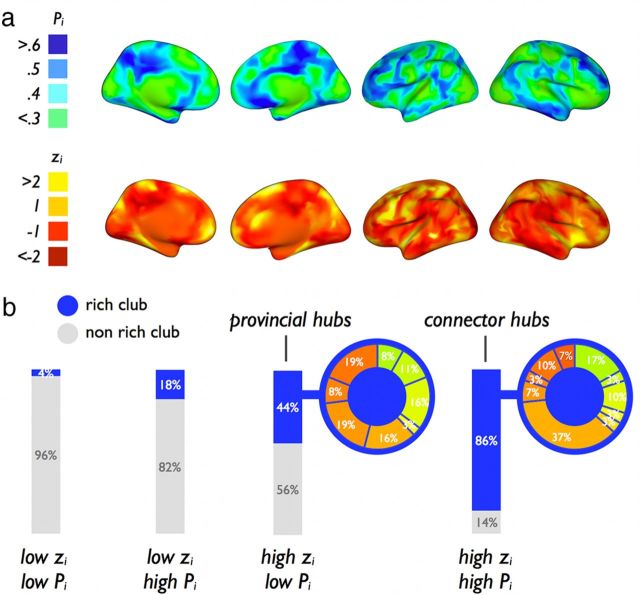
Figure 6a shows a surface plot of the Pi and zi values. Rich club nodes showed significantly higher within-module degree z-score zi, compared with nonrich club nodes (mean ± SD: rich club, 0.77 ± 1.26; nonrich club, −0.16 ± 0.84; p < 0.001; permutation testing, 10,000 permutations). In addition, rich club nodes showed a significantly higher participation index Pi, compared with nonrich club nodes (mean ± SD: rich club, 0.47 ± 0.21; nonrich club, 0.34 ± 0.25; p < 0.001, 10,000 permutations). Furthermore, rich club nodes showed on average a significantly more diverse functional neighborhood, as measured by the number of different RSNs to which an individual node was directly structurally connected, compared with nonrich club nodes (rich club, 3.36 ± 1.36; nonrich club, 2.27 ± 1.04, p < 0.001, 10,000 permutations).
Figure 6.
Participation index and within-module z-score of nodes. a, Structural–functional participation index Pi (top row) and the within-module degree degree z-score zi (bottom row) of each node of the network (maps are spatially smoothed for visualization). b, Categorizing the nodes of the network based on their Pi and zi score and the involvement of rich club and nonrich club nodes in each of these categories. Figure shows a strong involvement of rich club nodes in the “connector hubs” of the network (high Pi, high zi) expressing a high level of within-module connectivity and a diverse connectivity profile across RSNs.
Examination of the distribution of rich club and nonrich club nodes across the four classes revealed that 86% of the so-called “connector hubs” (high zi, high Pi class) comprised rich club members, compared with 14% of nonrich club nodes (Fig. 6b). The largest proportion of rich club “connector hubs” (37%) was found in the DMN, most of them located in the posterior part of the network. A total of 44% of the high zi, low Pi class comprised rich club nodes, indicating a moderate contribution of the rich club to “provincial hubs” (Fig. 6b). A large proportion of the rich club members in the salience network (19%), frontal network (19%), and DMN (16%, most of them located in the anterior portion) were found to be local provincial hubs.
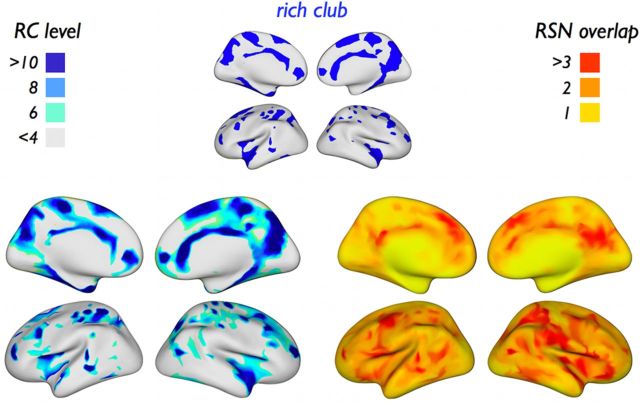
Relation between rich club level and local confluence of RSNs
Visual comparison between the rich club level per node of the network (Fig. 7, left), and the level of RSN overlap at each location on the brain surface (Fig. 7, right) shows that, in many cases, areas of RSN overlap or “confluence zones” (determined entirely on the basis of functional connectivity) were found to coincide with areas exhibiting a high rich club level (determined entirely on the basis of structural connectivity). Quantifying this overlap, rich club nodes showed a significantly higher RSN overlap count than nonrich club nodes (mean ± SD: rich club, 2.86 ± 0.94; nonrich club, 2.57 ± 0.92; p = 0.032, χ2 frequency test). Nodewise correlation (i.e., across all cortical parcels) between rich club level and RSN overlap score exhibited a modest significant correlation (r = 0.14, p < 0.001, linear regression).
Figure 7.
Rich club level and RSN overlap. Left, Rich club level projected on a reconstruction of the cortical surface. Right, Level of “RSN overlap” for each structural node of the network, expressing the number of RSNs in which a node is (proportionally) involved. Comparison shows that, in many cases, areas of RSN overlap coincide with regions that have a high rich club level (see Results). (Maps are spatially smoothed for visualization.)
Structural connections within and between functional RSNs
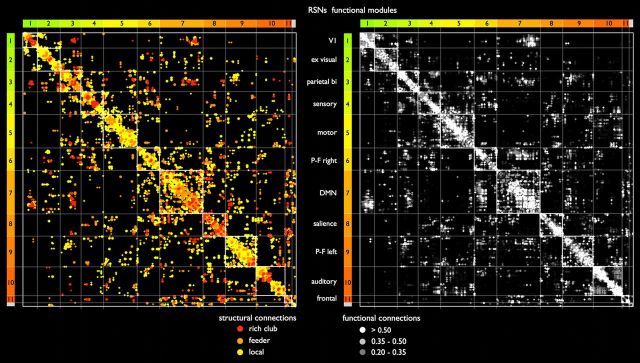
Connectivity matrices
A side-by-side comparison of the group-averaged structural connectivity and functional connectivity matrix, with network nodes ordered identically according to RSN assignment, revealed consistent overlap between the two connectivity modalities (Fig. 8). Comparison of the two connection matrices showed that relatively low levels of structural connectivity were found between visual and frontal RSNs and sensory and auditory networks (off-diagonal parts of the matrix), whereas relatively high structural connectivity existed between the DMN, salience processing, and bilateral parietal–frontal task-activation networks (central parts of the matrix).
Figure 8.
Structural and functional connectivity matrices. Side-by-side plot of the group-averaged structural and functional connectivity matrix. Network nodes in both the structural (left) and functional (right) matrix are arranged according to their “winner-take-all” assignment to each of the 11 functional RSNs. Existing entries within the matrix are marked by colored dots to allow the comparison of structural and functional connections across matrices. Colored bars to the left and top of the matrices indicate RSN assignment (compare with Fig. 4). RSNs are ordered according to their level of inter-RSN connectivity, with RSNs showing high levels of interconnectivity appearing closer to the center of the matrix. Left, Structural connections, color coded according to their connection class. Right, Connection strengths of strong positive functional couplings between nodes of the network depicted in grayscale.
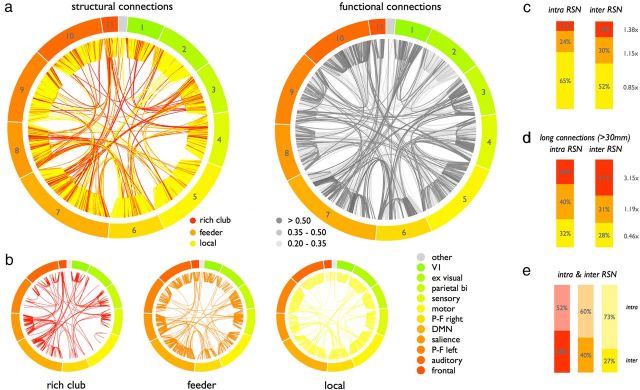
Inter-RSN versus intra-RSN links
Rich club and feeder connections were found to be highly represented among inter-RSN connections (Fig. 9a), expressing an important role of rich club and feeder connections in cross-linking different RSNs. Figure 9b shows the structural connections (as shown in Fig. 9a) divided up according to their structural connection class, illustrating a high proportional involvement of inter-RSN connections in the class of rich club and feeder connections. Quantifying this intermodular role, rich club, feeder, and local connections accounted for 18% (1.38 times their overall share of 13% connections), 30% (1.15 times), and 52% (0.85 times), respectively, of all inter-RSN connections, whereas they accounted for 11% (0.80 times their overall share of connections), 24% (0.92 times), and 65% (1.08 times), respectively, of all intra-RSN connections (Fig. 9c). Excluding connections crossing between spatially adjacent RSNs (examining only long-distance structural connections with a length >30 mm), the disproportional role of rich club connections in interlinking different RSNs became even more prominent, with rich club connections accounting for 41% (3.2 times more than their overall share) of all inter-RSN connections and 28% of all intra-RSN connections (Fig. 9d). Examining the contributions to inter-RSN and intra-RSN connections per connection class (Fig. 9e), 48% of all rich club connections linked rich club nodes located in different RSNs, in contrast to 40% of the feeder and 27% of the local connections. Examining the level of interconnectivity per RSN, the total number of inter-RSN connections was found to be the highest for the DMN and the salience processing network, with the left and right frontal–parietal network in third and fourth place. Mutual structural connectivity among those four central RSNs (i.e., the average number of structural inter-RSN connections between them) was found to be 7.12 times higher than the average interconnectivity between the other eight RSNs (e.g., motor to visual, visual to auditory). Furthermore, total connectivity (i.e., number of reconstructed tractography streamlines) between these eight more peripheral networks with the DMN, salience, and bilateral task-executive networks was dominated by rich club and feeder connections, which accounted for 76% of all connectivity strength between these two groups of RSNs.
Figure 9.
Rich club, feeder, and local intra-RSN and inter-RSN connectivity. a, Side-by-side plot of the long-distance (>30 mm, corresponding to the statistics presented in d) structural and functional connections arranged in the same node ordering. Left, Long-distance inter-RSN and intra-RSN connections, color-coded according to their connection class. Right, Connection strengths of strong positive functional couplings between all network nodes depicted in grayscale. b, Panels show the connections of a divided up according to their connection class. c, Distribution of rich club, feeder, and local connections over the two classes of intra-RSN and inter-RSN connections. d, Distribution of intra-RSN and inter-RSN connections, excluding connections crossing between spatially adjacent RSNs (examining only connections with a length >30 mm). e, Distribution of intra-RSN and inter-RSN connections split out according to connection class.
Connection usage in intra-RSN and inter-RSN communication
A total of 95% of all inter-RSN communication paths (i.e., paths of which the source and target nodes belonged to different RSNs) were found to travel through the rich club (i.e., across at least one rich club node), whereas 72% of all intra-RSN communication paths (i.e., source and target nodes were members of the same RSN) traveled through the rich club.
Inter-RSN and intra-RSN communication
Examining inter-RSN communication paths across the network in more detail, rich club inter-RSN connections were used on average 988 times, 2.9 times more than local inter-RSN connections (335 passes on average). Feeder inter-RSN connections (838 passes) were used 2.5 times more often than local inter-RSN connections. In contrast, on these inter-RSN communication paths, rich club intra-RSN connections were used (on average) 441 times, feeder intraconnections 620 times, and local intraconnections 310 times, indicating that rich club connections were passed only 1.42 times more often than local connections. A central role of rich club and feeder connections in inter-RSN communication was further illustrated by ranking the inter-RSN connections on the basis of the number of times they were used among all communication paths. Of the top 100 most-used inter-RSN network edges, 88% belonged to rich club (36%) and feeder connections (52%).
On intra-RSN communication paths (i.e., paths between nodes of the same RSN), rich club intra-RSN connections were used (on average) 39 times, feeder intraconnections 56 times, and local intraconnections 34 times, indicating that rich club connections were passed only 1.15 times more often than local connections.
Structural–functional modularity
RSNs defined a modular partitioning of the human brain, and these modules were used in combination with data on the structural connections of the network to compute the structural–functional modularity Qsc_fc. Qsc_fc levels (0.556, Fig. 10, blue line) indicated significant levels of modularity compared with the maximal level of modularity Qsc (0.78) achievable in the structural network, suggesting that RSNs partitioned the structural connection matrix quite well (see also Fig. 8).
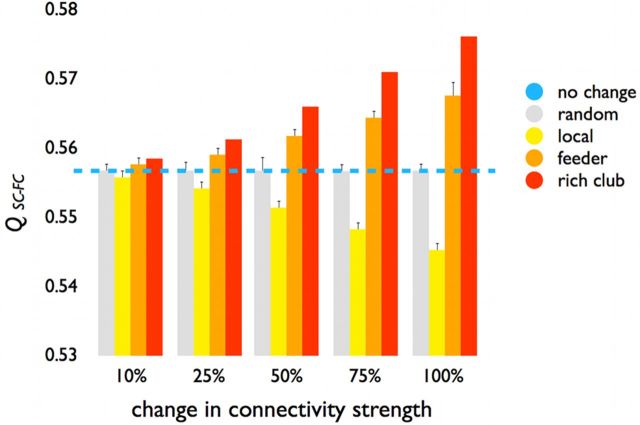
Figure 10.
Structural–functional modularity score. Reducing the weights of rich club and feeder connections (red bars, 10%–100% reduction) resulted in a gradual increase of the Qsc_fc metric, suggesting an increasingly modular partitioning of the brain. In contrast, reducing the weights of the local connections in equal proportions resulted in a decrease in Qsc_fc, indicating a progressively less modular organization of the brain's network. Gray bars represent the effects when the weights of 1000 random sets of connections are reduced (in equal proportions to the other conditions), showing no impact of random weight changes on Qsc_fc. Blue line indicates normal (i.e., 0%) level of Qsc_fc.
Vulnerability analysis
Uniformly reducing the weights of rich club connections by 10%, 25%, 50%, 75%, and 100% resulted in a gradual increase in Qsc_fc (Fig. 10, red bars). Reducing the weights of feeder connections also resulted in a gradual increase in Qsc_fc (Fig. 10, orange bars), but to a smaller extent compared with rich club connections. These findings suggest that rich club and feeder connections stimulate a less segregated or modular architecture, again underscoring their central role for linking segregated functional domains. In contrast, reducing the weights of the local connections by equal proportions resulted in a gradual decrease in Qsc_fc (Fig. 10, yellow bars), indicating that structural local connections tend to facilitate functional segregation and modularity. Reducing the weights by corresponding proportions across 1000 randomly selected sets of connections (i.e., a mixture of rich club, feeder, and local connections) had no effect on Qsc_fc (Fig. 10, gray bars).
Discussion
Our results provide five lines of evidence, which jointly support the notion that the structural rich club is essential for linking segregated functional RSNs of the human brain, thus forming an anatomical substrate for integrating functional communities of the human cortex. First, the structural rich club was found not to be restricted to a single functional RSN or a small set of RSNs. Instead, it involved high-degree hub nodes across all 11 RSNs, including nodes that were part of the default mode, salience, and parietal–frontal networks, as well as more unimodal systems, such as the motor, visual, and auditory networks (Figs. 4 and 5). Second, consistent with this observation, rich club nodes were found to be highly represented among the so-called connector hubs of the network (Fig. 6a), a type of node that has been suggested to be implicated in the integration of information across different parts of the brain network (Sporns et al., 2007; Bullmore and Sporns, 2009; Tomasi and Volkow, 2010; Zuo et al., 2011). Third, structural rich club nodes were predominantly located close to portions of the cortical surface where multiple RSNs overlapped, suggesting a high involvement of rich club members in the so-called functional “confluence zones” of the cerebral cortex, corresponding to locations that are associated with multiple RSNs. Previous studies have indeed demonstrated that such regions (e.g., portions of the medial parietal, superior frontal, and superior parietal cortex) are highly heterogeneous, comprising a large number of specialized areas that are involved in a wide variety of brain functions (Cavanna and Trimble, 2006; Vogt et al., 2006; Margulies et al., 2009; Leech et al., 2012). The heterogeneity of these regions (Fransson et al., 2011; Leech et al., 2012; Spreng et al., 2013) further supports our hypothesis of the cortical rich club, as a system, to involve a diverse set of functional networks and thus provide a key structural substrate for enabling the integration of information between segregated functional networks (Crossley et al., 2013). Fourth, analysis of the topological layout of the network's structural connections and their relationship to functional RSNs showed a disproportionally strong role of rich club and feeder connections in inter-RSN connectivity, with almost half of all structural inter-RSN connections found to be rich club or feeder edges (Fig. 9) (de Reus and van den Heuvel, 2013b). This stands in sharp contrast to the class of intra-RSN connections, which were found to be mainly dominated by local connections (Fig. 9). And fifth, inter-RSN rich club and feeder connections showed a strong involvement in shortest communication paths in the network, suggesting an important role for these connections in global communication across the network. Together, our findings support the notion of the rich club to form, as a system, an anatomical substrate that promotes communication and integration between segregated functional networks of the brain.
The notion of the rich club as a central integrator runs parallel to theories of cognition and consciousness that propose the existence of a distributed neural system as a basis for a “global workspace,” a core system in which segregated functional communities can share their information though means of neuronal interactions (Baars, 1989; Dehaene et al., 1998; Dehaene and Naccache, 2001). In concordance with the relations of the cortical rich club and functional modules as put forward in this paper, these theories hypothesize that the global workspace does not coincide with a single anatomical or functional module in the brain, but rather involves a widely distributed neural system of long-distance anatomical projections. Our findings might suggest that the rich club, characterized by a spatially distributed, yet topologically central organization, is a neurobiological substrate of such a global workspace, supporting the functional coupling and global binding of neural information. With recent graph theoretical advances allowing the study of fast temporal dynamics of functional networks (Bassett et al., 2012; Handwerker et al., 2012; Hutchison et al., 2012; Jones et al., 2012), it might be of particular interest for future studies to examine the structural rich club in the context of transient and dynamic binding of functional modules of the human brain.
While our findings are supportive of the notion that the rich club constitutes a structural skeleton cross-linking all functional communities of the brain, some RSNs appear to have a larger representation in the rich club than others. For example, the motor and extra-striate visual networks were found to be relatively underrepresented, whereas the DMN, in particular its posterior part, and the salience network were found to provide the largest contributions to the rich club (Fig. 5). Notably, inter-RSN connectivity between the DMN, the salience, and the left and right frontal–parietal network was found to be the strongest in the network, and the majority of structural connections linking the more peripheral RSNs (e.g., motor visual, sensory, auditory) to these four densely connected networks were found to comprise rich club and feeder connections. This observation puts forward a possible hierarchical model for the structural relations among RSNs (Fig. 11). The high density of structural connections between the DMN, salience, and task-executive networks suggests that the more cognitive, multimodal networks may act as a structural and functional core, to which other functional networks may be linked as satellites communicating to regions within other RSNs mainly through their representatives in the rich club (Fig. 11). Studies examining network interactions in the functional domain have indeed suggested a multimodal organization of functional connectivity in the human brain (Sepulcre et al., 2012) with a possible central position for the default mode and frontal–parietal task-activation networks (Spreng et al., 2013).
Figure 11.
Structural hierarchy of functional RSNs. Examination of structural inter-RSN connectivity revealed a disproportionately dense level of interconnectivity between the DMN, salience, and left and right parietal networks (see also Fig. 9a,b). This suggests a central role of more cognitive networks in a structural hierarchy of interconnectivity between RSNs, with more primary, unimodal RSNs connected as “spokes” or “satellite networks” to this functionally central core. The figure schematically illustrates such a hypothesized structural hierarchy of functional networks in the human brain.
A simulated reduction of the connectivity strength of rich club connections in a computational vulnerability analysis was shown to result in an increase in the structural–functional modular organization of the network (Fig. 10), which is in support of these connections promoting a less segregated or modular architecture of the brain's network. In this context, one might hypothesize that disease-related reductions in rich club connectivity might have a disproportional effect on brain functioning, in particular on higher-order cognitive processes that depend on global integration of information. Recent studies have indeed suggested that rich club organization may play a potential role in the pathogenesis of schizophrenia (Yu et al., 2013; van den Heuvel et al., 2013), a brain disorder long hypothesized as a disconnection syndrome involving decoupling of psychological brain processes (Friston, 1998; Andreasen, 1999; Stephan et al., 2006). Computational neural mass modeling may provide a powerful approach to further elucidate the dynamical effects of normal and abnormal rich club connectivity to the formation and integration of functional networks in the healthy and diseased brain (Loh et al., 2007; Rubinov and Bassett, 2011).
In our cross-model analysis, functional communities were inferred from resting-state fMRI, and anatomical connectivity on the macroscopic scale was inferred from diffusion imaging. Our results are subject to inherent limitations imposed by these imaging modalities (Johansen-Berg and Rushworth, 2009; Jones, 2010; Jbabdi and Johansen-Berg, 2011). For example, diffusion imaging is known to under-represent the density of long-distance white matter connections in the brain (de Reus and van den Heuvel, 2013a), when results are compared with observations of pathways derived from studies using anatomical tracers. However, analyses of datasets compiled from histological and tract tracing studies have also revealed rich club organization in the nematode Caenorhabditis elegans (Towlson et al., 2013), the cat (Zamora-Lopez et al., 2009), and the macaque (Harriger et al., 2012). Furthermore, the latter study (de Reus and van den Heuvel, 2013b) made the observation that the rich club interlinked all structural modules of the macaque cortex and suggested a pivotal role for rich club and feeder connections in generating global communication paths. Studies that further examine the rich club as an integrative system that bridges not only structural modules but also functional communities in nonhuman species would be of particular interest. These studies could use a cross-modal structural–functional design similar to the one adopted in the present paper, substituting diffusion imaging with weighted network data based on detailed tract-tracing reconstructions (Markov et al., 2011, 2012), combined with measurements of resting or spontaneous neural signal fluctuations from which functional modules and communities can be derived.
In conclusion, our findings suggest that the rich club forms an anatomical substrate for linking and integrating functional RSNs in human cortex. The anatomical infrastructure of the brain's rich club may be critically important for facilitating neuronal signaling and thus global binding of information between functionally segregated domains of the human brain.
Footnotes
M.P.v.d.H. was supported by a VENI Grant of the Netherlands Organization for Scientific Research. O.S. was supported by the J.S. McDonnell Foundation. The authors gratefully thank Marcel de Reus for providing the graph visualization tools for Fig 9a,b.
The authors declare no competing financial interests.
References
- Andersson JL, Skare S. A model-based method for retrospective correction of geometric distortions in diffusion-weighted EPI. Neuroimage. 2002;16:177–199. doi: 10.1006/nimg.2001.1039. [DOI] [PubMed] [Google Scholar]
- Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. A unitary model of schizophrenia: Bleuler's “fragmented phrene” as schizencephaly. Arch Gen Psychiatry. 1999;56:781–787. doi: 10.1001/archpsyc.56.9.781. [DOI] [PubMed] [Google Scholar]
- Baars BJ. A cognitive theory of consciousness. Cambridge: Cambridge UP; 1989. [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanna AE, Trimble MR. The precuneus: a review of its functional anatomy and behavioural correlates. Brain. 2006;129:564–583. doi: 10.1093/brain/awl004. [DOI] [PubMed] [Google Scholar]
- Chang LC, Jones DK, Pierpaoli C. RESTORE: robust estimation of tensors by outlier rejection. Magn Reson Med. 2005;53:1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- Chklovskii DB, Schikorski T, Stevens CF. Wiring optimization in cortical circuits. Neuron. 2002;34:341–347. doi: 10.1016/S0896-6273(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Colizza V, Flammini A, Serrano MA, Vespignani A. Detecting rich-club ordering in complex networks. Nat Physics. 2006;2:5. [Google Scholar]
- Collin G, Sporns O, Mandl RCW, van den Heuvel MP. Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb Cortex. 2013 doi: 10.1093/cercor/bht064. doi: 10.1093/cercor/bht064. Advance online publication. Retrieved April 3, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Vértes PE, Winton-Brown TT, Patel AX, Ginestet CE, McGuire P, Bullmore ET. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc Natl Acad Sci U S A. 2013;110:11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaene S, Naccache L. Towards a cognitive neuroscience of consciousness: basic evidence and a workspace framework. Cognition. 2001;79:1–37. doi: 10.1016/S0010-0277(00)00123-2. [DOI] [PubMed] [Google Scholar]
- Dehaene S, Kerszberg M, Changeux JP. A neuronal model of a global workspace in effortful cognitive tasks. Proc Natl Acad Sci U S A. 1998;95:14529–14534. doi: 10.1073/pnas.95.24.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. Estimating false positives and negatives in brain networks. Neuroimage. 2013a;70:402–409. doi: 10.1016/j.neuroimage.2012.12.066. [DOI] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. Rich club organization and intermodular communication in the cat connectome. J Neurosci. 2013b;33:12929–12939. doi: 10.1523/JNEUROSCI.1448-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Jr, Barch DM, Petersen SE, Schlaggar BL. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P, Aden U, Blennow M, Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state fMRI. Cereb Cortex. 2011;21:145–154. doi: 10.1093/cercor/bhq071. [DOI] [PubMed] [Google Scholar]
- Friston KJ. The disconnection hypothesis. Schizophr Res. 1998;30:115–125. doi: 10.1016/S0920-9964(97)00140-0. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2008;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. Periodic changes in fMRI connectivity. Neuroimage. 2012;63:1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriger L, van den Heuvel MP, Sporns O. Rich club organization of macaque cerebral cortex and its role in network communication. PLoS One. 2012;7:e46497. doi: 10.1371/journal.pone.0046497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2012 doi: 10.1002/hbm.22058. doi: 10.1002/hbm.22058. Advance online publication. Retrieved March 22, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodríguez EJ, Aleman-Gómez Y, Melie-García L. Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory. Neuroimage. 2008;40:1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Jbabdi S, Johansen-Berg H. Tractography: where do we go from here? Brain Connect. 2011;1:169–183. doi: 10.1089/brain.2011.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Rushworth MF. Using diffusion imaging to study human connectional anatomy. Annu Rev Neurosci. 2009;32:75–94. doi: 10.1146/annurev.neuro.051508.135735. [DOI] [PubMed] [Google Scholar]
- Jones DK. Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med. 2010;2:14. [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr Non-stationarity in the “resting brain's” modular architecture. PLoS One. 2012;7:e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol. 2006;2:e95. doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Varier S. Evolution and development of brain networks: from Caenorhabditis elegans to Homo sapiens. Network. 2011;22:143–147. doi: 10.3109/0954898X.2011.638968. [DOI] [PubMed] [Google Scholar]
- Leech R, Braga R, Sharp DJ. Echoes of the brain within the posterior cingulate cortex. J Neurosci. 2012;32:215–222. doi: 10.1523/JNEUROSCI.3689-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh M, Rolls ET, Deco G. A dynamical systems hypothesis of schizophrenia. PLoS Comput Biol. 2007;3:e228. doi: 10.1371/journal.pcbi.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci U S A. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Misery P, Falchier A, Lamy C, Vezoli J, Quilodran R, Gariel MA, Giroud P, Ercsey-Ravasz M, Pilaz LJ, Huissoud C, Barone P, Dehay C, Toroczkai Z, Van Essen DC, Kennedy H, Knoblauch K. Weight consistency specifies regularities of macaque cortical networks. Cereb Cortex. 2011;21:1254–1272. doi: 10.1093/cercor/bhq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey-Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, et al. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs270. doi: 10.1093/cercor/bhs270. Advance online publication. Retrieved Sept. 25, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. Fiber tracking: principles and strategies—a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan RK, Chatterjee N, Sinha S. Mesoscopic organization reveals the constraints governing Caenorhabditis elegans nervous system. PLoS One. 2010;5:e9240. doi: 10.1371/journal.pone.0009240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Bassett DS. Emerging evidence of connectomic abnormalities in schizophrenia. J Neurosci. 2011;31:6263–6265. doi: 10.1523/JNEUROSCI.0382-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Yeo TB, Liu H, Johnson KA. Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. J Neurosci. 2012;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kötter R. Identification and classification of hubs in brain networks. PLoS One. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN, Sepulcre J, Turner GR, Stevens WD, Schacter DL. Intrinsic architecture underlying the relations among the default, dorsal attention, and fronto-parietal control networks of the human brain. J Cogn Neurosci. 2013;25:74–86. doi: 10.1162/jocn_a_00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Baldeweg T, Friston KJ. Synaptic plasticity and dysconnection in schizophrenia. Biol Psychiatry. 2006;59:929–939. doi: 10.1016/j.biopsych.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Functional connectivity density mapping. Proc Natl Acad Sci U S A. 2010;107:9885–9890. doi: 10.1073/pnas.1001414107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towlson EK, Vértes PE, Ahnert SE, Schafer WR, Bullmore ET. The rich club of the C. elegans neuronal connectome. J Neurosci. 2013;33:6380–6387. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31:11. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized group clustering of resting-state fMRI data. PLoS One. 2008;3:e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RCW, Kahn RS, Hulshoff Pol HE. Functionally linked resting state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 2009;30:3127–3141. doi: 10.1002/hbm.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS, Goni J, Sporns O. High cost, high capacity backbone for global brain communication. Proc Natl Acad Sci U S A. 2012;109:11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, Goni J, Hulshoff Pol HE, Kahn RS. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 2013:1–10. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Vogt L, Laureys S. Cytology and functionally correlated circuits of human posterior cingulate areas. Neuroimage. 2006;29:452–466. doi: 10.1016/j.neuroimage.2005.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sui J, Liu J, Plis SM, Kiehl KA, Pearlson G, Calhoun VD. Disrupted correlation between low frequency power and connectivity strength of resting state brain networks in schizophrenia. Schizophr Res. 2013;143:165–171. doi: 10.1016/j.schres.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-López G, Zhou C, Kurths J. Graph analysis of cortical networks reveals complex anatomical communication substrate. Chaos. 2009;19 doi: 10.1063/1.3089559. 015117. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP. Network centrality in the human functional connectome. Cereb Cortex. 2012;22:1862–1875. doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]