Abstract
Nerve growth factor (NGF) is a target-derived neurotrophic growth factor that controls many aspects of sensory and sympathetic neuronal development. The identification of transcription factors and downstream target genes that mediate NGF-dependent neuronal differentiation and target field innervation is currently a major challenge. Here, we show that the Pea3 transcription factor family members Etv4 and Etv5 are expressed by developing TrkA-positive dorsal root ganglion (DRG) neurons during the period of target innervation. Real-time PCR assays indicated that Etv4 and Etv5 mRNAs are significantly induced by NGF in different neuronal cells, suggesting that they could be involved in the biological responses induced by this neurotrophin. Interestingly, distal axon application of NGF in compartmentalized cultures of rat DRG sensory neurons was sufficient to induce a significant increase in Etv4 and Etv5 mRNA expression. Pharmacological assays also revealed that activation of MEK/ERK (MAPK) pathway is required for Etv4 and Etv5 gene induction in response to NGF. Downregulation of Etv4 and Etv5 using small interference RNA knockdown experiments inhibited NGF-induced neurite outgrowth of rat sensory neurons, while overexpression of full-length Etv4 or Etv5 potentiated neuronal differentiation in response to this neurotrophin. Together, these data establish Etv4 and Etv5 as essential molecules of the transcriptional program linking neurotrophin signaling to sensory neuronal differentiation, and suggest that they can be involved in NGF-mediated target innervation.
Introduction
Sensory neuron subtypes in the dorsal root ganglion (DRG) could be characterized by the expression of different neurotrophin receptors. These receptors are essential for survival, target tissue innervation, as well as for the expression of neurotransmitters and ion channels, which define the functional characteristics of the different types of DRG neurons (Huang and Reichardt, 2003; Lallemend and Ernfors, 2012).
The majority of small-diameter DRG neurons, which respond to noxious stimuli and mediate pain sensation, express the nerve growth factor (NGF) receptor TrkA (Holtzman et al., 1992). These neurons project unmyelinated or thinly myelinated fibers to innervate the epidermis and depend on target-derived NGF for their development (Luo et al., 2007; Lallemend and Ernfors, 2012). The physiological relevance of NGF on peripheral neurons has been evidenced by the reduction in the number of sensory and sympathetic neurons observed in NGF mutant mice (Crowley et al., 1994). Moreover, analysis of Bax transgenic mice lacking TrkA revealed that NGF is essential not only for neuronal survival but also for the establishment of correct target innervation (Patel et al., 2000).
Target-derived NGF elicits both local and long-range effects, which require retrograde signaling from the distal axon (DA) to the cell body (CB) and involve the MEK/ERK (MAPK) and PI3K signaling pathway (Barker et al., 2002; Cosker et al., 2008; Ascano et al., 2012; Harrington and Ginty, 2013).
In mammals, peripheral neurotrophic signals have been shown to induce the expression of the Pea3 (polyoma enhancer activator 3) subfamily of ETS (E26 transformation specific) transcription factors, which comprise three members, namely Etv1, Etv4, and Etv5 (also named Er81, Pea3, and Erm, respectively). Several reports propose the different ETS proteins as effectors of the Ras/ERK (MAPK) pathways (Yordy and Muise-Helmericks, 2000; Oikawa and Yamada, 2003; de Launoit et al., 2006; Hollenhorst et al., 2011).
In particular, the three members of the Pea3 family have been described to be expressed in different subpopulations of DRG neurons (Chotteau-Lelièvre et al., 1997; Arber et al., 2000; Hagedorn et al., 2000; Paratore et al., 2002). Interestingly, peripheral neurotrophic signals have been shown to direct the expression of the ETS transcription factors Er81/Etv1 and Etv4 in DRG neurons and motor neuron pools respectively. In particular, Er81/Etv1 is expressed by the majority of DRG proprioceptive neurons in response to NT3 and mediates the formation of proprioceptive afferent-motor neuron connections (Arber et al., 2000; Patel et al., 2003). While the role of Er81/Etv1 in the development of DRG proprioceptive neurons is well established, the role of Etv4 and Etv5 in DRG development remains unknown.
Here, we show that Etv4 and Etv5 are expressed by developing TrkA+ DRG neurons, raising the possibility that they mediate the biological effects triggered by NGF on sensory neurons. Thus, our aim was to explore the role of these ETS transcription factors in the biological response induced by NGF in DRG neurons. The data presented in this study indicates that both Etv4 and Etv5 are induced by peripheral NGF and play an essential role as transcription factors linking distal NGF signaling to axonal growth of DRG sensory neurons.
Materials and Methods
Cell lines, recombinant proteins and inhibitors.
PC12 cells were grown in DMEM supplemented with 5% horse serum and 10% FBS (Invitrogen; Shirazi Fard et al., 2010). COS cells were grown in DMEM supplemented with 10% FBS as previously described (Paratcha et al., 2003). NGF and bFGF were purchased from Promega and R&D Systems respectively. MEK inhibitor PD98059 was from Sigma-Aldrich; the pan-caspase inhibitor N-Benzyloxycarbonyl-Val-Ala-Asp (O-Me) fluoromethyl ketone (Z-VAD-FMK) was obtained from R&D Systems; and the matrix metalloproteinase (MMP) inhibitor GM6001 was purchased from Calbiochem.
Real-time PCR.
The expression of Etv4, Etv5, TATA box binding protein (Tbp), TrkA, and MMP3 mRNAs were analyzed by real-time PCR. Total RNA was isolated from PC12 cells and DRG neurons treated with NGF for the indicated periods of time using RNA-easy columns (Quiagen). cDNA was synthesized using Multiscribe reverse transcriptase and random hexamers (Applied Biosystems). The cDNA was amplified using the following primer sets: TATA box binding protein (Tbp): forward, 5′-GGG GAG CTG TGA TGT GAA GT-3′; reverse, 5′-CCA GGA AAT AAT TCT GGC TCA-3′ (Ledda et al., 2007); rat Etv5: forward, 5′-GAA GTG CCT AAC TGC CAG TCA CCC-3′; reverse, 5′-GGC ACC ACG CAA GTG TCA TCG A-3′; rat Etv4: forward, 5′-ACT CAG ATG TCC CTG GAT GTG-3′; reverse, 5′-TCA CTC CAT CAC CTG AAG AGG-3′; rat TrkA: forward, 5′-CTG CCA GGA CAA ACA GAA CAC-3′; reverse, 5′-GTG GTT GGC TTC GTC TGA GTA-3′; rat MMP3: forward, 5′-GCT ATT CTG GCC ACC TTC TTC-3′; reverse, 5′ TCA TCA TCA CCA TAG GGA AGG-3′; rat MMP13: forward, 5′-GAC AGA TTC TTC TGG CGT CTG-3′; reverse, 5′-GAA GTT CTG GCC AAA AGG ACT-3′. The specificity of the primers was controlled by conventional PCR.
Real-time PCR was performed using the SYBR Green qPCR Master Mix (Invitrogen) on an ABI7500 sequence detection system (Applied Biosystems), according to the manufacturer's instructions. Reactions were performed in 25 μl volume. Nucleotides, TaqDNA polymerase, and buffer were included in the SYBR Green Master Mix (Invitrogen).
Cell transfection, plasmids, and pharmacological treatments.
COS cells were transfected with polyethylenimine (PEI from Polyscience). PC12 cells were transfected using X-tremeGENE (Roche) following manufacturer's instructions. In the indicated cases, the cells were transfected with the corresponding ETS-shRNA expressing green fluorescent protein (GFP; Etv4-shRNA-GFP and Etv5-shRNA-GFP) and selected with puromycin to enrich in transfected cells. PC12-Etv4-shRNA clones and PC12-Etv5-shRNA clones were generated by isolation of colonies transfected with the corresponding constructs and selected with puromycin. Transient transfection of primary DRG neurons was performed using Lipofectamine 2000 (Invitrogen) in 300 μl of DMEM/F12 serum-free medium containing 1 μg of total plasmid DNA per well in 24-well plates. For downregulation experiments, DRG sensory neurons were transfected with 1 μg of ETS-shRNA construct expressing GFP protein. For overexpression experiments, neurons or PC12 cells were cotransfected with HA-tagged-Etv4 or Flag-tagged-Etv5, HA-tagged-MMP3 or Flag-tagged-MMP13 (0.9 μg) constructs, and GFP expression vector (0.1 μg). Etv4-shRNA-GFP, Etv5-shRNA-GFP, HA-tagged-Etv4, Flag-tagged-Etv5, HA-tagged-MMP3, and Flag-tagged-MMP13 were purchased from Cellogenetics. The retroviral vector pRetro-U6G shRNA was used for expression of Etv4-shRNA and Etv5-shRNA targeting rat Etv4 and Etv5. HA-tagged-Etv4 and Flag-tagged-Etv5 were cloned in pRPCX1.0 vector. Plasmid encoding GFP was obtained from Clontech. Pretreatment with the specific MEK inhibitor PD98059 (50 μm) was done for 30 min at 37°C before NGF stimulation. Z-VAD-FMK (Neumar et al., 2003) was used at 50 nm. GM6001 was used at 2.5 μm.
Sensory neuron cultures.
DRG neurons from embryonic day (E) 15 Wistar rats of either sex (School of Pharmacy and Biochemistry, University of Buenos Aires) were prepared as previously described (Baudet et al., 2000). Briefly, the ganglia were dissociated with collagenase (Sigma-Aldrich), trypsin (Invitrogen), and DNase1 (Invitrogen), and then seeded onto plates coated with poly-ornithine (Sigma-Aldrich) and laminin (Sigma-Aldrich). The neurons were maintained in DMEM/F12 medium supplemented with 60 mg/ml penicillin, 100 mg/ml streptomycin, 2 mm glutamine (Invitrogen), 1 mg/ml BSA (Sigma-Aldrich), and NGF (50 ng/ml).
Compartmentalized cultures were performed as previously described (Campenot, 1994; Ledda et al., 2002). Briefly, 60 mm culture dishes were coated with rat-tail collagen, and parallel traces in the substrate were made by scratching the culture dish surface with a pin rake (Tyler Research Instruments). Subsequently, a Teflon divider (Tyler Research Instruments) was seated on top of the traces as described previously (Campenot, 1994). Dissociated E15 rat DRG neurons were plated in one compartment of a two-compartment chamber and maintained for 2 d in DMEM/F12 supplemented with 2 mm glutamine, 1 mg/ml BSA, and 100 ng/ml of NGF. After 2 d, the medium was replaced with DMEM/F12 containing 10–20 ng/ml of NGF in the CB compartment and 100 ng/ml of NGF in the side (DA) compartment. After 7 d, the cells were starved and then treated with NGF in the CB or DA compartment. Following 8 h of treatment, the CBs were lysed for RNA extraction, and analyzed for Etv4 and Etv5 expression.
Western blotting.
Cells were lysated at 4°C in buffer containing 0.5% Triton X-100, 1% octylglucoside plus protease, and phosphatase inhibitors. Protein lysates were clarified by centrifugation and analyzed by Western blotting as previously described (Ledda et al., 2008). The blots were scanned in a Storm 845 PhosphorImager (GE Healthcare Life Sciences).
The antibodies were obtained from various sources as follows: anti-Etv5 (H-100), anti-Etv4 (G-10), and anti-actin were from Santa Cruz Biotechnology; anti-TrkA was from R&D Systems; anti-Flag antibodies were from Sigma-Aldrich; anti-phospho-MAPK was from New England Biolabs; and anti-βIII-tubulin was from Promega.
Immunofluorescence.
Cryostat sections of rat embryos at different developmental stages and dissociated DRG neurons of either sex were fixed with 4% PFA, permeabilized with 0.25% Triton X-100, blocked with 10% donkey serum (Jackson ImmunoResearch), and then incubated with polyclonal anti-TrkA (dilution 1:200 on tissue sections and 1:1000 on cultured cells; R&D Systems), anti-Ret (dilution 1:200 on tissue sections; R&D Systems), anti-Etv4 (dilution 1:200 on tissue sections and 1:800 on cultured cells), and/or anti-Etv5 (dilution 1:200 on tissue sections and 1:800 on cultured cells; Santa Cruz Biotechnology) antibodies, anti-βIII-tubulin (dilution 1:3000; Promega), anti-Sox2 (dilution 1:400; Sigma-Aldrich). Anti-Er81/Etv1 (1:2000) was kindly provided by Dr. Silvia Arber (Basel University, Basel, Switzerland). Second antibodies were from Jackson ImmunoResearch. For each developmental stage, lumbar DRGs coming from 4–6 rats were isolated and embedded in optimum cutting temperature tissue-embedding medium. Ten-micrometer-thick serial cryosections were made through the entire DRG and counted every 30 μm. Photographs were obtained using an Olympus IX-81 inverted or an Olympus confocal microscope.
Neurite outgrowth assays.
For PC12 cell differentiation assays, the cells were transfected with the different ETV-shRNAs using X-tremeGENE reagent in complete medium. The next day, the cells were plated on coverslips coated with poly-d-lysine (Sigma-Aldrich) in 24-well plates and cultured in DMEM medium (control condition) or DMEM supplemented with NGF (50 ng/ml). After 72 h, the cells were fixed with 4% paraformaldehyde (PFA). The number of cells bearing neuritis longer than 1.5 or 2 CBs (as specified) was quantified relative to the total number of cells counted in at least 10 random fields of three different wells in each experiment. PC12 cell differentiation was evaluated in three independent experiments. The pictures were obtained using an Olympus IX-81 inverted microscope.
Neurite outgrowth assays were performed in dissociated cultures of E15 rat DRG neurons. Primary cultures were prepared as previously described (see above). Neurons were transfected with Etv4-shRNA-GFP, Etv5-shRNA-GFP, HA-tagged-Etv4, Flag-tagged-Etv5, HA-tagged-MMP3, Flag-tagged-MMP13, or control plasmid expressing GFP and cultured in the presence of NGF (50 ng/ml) for 24 or 36 h. Then, cells were fixed with 4% PFA and stained with anti-βIII-tubulin to identify neuronal cells. Neuronal survival was evaluated using the nuclear stain 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich). GFP-positive neurons containing fragmented or condensed nuclear staining were scored as apoptotic cells and not computed in the differentiation assays. Quantification of neurite length was performed using National Institutes of Health ImageJ software. The pictures were obtained using an Olympus IX-81 inverted microscope.
shRNA-mediated knockdown assays.
Rat Etv4-shRNA-GFP and Etv5-shRNA-GFP expression vectors were purchased from Cellogenetics. The retroviral vector pRetro-U6G shRNA was used for expression of Etv4-shRNA and Etv5-shRNA targeting rat Etv4 and Etv5. The target sequence of the Etv5-shRNA is 5′-GGATCACAGCAGCAAACATTT-3′, and corresponds to nucleotides 691–711 of rat Etv5 mRNA. The target sequence of the Etv4-shRNA is 5′-GCAGCAAATCTCCCGGAAATG-3′, and corresponds to nucleotides 324–344 of rat Etv4 mRNA. These regions were not homologous to other ETS transcription factors or other known genes determined by a BLAST search. The specificity of Etv4 and Etv5 knockdown were confirmed by real-time PCR, Western blot, and immunofluorescence.
Statistical analysis.
Statistical analysis was performed in GraphPad Prism 5.0. In the indicated cases, Student's t test or one-way ANOVA analysis followed by Dunnett or Newman–Keuls post-test was performed.
Results
Expression of Pea3 transcription factor family members during DRG development
Although the role of the ETS transcription factor Er81/Etv1 in DRG development is well established, the role of Etv4 and Etv5 in the peripheral nervous system development is still unclear. To address this, the expression of Etv4 and Etv5 mRNA was analyzed by real-time PCR in rat DRG tissue at different developmental stages (Fig. 1A). A developmental increase in Etv4 and Etv5 mRNA expression was detected between E15 and E17, a period in which sensory neurons encounter target-derived neurotrophins, such as NGF.
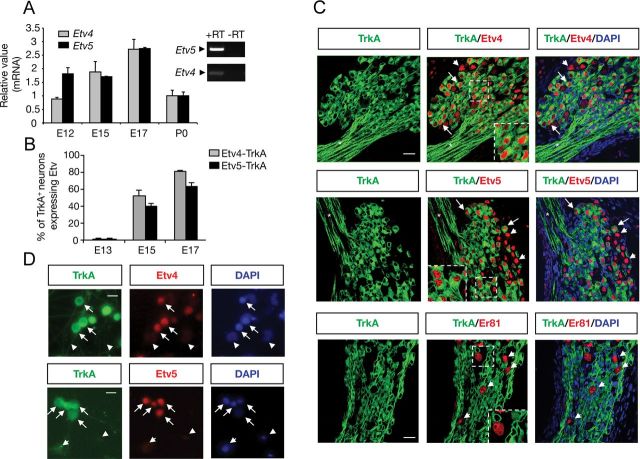
Figure 1.
Developmental expression and localization of Etv4 and Etv5 in DRG neurons. A, Quantitative analysis of developmental expression of Etv4 and Etv5 mRNA by real-time PCR in DRG neurons in E12, E15, E17, and newborn (P0) mice. Expression at each age was normalized to that of the housekeeping gene Tbp and is stated relative to the expression at postnatal day 0 (P0). Shown are averages ± SD of triplicate determinations. Right, Etv4 and Etv5 mRNA expression by PCR in DRG. Control samples without reverse transcriptase (−RT) were included. B, Quantitative analysis of developmental coexpression of either Etv4 or Etv5 with TrkA in lumbar DRG sections. The histogram shows the average ± SEM. C, Immunofluorescence in E16 DRG sections. Top, Staining for Etv4 (red). Middle, Staining for Etv5 (red). Bottom, Staining for Er81/Etv1 (red). Green indicates staining for TrkA. High-magnification images of boxed areas are also shown. Arrows indicate individual cells showing Etv4-TrkA or Etv5-TrkA coexpression. Arrowheads indicate cells expressing Etv4, Etv5, or Er81 in the absence of TrkA staining. Asterisk indicates fibers exiting the ganglion labeled with TrkA but lacking ETS immunoreactivity. Scale bar, 30 μm. D, Localization by immunofluorescence of Etv4 (top, red), Etv5 (bottom, red), and TrkA (green) in DRG-dissociated neurons. Arrows indicate cells showing Etv4–TrkA or Etv5–TrkA coexpression. Arrowheads indicate cells negative for TrkA and the different ETS. Scale bars, 15 μm.
To determine whether these ETS transcription factors could play an in vivo role in NGF/TrkA signaling, we examined the expression pattern of Etv4, Etv5, and the NGF receptor TrkA in transverse sections of DRGs by immunofluorescence using specific antibodies. Quantitative assessment of Etv4 and Etv5 expression in TrkA-positive neurons is shown in Figure 1B. Coexpression of either Etv4 or Etv5 with TrkA was not observed at early developmental stages. At E13, Etv4 and Etv5 were detected in cells negative for TrkA and positive for the migratory neural crest cell marker Sox2 (Wakamatsu et al., 2000). From E15 to E17, the expression of the two transcription factors were detected in a large number of cells expressing TrkA (Fig. 1B,C). Quantitative determinations (average ± SEM) indicate that at E17, 80.7 ± 1.5% of TrkA-positive neurons expressed Etv4 and 63.3 ± 4.1% of TrkA-positive neurons expressed Etv5. Furthermore, at this developmental stage, the majority of the cells positive for Etv5 and TrkA also expressed Etv4 (percentage of cells positive for TrkA, Etv4, and Etv5: 61.9 ± 3.0%; average ± SEM).
Our analysis also revealed that ∼80% of the Etv4 and Etv5 are localized in TrkA-positive neurons, supporting a relevant contribution of these factors to the biological response triggered by NGF. It is known that many TrkA-positive neurons at E16 begin to coexpress Ret (Luo et al., 2007). Interestingly, we observed that at E17 Etv4 and Etv5 were expressed by the majority of Ret-positive cells. In addition, we found that 8.7 ± 1.1% (average ± SEM) of Etv4-expressing cells and 10.4 ± 2.3% of Etv5-expressing cells were negative for TrkA and Ret. These DRG cells expressing Etv4 and Etv5 in non-TrkA/Ret cells were also negative for the glial cell marker S100β, indicating that they were neurons (data not shown).
In agreement with previous results, expression of these transcription factors was not detected in the nuclei of presumptive Schwann cells surrounding fibers exiting the DRG (Paratore et al., 2002; Fig. 1C). The expression of Er81/Etv1 was also evaluated in vivo in DRG at E16. As shown in Figure 1B (bottom), and in agreement with previous results, Er81/Etv1 was excluded from TrkA-positive neurons and only detected in few TrkA-negative cells with large nuclei.
Primary dissociated DRG neurons obtained from E15 rats and maintained in the presence of NGF for 48 h, were also assessed by immunofluorescence using either anti-Etv5 or anti-Etv4, and anti-TrkA antibodies. In agreement with the in vivo results, the staining revealed a striking coexpression of the two transcription factors with the NGF receptor TrkA in DRG cultured neurons (Fig. 1D). Antibody specificity was confirmed by knockdown of Etv4 and Etv5 expression by immunoblot (see Fig. 4D) and immunofluorescence (data not shown).
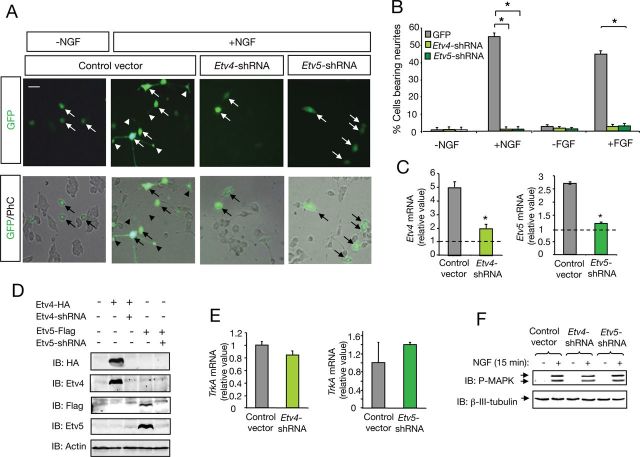
Figure 4.
Etv4 and Etv5 are independently required for neuronal differentiation induced by NGF. A, Photomicrographs show PC12 cells transfected with control GFP expression vector (Control), Etv4-shRNA, or Etv5-shRNA plasmids, each expressing GFP. After 72 h of NGF treatment (50 ng/ml), the cells were fixed. Arrows indicate neuronal CBs and arrowheads denote neurite tips. Scale bar, 20 μm. B, The histogram shows the quantification of the relative number of GFP-positive PC12 cells bearing neurites longer than 1.5 CB diameters after 72 h of treatment with NGF (50 ng/ml) or FGF (25 ng/ml). The results are presented as average ± SD of a representative experiment performed in triplicate. *p < 0.001 (ANOVA followed by Student–Newman–Keuls). The experiment was repeated at least three times with similar results. C, Etv4 and Etv5 mRNA levels were analyzed by real-time PCR from PC12 cells transfected with control vector (Ctrl), Etv4-shRNA, or Etv5-shRNA. Transfected cells were enriched by puromycin treatment to increase the population of cells containing shRNA constructs. Cells were maintained in control conditions (nontreated) or stimulated 8 h with NGF. Histogram shows fold changes relative to control cells (nontreated cells, dotted line). Quantitative analysis is shown as average ± SD of triplicate. *p < 0.05 (Student's test). D, Etv4 and Etv5 protein levels were analyzed by immunoblotting (IB) in COS cells transfected with HA-tagged-Etv4, Flag-tagged-Etv5 alone, or together with Etv4-shRNA or Etv5-shRNA respectively. Downregulation of Etv4 was probed using anti-HA and Etv4 antibodies. Downregulation of Etv5 was probed using anti-Flag and Etv5 antibodies. Reprobing with anti-actin is shown as loading control. E, TrkA mRNA levels were analyzed by real-time PCR from PC12 cells transfected with control vector, Etv4-shRNA, or Etv5-shRNA constructs. Quantitative analysis is shown as averages ± SD of triplicate determinations. The levels of Etv4, Etv5, and TrkA mRNAs were normalized using the expression of the housekeeping gene Tbp. F, MAPK activation (P-MAPK) in cell lysates of PC12 cells and PC12 cells transfected with Etv4-shRNA or Etv5-shRNA treated with NGF for 10 min and detected by IB. Reprobing of the same blot with βIII-tubulin is shown.
The expression of Etv4 and Etv5 in TrkA+ sensory neurons, together with their upregulation during the period of NGF-dependent tissue invasion, suggested that these factors could mediate axonal growth and target innervation in response to this neurotrophin during embryonic development.
NGF induces Etv4 and Etv5 expression through a MAPK-dependent pathway
It is known that NGF signaling through TrkA is essential for the expression of numerous genes involved in survival, differentiation, axonal outgrowth, and target innervation of cutaneous sensory neurons (Chen et al., 1999; Huang and Reichardt, 2001, 2003). Interestingly, Etv4 and Etv5 have been involved in the expression control of a number of genes important for neuronal outgrowth (Oikawa and Yamada, 2003; de Launoit et al., 2006; Hollenhorst et al., 2011).
To address whether Etv4 and Etv5 are NGF-regulated genes, we examined the mRNA expression pattern of these two ETS transcription factors in response to NGF in dissociated DRG neurons and PC12 cells, two well established models responsive to NGF. DRG-derived neurons from E15 rat embryos were cultured and exposed to this neurotrophin for different times. Analysis by real-time PCR revealed a significant induction of Etv4 and Etv5 mRNA in DRG neurons exposed to NGF compared with untreated controls (Fig. 2A). This finding indicates that the expression of Etv4 and Etv5 in developing neurons is regulated by NGF. The expression pattern of the two ETS mRNAs in response to NGF was also evaluated in PC12 cells. This quantitative analysis also revealed a significant induction of Etv4 and Etv5 mRNA in this neuronal cell line (Fig. 2B).
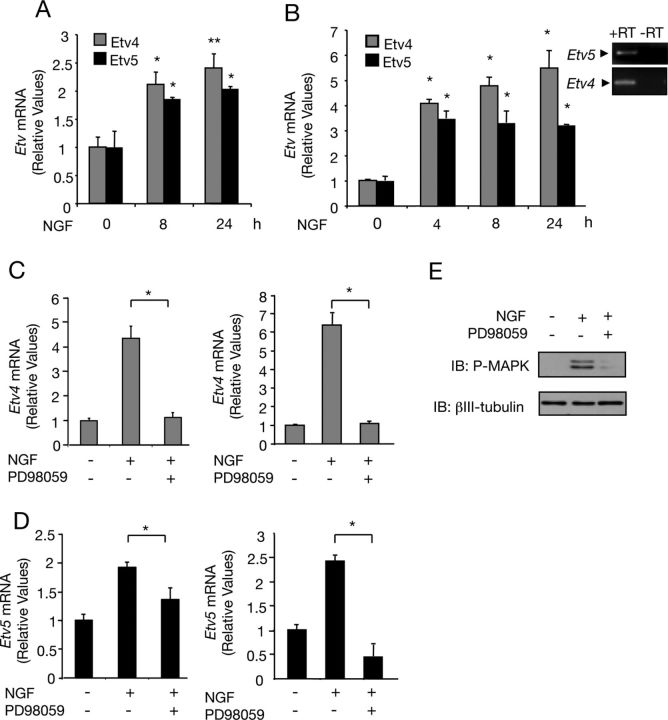
Figure 2.
NGF signaling induces Etv4 and Etv5 gene expression in neuronal cells. A, B, Analysis of Etv4 and Etv5 mRNA expression by real-time PCR in DRG-derived neuronal cultures (A) and PC12 cells (B) treated with NGF (50 ng/ml) for the indicated times. The levels of Etv4 and Etv5 mRNA were normalized using the expression of the housekeeping gene Tbp. Shown are averages ± SD of triplicate determinations. A, *p < 0.01, **p < 0.001; B, *p < 0.001, NGF-treated versus control (nonstimuated) group (ANOVA followed by Dunnett's test). B, Top right, Etv4 and Etv5 mRNA by PCR in PC12 cells. C, D, Analysis of Etv4 (C) and Etv5 (D) mRNA expression by real-time PCR in DRG (left) and PC12 (right) cells treated first with the specific MEK inhibitor PD98059 (50 μm) for 30 min, and then stimulated with NGF (50 ng/ml) for 10 min. The levels of Etv4 and Etv5 mRNA were normalized using the expression of the housekeeping gene Tbp. Shown are averages ± SD of triplicate determinations. *p < 0.01 (ANOVA followed by Student–Neuman–Keuls). E, The inhibition of the MAPK by PD98059 was tested measuring MAPK activation (P-MAPK) by immunoblotting (IB) of PC12 cells stimulated for 10 min with NGF (50 ng/ml). Reprobing was done with antibodies against βIII-tubulin.
NGF activates different signaling pathways to trigger its biological effects, including the MEK/ERK (MAPK) signaling cascade (Markus et al., 2002; Zhong et al., 2007). Moreover, different reports indicate that ETS proteins act as effectors of the MEK/ERK (MAPK) pathway (Wasylyk et al., 1998; Yordy and Muise-Helmericks, 2000). Using a pharmacological approach, we investigated whether NGF-induced Etv4 and Etv5 gene expression was mediated by this signaling pathway. To this end, DRG and PC12 cells were pretreated with the MEK inhibitor PD98059 (Alessi et al., 1995) and then stimulated with NGF. The mRNA level of the two ETS transcription factors was determined by real-time PCR. As shown in Figure 2C,D, the MEK inhibitor blocked NGF-induced gene expression of the Etv4 and Etv5 transcription factors, demonstrating that MEK/ERK (MAPK) pathway activation was required for NGF induction of the two ETS transcription factors. The inhibitory activity of PD98059 was confirmed by measuring MAPK activation by immunoblotting of PC12 total lysates obtained from cells maintained in the presence or absence of PD98059 and stimulated for 10 min with NGF (Fig. 2E).
NGF-retrograde signaling induces Etv4 and Etv5 mRNA transcription
During development, sensory neurons extend axons across long distances to innervate postsynaptic target cells. It is known that NGF released by target tissues support neuronal growth and survival of specific DRG neuronal subpopulations that express TrkA receptors. The mechanisms by which the information triggered by NGF at the nerve terminal is retrogradely transported to the CB and nucleus have been under discussion for several years. During the last decade, several experiments have demonstrated that retrograde transport of the signals originated by target-derived NGF is required for gene transcription, survival response, axonal growth, and connectivity (Riccio et al., 1997; Ginty and Segal, 2002; Ye et al., 2003; Harrington and Ginty, 2013). To determine whether the induction of Etv4 and Etv5 could be triggered by TrkA activation in DAs, we cultured E15 DRG sensory neurons in compartmentalized chambers, allowing growth of neurites to the distal compartment (see Fig. 3A). After starvation, DA or CB compartments were stimulated with saturating NGF concentration for 8 h and the levels of Etv4 and Etv5 mRNA were measured by real-time PCR. Our results revealed that either CB or DA stimulation with NGF induced a significant increase in the levels of Etv4 and Etv5 mRNAs, indicating that retrograde propagation of TrkA signaling is sufficient to induce the transcription of both ETS genes (Fig. 3), suggesting a possible role of these factors in NGF-dependent target innervation. As DRG nociceptive TrkA+ neurons critically depend on NGF secreted by peripheral tissues for their survival, axonal growth, and differentiation, this finding establishes Etv4 and Etv5 as critical mediators of the transcriptional response linking target-derived NGF to axonal growth.
Figure 3.
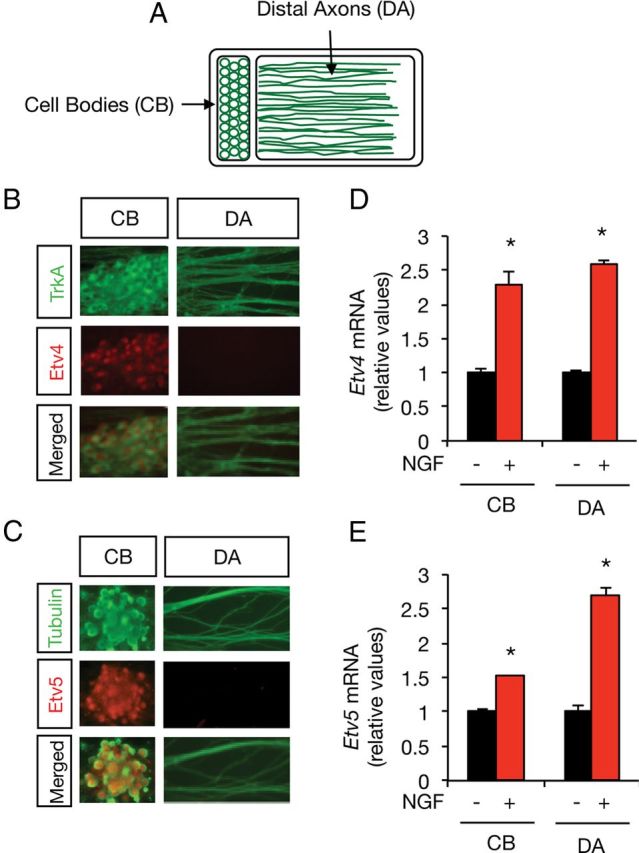
Etv4 and Etv5 gene expression is regulated by DA exposure to NGF. A, Schematic representation of compartmentalized cultures of DRG-derived neurons showing CB and DA compartments. B, C, Sensory neurons were grown in compartmentalized chambers for 7 d in the presence of distal NGF (100 ng/ml) and immunostained with anti-Etv4 (red) and anti-TrkA (green; B), or anti-Etv5 (red) and anti-βIII-tubulin (green; C). Note specific staining of Etv4 and Etv5 in the CB compartment. D, E, Analysis of Etv4 and Etv5 mRNA levels after stimulation with NGF on DAs or CBs. DRG neurons were grown in compartmentalized chambers for 7 d. After 16 h of starving, the DA or the CB compartment was stimulated for 8 h with NGF (100 ng/ml). Total RNA was prepared from the CB compartment and the expression of Etv4 and Etv5 mRNA was analyzed by real-time PCR. The levels of Etv4 and Etv5 mRNA were normalized using the expression of the housekeeping gene Tbp. Shown are averages ± SD of triplicate determinations. D, CB: *p < 0.001 (Student's t test, t = 10.55); DA: *p < 0.001 (Student's t test, t = 36.33); E, CB: *p < 0.001 (Student′s t test, t = 16.45); DA: *p < 0.001(Student′s t test, t = 22.9).
Etv4 and Etv5 are independently required for NGF-dependent neurite outgrowth
Previous work showed that NGF-induced axonal growth of sensory neurons requires MEK/ERK (MAPK) signaling. To investigate whether Etv4 and/or Etv5 play a role in NGF-induced cell differentiation, PC12 cells were transiently transfected with different vectors containing specific shRNA directed to the rat Etv4 or Etv5 mRNA sequence. These shRNA vectors were able to abolish the levels of ectopically expressed Etv4 and Etv5 protein in COS cells (Fig. 4D). PC12 transfected with Etv4-shRNA, Etv5-shRNA expressing GFP vectors, or GFP expressing control plasmid were maintained in the presence of NGF during 72 h and then analyzed for neuronal differentiation. In cells transfected with control plasmids, NGF treatment induced neurite outgrowth and growth cones were present at the tips of neurites. In contrast, downregulation of Etv4 and Etv5 by RNA interference abrogated NGF-induced neurite outgrowth (Fig. 4A,B). No effect on cell survival was observed in the different groups. Similar results were obtained in three stable PC12 clones overexpressing each ETS-shRNA construct (data not shown). Downregulation of the two transcription factor mRNAs in PC12 cells was controlled by real-time PCR (Fig. 4C). Since Etv4 and Etv5 have been described as mediators of fibroblast growth factor (FGF) signaling (Mao et al., 2009; Zhang et al., 2009; Znosko et al., 2010), we tested their requirement for the differentiation triggered by this trophic factor in PC12 cells. In agreement with the results obtained with NGF, a complete abrogation of neurite outgrowth in response to FGF was observed (Fig. 4B).
The abrogation of the morphological differentiation induced by NGF in PC12 cells prompted us to examine whether downregulation of the ETS leads to a reduction of TrkA receptor and its downstream signaling effector MAPK. To test this idea, we analyzed the expression of the NGF receptor TrkA by real-time PCR and the NGF-dependent activation of MAPK by immunoblot in cells transfected with Etv4-shRNA or Etv5-shRNA constructs. Transfected cells were enriched by puromycin treatment to increase the population of cells expressing each ETS-shRNA construct. No difference was detected in the levels of TrkA mRNA in PC12 cells transfected with control, Etv4-shRNA, or Etv5-shRNA vectors (Fig. 4E). In addition, similar levels of MAPK activation in response to NGF were observed in all conditions (Fig. 4F), indicating that the complete abrogation of NGF-dependent differentiation was not due to reduced expression of NGF receptor TrkA or to altered MAPK signaling.
Together, these results indicate that both Etv4 and Etv5 transcription factors are necessary for NGF-triggered differentiation, but this is not mediated by the regulation of TrkA expression/MAPK signaling.
Etv4 and Etv5 are involved in axonal growth of DRG sensory neurons in response to NGF
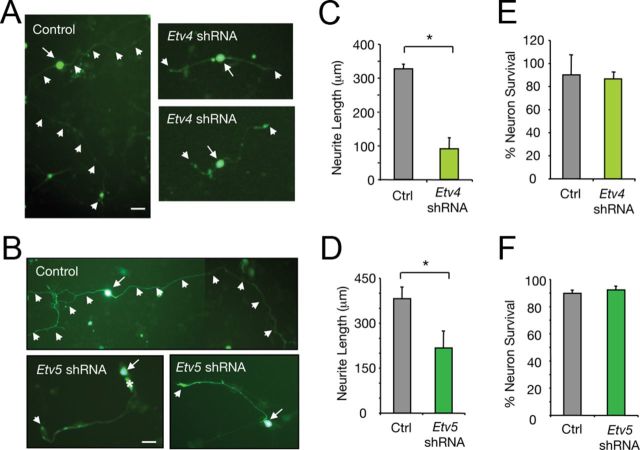
The results obtained in PC12 cells lead us to examine the physiological significance of Etv4 and Etv5 as mediators of NGF signaling in primary neurons. To this end, we introduced Etv4-shRNA and Etv5-shRNA constructs expressing GFP into sensory neurons isolated from E15 DRG. At this developmental stage, the majority of DRG-derived neurons express high levels of TrkA and respond to NGF. After transfection, the cells were maintained in the presence of NGF for 36 h, fixed, and analyzed for neuronal differentiation. In agreement with the results obtained in PC12 cells, the neurite outgrowth stimulated by NGF was significantly reduced in DRG neuronal cells transfected with Etv4-shRNA or Etv5-shRNA. While Etv5 downregulation resulted in a significant decrease in the neurite length, a stronger inhibition of neurite outgrowth was observed in the presence of Etv4-shRNA (Fig. 5). To exclude the possibility that the effects seen with Etv4-shRNA or Etv5-shRNA were due to apoptosis, the morphology of the nuclei was assessed using the nuclear staining DAPI. Survival was similar in control and shRNA-transfected neurons (Fig. 5E,F). Thus, our findings demonstrate that Etv4 and Etv5 are required for sensory neuron axonal growth in response to NGF.
Figure 5.
Etv4 and Etv5 are involved in NGF-induced sensory neuron differentiation. A, B, Dissociated DRG neurons transfected with control, Etv4-shRNA (A), or Etv5-shRNA (B) expressing GFP constructs and maintained in the presence of NGF (50 ng/ml). After 36 h in culture, neurons were fixed and stained with anti-βIII-tubulin antibodies (data not shown). Scale bars, 20 μm. Arrows indicate neuronal CBs and arrowheads indicate neurite trajectory and neurite tips. C, D, Histograms show the inhibition of neurite outgrowth in DRG neurons by knockdown of Etv4 (C) or Etv5 (D) expression. The length of the longest neurite was measured. Results are shown as the average ± SD of a representative experiment performed in triplicate. C, *p = 0.0135 (Student's t test, t = 4.2); D, *p = 0.0003 (Student's t test, t = 11.6). The experiment was repeated at least two times with similar results. E, F, Histograms showing the survival of DRG neurons transfected with control, Etv4-shRNA (E) or Etv5-shRNA (F). Neuronal survival was evaluated using DAPI for the nuclear staining. GFP-positive neurons containing fragmented or condensed nuclear staining were scored as apoptotic cells. The results are averages ± SD of a representative experiment performed in triplicate.
Etv4 and Etv5 overexpression potentiates neuronal differentiation in response to NGF
Our data indicating that knockdown of Etv4 and Etv5 restricts neurite outgrowth induced by NGF prompted us to analyze whether overexpression of Etv4 or Etv5 is sufficient to promote neurite outgrowth. For this purpose, PC12 cells were transfected with control, HA-tagged-Etv4, or Flag-tagged-Etv5 plasmids in combination with an enhanced GFP expression vector. Cells were maintained in the presence of NGF for 72 h and analyzed for neurite outgrowth response. Consistent with a role of Etv4 and Etv5 in the control of TrkA-mediated downstream signaling, a significant potentiation of PC12 cell differentiation was observed in cells overexpressing each transcription factor. This effect was only evidenced in the presence of NGF (Fig. 6A,B). In agreement with this, Etv4 and Etv5 overexpression in sensory DRG neurons also potentiated axonal growth in the presence of NGF (Fig. 6C--E). Similar results were observed in the presence of the apoptotic inhibitor Z-VAD-FMK (50 nm), indicating that this effect is independent of neuronal survival (Fig. 6F). Together, the data indicate that either Etv4 or Etv5 overexpression potentiates the morphological differentiation triggered by NGF. However, experiments in PC12 and DRG neurons revealed that overexpression of either Etv4 or Etv5 is not sufficient to induce maximal outgrowth response in the absence of NGF (Fig. 6B,F). Only overexpression of Etv4 in DRG neurons revealed a marginal, but consistent, increase in neurite outgrowth in the absence of NGF (Fig. 6F). Thus, post-translational modifications or the synthesis of other transcriptional components induced by NGF are required for maximal NGF-induced morphological differentiation.
Figure 6.
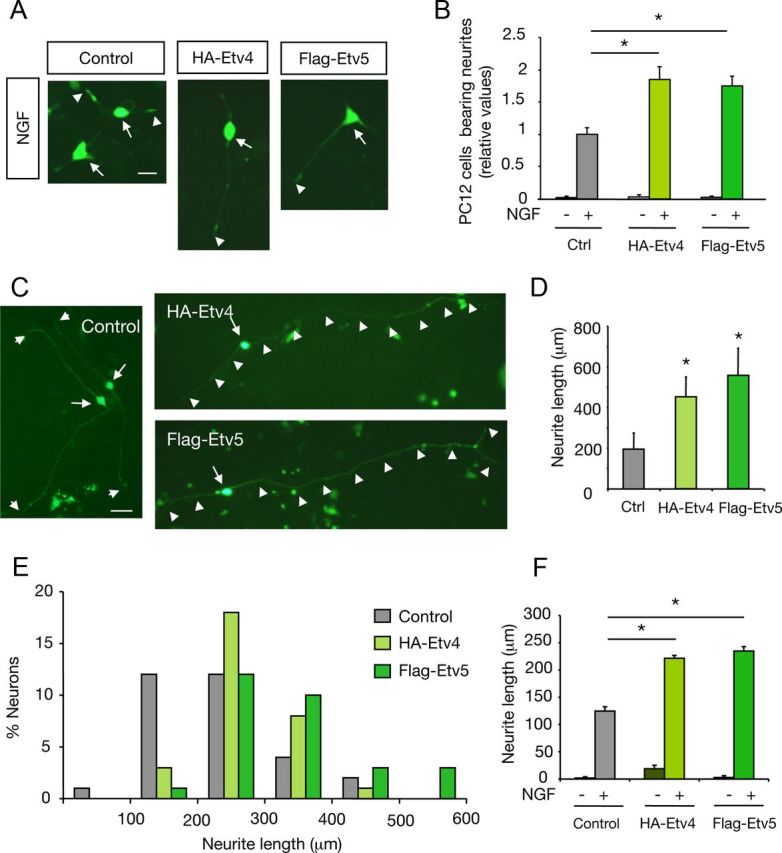
Overexpression of Etv4 and Etv5 potentiates PC12 cell differentiation in response to NGF. A, Photomicrographs show PC12 cells transfected with control vector, HA-tagged Etv4 (HA-Etv4), or Flag-tagged Etv5 (Flag-Etv5) constructs together with a GFP expression vector. After 72 h maintained in the presence of NGF (50 ng/ml), cells were fixed and analyzed. Arrows indicate neuronal CBs and arrowheads denote neurite tips. Scale bar, 15 μm. B, The histogram shows the quantification of the relative number of GFP-positive neurite-bearing cells longer than two CB diameters in the different experimental conditions. The results are shown as averages ± SD of a representative experiment performed in triplicate. *p < 0.01 (ANOVA followed by Student–Newman–Keuls). The experiment was repeated three times with similar results. C, Photomicrographs show DRG cells transfected with control vector, HA-Etv4, or Flag-Etv5 constructs together with GFP expression vector. After 24 h maintained in the presence of NGF (50 ng/ml), cells were fixed and analyzed. Arrows indicate neuronal CBs and arrowheads indicate the neurite trajectory and neurite tips. D, The histogram shows quantification of neurite length. The longest neurite was measured. Results are shown as the average ± SD of a representative experiment performed in triplicate. The experiment was repeated three times with similar results. p < 0.01 (ANOVA, followed by Dunnett's test). E, The histogram shows the distribution of neurons carrying neurites classified in different length categories after transfection with control, HA-Etv4, or Flag-Etv5 vectors. F, Histogram showing the quantification of neurite outgrowth in DRG neurons overexpressing either HA-Etv4 or Flag-Etv5 constructs together with GFP expression vector cultured in the presence of the apoptosis inhibitor Z-VAD-FMK (50 nm). The cells were maintained as indicated in the absence or presence of NGF during 24 h, fixed, and analyzed. The longest neurite was measured. Results are shown as the average ± SEM of a representative experiment performed in triplicate *p < 0.001 HA-Etv4 and Flag-Etv5 versus control in the presence of NGF (ANOVA, followed by Student–Newman–Keuls).
Etv4 and Etv5 mediate NGF-dependent MMP3 and MMP13 gene expression
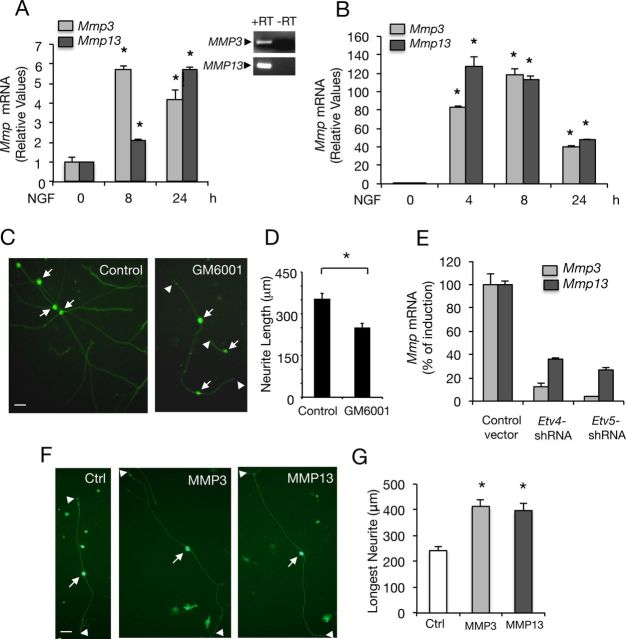
ETS family proteins regulate gene expression of several genes by functional interaction with other transcription factors (Hollenhorst et al., 2011). Among the genes that have been described to be regulated by Pea3 transcription factors are several members of the MMP family (i.e., MMP3, MMP10, and MMP13), which have been implicated in extracellular matrix remodelling that occurs during normal development and in pathological conditions (Page-McCaw et al., 2007). In particular, MMP3 (also named stromelysin-1) has been reported to be highly induced by NGF and to mediate axonal growth and growth cone invasiveness through extracellular matrix barriers (Nordstrom et al., 1995). In agreement with this, we observed a significant induction of MMP3 and MMP13 (also named collagenase 3) in PC12 cells and DRG neurons in response to NGF (Fig. 7A,B). In addition, NGF-mediated neurite outgrowth was blocked in PC12 cells (data not shown) and DRG neurons in the presence of the MMP inhibitor GM6001 (Fig. 7C,D).
Figure 7.
NGF regulates MMP3 and MMP13 expression through Etv4/Etv5-dependent transcription. A, B, Quantitative analysis of Mmp3 and Mmp13 mRNA expression by real-time PCR in DRG neurons (A) and PC12 cells (B) treated with NGF (50 ng/ml) during the indicated times. The levels of Mmp3 and Mmp13 mRNA were normalized using the expression of the housekeeping gene Tbp. Shown are averages ± SD of triplicate determinations. *p < 0.001 NGF-treated versus control (nonstimulated) group (ANOVA followed by Dunnett's test). A, Top, Expression of Mmp3 and Mmp13 examined by PCR in PC12 cells treated with NGF (50 ng/ml). Control sample without reverse transcriptase (−RT) was included. C, Photomicrographs show DRG cells grown in the presence of NGF in control conditions or in the presence of the MMP inhibitor GM6001 (2.5 μm) for 36 h. Scale bar, 30 μm. D, Quantification of neurite outgrowth observed in C. *p = 0.0001 (Student's t test, t = 14.78). E, Mmp3 and Mmp13 mRNA levels were analyzed by real-time PCR from PC12 cells transfected with control vector, Etv4-shRNA, or Etv5-shRNA followed by 8 h of treatment with NGF. The levels of Mmp3 and Mmp13 mRNA were normalized using the expression of the housekeeping gene Tbp. Transfected cells were enriched by puromycin treatment to increase the population of cells containing shRNA constructs. Percentage of Mmp3 and Mmp13 mRNA induction in the presence of NGF relative to untreated conditions is indicated for each experimental group. F, Photomicrographs show DRG cells transfected with control vector, HA-MMP3 or Flag-MMP13 constructs together with GFP expression vector. After 24 h maintained in the presence of NGF (50 ng/ml), cells were fixed and analyzed. Scale bars, 30 μm. Arrows indicate neuronal CBs and arrowheads indicate the neurite tips. G, The histogram shows quantification of neurite length. The longest neurite was measured. Results are shown as average ± SEM of a representative experiment performed in triplicate. The experiment was performed in duplicate with similar results. *p < 0.005 (ANOVA followed by Dunnett's test).
Given that conserved Pea3 elements have been described to be present in the promoter region of the MMP3 and MMP13 genes (Yan and Boyd, 2007), we next analyzed whether Etv4 and Etv5 could mediate NGF-dependent MMP3 and MMP13 induction. To this purpose, the levels of Mmp3 and Mmp13 mRNA were measured by real-time PCR in PC12 cells transfected with Etv4-shRNA or Etv5-shRNA, selected by puromycin treatment to increase the population of cells expressing each ETS-shRNA construct and treated with NGF. As shown in Figure 7E, a significant decrease in the levels of Mmp3 and Mmp13 mRNA induction were observed in response to NGF upon knockdown of Etv4 or Etv5, indicating that these two transcription factors are involved in NGF-dependent Mmp3 and Mmp13 gene expression. Moreover, overexpression of HA-tagged-MMP3 and Flag-tagged-MMP13 in DRG cells was able to induce a significant increase in NGF-mediated neurite outgrowth (Fig. 7F,G).
Altogether, these data establish that both Etv4 and Etv5 transcription factors mediate NGF-dependent MMP3 and MMP13 expression, two metalloproteinases involved in the neurite outgrowth response induced by this neurotrophin.
Discussion
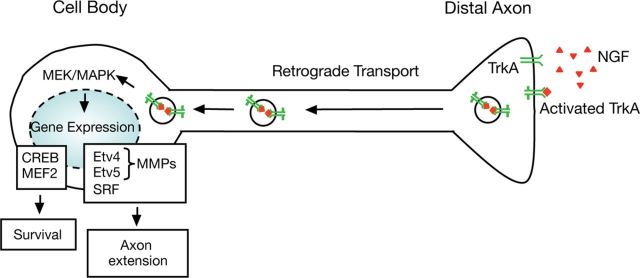
In this study, we provide evidence that the ETS transcription factors Etv4 and Etv5 are critical mediators of retrograde NGF signaling, gene expression, and axonal growth of DRG sensory neurons. We also report that NGF-induced Etv4 and Etv5 expression requires activation of MEK/ERK (MAPK) signaling pathway and mediates the expression of MMP3 and MMP13, two enzymes involved in neurite invasiveness. These findings lead us to propose a model in which target-derived NGF and TrkA retrograde signaling promote axonal growth of sensory DRG neurons through the control of a transcriptional program mediated by Etv4 and Etv5 (Fig. 8).
Figure 8.
Model describing Etv4 and Etv5 induction following distal NGF stimulation. NGF binds TrkA at axon terminals and forms signaling endosomes that are transported to the CB. There, TrkA activates MEK/ERK signaling pathway, inducing diverse transcription factors, which are involved in survival and axonal extension. Among them are Etv4 and Etv5, which mediate MMP (i.e., MMP3 and MMP13) expression and axonal growth.
ETS gene expression in sensory neuron subpopulations
It is well known that neurotrophin signaling is essential for survival, neuronal specification, axonal growth, and terminal arborization of different DRG neuronal subpopulations (Bibel and Barde, 2000; Markus et al., 2002; Huang and Reichardt, 2003). However, little is known about the transcriptional programs induced by the neurotrophic factors acting on these neuronal populations.
In recent years, Pea3 transcription factors Er81/Etv1, Etv4, and Etv5 have been shown to be expressed by different populations of DRG cells. It is well established that Er81/Etv1 is expressed by proprioceptive neurons after axons have entered the periphery (Lin et al., 1998; Arber et al., 2000). However, Etv4 and Etv5 expression has not been characterized in detail. It has been described that at early developmental stages all neurons expressing the NT3 receptor TrkC express Er81/Etv1. Regarding Etv4, it has been shown to be expressed in a subpopulation of proprioceptive neurons and other DRG sensory neurons that have not been well characterized yet (Arber et al., 2000). Both, Etv1/Er81 and Etv4 are not only expressed in proprioceptive DRG neurons, but are also expressed in nonoverlapping pools of spinal motor neurons (Lin et al., 1998; Arber et al., 2000). Interestingly, Etv4 expression can be induced by the neurotrophic factor GDNF in a subpopulation of motor neurons, but not in DRG neurons. In contrast to Etv1/Er81 and Etv4, the third member of the Pea3 subfamily, Etv5, is expressed in DRG cells but not in motor neurons (Lin et al., 1998; Hagedorn et al., 2000; Paratore et al., 2002). Like Etv4, the population of sensory neurons expressing Etv5 have not been characterized in detail yet. In this work, we show that Etv4 and Etv5 are expressed in TrkA+ sensory neurons. Interestingly, this expression is coincident with the period of sensory axonal growth in response to peripheral NGF, indicating that these two transcriptional factors could mediate the biological effects triggered by this neurotrophin.
We found that the majority of TrkA+ neurons express Etv4 and Etv5 with markedly variable levels among them. However, Etv4 and Etv5 were not exclusively expressed in NGF-responsive neurons. They were also expressed in Ret-positive neurons and in a small subpopulation of neurons negative for TrkA and Ret. In addition, neither Etv4 nor Etv5 was detected in glial cells surrounding DRG exiting fibers. In agreement with previous results, we did not detect Etv1/Er81 expression in TrkA+ cells (Arber et al., 2000; Fig. 1). From a developmental point of view, the expression of variable levels of Etv4 and Etv5 in TrkA+ neurons may result in a diverse NGF-dependent transcriptional repertoire, thereby defining different TrkA neuronal subtypes.
Etv4/Etv5 are induced by NGF-TrkA-MEK/ERK signaling pathway
Target-derived NGF is essential for neuronal survival as well as for the development of cutaneous sensory neuron innervation (Zweifel et al., 2005). The evidence that NGF plays a crucial role in development of sensory projections in a manner independent of its role in regulating neuronal survival came from the analysis of Bax/NGF and Bax/TrkA double-mutant mice. These mice show severe defects in the development of peripheral projections, which involves the impaired branching and innervation of cutaneous target tissues (Patel et al., 2000).
The main mechanism through which target-derived NGF retrograde signal is propagated from the DA to the CB is through neurotrophin-induced internalization of TrkA receptors (Barker et al., 2002; Cosker et al., 2008; Harrington and Ginty, 2013). Although recent work has established many of the cellular and molecular events that underlie retrograde signaling, the identification of transcription factors and downstream target genes that mediate NGF-dependent axonal growth and target field innervation are still under intensive analysis.
The expression pattern of Etv4 and Etv5 at the time when DRG sensory axons invade their peripheral tissues in response to NGF raised the question of whether their induction might be regulated by this neurotrophin. We found that NGF regulates Etv4 and Etv5 gene expression and axonal growth in DRG neurons through activation of MEK/ERK (MAPK) signaling pathway. We also show that DA stimulation of TrkA+ DRG neurons is sufficient to upregulate Etv4 and Etv5 expression, probably through ERK5 activation. The mitogen-activated protein kinase ERK5, but not ERK1/2, is the ERK family member that has been shown to be activated in the CB compartment upon DA stimulation with NGF. However, both ERK5 and ERK1/2 are inhibited by the MEK inhibitor PD98059 (Watson et al., 2001). Thus, we propose that these two ETS factors may act as key nuclear molecules linking NGF retrograde signaling to transcriptional programs associated with axonal growth (Fig. 8).
The ETS transcription factors of the Pea3 group share a highly conserved ETS domain and two conserved transactivating domains. In most cases, the ETS transcription factors behave as transcriptional activators. These proteins regulate transcription of multiple genes through protein–protein interactions with other cellular partners present at defined times and cellular conditions, as well as through specific protein modifications, such as phosphorylation and acetylation (Li et al., 2000; Verger and Duterque-Coquillaud, 2002). Interactions with various other transcriptional coregulatory partners determine the specificity of the ETS for different target gene promoter regions. Therefore, the function of ETS family proteins has to be considered in combination with other cellular proteins since the function of the same ETS differs in the different cellular contexts. Several studies have identified many transcription factors as mediators of NGF-dependent response in sensory neurons, such as CREB, NFAT (nuclear factor of activated T cells), MEF2, and SRF (serum response factor) among others, which control distinct aspects of the biological response induced by NGF (Lonze and Ginty, 2002; Graef et al., 2003; Wickramasinghe et al., 2008; Pazyra-Murphy et al., 2009). Thus, understanding the functional interaction and specific developmental outputs generated by them is one of the most challenging tasks in the neurotrophin field.
NGF-induced Etv4 and Etv5 expression is necessary for sensory axonal growth
In this paper we describe that downregulation of Etv4 or Etv5 significantly reduces DRG axonal growth in response to NGF, while overexpression of either Etv4 or Etv5 potentiates neurite outgrowth induced by NGF. Interestingly, previous experiments showed that precocious expression in DRG neurons of a fusion product between the amino-terminal domain of Ewing sarcoma gene and the Pea3 DNA binding domain was able to promote neurotrophin-independent survival and outgrowth of sensory neurons (Hippenmeyer et al., 2005). However, our study revealed that overexpression of the ETS transcription factors in the absence of NGF is not enough to trigger the differentiation program, indicating that the NGF signal is necessary not only to induce Etv4 and Etv5 expression but also to activate these factors or to induce the expression of other protein partners that can interact with them.
One important issue is the identity of downstream targets of Etv4 and Etv5 that could participate in NGF-dependent axonal outgrowth. Candidates include several MMPs, which are enzymes that have been described to be regulated by ETS transcription factors (Trojanowska, 2000; Oikawa and Yamada, 2003; de Launoit et al., 2006). MMPs are a group of enzymes involved in the degradation of the extracellular matrix (ECM). Interestingly, NGF has been reported to regulate the expression of different MMPs, such as MMP3 and MMP10 (Nordstrom et al., 1995; Chen et al., 2008). In agreement with previous results showing that MMP3 is highly induced by NGF in PC12 cells, we observed that this MMP as well as MMP13 is upregulated in DRG neurons upon NGF treatment. Moreover, we found that downregulation of either Etv4 or Etv5 reduces NGF-dependent MMP3 and MMP13 expression. This finding is supported by the presence of Pea3 binding sites in the promoter region of these MMPs (Yan and Boyd, 2007, Tardif et al., 2004). Moreover, gain-of-function assays indicated that overexpression of MMP3 and MMP13 can potentiate DRG neurite outgrowth in response to NGF. Even though we provide evidence that MMP3 and MMP13 expression is regulated by Etv4 and Etv5 transcription factors in response to NGF, we consider the possibility that other target genes might be also involved in sensory neurite outgrowth. Therefore, other MMPs, such as MMP10, which also contain Pea3 sites in their promoter region and are upregulated by NGF, can also participate in the biological processes triggered by this neurotrophin (Chen et al., 2008). Future work is necessary to reveal whether NGF-induced MMPs can reach DAs to regulate growth and connectivity in response to NGF.
Pea3 transcription factors are expressed during normal development and in oncogenic processes that involve remodelling of the ECM (Lu et al., 2009; Zhang et al., 2009; Oh et al., 2012). Our results indicating a key role for Etv4 and Etv5 in normal axonal growth during development, suggest that they could also be involved in axonal regeneration following acute nerve injury. Therefore, we believe that the precise understanding of the role of Etv4 and Etv5 in the complex molecular network that controls axonal growth in response to extracellular signals will be important for the design of new therapies for repair and regeneration of the nervous system.
Footnotes
This work was supported by the Argentine Agency for Promotion of Science and Technology (ANPCyT-PICT-PRH2008-0282, PICT2010-1523, and PICT2010-1012), the University of Buenos Aires (UBACyT-2011-2013-FL), the Swedish Royal Academy of Science (2006), and the Åke-Wieberg Foundation (2007). G.P. and F.L. were supported by an Independent Research Career Position from the Argentine Medical Research Council (CONICET). G.P. is additionally supported by a J. S. Guggenheim Memorial Foundation Fellowship Award. P.F. and D.I. were supported by a fellowship from CONICET and F.C.A. was supported by a fellowship from the ANPCyT. We thank Dr. Silvia Arber (Basel University, Basel, Switzerland) for anti-Er81 antibody. We also thank Dr. Helena Mira and Dr. Marçal Vilar for comments on the manuscript, Dr. María Clara Gravielle and Dr. Pablo Iribarren for technical advice, Andrea Pecile and Manuel Ponce for animal care, Roux-Ocefa for reagent supply, and Innova-t and UBATEC for research grant administration.
References
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/S0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Ascano M, Bodmer D, Kuruvilla R. Endocytic trafficking of neurotrophins in neural development. Trends Cell Biol. 2012;22:266–273. doi: 10.1016/j.tcb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker PA, Hussain NK, McPherson PS. Retrograde signaling by the neurotrophins follows a well-worn trk. Trends Neurosci. 2002;25:379–381. doi: 10.1016/S0166-2236(02)02199-9. [DOI] [PubMed] [Google Scholar]
- Baudet C, Mikaels A, Westphal H, Johansen J, Johansen TE, Ernfors P. Positive and negative interactions of GDNF, NTN and ART in developing sensory neuron subpopulations, and their collaboration with neurotrophins. Development. 2000;127:4335–4344. doi: 10.1242/dev.127.20.4335. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Campenot RB. NGF and the local control of nerve terminal growth. J Neurobiol. 1994;25:599–611. doi: 10.1002/neu.480250603. [DOI] [PubMed] [Google Scholar]
- Chen L, Maures TJ, Jin H, Huo JS, Rabbani SA, Schwartz J, Carter-Su C. SH2B1beta (SH2-Bbeta) enhances expression of a subset of nerve growth factor-regulated genes important for neuronal differentiation including genes encoding urokinase plasminogen activator receptor and matrix metalloproteinase 3/10. Mol Endocrinol. 2008;22:454–476. doi: 10.1210/me.2007-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Chang YC, Hsieh ST. Trophic interactions between sensory nerves and their targets. J Biomed Sci. 1999;6:79–85. doi: 10.1007/BF02256438. [DOI] [PubMed] [Google Scholar]
- Chotteau-Lelièvre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene. 1997;15:937–952. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- Cosker KE, Courchesne SL, Segal RA. Action in the axon: generation and transport of signaling endosomes. Curr Opin Neurobiol. 2008;18:270–275. doi: 10.1016/j.conb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, McMahon SB, Shelton DL, Levinson AD. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Ginty DD, Segal RA. Retrograde neurotrophin signaling: Trk-ing along the axon. Curr Opin Neurobiol. 2002;12:268–274. doi: 10.1016/S0959-4388(02)00326-4. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/S0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Hagedorn L, Paratore C, Brugnoli G, Baert JL, Mercader N, Suter U, Sommer L. The Ets domain transcription factor Erm distinguishes rat satellite glia from Schwann cells and is regulated in satellite cells by neuregulin signaling. Dev Biol. 2000;219:44–58. doi: 10.1006/dbio.1999.9595. [DOI] [PubMed] [Google Scholar]
- Harrington AW, Ginty DD. Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14:177–187. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Li Y, Parada LF, Kinsman S, Chen CK, Valletta JS, Zhou J, Long JB, Mobley WC. p140trk mRNA marks NGF-responsive forebrain neurons: evidence that trk gene expression is induced by NGF. Neuron. 1992;9:465–478. doi: 10.1016/0896-6273(92)90184-F. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Lallemend F, Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–381. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Ledda F, Paratcha G, Ibañez CF. Target-derived GFRalpha1 as an attractive guidance signal for developing sensory and sympathetic axons via activation of Cdk5. Neuron. 2002;36:387–401. doi: 10.1016/S0896-6273(02)01002-4. [DOI] [PubMed] [Google Scholar]
- Ledda F, Paratcha G, Sandoval-Guzmán T, Ibañez CF. GDNF and GFRalpha1 promote formation of neuronal synapses by ligand-induced cell adhesion. Nat Neurosci. 2007;10:293–300. doi: 10.1038/nn1855. [DOI] [PubMed] [Google Scholar]
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28:39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei H, Watson DK. Regulation of Ets function by protein-protein interactions. Oncogene. 2000;19:6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/S0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/S0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, Arber S, Hassell J, MacNeil L, Hoshi M, Jain S, Asai N, Takahashi M, Schmidt-Ott KM, Barasch J, D'Agati V, Costantini F. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A, Patel TD, Snider WD. Neurotrophic factors and axonal growth. Curr Opin Neurobiol. 2002;12:523–531. doi: 10.1016/S0959-4388(02)00372-0. [DOI] [PubMed] [Google Scholar]
- Neumar RW, Xu YA, Gada H, Guttmann RP, Siman R. Cross-talk between calpain and caspase proteolytic systems during neuronal apoptosis. J Biol Chem. 2003;278:14162–14167. doi: 10.1074/jbc.M212255200. [DOI] [PubMed] [Google Scholar]
- Nordstrom LA, Lochner J, Yeung W, Ciment G. The metalloproteinase stromelysin-1 (transin) mediates PC12 cell growth cone invasiveness through basal laminae. Mol Cell Neurosci. 1995;6:56–68. doi: 10.1006/mcne.1995.1006. [DOI] [PubMed] [Google Scholar]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/S0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratcha G, Ledda F, Ibanez CF. The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell. 2003;113:867–870. doi: 10.1016/S0092-8674(03)00435-5. [DOI] [PubMed] [Google Scholar]
- Paratore C, Brugnoli G, Lee HY, Suter U, Sommer L. The role of the Ets domain transcription factor Erm in modulating differentiation of neural crest stem cells. Dev Biol. 2002;250:168–180. doi: 10.1006/dbio.2002.0795. [DOI] [PubMed] [Google Scholar]
- Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–357. doi: 10.1016/S0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Patel TD, Kramer I, Kucera J, Niederkofler V, Jessell TM, Arber S, Snider WD. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003;38:403–416. doi: 10.1016/S0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- Pazyra-Murphy MF, Hans A, Courchesne SL, Karch C, Cosker KE, Heerssen HM, Watson FL, Kim T, Greenberg ME, Segal RA. A retrograde neuronal survival response: target-derived neurotrophins regulate MEF2D and bcl-w. J Neurosci. 2009;29:6700–6709. doi: 10.1523/JNEUROSCI.0233-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- Shirazi Fard S, Kele J, Vilar M, Paratcha G, Ledda F. Tiam1 as a signaling mediator of nerve growth factor-dependent neurite outgrowth. PLoS One. 2010;5:e9647. doi: 10.1371/journal.pone.0009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif G, Reboul P, Pelletier JP, Martel-Pelletier J. Ten years in the life of an enzyme: the story of the human MMP-13 (collagenase-3) Mod Rheumatol. 2004;14:197–204. doi: 10.1007/s10165-004-0292-7. [DOI] [PubMed] [Google Scholar]
- Trojanowska M. Ets factors and regulation of the extracellular matrix. Oncogene. 2000;19:6464–6471. doi: 10.1038/sj.onc.1204043. [DOI] [PubMed] [Google Scholar]
- Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Maynard TM, Weston JA. Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development. 2000;127:2811–2821. doi: 10.1242/dev.127.13.2811. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/S0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Watson FL, Heerssen HM, Bhattacharyya A, Klesse L, Lin MZ, Segal RA. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat Neurosci. 2001;4:981–988. doi: 10.1038/nn720. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe SR, Alvania RS, Ramanan N, Wood JN, Mandai K, Ginty DD. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58:532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Boyd DD. Regulation of matrix metalloproteinase gene expression. J Cell Physiol. 2007;211:19–26. doi: 10.1002/jcp.20948. [DOI] [PubMed] [Google Scholar]
- Ye H, Kuruvilla R, Zweifel LS, Ginty DD. Evidence in support of signaling endosome-based retrograde survival of sympathetic neurons. Neuron. 2003;39:57–68. doi: 10.1016/S0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell. 2009;16:607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Li X, McNamee C, Chen AP, Baccarini M, Snider WD. Raf kinase signaling functions in sensory neuron differentiation and axon growth in vivo. Nat Neurosci. 2007;10:598–607. doi: 10.1038/nn1898. [DOI] [PubMed] [Google Scholar]
- Znosko WA, Yu S, Thomas K, Molina GA, Li C, Tsang W, Dawid IB, Moon AM, Tsang M. Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development. Dev Biol. 2010;342:11–25. doi: 10.1016/j.ydbio.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]