Abstract
Colorectal cancer (CRC) is the third most prevalent cancer and second in terms of mortality. Emerging evidence from recent studies suggests a potential role of Fusobacterium nucleatum in the development of CRC. In this article, we review studies from different geographical regions examining the association between F. nucleatum and CRC, the detection methods and the tumorigenic mechanisms. Furthermore, we discuss the potential clinical impact of F. nucleatum in CRC and suggest future study directions.
Keywords: colorectal cancer, CRC, Fusobacterium nucleatum, F. nucleatum, tumorigenic mechanisms
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and second in terms of mortality (1). Over 95% of CRC are adenocarcinomas, and the majority develop via the adenoma-carcinoma sequence (2). Several risk factors are associated with CRC, such as genetic mutations on tumor suppressor genes and oncogenes, older age, diet, and chronic inflammation (3). In addition to these established risk factors, increasing evidence has linked CRC with some bacterial species in the gastrointestinal tract such as Fusobacterium nucleatum.
This article will review studies examining the association between F. nucleatum and CRC from different geographical regions. The potential clinical impact of F. nucleatum in CRC and the tumorigenic mechanisms of F. nucleatum in CRC will also be discussed.
Fusobacterium Nucleatum
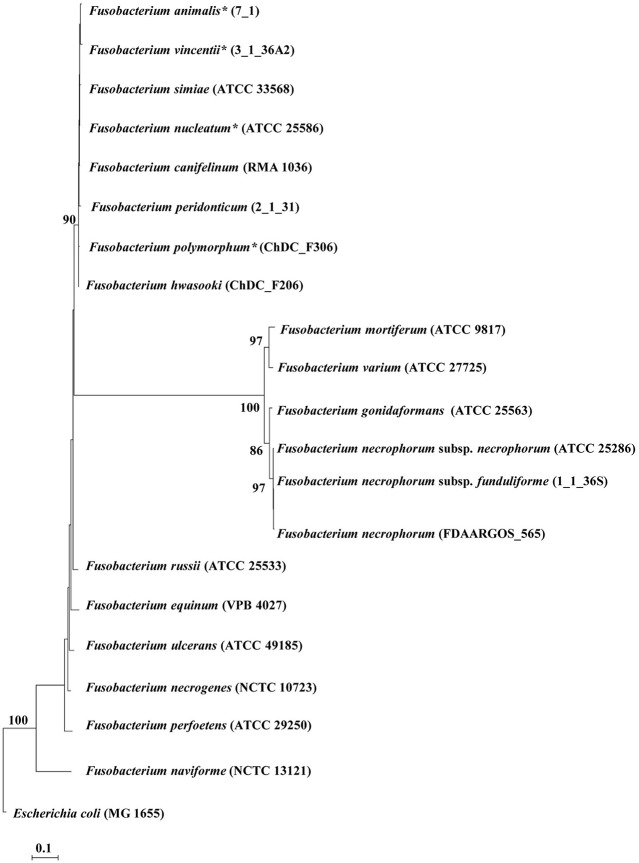
Fusobacterium is a genus of the Fusobacteriaceae family, containing bacterial species isolated from both human and animal sources (4–6). F. nucleatum, a species of the Fusobacterium genus, previously contained four subspecies including F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. nucleatum subsp. vincentii, and F. nucleatum subsp. animalis (7). A recent study by Kook et al. compared the genomes of the F. nucleatum subspecies with other Fusobacterium species and their study strongly suggested that these four subspecies should be reclassified as species F. nucleatum, F. polymorphum, F. vincentii, and F. animalis, respectively (6). The Fusobacterium genus currently contains 20 species and subspecies (Figure 1).
Figure 1.
The phylogenetic tree generated based on 16S rRNA sequences of Fusobacterium species. The phylogenetic tree was generated using the maximum likelihood method. Bootstrap values were generated from 1,000 replicates and values of more than 70 were indicated. Escherichia coli MG1655 was included as an out group. Updated Fusobacterium species from previous subspecies are indicated by asterisks (*) (6). The 16S rRNA sequences of Fusobacterium species and Escherichia coli MG1655 were obtained from the National Center Biotechnology Information database.
Within the Fusobacterium genus, most infections in humans are caused mainly by two species, F. necrophorum and F. nucleatum. F. necrophorum is a Gram-negative anaerobic bacterium with a rod shape and non-spore forming. F. necrophorum is found in the gastrointestinal tract of humans and animals and also in female urogenital tract and is the causative agent for Lemierre's syndrome (8–14). F. nucleatum is an oral Gram-negative anaerobe, with a spindle-rod shape, non-motile, and non-spore forming. F. nucleatum is associated with periodontitis (15–17). Furthermore, recent studies have linked F. nucleatum with human CRC.
Studies Examining the Associations Between F. nucleatum and CRC and Pre-Cancerous Lesions From Different Geographical Regions
Studies From Asia
Studies investigating F. nucleatum in CRC in Asia were reported from China and Japan. Using quantitative polymerase chain reaction (qPCR), Yan et al. examined the level of F. nucleatum relative to beta-actin in 280 stage III/IV Chinese CRC patients from Shanghai and found that the relative level of F. nucleatum in tumor tissues was significantly higher than the paired adjacent normal tissues (0.1092 ± 0.215 vs. 0.0245 ± 0.0553, P < 0.001; Table 1) (18). This study also found that a higher level of F. nucleatum was associated with tumor invasion, and patients with a low level of F. nucleatum had a significantly better cancer-specific survival and a disease-free survival (18).
Table 1.
Studies examining the association between Fusobacterium nucleatum and CRC using colorectal tissues and fecal samples.
| References | Population | Samples (N) | Results | Detection methods |
|---|---|---|---|---|
| Yan et al. (18) | China | CRC (280), adjacent normal tissues (280) | •F. nucleatum level was significantly higher in CRC tissues than in adjacent normal tissues (CRC vs. normal: 0.1092 ± 0.215 vs. 0.0245 ± 0.0553, P < 0.001). | Quantitative reverse transcription PCR using F. nucleatum specific primers |
| Li et al. (19) | China | CRC (101), adjacent normal tissues (101) | •The median abundance of F. nucleatum was significantly greater in the tumor samples [0.242 (0.178, 0.276)] than that in the matched normal controls [0.050 (0.023, 0.067)] (P < 0.001). | Fluorescent quantitative PCR using primer and probe sequences for F. nucleatum and fluorescence in situ hybridization (FISH) analysis |
| Wei et al. (20) | China | CRC (180), adjacent normal tissues (180) | •F. nucleatum between non-survival group and survival group (5.66 vs. 1.08%) and F. nucleatum (5.10 vs. 1.08%) exhibited a greater abundance in the recurrence group than in survival group, however, was not statistically significant. •High abundance of F. nucleatum was significantly correlated with the worse depth of invasion (P = 0.015). |
Sequencing of V4 region of bacterial 16S rRNA gene |
| Yu et al. (21) | China | Cohort 1: CRC with recurrence (16), CRC without recurrence (15) Cohort 2: CRC with recurrence (48), CRC without recurrence (44) Cohort 3: CRC with recurrence (87), CRC without recurrence (86) | •In cohort 1, Fusobacterium was significantly enriched in recurrent tissues than non-recurrent tissues. Using qPCR, they also found that the relative abundance of F. nucleatum was significantly higher in recurrent CRC than non-recurrent tissues (P < 0.01). •In cohort 2, they also found that the relative abundance of F. nucleatum was significantly higher in tumor tissues, as compared to para-tumor tissues in both CRC patients with and without recurrence (P < 0.05). Furthermore, the F. nucleatum abundance in tumor tissues of CRC patients with recurrence was significantly higher than that in tumor tissues of CRC patients without recurrence (P < 0.01). •In cohort 3, F. nucleatum had a significantly higher abundance in recurrent CRC tissues than non-recurrent CRC tissues (P < 0.01). |
Sequencing using 16S rRNA gene and quantitative real-time PCR using primers specific for F. nucleatum |
| Liang et al. (22) | China | Cohort 1: CRC (170), healthy controls (200)Cohort 2: CRC (33), healthy controls (36) | •The median abundance of F. nucleatum was significantly higher in CRC (0.0288 vs. 8.1E-6) than controls for Cohort 1 (P < 0.0001). For Cohort 2, abundance was also significantly higher in CRC than controls (P < 0.01). •The occurrence rate of F. nucleatum in CRC was 98.2% whereas in control 72% (P < 0.0001). |
Duplex quantitative PCR targeting 16S rRNA gene (TaqMan method) |
| Wong et al. (23) | China | CRC (104), advanced adenoma (103) and healthy controls (102) | •In comparison to healthy controls, patients with CRC had a significantly higher abundance of F. nucleatum (132-fold, P < 0.001) •In comparison to healthy controls, patients with advanced adenoma had a significantly higher level of F. nucleatum (3.8-fold, P = 0.022). |
Species specific quantitative real-time PCR (SYBR Green method) |
| Yamaoka et al. (24) | Japan | CRC (100), matched normal mucosa (72) | •The detection rates of F. nucleatum were 63.9% (46/72) in normal-appearing mucosal tissues and 75.0% (75/100) in CRC tissue samples and this was statistically insignificant. •The median copy number of F. nucleatum was 0.4 copies/ng DNA in the normal-appearing colorectal mucosa in patients with colorectal cancer and 1.9 copies/ng DNA in the colorectal cancer tissues (P = 0.0031). |
Droplet digital PCR using primer targeting F. nucleatum |
| Ito et al. (25) | Japan | CRC (511), premalignant lesions: serrated lesions (343) and non-serrated adenomas (122) | •F. nucleatum positivity was significantly higher in CRCs (56%) than in premalignant lesions of any histological type (24% in hyperplastic polyps, 35% in sessile serrated adenomas, 30% in traditional serrated adenomas and 33% in non-serrated adenomas, P < 0.0001). | Quantitative PCR using primer specific for F. nucleatum (TaqMan method) |
| Suehiro et al. (26) | Japan | CRC stage I to IV (158), colorectal advanced adenoma/carcinoma in situ (19), colorectal non-advanced adenomas (11), healthy controls (60) | •The median copy numbers of F. nucleatum were 17.5 in the control group, 311 in the non-advanced adenoma group, 122 in the advanced adenoma/CIS group and 317 in the CRC group. •F. nucleatum level was significantly higher in the non-advanced adenoma group (P = 0.0147), the advanced adenoma/CIS group (P = 0.0060) and the CRC group (P < 0.0001) than the control group. |
Droplet digital PCR using sequences of the F. nucleatum primer and probe set |
| Flanagan et al. (27) | Ireland, Czech Republic and Germany (European cohorts) | CRC (122), matched normal controls (122) | •In all three European cohorts, the abundance of F. nucleatum in tumor tissues were significantly higher as compared to the normal tissues (Czech Republic 266-fold P = 0.002, Germany 43-fold P = 0.0001 and Ireland 9-fold P = 0.006 respectively). •Significantly higher levels of F. nucleatum in tumor tissue (pooled cohorts RQNormal 2−19 vs. RQTumour 2−10, P < 0.0001) were found. The average F. nucleatum levels were increased by over 45-fold in the pooled cohorts. |
Quantitative real-time PCR using primers specific for Fusobacterium (SYBR Green method) |
| Bundgaard-Nielsen et al. (28) | Denmark | CRC (99), paired normal controls (99), CRA (96) and diverticular disease of the colon (104) | •F. nucleatum was detected higher in tumor tissues compared to adenoma (29.3 vs. 3.0%, P < 0.001). •Detection of F. nucleatum did not result in significant changes in survival or disease-free survival rates of patients within a 5-year period. |
Quantitative real-time PCR using primers specific for F. nucleatum (SYBR Green method) |
| Eklöf et al. (29) | Sweden | CRC (39), dysplasia (134), controls (66) | •The abundance of F. nucleatum was significantly higher in fecal samples from patients with CRC than samples from dysplasia and controls (P < 0.001). •F. nucleatum was detected in 27 (69.2%) CRC, 27 (20.1%) dysplasia, and 15 (24.3%) controls (P < 0.001). |
Quantitative real-time PCR using a FAM-labeled probe specific for F. nucleatum 16S rRNA gene |
| Russo et al. (30) | Italy | CRC (10), healthy controls (10) | •No significant difference between stool samples of healthy subjects of CRC patients was observed. However, it was found that F. nucleatum abundance was higher in saliva samples than stool samples in both healthy subjects (P < 0.002) and CRC patients (P < 0.01). | Quantitative real-time PCR using species-specific primers targeting 16S rRNA sequence |
| Castellarin et al. (31) | Canada | CRC (99), matched normal (99) | •RNA-Seq showed that F. nucleatum had the highest hits overall (21% of all alignments) and 9/11 subjects showed at least 2-fold higher read counts in tumor relative to corresponding control tissue. •qPCR showed that the mean overall abundance of Fusobacterium was found to be 415 times greater in the tumor samples than in the matched normal samples using qPCR (P < 2.52E-6). |
RNA-Seq; quantitative PCR using primer/probe targeting Fusobacterium DNA (TaqMan method) |
| Mima et al. (32) | United States | CRC (598), adjacent non-tumor tissues (558) | •F. nucleatum was detected significantly higher in CRC tissues (13%, 76/598) than in non-tumor tissues (3.4%, 19/558), (P < 0.001). •In the 558 pairs of CRC and adjacent non-tumor tissues, the amount of F. nucleatum DNA in tumor tissues was significantly higher than in adjacent non-tumor tissues (P < 0.0001). |
Quantitative PCR using primer targeting the nusG gene of F. nucleatum (TaqMan method) |
| Mima et al. (33) | United States | CRC (1102) | •F. nucleatum DNA was detected in 13% (138/1102) CRC tissues. The proportion of F. nucleatum-high colorectal cancers gradually increased from rectal cancers (2.5% 4/157) to cecal cancers (11% 19/178), with a statistically significant linear trend along with all subsites (P < 0.0001). | Quantitative PCR using primer targeting the nusG gene of F. nucleatum (TaqMan method) |
| Proença et al. (34) | Brazil | CRC (43), adjacent normal tissue (N-CRC 43), CRA (27), matched adjacent normal tissue (N-CRA 27) | •A significant increase in bacterial DNA was found for both CRA and CRC tissues compared to the respective normal adjacent tissues (P = 0.0002). The quantity of F. nucleatum was 24.84 times greater in CRC samples than in CRA samples (P < 0.0001). | Quantitative real-time PCR using nusG gene of F. nucleatum (TaqMan method) |
| Fukugaiti et al. (35) | Brazil | CRC (7), healthy controls (10) | •The level of F. nucleatum was significantly higher in fecal samples from patients with CRC than healthy patients (6.2 ±1.5 vs. 4.0 ± 1.5, P < 0.01). | Quantitative real-time PCR using species-specific primers (SYBR Green method) |
For studies from each country, studies using CRC tissues were listed first, followed by studies using fecal samples. CRC, colorectal cancer; CRA, colorectal adenoma; CIS, carcinoma in situ; RQ, relative quantification.
A study by Li et al. using TaqMan probe-based qPCR examined F. nucleatum in cancer and adjacent normal tissues of 101 patients with CRC (19). They found that the median abundance of F. nucleatum in CRC tissues was 0.242 (0.178, 0.276), which was significantly higher than that in normal tissues, 0.05 (0.023, 0.067; P < 0.001) (19). In this study, Li et al. also found that the lymph node metastases was more frequently seen in patients with high abundance of F. nucleatum (19).
Wei et al. established libraries of 16S rRNA gene V4 region amplicon prepared from amplification of tumor samples from 180 Chinese patients with CRC stages I-IV from Qingdao (20). This study divided the patients into four groups including non-survival, recurrence, survival, and unknown. They found a greater abundance of F. nucleatum in the recurrence group than in the survival group and found that the high abundance of F. nucleatum significantly correlated with the worse depth of invasion (P = 0.015).
Yu et al. examined three cohorts of patients with CRC from a hospital in Shanghai, China (21). Fresh intestinal tissues were collected from patients of cohort 1 (31 patients) and subjected for microbiota analysis using 16S RNA sequencing, and it was found that Fusobacterium genus was enriched in recurrent CRC patients. Cohort 2 contained 44 recurrent CRC patients and 48 non-recurrent CRC patients, and formalin-fixed paraffin-embedded tissues were subjected for analysis using qPCR method. A significantly higher abundance of F. nucleatum in tumor tissues was observed, as compared to para-tumor tissues in both CRC patients with and without recurrence (P < 0.05). Furthermore, the abundance of F. nucleatum in tumor tissues of CRC patients with recurrence was significantly higher than that in tumor tissues of CRC patients without recurrence (P < 0.01). The higher level of F. nucleatum was strongly associated with shorter recurrence free survival. Cohort 3 contained 173 patients with CRC, and formalin-fixed paraffin-embedded tissues were used for qPCR analysis of F. nucleatum. Patients were divided into high and low F. nucleatum groups and an association between a higher level of F. nucleatum and shorter recurrence free survival was again found (21).
In addition to colonic tissue samples, fecal samples were also used to detect F. nucleatum. Liang et al. examined the abundance of five bacterial species in fecal samples of two independent Chinese cohorts using duplex qPCR targeting 16S rRNA gene (22). One cohort was from Hong Kong consisting of 170 patients with CRC and 200 healthy controls. The second cohort was from Shanghai consisting of 33 patients with CRC and 36 healthy controls. They found that the relative abundances of F. nucleatum, Clostridium hathewayi, and one unidentified species were higher in patients with CRC than in healthy controls for Hong Kong cohort (P < 0.0001) and the relative abundance of F. nucleatum was significantly higher in CRC patients than healthy controls for Shanghai cohort (P = 0.01) (22). Wong et al. examined the abundance of F. nucleatum, Peptostreptococcus anaerobius and Parvimonas micra relative to the total bacterial DNA in fecal samples collected from 104 patients with CRC, 103 patients with high grade adenoma and 102 controls in Hong Kong using qPCR SYBR Green method (23). This study found that the relative abundance of F. nucleatum in patients with CRC and advanced adenoma was 132-fold and 3.8-fold higher compared to controls (P < 0.001 and P = 0.022, respectively) (23).
The above studies examining the association between F. nucleatum and CRC in Chinese patients were based on F. nucleatum abundance. None of these studies reported the positive detection rate of F. nucleatum in the samples studied, it is therefore not clear how many patients with CRC were positive for F. nucleatum in the intestinal tissues or fecal samples.
A number of studies examining F. nucleatum in CRC were reported from Japan. Using digital PCR targeting F. nucleatum, Yamaoka at al. found that the median copy number of F. nucleatum in cancer mucosa of patients with CRC was significantly higher than that in normal-appearing mucosal tissues of these patients (1.9 copy number/ng vs. 0.4 copy number/ng DNA, P = 0.0031; Table 1) (24). After separating the CRC patients based on their disease stage, they found that the copy numbers of F. nucleatum in stage IV (not those in stages I to III) was significantly higher than that in the normal-appearing mucosa (P = 0.0016). This study also reported that F. nucleatum is commonly present in both mucosal tissues of CRC and normal-appearing tissues (75/100, 75% vs. 46/72, 63.95%) and the prevalence was not statistically different. Using F. nucleatum specific TaqMan qPCR method, Ito et al. examined tumor tissues from 511 Japanese CRC patients and 465 premalignant lesions including 343 serrated lesions and 122 non-serrated adenomas (25). They found that the F. nucleatum positivity was 56% in patients with CRC, which was significantly higher than in any of the premalignant lesions including 24% in hyperplastic polys, 35% in sessile serrated adenomas, 30% in traditional serrated adenomas (P < 0.0001). In contrast to other studies, the association between F. nucleatum and CRC reported by Ito et al. was based on prevalence rather than abundance. They also found that F. nucleatum was more frequently detected in premalignant lesions with high CpG Island methylator phenotype (CIMP) than in lesions with low or absent CIMP (46/108, 43 vs. 96/357, 27; P = 0.0023) (25).
Suehiro et al. also used digital PCR targeting F. nucleatum and found that in fecal samples, the median copy number of F. nucleatum was 317 in CRC group (158 patients), 122 in high grade adenoma/carcinoma in situ group (19 patients) and 311 in low grade adenoma group (11 patients). These were all significantly higher than the median copy number (17.5) in healthy controls (60 individuals) (26).
Studies From Europe
A study from Flanagan et al. examined F. nucleatum in tumor and matched normal tissues from 122 patients with CRC from three European cohorts (Czech Republic, Germany, and Ireland; Table 1) (27). They found that in all three cohorts, the abundance of F. nucleatum in tumor tissues were significantly higher as compared to the normal tissues (P = 0.002, P = 0.0001 and P = 0.006 for Czech Republic, Germany, and Ireland, respectively) (27). They also examined F. nucleatum in 52 Irish patients with colorectal adenoma (CRA) and found a significantly higher level of F. nucleatum in adenoma tissues with high-grade dysplasia as compared to matched normal tissues (P = 0.015) (27).
A study from Denmark by Bundgaard-Nielsen et al. used tumor tissues, matched normal tissues, tissues from patients with adenoma and patients with diverticular disease to examine the association between F. nucleatum and CRC (Table 1) (28). By using qPCR, they have found that the abundance of F. nucleatum in tumor tissues were significantly higher as compared to adenoma tissues (29.3 vs. 3.0%, P < 0.001), however, when comparing to normal tissues, no significant difference was observed (28). This study also demonstrated that F. nucleatum did not affect the risk of death or the risk of developing new adenomas or CRC in patients with CRC, adenoma or diverticular disease (28).
Using fecal samples, Eklöf et al. from Sweden detected F. nucleatum in 69.2% of patients with CRC, 20.1% of patients with dysplasia and 24.3% of controls (P < 0.001; Table 1). They also found that F. nucleatum was significantly more abundant in fecal samples from patients with CRC than samples from dysplasia and controls (P < 0.001) (29). A study from Italy by Russo et al. did not find any significant difference between fecal samples from patients with CRC and healthy individuals (Table 1) (30). However, the sample size in that study was small (10 patients and 10 controls) (30).
Studies From North America
Castellarin et al. compared the levels of F. nucleatum in tumor tissues and matched normal tissues from 99 Canadian patients and found a significantly higher level of F. nucleatum in tumor tissues as compared to normal tissues (P = 2.52E−6; Table 1) (31). The mean overall abundance of F. nucleatum was found to be 415 times greater in the tumor samples than in the matched normal tissues. They also found that patients with high-relative-abundance of F. nucleatum in tumor tissues relative to matched control tissues were significantly more likely to have regional lymph node metastases (P = 0.0035). Mima et al. from the United States examined F. nucleatum in tumor tissues of 598 patients with CRC and 558 adjacent non-tumor tissues; they found that both the positivity and the amount of F. nucleatum in tumor tissues were significantly higher than in non-tumor tissues (Table 1) (32). However, the detection rates of F. nucleatum in tumor and control tissues in the study from Mima et al. were 13 vs. 3.4%, which were dramatically lower than those reported from Asian countries. It is not clear whether this difference was due to different target genes used in the detection methods or ethnic background. In the following study, Mima et al. examined tumor tissues from 1,102 patients with CRC, found that the proportion of F. nucleatum-high cancers gradually increased from rectal cancers (2.5%; 4/157) to cecal cancers (11%; 19/178) (33). This is an interesting observation, given that F. nucleatum is an oral bacterium, it may have more opportunities to encounter with tumors at the cecum than those at the rectum.
Studies From South America
A study from Brazil conducted by Proença et al. examined F. nucleatum in tumor tissues of 43 patients with CRC, 27 patients with CRA and matched adjacent normal tissues (Table 1) (34). A significant increase in F. nucleatum DNA was found in both CRA and CRC tissues compared to the respective normal adjacent tissues (P = 0.0002). Furthermore, the quantity of F. nucleatum was 24.84 times greater in CRC samples than in CRA samples (P < 0.0001). Fukugaiti et al. used fecal samples from seven patients with CRC and 10 healthy controls in Brazil to examine the level of F. nucleatum and found that F. nucleatum was detected at a significantly higher level in patients with CRC than healthy controls (6.2 ± 1.5 vs. 4.0 ± 1.5, P < 0.01) (35).
Potential Clinical Implications of F. nucleatum in CRC
Two studies examined the use of F. nucleatum as a potential biomarker for detection of CRC: The significantly higher abundance of F. nucleatum in fecal samples of CRC patients when compared to healthy controls suggests that F. nucleatum is a potential biomarker for the early detection of CRC. In a study by Liang et al., DNA was first extracted from fecal samples collected from patients with CRC and healthy individuals, and duplex qPCR was performed using primers targeting F. nucleatum (22). Liang et al. reported a sensitivity of 77.7% and specificity of 79.5% of using F. nucleatum in fecal samples to discriminate CRC from controls (22). In the study from Wong et al., DNA was extracted from fecal samples of patients with CRC and high grade adenoma, and quantitative real-time PCR was performed using primers specific for F. nucleatum (23). Overall, the sensitivities of using F. nucleatum in fecal samples as a marker in detecting CRC and high grade adenoma were 73.1 and 15.5%, respectively, (23).
A number of studies also suggested that F. nucleatum is a potential therapeutic target for interfering with the progression of CRC: In the study from Yamaoka at al., the high abundance of F. nucleatum was found to be associated with stage IV of CRC, not the stages of I to III CRC (24). Yan et al. and Wei et al. found that F. nucleatum was adversely associated with CRC patient survival (18, 20). Castellarin et al. found that patients with high relative abundance of F. nucleatum in tumor tissues relative to matched control tissues were more likely to have regional lymph node metastases (31). These findings provide initial evidence that high levels of F. nucleatum may promote the progression of CRC. If these findings are confirmed, reducing F. nucleatum may be a potential strategy to alleviate the progression of CRC.
Tumorigenic Mechanisms of F. nucleatum in CRC
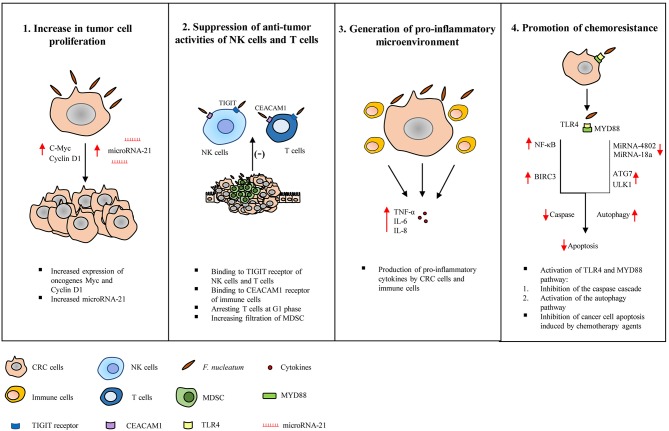
A number of studies have suggested the following mechanisms by which F. nucleatum may contribute to the development and progression of CRC, these mechanisms are summarized in Figure 2.
Figure 2.
Tumorigenic mechanisms of Fusobacterium nucleatum in colorectal cancer. Studies have suggested the following tumorigenic mechanisms of F. nucleatum in colorectal cancer (CRC). (1) Increase in tumor cell proliferation: F. nucleatum increases the expression of oncogenes such as c-Myc and Cyclin D1 and microRNA-21, directly promoting tumor growth. (2) Suppression of the anti-tumor activity of natural killer (NK) and T cells: The activity of NK cells and T cells are suppressed by F. nucleatum through interaction with the T cell immunoglobulin and ITIM domain (TIGIT) receptor expressed on NK cells and T cells, binding and activating the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) 1, arresting T cells at G1 phase and increasing the infiltration of myeloid-derived suppressor cells. (3) Generation of pro-inflammatory microenvironment: F. nucleatum induces pro-inflammatory cytokines production by cancer cells and immune cells. (4) Promotion of chemoresistance: F. nucleatum promotes chemoresistance to 5-fuorouracil (5-FU) by up-regulating baculoviral inhibitor of apoptosis protein repeat 3 (BIRC3) in CRC cells via Toll-like receptor 4 (TLR4)/Nuclear factor-kappa B (NF-κB) pathway, which results in inhibition of cancer cell apoptosis and reduced chemosensitivity to 5-FU. F. nucleatum also promotes chemoresistance by modulating autophagy. CRC cells infected with F. nucleatum activate the TLR4 and MYD88 innate immune signaling pathway, causing the loss of microRNAs miR-18a and miR-4802, and up-regulating autophagy elements, ULK1 and ATG7, which ultimately leads to inhibition of cancer cell apoptosis.
Direct Promotion of the Growth of CRC Cells
Using CRC cell line models, several studies revealed that F. nucleatum was able to directly increase the growth of cancer cells. Rubinstein et al. showed that F. nucleatum stimulated the growth of several CRC cell lines such as HT-29 cells but did not stimulate the growth of the non-CRC cells such as HEK293 (36). By deletion of the fadA gene, this study demonstrated that FadA is a key virulence factor in promoting CRC cell growth by F. nucleatum (36). Rubinstein et al. also demonstrated that the cell-adhesion molecule E-cadherin acts as a receptor for FadA and binding of FadA to E-cadherin led to increased expression of transcription factor Nuclear factor-kappa B (NF-κB) and oncogenes such as c-Myc and Cyclin D1 (36). Another interesting finding from this study was that increased tumor growth and inflammatory responses caused by F. nucleatum were differentially regulated, further supporting a direct cancer promoting effect by F. nucleatum (36). A recent study conducted by the same group also reported that FadA can upregulate Wnt/β-catenin modulator Annexin A1 expression through E-cadherin (37).
Yang et al. demonstrated that F. nucleatum increases proliferation of CRC cell lines by up-regulating the expression of microRNA-21 (38). MicroRNA-21 functions as an oncogene, which suppresses the expression of tumor suppressor genes (38). This study further demonstrated that the increased expression of microRNA-21 by F. nucleatum was through binding of NF-κB to microRNA-21 promoter and the NF-κB pathway was activated through Toll-like receptor 4 (TLR4) (38).
Suppression of Host Anti-tumor Immunity
Cytotoxic T cells and natural killer (NK) cells are the major effector cells to remove cancerous and pre-cancerous cells (39, 40). Shenker et al. demonstrated that F. nucleatum suppresses human T cell responses to mitogens and antigens by arresting cells in the G1 phase of the cell cycle (41, 42). Kaplan et al. showed that F. nucleatum outer membrane proteins Fap2 and RadD induce cell death in Jurkat cells (immortalized human lymphocytes) (43). Gur et al. further demonstrated that F. nucleatum prevents tumor cells from NK cell killing, via Fap2 protein binding to human inhibitory receptor T cell immunoglobulin and ITIM domain (TIGIT) on NK cells (44). They also showed that tumor-infiltrating lymphocytes expressed TIGIT and T cell activities were also inhibited by F. nucleatum via Fap2 (44). In addition, Fap2 also mediates F. nucleatum enrichment by interacting with Gal-GalNAc, a polysaccharide that is found to be overexpressed in CRC (45).
A recent study by Gur et al. demonstrated that an unidentified ligand of F. nucleatum inhibits NK cell and T cell function by binding to carcinoembryonic antigen-related cell adhesion molecule (CEACAM) 1 (46). CEACAM1 functions as an inhibitory receptor on various immune cell subsets (47). CEACAM1 is also expressed on the surface of tumors, and is considered to be a biomarker associated with tumor progression, metastasis and poor prognosis (48). It was previously shown that CEACAM1 expression is higher with more advanced stages of CRC, particularly in metastatic colon cancer, suggesting its role in CRC progression (49).
Mima et al. observed an inverse association between the amount of F. nucleatum and CD3+ T cell density in CRC tissues (32). Kostic et al. showed that F. nucleatum fed APCMin/+ mice had a significantly higher number of colonic tumors as compared to control groups without causing enteritis (50). Furthermore, this study found increased filtration of CD11b+ myeloid-derived suppressor cells (MDSC) in the tumors of APCMin/+ mice fed with F. nucleatum as compared to control APCMin/+ mice, while the numbers of CD3+/CD4+ and CD3+/CD8+ T lymphocytes were not affected. MDSCs are known to suppress T cells and play an important role in promoting tumor progression (51).
Microsatellite instability (MSI) is a form of genetic instability caused by alterations in the DNA mismatch repair system and ~15% of colorectal cancers display MSI (52). A number of studies have shown the association between high MSI and a great amount of F. nucleatum in CRC tissues (32, 50, 53, 54). A recent study by Hamada et al. has shown an association between F. nucleatum with the immune response to CRC and tumor MSI status (53). The study found that the presence of F. nucleatum was negatively associated with TIL in MSI-high tumors whereas, positively associated with TIL in non–MSI-high tumors (53). Hence, F. nucleatum may promote immune evasion in MSI-high colorectal carcinomas by inducing suppressive effects on adaptive anti-tumor immune responses in MSI-high CRC. However, further studies are required to discover the exact mechanism of how F. nucleatum affects the epigenetic changes in CRC. Collectively, these studies have shown that colonization of F. nucleatum in CRC and pre-cancerous tissues may reduce host ability in eliminating cancer and precancerous cells by direct inhibition of immune effector cells or modulating tumor-immune microenvironment.
Inducing Inflammation
It is well-known that chronic inflammation induced by persistent microbes, such as Helicobacter pylori and hepatitis B and C, promotes cancer development and progression (55, 56). Chronic inflammation contributes to tumorigenesis through multiple mechanisms such as causing DNA damage, increasing mutation rates and damaging repair enzymes (57). Kostic et al. found that tumor tissues in F. nucleatum fed APCMin/+ mice had increased expression of cytokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-6 and IL-8, which were similar to what has been observed in human CRC (50). Several other studies also showed that F. nucleatum induced production of pro-inflammatory cytokines and activation of NF-κB pathway (58, 59). NF-κB activates genes that control cell survival, proliferation and angiogenesis (60). Collectively, these data suggest that F. nucleatum may contribute to CRC development and progression via generating a pro-inflammatory microenvironment.
Promoting Chemoresistance
Studies have shown that gut microbiota can influence the efficacy of anti-tumor immunotherapy drugs and have an impact on the treatment of CRC (61–66). Using colon cancer cell lines, Zhang et al. demonstrated that F. nucleatum infection reduced chemosensitivity of CRC cells to 5-fluorouracil (5-FU), a chemotherapeutic drug commonly used in treating patients with advanced CRC (67). This study showed that F. nucleatum infection up-regulated baculoviral inhibitor of apoptosis protein repeat 3 (BIRC3), a member of the inhibitor of apoptosis proteins (IAPs), which inhibits apoptosis by directly inhibiting the caspase cascade (67–69). IAPs are also known to promote the survival of tumor cells and induce chemoresistance (70, 71). Studies have reported that overexpression of BIRC3 is associated with chemoresistance in the treatment of CRC (72, 73). Using the same cell line models, Zhang et al. further demonstrated that BIRC3 expression in CRC cells infected with F. nucleatum is regulated via TLR4/NF-κB pathway and reduced the chemosensitivity of CRC cells to 5-FU. Consistent with studies using the cell line models, F. nucleatum also induced chemoresistance of CRC cells in response to 5-FU in a mouse model. It was found that a high abundance of F. nucleatum correlated with chemoresistance in advanced CRC patients receiving 5-FU-based chemotherapy, suggesting that F. nucleatum may be used as a target to improve the therapeutic response of advanced CRC patients receiving 5-FU-based chemotherapy (67).
Yu et al. has found that F. nucleatum promoted chemoresistance to CRC cells by modulating autophagy (21). By using colon cancer cell lines, they showed that F. nucleatum induced CRC resistance to chemotherapeutic drugs, Oxaliplatin and 5-FU, via TLR4 and MYD88 innate immune signaling pathway (21). By acting on this pathway, F. nucleatum downregulated microRNAs miR-18a and miR-4802, and upregulated autophagy elements such as ULK1 and ATG7, leading to inhibition of cancer cell apoptosis and enhanced chemoresistance (21). Tissues from patients with CRC and normal subjects were also examined, which showed that a higher F. nucleatum abundance, upregulated expression of ULK1, and ATG7 and loss of miR18a and miRA-4802 were associated with disease recurrence (21). Collectively, the study suggests that F. nucleatum plays a role in chemoresistance.
Discussion and Future Work
Studies from different geographical regions reported the association between F. nucleatum and CRC using molecular detection methods. In most of these studies, the association was based on comparison of F. nucleatum abundance in CRC tissue sample with matched normal tissues. A few studies also reported the association based on the prevalence of F. nucleatum in CRC tissues or fecal samples as compared to controls. The prevalence of F. nucleatum in CRC tissues reported varied greatly, while the study by Mima et al. from the United States reported the detection rate of 13%, a study by Yamaoka et al from Japan detected F. nucleatum in 75% CRC tissues (24, 32, 33). It is not clear whether such a great difference was due to different detection methods used. Furthermore, a study from Denmark did not find an association between F. nucleatum and CRC and the CRC progression (28). A standardized method is required to generate reproducible and consistent data as well as to clarify whether the differences between the detection rate of F. nucleatum is due to ethnicity.
A higher level of F. nucleatum in fecal samples of patients with CRC and adenoma was detected as compared to healthy controls (23, 26). This may be due to increased shedding of F. nucleatum from the neoplastic tissues. Another possibility is that more F. nucleatum has been transported from the oral cavity to the intestinal tract in patients with CRC than healthy controls. This view is supported by a recent study by Komiya et al. showing that patients with CRC had identical strains in their colorectal cancer and oral cavity (74). However, examinations of the abundance of F. nucleatum in saliva samples of patients with CRC and healthy controls reported inconsistent results. While Guven et al. reported a significantly higher amount of F. nucleatum in saliva samples of patients with CRC than in controls, Russo et al. did not observe such a difference (30, 75). In contrast, Russo et al. detected a high prevalence of F. nucleatum in saliva samples of both patients with CRC (P < 0.01) and healthy controls (P < 0.002) than in stool samples showing that F. nucleatum is commonly present in the human oral cavity (30). Thus, whether oral F. nucleatum directly contributes to the increased F. nucleatum in the fecal samples of CRC remains to be investigated.
Several studies also suggest that high levels of F. nucleatum may promote the progression of CRC (27, 36, 37). If these mechanisms are confirmed, reducing or eliminating F. nucleatum may be a potential strategy to alleviate the progression of CRC, and the use of antibiotics might be one of the therapeutic interventions. However, a study from Cao et al. showed that the long-term use of antibiotics is associated with an increased risk of developing colorectal adenoma, although this study did not specify the antibiotics that were being used (76). Furthermore, whether these antibiotics could change the composition of microbiota in the gastrointestinal tract, and how this may affect the progression of CRC is unclear. Therefore, further studies are required to evaluate the efficacy of different antibiotics in reducing or eliminating F. nucleatum from the gastrointestinal tract and their relationship with CRC development and progression. Another possible therapeutic strategy is through microbiota manipulation. Bacterial species that are positively associated with a better prognosis of CRC in patients with a low level of F. nucleatum could be considered as potential candidates, which may be used as probiotic bacterial species to reduce the level of F. nucleatum. The use of fecal F. nucleatum as a biomarker for detection of CRC was suggested by two studies, which remains to be verified by additional studies (22, 23).
In conclusion, despite recent interesting findings in the field of F. nucleatum and CRC, whether this bacterium can be used as a CRC detection marker or a therapeutic target for intervention in tumor progression still need further investigation. Furthermore, F. nucleatum strains should be isolated from patients with CRC and healthy controls to investigate whether there are tumor-associated virulence factors.
Author Contributions
SL played a major role in writing the manuscript. LZ, FL, SR, and CL provided important feedback and helped in editing the manuscript. All authors have approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work is supported by Science Silverstar awarded to LZ from the University of New South Wales (Grant No: PS49348).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Thrumurthy SG, Thrumurthy SSD, Gilbert CE, Ross P, Haji A. Colorectal adenocarcinoma: risks, prevention and diagnosis. BMJ. (2016) 354:i3590. 10.1136/bmj.i3590 [DOI] [PubMed] [Google Scholar]
- 3.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. (2009) 22:191–7. 10.1055/s-0029-1242458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krieg NR, Holt JG. Bergey's Manual of Systematic Bacteriology 1 ed Baltimore, MD: Williams & Wilkins; (1984). p. 964. [Google Scholar]
- 5.Cho E, Park S-N, Lim YK, Shin Y, Paek J, Hwang CH, et al. Fusobacterium hwasookii sp. nov., isolated from a human periodontitis lesion. Curr Microbiol. (2015) 70:169–75. 10.1007/s00284-014-0692-7 [DOI] [PubMed] [Google Scholar]
- 6.Kook J-K, Park S-N, Lim YK, Cho E, Jo E, Roh H, et al. Genome-based reclassification of Fusobacterium nucleatum subspecies at the species level. Curr Microbiol. (2017) 74:1137–47. 10.1007/s00284-017-1296-9 [DOI] [PubMed] [Google Scholar]
- 7.Gharbia SE, Shah HN. Heterogeneity within Fusobacterium nucleatum, proposal of four subspecies. Lett Appl Microbiol. (1990) 10:105–8. 10.1111/j.1472-765X.1990.tb00276.x [DOI] [Google Scholar]
- 8.Riordan T. Human infection with Fusobacterium necrophorum (Necrobacillosis), with a focus on Lemierre's syndrome. Clin Microbiol Rev. (2007) 20:622–59. 10.1128/CMR.00011-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemierre A. On certain septicaemias due to anaerobic organisms. Lancet. (1936) 227:701–3. 10.1016/S0140-6736(00)57035-4 [DOI] [Google Scholar]
- 10.Aliyu SH, Yong PFK, Newport MJ, Zhang H, Marriott RK, Curran MD, et al. Molecular diagnosis of Fusobacterium necrophorum infection (Lemierre's syndrome). Eur J Clin Microbiol Infect Dis. (2005) 24:226–9. 10.1007/s10096-005-1298-6 [DOI] [PubMed] [Google Scholar]
- 11.Hagelskjaer LH, Prag J, Malczynski J, Kristensen JH. Incidence and clinical epidemiology of necrobacillosis, including Lemierre's syndrome, in Denmark 1990–1995. Eur J Clin Microbiol Infect Dis. (1998) 17:561–5. 10.1007/BF01708619 [DOI] [PubMed] [Google Scholar]
- 12.Batty A, Wren MWD, Gal M. Fusobacterium necrophorum as the cause of recurrent sore throat: comparison of isolates from persistent sore throat syndrome and Lemierre's disease. J Infect. (2005) 51:299–306. 10.1016/j.jinf.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 13.Jousimies-Somer H, Savolainen S, Mäkitie A, Ylikoski J. Bacteriologic findings in peritonsillar abscesses in young adults. Clin Infect Dis. (1993) 16(Suppl_4):S292–S8. 10.1093/clinids/16.Supplement_4.S292 [DOI] [PubMed] [Google Scholar]
- 14.Jensen A, Kristensen LH, Prag J. Detection of Fusobacterium necrophorum subsp. funduliforme in tonsillitis in young adults by real-time PCR. Clin Microbiol Infect. (2007) 13:695–701. 10.1111/j.1469-0691.2007.01719.x [DOI] [PubMed] [Google Scholar]
- 15.Moore WEC, Moore LVH. The bacteria of periodontal diseases. Periodontology 2000. (1994) 5:66–77. 10.1111/j.1600-0757.1994.tb00019.x [DOI] [PubMed] [Google Scholar]
- 16.Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. (1996) 9:55–71. 10.1128/CMR.9.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. (2005) 187:5330–40. 10.1128/JB.187.15.5330-5340.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. (2017) 10:5031–46. 10.2147/OTT.S145949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y-Y, Ge Q-X, Cao J, Zhou Y-J, Du Y-L, Shen B, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in chinese patients. World J Gastroenterol. (2016) 22:3227–33. 10.3748/wjg.v22.i11.3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients' survival? a pilot study on relevant mechanism. Oncotarget. (2016) 7:46158–72. 10.18632/oncotarget.10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–56.e16. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for non-invasive diagnosis of colorectal cancer. Clin Cancer Res. (2017) 23:2061 10.1158/1078-0432.CCR-16-1599 [DOI] [PubMed] [Google Scholar]
- 23.Wong SH, Kwong TNY, Chow T-C, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. (2017) 66:1441–8. 10.1136/gutjnl-2016-312766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. (2018) 53:517–24. 10.1007/s00535-017-1382-6 [DOI] [PubMed] [Google Scholar]
- 25.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. (2015) 137:1258–68. 10.1002/ijc.29488 [DOI] [PubMed] [Google Scholar]
- 26.Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, et al. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. (2016) 54:86–91. 10.1177/0004563216643970 [DOI] [PubMed] [Google Scholar]
- 27.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. (2014) 33:1381–90. 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- 28.Bundgaard-Nielsen C, Baandrup UT, Nielsen LP, Sørensen S. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer. (2019) 19:399. 10.1186/s12885-019-5571-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. (2017) 141:2528–36. 10.1002/ijc.31011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo E, Bacci G, Chiellini C, Fagorzi C, Niccolai E, Taddei A, et al. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front Microbiol. (2018) 8:2699. 10.3389/fmicb.2017.02699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. (2012) 22:299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in colorectal carcinoma. JAMA Oncol. (2015) 1:653–61. 10.1001/jamaoncol.2015.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol. (2016) 7:e200–e. 10.1038/ctg.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proença MA, Biselli JM, Succi M, Severino FE, Berardinelli GN, Caetano A, et al. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. (2018) 24:5351–65. 10.3748/wjg.v24.i47.5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. (2015) 46:1135–40. 10.1590/S1517-838246420140665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. (2013) 14:195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. (2019) 20:e47638. 10.15252/embr.201847638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-Like Receptor 4 signaling to Nuclear Factor–κB, and up-regulating expression of microRNA-21. Gastroenterology. (2017) 152:851–66.e24. 10.1053/j.gastro.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Lostao L, Anel A, Pardo J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res. (2015) 21:5047. 10.1158/1078-0432.CCR-15-0685 [DOI] [PubMed] [Google Scholar]
- 40.Marcus A, Gowen BG, Thompson TW, Iannello A, Ardolino M, Deng W, et al. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. (2014) 122:91–128. 10.1016/B978-0-12-800267-4.00003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shenker BJ, DiRienzo JM. Suppression of human peripheral blood lymphocytes by Fusobacterium nucleatum. J Immunol. (1984) 132:2357. [PubMed] [Google Scholar]
- 42.Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. (1995) 63:4830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. (2010) 78:4773–8. 10.1128/IAI.00567-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. (2015) 42:344–55. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. (2016) 20:215–25. 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. (2019) 8:e1581531. 10.1080/2162402X.2019.1581531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. (2006) 6:433–46. 10.1038/nri1864 [DOI] [PubMed] [Google Scholar]
- 48.Wang N, Feng Y, Wang Q, Liu S, Xiang L, Sun M, et al. Neutrophils infiltration in the tongue squamous cell carcinoma and its correlation with CEACAM1 expression on tumor cells. PloS ONE. (2014) 9:e89991. 10.1371/journal.pone.0089991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arabzadeh A, Beauchemin N. Stromal CEACAM1 expression regulates colorectal cancer metastasis. Oncoimmunology. (2012) 1:1205–7. 10.4161/onci.20735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor immune microenvironment. Cell Host Microbe. (2013) 14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. (2012) 12:253–68. 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nojadeh JN, Behrouz Sharif S, Sakhinia E. Microsatellite instability in colorectal cancer. EXCLI J. (2018) 17:159–68. 10.17179/excli2017-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. (2018) 6:1327. 10.1158/2326-6066.CIR-18-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hale VL, Jeraldo P, Chen J, Mundy M, Yao J, Priya S, et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. (2018) 10:78. 10.1186/s13073-018-0586-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamb A, Chen L-F. Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem. (2013) 114:491–7. 10.1002/jcb.24389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ringelhan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans Royal Soc B. (2017) 372:20160274. 10.1098/rstb.2016.0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. (2010) 140:883–99. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubenstein JH, Shaheen NJ. Epidemiology, diagnosis, and management of esophageal adenocarcinoma. Gastroenterology. (2015) 149:302–17.e1. 10.1053/j.gastro.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through toll-like-receptor-independent mechanism. Int Endod J. (2013) 47:550–9. 10.1111/iej.12185 [DOI] [PubMed] [Google Scholar]
- 60.Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. (2018) 11:2063–73. 10.2147/OTT.S161109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. (2015) 350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. (2015) 350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zitvogel L, Galluzzi L, Viaud S, Vétizou M, Daillère R, Merad M, et al. Cancer and the gut microbiota: an unexpected link. Sci Transl Med. (2015) 7:271. 10.1126/scitranslmed.3010473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Routy B, Gopalakrishnan V, Daillere R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anti-cancer immunosurveillance and general health. Nat Rev Clin Oncol. (2018) 15:382–96. 10.1038/s41571-018-0006-2 [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Ma R, Liu F, Lee SA, Zhang L. Modulation of gut microbiota: a novel paradigm of enhancing the efficacy of programmed death-1 and programmed death Ligand-1 blockade therapy. Front Immunol. (2018) 9:374. 10.3389/fimmu.2018.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coleman OI, Nunes T. Role of the microbiota in colorectal cancer: updates on microbial associations and therapeutic implications. Biores Open Access. (2016) 5:279–88. 10.1089/biores.2016.0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, et al. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. (2019) 38:14. 10.1186/s13046-018-0985-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang CY, Cusack JC, Liu R, Baldwin AS. Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med. (1999) 5:412–7. 10.1038/7410 [DOI] [PubMed] [Google Scholar]
- 69.Park SM, Yoon JB, Lee TH. Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. (2004) 566:151–6. 10.1016/j.febslet.2004.04.021 [DOI] [PubMed] [Google Scholar]
- 70.Deveraux QL, Reed JC. IAP family proteins–suppressors of apoptosis. Genes Dev. (1999) 13:239–52. 10.1101/gad.13.3.239 [DOI] [PubMed] [Google Scholar]
- 71.Miura K, Karasawa H, Sasaki I. cIAP2 as a therapeutic target in colorectal cancer and other malignancies. Expert Opin Ther Targets. (2009) 13:1333–45. 10.1517/14728220903277256 [DOI] [PubMed] [Google Scholar]
- 72.Krajewska M, Kim H, Kim C, Kang H, Welsh K, Matsuzawa S, et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res. (2005) 11:5451–61. 10.1158/1078-0432.CCR-05-0094 [DOI] [PubMed] [Google Scholar]
- 73.Karasawa H, Miura K, Fujibuchi W, Ishida K, Kaneko N, Kinouchi M, et al. Down-regulation of cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in human colon cancer cells. Cancer Sci. (2009) 100:903–13. 10.1111/j.1349-7006.2009.01112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. (2018) 68:1335–7. 10.1136/gutjnl-2018-316661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guven DC, Dizdar O, Alp A, Akdogan Kittana FN, Karakoc D, Hamaloglu E, et al. Analysis of Fusobacterium nucleatum, Streptococcus gallolyticus and Porphyromonas gingivalis in saliva in colorectal cancer patients and healthy controls. J Clin Oncol. (2018) 36(suppl 15):e15617–e. 10.1200/JCO.2018.36.15_suppl.e15617 [DOI] [Google Scholar]
- 76.Cao Y, Wu K, Mehta R, Drew DA, Song M, Lochhead P, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut. (2018) 67:672. 10.1158/1538-7445.CRC16-A24 [DOI] [PMC free article] [PubMed] [Google Scholar]