Abstract
Olfactory ensheathing cells (OECs) have neuro-restorative properties in animal models for spinal cord injury, stroke, and amyotrophic lateral sclerosis. Here we used a multistep screening approach to discover genes specifically contributing to the regeneration-promoting properties of OECs. Microarray screening of the injured olfactory pathway and of cultured OECs identified 102 genes that were subsequently functionally characterized in cocultures of OECs and primary dorsal root ganglion (DRG) neurons. Selective siRNA-mediated knockdown of 16 genes in OECs (ADAMTS1, BM385941, FZD1, GFRA1, LEPRE1, NCAM1, NID2, NRP1, MSLN, RND1, S100A9, SCARB2, SERPINI1, SERPINF1, TGFB2, and VAV1) significantly reduced outgrowth of cocultured DRG neurons, indicating that endogenous expression of these genes in OECs supports neurite extension of DRG neurons. In a gain-of-function screen for 18 genes, six (CX3CL1, FZD1, LEPRE1, S100A9, SCARB2, and SERPINI1) enhanced and one (TIMP2) inhibited neurite growth. The most potent hit in both the loss- and gain-of-function screens was SCARB2, a protein that promotes cholesterol secretion. Transplants of fibroblasts that were genetically modified to overexpress SCARB2 significantly increased the number of regenerating DRG axons that grew toward the center of a spinal cord lesion in rats. We conclude that expression of SCARB2 enhances regenerative sprouting and that SCARB2 contributes to OEC-mediated neuronal repair.
Introduction
In mammals, functional regeneration of injured nerve tracts is very limited in the CNS, but olfactory neurons that originate in the periphery do have a remarkable capacity to regrow axons into the CNS. In the olfactory neuroepithelium (OE), neurogenesis is preserved (Moulton, 1974; Graziadei and Graziadei, 1979). New olfactory neurons extend axons through the lamina propria to enter the CNS to form the olfactory nerve layer (ONL) around the olfactory bulb. The olfactory axons that enter the CNS contain a specialized type of glia cell, the olfactory ensheathing cell (OEC) (Raisman, 1985; Doucette, 1991).
In the 1990s, it was shown that OECs stimulate axon regeneration in the injured spinal cord after various types of lesions (Ramon-Cueto and Nieto-Sampedro, 1994; Li et al., 1997; Ramon-Cueto et al., 1998) can remyelinate axons (Franklin et al., 1996; Imaizumi et al., 2000; Radtke et al., 2004) and stimulate neural tissue repair and angiogenesis (Lakatos et al., 2003; Plant et al., 2003; Ramer et al., 2004; Sasaki et al., 2004; Richter et al., 2005).
OECs contribute to regeneration by expressing cell adhesion molecules (NCAM, L1, N-cadherin) and by secreting a variety of signaling molecules, including neurotrophic factors (Boruch et al., 2001; Woodhall et al., 2001; Lipson et al., 2003; Santos-Silva et al., 2007), and extracellular matrix molecules (e.g., laminin, clusterin, matrix metalloprotease 2 (MMP2), SPARC, and plasminogen activator inhibitor-1) (Doucette, 1990; Tisay and Key, 1999; Fairless et al., 2005; Pastrana et al., 2006; Au et al., 2007; Liu et al., 2010; Simon et al., 2011). Neurites of cultured primary olfactory neurons are enfolded by OECs (Ramon-Cueto et al., 1993), which promotes their extension (Kafitz and Greer, 1997; Tisay and Key, 1999). OECs provide neurotrophic support to adult retinal ganglion cells, embryonic, postnatal, and adult dorsal root ganglion neurons (DRG), postnatal cerebral cortical neurons, and postnatal hippocampal neurons (Goodman et al., 1993; Lipson et al., 2003; Chung et al., 2004; Leaver et al., 2006; Au et al., 2007; Pellitteri et al., 2009; Runyan and Phelps, 2009).
Large-scale transcriptional profiling (Vincent et al., 2005; Pastrana et al., 2006; Ruitenberg et al., 2006; Franssen et al., 2008; Guerout et al., 2010; Roet et al., 2011) and proteomics (Boyd et al., 2006; Au et al., 2007; Jahed et al., 2007; Liu et al., 2010) have elucidated molecular differences between early- and late-passage OECs (Pastrana et al., 2006), lamina propria OECs (Guerout et al., 2010), and Schwann cells (SCs) (Vincent et al., 2005; Franssen et al., 2008). However, systematic insight into the molecular mechanisms underlying the neuro-restorative capacity of OECs remained elusive.
In this study, a multilevel screening strategy was used to systematically identify genes underlying the neurite outgrowth-promoting properties of OECs, starting with two microarray studies: (1) on regeneration of the ONL after peripheral deafferentiation and (2) on cultured OECs. siRNA-mediated loss- and lentiviral-mediated gain-of-function cellular screening identified SCARB2 among the most potent genes to modify outgrowth of DRG neurons. Gene therapeutic intervention demonstrated that SCARB2 significantly enhanced the regeneration of lesioned afferent adult rat dorsal column fibers in vivo. SCARB2 is a potent enhancer of regenerative sprouting and represents a newly identified contributor of OEC-mediated neurorepair.
Materials and Methods
Animals and surgical procedures
All animal experimental procedures were approved by the animal welfare committee of the Royal Netherlands Academy of Sciences. Animals were housed under standard conditions with ad libitum access to food and water. Adult female Fischer (F344) rats (180–220 g, 8–10 weeks of age; Harlan) were used for gene expression profiling of the ONL, the dorsal column lesion experiment, and for the primary cell cultures. For the primary cell cultures, rats were deeply anesthetized with CO2 and decapitated. Embryonic DRG neuron cultures were prepared from embryonic day 15 (E15) embryos derived from timed-pregnant female Wistar rats (Harlan). OECs, skin fibroblasts, and adult and embryonic DRG neuron cultures were prepared as described below.
Olfactory epithelium lesion.
A total of 72 animals were used for transcriptional profiling of the ONL after lesioning of the olfactory epithelium. To lesion the primary sensory neurons in the olfactory neuroepithelium, animals were anesthetized lightly with 5% isoflurane and immediately given intranasal injections of 50 μl of 0.7% Triton X-100 (Sigma-Aldrich) in 0.1 m sodium phosphate buffer, pH 7.4, in each nostril. This treatment results in peripheral deafferentiation of the olfactory bulb and is based on a procedure previously documented for lesioning of the olfactory epithelium in mice (Rochel and Margolis, 1980; Nadi et al., 1981; Verhaagen et al., 1990). Nonlesioned animals (t = 0) were included as controls. Lesioned animals were killed at 1, 3, 6, 10, 15, 20, 30, and 60 d post lesion (dpl) (n = 27 to confirm the effectiveness of the lesion procedure, 3 per time point; n = 45 for microarrays, n = 5 per time point). Animals were deeply anesthetized using 0.11 ml Nembutal/100 g body weight (sodium pentobarbital; Sanofi Sante). To study the effectiveness of the lesion, animals were perfused with 4% PFA in PBS, pH 7.4, and processed for immunohistochemistry. The extent of the lesion was quantified as described below. For gene expression profiling, the ONL was dissected and RNA was isolated as described below.
Dorsal column lesion.
A total of 34 animals were used for the dorsal column lesion experiment. Of these, 2 were used to prepare the skin fibroblast cultures and 32 rats underwent surgery and received transplants of fibroblasts that were genetically modified to express GFP (n = 8), S100A9 (n = 8), SCARB2 (n = 8), or SERPINI1 (n = 8). Fibroblasts that were isolated and transduced as described below were thawed and cultured on poly-l-lysine (PLL; Sigma-Aldrich)-coated dishes. After 2 weeks, cells were harvested by trypsinization, washed with DMEM, and centrifuged (3 min at 290 × g) 3 times. The pellets were resuspended in DMEM, cells were counted, and the concentration was adjusted to 1.0 × 105 cells/μl, and cells were kept at 4°C until needed for implantation.
Implantation of fibroblasts into a dorsal column lesion was performed as follows. Rats were anesthetized with an intramuscular injection of ketamine (100 mg/kg bodyweight; Nimatek) and xylazine (5 mg/kg body weight; Rompun, Bayer). The dorsal column was lesioned at thoracic vertebra 8 (T8) as previously described (Hendriks et al., 2007). The rats were immobilized with a spinal cord fixator, after which a laminectomy at T8 exposed the spinal cord. Subsequently, a bilateral lesion (1.2 mm deep) of the dorsal columns was made using a microknife (a 25-gauge needle with a file-sharpened 1.2 mm tip bended 45°). Immediately after the lesion was made, fibroblasts were carefully injected as previously described (Ruitenberg et al., 2002; Hendriks et al., 2007). Each animal received three injections of a 1 μl suspension of fibroblasts containing 1.0 × 105 cells/μl. One injection was placed directly in the lesion, one injection 1 mm rostral from the lesion, and one 1 mm caudal from the lesion. Injections were conducted at a depth of 0.8 mm from the dorsal surface of the spinal cord and consisted of pressure injections (syringe pump, Harvard Apparatus; injection speed 0.2 μl/min). After infusions, muscle and skin were closed in separate layers. While rats were still under anesthesia, the sciatic nerve of the left hindlimb was exposed at mid thigh level and crushed for 30 s by closing a hemostatic forceps with serrated jaws, after which the skin was closed. Animals received postoperative care, including 3 d subcutaneous injections with the analgesic Temgesic (0.03 ml/100 g body weight buprenorphine hydrochloride; Schering-Plough). One rat of both the SCARB2 group and the SERPINI1 group did not survive the surgery. One rat of the SCARB2 group was taken out of the experiment after deliberation with members of the animal welfare committee because it met established humane endpoint criteria. One rat of the SERPINI1 group died prematurely of unknown causes.
Cholera toxin B subunit (CTB) tracer injection.
After 67 d, a CTB tracer was injected in the sciatic nerve. Anesthesia was induced in all rats with 5% isoflurane and maintained at 2.5%. The sciatic nerve of the left hindpaw was exposed, and each of the two branches was injected with 1 μl of CTB solution (#104; List Biological Laboratories; 10 mg/ml) mixed with 0.05 μl 1% fast blue (EMS) in PBS using a Hamilton syringe, after which the skin was closed again. Three days later, animals were deeply anesthetized using 0.11 ml Nembutal/100 g body weight (sodium pentobarbital; Sanofi Sante) and subsequently perfused with 4% PFA in PBS.
Microarray analysis and candidate gene selection
RNA isolation and microarray hybridization.
From each animal, both olfactory bulbs were dissected, and each bulb was cut into 400-μm-thick slices with a McIlwain tissue chopper. The ONL was manually dissected from these tissue slices using a laser dissection microscope. Tissue from the ONL of two bulbs was pooled, and RNA was isolated from the ONLs with the Rneasy Mini kit (QIAGEN) according to the manufacturer's protocol. For microarray analysis, equal amounts of RNA from five individual animals was pooled per time point, amplified, and labeled with Cy3-CTP and Cy5-CTP using the Agilent Low RNA Input Fluorescent Linear Amplification kit (5184–3523) according to the manufacturer's guidelines (Agilent Technologies). A D-optimal (Montgomery, 2001) and admissible (Glonek and Solomon, 2004) design was created. Each pool was measured in quadruplicate over four arrays with a balanced dye swap. A total of 18 microarrays was used. Hybridization was performed with the Agilent In Situ Hybridization Kit (G2559A, Agilent Technologies) according to the manufacturer's protocol. Microarrays were scanned with an Agilent microarray scanner (G2505A), and Agilent Feature Extraction software was used to extract the intensity data (Agilent Technologies). Per probe, local background and foreground levels were obtained for both red and green channels. We also used a second microarray dataset that has been published previously (Franssen et al., 2008a). In this study, the transcriptome of cultured OECs was compared with that of cultured SCs or the intact ONL from adult Fischer 344 rats. In this study, RNA hybridization and microarray hybridization were performed as described above. The data from 57 microarray probes were verified by quantitative real-time PCR (qPCR) (data not shown). RNA was derived from the same RNA samples that had been used for the microarray study.
Data analysis and selection of candidate genes.
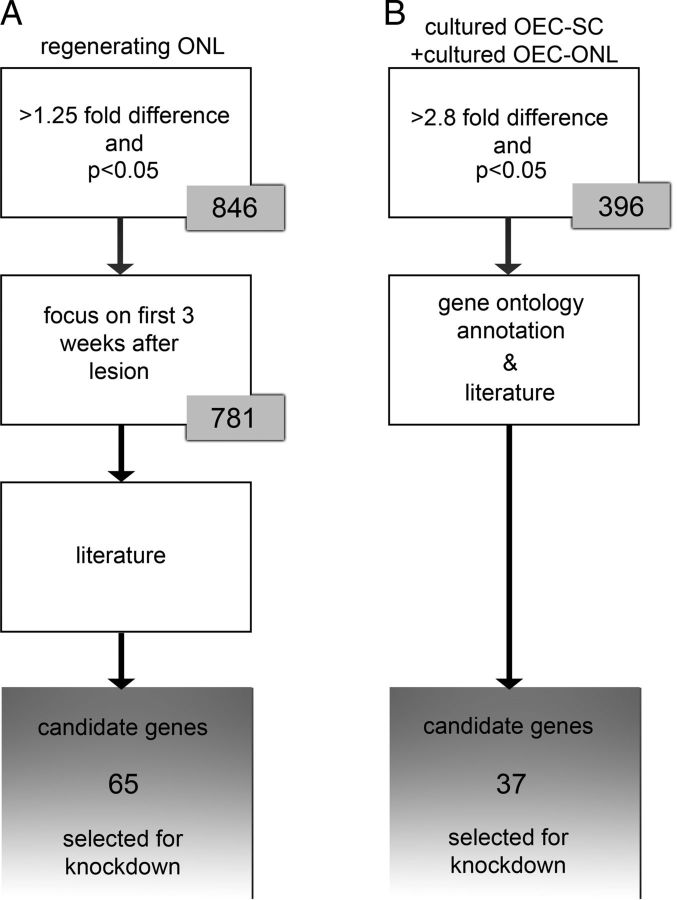
Data analysis was performed in R (http://www.r-project.org), using the packages marray and limma. Internal normalization of the arrays was based on Loess, and global normalization was based on the median absolute deviation. Normalized data were modeled in limma. For the regenerating olfactory nerve layer, differential gene expression was calculated at each time point relative to day 0 (nonlesioned control). Uncorrected p values were corrected for multiple testing using the Benjamini-Hochberg correction algorithm. Gene expression profiles of regulated genes were subjected to a trend clustering in R, which was based on Pearson's noncentered correlation. Genes that were exclusively regulated at 1 or 60 dpl were left out of the clustering. Clustering was done based on a correlation threshold of 90% on all gene expression profiles. The first 3 weeks after the lesion is the postlesion period in which the newly formed primary olfactory neurons extend their axons through the ONL toward the glomeruli. A total of 65 genes that were at least 1.25-fold upregulated with an adjusted p < 0.05 within the first 3 weeks after the lesion were selected by means of an extensive literature study to reveal genes that had the greatest potential to be involved in various aspect of neurite outgrowth, including neurite outgrowth per se, cellular adhesion, modification of the extracellular environment, cell migration, and cell growth. To compare cultured OECs with cultured SCs or the intact ONL, the differential expression was calculated for each gene as described previously (Franssen et al., 2008a). Three selection criteria were applied. First, only genes that were 2.8-fold higher expressed in OECs with an adjusted p < 0.05 were included. Second, the web-based tool eGOn (http://www.genetools.microarray.ntnu.no/common/intro.php) (Beisvag et al., 2006) was used to select genes with a gene ontology (GO) annotation as listed in Table 1. A third and parallel selection criterion was based on an extensive literature study as indicated above. This resulted in the selection of 37 genes. We predicted that, based on these criteria, we would select for genes that would potentially be involved in stimulating axon outgrowth. Figure 1 shows a schematic representation of the entire selection procedure, with the selected genes shown in Table 2.
Table 1.
Gene ontology classes including their children that were used for selection of target genesa
| Gene ontology classes | Characteristic |
|---|---|
| Biological process | |
| GO:0009653 | Anatomical structure morphogenesis |
| GO:0007155 | Cell adhesion |
| GO:0007154 | Cell communication |
| GO:0016049 | Cell growth |
| GO:0006928 | Cell motility |
| GO:0016043 | Cellular component organization and biogenesis |
| GO:0040007 | Growth |
| GO:0040011 | Locomotion |
| GO:0050896 | Response to stimulus |
| GO:0009888 | Tissue development |
| Cellular component | |
| GO:0009986 | Cell surface |
| GO:0031012 | Extracellular matrix |
| GO:0005576 | Extracellular region |
| GO:0016020 | Membrane |
aTo select genes involved in the “stimulation of neurite outgrowth,” all target genes belonging to the represented gene ontology classes or to one of the subordinate branches were selected.
Figure 1.
Schematic representation of selection criteria used to refine the number of target genes. The number of selected genes in each step is represented. A, Selection criteria used for gene expression analysis of the regenerating ONL after lesion. First, genes >1.25-fold upregulated after injury were included. p < 0.05. Second, only genes differentially expressed in the first 3 weeks after the lesion were selected. Third, genes that could be involved in “axon outgrowth” based on their function as documented in the literature were selected. This resulted in a list of 65 target genes. B, Selection criteria used for gene expression analysis of cultured OECs. Similar selection criteria were applied to the comparisons of cultured OEC-SC and the cultured OEC-intact ONL. First, only genes higher expressed in OEC compared with SCs, as well compared with the intact ONL, were included (>2.8-fold expression difference; p < 0.05). Second, genes that could be involved in “axon outgrowth” were selected based on their GO annotation (Table 1). Third, in parallel to the GO analysis, an extensive literature study was used to further select those genes that, based on their proposed or known function in neurite outgrowth per se, cellular adhesion, modification of the extracellular environment, cell migration, and cell growth, had the greatest potential to be involved in various aspects of axon outgrowth. This resulted in a list of 37 genes that are higher expressed in OECs than SCs and/or the intact ONL. In total, 102 genes were selected for siRNA-mediated knockdown.
Table 2.
The 102 selected genes for the siRNA screens of the microarray analysis in the olfactory pathway after peripheral deafferentiation and of cultured OECs
| Gene symbol | Accession number | Gene name | siRNA screena | Fold change in gene expression (days after lesion) |
p value of fold change |
dpl 30 | dpl 60 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 10 | 15 | 20 | 30 | 60 | dpl 1 | dpl 3 | dpl 6 | dpl 10 | dpl 15 | dpl 20 | ||||||
| ACP5 | NM_019144 | Acid phosphatase 5, tartrate resistant | 1 | 1.05 | 1.24 | 1.38 | 1.45 | 1.46 | 1.08 | 1.04 | 1.16 | 0.28 | 0.00 | 0.00 | 0.00 | 0.00 | 0.08 | 0.43 | 0.01 |
| ADAMTS1 | NM_024400 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 2 | 1.42 | 1.04 | 1.30 | 1.04 | −0.49 | 1.07 | 1.11 | 1.21 | 0.02 | 0.75 | 0.05 | 0.74 | 0.84 | 0.57 | 0.45 | 0.16 |
| ADM | NM_012715 | Adrenomedullin | 2 | −1.19 | −1.08 | 1.01 | 1.14 | −0.45 | −1.07 | −1.14 | 1.05 | 0.32 | 0.66 | 0.94 | 0.43 | 0.49 | 0.69 | 0.47 | 0.80 |
| α 7A integrin | AA875092 | Integrin, α 7 | 2 | −1.19 | 1.10 | 1.06 | 1.02 | 1.13 | 1.27 | 1.12 | −1.04 | 0.01 | 0.14 | 0.33 | 0.76 | 0.04 | 0.00 | 0.09 | 0.50 |
| ANXA3 | NM_012823 | Annexin A3 | 1 | 1.73 | 1.21 | 1.51 | 1.37 | 1.21 | 1.19 | −1.01 | 1.41 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.79 | 0.00 |
| ANXA5 | NM_013132 | Annexin A5 | 2 | −1.29 | −1.10 | −1.01 | 1.09 | −0.47 | −1.12 | −1.13 | −1.04 | 0.00 | 0.16 | 0.83 | 0.15 | 0.79 | 0.07 | 0.08 | 0.51 |
| APLN | NM_031612 | Apelin | 2 | 1.92 | 1.12 | 1.39 | 1.24 | 1.16 | 1.02 | 1.04 | 1.31 | 0.00 | 0.30 | 0.00 | 0.04 | 0.15 | 0.81 | 0.72 | 0.02 |
| BGN | NM_017087 | Biglycan | 2 | −1.19 | 1.07 | 1.06 | 1.02 | 1.09 | 1.15 | 1.07 | −1.10 | 0.01 | 0.26 | 0.28 | 0.67 | 0.13 | 0.02 | 0.28 | 0.13 |
| CCK | NM_012829 | Cholecystokinin | 1 | 2.02 | 1.07 | 1.28 | 1.32 | 1.64 | −1.04 | 1.17 | 1.33 | 0.00 | 0.19 | 0.00 | 0.00 | 0.00 | 0.46 | 0.01 | 0.00 |
| CD44 | NM_012924 | CD44 molecule (Indian blood group) | 2 | 1.48 | 1.10 | 1.27 | 1.15 | 1.16 | 1.11 | 1.01 | 1.20 | 0.00 | 0.41 | 0.03 | 0.17 | 0.16 | 0.31 | 0.96 | 0.10 |
| CD9 | AW253649 | CD9 molecule | 2 | −1.16 | 1.02 | −1.07 | −1.03 | −2.04 | 1.02 | −1.06 | −1.18 | 0.44 | 0.93 | 0.69 | 0.86 | 0.68 | 0.93 | 0.78 | 0.40 |
| CDH3 | AI010270 | Cadherin 3, type 1, P-cadherin (placental) | 2 | −1.20 | −1.02 | −1.08 | −1.09 | −2.13 | 1.01 | 1.18 | 1.22 | 0.42 | 0.94 | 0.72 | 0.68 | 0.72 | 0.96 | 0.46 | 0.37 |
| CDHR1 | NM_053572 | Cadherin-related family member 1 | 2 | 2.78 | 1.38 | 2.07 | 1.29 | 1.57 | −1.05 | 1.17 | 1.37 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.49 | 0.06 | 0.00 |
| CNN1 | NM_031747 | Calponin 1, basic, smooth muscle | 1 | 1.01 | −1.03 | 1.04 | 1.11 | 1.01 | −1.06 | −1.09 | 1.01 | 0.84 | 0.63 | 0.44 | 0.07 | 0.89 | 0.29 | 0.18 | 0.88 |
| CNTF | NM_013166 | Ciliary neurotrophic factor | 2 | −1.09 | 1.06 | 1.03 | −1.01 | −2.02 | 1.00 | −1.07 | 1.17 | 0.09 | 0.21 | 0.52 | 0.73 | 0.69 | 0.91 | 0.15 | 0.00 |
| CTGF | NM_022266 | Connective tissue growth factor | 1 | 1.26 | 1.27 | 1.40 | 1.27 | 1.13 | −1.08 | −1.08 | 1.31 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.17 | 0.24 | 0.00 |
| CTHRC1 | NM_172333 | Collagen triple helix repeat containing 1 | 2 | −1.17 | −1.06 | −1.02 | −1.01 | −2.01 | 1.14 | 1.19 | 1.19 | 0.29 | 0.70 | 0.85 | 0.94 | 0.70 | 0.31 | 0.23 | 0.24 |
| CTSC | NM_017097 | Cathepsin C | 2 | −1.05 | 1.58 | 1.82 | 1.51 | 1.27 | 1.43 | 1.04 | 1.34 | 0.80 | 0.05 | 0.01 | 0.05 | 0.23 | 0.08 | 0.86 | 0.18 |
| CTSL2 | NM_013156 | Cathepsin L2 | 2 | 1.30 | 1.16 | 1.28 | 1.28 | 1.16 | 1.10 | 1.00 | 1.27 | 0.00 | 0.02 | 0.00 | 0.00 | 0.01 | 0.07 | 0.94 | 0.00 |
| CX3CL1 | NM_134455 | Chemokine (C-X3-C motif) ligand 1 | 2 | 2.14 | 1.25 | 1.89 | 1.43 | 1.49 | 1.20 | 1.30 | 1.44 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 |
| CYR61 | NM_031327 | Cysteine-rich, angiogenic inducer, 61 | 2 | 1.03 | 1.02 | 1.20 | 1.08 | −0.47 | −1.03 | −1.15 | 1.06 | 0.85 | 0.86 | 0.16 | 0.52 | 0.42 | 0.81 | 0.32 | 0.68 |
| DBC1 | NM_080482 | Deleted in bladder cancer 1 | 2 | 1.76 | 1.06 | 1.34 | 1.03 | 1.09 | −1.24 | −1.09 | 1.16 | 0.00 | 0.55 | 0.01 | 0.72 | 0.39 | 0.03 | 0.41 | 0.17 |
| EGR1 | NM_012551 | Early growth response 1 | 1 | 1.60 | 1.25 | 2.62 | 1.26 | 1.24 | 1.39 | 1.56 | 1.24 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 |
| ENPP3 | NM_019370 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | 1 | −1.05 | −1.04 | −1.02 | 1.02 | −0.49 | −1.02 | −1.01 | −1.11 | 0.52 | 0.56 | 0.80 | 0.72 | 0.57 | 0.75 | 0.88 | 0.17 |
| F2R | NM_012950 | Coagulation factor II (thrombin) receptor | 1 | 1.07 | −1.04 | −1.02 | 1.07 | 1.01 | −1.07 | −1.05 | 1.01 | 0.31 | 0.52 | 0.78 | 0.26 | 0.86 | 0.28 | 0.50 | 0.89 |
| FEN1 | NM_053430 | Flap structure-specific endonuclease 1 | 1 | 1.69 | 1.15 | 1.32 | 1.16 | 1.10 | 1.00 | 1.06 | 1.21 | 0.00 | 0.01 | 0.00 | 0.00 | 0.03 | 0.92 | 0.18 | 0.00 |
| FEZ1 | NM_031066 | Fasciculation and elongation protein ζ 1 (zygin I) | 2 | 1.12 | 1.12 | −1.00 | −1.11 | 1.07 | −1.03 | 1.05 | 1.05 | 0.36 | 0.39 | 0.98 | 0.38 | 0.54 | 0.81 | 0.68 | 0.70 |
| FGFRL1 | BQ205363 | Fibroblast growth factor receptor-like 1 | 1 | 1.27 | 1.13 | 1.44 | 1.30 | 1.27 | 1.17 | 1.15 | 1.17 | 0.00 | 0.05 | 0.00 | 0.00 | 0.00 | 0.01 | 0.02 | 0.01 |
| FZD1 | NM_021266 | Frizzled homolog 1 (Drosophila) | 2 | −1.01 | 1.11 | 1.12 | 1.08 | 1.03 | 1.21 | 1.08 | 1.21 | 0.92 | 0.42 | 0.32 | 0.52 | 0.79 | 0.11 | 0.55 | 0.14 |
| GADD45G | BF419904 | Growth arrest and DNA-damage-inducible, γ | 1 | 2.03 | 1.22 | 1.84 | 1.28 | 1.10 | 1.09 | 1.07 | 1.31 | 0.00 | 0.06 | 0.00 | 0.02 | 0.30 | 0.36 | 0.53 | 0.02 |
| GFRA1 | NM_012959 | GDNF family receptor α 1 | 2 | 1.31 | −1.01 | 1.09 | −1.03 | −2.05 | 1.01 | −1.02 | 1.09 | 0.00 | 0.93 | 0.19 | 0.60 | 0.73 | 0.91 | 0.77 | 0.25 |
| GLIPR1 | AI012463 | GLI pathogenesis-related 1 | 1 | 1.11 | 1.18 | 1.44 | 1.28 | 1.11 | 1.09 | −1.01 | 1.15 | 0.22 | 0.07 | 0.00 | 0.01 | 0.20 | 0.28 | 0.94 | 0.12 |
| GPC1 | NM_030828 | Glypican 1 | 2 | 1.10 | 1.01 | 1.04 | 1.10 | 1.15 | 1.00 | 1.03 | −1.03 | 0.14 | 0.88 | 0.46 | 0.12 | 0.03 | 0.95 | 0.68 | 0.58 |
| GPNMB | NM_133298 | Glycoprotein (transmembrane) nmb | 2 | 1.81 | 1.77 | 7.01 | 3.27 | 1.98 | 1.41 | −1.15 | 1.35 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.28 | 0.03 |
| ICAM1 | NM_012967 | Intercellular adhesion molecule 1 | 1 | 1.45 | 1.10 | 1.64 | 1.28 | 1.27 | 1.26 | 1.15 | 1.02 | 0.00 | 0.23 | 0.00 | 0.01 | 0.01 | 0.01 | 0.10 | 0.83 |
| IGF2R | NM_012756 | Insulin-like growth factor 2 receptor | 1 | 1.04 | 1.06 | 1.07 | 1.08 | 1.05 | −1.07 | −1.06 | 1.14 | 0.56 | 0.40 | 0.23 | 0.18 | 0.40 | 0.23 | 0.39 | 0.06 |
| IGFBP2 | NM_013122 | Insulin-like growth factor binding protein 2, 36 kDa | 1 | −1.27 | 1.01 | 1.28 | 1.80 | 1.36 | 1.11 | −1.11 | −1.09 | 0.01 | 0.91 | 0.00 | 0.00 | 0.00 | 0.14 | 0.20 | 0.25 |
| IGFBP3 | NM_012588 | Insulin-like growth factor binding protein 3 | 1 | −1.11 | −1.57 | −1.69 | −1.66 | −3.16 | −1.55 | −1.08 | −1.26 | 0.41 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.52 | 0.08 |
| IGSF1 | NM_175763 | Ig superfamily, member 1 | 2 | 1.52 | 1.18 | 1.33 | 1.32 | 1.25 | −1.05 | −1.07 | 1.26 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.33 | 0.18 | 0.00 |
| ITIH3 (PAIHC3) | NM_017351 | Inter-α (globulin) inhibitor H3 | 2 | 1.82 | 1.59 | 2.20 | 1.53 | 1.38 | 1.03 | 1.03 | 1.40 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.75 | 0.80 | 0.00 |
| LEPRE1 | NM_053667 | Leucine proline-enriched proteoglycan (leprecan) 1 | 1 | −1.08 | 1.06 | 1.00 | 1.03 | 1.05 | 1.05 | 1.05 | 1.02 | 0.09 | 0.19 | 0.96 | 0.49 | 0.26 | 0.24 | 0.26 | 0.67 |
| LGALS3 | NM_031832 | Lectin, galactoside-binding, soluble, 3 | 1 | 3.22 | 1.33 | 2.94 | 1.85 | 1.31 | 1.26 | 1.06 | 1.22 | 0.00 | 0.04 | 0.00 | 0.00 | 0.03 | 0.05 | 0.62 | 0.13 |
| LPL | NM_012598 | Lipoprotein lipase | 2 | 1.26 | 1.11 | 1.42 | 1.45 | 1.08 | 1.03 | −1.11 | 1.23 | 0.00 | 0.04 | 0.00 | 0.00 | 0.10 | 0.46 | 0.04 | 0.00 |
| LTBP2 | NM_021586 | Latent-transforming growth factor β binding protein 2 | 2 | −1.09 | −1.10 | −1.03 | −1.01 | −2.02 | −1.11 | −1.10 | −1.12 | 0.12 | 0.09 | 0.50 | 0.81 | 0.13 | 0.06 | 0.11 | 0.05 |
| LYN | NM_030857 | v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog | 1 | 1.46 | 1.14 | 1.32 | 1.16 | 1.07 | 1.11 | 1.07 | 1.07 | 0.00 | 0.05 | 0.00 | 0.02 | 0.23 | 0.07 | 0.28 | 0.24 |
| MGP | NM_012862 | Matrix Gla protein | 2 | 1.16 | 1.09 | 1.38 | 1.39 | 1.19 | −1.13 | −1.12 | 1.26 | 0.10 | 0.33 | 0.00 | 0.00 | 0.05 | 0.13 | 0.21 | 0.02 |
| MME | NM_012608 | Membrane metallo-endopeptidase | 1 | −1.18 | 1.00 | −1.04 | 1.05 | −0.48 | −1.06 | −1.01 | 1.12 | 0.05 | 0.98 | 0.55 | 0.50 | 0.28 | 0.43 | 0.88 | 0.15 |
| MSLN | NM_031658 | Mesothelin | 1 | −1.58 | −1.09 | 1.01 | 1.34 | 1.15 | −1.05 | −1.25 | −1.13 | 0.00 | 0.21 | 0.80 | 0.00 | 0.03 | 0.43 | 0.00 | 0.06 |
| NCAM1 | NM_031521 | Neural cell adhesion molecule 1 | 1 | 1.05 | 1.09 | 1.10 | −1.06 | 1.30 | 1.54 | 1.44 | 1.10 | 0.42 | 0.15 | 0.08 | 0.25 | 0.00 | 0.00 | 0.00 | 0.11 |
| NDRG4 | NM_031967 | NDRG family member 4 | 1 | 2.28 | 1.18 | 1.62 | 1.12 | 1.41 | −1.11 | 1.10 | 1.25 | 0.00 | 0.01 | 0.00 | 0.03 | 0.00 | 0.05 | 0.10 | 0.00 |
| NGFR | NM_012610 | Nerve growth factor receptor | 1 | −1.08 | −1.05 | −1.12 | −1.09 | −2.13 | −1.12 | −1.23 | −1.11 | 0.48 | 0.67 | 0.25 | 0.40 | 0.20 | 0.28 | 0.08 | 0.34 |
| NID2 | BG671163 | Nidogen 2 (osteonidogen) | 2 | −1.16 | −1.01 | 1.03 | 1.20 | 1.06 | 1.13 | 1.12 | 1.13 | 0.01 | 0.87 | 0.55 | 0.00 | 0.20 | 0.02 | 0.05 | 0.03 |
| NPTX1 | NM_153735 | Neuronal pentraxin I | 1 | 1.81 | 1.10 | 1.30 | 1.12 | 1.41 | 1.03 | 1.16 | 1.37 | 0.00 | 0.51 | 0.05 | 0.36 | 0.02 | 0.83 | 0.29 | 0.03 |
| NRP1 | NM_145098 | Neuropilin 1 | 1 | 1.13 | 1.16 | 1.54 | 1.48 | 1.26 | 1.05 | 1.07 | 1.04 | 0.07 | 0.03 | 0.00 | 0.00 | 0.00 | 0.37 | 0.28 | 0.50 |
| NRTN | BF407571 | Neurturin | 1 | 1.52 | 1.18 | 1.38 | 1.23 | 1.17 | 1.16 | 1.07 | 1.14 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.22 | 0.02 |
| NRXN1 | NM_021767 | Neurexin 1 | 2 | 1.60 | 1.12 | 1.21 | 1.00 | 1.07 | −1.27 | −1.10 | 1.00 | 0.00 | 0.20 | 0.03 | 0.96 | 0.37 | 0.01 | 0.29 | 0.97 |
| NTRK2 | NM_012731 | Neurotrophic tyrosine kinase, receptor, type 2 | 1 | 1.89 | 1.02 | 1.17 | 1.15 | 1.21 | −1.11 | −1.01 | 1.36 | 0.00 | 0.58 | 0.00 | 0.00 | 0.00 | 0.01 | 0.74 | 0.00 |
| NTSR2 | NM_022695 | Neurotensin receptor 2 | 1 | 1.44 | 1.12 | 1.37 | 1.38 | 1.25 | 1.05 | −1.04 | 1.17 | 0.00 | 0.07 | 0.00 | 0.00 | 0.00 | 0.39 | 0.56 | 0.02 |
| OLFM1 | NM_053573 | Olfactomedin 1 | 2 | 1.60 | −1.07 | 1.21 | −1.00 | 1.10 | −1.05 | 1.04 | 1.07 | 0.00 | 0.18 | 0.00 | 0.95 | 0.05 | 0.26 | 0.38 | 0.15 |
| PCOLCE | NM_019237 | Procollagen C-endopeptidase enhancer | 1 | −1.26 | 1.00 | 1.11 | 1.42 | 1.16 | 1.10 | −1.04 | −1.05 | 0.00 | 0.96 | 0.03 | 0.00 | 0.00 | 0.05 | 0.37 | 0.32 |
| PENK | NM_017139 | Proenkephalin | 1 | 1.96 | 1.09 | 1.46 | 1.36 | 1.48 | 1.04 | 1.18 | 1.27 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.29 | 0.00 | 0.00 |
| PLUNC | NM_172031 | Palate, lung, and nasal epithelium associated | 2 | −1.08 | 3.23 | 1.20 | −1.20 | 1.27 | −1.34 | 2.69 | 1.30 | 0.63 | 0.00 | 0.25 | 0.26 | 0.15 | 0.07 | 0.00 | 0.14 |
| PMP22 | NM_017037 | Peripheral myelin protein 22 | 1 | −1.01 | 1.15 | 1.13 | 1.12 | 1.16 | 1.08 | 1.15 | 1.09 | 0.84 | 0.06 | 0.07 | 0.10 | 0.04 | 0.24 | 0.06 | 0.23 |
| PTK2B | NM_017318 | PTK2B protein tyrosine kinase 2 β | 1 | 1.61 | 1.02 | 1.29 | −1.00 | 1.04 | −1.01 | 1.15 | 1.19 | 0.00 | 0.66 | 0.00 | 0.94 | 0.44 | 0.89 | 0.03 | 0.01 |
| PTPN6 | NM_053908 | Protein tyrosine phosphatase, nonreceptor type 6 | 1 | 1.40 | 1.43 | 1.87 | 1.56 | 1.26 | 1.17 | 1.03 | 1.09 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.60 | 0.16 |
| PTPRC | NM_001109887 | Protein tyrosine phosphatase, receptor type, C | 2 | 1.38 | 1.29 | 1.58 | 1.37 | 1.10 | 1.04 | −1.06 | 1.26 | 0.00 | 0.00 | 0.00 | 0.00 | 0.07 | 0.42 | 0.31 | 0.00 |
| PTPRF | NM_019249 | Protein tyrosine phosphatase, receptor type, F | 2 | 1.31 | 1.07 | 1.15 | 1.14 | 1.08 | −1.14 | −1.03 | 1.00 | 0.00 | 0.25 | 0.01 | 0.02 | 0.15 | 0.02 | 0.64 | 0.95 |
| PTPRO | NM_017336 | Protein tyrosine phosphatase, receptor type, O | 1 | 1.86 | 1.22 | 1.49 | 1.32 | 1.29 | 1.05 | 1.16 | 1.18 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.18 | 0.00 | 0.00 |
| QKI | AA891889 | Quaking homolog, KH domain RNA binding (mouse) | 2 | −1.26 | 1.08 | 1.12 | 1.19 | 1.36 | 1.52 | 1.43 | −1.24 | 0.02 | 0.43 | 0.18 | 0.05 | 0.00 | 0.00 | 0.00 | 0.03 |
| RAMP3 | NM_020100 | Receptor (G protein-coupled) activity modifying protein 3 | 1 | −1.08 | −1.01 | −1.03 | −1.04 | −2.06 | −1.00 | −1.03 | −1.06 | 0.12 | 0.81 | 0.44 | 0.33 | 0.41 | 0.90 | 0.46 | 0.18 |
| RARA | NM_031528 | Retinoic acid receptor, α | 2 | −1.29 | −1.09 | −1.01 | 1.02 | 1.04 | 1.16 | −1.01 | 1.10 | 0.00 | 0.09 | 0.84 | 0.59 | 0.40 | 0.00 | 0.77 | 0.05 |
| RARRES1 | AA819288 | Retinoic acid receptor responder (tazarotene induced) 1 | 1 | 1.63 | 1.76 | 2.99 | 2.23 | 1.51 | 1.30 | 1.05 | 1.43 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.65 | 0.00 |
| RND1 | NM_001013222 | Rho family GTPase 1 | 2 | 1.51 | 1.00 | 1.64 | 1.04 | 1.09 | 1.08 | 1.12 | 1.14 | 0.00 | 0.98 | 0.00 | 0.46 | 0.08 | 0.14 | 0.06 | 0.03 |
| S100A9 | NM_053587 | S-100 calcium binding protein A9 | 1 | 1.12 | 1.01 | 1.97 | 1.02 | 1.01 | −1.03 | −1.05 | 1.10 | 0.20 | 0.88 | 0.00 | 0.82 | 0.86 | 0.67 | 0.59 | 0.26 |
| SCARB2 | NM_054001 | Scavenger receptor class B, member 2 | 1 | 1.67 | 1.11 | 1.26 | 1.13 | 1.02 | 1.05 | 1.06 | 1.14 | 0.00 | 0.03 | 0.00 | 0.01 | 0.64 | 0.20 | 0.19 | 0.01 |
| SERPINF1 (PEDF) | AI179984 | Serpin peptidase inhibitor, clade F (α-2 anti-plasmin, pigment epithelium-derived factor), member 1 | 2 | −1.14 | −1.08 | −1.10 | 1.21 | 1.14 | −1.04 | −1.01 | 1.10 | 0.01 | 0.10 | 0.02 | 0.00 | 0.00 | 0.28 | 0.79 | 0.03 |
| SERPINI1 | NM_053779 | Serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 1 | 2.05 | 1.20 | 1.41 | 1.31 | 1.34 | −1.13 | 1.08 | 1.36 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.06 | 0.27 | 0.00 |
| SGCG | AA850867 | Sarcoglycan, γ (35 kDa dystrophin-associated glycoprotein) | 2 | 1.33 | 1.11 | 1.52 | 1.13 | 1.16 | 1.06 | −1.03 | 1.12 | 0.17 | 0.61 | 0.04 | 0.51 | 0.43 | 0.75 | 0.90 | 0.56 |
| SKAP2 | NM_130413 | src kinase-associated phosphoprotein 2 | 1 | 1.25 | 1.23 | 1.44 | 1.30 | 1.13 | 1.02 | −1.05 | 1.15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.21 | 0.00 |
| SLC7A5 | NM_017353 | Solute carrier family 7 (cationic amino acid transporter, y + system), member 5 | 1 | 1.07 | 1.17 | 1.14 | 1.02 | 1.11 | 1.23 | 1.13 | 1.22 | 0.79 | 0.52 | 0.56 | 0.92 | 0.65 | 0.35 | 0.61 | 0.42 |
| SNAP25 | NM_030991 | Synaptosomal-associated protein, 25 kDa | 2 | 1.89 | 1.04 | 1.34 | −1.07 | 1.14 | −1.23 | 1.03 | 1.29 | 0.00 | 0.69 | 0.00 | 0.42 | 0.16 | 0.03 | 0.73 | 0.02 |
| SOD3 | NM_012880 | Superoxide dismutase 3, extracellular | 2 | −1.17 | 1.06 | 1.04 | 1.04 | 1.03 | 1.13 | 1.15 | 1.06 | 0.00 | 0.21 | 0.31 | 0.37 | 0.45 | 0.01 | 0.01 | 0.18 |
| SPARC | NM_012656 | Secreted protein, acidic, cysteine-rich (osteonectin) | 2 | 1.06 | 1.13 | 1.05 | 1.14 | 1.04 | −1.03 | 1.05 | −1.22 | 0.27 | 0.03 | 0.25 | 0.01 | 0.38 | 0.53 | 0.35 | 0.00 |
| SPOCK2 | BG381644 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 | 2 | 1.48 | 1.26 | 1.21 | 1.14 | 1.33 | 1.28 | 1.20 | 1.15 | 0.00 | 0.01 | 0.01 | 0.07 | 0.00 | 0.00 | 0.03 | 0.07 |
| SPP1 | NM_012881 | Secreted phosphoprotein 1 | 2 | 1.94 | 1.33 | 1.55 | 1.18 | 1.19 | 1.15 | 1.02 | 1.24 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.64 | 0.00 |
| STMN1 | NM_017166 | Stathmin 1 | 1 | 1.42 | 1.04 | 1.15 | 1.01 | 1.08 | 1.00 | 1.02 | 1.07 | 0.00 | 0.40 | 0.00 | 0.79 | 0.10 | 0.90 | 0.70 | 0.17 |
| SUSD3 | AA866228 | Sushi domain containing 3 | 2 | −1.34 | 1.29 | 1.34 | 1.46 | 1.16 | 1.16 | 1.09 | 1.22 | 0.01 | 0.02 | 0.00 | 0.00 | 0.11 | 0.10 | 0.37 | 0.05 |
| TEC | BE097141 | tec protein tyrosine kinase | 2 | −1.29 | 1.10 | 1.05 | 1.10 | 1.10 | 1.16 | 1.13 | 1.18 | 0.02 | 0.32 | 0.57 | 0.29 | 0.27 | 0.10 | 0.20 | 0.10 |
| TGFB2 | BF420705 | Transforming growth factor, β 2 | 1 | −1.04 | 1.06 | 1.33 | 1.17 | 1.23 | 1.18 | 1.14 | −1.03 | 0.64 | 0.50 | 0.00 | 0.06 | 0.02 | 0.05 | 0.14 | 0.72 |
| THBS2 | BF559836 | Thrombospondin 2 | 2 | −1.35 | 1.18 | 1.20 | 1.36 | 1.34 | 1.57 | 1.14 | 1.46 | 0.01 | 0.13 | 0.07 | 0.01 | 0.01 | 0.00 | 0.24 | 0.00 |
| TIMP2 | NM_021989 | TIMP metallopeptidase inhibitor 2 | 1 | −1.16 | 1.18 | 1.39 | 1.47 | 1.20 | 1.03 | −1.10 | 1.03 | 0.15 | 0.11 | 0.00 | 0.00 | 0.06 | 0.75 | 0.34 | 0.75 |
| TINAGL1 (LCN7) | NM_053582 | Tubulointerstitial nephritis antigen-like 1 | 2 | 1.00 | 1.07 | −1.01 | 1.08 | −0.47 | −1.08 | −1.07 | −1.07 | 0.99 | 0.38 | 0.91 | 0.28 | 0.63 | 0.26 | 0.40 | 0.37 |
| TNC | BM387134 | Tenascin C | 1 | −1.64 | −1.07 | −1.12 | −1.06 | 1.11 | 1.35 | 1.38 | 1.27 | 0.00 | 0.11 | 0.01 | 0.11 | 0.02 | 0.00 | 0.00 | 0.00 |
| TPBG | NM_031807 | Trophoblast glycoprotein | 2 | 1.73 | 1.25 | 1.32 | 1.19 | 1.30 | 1.10 | 1.14 | 1.19 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.14 | 0.06 | 0.02 |
| TRH | NM_013046 | Thyrotropin-releasing hormone | 1 | 2.08 | 1.28 | 1.42 | 1.36 | 1.45 | 1.18 | 1.26 | 1.30 | 0.00 | 0.04 | 0.00 | 0.01 | 0.00 | 0.10 | 0.05 | 0.02 |
| TYROBP | AW915488 | TYRO protein tyrosine kinase binding protein | 1 | 1.29 | 1.64 | 2.27 | 1.71 | 1.49 | 1.36 | 1.12 | 1.24 | 0.20 | 0.02 | 0.00 | 0.01 | 0.04 | 0.09 | 0.57 | 0.27 |
| UPK1B | BE096378 | Uroplakin 1B | 2 | −1.57 | −1.02 | −1.06 | 1.32 | 1.16 | 1.09 | −1.10 | −1.06 | 0.00 | 0.85 | 0.62 | 0.03 | 0.19 | 0.44 | 0.46 | 0.65 |
| VAV1 | NM_012759 | vav 1 guanine nucleotide exchange factor | 2 | 1.37 | 1.61 | 2.07 | 1.62 | 1.26 | 1.17 | 1.00 | 1.15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.95 | 0.05 |
| VGF | NM_030997 | VGF nerve growth factor inducible | 1 | 1.87 | 1.14 | 1.36 | 1.33 | 1.78 | 1.14 | 1.15 | 1.49 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.05 | 0.07 | 0.00 |
| VIP | AI412212 | Vasoactive intestinal peptide | 1 | 1.68 | 1.12 | 1.20 | 1.07 | 1.28 | 1.04 | 1.04 | 1.17 | 0.00 | 0.31 | 0.09 | 0.51 | 0.03 | 0.73 | 0.71 | 0.16 |
| WAS | AW534613 | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | 1 | 1.21 | 1.55 | 1.47 | 1.56 | 1.27 | 1.11 | 1.04 | 1.24 | 0.06 | 0.00 | 0.00 | 0.00 | 0.02 | 0.25 | 0.66 | 0.04 |
| BM385941 | Transcribed locus | 1 | 1.25 | 1.11 | 1.32 | 1.26 | 1.44 | 1.11 | 1.13 | 1.12 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.07 | 0.06 | 0.07 | |
aThis column shows whether the gene is tested in phase 1 or phase 2 of the siRNA screen. For each gene, the relative fold change in gene expression for the time points 1, 3, 6, 10, 15, 20, 30, and 60 dpl compared with the gene expression in nonlesioned animals is presented together with the calculated p values for each time point. A p value of 0.00 indicates a p value < 0.005.
For target genes selected based on the gene expression analysis of the regenerating olfactory nerve, we verified whether they were indeed expressed in purified cultured OECs. Genes with a foreground expression level >2 times background in the microarray analysis of the cultured OECs were included as genes being expressed in cultured OECs. Only target genes that showed expression on >2 separate arrays (96 genes) or genes for which expression in cultured OECs could be determined with qPCR (6 genes) were used for the siRNA-mediated loss of function neurite outgrowth assays.
Cell cultures
OECs.
For each OEC culture, olfactory bulbs were dissected and placed in Leibovitz-15 medium (L15; Invitrogen). Meninges were removed using forceps. The olfactory nerve and glomerular layers were isolated manually from the rest of the bulbs using a dissection microscope and incubated in Ca2+- and Mg2+-free Hanks buffered salt solution (Invitrogen) containing 0.1% trypsin (Invitrogen) for 10 min at 37°C. Trypsinization was stopped by adding Dulbecco's modified Eagle's/Ham's F12 medium (DMEM/F12, Invitrogen), supplemented with 10% FCS (Invitrogen) and 1% penicillin/streptomycin (PS; Invitrogen) (DF-10S). The tissue was washed in DF-10S and triturated in DF-10S until a homogeneous cell suspension was obtained. Cells were plated in DF-10S in 25 cm2 flasks precoated for 2 h at 37°C with 20 μg/ml PLL. After 1 week, p75-positive OECs were purified from the cultures by immunopanning, as described previously (Ramon-Cueto et al., 1998; Franssen et al., 2008). Petri dishes (100 mm) were incubated with anti-mouse IgG, Fc-specific (Jackson ImmunoResearch Laboratories; 1:1000) at 4°C overnight followed by another incubation at 4°C overnight with p75 monoclonal antibody (gift of Dr. P Wood) diluted 1:2.5 in PBS, pH 7.3. Dishes were blocked with 0.5% BSA (Roche) in PBS for 1 h at room temperature (RT). Cells were seeded onto three antibody-coated dishes and left for 5 min at 37°C. Unbound cells were removed by washing six times with DMEM/F12. Bound cells were scraped off from the dishes and seeded onto 2 other antibody-coated dishes. Incubation on antibody-coated dishes was repeated to diminish the number of contaminating cells. Purified cultures were plated on PLL-coated 25 cm2 flasks. After 2 d, medium was changed in DF-10S containing 2 μm forskolin (Sigma-Aldrich) and 20 μg/ml pituitary extract (PEX; Sigma-Aldrich). For siRNA phase 2, OECs were frozen after 4 d of culturing in DMEM/F12 containing 30% FCS, PS, forskolin, PEX, and 10% DMSO and stored in liquid nitrogen.
Skin fibroblasts.
Fibroblasts were obtained from 1 cm2 skin biopsies of the shaved abdominal region of Fischer 344 rats. The skin biopsies were placed in Petri dishes containing D-10S for 2–3 weeks. D-10S was replaced every 7 d. When the fibroblasts that migrated from the skin biopsies reached ∼80% confluency, they were split 1:3. When these split cultures again reached 80% confluency, they were frozen in DMEM containing 30% FCS, PS, and 10% DMSO and stored in liquid nitrogen.
Embryonic DRG neurons.
Embryonic DRGs were dissected from E15 Wistar rat embryos. All internal organs were removed from the embryos, and the vertebral column was cut ventrally to expose the spinal cord. The spinal cord, including all embryonic DRGs, was carefully raised and removed from the embryo. Embryonic DRGs were detached from the spinal cord, stripped from nerve roots, and incubated in 500 μl Hanks buffered salt solution, containing 0.125% trypsin for 30 min at 37°C. Trypsinization was stopped by adding an equal volume of DF-10S. Cells were triturated, centrifuged, and washed with DF-10S.
Adult DRG neurons.
Adult DRGs were obtained from Fischer 344 rats. After the spinal column was removed and cut open dorsally, the spinal cord was removed, leaving the adult DRG visible. Adult DRGs were carefully detached from peripheral nerves and placed into cold L15 medium. Adult DRGs were then digested for 30 min at 37°C in L15 containing 0.25% collagenase (Invitrogen), triturated, digested again for 30 min at 37°C, and triturated again. Chunks were allowed to sink, after which the cell suspension was layered carefully on 1 ml PBS containing 15% BSA in 1.5 ml Eppendorf tubes and centrifuged at 2800 rpm for 4 min in a tabletop centrifuge to enrich cultures for neurons. The pellet was triturated in Neurobasal medium (Invitrogen) containing Glutamax (Invitrogen), PS, B27 (Invitrogen), and the concentration of cells was determined.
siRNA transfection
The siRNA screen that was performed to test the effects of the candidate genes on neurite outgrowth consisted of 2 phases. The first phase was conducted with OECs that were cultured for 3.5 weeks (Phase 1). The second phase was performed with OECs that were stored in liquid nitrogen after 11 d in culture, and thawed and cultured for another 2 weeks (Phase 2). OECs were plated in PLL-coated 96-well plates at a density of 8500 cells/well. The next day, cells were transfected with siRNA pools (siGENOME SMART pools; Dharmacon) consisting of 4 siRNA duplexes per gene to a final concentration of 100 nm. The transfection was performed according to the manufacturer's protocol, using DharmaFECT 3 (Dharmacon). In short, transfection mix was prepared by mixing siRNA and DharmaFECT 3, both diluted in serum-free medium. After incubation for 20 min at RT, prewarmed (37°C) medium containing 0.5% FCS was added. Culture medium was removed from the cells and replaced by transfection mix to a final volume of 50 μl per well. After 4 h, the transfection mix was removed and replaced by culture medium. Individual siRNAs from the SMART pools were tested at a concentration of 100 nm each.
Determination of transfection efficiency.
OECs were plated on PLL-coated coverslips at a density of 5.0 × 104 cells per well and transfected the next day with siGLO at a final concentration of 100 nm. Untreated cells were used as control. To determine the transfection efficiency, cells were fixed at 2 d after transfection with 4% PFA for 20 min. Cell nuclei were stained with Hoechst 33258 (Bio-Rad) and mounted in Mowiol (0.1 m Tris-HCl, pH 8.5, 25% glycerol, Sigma; 10% Mowiol 4–88 Reagent, EMD Biosciences; and 0.1% 1,4-diazabicyclo-[2.2.2]octane, Sigma). Transfection efficiency was determined by counting the number of cells positive for the fluorescent marker siGlo as percentage of all Hoechst-positive cells.
Determination of knockdown efficiency by quantitative PCR
To determine the knockdown efficiency, OECs were plated in PLL-coated 24-well plates at a density of 1.0 × 105 cells per well. OECs were transfected the next day with siRNA pools directed to the candidate genes or siGLO at a final concentration of 100 nm. Three days later, cells were lysed, RNA was isolated, and cDNA was synthesized as described above. qPCR was performed on the ABI PRISM 7300 Sequence Detection System (Applied Biosystems) using a SYBR Green PCR reagent kit (Applied Biosystems). Data were normalized against β-actin, EF1α, and GAPDH, which were selected based on stability of expression with use of Genorm (http://medgen.ugent.be/∼jvdesomp/genorm/).
Quantification of protein expression by Western blot analysis
The effect of siRNA-mediated knockdown on the expression of LEPRE1, SCARB2, and SERPINI1 protein in OECs and of lentiviral-mediated overexpression of SCARB2 in fibroblasts was investigated by Western blot analysis. OECs and transduced fibroblasts were thawed and cultured on PLL-coated 6 cm plates in DF-10S containing forskolin and PEX or D-10S, respectively. After ∼1 week, cells were plated in 24-well plates at a density of 1.0 × 105 per well. The next day, OECs were transfected with siRNA pools or with siGlo as described in siRNA transfection. After 3 d, chondroitinase ABC (ChABC; 100332–1A; Seikagaku) was added to the culture medium of the OECs that were transfected with siRNA pools targeting LEPRE1 and its siGlo control conditions at a final concentration of 0.2 U/ml. Treatment of the cells with ChABC is necessary to optimize antibody binding to the protein core by removal of glycosaminoglycan chains (Wassenhove-McCarthy and McCarthy, 1999). After 3 h, the medium of all conditions was harvested and cells were lyzed in RIPA buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and Complete Protease Inhibitor Cocktail, Roche). Protein concentration was determined by using the bicinchoninic acid protein assay kit (Pierce Thermo Scientific). Blots were labeled with the appropriate primary antibodies for β-actin (A5316; Sigma-Aldrich; 1:2000), LEPRE1 (kind gift of professor Kevin McCarthy; LSUHSC-S; 1:100), SCARB2 (AF1888; R&D Systems; 1:500), and SERPINI1 (ab33077; Abcam; 1–250) followed by secondary IR-dye 800CW antibody (Odyssey, Licor; 1:2000) for the candidate genes and CY-5 (Jackson ImmunoResearch Laboratories; 1:2000) for β-actin detection. The β-actin signal was used to normalize the final protein quantifications.
Lentiviral vector production
DNA containing the coding sequence for ADAMTS1, APLN, BM_385941, CX3CL1, FZD1, GPNMB, LEPRE1, MSLN, S100A9, SCARB2, SERPINI1, and STMN1 was obtained as follows. RNA was isolated from either cultured OECs or rat cerebellum, and cDNA was synthesized with a QuantiTect Reverse Transcription Kit (QIAGEN). cDNA was amplified by PCR (Phusion DNA polymerase; Finnzymes) using cDNA primers. After agarose gel electrophoresis, the appropriate bands were isolated and purified with a Nucleospin Extract II kit (Macherey-Nagel). For NRXN1, PTPRC, RND1, SCARB1, SPP1, TIMP2, and VAV1 plasmids with a cDNA encoding the complete coding region of the gene were obtained from Imagenes. Clones were inoculated overnight at 37°C in Difco Luria-Bertani Broth, Miller (LB; BD Biosciences) in the presence of the appropriate antibiotics, and plasmid DNA was isolated with a Nucleospin plasmid quickpure kit (Macherey-Nagel). The coding sequence of each of the 19 genes was amplified by PCR using gateway primers. After electrophoresis, the appropriate bands were isolated and recombined with the pDONR221 plasmid from the Gateway Vector Conversion System (Invitrogen) using the BP Clonase II enzyme mix overnight at 25°C. The recombination product was transformed in One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen). Colonies were used for inoculation and grown overnight at 37°C in Luria-Bertani Broth with 50 μg/ml kanamycin (Invitrogen), plasmid DNA was purified, and the coding sequence of each gene was sequenced.
The LV-destination plasmids containing the candidate genes were constructed by replacing the GFP sequence from the transfer vector pRRLsin-PPthCMV-GFP-wpre with Reading Frame (Rf) Cassette B (RfB) of the Gateway Vector Conversion System (Invitrogen) followed by an IRES-GFP sequence. pDonor plasmids containing validated candidate gene sequences were recombined overnight at 25°C with the LV-destination vector using LR Clonase II (Invitrogen) following gateway manual instructions. The recombination product was transformed in DH5-α E. coli (Invitrogen) or One Shot TOP10 Chemically Competent E. coli. Plasmids were isolated and confirmed by restriction analysis.
Self-inactivating lentiviral vectors were produced as described previously (Naldini et al., 1996; Dull et al., 1998; Hendriks et al., 2007). Stocks were generated for all LV constructs by a polyethylenimine (linear; Polysciences) based cotransfection with 3 plasmids [the transfer GFP or destination vectors (25 μg), the VSV-G envelope protein vector pMD.G.2 (8.75 μg), and the viral core packaging construct pCMVdeltaR8.74 (16.25 μg)] into 1.13 × 107 human embryonic kidney (HEK) 293T cells per 15 cm dish in Iscove's modified Dulbecco's medium (Invitrogen) containing 10% FCS, 1% PS, and 1× Glutamax (Invitrogen). After 16 h incubation at 37°C, the medium was replaced by Iscove's modified Dulbecco's medium containing 2% FCS, 1% PS, and 1× Glutamax. To obtain high titer LV stocks, the medium was harvested after 24 h, filtered through 0.20 μm pores, and ultracentrifuged for 2.5 h at 20,000 rpm in a SW-28 rotor (Beckman Coulter). The viral pellet was resuspended in PBS, pH 7.4, and the viral vector stocks were stored in aliquots at −80°C. To determine the number of transducing particles in the LV stocks, HEK 293T cells were transduced and after 48 h the number of GFP-expressing cells was counted. Titers were expressed as transducing units per milliliters.
LV transductions and validation of overexpression by quantitative PCR or immunocytochemistry
Fibroblasts, OECs, or HEK 293T cells were plated in 24-well plates at a density of 1.0 × 105 cells in 0.5 ml per well. Cells were transduced the next day with the appropriate LV vectors at an multiplicity of infection of 100 resulting in 100% transduction. Medium was replaced after 16 h. For the fibroblasts, the transduction procedure was repeated 8 h later and again the next day to obtain near 100% transduction efficiency. For validation of LV-mediated overexpression, cells were lysed 3 d after the virus was removed, RNA was isolated, cDNA was synthesized, and qPCR was performed to determine expression levels. Overexpression in fibroblasts was confirmed by immunocytochemistry for FZD1, SCARB2, and SERPINI1. Transduced fibroblasts were plated on PLL-coated coverslips at a density of 1 × 103 cells/coverslip. After 24 h, fibroblasts were fixed with PFA for 30 min and labeled for GFP (AB16901; Millipore Bioscience Research Reagents; 1:1000), followed by a Dyelight-488 (Jackson ImmunoResearch Laboratories; 1:800) secondary antibody and labeled for FZD1 (AF1120; R&D Systems; 1:50), SCARB2 (AF1888; R&D Systems; 1:500), or SERPINI1 (ab33077; Abcam; 1:500) followed by a biotinylated (Vector Laboratories; 1:400) secondary antibody and a Cy3-Streptavidine (Jackson ImmunoResearch Laboratories; 1:800) tertiary antibody.
Bioassays
Loss of function bioassay: cocultures of OECs and embryonic DRG neurons.
OECs were plated in PLL-coated 96-well plates and transfected with siRNA pools, each targeting an individual candidate gene as described above. After 3 d, the monolayer of the feeding layer in each well was examined by phase-contrast microscopy, and transfection efficiencies were estimated in OECs transfected with the fluorescent indicator siGlo. Next, dissociated embryonic DRG neurons were plated onto the feeding layer of OECs at a density of 6000 cells/well in DF-10S containing forskolin and PEX. For Phase 1, cultures were fixed with 4% PFA for 20 min at 2, 4, 6, and 8 h after plating the embryonic DRG. For Phase 2, cultures were fixed after 8 h. For each experiment, one well was labeled for p75 (G323A; Promega; 1:500) followed by an Alexa-488 secondary antibody (Invitrogen) combined with Hoechst 33258 (Invitrogen; 1:1000) to determine the purity of the cultures. Experiments with OEC cultures containing <95% p75-positive cells were discarded. Neurites of embryonic DRG neurons were labeled with βIII-tubulin (MMS-435P; Covance; 1:500) followed by an Alexa-488 (Invitrogen) secondary antibody. Total neurite length per neuron was measured on a Cellomics ArrayScan VTI HCS Reader (Thermo Fischer). A maximum of 40 fields per well was imaged (10× magnification), containing ∼500 neurons per well in total. Using the Neuronal Profiling 3.5 Bioapplication (Thermo Fischer), neurites were traced and neurite length per neuron was determined. The bioassays included at least 3 wells per condition. For Phase 1, the conditions with siRNA pools that showed a significant difference in the first experiment were repeated twice. The results were analyzed for statistically significant differences via a two-way ANOVA to test for effects of gene and interaction of gene and time (gene × time). ANOVA was followed by a Bonferroni post hoc test to find significance between the different groups, and per experiment, differences were considered statistically significant if p < 0.05. A gene was considered to have an effect when the results of at least two of three independent experiments were statistically significant. For Phase 2, all siRNA pools conditions were tested with at least three independent siRNA transfections with OECs obtained from at least two independent cultures. Only neurons with a total neurite length of >10 μm were included in the analysis. For each well, the average total neurite length per neuron was calculated. All wells were then normalized to the average total neurite length per neuron in the siGlo condition. The normalized averages per well of the 8 h time point for all genes of all biological replicates were analyzed together using the Student's t test followed by a Bonferroni correction for multiple testing, and significance was set at p < 0.05. To measure the effects of 50 ng/ml NGF on embryonic DRG neurite length in a coculture with OECs or fibroblasts, we applied the same method as described above without transfections. Fibroblast-embryonic DRG cultures were fixed after 21 h.
Gain-of-function bioassay: cocultures of LV transduced fibroblasts or OECs and DRGs.
Fibroblasts or OECs were thawed and cultured on PLL-coated plates in D-10S (fibroblasts) and DF-10S containing forskolin and PEX (OECs). After 2 d, cells were transduced as described above and replated to expand the numbers of cells. After 1 week, the transduced cells were plated at a density of 2.5 × 104 cells per well in 24-well plates. Batches of transduced cells were frozen for future experiments, including the analysis of transgene expression. After 3–4 d, culture medium was replaced by 0.5 ml/well of Neurobasal containing Glutamax, PS, and B27. Two days later, 0.1 ml of the same medium containing 500 dissociated adult DRG was added to each well. Cultures were fixed with 4% PFA for 30 min after 27 h for OECs and after 40 h for fibroblasts and labeled for βIII-tubulin (1:500) followed by an Alexa-594 secondary antibody (Invitrogen; 1:800). Tiled images were created using a 5× objective. The length of the longest neurite/cell of a minimum of 80 neurons per well was measured blindly using NIH Image software (ImageJ) with the NeuronJ plugin to determine the mean neurite length per well. For each candidate gene tested with fibroblasts, at least 2 wells were measured per experiment, and the averages of each well were normalized to the average of all wells of the GFP condition. Each gene was tested at least 3 times with batches of fibroblasts that were prepared by at least two independent transductions and using at least two different adult DRG donor rats. The normalized averages per well for each gene were compared with the normalized averages of the GFP condition with SPSS using a one-way nested ANOVA using SPSS Statistics 17.0. Significance was set at p < 0.05. To measure the effects of NGF on dissociated embryonic and adult DRG neurite length, 50 ng/ml NGF was added to the cocultures or fibroblasts were transduced with LV-NGF (Tannemaat et al., 2007) before adding DRGs. Total neurite length was measured as described above. Fibroblast-dissociated embryonic DRG cultures were fixed after 21 h.
Immunohistochemistry
The olfactory epithelium, olfactory bulbs, and frontal pole of the cortex were dissected en bloc. All tissue used for immunohistochemistry was postfixed overnight at 4°C, incubated in 250 mm ethylenediaminetetraacetic acid in PBS for 24 h at 4°C, incubated in 30% sucrose in PBS for 24 h at 4°C, mounted in Tissue-tek (Sakura Finetek), snap-frozen in dry-ice-cooled isopentane, and stored at −80°C. The following primary antibodies were used: anti-GAP43 (#9527, gift from P.N.E. De Graan, Utrecht University, Utrecht, The Netherlands; 1:2000), anti-p75 (MAB365, Millipore Bioscience Research Reagents; 1:50) and anti-Gfap (Z-0334, Dako; 1:2000), anti-SCARB2 (NBP1–77326, Novus Biologicals; 1:250), and anti-SERPINI1 (ab33077, Abcam; 1:50). Except for the CTB stainings (see below), sections were incubated overnight at 4°C with primary antibodies. CX3CL1 (ab25088, Abcam) and S100A9 (sc-8115, Santa Cruz Biotechnology) labeling was not successful. Subsequently, the sections were incubated at RT with secondary antibodies (1:800) that were either fluorescently labeled or biotinylated for diaminobenzidine staining with the VectaStain ABC Elite kit (Vector Laboratories).
Effect of the lesioning procedure on the olfactory epithelium.
The effect of the lesion on the epithelium was analyzed by examining 20-μm-thick transversal cryostat sections of the olfactory epitheliobulbar complex from PFA-perfused animals. These sections were stained with hematoxylin-eosin (Merck), dehydrated and embedded in Entallan (Merck), or immunostained for GAP43 to visualize immature primary olfactory neurons and olfactory marker protein (OMP) to detect mature olfactory neurons. The efficacy of the lesioning procedure on the olfactory epithelium was determined as follows. Normal olfactory epithelium was defined as epithelium in which the GAP43-positive staining was located in a thin band of 1 or 2 cell bodies thick in the lower one-fifth of the height of the epithelium, whereas the upper portion of the epithelium is filled with OMP-positive cells (Verhaagen et al., 1989). Lesioned olfactory neuroepithelium was defined as epithelium in which OMP-positive cells had essentially vanished (this is usually the case during the first 1–3 postlesion days). To corroborate the completeness of the lesion, the epithelium of rats (n = 3) killed at 3 dpl was imaged in horizontal sections and outlined in ImagePro (Media Cybernetics). The percentage of damaged epithelium was determined in triplicate sections at 6 or 7 evenly spaced planes.
Immunohistochemical labeling of CTB.
To examine the regeneration of the dorsal column fibers, animals were killed 10 weeks after the lesion and 3 d after CTB injection and CTB signal was visualized and quantified as follows. The spinal cords were cut in 20 μm sections in sagittal orientation in 2 series. Sections were postfixed for 15 min with 4% PFA in PBS, incubated in TBS with 0.3% H2O2 for 20 min, incubated in TBS containing 0.1% SDS (MP Biomedicals), blocked with TBS containing 2% horse serum (Invitrogen) and 0.3% Triton X-100 (blocking buffer) for 45 min and incubated at 4°C with goat-anti-CTB (#703, List Biological Laboratories; 1:80,000) and rabbit-anti-GFAP (Z0334; Dako; 1:1.000) in blocking buffer for 72 h. Next, sections were incubated for 2 h at RT with biotinylated horse-anti-goat (BA-9500; Vector Laboratories; 1:400) and donkey-anti-rabbit-Dylight-649 (Jackson ImmunoResearch Laboratories; 1:800) in blocking buffer and then incubated in ABC reagent (Vectastain elite; Vector Laboratories; 1:400) in TBS at RT. After 1.5 h, sections were incubated in TBS containing 0.1% Tween 20 (P137–9; Sigma-Aldrich) for 5 min, in biotinyl tyramide reagent (NEL700A001KT; PerkinElmer; 1:400) in TBS containing 0.001% H2O2 at RT for 15 min and with streptavidin-Cy3 (Jackson ImmunoResearch Laboratories; 1:800) at RT for 1 h and mounted in Mowiol.
Quantification of CTB-labeled fibers
Quantification of fibers.
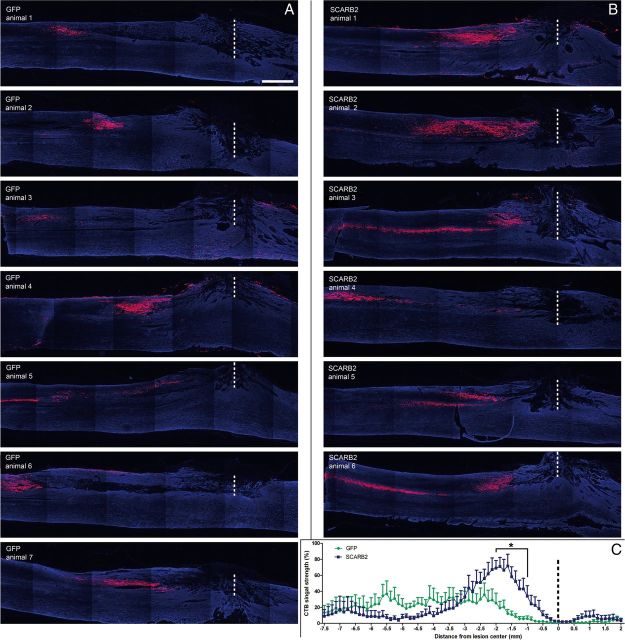
For quantification of CTB-positive fibers in and around the lesion area every fourth section of all sections containing gray matter was analyzed blindly. The lesion center was determined based on GFAP expression for each section at 100-fold magnification and was set at 0. Fibers were counted at 400-fold magnification by crossing lines perpendicular to the direction of the spinal cord at −1.5, −1, −0.5, 0, 0.5, 1, and 1.5 mm from the lesion center for each section that contained lesioned tissue. Fiber counts of all sections of each animal were then combined for each distance from the lesion center and multiplied by 4. For all groups, the total number of fibers crossing each distance was averaged and averages were compared using the Student's t test.
CTB-signal quantification.
For quantification of the intensity of the CTB signal in the dorsal column, it was first determined which section of each individual animal had most CTB signal in the dorsal column. Of this section together with the two adjacent CTB-stained sections, fluorescence photographs were acquired at 50-fold magnification with a charge-coupled device camera (CoolSnap fx; Photometrics) with the same exposure times for all spinal cords. All spinal cord images were blindly aligned to the lesion center based on GFAP staining to allow comparison. Measured intensities were linearly mapped into pixels with 255 representative gray values using Image-pro software. The background of the red channel (containing the CTB signal) was set at gray value 50 for all images. Projections into the gray matter were blindly excluded from analysis. All images were divided in 77 segments (1–77) of equal size in caudal to rostral direction. In each segment, the number of pixels higher than background was quantified and the pixel values of the same segment number of the three sections of each animal were combined. Of each animal, the combined segment with the highest value was set at 100%, and values of other segments were expressed as a percentage of this value. Segments of the GFP and treatment groups were compared using the Student's t test. Significance was set at p < 0.05.
Results
Peripheral deafferentation of the olfactory bulb induces neuroregeneration
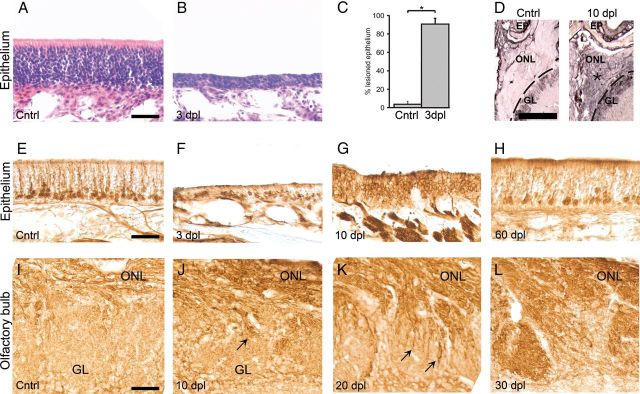
We first induced a lesion of the neuroepithelium to examine the temporal pattern of reinnervation of the ONL and activation of supporting OECs. Olfactory epithelia of rats were lesioned by infusion of Triton X-100 and assessed by examining the epithelium as well as the ONL at 1, 3, 6, 10, 20, 30, and 60 dpl. The effect of the lesion on the epithelium was clearly visible macroscopically at 3 d after the lesion. At 3 dpl, exudate was present throughout the pharynges, and the epithelium had become very thin compared with control epithelium (Fig. 2A,B). Quantitative analysis of the extent of the lesion at 3 dpl revealed that 90.4 ± 6.6% of the epithelium was successfully lesioned (Fig. 2C). The thickness of the ONL had decreased slightly after the lesion, and p75 expression in the ONL was highly increased (Fig. 2D). This increase in p75 expression is consistent with previously reported results in mice (Turner and Perez-Polo, 1993). At 3 dpl, occasional GAP43-labeled cells were visible in the remaining thin layer of epithelium that remained after the lesion (Fig. 2E,F). At 10 dpl, the thickness of the epithelium had strongly increased and the ONL was almost completely filled with newly born GAP43-positive neurons (Fig. 2G). Thereafter, the number of GAP43-positive neurons declined, whereas the number of OMP-positive neurons increased over time (data not shown), and at 60 dpl, GAP43-positive neurons became restricted to the lower one-fifth of the regenerated epithelium, a pattern comparable with the unlesioned control epithelium (Fig. 2H). In the olfactory bulb, new GAP43-positive axons had started to grow through the ONL and entered the glomeruli ∼10 dpl (Fig. 2I,J). The number of ingrowing axons had strongly increased at 20 dpl (Fig. 2K), and at 30 dpl the glomeruli were completely filled with new axons (Fig. 2L). Together, these observation are consistent with previous work in mice using chemical deafferentation of the bulb (Verhaagen et al., 1990; Schwob et al., 1999) and corroborated that a vigorous neuroregenerative response occurs after peripheral deafferentation of the rat olfactory bulb.
Figure 2.
Effects of the TX-100 lesion on the olfactory epithelium and the olfactory nerve layer. A, B, In contrast to undamaged epithelium, at 3 dpl, the epithelium was much thinner than normal epithelium. Sections were hematoxylin-eosin stained. C, At 3 dpl, ∼90% of the epithelium was damaged (*p < 0.05). D, There was increased expression of p75 in the ONL and glomerular layer (GL) in the olfactory bulb at 10 dpl. GAP43-staining of the epithelium was strongly diminished at 3 dpl and was located in close proximity to the lumen (E,F). At 10 dpl, GAP43-positive cells reappeared in great numbers and the epithelium was much thicker again (G). At 60 dpl, the number of GAP43-positive cells returned to normal levels, and the epithelium was comparable with control sections (H). The ONL and GL of a control animal (I). J, At 10 dpl, the first GAP43-positive olfactory axons (arrow) grow from the ONL toward the glomeruli. At 20 dpl, there is massive ingrowth of GAP43-positive axons into the GL (arrows) (K); and at 30 dpl, the glomeruli are completely filled with GAP43-positive axons (L). Scale bars: A, E, I, 50 μm; D, 400 μm.
A total of 846 genes are regulated in the ONL after peripheral deafferentation of the olfactory bulb
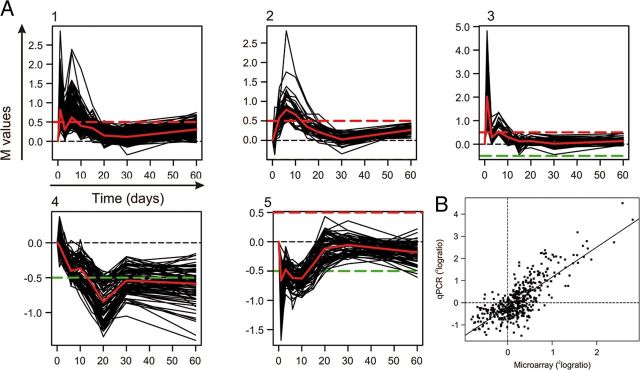
To obtain insight in the molecular mechanisms underlying successful regeneration of olfactory axons along the OECs, gene expression changes in the OEC-containing ONL were profiled after peripheral deafferentation of the olfactory bulb. Microarray analysis was conducted on the ONL of intact, nonlesioned animals and on animals at 1, 3, 6, 10, 15, 20, 30, and 60 dpl. Clustering of genes with >90% similarity in expression changes over time revealed five prominent expression profiles (Fig. 3A, 1–5). Genes in profiles 1–3 show upregulation followed by a decline in expression over time, whereas genes in profiles 4 and 5 exhibit a “down-up” pattern of expression. A total of 846 genes were differentially expressed at one or more time points after the lesion (>1.25 and an adjusted p < 0.05), and changes in expression of a random selection of 60 of these 847 genes were validated by qPCR (Fig. 3B). The differential expression measured by qPCR showed a linear correlation of R2 = 0.61 with the microarray data, and the qPCR revealed on average 35% higher expression levels. The majority of regulated genes (781 in total) were differentially expressed in the first 3 weeks after the lesion, which is the time period in which most regenerative axonal growth occurs through the ONL toward the glomeruli in the olfactory bulb.
Figure 3.
Gene expression profiling on the regenerating ONL. A, The five major profiles of significantly regulated genes that are clustered with other genes that have a similar expression profile (>90% correlation). The differential expression, M, is expressed as 2log(x), in which × is the differential expression at a post lesion time point compared with the expression at day 0. The red and green dashed lines indicate M = 0.5 or −0.5, respectively. The expression profile in red in each cluster represents the median of the cluster. The genes that were selected for the siRNA screen were predominantly present in cluster 1. B, A validation of microarray data by qPCR. A group of 57 microarray probes that detected differential expression were validated by qPCR. The scatter plot shows the relationship between qPCR and microarray data per time point after the lesion per gene (i.e., 8 data points per gene). The linear regression of the data points was as follows: y = 1.35x − 0.19 (R2 = 0.61).
Loss-of-function cellular screening
We next set out to identify genes that contribute to the neurite outgrowth-promoting properties of OECs. Genes that are upregulated in the ONL shortly after the lesion and during the ingrowth of olfactory nerve fibers in the ONL are potentially involved in supporting axon growth. Sixty-five genes that were upregulated in the first 3 weeks in our microarray study together with an additional set of 37 genes that were selected based on high expression in cultured OECs compared with either the intact ONL or cultured SCs were selected for a functional siRNA screen (Franssen et al., 2008a). All genes were selected according to the criteria specified in Materials and Methods. The selected genes are shown in Table 2 together with the relative fold change in gene expression after peripheral deafferentiation of the olfactory pathway. Together, these 102 genes represent a subset of all differentially expressed genes. Each gene was knocked down by transfection with siRNA duplex pools (four independent duplexes per gene target) in a confluent layer of cultured OECs (Fig. 4A). Transfection and knockdown efficiencies were tested in OECs using the fluorescent control siRNA siGlo and a siRNA pool targeting Lamin A/C. At 2 d after transfection, 75% of the OECs was transfected based on fluorescence (Fig. 4B), and the knockdown efficiency of lamin A/C was 83% after 2 d and 66% after 7 d based on qPCR measurements.
Figure 4.
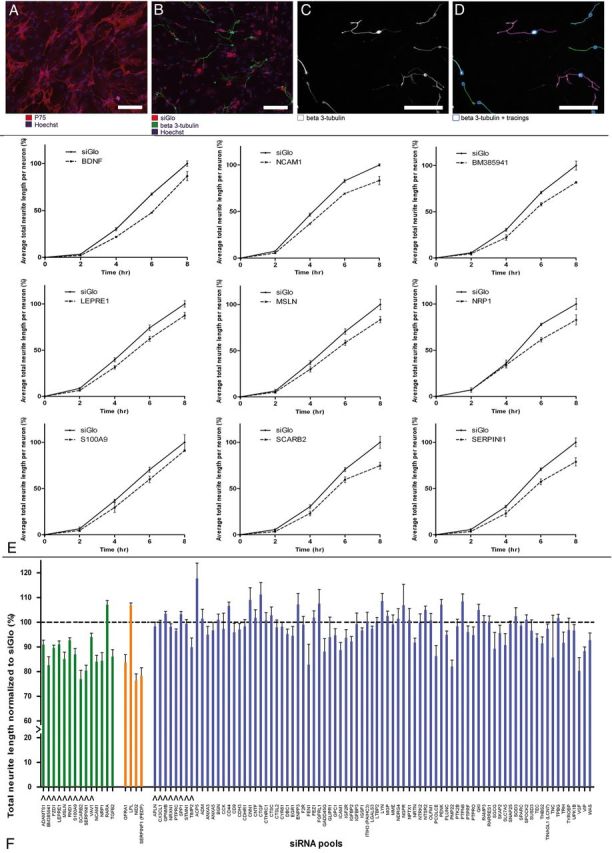
Functional study of candidate genes in siRNA-mediated outgrowth screen. A, OEC feeding layer in a 96-well plate visualized by p75 staining (red) and Hoechst (blue). B, DRGs, labeled for β3-tubulin (green), were plated onto an OEC feeding layer (Hoechst; blue), which was transfected with siGlo (red). C, D, Automated tracings of embryonic DRG cell bodies and neurites, labeled for β3-tubulin (white) by Cellomics technology. Scale bar, 100 μm. E, BDNF and the 8 genes that had a significant effect on neurite outgrowth in siRNA screen Phase 1. Graphs represent total neurite length of embryonic DRGs cultured on OECs transfected with an siRNA pool against BDNF, NCAM1, BM385941, LEPRE1, MSLN, NRP1, S100A9, SCARB2, and SERPIN1. Embryonic DRGs were incubated on the transfected OEC feeding layer for 2, 4, 6, and 8 h. Values are normalized to the siGlo condition. All graphs are the combined result of three independent experiments. F, Graph presenting combined results from siRNA screen Phase 1 and Phase 2. Total neurite length is shown of embryonic DRGs cultured for 8 h on OECs transfected with an siRNA pool against one of all 102 candidate genes. Neurite length was normalized to the siGlo condition (dotted line). Data are presented as the normalized mean ± SEM of the average neurite length. S100A9 was included as an extra gene in Phase 2 to confirm the results obtained for this gene in Phase 1 of the screen. Green and orange bars represent the 18 genes for which knockdown significantly affected neurite length in Phase 1 and 2 of the screen (p < 0.05). Knockdown of 16 genes resulted in a significant reduction of total neurite length, and knockdown of two genes resulted in a significant increase of total neurite length. Orange bars represent genes for which knockdown disrupted the OEC feeding layer. Blue bars represent the 84 genes that had no significant effect on total neurite length. Arrows under the bars indicate the genes that were selected for LV-mediated overexpression experiments.
In the screen, dissociated embryonic DRG neurons were plated on top of the transfected OECs 72 h after transfection (Fig. 4B–D), and neurite outgrowth was monitored using automated high-content microscopy. To determine the optimal time point for analysis of knockdown effects, we initially screened 52 genes at 2, 4, 6, or 8 h after plating of the embryonic DRG neurons on the layer of transfected OECs. Knockdown of eight genes (BM385941, LEPRE1, NCAM1, MSLN, NRP1, S100A9, SCARB2, and SERPINI1) resulted in a significant reduction of neurite outgrowth (p < 0.05) (Fig. 4E). Knockdown of BDNF was used as a positive control because BDNF is expressed in OECs (Woodhall et al., 2001), and it has been shown to actively promote neurite ougrowth of embryonic DRG neurons (Ruitenberg et al., 2003). Indeed, knockdown of the BDNF transcripts reduced DRG outgrowth substantially, and the reduction in neurite outgrowth observed for the eight hits was comparable to the reduction of neurite outgrowth after knockdown of BDNF (Fig. 4E). NCAM1 was previously shown to be involved in neurite outgrowth stimulation by OECs (Doucette, 1990; Aoki et al., 1999) and thus served as a positive control for the screening approach.
Because the effects on neurite outgrowth were always strongest at the 8 h time point, the remaining 50 genes were only screened at this time point. Figure 4F shows the normalized neurite lengths of DRG neurons after knockdown in OECs of all 102 genes at the 8 h time point. In total, knockdown of 16 genes (ADAMTS1, BM385941, FZD1, GFRA1, LEPRE1, NCAM1, NID2, NRP1, MSLN, RND1, S100A9, SCARB2, SERPINI1, SERPINF1, TGFB2, and VAV1) significantly decreased neurite length, and knockdown of two genes (LPL and RARA) significantly increased neurite length (p < 0.05) (Fig. 4F, green and orange bars, respectively). Knockdown of four genes (GFRA1, LPL, NID2, and SERPINF1) also had a visible effect on the integrity of the OEC feeding layer, suggesting that they may be involved in OEC cell–cell signaling and adhesion (Fig. 4F, orange bars).
Validation of knockdown efficiency
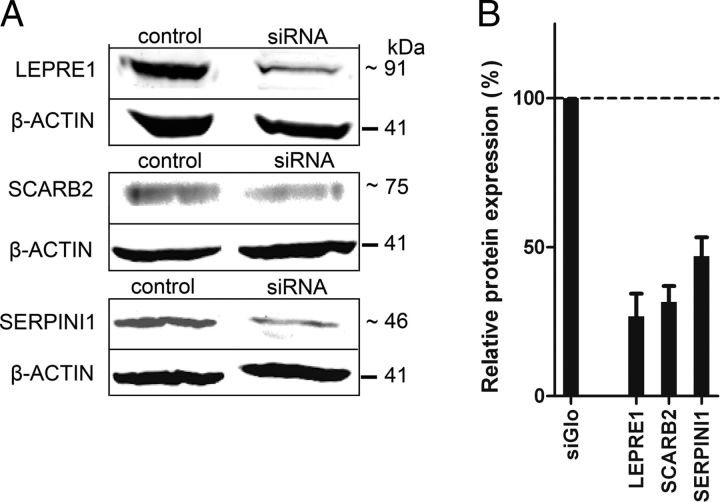
Knockdown efficiencies of siRNA pools were determined by qPCR for ADAMTS1, BM385941, FZD1, LEPRE1, MSLN, NRP1, RND1, S100A9, SCARB2, SERPINI1, and VAV1, which showed a significant decrease of neurite outgrowth after knockdown without affecting the feeding layer of OECs (data not shown). mRNA levels after knockdown varied from 21% of endogenous levels (for RND1) to 64% of endogenous levels (for SCARB2). For three genes (LEPRE1, SCARB2, and SERPINI1), knockdown efficiencies were also measured at the protein level on Western blots (Fig. 5A,B), and reduced levels of 73.2%, 68.4%, and 53.0% on average were observed, respectively, indicating that reduced transcript levels indeed result in lowered protein levels.
Figure 5.
Western blot confirms siRNA knockdown in olfactory ensheathing glial cells. A, Knockdown at the protein level for SCARB2, LEPRE1, and SERPINI1 analyzed by Western blot after transfection of cultured OECs with siRNA pools. B, The protein level of LEPRE1, SCARB2, and SERPINI1 after transfections with siRNA pools was compared with the endogenous expression in siGlo-transfected OECs by normalizing to the expression of β-actin as described in Materials and Methods. The protein levels are as follows: 26.8%, 31.6%, and 47% of endogenous levels, which indicates a knockdown of 73.2%, 68.4%, and 53.0% for LEPRE1, SCARB2, and SERPINI1, respectively. Data are presented as the normalized mean ± SEM of 3 biological replicates for LEPRE1, 2 technical replicates for SCARB2, and 2 biological replicates for SERPINI1.
The rationale behind using pools of four different siRNAs targeting the same gene is that the concentrations of each individual siRNA can be lowered, thereby diminishing the chance of a potential off-target effect of one of the siRNAs. To assess the efficacy of knockdown in OECs, we also measured the knockdown efficiencies of the single siRNAs that make up the siRNA pools for seven genes (BM385941, LEPRE1, MSLN, NRP1, S100A9, SCARB2, and SERPINI1; data not shown). All single siRNAs showed a certain degree of knockdown with mRNA levels varying from 7% of endogenous levels (for S100A9 #2) to 77% of endogenous levels (for SCARB2 #2). Thus, single siRNAs likely contributed to the knockdown observed with the siRNA pool.
Gain-of-function cellular screening
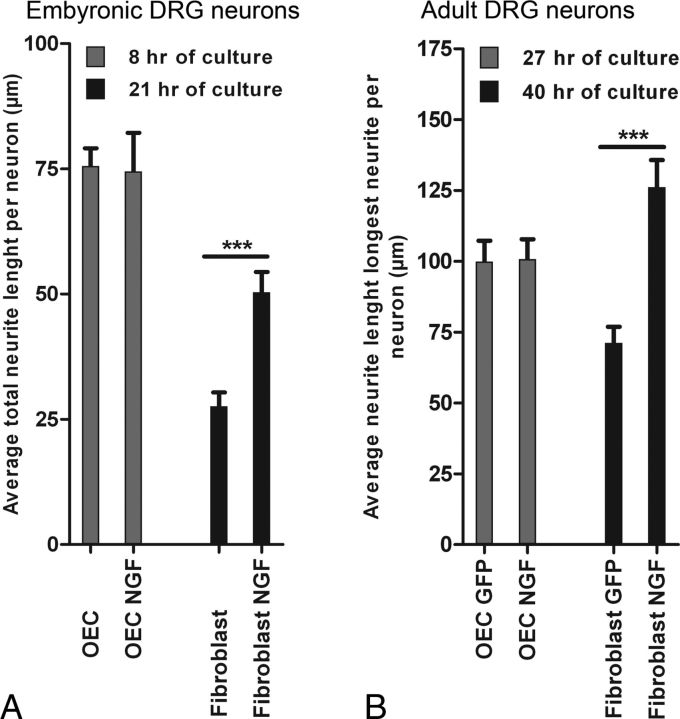
We next performed gain-of-function screening using lentiviral vector-mediated overexpression of a selected set of candidate genes. Initial experiments showed that neurite outgrowth of embryonic DRG neurons on a feeding layer of OECs was robust and could not be further enhanced by NGF (Fig. 6A). Therefore, we adapted the bioassay to facilitate gain-of-function screening and to identify growth-promoting effects reliably. We replaced OECs with skin fibroblasts, which are less capable of supporting neurite growth, and we used adult DRG neurons, which have less endogenous growth capacity compared with embryonic DRG neurons. Indeed, by doing so, we could easily detect the growth-promoting effects of lentiviral vector-directed overexpression of NGF in fibroblast on adult DRG neurons (Fig. 6B).
Figure 6.
NGF increases neurite length of dissociated DRGs on fibroblasts but not on OECs. A, Dissociated embryonic DRG neurons plated onto OECs did not show increased total neurite length when cocultured in the presence of 50 ng/ml NGF after 8 h. Dissociated embryonic DRG on skin fibroblasts showed a significant (***p < 0.001) increase in total neurite length when cultured in the presence of NGF (27.6–50.4 μm). These results are an average of two independent experiments. Data are presented as the normalized mean ± SEM of the average neurite length. B, Dissociated adult DRG neurons cultured for 27 h onto OECs transduced by LV-NGF did not show increased outgrowth compared with adult DRG on OECs transduced by LV-GFP. Dissociated adult DRGs cultured for 40 h on skin fibroblasts transduced by LV-NGF showed a significant (***p < 0.001) increase in neurite length compared with adult DRGs on fibroblasts transduced by LV-GFP (GFP, 71.1 μm; NGF, 126.2 μm). Data are presented as the normalized mean ± SEM of the average neurite length.
Not all genes that were tested in our siRNA screen were also selected for overexpression. First, genes that showed an increase in neurite length after knockdown were excluded (LPL and RARA). Second, genes for which knockdown clearly affected the consistency of the OEC feeding layer were excluded (ITGA7, GFRA1, NID2, and SERPINF1; Fig. 4F, orange bars). Finally, genes that were already extensively studied in the context of neural repair by others were also not selected (NCAM1, NRP1, and TGFB2) (Miragall et al., 1988; Doherty and Walsh, 1991; Takagi et al., 1995; Moon and Fawcett, 2001; De Winter et al., 2002a; Day et al., 2005). This resulted in the following selection of 10 genes for overexpression: ADAMTS1, BM385941, FZD1, LEPRE1, MSLN, RND1, S100A9, SCARB2, SERPINI1, and VAV1. In addition, eight genes were selected for overexpression for which knockdown in OECs had no significant effect on neurite outgrowth but that were significantly upregulated in the ONL within the first 3 weeks after a lesion of the olfactory epithelium: APLN, CX3CL1, GPNMB, NRX1, PTPRC, SPP1, STMN1, and TIMP2. In total, LV overexpression vectors were generated for 18 genes.
Transduction of fibroblasts with each of these 18 LV vectors resulted in an increase of relative mRNA expression levels varying from 1.4 to 1.1 × 106-fold of control levels (Fig. 7A). The large differences in relative expression level changes result from differences in endogenous expression levels for the different target genes. This overexpression resulted in higher protein levels as was confirmed for FZD1, SCARB2, and SERPINI1 (Fig. 7B–D). Protein levels of SCARB2 in overexpressing fibroblasts were ∼4.1- and 2.7-fold higher than endogenous levels in fibroblasts and OECs, respectively (Fig. 7E,F).
Figure 7.
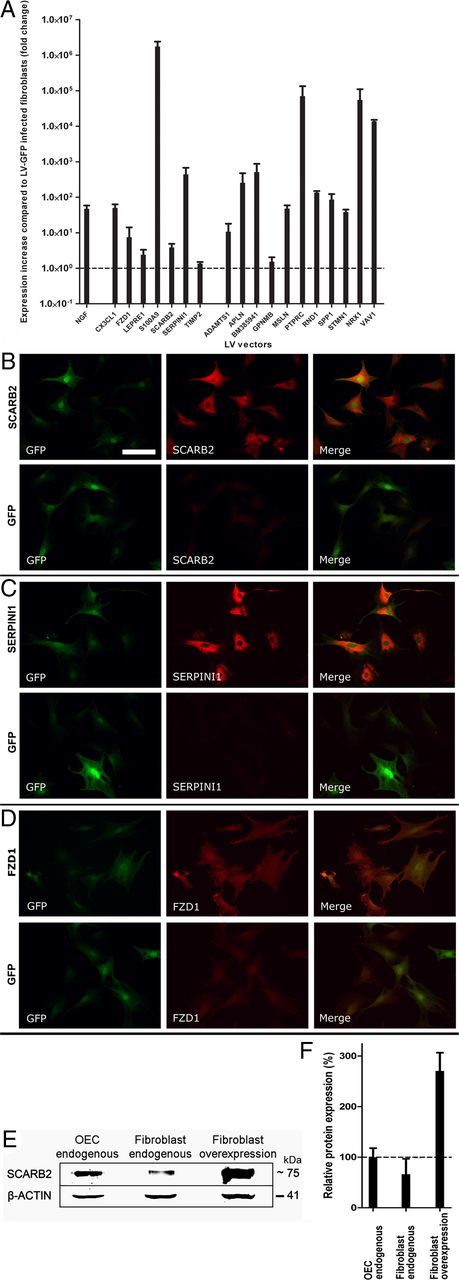
Validation of LV-mediated overexpression in skin fibroblasts by qPCR and immunocytochemistry. A, Relative gene expression after transduction of fibroblasts with either LV-NGF or 1 of the 18 constructed LV vectors containing the candidate gene sequences. Gene expression is presented relative to the endogenous expression levels in LV-GFP-transduced fibroblasts (dotted line). Differences in relative expression levels between candidate genes can largely be explained by differences in endogenous expression levels. The value for each condition is the average of at least two separate transduction experiments. B–D, Fibroblasts were transduced with LV-SCARB2 (B), LV-SERPINI1 (C), and LV-FZD1 (D). LV-GFP was used as control. Cells were stained for GFP (green) and SCARB2, SERPINI1, and FZD1, respectively (red). Similar exposure times were used for SCARB2, SERPINI1, and FZD1 stainings for cells transduced with LV vectors containing the respective gene and LV-GFP-transduced cells. Scale bar, 100 μm. E, Western blot of endogenous protein levels for SCARB2 in OECs and fibroblasts and SCARB2 protein level in overexpressing fibroblasts. F, Quantification of the protein levels shown in E after background correction and normalization to the expression of β-actin. Data are presented as the mean ± SEM of two technical replicates normalized to the endogenous levels in OECs. SCARB2 levels were 66% and 271% for endogenous and overexpressing fibroblasts, respectively.
Next, the effect of the overexpression of the candidate genes on neurite outgrowth was measured. To accurately trace the complex neurite outgrowth patterns of adult DRG neurons, we adopted an established method in which the longest neurite of each adult DRG neuron is traced (Gao et al., 2004; Cao et al., 2006). Overexpression of six candidate genes resulted in significantly increased neurite length as follows: CX3CL1 (128%; p < 0.05), FZD1 (118%; p < 0.001), LEPRE1 (115%, p < 0.05), S100A9 (152%, p < 0.01), SCARB2 (123%, p < 0.05), and SERPINI1 (135%, p < 0.01) (Fig. 8A). Overexpression of TIMP2 resulted in a significant decrease in neurite length (90%, p < 0.05). Overexpression of NGF resulted in a significant increase in the average neurite length (160%, p < 0.001) compared with fibroblasts expressing GFP (Fig. 8B). Overexpression of the other 11 candidate genes had no significant effect on neurite length.
Figure 8.
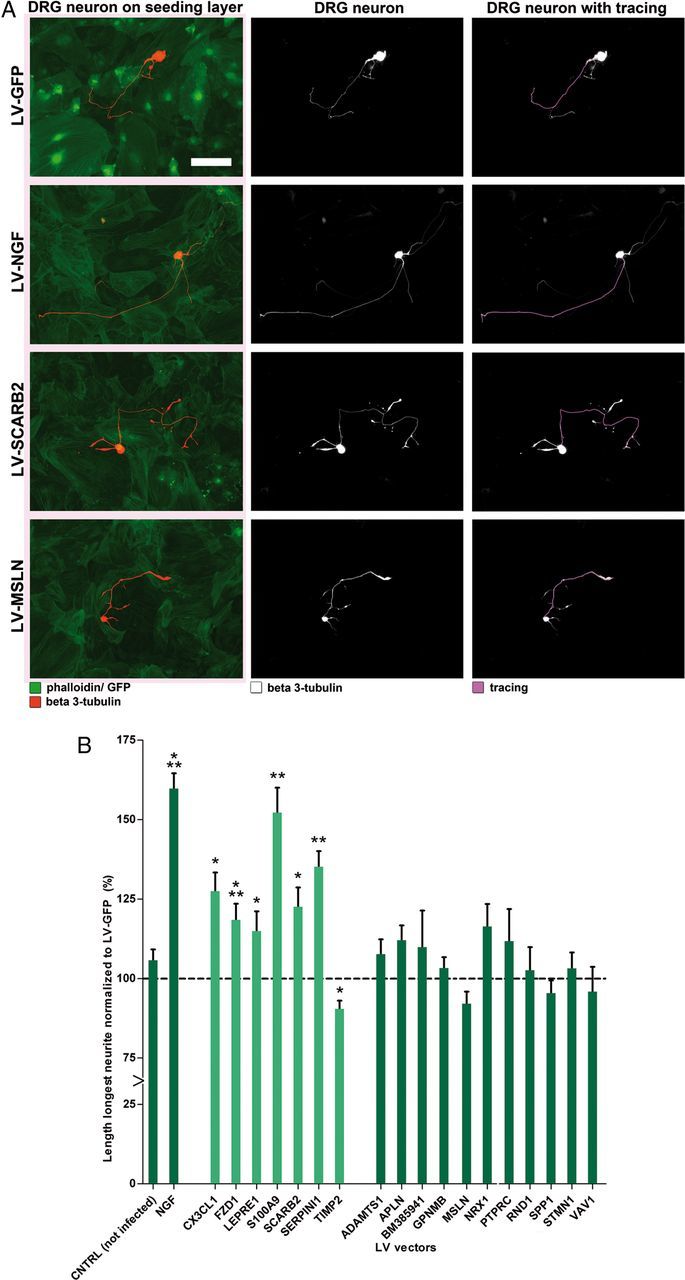
Overexpression of six candidate genes in a feeding layer of skin fibroblasts increases neurite length of dissociated adult DRG neurons. A, Representative pictures of adult DRG neurons cultured for 40 h on a feeding layer of fibroblasts expressing GFP, NGF, SCARB2, or MSLN, respectively. Fibroblast feeding layer is labeled with phalloidin (green; left images) and adult DRG are labeled with β3-tubulin (red, left; and white, middle and right). The longest neurite of each neuron was traced using cellomics technology (purple; right). B, Average neurite length of adult DRG neurons on nontransduced fibroblasts, fibroblasts overexpressing NGF, or overexpressing one of the 18 candidate genes. The average length of the longest neurite in each condition was normalized to the average length on a LV-GFP transduced feeding layer (dotted line). Adult DRG cultured on nontransduced fibroblasts showed comparable outgrowth to DRG on LV-GFP transduced fibroblasts. Overexpression of NGF (positive control) and six of the candidate genes resulted in a significant increase in neurite length as follows: NGF (160%, p < 0.001), CX3CL1 (128%; *p < 0.05), FZD1 (118%; ***p < 0.001), LEPRE1 (115%, *p < 0.05), S100A9 (152%, **p < 0.01), SCARB2 (123%, *p < 0.05), and SERPINI1 (135%, **p < 0.01). Overexpression of TIMP2 resulted in a significant decrease in length (90%, *p < 0.05). Overexpression of the other 11 candidate genes had no significant effect on neurite length. Data are presented as the normalized mean ± SEM of the average neurite length. The value for each condition is the average of the wells of at least three different outgrowth experiments, each with a minimum of 2 wells per condition and 75 neurons per well. Scale bar, 100 μm.
SCARB2 and SERPINI1 protein expression is increased in the ONL after a lesion
Protein expression and localization of SCARB2 and SERPINI1 were examined in the ONL at 6 dpl, a time point at which both genes were significantly upregulated (Fig. 9A,B). SCARB2 immunoreactivity was increased in the ONL after deafferentation and was observed in numerous OECs that aligned in typical olfactory nerve bundles (Fig. 9C,D). In many OECs, SCARB2 displayed the typical punctuate intracellular staining as observed previously in COS, HeLa cells, and MDCK cells (Kuronita et al., 2002; Eckhardt et al., 2004). An increase of SERPINI1 labeling was mainly observed in OECs located in the inner portion of the ONL closely associated with the glomerular layer at 6 dpl, in a position where OECs guide newly formed axons toward the glomeruli (Fig. 9E,F).
Figure 9.
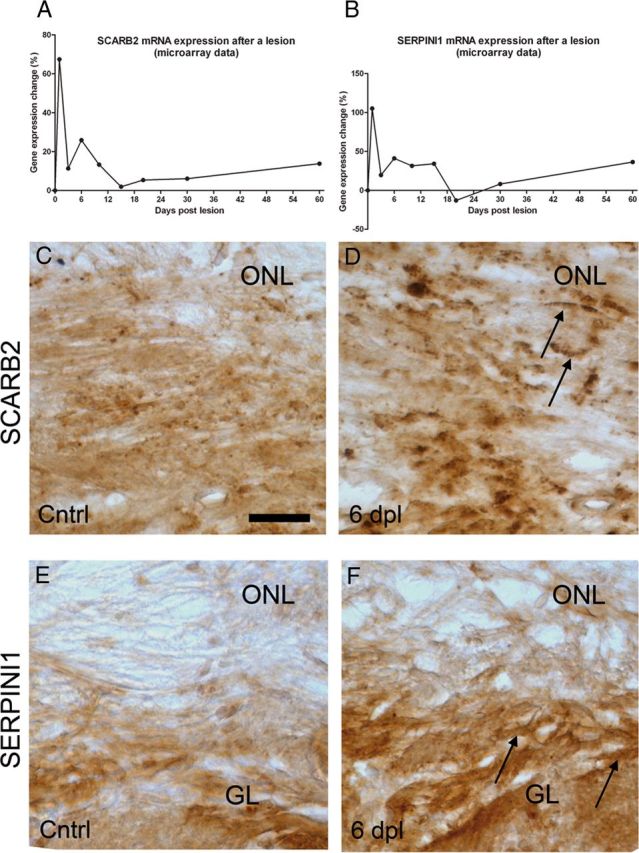
SCARB2 and SERPINI1 protein expression is increased in the ONL after a lesion. A, B, Gene expression of SCARB2 and SERPINI1 in the ONL after a lesion of the olfactory epithelium. C, D, SCARB2 expression is increased in the ONL at 6 dpl. SCARB2 staining can be observed diffused throughout the ONL, in cells with an immune-like morphology and in endosomal-like structures in bipolar OEC-like cells (arrows). E, F, SERPINI1 protein expression is increased in the ONL at 6 dpl. SERPINI1 staining is mainly observed in the transition phase from the ONL to the GL (arrows). This is the region where OECs guide newly formed axons toward their targets in the glomeruli. Scale bar, 25 μm.
Ex vivo gene delivery of SCARB2 in the lesioned dorsal column significantly increases axonal regenerative sprouting in and around the lesion center
Of all genes screened so far, knockdown and overexpression of SERPINI1, S100A9, and SCARB2 had strong and opposite effects on neurite outgrowth in the coculture assays. Therefore, the growth-promoting effects of overexpression of these three genes were investigated in vivo. The dorsal columns of adult rats were transected, and fibroblasts overexpressing SCARB2, S100A9, SERPINI1, or GFP were implanted in and around the lesioned area concomitant to the transection. Regenerating ascending sensory fibers in the dorsal column were visualized by a CTB-tracer injection in the sciatic nerve and quantified. The number of fibers in the SCARB2 group was ∼4-fold higher than in the GFP group from 1.5 mm before to 1 mm after the lesion (Fig. 10A,B). The S100A9 and SERPINI1-treated animals had a similar number of fibers as the GFP group. In contrast to the GFP group, most CTB-labeled fibers of the rats that received ex vivo gene delivery of SCARB2 were found more proximal to the lesion center (Fig. 11A–C). The CTB signal of the SCARB2 group was detected in a significantly larger area than the GFP group in the GFAP-positive area from 1 to 2 mm caudal of the lesion center. However, no significant difference was observed in the size of the GFAP-negative area between the SCARB2 and the GFP group (data not shown). There were no apparent morphological differences between the fibers of the SCARB2 and GFP group.
Figure 10.
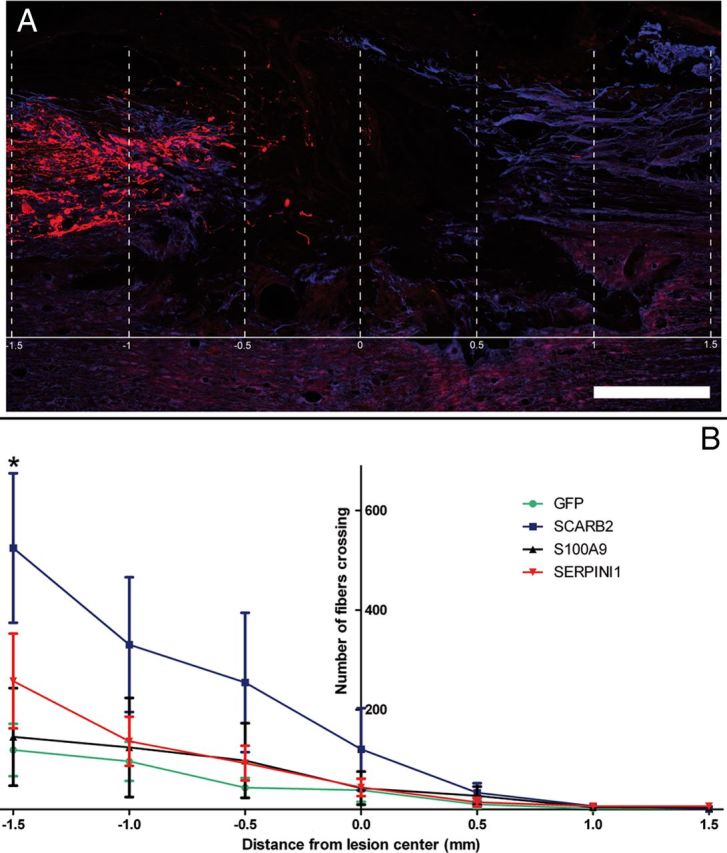
Implantation of fibroblasts overexpressing SCARB2 in a lesioned dorsal column leads to an increase of axons in and around the lesion. A, Sagittal section of an animal of the SCARB2 group. Fibers were counted at −1.5, −1, −0.5, 0, 0.5, 1, and 1.5 mm from the lesion center. B, Significantly more fibers were present around the lesion site in the SCARB2 group than in the GFP group (*p < 0.05). Scale bar, 500 μm.
Figure 11.
Implantation of fibroblasts overexpressing SCARB2 in a lesioned dorsal column leads to an increase of CTB-positive fibers in proximity of the lesion. A, B, Sections showing the largest area of CTB staining (red) in the dorsal column of each rat implanted with fibroblasts expressing GFP (A) or SCARB2 (B). Astrocytes in spinal tissue are stained for GFAP (blue). The lesion center is indicated by a white dotted line. C, Quantification of CTB signal at different distances from the lesion center. The graph shows the average normalized CTB signal at different distances from the lesion center of all animals of the GFP and the SCARB2 groups. The CTB signal is significantly higher in the SCARB2 group from 2 to 1 mm caudal to the lesion center (*p < 0.05). Scale bar, 1 mm.
SCARB2 is a splice variant of SCARB1, and both proteins are involved in the uptake and secretion of cholesterol (Webb et al., 1997; Eckhardt et al., 2004). However, SCARB1 overexpression did not result in a significant increase of neurite length as was observed for SCARB2 in vitro (data not shown). Thus, the observed effect of SCARB2 on regenerative sprouting is most likely specific for SCARB2.
Discussion
A multistep screening strategy was used to identify genes involved in the neurite outgrowth-promoting properties of OECs. We selected 102 candidate genes after transcriptional profiling of either cultured OECs (Franssen et al., 2008a) or of the ONL after peripheral deafferentiation (this study). The involvement of these genes in neurite outgrowth was assessed in cocultures of OECs and primary DRG neurons using selective siRNA-mediated knockdown in OECs. Sixteen genes were identified (ADAMTS1, BM385941, FZD1, GFRA1, LEPRE1, NCAM1, NID2, NRP1, MSLN, RND1, S100A9, SCARB2, SERPINI1, SERPINF1, TGFB2, and VAV1) that significantly reduced neurite outgrowth after knockdown, indicating their importance for neurite growth support. A gain-of-function screen of 18 genes (10 genes that were identified as positive hits in the knockdown screen and 8 additional genes) identified 6 genes (CX3CL1, FZD1, LEPRE1, S100A9, SCARB2, and SERPINI1) that enhanced, and one gene (TIMP2) that inhibited, neurite outgrowth. Because S100A9, SCARB2, and SERPINI1 were the most effective in the loss- and gain-of-function screens, the effects of these genes on the regeneration of injured afferent DRG axons were tested in vivo. Ex vivo gene delivery of SCARB2 significantly enhanced the number of regenerating DRG axonal sprouts that grew toward the center of a thoracic lesion of the dorsal funiculus of the rat spinal cord.
In the cellular screen, 11 of the genes that modulate neurite outgrowth encode secreted proteins, including structural extracellular matrix (LEPRE1, NID1) and matricellular proteins (ADAMTS1, MSLN, SERPINI1, SERPINF1, and TIMP2), immune-modulatory factors (CX3CL1, S100A9, and TGFB2), and a protein involved in lipid metabolism (LPL). The other 9 genes are transmembrane receptors (GFRA1, FZD1, NRP1, RARA, and SCARB2, which is also involved in lipid metabolism), a cell adhesion molecule (NCAM1), or intracellular signaling molecules (BM385941, RND1, and VAV1). Together, these data suggest major roles for three cellular mechanisms in the growth-promoting properties of OECs: (1) organization of the basement membrane, (2) modulation of the extracellular matrix, and (3) lipid transfer.
The basement membrane
LEPRE1, a leucine-proline rich chondroitin sulfate proteoglycan, and NID2, an adhesive protein that binds collagen type I, IV, and laminin and is important for the formation of neuromuscular junctions, are both basement membrane-associated proteins (Kohfeldt et al., 1998; Wassenhove-McCarthy and McCarthy, 1999; Fox et al., 2008). OECs were reported to express three other major neurite growth-supporting molecular components of the basement membrane (i.e., collagen type IV, fibronectin, and laminin) (Laurie and Leblond, 1983; Lein et al., 1991; Ramon-Cueto and Nieto-Sampedro, 1992; Doucette, 1996; Kafitz and Greer, 1997). In the primary olfactory pathway, layers of interconnected OECs form a permissive pathway for growing olfactory axons (Field et al., 2003; Li et al., 2005a, b). The results of the bioassay indicate that the OEC expressed basement membrane-associated proteins LEPRE1 and NID2 also contribute to the formation of a neurite growth permissive substrate.
The extracellular matrix
ADAMTS1 (a matrix metalloprotease), SERPINF1, SERPINI1, and TIMP2 (three protease inhibitors) are all involved in modification of the extracellular matrix (Yasui et al., 2003; Miranda and Lomas, 2006; Pastrana et al., 2006a; Yamagishi et al., 2009; Esselens et al., 2010). Overexpression of the MMP2 inhibitor TIMP2 inhibited neurite outgrowth. This supports the previous observation that MMP2 is a metalloprotease important for the outgrowth-promoting properties of OECs (Pastrana et al., 2006a). ADAMTS1 can cleave proteins, such as versican, and ADAMTS1 activity releases the repulsive axon guidance molecule SEMA3C, an inhibitory protein present in the neural scar, from the extracellular matrix (Steup et al., 2000; De Winter et al., 2002b; Russell et al., 2003; Esselens et al., 2010). MSLN is a glycoprotein, which stimulates the invasion and adhesion of cancer cells (Chang and Pastan, 1996; Rump et al., 2004; Uehara et al., 2008). Thus, ADAMTS1 and MSLN expressed by OECs could make the extracellular matrix more permissive for regenerating axons.
OECs express at least six members of the serine protease inhibitor gene family (SERPINB6b, SERPINE1, SERPINF1, SERPING1, SERPINH1, and SERPINI1) (Franssen et al., 2008; Simon et al., 2011). Here we show that the two serine protease inhibitors, SERPINI1 and SERPINF1, modify neurite outgrowth of cocultured DRG neurons. SERPINI1 is an inhibitor of tissue-type plasminogen activator (PLAT), a protease involved in fibrinolysis. PLAT and several other members of the fibrinolysis system, including SERPINE1, SERPINF1, PAR1, and thrombomodulin, have been implicated in neural repair (Tanimoto et al., 2006; Yamagishi et al., 2009; Roet et al., 2011; Simon et al., 2011). PLAT, expressed in neurons and glia, plays a prominent role in regulating neuroplasticity and synaptogenesis (Gveric et al., 2003; Miranda and Lomas, 2006; Samson and Medcalf, 2006; Borges et al., 2010). PLAT can degrade the axon growth-supporting extracellular matrix proteins laminin, fibrin, and fibronectin, and PLAT-deficient mice show a decrease in size of the injury area and increased functional outcome after spinal cord injury (Alexander and Werb, 1989; Abe et al., 2003). The effects found for SERPINI1 in the in vitro assays could be related to a role in extracellular matrix remodeling by inhibition of PLAT. However, the activity of SERPINI1 as a PLAT inhibitor was not investigated in the context of the experiments described in this study. No positive effects on afferent dorsal column regeneration were observed after ex vivo gene delivery of SERPINI1 in the lesioned area. The absence of an effect of SERPINI1 in vivo highlights that it is difficult to directly translate observations made in bioassays to the actual complex cellular microenvironment of the injured nervous system. Taken together, OECs appear to actively regulate the outgrowth-promoting properties of the extracellular matrix by coexpression of matrix molecules, proteases, and protease inhibitors.
Lipid transfer
SCARB2 expression is upregulated in the ONL immediately after a lesion and remains high during the ingrowth of newly formed axons. Knockdown of SCARB2 in OECs resulted in the strongest decrease of neurite length of cocultured embryonic DRG neurons. Overexpression of SCARB2 increased neurite outgrowth of adult DRG neurons. Moreover, ex vivo gene delivery of SCARB2 after a thoracic lesion of the dorsal funiculus significantly enhanced the number of DRG axons in proximity of the lesion site. Although the in vitro results presented in this study show a proregenerative effect of SCARB2 on adult DRG neurites, it is also possible that the observed effects are the result of a reduction of axonal die-back in combination with axonal sprouting. A number of axons did grow through the lesion site; however, it was not possible to associate these with the implanted fibroblasts. The bulk of the CTB labeling in the SCARB2 group was observed at 0.5–2 mm distance of the lesion center, approximately at the border of the GFAP-negative area of the scar. Repulsive molecules that are present after a spinal cord lesion, such as the myelin inhibitors, Nogo, MAG, and OMpg, and scar-associated inhibitors, including semaphorins, chondroitin sulfate proteoglycans, and collagens (Niclou et al., 2006; Fawcett et al., 2012; Lee and Zheng, 2012), may counteract the growth permissive environment created by SCARB2 expression. Interestingly, a recent study suggested that scavenger receptors can play a role in myelin clearance (Hendrickx et al., 2013). It is therefore possible that SCARB2 is involved in the removal of myelin-associated inhibitors.
SCARB2 is a cargo receptor and splice variant of SCARB1 (Webb et al., 1997) and belongs to the scavenger receptor class B family. Both proteins mediate the uptake of cholesterol to cells as well as cholesterol efflux by high-affinity binding of high-density lipoprotein (HDL) (Ji et al., 1997; Krieger and Kozarsky, 1999; Eckhardt et al., 2004; Mulcahy et al., 2004). SCARB2 is highly expressed in many neuroblastoma cell lines and in primary neurons (Thilakawardhana et al., 2005; Yamayoshi et al., 2012). In this study, only overexpression of SCARB2, and not SCARB1, enhanced neurite outgrowth significantly. SCARB1 and SCARB2 are present in caveolae at the cell surface, but SCARB2 is predominantly expressed intracellularly (Eckhardt et al., 2004). This is determined by its highly conserved cytoplasmic C terminus, which also enables rapid clathrin-dependent endocytosis of HDL to a perinuclear region and in the endosomal recycling compartment (Webb et al., 1997; Eckhardt et al., 2004, 2006).
There is accumulating evidence that local lipid transfer mechanisms are important for rapid axon membrane biosynthesis during regeneration after a nerve injury (Boyles et al., 1989; Jurevics et al., 1998; Li et al., 2010). Damage to the peripheral nervous system or CNS leads to the synthesis of proteins involved in lipid and cholesterol metabolism (Dawson et al., 1986; Ignatius et al., 1986; Mahley, 1988; Seitz et al., 2003). Pharmacological or genetic interference with local cholesterol reutilization after a peripheral nerve lesion results in a decrease of axonal outgrowth and a reduction of axon diameter (Goodrum et al., 2000). Glia secrete cholesterol-containing lipoproteins, which can strongly enhance neurite outgrowth and synapse formation of cultured retinal ganglion cells and DRG neurons (Handelmann et al., 1992; Mauch et al., 2001; Hayashi et al., 2004). Lipoprotein-stimulated neurite growth is mediated by the low density lipid receptor family on neuronal processes (Hayashi et al., 2004) and may explain the increased dorsal column axon outgrowth that we observed after ex vivo gene delivery of SCARB2 in the lesioned rat spinal cord.
SCARB2 is also known as a lysosomal protein, hence its alternative name, lysosomal integral membrane protein type 2. SCARB2 mediates transport of the lysosomal hydrolase β-glucocerebrosidase from the endoplasmatic reticulum to the lysosome (Reczek et al., 2007). β-Glucocerebrosidase transport and cholesterol secretion are functionally linked because treatment with a β-glucocerebrosidase inhibitor reduces APOA-I-mediated cholesterol efflux in fibroblasts (Glaros et al., 2005).
SCARB2 mutations have been linked to electrophysiological abnormalities that indicate a demyelinating neuropathy (Costello et al., 2009; Dibbens et al., 2011). SCARB2 deficiency in mice causes impairment in membrane transport processes and results in peripheral demyelination and degeneration of the spiral ganglia in the cochlea (Gamp et al., 2003). SCs from SCARB2 knock-out mice show a downregulation of peripheral myelin proteins and an upregulation of lysosomal enzymes (Gamp et al., 2003). In line with its function as cargo receptor, in cardiac myocytes SCARB2 interacts with and determines the cellular distribution of N-cadherin (CDH2) (Schroen et al., 2007), an important mediator of axonal outgrowth and myelination.
In conclusion, the positive effects of SCARB2 on regenerative sprouting of axons could result from increased cholesterol secretion and HDL recycling. Moreover, SCARB2 might modulate the function of CDH2, a protein that supports axonal growth and facilitates the onset of myelination. Regulation of SCARB2 expression provides a new opportunity to enhance neural repair after an injury. Future experiments will aim at deciphering the underlying molecular mechanisms that underlie the regenerative effects of SCARB2.
Footnotes
This work was supported by SenterNovem and The Netherlands Organization for Scientific Research (ZonMW investment 40-00506-98-10011). We thank Matthew R. Mason (Netherlands Institute for Neuroscience) and Edmund R. Hollis (University of California at San Diego) for advice on CTB labeling and Koen Bossers (Netherlands Institute for Neuroscience) for help with the microarray analysis. All information on primers and image clones used can be made available by the first author to colleagues that are interested.
The authors declare no competing financial interests.
References
- Abe Y, Nakamura H, Yoshino O, Oya T, Kimura T. Decreased neural damage after spinal cord injury in tPA-deficient mice. J Neurotrauma. 2003;20:43–57. doi: 10.1089/08977150360517173. [DOI] [PubMed] [Google Scholar]
- Alexander CM, Werb Z. Proteinases and extracellular matrix remodeling. Curr Opin Cell Biol. 1989;1:974–982. doi: 10.1016/0955-0674(89)90068-9. [DOI] [PubMed] [Google Scholar]
- Aoki K, Nakahara Y, Yamada S, Eto K. Role of polysialic acid on outgrowth of rat olfactory receptor neurons. Mech Dev. 1999;85:103–110. doi: 10.1016/S0925-4773(99)00092-1. [DOI] [PubMed] [Google Scholar]
- Au E, Richter MW, Vincent AJ, Tetzlaff W, Aebersold R, Sage EH, Roskams AJ. SPARC from olfactory ensheathing cells stimulates Schwann cells to promote neurite outgrowth and enhances spinal cord repair. J Neurosci. 2007;27:7208–7221. doi: 10.1523/JNEUROSCI.0509-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisvag V, Jünge FK, Bergum H, Jølsum L, Lydersen S, Günther CC, Ramampiaro H, Langaas M, Sandvik AK, Laegreid A. GeneTools: application for functional annotation and statistical hypothesis testing. BMC Bioinformatics. 2006;7:470. doi: 10.1186/1471-2105-7-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges VM, Lee TW, Christie DL, Birch NP. Neuroserpin regulates the density of dendritic protrusions and dendritic spine shape in cultured hippocampal neurons. J Neurosci Res. 2010;88:2610–2617. doi: 10.1002/jnr.22428. [DOI] [PubMed] [Google Scholar]
- Boruch AV, Conners JJ, Pipitone M, Deadwyler G, Storer PD, Devries GH, Jones KJ. Neurotrophic and migratory properties of an olfactory ensheathing cell line. Glia. 2001;33:225–229. doi: 10.1002/1098-1136(200103)33:3<225::AID-GLIA1021>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Boyd JG, Jahed A, McDonald TG, Krol KM, Van Eyk JE, Doucette R, Kawaja MD. Proteomic evaluation reveals that olfactory ensheathing cells but not Schwann cells express calponin. Glia. 2006;53:434–440. doi: 10.1002/glia.20299. [DOI] [PubMed] [Google Scholar]
- Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J Clin Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Gao Y, Bryson JB, Hou J, Chaudhry N, Siddiq M, Martinez J, Spencer T, Carmel J, Hart RB, Filbin MT. The cytokine interleukin-6 is sufficient but not necessary to mimic the peripheral conditioning lesion effect on axonal growth. J Neurosci. 2006;26:5565–5573. doi: 10.1523/JNEUROSCI.0815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996;93:136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RS, Woodhouse A, Fung S, Dickson TC, West AK, Vickers JC, Chuah MI. Olfactory ensheathing cells promote neurite sprouting of injured axons in vitro by direct cellular contact and secretion of soluble factors. Cell Mol Life Sci. 2004;61:1238–1245. doi: 10.1007/s00018-004-4026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello DJ, Chiappa KH, Siao P. Progressive myoclonus epilepsy with demyelinating peripheral neuropathy and preserved intellect: a novel syndrome. Arch Neurol. 2009;66:898–901. doi: 10.1001/archneurol.2009.131. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Schechter N, Williams DL. Induction of rat E and chicken A-I apolipoproteins and mRNAs during optic nerve degeneration. J Biol Chem. 1986;261:5681–5684. [PubMed] [Google Scholar]
- Day WA, Koishi K, Nukuda H, McLennan IS. Transforming growth factor-β 2 causes an acute improvement in the motor performance of transgenic ALS mice. Neurobiol Dis. 2005;19:323–330. doi: 10.1016/j.nbd.2005.01.010. [DOI] [PubMed] [Google Scholar]
- De Winter F, Holtmaat AJ, Verhaagen J. Neuropilin and class 3 semaphorins in nervous system regeneration. Adv Exp Med Biol. 2002a;515:115–139. doi: 10.1007/978-1-4615-0119-0_10. [DOI] [PubMed] [Google Scholar]
- De Winter F, Oudega M, Lankhorst AJ, Hamers FP, Blits B, Ruitenberg MJ, Pasterkamp RJ, Gispen WH, Verhaagen J. Injury-induced class 3 semaphorin expression in the rat spinal cord. Exp Neurol. 2002b;175:61–75. doi: 10.1006/exnr.2002.7884. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Karakis I, Bayly MA, Costello DJ, Cole AJ, Berkovic SF. Mutation of SCARB2 in a patient with progressive myoclonus epilepsy and demyelinating peripheral neuropathy. Arch Neurol. 2011;68:812–813. doi: 10.1001/archneurol.2011.120. [DOI] [PubMed] [Google Scholar]
- Doherty P, Walsh FS. The contrasting roles of N-CAM and N-cadherin as neurite outgrowth-promoting molecules. J Cell Sci Suppl. 1991;15:13–21. doi: 10.1242/jcs.1991.supplement_15.3. [DOI] [PubMed] [Google Scholar]
- Doucette R. Glial influences on axonal growth in the primary olfactory system. Glia. 1990;3:433–449. doi: 10.1002/glia.440030602. [DOI] [PubMed] [Google Scholar]
- Doucette R. PNS-CNS transitional zone of the first cranial nerve 1. J Comp Neurol. 1991;312:451–466. doi: 10.1002/cne.903120311. [DOI] [PubMed] [Google Scholar]
- Doucette R. Immunohistochemical localization of laminin, fibronectin and collagen type IV in the nerve fiber layer of the olfactory bulb. Int J Dev Neurosci. 1996;14:945–959. doi: 10.1016/S0736-5748(96)00042-1. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt ER, Cai L, Sun B, Webb NR, van der Westhuyzen DR. High density lipoprotein uptake by scavenger receptor SR-BII. J Biol Chem. 2004;279:14372–14381. doi: 10.1074/jbc.M313793200. [DOI] [PubMed] [Google Scholar]
- Eckhardt ER, Cai L, Shetty S, Zhao Z, Szanto A, Webb NR, Van der Westhuyzen DR. High density lipoprotein endocytosis by scavenger receptor SR-BII is clathrin-dependent and requires a carboxyl-terminal dileucine motif. J Biol Chem. 2006;281:4348–4353. doi: 10.1074/jbc.M513154200. [DOI] [PubMed] [Google Scholar]
- Esselens C, Malapeira J, Colomé N, Casal C, Rodríguez-Manzaneque JC, Canals F, Arribas J. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J Biol Chem. 2010;285:2463–2473. doi: 10.1074/jbc.M109.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairless R, Frame MC, Barnett SC. N-cadherin differentially determines Schwann cell and olfactory ensheathing cell adhesion and migration responses upon contact with astrocytes. Mol Cell Neurosci. 2005;28:253–263. doi: 10.1016/j.mcn.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Fawcett JW, Schwab ME, Montani L, Brazda N, Müller HW. Defeating inhibition of regeneration by scar and myelin components. Handb Clin Neurol. 2012;109:503–522. doi: 10.1016/B978-0-444-52137-8.00031-0. [DOI] [PubMed] [Google Scholar]
- Field P, Li Y, Raisman G. Ensheathment of the olfactory nerves in the adult rat 1. J Neurocytol. 2003;32:317–324. doi: 10.1023/B:NEUR.0000010089.37032.48. [DOI] [PubMed] [Google Scholar]
- Fox MA, Ho MS, Smyth N, Sanes JR. A synaptic nidogen: developmental regulation and role of nidogen-2 at the neuromuscular junction. Neural Dev. 2008;3:24. doi: 10.1186/1749-8104-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ, Gilson JM, Franceschini IA, Barnett SC. Schwann cell-like myelination following transplantation of an olfactory bulb-ensheathing cell line into areas of demyelination in the adult CNS. Glia. 1996;17:217–224. doi: 10.1002/(SICI)1098-1136(199607)17:3<217::AID-GLIA4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Franssen EH, De Bree FM, Essing AH, Ramon-Cueto A, Verhaagen J. Comparative gene expression profiling of olfactory ensheathing glia and Schwann cells indicates distinct tissue repair characteristics of olfactory ensheathing glia. Glia. 2008;56:1285–1298. doi: 10.1002/glia.20697. [DOI] [PubMed] [Google Scholar]
- Gamp AC, Tanaka Y, Lüllmann-Rauch R, Wittke D, D'Hooge R, De Deyn PP, Moser T, Maier H, Hartmann D, Reiss K, Illert AL, von Figura K, Saftig P. LIMP-2/LGP85 deficiency causes ureteric pelvic junction obstruction, deafness and peripheral neuropathy in mice. Hum Mol Genet. 2003;12:631–646. doi: 10.1093/hmg/ddg062. [DOI] [PubMed] [Google Scholar]
- Gao Y, Deng K, Hou J, Bryson JB, Barco A, Nikulina E, Spencer T, Mellado W, Kandel ER, Filbin MT. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron. 2004;44:609–621. doi: 10.1016/j.neuron.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Glaros EN, Kim WS, Quinn CM, Wong J, Gelissen I, Jessup W, Garner B. Glycosphingolipid accumulation inhibits cholesterol efflux via the ABCA1/apolipoprotein A-I pathway: 1-phenyl-2-decanoylamino-3-morpholino-1-propanol is a novel cholesterol efflux accelerator. J Biol Chem. 2005;280:24515–24523. doi: 10.1074/jbc.M413862200. [DOI] [PubMed] [Google Scholar]
- Glonek GF, Solomon PJ. Factorial and time course designs for cDNA microarray experiments. Biostatistics. 2004;5:89–111. doi: 10.1093/biostatistics/5.1.89. [DOI] [PubMed] [Google Scholar]
- Goodman MN, Silver J, Jacobberger JW. Establishment and neurite outgrowth properties of neonatal and adult rat olfactory bulb glial cell lines. Brain Res. 1993;619:199–213. doi: 10.1016/0006-8993(93)91613-W. [DOI] [PubMed] [Google Scholar]
- Goodrum JF, Brown JC, Fowler KA, Bouldin TW. Axonal regeneration, but not myelination, is partially dependent on local cholesterol reutilization in regenerating nerve. J Neuropathol Exp Neurol. 2000;59:1002–1010. doi: 10.1093/jnen/59.11.1002. [DOI] [PubMed] [Google Scholar]
- Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals: I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- Guérout N, Derambure C, Drouot L, Bon-Mardion N, Duclos C, Boyer O, Marie JP. Comparative gene expression profiling of olfactory ensheathing cells from olfactory bulb and olfactory mucosa. Glia. 2010;58:1570–1580. doi: 10.1002/glia.21030. [DOI] [PubMed] [Google Scholar]
- Gveric D, Herrera B, Petzold A, Lawrence DA, Cuzner ML. Impaired fibrinolysis in multiple sclerosis: a role for tissue plasminogen activator inhibitors. Brain. 2003;126:1590–1598. doi: 10.1093/brain/awg167. [DOI] [PubMed] [Google Scholar]
- Handelmann GE, Boyles JK, Weisgraber KH, Mahley RW, Pitas RE. Effects of apolipoprotein E, β-very low density lipoproteins, and cholesterol on the extension of neurites by rabbit dorsal root ganglion neurons in vitro. J Lipid Res. 1992;33:1677–1688. [PubMed] [Google Scholar]
- Hayashi H, Campenot RB, Vance DE, Vance JE. Glial lipoproteins stimulate axon growth of central nervous system neurons in compartmented cultures. J Biol Chem. 2004;279:14009–14015. doi: 10.1074/jbc.M313828200. [DOI] [PubMed] [Google Scholar]
- Hendrickx DA, Koning N, Schuurman KG, van Strien ME, van Eden CG, Hamann J, Huitinga I. Selective upregulation of scavenger receptors in and around demyelinating areas in multiple sclerosis. J Neuropathol Exp Neurol. 2013;72:106–118. doi: 10.1097/NEN.0b013e31827fd9e8. [DOI] [PubMed] [Google Scholar]
- Hendriks WT, Eggers R, Verhaagen J, Boer GJ. Gene transfer to the spinal cord neural scar with lentiviral vectors: predominant transgene expression in astrocytes but not in meningeal cells. J Neurosci Res. 2007;85:3041–3052. doi: 10.1002/jnr.21432. [DOI] [PubMed] [Google Scholar]
- Ignatius MJ, Gebicke-Harter PJ, Skene JH, Schilling JW, Weisgraber KH, Mahley RW, Shooter EM. Expression of apolipoprotein E during nerve degeneration and regeneration. Proc Natl Acad Sci U S A. 1986;83:1125–1129. doi: 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Lankford KL, Burton WV, Fodor WL, Kocsis JD. Xenotransplantation of transgenic pig olfactory ensheathing cells promotes axonal regeneration in rat spinal cord. Nat Biotechnol. 2000;18:949–953. doi: 10.1038/79432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed A, Rowland JW, McDonald T, Boyd JG, Doucette R, Kawaja MD. Olfactory ensheathing cells express smooth muscle alpha-actin in vitro and in vivo. J Comp Neurol. 2007;503:209–223. doi: 10.1002/cne.21385. [DOI] [PubMed] [Google Scholar]
- Ji Y, Jian B, Wang N, Sun Y, Moya ML, Phillips MC, Rothblat GH, Swaney JB, Tall AR. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem. 1997;272:20982–20985. doi: 10.1074/jbc.272.34.20982. [DOI] [PubMed] [Google Scholar]
- Jurevics H, Bouldin TW, Toews AD, Morell P. Regenerating sciatic nerve does not utilize circulating cholesterol. Neurochem Res. 1998;23:401–406. doi: 10.1023/A:1022469803426. [DOI] [PubMed] [Google Scholar]
- Kafitz KW, Greer CA. Role of laminin in axonal extension from olfactory receptor cells. J Neurobiol. 1997;32:298–310. doi: 10.1002/(SICI)1097-4695(199703)32:3<298::AID-NEU4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kohfeldt E, Sasaki T, Göhring W, Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J Mol Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- Krieger M, Kozarsky K. Influence of the HDL receptor SR-BI on atherosclerosis. Curr Opin Lipidol. 1999;10:491–497. doi: 10.1097/00041433-199912000-00003. [DOI] [PubMed] [Google Scholar]
- Kuronita T, Eskelinen EL, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J Cell Sci. 2002;115:4117–4131. doi: 10.1242/jcs.00075. [DOI] [PubMed] [Google Scholar]
- Lakatos A, Barnett SC, Franklin RJ. Olfactory ensheathing cells induce less host astrocyte response and chondroitin sulphate proteoglycan expression than Schwann cells following transplantation into adult CNS white matter. Exp Neurol. 2003;184:237–246. doi: 10.1016/S0014-4886(03)00270-X. [DOI] [PubMed] [Google Scholar]
- Laurie GW, Leblond CP. What is known of the production of basement membrane components. J Histochem Cytochem. 1983;31:159–163. doi: 10.1177/31.1A_Suppl.6186721. [DOI] [PubMed] [Google Scholar]
- Leaver SG, Harvey AR, Plant GW. Adult olfactory ensheathing glia promote the long-distance growth of adult retinal ganglion cell neurites in vitro. Glia. 2006;53:467–476. doi: 10.1002/glia.20311. [DOI] [PubMed] [Google Scholar]
- Lee JK, Zheng B. Role of myelin-associated inhibitors in axonal repair after spinal cord injury. Exp Neurol. 2012;235:33–42. doi: 10.1016/j.expneurol.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein PJ, Higgins D, Turner DC, Flier LA, Terranova VP. The NC1 domain of type IV collagen promotes axonal growth in sympathetic neurons through interaction with the α 1 β 1 integrin. J Cell Biol. 1991;113:417–428. doi: 10.1083/jcb.113.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FQ, Fowler KA, Neil JE, Colton CA, Vitek MP. An apolipoprotein E-mimetic stimulates axonal regeneration and remyelination after peripheral nerve injury. J Pharmacol Exp Ther. 2010;334:106–115. doi: 10.1124/jpet.110.167882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Field PM, Raisman G. Repair of adult rat corticospinal tract by transplants of olfactory ensheathing cells. Science. 1997;277:2000–2002. doi: 10.1126/science.277.5334.2000. [DOI] [PubMed] [Google Scholar]
- Li Y, Field PM, Raisman G. Olfactory ensheathing cells and olfactory nerve fibroblasts maintain continuous open channels for regrowth of olfactory nerve fibres. Glia. 2005a;52:245–251. doi: 10.1002/glia.20241. [DOI] [PubMed] [Google Scholar]
- Li Y, Li D, Raisman G. Interaction of olfactory ensheathing cells with astrocytes may be the key to repair of tract injuries in the spinal cord: the ‘pathway hypothesis.’. J Neurocytol. 2005b;34:343–351. doi: 10.1007/s11068-005-8361-1. [DOI] [PubMed] [Google Scholar]
- Lipson AC, Widenfalk J, Lindqvist E, Ebendal T, Olson L. Neurotrophic properties of olfactory ensheathing glia. Exp Neurol. 2003;180:167–171. doi: 10.1016/S0014-4886(02)00058-4. [DOI] [PubMed] [Google Scholar]
- Liu Y, Teng X, Yang X, Song Q, Lu R, Xiong J, Liu B, Zeng N, Zeng Y, Long J, Cao R, Lin Y, He Q, Chen P, Lu M, Liang S. Shotgun proteomics and network analysis between plasma membrane and extracellular matrix proteins from rat olfactory ensheathing cells. Cell Transplant. 2010;19:133–146. doi: 10.3727/096368910X492607. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mauch DH, Nägler K, Schumacher S, Göritz C, Müller EC, Otto A, Pfrieger FW. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- Miragall F, Kadmon G, Husmann M, Schachner M. Expression of cell adhesion molecules in the olfactory system of the adult mouse: presence of the embryonic form of N-CAM. Dev Biol. 1988;129:516–531. doi: 10.1016/0012-1606(88)90397-1. [DOI] [PubMed] [Google Scholar]
- Miranda E, Lomas DA. Neuroserpin: a serpin to think about. Cell Mol Life Sci. 2006;63:709–722. doi: 10.1007/s00018-005-5077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC. Design and analysis of experiments. 5th ed. New York: Wiley; 2001. [Google Scholar]
- Moon LD, Fawcett JW. Reduction in CNS scar formation without concomitant increase in axon regeneration following treatment of adult rat brain with a combination of antibodies to TGFβ1 and β2. Eur J Neurosci. 2001;14:1667–1677. doi: 10.1046/j.0953-816x.2001.01795.x. [DOI] [PubMed] [Google Scholar]
- Moulton DG. Dynamics of cell populations in the olfactory epithelium. Ann N Y Acad Sci. 1974;237:52–61. doi: 10.1111/j.1749-6632.1974.tb49843.x. [DOI] [PubMed] [Google Scholar]
- Mulcahy JV, Riddell DR, Owen JS. Human scavenger receptor class B type II (SR-BII) and cellular cholesterol efflux. Biochem J. 2004;377:741–747. doi: 10.1042/BJ20030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadi NS, Head R, Grillo M, Hempstead J, Grannot-Reisfeld N, Margolis FL. Chemical deafferentation of the olfactory bulb: plasticity of the levels of tyrosine hydroxylase, dopamine and norepinephrine. Brain Res. 1981;213:365–377. doi: 10.1016/0006-8993(81)90241-9. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Niclou SP, Franssen EH, Ehlert EM, Taniguchi M, Verhaagen J. Meningeal cell-derived semaphorin 3A inhibits neurite outgrowth. Mol Cell Neurosci. 2003;24:902–912. doi: 10.1016/S1044-7431(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Niclou SP, Ehlert EM, Verhaagen J. Chemorepellent axon guidance molecules in spinal cord injury. J Neurotrauma. 2006;23:409–421. doi: 10.1089/neu.2006.23.409. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Moreno-Flores MT, Gurzov EN, Avila J, Wandosell F, Diaz-Nido J. Genes associated with adult axon regeneration promoted by olfactory ensheathing cells: a new role for matrix metalloproteinase 2. J Neurosci. 2006;26:5347–5359. doi: 10.1523/JNEUROSCI.1111-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellitteri R, Spatuzza M, Russo A, Zaccheo D, Stanzani S. Olfactory ensheathing cells represent an optimal substrate for hippocampal neurons: an in vitro study. Int J Dev Neurosci. 2009;27:453–458. doi: 10.1016/j.ijdevneu.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Plant GW, Christensen CL, Oudega M, Bunge MB. Delayed transplantation of olfactory ensheathing glia promotes sparing/regeneration of supraspinal axons in the contused adult rat spinal cord. J Neurotrauma. 2003;20:1–16. doi: 10.1089/08977150360517146. [DOI] [PubMed] [Google Scholar]
- Radtke C, Akiyama Y, Brokaw J, Lankford KL, Wewetzer K, Fodor WL, Kocsis JD. Remyelination of the nonhuman primate spinal cord by transplantation of H-transferase transgenic adult pig olfactory ensheathing cells. FASEB J. 2004;18:335–337. doi: 10.1096/fj.03-0214fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisman G. Specialized neuroglial arrangement may explain the capacity of vomeronasal axons to reinnervate central neurons. Neuroscience. 1985;14:237–254. doi: 10.1016/0306-4522(85)90176-9. [DOI] [PubMed] [Google Scholar]
- Ramer LM, Au E, Richter MW, Liu J, Tetzlaff W, Roskams AJ. Peripheral olfactory ensheathing cells reduce scar and cavity formation and promote regeneration after spinal cord injury. J Comp Neurol. 2004;473:1–15. doi: 10.1002/cne.20049. [DOI] [PubMed] [Google Scholar]
- Ramón-Cueto A, Nieto-Sampedro M. Glial cells from adult rat olfactory bulb: immunocytochemical properties of pure cultures of ensheathing cells. Neuroscience. 1992;47:213–220. doi: 10.1016/0306-4522(92)90134-N. [DOI] [PubMed] [Google Scholar]
- Ramón-Cueto A, Nieto-Sampedro M. Regeneration into the spinal cord of transected dorsal root axons is promoted by ensheathing glia transplants. Exp Neurol. 1994;127:232–244. doi: 10.1006/exnr.1994.1099. [DOI] [PubMed] [Google Scholar]
- Ramón-Cueto A, Pérez J, Nieto-Sampedro M. In vitro enfolding of olfactory neurites by p75 NGF receptor positive ensheathing cells from adult rat olfactory bulb. Eur J Neurosci. 1993;5:1172–1180. doi: 10.1111/j.1460-9568.1993.tb00971.x. [DOI] [PubMed] [Google Scholar]
- Ramón-Cueto A, Plant GW, Avila J, Bunge MB. Long-distance axonal regeneration in the transected adult rat spinal cord is promoted by olfactory ensheathing glia transplants. J Neurosci. 1998;18:3803–3815. doi: 10.1523/JNEUROSCI.18-10-03803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Schwake M, Schröder J, Hughes H, Blanz J, Jin X, Brondyk W, Van Patten S, Edmunds T, Saftig P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Richter MW, Fletcher PA, Liu J, Tetzlaff W, Roskams AJ. Lamina propria and olfactory bulb ensheathing cells exhibit differential integration and migration and promote differential axon sprouting in the lesioned spinal cord. J Neurosci. 2005;25:10700–10711. doi: 10.1523/JNEUROSCI.3632-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochel S, Margolis FL. The response of ornithine decarboxylase during neuronal degeneration and regeneration in olfactory epithelium. J Neurochem. 1980;35:850–860. doi: 10.1111/j.1471-4159.1980.tb07082.x. [DOI] [PubMed] [Google Scholar]
- Roet KC, Bossers K, Franssen EH, Ruitenberg MJ, Verhaagen J. A meta-analysis of microarray-based gene expression studies of olfactory bulb-derived olfactory ensheathing cells. Exp Neurol. 2011;229:10–45. doi: 10.1016/j.expneurol.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Ruitenberg MJ, Plant GW, Christensen CL, Blits B, Niclou SP, Harvey AR, Boer GJ, Verhaagen J. Viral vector-mediated gene expression in olfactory ensheathing glia implants in the lesioned rat spinal cord. Gene Ther. 2002;9:135–146. doi: 10.1038/sj.gt.3301626. [DOI] [PubMed] [Google Scholar]
- Ruitenberg MJ, Plant GW, Hamers FP, Wortel J, Blits B, Dijkhuizen PA, Gispen WH, Boer GJ, Verhaagen J. Ex vivo adenoviral vector-mediated neurotrophin gene transfer to olfactory ensheathing glia: effects on rubrospinal tract regeneration, lesion size, and functional recovery after implantation in the injured rat spinal cord. J Neurosci. 2003;23:7045–7058. doi: 10.1523/JNEUROSCI.23-18-07045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruitenberg MJ, Vukovic J, Sarich J, Busfield SJ, Plant GW. Olfactory ensheathing cells: characteristics, genetic engineering, and therapeutic potential. J Neurotrauma. 2006;23:468–478. doi: 10.1089/neu.2006.23.468. [DOI] [PubMed] [Google Scholar]
- Rump A, Morikawa Y, Tanaka M, Minami S, Umesaki N, Takeuchi M, Miyajima A. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–9198. doi: 10.1074/jbc.M312372200. [DOI] [PubMed] [Google Scholar]
- Runyan SA, Phelps PE. Mouse olfactory ensheathing glia enhance axon outgrowth on a myelin substrate in vitro. Exp Neurol. 2009;216:95–104. doi: 10.1016/j.expneurol.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DL, Doyle KM, Ochsner SA, Sandy JD, Richards JS. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem. 2003;278:42330–42339. doi: 10.1074/jbc.M300519200. [DOI] [PubMed] [Google Scholar]
- Samson AL, Medcalf RL. Tissue-type plasminogen activator: a multifaceted modulator of neurotransmission and synaptic plasticity. Neuron. 2006;50:673–678. doi: 10.1016/j.neuron.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Santos-Silva A, Fairless R, Frame MC, Montague P, Smith GM, Toft A, Riddell JS, Barnett SC. FGF/heparin differentially regulates Schwann cell and olfactory ensheathing cell interactions with astrocytes: a role in astrocytosis. J Neurosci. 2007;27:7154–7167. doi: 10.1523/JNEUROSCI.1184-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lankford KL, Zemedkun M, Kocsis JD. Identified olfactory ensheathing cells transplanted into the transected dorsal funiculus bridge the lesion and form myelin. J Neurosci. 2004;24:8485–8493. doi: 10.1523/JNEUROSCI.1998-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroen B, Leenders JJ, van Erk A, Bertrand AT, van Loon M, van Leeuwen RE, Kubben N, Duisters RF, Schellings MW, Janssen BJ, Debets JJ, Schwake M, Høydal MA, Heymans S, Saftig P, Pinto YM. Lysosomal integral membrane protein 2 is a novel component of the cardiac intercalated disc and vital for load-induced cardiac myocyte hypertrophy. J Exp Med. 2007;204:1227–1235. doi: 10.1084/jem.20070145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Ring G, Iwema CL, Mezza RC. Reinnervation of the rat olfactory bulb after methyl bromide-induced lesion: timing and extent of reinnervation. J Comp Neurol. 1999;412:439–457. doi: 10.1002/(SICI)1096-9861(19990927)412:3<439::AID-CNE5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Seitz A, Kragol M, Aglow E, Showe L, Heber-Katz E. Apolipoprotein E expression after spinal cord injury in the mouse. J Neurosci Res. 2003;71:417–426. doi: 10.1002/jnr.10482. [DOI] [PubMed] [Google Scholar]
- Simón D, Martín-Bermejo MJ, Gallego-Hernández MT, Pastrana E, García-Escudero V, García-Gómez A, Lim F, Díaz-Nido J, Avila J, Moreno-Flores MT. Expression of plasminogen activator inhibitor-1 by olfactory ensheathing glia promotes axonal regeneration. Glia. 2011;59:1458–1471. doi: 10.1002/glia.21189. [DOI] [PubMed] [Google Scholar]
- Steup A, Lohrum M, Hamscho N, Savaskan NE, Ninnemann O, Nitsch R, Fujisawa H, Püschel AW, Skutella T. Sema3C and netrin-1 differentially affect axon growth in the hippocampal formation. Mol Cell Neurosci. 2000;15:141–155. doi: 10.1006/mcne.1999.0818. [DOI] [PubMed] [Google Scholar]
- Takagi S, Kasuya Y, Shimizu M, Matsuura T, Tsuboi M, Kawakami A, Fujisawa H. Expression of a cell adhesion molecule, neuropilin, in the developing chick nervous system. Dev Biol. 1995;170:207–222. doi: 10.1006/dbio.1995.1208. [DOI] [PubMed] [Google Scholar]
- Tanimoto S, Kanamoto T, Mizukami M, Aoyama H, Kiuchi Y. Pigment epithelium-derived factor promotes neurite outgrowth of retinal cells. Hiroshima J Med Sci. 2006;55:109–116. [PubMed] [Google Scholar]
- Tannemaat MR, Boer GJ, Verhaagen J, Malessy MJ. Genetic modification of human sural nerve segments by a lentiviral vector encoding nerve growth factor. Neurosurgery. 2007;61:1286–1294. doi: 10.1227/01.neu.0000306108.78044.a2. discussion 1294–1296. [DOI] [PubMed] [Google Scholar]
- Thilakawardhana S, Everett DM, Murdock PR, Dingwall C, Owen JS. Quantification of apolipoprotein E receptors in human brain-derived cell lines by real-time polymerase chain reaction. Neurobiol Aging. 2005;26:813–823. doi: 10.1016/j.neurobiolaging.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Tisay KT, Key B. The extracellular matrix modulates olfactory neurite outgrowth on ensheathing cells. J Neurosci. 1999;19:9890–9899. doi: 10.1523/JNEUROSCI.19-22-09890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CP, Perez-Polo JR. Expression of p75NGFR in the olfactory system following peripheral deafferentation. Neuroreport. 1993;4:1023–1026. doi: 10.1097/00001756-199308000-00007. [DOI] [PubMed] [Google Scholar]
- Uehara N, Matsuoka Y, Tsubura A. Mesothelin promotes anchorage-independent growth and prevents anoikis via extracellular signal-regulated kinase signaling pathway in human breast cancer cells. Mol Cancer Res. 2008;6:186–193. doi: 10.1158/1541-7786.MCR-07-0254. [DOI] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher AB, Gispen WH, Margolis FL. The expression of the growth associated protein B50/GAP43 in the olfactory system of neonatal and adult rats. J Neurosci. 1989;9:683–691. doi: 10.1523/JNEUROSCI.09-02-00683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher AB, Grillo M, Khew-Goodall YS, Gispen WH, Margolis FL. Neuroplasticity in the olfactory system: differential effects of central and peripheral lesions of the primary olfactory pathway on the expression of B-50/GAP43 and the olfactory marker protein. J Neurosci Res. 1990;26:31–44. doi: 10.1002/jnr.490260105. [DOI] [PubMed] [Google Scholar]
- Vincent AJ, Taylor JM, Choi-Lundberg DL, West AK, Chuah MI. Genetic expression profile of olfactory ensheathing cells is distinct from that of Schwann cells and astrocytes. Glia. 2005;51:132–147. doi: 10.1002/glia.20195. [DOI] [PubMed] [Google Scholar]
- Wassenhove-McCarthy DJ, McCarthy KJ. Molecular characterization of a novel basement membrane-associated proteoglycan, leprecan. J Biol Chem. 1999;274:25004–25017. doi: 10.1074/jbc.274.35.25004. [DOI] [PubMed] [Google Scholar]
- Webb NR, de Villiers WJ, Connell PM, de Beer FC, van der Westhuyzen DR. Alternative forms of the scavenger receptor BI (SR-BI) J Lipid Res. 1997;38:1490–1495. [PubMed] [Google Scholar]
- Woodhall E, West AK, Chuah MI. Cultured olfactory ensheathing cells express nerve growth factor, brain-derived neurotrophic factor, glia cell line-derived neurotrophic factor and their receptors. Brain Res Mol Brain Res. 2001;88:203–213. doi: 10.1016/S0169-328X(01)00044-4. [DOI] [PubMed] [Google Scholar]
- Yamagishi S, Matsui T, Nakamura K, Takenaka K. Administration of pigment epithelium-derived factor prolongs bleeding time by suppressing plasminogen activator inhibitor-1 activity and platelet aggregation in rats. Clin Exp Med. 2009;9:73–76. doi: 10.1007/s10238-008-0010-4. [DOI] [PubMed] [Google Scholar]
- Yamayoshi S, Iizuka S, Yamashita T, Minagawa H, Mizuta K, Okamoto M, Nishimura H, Sanjoh K, Katsushima N, Itagaki T, Nagai Y, Fujii K, Koike S. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol. 2012;86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui N, Mori T, Morito D, Matsushita O, Kourai H, Nagata K, Koide T. Dual-site recognition of different extracellular matrix components by anti-angiogenic/neurotrophic serpin, PEDF. Biochemistry. 2003;42:3160–3167. doi: 10.1021/bi0206558. [DOI] [PubMed] [Google Scholar]