Abstract
Axon regeneration after injury requires the extensive reconstruction, reorganization, and stabilization of the microtubule cytoskeleton in the growth cones. Here, we identify KIF3C as a key regulator of axonal growth and regeneration by controlling microtubule dynamics and organization in the growth cone. KIF3C is developmentally regulated. Rat embryonic sensory axons and growth cones contain undetectable levels of KIF3C protein that is locally translated immediately after injury. In adult neurons, KIF3C is axonally transported from the cell body and is enriched at the growth cone where it preferentially binds to tyrosinated microtubules. Functionally, the interaction of KIF3C with EB3 is necessary for its localization at the microtubule plus-ends in the growth cone. Depletion of KIF3C in adult neurons leads to an increase in stable, overgrown and looped microtubules because of a strong decrease in the microtubule frequency of catastrophes, suggesting that KIF3C functions as a microtubule-destabilizing factor. Adult axons lacking KIF3C, by RNA interference or KIF3C gene knock-out, display an impaired axonal outgrowth in vitro and a delayed regeneration after injury both in vitro and in vivo. Murine KIF3C knock-out embryonic axons grow normally but do not regenerate after injury because they are unable to locally translate KIF3C. These data show that KIF3C is an injury-specific kinesin that contributes to axon growth and regeneration by regulating and organizing the microtubule cytoskeleton in the growth cone.
Introduction
Injury to an axon triggers a series of complex anatomical and molecular changes, including retraction, terminal enlargement formation, growth cone reconstruction, and finally axon extension (Bradke et al., 2012). All of these steps involve the dynamic microtubule cytoskeleton and controlled delivery of cargos transported via microtubules. The microtubules extending into the periphery of the growth cone are considered to be particularly dynamic and involved in the processes of growth cone turning and path-finding (Conde and Cáceres, 2009; Lowery and Van Vactor, 2009). Growth cone regeneration requires precise reconstruction and remodeling of the axonal microtubule cytoskeleton (Ertürk et al., 2007; Bradke et al., 2012). However, the factors that control microtubule restructuring in mammalian growth cones and axonal regeneration after injury have received less attention. Furthermore, it is still unclear by what mechanism the cellular machinery at the growth cone or injured axon tip integrates and coordinates microtubule stabilization and remodeling during axon growth and regeneration. The kinesin superfamily of microtubule molecular motors can affect microtubule dynamics and structure either by directly acting as microtubule organizers through their interaction with microtubules, promoting polymerization/depolymerization or alignment, or indirectly, through their role in the selective transport of molecules to the axon tips (Wu et al., 2006; Hirokawa et al., 2009; Liu et al., 2010; Mattie et al., 2010; Daire and Poüs, 2011; Drummond, 2011; Su et al., 2012). Thus, kinesins appear as robust candidates for a role in the coordination and restructuring of the microtubule network at the growth cone or axon tip after injury.
We previously found that mRNAs encoding various motor proteins were enriched in embryonic sensory axons and growth cones, and their presence in axons is developmentally regulated (Gumy et al., 2011). Kinesin-2 family member KIF3C is particularly interesting because it is expressed almost exclusively in the nervous system, being strongly upregulated at the mRNA level during sensory neuron differentiation and localized to the tip of outgrowing neurites (Muresan et al., 1998; Yang and Goldstein, 1998; Navone et al., 2001; Yang et al., 2001; Laketa et al., 2007). This led us to examine whether KIF3C might play a specific role during axon regeneration after injury. Here, we show that KIF3C contributes to sensory axon regeneration by regulating microtubule dynamics and organization in the growth cone. Localization of KIF3C to the plus-ends of microtubules in the growth cone is achieved by binding to tyrosinated microtubules and by interacting with EB3. We found that KIF3C functions as a microtubule-destabilizing factor and that its localization at the tips of growing microtubules in the growth cone is necessary for the maintenance of microtubule dynamic behavior at the axonal growth cone. Importantly, we show that loss of KIF3C function adversely affects axon growth in adult dorsal root ganglia (DRG) neurons and impairs regeneration after injury in both embryonic and adult DRG neurons in vitro and in vivo.
Materials and Methods
Neuronal cultures and transfections
DRG explants.
Embryonic and adult DRG explants and neurons were isolated as previously described (Gumy et al., 2008, 2011). Briefly, DRG explants dissected from adult female Sprague Dawley rats (3–5 months old) or E16 rat embryos or from adult KIF3C−/− or wild-type (WT) mice (3–5 months old) or E15 KIF3C−/− or WT mice were plated onto Permanox chamber slides (Lab-Tek), which had been coated with poly-d-lysine (20 μg/ml, Sigma) and laminin (1 μg/ml; Sigma). Explants were cultured in DMEM containing ITS+ (1:100, BD Biosciences), penicillin-streptomycin-fungizone (1×; Sigma), and 20 ng/ml of NGF (Sigma) or 100 ng/ml for embryonic cultures. Explants were kept at 37°C in 7% CO2. Axons from the explants were axotomized using a pulled Pasteur pipette.
Dissociated DRG neurons.
Dissociated DRG neurons were isolated from adult female Sprague Dawley rats (3–5 months old) or E16 rat embryos or from adult KIF3C−/− or WT mice (3–5 months old). The neurons were dissociated with collagenase type IV (Sigma) and 0.1% trypsin (Sigma) and centrifuged through a 15% BSA density gradient. Dissociated neurons were plated on coverslips coated with poly-d-lysine (20 μg/ml) and 1 μg/ml laminin, and cultured in dissociated DRG culture medium (DMEM; Sigma), 1% FBS (Invitrogen), penicillin-streptomycin-fungizone (1×; Sigma), and NGF (20 ng/ml or 100 ng/ml for E16 neurons; Sigma), and kept at 37°C in 7% CO2. For experiments requiring taxol treatment (Cytoskeleton), this was added 24 h after plating to the neurons at a concentration of 100 nm for 3 h. Control neurons were treated with DMSO.
Compartmentalized embryonic DRG cultures and axotomy.
Separation of the DRG axon and cell body compartments was achieved using Campenot chambers (Tyler Research). Cultures were set up following an established protocol (Pazyra-Murphy and Segal, 2008) and using permanox chamber slides (Nalge Nunc International). After 24 h at 37°C to test for leakage, watertight chambers were coated with 1 μg/ml laminin in both the somatic and axonal compartment for at least 3 h at 37°C. Dissociated cells were then plated into the middle compartment and allowed to grow axons into the outer compartments. Normal DRG medium was used for culturing. After 4 d in culture, control medium or medium containing cycloheximide (25 μm; Calbiochem) was replaced in the cell body compartment, the axonal compartment or both for 1 h. Next, the axons were cut and 1 h later fixed in 4% PFA for immunostainings.
DRG neuron transfection.
Dissociated DRG neurons were transfected using a Microporator (Invitrogen), which electroporates within a micropipette tip. Approximately 1 × 105 cells were transfected per reaction, in a volume of 10 μl. Transfected cells were plated and cultured as described above, but without antibiotics for the first 24 h after electroporation.
Cortical neuron culture and transfection.
Primary cortical neurons were isolated from E18 rat brain (Banker and Goslin, 1998). Cells (1 × 106) were transfected using the Amaxa Rat Neuron Nucleofector kit (Lonza) with 3 μg of plasmid DNA and plated in 6-well plates (5 × 105 cells/well) coated with poly-l-lysine (37.5 μg/ml) and laminin (5 μg/ml) containing DMEM supplemented with 10% FBS as described previously (Kaech and Banker, 2006). Cells were allowed to recover and adhere to the surface at 37°C in 5% CO2; after 4 h, the medium was replaced with Neurobasal medium supplemented with 2% B27, 0.5 mm glutamine, 15.6 μm glutamate, and 1% penicillin/streptomycin. Cells were grown for 4 d at 37°C in 5% CO2 before lysis.
DNA expression constructs.
For transfection, the following mammalian expression plasmids have been described: EB3-GFP (Stepanova et al., 2003), GST-EB3 and GST-EB3ΔC (Komarova et al., 2009), MACF43-GFP (Honnappa et al., 2009), mRFP-KIF3C, and the dominant-negative form lacking the motor domain (MD) mRFP-KIF3CDN (Davidovic et al., 2007). The TagRFP-EB3 construct and the GFP-KIF3C expression construct and its deletion mutants were generated by cloning and PCR-based strategies. GFP-KIF3C-SxNN mutation was introduced by overlapping PCR.
Short hairpin RNAs.
For transfection the following shRNA has been described, pSuper EB1/EB3 (Jaworski et al., 2009). Suppression of KIF3C expression was performed by shRNA interference using two specific GFP-tagged pSuper shRNA vectors purchased from Oligoengine targeted to different regions of the rat KIF3C mRNA sequence. The KIF3C pSuper shRNA-1 sequence corresponds to 5′-CGAACCGAGCCAAGAACAUUCUCUUGAAAUGUUCUUGGCUCGGUUCGGGG-3′, and the KIF3C shRNA-2 sequence corresponds to 5′-GCUGCCCAAGACCUUCACUUCUCUUGAAAGUGAAGGUCUUGGGCAGCGGG-3′. For control experiments, a GFP-tagged or empty pSuper shRNA vector containing a mammalian scramble sequence was used. shRNA vectors targeting KIF3C were transfected into dissociated DRG neurons using the Microporator.
In vitro experiments: immunohistochemistry
Dissociated adult DRG neurons were fixed in 4% PFA for 20 min. The cultures were permeabilized in 0.1% Triton X-100 in PBS for 15 min at room temperature and incubated with the primary antibody diluted in PBS with 10% normal goat serum for 30 min at room temperature. The primary antibody dilution was as follows: monoclonal anti-β-tubulin isotype III 1:400 (Sigma), polyclonal anti-KIF3C, 1:250 (Cytoskeleton), rat monoclonal anti-tyrosinated tubulin 1:200 (clone YL1/2; Abcam), monoclonal anti-acetylated tubulin, 1:400 (clone 6-11B-1; Abcam). To visualize EB3-positive microtubules, dissociated neurons were fixed in ice-cold methanol followed by 4% PFA for 20 min. The primary antibody dilution was as follows: polyclonal anti-EB3 (1:500; Jaworski et al., 2009). The cultures were washed three times in PBS and incubated for 30 min at room temperature with the secondary antibody Alexa Fluor 488 or Alexa Fluor 568 (1:500; Invitrogen) diluted in PBS. Coverslips were mounted with Fluoromount G mounting medium (Southern Biotech).
Microscopy
Standard fluorescent microscopy was performed using a Leica DM6000B with a Leica DFC350 FX CCD camera or a Leica AF7000 with a Hamamatsu EM CCD C9100 camera and Leica LAS AF software. Images for presentation were processed in their entirety with Leica and ImageJ software. Images acquired for quantification were analyzed raw.
Live cell imaging of adult DRG growth cones.
Total internal reflection fluorescence microscopy was performed as described previously (Jaworski et al., 2009; Kapitein et al., 2010) on an inverted research microscope (Nikon Eclipse TE2000E; Nikon) with a CFI Apo TIRF 100×, 1.49 numerical aperture (NA), oil objective (Nikon), and using a DualView (Photometrics) to project the RFP and GFP images side-by-side onto the EMCCD camera (Photometrics Evolve 512).
Live cell imaging of MACF43-GFP in adult DRG growth cones was performed on an inverted research spinning-disk microscope Nikon Eclipse Ti-E (Nikon) with perfect focus system (Nikon), equipped with Nikon CFI Apo TIRF 100× 1.49 NA oil objective (Nikon), Photometrics Evolve 512 EMCCD (Roper Scientific), and controlled with MetaMorph 7.7 software (Molecular Devices). The microscope's magnified image was projected onto the chip of the 16-bit Evolve 512 camera with intermediate lens 2.5× at final magnification of 0.063 μm/pixel. To keep cells at 37°C, we used stage top incubator (model INUBG2E-ZILCS; Tokai Hit).
Time-lapse microscopy of injured axons.
DRG explants from KIF3C−/− and control embryonic and adult mice were cultured as described above in L15 medium (Sigma) instead of DMEM to prevent pH fluctuations. Proximal and distal axonal segments from axotomized axons were observed for 1 h to fully characterize axonal behavior after injury. All time-lapse analyses were performed on a heated stage and CO2 chamber using a digital camera (Nikon) and the Lucia Imaging Software (Nikon). Frames were taken every 2 min.
Analysis of microtubule dynamics
Movies of growth cones labeled with MACF43-GFP and acquired on the spinning disk confocal microscope were imported into ImageJ software. Maximum intensity projections of those movies contained easily visible tracks corresponding to the microtubules growth episodes marked by MACF43 comets (see Fig. 9C). Kymographs were built along those curves. On the kymographs, single growth episodes were distinguished as bright tilted straight lines (see Fig. 9D). The length and the duration of each growth episode were measured as horizontal and vertical projections of those lines, respectively. The growth velocity was calculated as a ratio of those values. The reported mean catastrophe frequency is equal to the inverse average growth time. The relative SE for the catastrophe frequency is equal to the relative SE of growth time according to the propagation of uncertainties (Taylor, 1997).
Figure 9.

KIF3C regulates microtubule dynamics and organization in the growth cone. A, Immunofluorescence images of control or KIF3C knockdown growth cones immunostained for TUBB3. High magnification of the boxed regions shows the arrangement of microtubules within the periphery of the growth cones. Scale bar, 10 μm. B, Quantification of the percentage of control and KIF3C knockdown growth cones containing looped microtubules (n = 102 and n = 121, respectively, compiled from three independent experiments). Error bars indicate SD. ***p < 0.001. C, Spinning-disk confocal live-cell imaging of adult DRG growth cones cotransfected with control, Kif3c shRNA, or mRFP-Kif3c and MACF43-GFP constructs. Images were collected for 100 s with a 0.2 s interval between frames. Left to right, Single frame of the MACF43-GFP channel; maximum intensity projection of the MACF43-GFP channel >500 frames (100 s). D, Representative kymographs illustrating the growth of MACF43-GFP-decorated microtubule tips in control, KIF3C-depleted, and KIF3C-overexpressing growth cones. Sudden shrinkage events are seen in the control and mRFP-KIF3C recordings, whereas the KIF3C knockdown shows steady elongation with no catastrophes. E–G, Quantification of the duration of growth, frequency of catastrophes, and the instantaneous growth velocity (growth rate) in control, KIF3C-depleted, and KIF3C-overexpressing growth cones. Fifty to 150 growth episodes in 10 growth cones were analyzed in each condition. Data for mRFP control and control shRNA showed no significant differences and were pooled. Error bars indicate SEM. **p < 0.01. ***p < 0.001.
Analysis of DRG neurite outgrowth and growth cone size
Images of dissociated DRG neurons were acquired using a Leica DM6000 microscope. ImageJ software was used to analyze the images. The drawing tool was used to trace and measure the length of the longest neurite per cell from the axon hillock to the growth cone. The same method was used to trace around the growth cone to obtain the area and perimeter in micrometers. Growth cones containing looped microtubules were scored as previously reported (Purro et al., 2008). Briefly, growth cone containing looped microtubules were those with at least 30% of their microtubules bent at an angle of at least 90°.
Quantification of immunofluorescence
All images were taken at the same settings for light and exposure with parameters adjusted so that the pixel intensities were below saturation. Images were analyzed by ImageJ analysis software. For quantification of intensity levels of tyrosinated and acetylated microtubules, the length of an axon or the whole area of a growth cone was traced using the software and the average pixel intensity per unit area was calculated. The intensity of the background was similarly measured in areas adjacent to the axons and subtracted from the axon value giving a final intensity value per axon. For quantification of intensity levels of KIF3C, the neuronal cell body was traced and the pixel intensity per unit area was calculated. The intensity of the background was similarly measured in areas near the cell body and subtracted from the initial value giving a final intensity value per cell body.
Western blotting
Samples were eluted in SDS sample buffer, equally loaded onto SDS-PAGE gels, and subjected to Western blotting on polyvinylidene difluoride membrane. Blots were blocked with 2% BSA/0.05% Tween 20 in PBS and incubated with the primary antibodies at 4°C overnight. Blots were washed with 0.05% Tween 20 in PBS three times for 10 min at room temperature and incubated with either anti-rabbit or anti-mouse IgG antibody conjugated to horseradish peroxidase (Dako). Blots were developed with enhanced chemiluminescent Western blotting substrate (Pierce).
Antibodies used for Western blotting
Antibodies used for Western blotting included the following: rabbit polyclonal anti-KIF3C 1:500 (Cytoskeleton), mouse monoclonal anti-α-tubulin 1:5000 (clone B5121, Sigma), mouse monoclonal anti-EB3 1:1000 (BD Biosciences), and rabbit polyclonal anti-GFP 1:5000 (Abcam).
Pull-down assays
GST pull-down assays.
GST-tagged fusions of EB3 and EB3 ΔC acidic tail were described previously (Komarova et al., 2009). GST fusion proteins were induced in BL21 Escherichia coli, as described previously (Smith and Johnson, 1988) and purified on glutathione-Sepharose 4B beads (GE Healthcare), according to the instructions of the manufacturer. GFP-KIF3C fragment constructs were expressed in HEK293 cells for 24 h; lysates were prepared in a lysis buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.5% Triton X-100, and protease inhibitors (Complete; Roche) and mixed with GST-fusion proteins. After incubation for 2 h on a rotating wheel at 4°C, beads were separated from the supernatant by centrifugation and washed several times with wash buffer containing 20 mm Tris, pH 8, 150 mm NaCl, 1 mm DTT, and 0.05% Triton X-100. Beads were resuspended in 2× Laemmli buffer and boiled at 95°C for 5 min. The proteins retained on the beads were analyzed by Western blotting as described above.
GFP pull-down.
GFP-KIF3C fragments were cotransfected with TagRFP-EB3 into HEK293T cells. One day after transfection, cells were lysed in a buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, and protease inhibitors (Complete; Roche) and purified with GFP-Trap magnetic beads (Chromotek) according to the manufacturer's instructions. The proteins retained on the beads were analyzed by Western blotting (TagRFP-EB3) using a rabbit polyclonal anti-EB3 antibody.
In vitro microtubule binding protein assay
Microtubule-binding assays were performed with the Microtubule Associated Protein spin-down biochemical assay kit (Cytoskeleton). Briefly, for each assay, 100 μg of pure tubulin (isolated from bovine brain; Cytoskeleton) was incubated at 35°C for 20 min in 20 μl of buffer containing 1 mm GTP, 5% glycerol to assemble the microtubules. In a cohort, carboxypeptidase A (10 μg/ml, Sigma) was added during polymerization to generate detyrosinated microtubules. Immediately after incubation, the assembled tyrosinated and detyrosinated microtubules were stabilized in 200 μl of warm buffer + 20 μm taxol. Then, the tyrosinated and detyrosinated microtubules (4 μm tubulin) were incubated with human KIF3C MD (100 μg/ml; Cytoskeleton) in a total volume of 50 μl at room temperature for 30 min. The reaction mixtures were then centrifuged through a 50% glycerol cushion containing buffer and taxol at 100,000 × g at 25°C for 15 min. The pellet was resolved on 10% SDS-PAGE, and the presence of KIF3C MD was detected by immunoblot analysis. Total tubulin abundance was determined by amido black staining and densitometry. This assay was repeated in the presence of 100 mm KCl.
Statistical analysis
Statistical analysis was performed using two-way ANOVA followed by calculation of statistical significance using two-tailed unpaired t test or Mann–Whitney test for analysis of microtubule dynamics parameters.
In vivo experiments: mutant mice
KIF3C−/− mice (B6; 129-KIF3Ctm3Gsn/Mmcd) were obtained from the Mutant Mouse Regional Resource Center at University California Davis and were originally donated by L.S.B Goldstein (Yang et al., 2001).
Animals
Adult male WT mice (20–30 g, n = 18) and KIF3C KO mice (20–30 g, n = 18) were used in this surgical study. All procedures were performed in accordance with the United Kingdom Home Office regulations (Animals Scientific Procedures Act, 1986).
Surgery
Mice were anesthetized with 5% isoflurane (Abbot Laboratories) in oxygen and nitrous oxide and maintained at 2% during the procedure. After left hindlimb incision, the sciatic nerve was exposed, from the sciatic notch to the trifurcation point. The nerve was crushed just distal to the sciatic notch using no. 5 Dumont forceps (Fine Science Tools) for 20 s, and the crush site marked with charcoal powder.
Tissue preparation
Mice were killed at 3, 5, and 8 d after sciatic nerve crush (N = 6 per group). Mice received a terminal intraperitoneal injection of pentobarbitone and transcardially perfused with 10 ml of saline followed by 50 ml of chilled PFA (4% in 0.1 m phosphate buffer, pH 7.4). Sciatic nerves were dissected, postfixed in 4% PFA for 2 h at 4°C, and stored in 20% sucrose (in 0.1 m phosphate buffer) for 2 d. Tissue was the blocked in OCT embedding compound and stored at −80°C before sectioning. Longitudinal sections of sciatic nerve were cut on a cryostat (Leica) at 8 μm thickness and mounted on superfrost Plus slides (VWR).
Immunohistochemistry
Slides were removed from −80°C, left for 2 h at room temperature, and washed twice for 10 min each in 1× PBS. Slides were then incubated for 1 h in 10% normal donkey serum with 2% BSA (diluted in 1× PBS), followed by α-rabbit GAP43 (1:1000 in 10% normal donkey serum; Abcam). Slides were incubated in a humid chamber at room temperature for 24 h. Slides were washed in 1× PBS + 0.2% Triton X-100, three times for 5 min each. Donkey α-rabbit Alexa Fluor 488 (1:1000 diluted in 1× PBS; Invitrogen) was then applied to the slides for 2 h. Slides were washed 3 times for 5 min each and mounted with Fluorsave (Merck).
Microscopy
Within sections of the sciatic nerve, three areas distal to the crush site were analyzed for axon count (2, 4, and 6 mm, at each time point, three sections per animal). Focusing through the tissue section using the z-plane on a Leica CRT 6000 fluorescence microscope (Leica), the number of GAP-43-positive axons within the nerve crossing these three distances was counted. Photomicrographs for figure illustration were taken at 20× magnification.
Statistical analysis
Results are expressed as mean ± SEM, with statistical comparisons between WT and KIF3C KO groups made using two-tailed Student's t test. The levels of significance are indicated above each data point as follows: *p < 0.05; **p < 0.005; ***p < 0.001.
Results
KIF3C is present in adult, but not embryonic, sensory axon growth cones
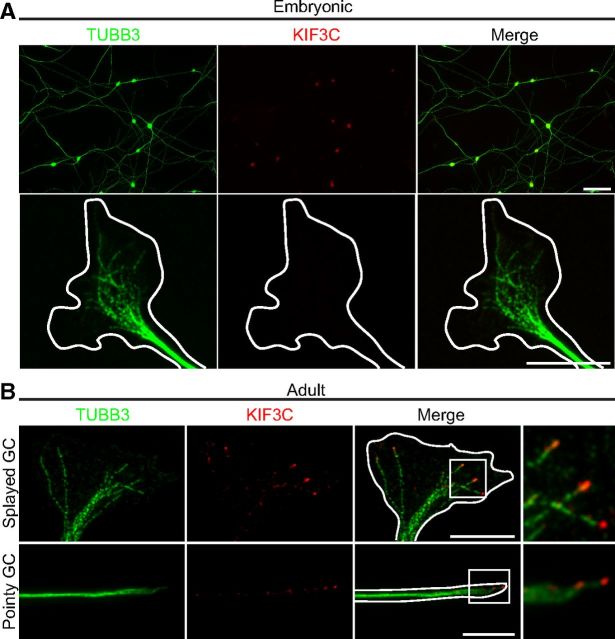
The localization of endogenous KIF3C protein during axon growth was characterized in embryonic and adult DRG neurons by immunostaining. In embryonic neurons, KIF3C expression in the axons and growth cones was almost undetectable despite the fact that the protein was being expressed in the cell body (Fig. 1A). However, in adult DRG neurons, KIF3C was enriched in both splayed and pointy growth cones (Fig. 1B) and decorating the plus-ends of a subset of microtubules (Fig. 1B, boxed regions). To explore the difference in KIF3C expression between embryonic and adult DRG neurons, we asked whether axotomy would influence the presence and localization of KIF3C in the injured axon tip.
Figure 1.
KIF3C decorates the plus-ends of microtubules in adult DRG growth cones. A, Immunostaining of KIF3C and tubulin-β-III (TUBB3) in embryonic (E16) dissociated DRG neurons cultured for 2 d. KIF3C is enriched in the cell body, but hardly any KIF3C is detected in the axons (scale bar: top panels, 100 μm) or growth cones (scale bar: bottom panels, 10 μm). B, Immunostaining of KIF3C and tubulin-β-III (TUBB3) in adult DRG growth cones. KIF3C is enriched at the tip of microtubules in both pointy and splayed growth cones (GC). High magnification of boxed regions shows KIF3C at the plus-ends of microtubules. Scale bar, 10 μm.
KIF3C is an injury-associated kinesin that is locally translated in embryonic axons
The formation of a new growth cone after axotomy has many differences compared with axonogenesis from the cell body. Cutting the axon causes extracellular fluids to flood in and initiates many local signaling events. To examine KIF3C localization in injured embryonic axons, E16 DRG explants were cultured for 3 d and then axotomized, following which almost all axons regenerated a growth cone within 1 h (Chierzi et al., 2005; Verma et al., 2005). The cultures were fixed 30 min after axotomy and immunostained for KIF3C. Surprisingly, we found that axotomy triggered the enrichment of KIF3C in the newly formed growth cones (Fig. 2A). Thus, KIF3C localization to embryonic growth cones is injury-associated. Adult sensory axons also regenerate in vitro after axotomy within 1 h. We examined the localization of Kif3C in these regenerating growth cones and found it enriched at the newly formed growth cone (Fig. 2B). Because embryonic axons contain undetectable levels of KIF3C protein but the mRNA of KIF3C is present, we asked whether axotomy induced the local translation of KIF3C at the proximal cut end (Gumy et al., 2011). Compartmentalized embryonic DRG cultures were established using Campenot chambers, which allow the separation of the cell body from the axonal compartment. Under control conditions, KIF3C readily localized to the newly formed growth cone after the axons were cut (Fig. 3A,B). However, when the protein synthesis inhibitor cycloheximide was added to both the cell body and axonal compartment, KIF3C did not localize anymore to the growth cones (Fig. 3A,B). To determine whether KIF3C was being synthesized in the cell body, in the axons, or in both after axotomy, cycloheximide was added to either the cell body compartment or the axonal compartment. Surprisingly, cycloheximide added to the cell body compartment did not prevent KIF3C localization to the growth cones after injury (Fig. 3A,C). However, cycloheximide added to the axonal compartment significantly abolished the enrichment of KIF3C in the growth cones demonstrating that KIF3C is locally translated in embryonic axons after axotomy (Fig. 3A,C). The lack of KIF3C in embryonic axons and growth cones outgrowing from the cell body and its presence after axotomy highlights the differences in mechanisms between the two processes and shows that adult and embryonic neurons share common mechanisms of KIF3C localization after axotomy.
Figure 2.

KIF3C is enriched in the newly formed embryonic and adult growth cone after axotomy. A, Immunostaining of KIF3C and TUBB3 in embryonic axons 30 min after axotomy. KIF3C is enriched at the newly formed growth cone of the regenerating axons. Scale bar, 10 μm. B, Immunostaining of KIF3C and TUBB3 in adult axons 1 h after injury. KIF3C is localized to the new growth cone. Scale bar, 20 μm.
Figure 3.
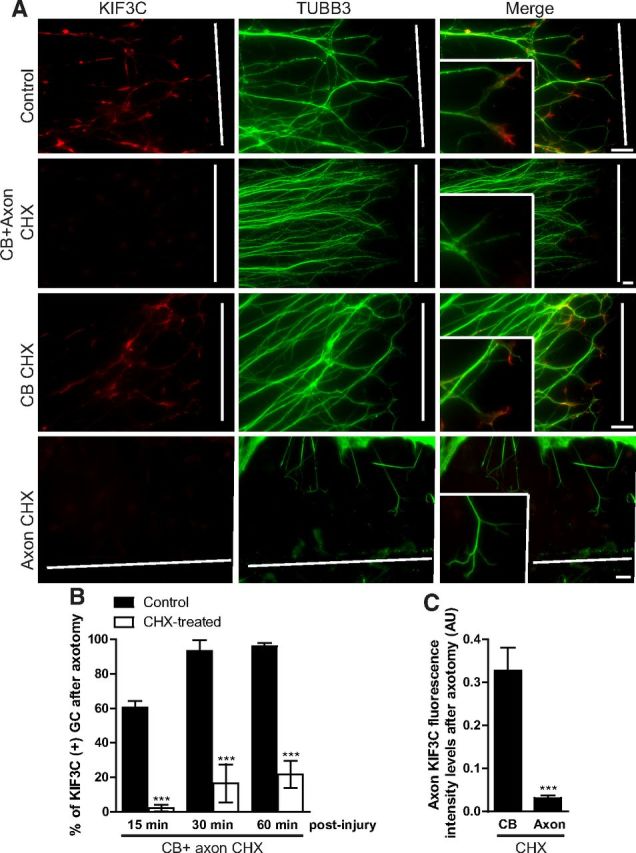
KIF3C is locally translated in embryonic axons after axotomy. A, Immunostaining of axotomized embryonic DRG axons cultured in Campenot chambers to separate the cell body (CB) from the axonal (Axon) compartment. The cultures were treated for 1 h with control medium or with the protein synthesis inhibitor cycloheximide (25 μm) (CHX), which was added to the CB and the axonal compartment, the CB compartment alone, or the axonal compartment alone. The white lines indicate the site of injury. B, Quantification of the percentage of growth cones (GC) enriched with KIF3C at different time points after axotomy. CHX was added to both the somatic and axonal compartments (30 axons were analyzed per condition and time point). C, Quantification of KIF3C fluorescence intensity levels in the growth cone of axotomized embryonic DRG axons. CHX was added either to the somatic or the axonal compartment (n = 30 and n = 30, compiled from three independent experiments). Error bars indicate SD. ***p < 0.001. Scale bar, 20 μm.
EB3 targets KIF3C to the plus-ends of microtubules in the growth cone
KIF3C is found at the tip of some, but not all, microtubules in growth cones. We therefore asked whether these decorated microtubules were specialized dynamic microtubule plus-ends. The plus-ends of dynamic growing microtubules are marked by End-Binding protein 3 (EB3) (Stepanova et al., 2003), so adult DRG neurons were transfected with a GFP-tagged cDNA construct encoding this protein (EB3-GFP). Analysis of images of fixed growth cones revealed that 98 ± 1.1% of endogenous KIF3C protein colocalized with EB3-labeled microtubule plus-ends, whereas only 64 ± 12.4% of EB3-labeled microtubule plus-ends colocalized with KIF3C in pointy and splayed growth cones combined (n = 287 and n = 302, respectively, compiled from three independent experiments) (Fig. 4A). The ability of KIF3C to associate with EB3-positive microtubule plus-ends was further confirmed by simultaneous dual-color TIRF live-imaging of mRFP tagged full-length KIF3C (mRFP-KIF3C) and EB3-GFP in DRG growth cones (Fig. 4B). mRFP-KIF3C was observed on a subset of EB3-GFP comets located at the periphery of the growth cone (Fig. 4B, arrows), whereas EB3-GFP comets in the central region of the growth cone did not coincide with mRFP-KIF3C (Fig. 4B, arrowheads). Analysis of the position of the maximum fluorescence intensity value of mRFP-KIF3C and EB3-GFP along the comet trajectory obtained from the time-lapse recording showed that both proteins move together (Fig. 4C). To determine whether EB3 targeted KIF3C to the microtubule plus-ends, we knocked down EB1 and EB3 expression in adult DRG neurons using a previously published shRNA that targeted mRNA regions of EB1 and EB3 (Jaworski et al., 2009) (Fig. 4D). As a control, we used an shRNA vector that contained a mammalian scramble sequence that does not target any specific mRNA. Neurons were cotransfected with an empty vector expressing GFP to define transfected cells. Interestingly, knockdown of EB1 and EB3 impaired KIF3C localization at the microtubule tips (Fig. 4D,E). However, it did not prevent the enrichment of KIF3C in the growth cone, suggesting that EB3 is required for localizing KIF3C at the microtubule plus-ends in the growth cone (Fig. 4D,E). We further explored the possibility that an interaction between KIF3C and EB3 mediated the localization of KIF3C at the microtubule tips. We performed GFP pull-down assays in Hek293T cells with GFP-KIF3C full-length (FL), GFP-KIF3C motor domain (MD), GFP-KIF3C tail domain (T), GFP-KIF3C–FL-mut where a putative EB3 binding site on the MD was mutated from SxIP to SxNN (Honnappa et al., 2009), GFP-KIF3C-MD-mut and tagRFP-EB3 (Fig. 5A). Our results show that GFP-KIF3C-FL, GFP-KIF3C-FL-mut, and GFP-KIF3C-T interacted with EB3 (Fig. 5B). Interestingly, the MD of KIF3C failed to interact with EB3. These data suggest that KIF3C interacts with EB3 through its tail domain and that the SxIP motif found in the MD does not bind to EB3 (Fig. 5B). To further validate these results, we performed GST-EB3 and GST-EB3ΔC pull-down assays with GFP-KIF3C-MD and GFP-KIF3C-T (Fig. 5C). The data confirmed our previous findings showing that KIF3C interacts with EB3 through its tail domain.
Figure 4.
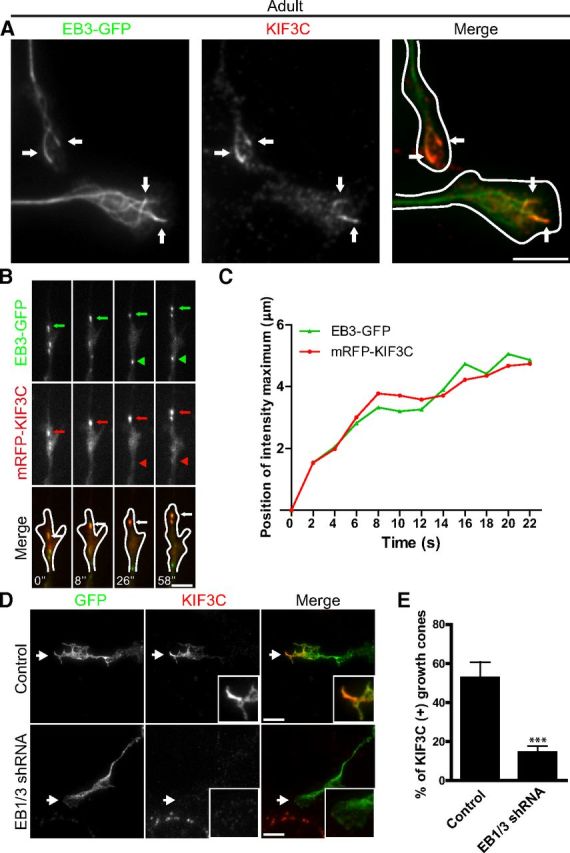
EB3 targets KIF3C to the plus-end of microtubules. A, Immunofluorescence image of an adult DRG growth cone showing that endogenous KIF3C protein accumulates at the plus-end of a subset of microtubules decorated with EB3-GFP comets (arrows). Scale bar, 5 μm. B, Simultaneous imaging of mRFP-KIF3C (red in overlay) and EB3-GFP (green in overlay) in transfected adult DRG growth cones. Selected frames from a time-lapse movie are shown; time is indicated at the bottom of the panel. A representative EB3-GFP comet (green arrows) that colocalizes with mRFP-KIF3C dots (red arrows) is indicated. An EB3-GFP comet that does not colocalize with mRFP-KIF3C is highlighted by arrowheads. Scale bar, 5 μm. C, The positions of the maximum fluorescence intensity values of EB3-GFP and mRFP-KIF3C along the moving comet track, which is indicated in Figure 3B (green and red arrows, respectively). The position of EB3-GFP in the first frame of the time-lapse was taken as a starting point. The plot shows correlated movement of EB3-GFP and mRFP-KIF3C. D, Immunofluorescence images of control and EB1/3-depleted growth cones stained for endogenous KIF3C. EB1/3 knockdown impairs the localization of KIF3C at the tip of microtubules in the growth cone (n = 282 and n = 288, respectively, compiled from three independent experiments). Scale bar, 5 μm. E, The quantification of the percentage of growth cones with microtubule tips decorated with KIF3C. Error bars indicate SD. ***p < 0.001.
Figure 5.
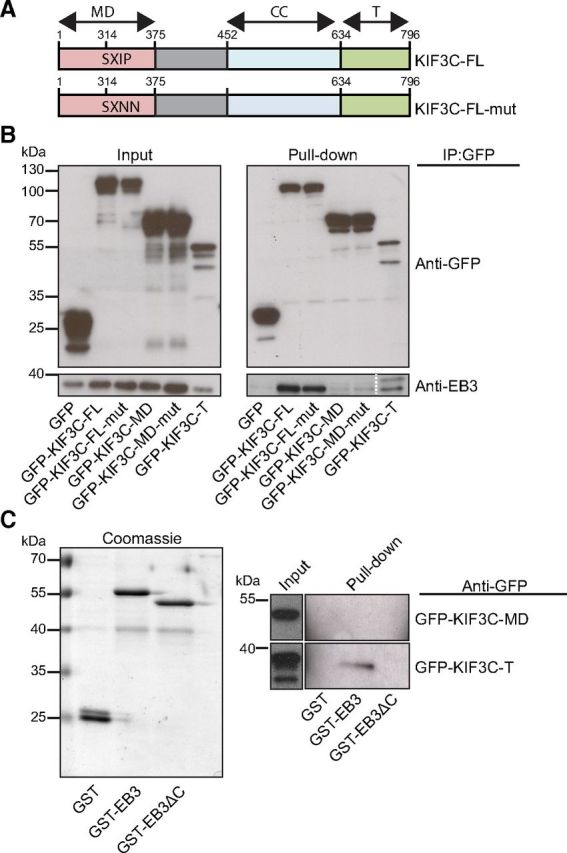
KIF3C interacts with EB3. A, Schematic overview of full-length KIF3C and full-length KIF3C with two mutations in the SXIP motif binding site for EB3. CC, Coiled-coil. B, GFP pull-down assay was performed with HEK293T cells extracts coexpressing the indicated GFP-KIF3C fragments and mTagRFP-EB3 and analyzed by Western blotting with the indicated antibody. C, GST-EB3 and GST-EB3ΔC pull-down assays were performed with HEK293T cells coexpressing the indicated GFP-tagged KIF3C fragment and analyzed by Western blotting with the indicated antibody.
Most proteins that localize to the plus-tips of microtubules bind the C terminus of EB3 through their SxIP motif. However, we found that KIF3C does not interact with EB3 via its putative SxIP motif. Moreover, the fact that GFP-KIF3C-T interacts with full-length EB3, but not with EB3ΔC (EB3 without the C-terminus), suggests that the interaction between KIF3C and EB3 is either a noncanonical direct interaction or most likely mediated by an additional factor containing an SxIP motif. Future work will need to determine the identity of this SxIP-containing factor. Thus, KIF3C requires EB3 for it to localize to the plus-ends of microtubules in the growth cone.
Microtubule tyrosination contributes to the enrichment of KIF3C in the growth cones
In adult sensory neurons, KIF3C is enriched at the growth cone where it also localizes to a subset of EB3-decorated microtubules at the growth cone periphery. Binding to EB3 cannot therefore be the only factor that determines KIF3C growth cone localization. Kinesins bind to the microtubule lattice via their MD (Vale and Fletterick, 1997), and microtubule post-translational modifications can regulate the interaction and localization of kinesin-1 on neuronal microtubules (Witte et al., 2008; Konishi and Setou, 2009; Hammond et al., 2010). Furthermore, recent studies have demonstrated that axon injury induces a decrease in tubulin acetylation and a significant increase in tubulin tyrosination at the injury site of embryonic DRG axons, and that tyrosinated tubulin is found predominantly in the growth cone of axons (Gordon-Weeks and Lang, 1988; Cho and Cavalli, 2012). We therefore asked whether tubulin tyrosination might contribute to the enrichment of KIF3C in embryonic growth cones after injury and in adult growth cones. We examined whether KIF3C colocalized with microtubule modifications associated with stable (acetylation) or with dynamic (tyrosination) microtubules in adult DRG axons (Fig. 6A). Coimmunostaining of tyrosinated and acetylated tubulin showed that the growth cones contained higher levels of tyrosinated (dynamic) microtubules compared with the axon, whereas acetylated (stable) microtubules were almost undetectable in the growth cone (Fig. 6A,B). KIF3C staining was also enriched at the growth cone and colocalized with tyrosinated but not with acetylated microtubules (Fig. 6A,B). The levels of acetylated microtubules were highest in the middle and proximal axonal segments near the cell body as reported previously (Robson and Burgoyne, 1989) (Fig. 6A,B). To determine whether the colocalization of KIF3C with tyrosinated microtubules was the result of their preferential binding, we performed an in vitro cosedimentation assay using tyrosinated or detyrosinated brain-derived microtubules and a purified KIF3C protein MD (Fig. 6C,D). At saturation, ∼50% less KIF3C MD was bound to detyrosinated microtubules compared with tyrosinated microtubules (Fig. 6E). This result was corroborated when the assay was conducted under stringent conditions, in the presence of 100 mm KCl (Fig. 6F,G), which triggers the dissociation of most microtubule-associated proteins from microtubules, indicating that KIF3C preferentially binds to tyrosinated microtubules in the absence of other microtubule-associated proteins.
Figure 6.
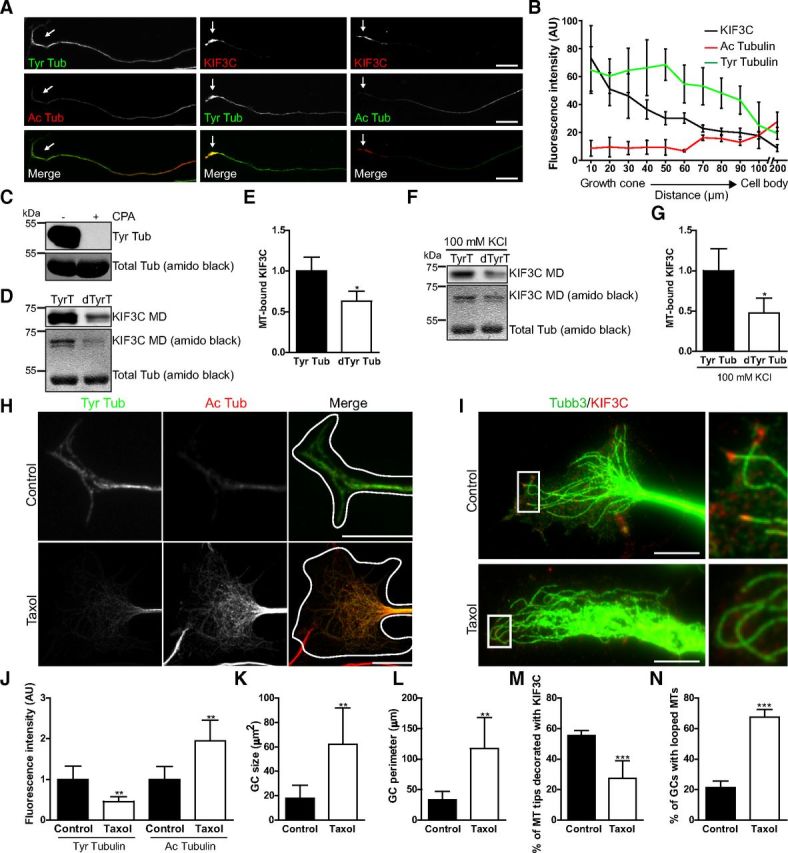
Tubulin tyrosination contributes to localizing KIF3C to the tip of growing microtubules in the growth cone. A, Immunostaining of tyrosinated (dynamic) and acetylated (stable) microtubules and KIF3C in three different adult DRG axons and growth cones. The growth cones (arrows) contain hardly detectable levels of acetylated microtubules but are enriched in tyrosinated microtubules and in KIF3C. Scale bar, 20 μm. B, Quantification of fluorescence intensity levels of tyrosinated, acetylated microtubules and KIF3C along adult DRG axons (n = 30 axons, respectively, compiled from three independent experiments). C, Western blot shows tyrosinated tubulin in the presence (+) or absence (−) of carboxypeptidase A (CPA). CPA is capable of detyrosinating the entire pool of tubulin. D, Immunoblot and amido black staining show the amount of human KIF3C MD that is bound to tyrosinated or detyrosinated microtubules. E, Quantification of the levels of KIF3C MD that bind to tyrosinated or detyrosinated microtubules based on the immunoblot (n = 8). F, Immunoblot and amido black staining show the amount of human KIF3C MD that is bound to tyrosinated or detyrosinated microtubules in the presence of 100 mm KCl. G, Quantification of the levels of KIF3C MD that bind to tyrosinated or detyrosinated microtubules in the presence of 100 mm KCl based on the immunoblot (n = 8). H, Immunostaining of tyrosinated and acetylated microtubules in adult DRG growth cones treated with taxol (100 nm) for 3 h. Scale bar, 10 μm. I, Immunostaining of KIF3C and TUBB3 in adult DRG growth cones treated with taxol (100 nm) for 3 h. Taxol releases KIF3C from the microtubule plus-ends tips and induces microtubule looping (enlarged boxed regions). Scale bar, 10 μm. J, Quantification of the fluorescence intensity levels of tyrosinated and acetylated microtubules in control and taxol-treated growth cones (n = 32 and n = 35, respectively, compiled from three independent experiments). K, Quantification of the effect of taxol on growth cone size (n = 105 and n = 99, respectively, compiled from three independent experiments). L, Quantification of the effect of taxol treatment on growth cone perimeter (n = 105 and n = 99, respectively, compiled from three independent experiments). M, Quantification of the percentage of microtubule tips decorated with KIF3C before and after taxol treatment (n = 289 and n = 305, respectively, compiled from three independent experiments). N, Quantification of the effect of taxol on the percentage of growth cones with looped microtubules (n = 255 and n = 305, respectively, compiled from three independent experiments). Error bars indicate SD. *p < 0.05. **p < 0.01. ***p < 0.001.
To confirm that KIF3C was preferentially bound to tyrosinated microtubules in cultured living neurons, we treated adult DRG neurons with taxol (100 nm), a microtubule-stabilizing drug that induces changes in the microtubule modification patterns (Robson and Burgoyne, 1989; Hammond et al., 2010) (Fig. 6H). After 3 h of taxol treatment, the fluorescence intensity levels of tyrosinated microtubules in the growth cones of adult DRG neurons had decreased compared with controls, whereas there was a simultaneous increase in the levels of acetylated microtubules (Fig. 6J), consistent with increased microtubule stabilization. In addition, taxol induced significant KIF3C removal from the microtubule tips with a concomitant increase in the number of looped microtubules and an increase in growth cone area and circumference (Fig. 6I,K–N). These results indicate that the high levels of tubulin tyrosination in the adult DRG distal axon contribute to enriching KIF3C to the growth cone and to localizing it to the plus-ends of growing microtubules.
KIF3C is required for axon outgrowth in adult, but not embryonic, sensory axons
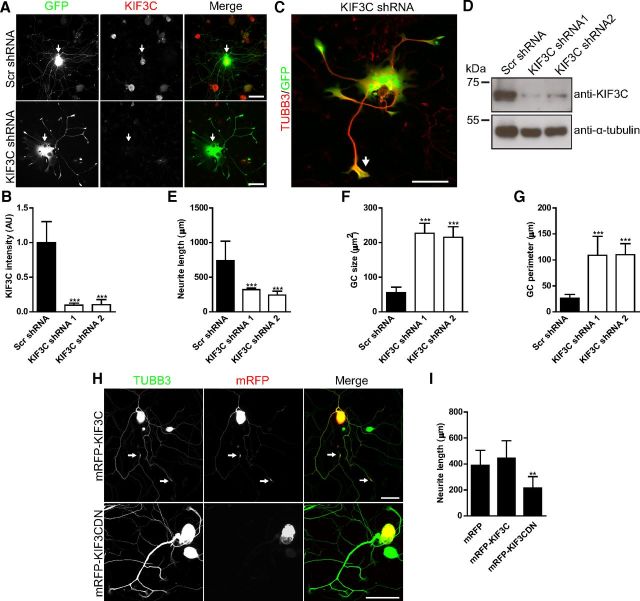
To establish whether KIF3C plays a functional role during neurite outgrowth, we knocked down KIF3C expression using two independent shRNA vectors that targeted different regions of the KIF3C mRNA. As a control, we used an shRNA vector that contained a mammalian scramble sequence that does not target any specific mRNA. All vectors expressed GFP to define transfected cells. As might be predicted from the lack of developmental phenotype in the KIF3C knock-out mouse and from the lack of KIF3C protein in embryonic sensory axons, knockdown of KIF3C had no effect on embryonic DRG neuron morphology or axon length, suggesting that KIF3C is not necessary for normal embryonic axon growth (Fig. 7A,C). However, overexpression of KIF3C considerably impaired growth of embryonic axons, leading to very short neurites (Fig. 7B,C).
Figure 7.

KIF3C is dispensable for embryonic DRG neuronal development. A, Fluorescence images of control (Scr shRNA) and KIF3C-depleted (KIF3C shRNA 1) embryonic DRG neurons. GFP is used to visualize the transfected neurons. Scale bar, 100 μm. B, Fluorescence images of embryonic DRG neurons transfected with mRFP alone or mRFP-KIF3C and immunostained for TUBB3. C, Quantification of neurite length in embryonic DRG neurons transfected with control, KIF3C knockdown, or KIF3C overexpressing constructs (n = 108, n = 101, and n = 107, respectively, compiled from three independent experiments). Scale bar, 100 μm. Error bars indicate SD. ***p < 0.001.
Adult DRG neurons transfected with KIF3C shRNA 1 or KIF3C shRNA 2 contained significantly lower levels of KIF3C protein in the cell body compared with the scramble controls, indicating that the shRNA-KIF3C constructs significantly knocked down KIF3C expression (∼85% knockdown; Fig. 8A,B). Significant KIF3C knockdown was further corroborated by Western blotting of cortical neurons transfected with the scramble control and the two KIF3C shRNA constructs (Fig. 8D). Strikingly, after KIF3C knockdown, adult DRG axons showed aberrant morphology with enlarged and flattened cell bodies and thick and short neurites, whereas scramble controls were unaffected (Fig. 8A,C).
Figure 8.
KIF3C knockdown in adult neurons impairs neuronal morphology and neurite outgrowth. A, Immunofluorescence images of control (Scr shRNA) and KIF3C-depleted (KIF3C shRNA) adult DRG neurons stained for KIF3C. Arrows indicate KIF3C staining in the cell bodies. Scale bar, 100 μm. B, Quantification of KIF3C fluorescence intensity levels in the cell bodies of adult DRG neurons transfected with control or KIF3C knockdown (KIF3C shRNA 1 or KIF3C shRNA 2) constructs (n = 98, n = 97, and n = 75, respectively, compiled from three independent experiments). C, Immunofluorescence image of an adult DRG neuron depleted of KIF3C and immunostained for TUBB3. The arrows indicate the enlarged growth cones. Scale bar, 100 μm. D, Cortical neurons were transfected with the indicated shRNAs and were lysed 24 h after transfection. Whole-cell lysates were probed for KIF3C or α-tubulin. E, Quantification of neurite length in adult DRG neurons transfected with control or KIF3C knockdown constructs (n = 114, n = 124, and n = 102, respectively, compiled from three independent experiments). F, Quantification of growth cone size in control and KIF3C knockdown neurons (n = 120 and n = 144, respectively, compiled from three independent experiments). G, Quantification of growth cone perimeter in KIF3C knockdown neurons (n = 120 and n = 144, respectively, compiled from three independent experiments). H, Immunofluorescence images of adult DRG neurons transfected with mRFP-KIF3C or mRFP-KIF3C dominant-negative (DN) cDNA and immunostained for TUBB3. mRFP-KIF3C accumulates at the growth cones as observed with the endogenous protein (arrows). Scale bar, 100 μm. I, Quantification of neurite length in control (mRFP), mRFP-KIF3C, or mRFP-KIF3CDN-transfected neurons (n = 158, n = 166, and n = 165, respectively, compiled from three independent experiments). Error bars indicate SD. **p < 0.01. ***p < 0.001.
Quantification of neurite length after 3 d in culture revealed that KIF3C knockdown neurites were significantly shorter than control ones (Fig. 8E). In addition, KIF3C knockdown adult DRG neurons displayed enlarged growth cones with a significantly increased area and perimeter compared with controls (Fig. 8F,G). Interestingly, overexpression of mRFP-KIF3C had no obvious effect on neuron morphology or neurite growth; however, adult DRG neurons transfected with a cDNA expressing a dominant-negative version of KIF3C that lacks the microtubule binding MD and accumulates in the cell body (mRFP-KIF3CDN) displayed a very similar phenotype to KIF3C knockdown neurons consisting of short, thick neurites with enlarged growth cones (Fig. 8H,I). These data demonstrate that enrichment of KIF3C in the growth cone is required for the maintenance of neuronal morphology and axon outgrowth in adult DRG neurons.
KIF3C is a microtubule-destabilizing factor that regulates microtubule dynamics and organization in the growth cone
We next asked whether the abnormal axonal and growth cone morphology caused by KIF3C knockdown was a direct consequence of changes in the microtubule cytoskeleton. A more detailed analysis of the microtubules in the growth cones revealed that a high proportion of adult KIF3C knockdown growth cones contained looped microtubules (Fig. 9A, enlarged box, B). This observation correlates with our results obtained in neurons treated with taxol where tyrosinated microtubule levels decreased and with the finding that looped microtubules are also found in pausing and static growth cones (Purro et al., 2008). We next examined microtubule dynamics between control, KIF3C-depleted, and KIF3C-overexpressing adult sensory growth cones. As a marker of growing microtubule ends, we used the dimeric version of the C-terminal 43-amino acid-long microtubule plus-end tracking fragment of the spectraplakin MACF2 (MACF43-GFP) (Honnappa et al., 2009). Microtubule plus-end tracking behavior of MACF43-GFP was observed in control, KIF3C-depleted, and KIF3C overexpressing growth cones (Fig. 9C). Strikingly, in the absence of KIF3C, microtubules in the growth cone experienced almost persistent growth because of a strong decrease in the frequency of catastrophes (Fig. 9D–F). Microtubule instantaneous growth rate was also reduced, suggesting that KIF3C might also play a role in the polymerization of microtubules (Fig. 9G). Overexpression of KIF3C had no significant effects on microtubule dynamics compared with controls (Fig. 9D–G), in line with its negligible effect on neuronal morphology. Thus, these data demonstrate that, in the growth cone, KIF3C functions as a microtubule-destabilizing factor that participates in the regulation of microtubule dynamics. Removal of KIF3C leads to stable overgrown and looped microtubules. Our data suggest that the abnormal axonal and growth cone phenotype observed in neurons lacking KIF3C results from impairments in microtubule dynamics and organization.
KIF3C knock-out adult neurons show impaired neurite outgrowth and increased levels of stable microtubules in the growth cone
We next investigated whether the changes in growth cone morphology and axon growth that we observed in KIF3C knockdown neurons occurred in adult DRG neurons from a KIF3C knock-out (KO) mouse, where the KIF3C gene had been completely deleted (Yang et al., 2001). After 1 d in culture, neurites from adult DRG KIF3C KO neurons had a similar appearance to those of the WT control (Fig. 10A). However, quantification of neurite length showed that KIF3C KO neurites were significantly shorter (Fig. 10A,B). After 2 d in culture, ∼40% of KIF3C KO neurons displayed significant aberrant morphology similar to the KIF3C knockdown neurons. Flattened cell bodies with short, thick neurites and enlarged growth cones were observed (Fig. 10A–D). After 3 d in culture, ∼60% of KIF3C KO DRG neurons displayed this phenotype. To test whether the abnormal morphology was specifically the result of a lack of KIF3C protein, we transfected KIF3C KO DRG neurons with mRFP-KIF3C using cDNA-expressing mRFP as control (Fig. 10E). Expression of mRFP-KIF3C protein in KIF3C KO neurons rescued the abnormal neuronal and growth cone morphology and restored neurite length, whereas overexpression of mRFP alone did not rescue the KIF3C KO phenotype (Fig. 10E,F).
Figure 10.
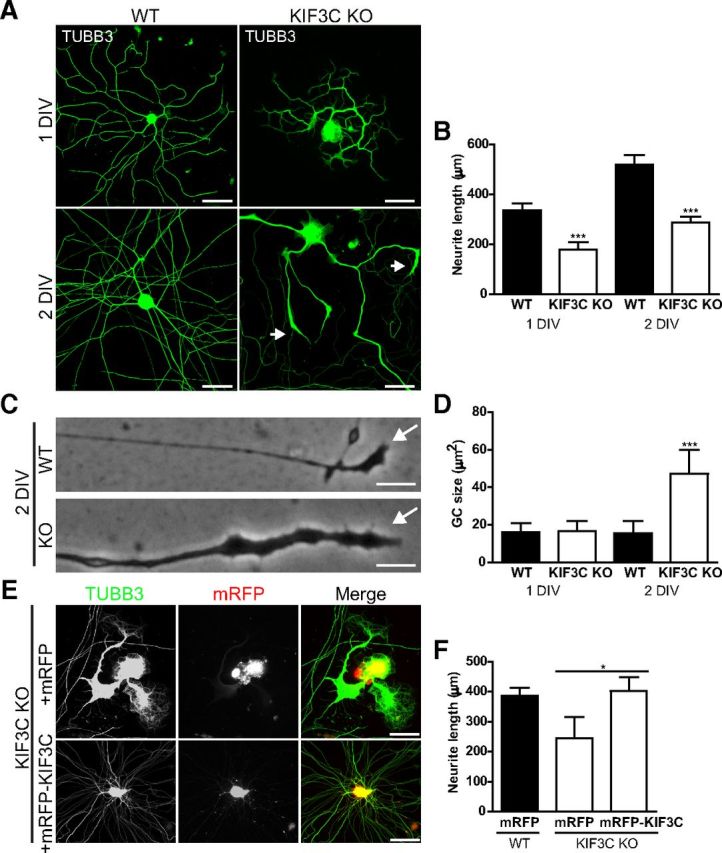
Adult KIF3C KO DRG neurons display morphological abnormalities and impaired neurite outgrowth. A, Immunostaining of TUBB3 in WT and KIF3C KO adult DRG neurons at 1 or 2 d in vitro (DIV). The arrows indicate enlarged growth cones of a KIF3C KO DRG neuron at 2 DIV. Scale bar, 100 μm. B, Quantification of neurite length in WT and KIF3C KO at 1 (n = 372 and n = 217, respectively) and 2 DIV (n = 310 and n = 201, respectively). C, Phase-contrast images of WT and KIF3C KO growth cones and axons. The arrows indicate the tip of the growth cones. Scale bar, 10 μm. D, Quantification of the growth cone size of WT and KIF3C KO DRG neurons at 1 or 2 DIV (n = 105 and n = 95, respectively). E, Immunofluorescence images of KIF3C KO DRG neurons transfected with mRFP or mRFP-KIF3C cDNAs and immunostained for TUBB3. Scale bar, 100 μm. F, Quantification of neurite length in WT DRG neurons transfected with mRFP (n = 56) or KIF3C KO DRG neurons transfected with mRFP or mRFP-KIF3C (n = 59 and n = 48, respectively). Error bars indicate SD. *p < 0.05. ***p < 0.001.
To examine whether the observed impairment in axon growth was associated with changes in microtubule stability, as suggested by our previous experiments, KIF3C KO cultures were stained with the respective antibodies, and the ratio of acetylated to tyrosinated microtubules was measured (Fig. 11A,B). Interestingly, growth cones of adult KIF3C KO neurons contained higher levels of acetylated (stable) microtubules, which were already detected in neurons cultured for 1 d before they showed morphological changes (Fig. 11A,B). By 2 d, the ratio of Ac/Tyr microtubules was increased by 45% in the growth cones where looped microtubules were also observed (Fig. 11A,B). To obtain a more direct quantification of the levels of dynamic microtubules in the growth cone, we examined the number of endogenous EB3 comets (Stepanova et al., 2003) (Fig. 11C). After 2 d in vitro, KIF3C KO growth cones contained significantly fewer EB3 comets compared with controls (a reduction of ∼70% despite the larger growth cones in KIF3C KO neurons), demonstrating a specific decrease in the number of growing microtubule plus-ends in KIF3C KO adult growth cones (Fig. 11C,D). These data further confirm our findings that KIF3C is required for regulating microtubule dynamics and organization in the growth cones.
Figure 11.
Adult KIF3C KO DRG neurons contain increased levels of stable microtubules in the growth cone. A, Immunostaining of tyrosinated and acetylated microtubules in WT or KIF3C KO DRG neurons at 1 or 2 DIV. Scale bar, 10 μm. B, Quantification of the ratio of fluorescence intensity of acetylated versus tyrosinated microtubules in the growth cones of WT or KIF3C KO DRG neurons (n = 55 and n = 65, respectively). C, Immunostaining of endogenous EB3 and TUBB3 in WT and KIF3C KO DRG growth cones. Arrows indicate EB3 comets. Scale bar, 5 μm. D, Quantification of the number of EB3 comets per area unit (μm2) in WT or KIF3C KO growth cones (n = 275 and n = 252, respectively). Error bars indicate SD. ***p < 0.001.
KIF3C is required for regeneration of both embryonic and adult axons after injury
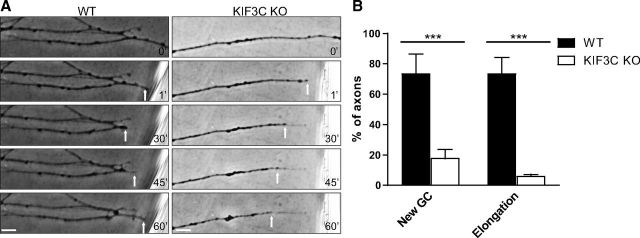
In this study, we found that, although KIF3C is undetectable in normally growing embryonic sensory axons, injury triggered the enrichment of KIF3C at the embryonic growth cone. Thus, to investigate the role of KIF3C in the regeneration of embryonic axons, WT and KIF3C KO E16 DRG neurons were axotomized as previously described and immediately photographed after injury, and the subsequent events were recorded by time-lapse video microscopy for 1 h (Fig. 12A) (Chierzi et al., 2005; Verma et al., 2005). Previous experiments have shown that, if DRG axons regenerate, they will do so within 1 h (Chierzi et al., 2005; Verma et al., 2005). We found that the percentage of KIF3C KO axons that elongated forward and that reformed new growth cones was significantly lower compared with WT axons (Fig. 12A,B). Together, these data demonstrate that, although KIF3C is dispensable during developmental growth, the axotomy-induced appearance of KIF3C in the axon stump in embryonic axons is required for regeneration.
Figure 12.
KIF3C is required for embryonic axon regeneration after injury A, Successive frames from a time-lapse recording illustrating the behavior of a WT or KIF3C KO embryonic axon after axotomy in vitro. Scale bar, 20 μm. B, The percentage of WT or KIF3C KO embryonic axons that newly reformed growth cones or elongated forward after axotomy (n = 30 and n = 30, respectively, from three independent experiments). Error bars indicate SD. ***p < 0.001.
We next assessed the physiological importance of KIF3C in adult axon regeneration. To examine this, we used the in vitro axotomy assay mentioned above, and the subsequent events were recorded by time-lapse video microscopy for 1 h (Fig. 13A). We analyzed four parameters to determine the regenerative ability of the adult axons: (1) the percentage of axons that retracted after injury, (2) the percentage of axons that reformed a growth cone, (3) the percentage of axons that elongated forward after the retraction period, and (4) the percentage of axons distal to the cut that degenerated after injury. Both WT and KIF3C KO neurons reformed growth cones at a similar rate after injury (Fig. 13B). However, the percentage of KIF3C KO axons that elongated forward was significantly lower compared with WT axons (Fig. 13C). Interestingly, most of the WT axons retracted after injury, but only a small percentage of KIF3C KO axons retracted (Fig. 13D). In addition, a lower percentage of KIF3C KO distal axons disconnected from the cell body degenerated after injury compared with controls (Fig. 13E). Thus, although KIF3C KO-injured axons were capable of reforming a growth cone, they failed to elongate or grow as efficiently as WT axons, which suggests that the axonal and growth cone microtubule cytoskeleton is less dynamic.
Figure 13.
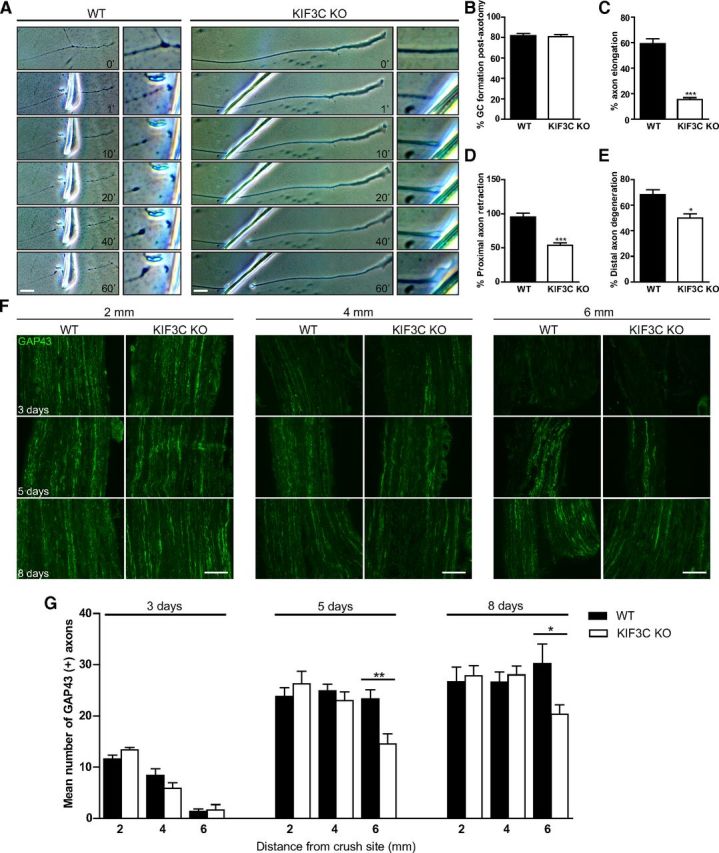
KIF3C is necessary for adult axon regeneration after injury. A, Successive frames from a time-lapse recording illustrating the behavior of a WT or KIF3C KO axon after axotomy in vitro. The enlarged box represents the site of axotomy at higher magnification. Scale bar, 20 μm. B, The percentage of WT or KIF3C KO axons that regrew growth cones after axotomy (n = 30 and n = 30, respectively, from three independent experiments). C, The percentage of WT or KIF3C KO axons that elongated after axotomy (n = 30 and n = 30, respectively). D, The percentage of WT or KIF3C KO axons that retracted after axotomy (n = 30 and n = 30, respectively). E, The percentage of WT or KIF3C KO distal axons that degenerated after axotomy (n = 30 and n = 30, respectively). Error bars indicate SD. *p < 0.05. ***p < 0.001. F, Representative photomicrographs of GAP43-positive axons in longitudinal cryosections from WT and KIF3C KO sciatic nerves, 3, 5, and 8 d after crush injury. Distance from crush site is 2, 4, and 6 mm. Scale bar, 100 μm. G, GAP43 axon count at 3, 5, and 8 d after sciatic nerve crush injury. Axon counts were taken at 2, 4, and 6 mm from the crush site. Error bars indicate SEM. *p < 0.05. **p < 0.01.
Having shown defective axon regeneration in vitro, we asked whether KIF3C KO axons regenerate normally after a sciatic nerve crush injury in vivo. The sciatic nerves of WT and KIF3C KO mice were crushed, and the time course of axonal regeneration was analyzed at 3, 5, and 8 d after injury. The extent of axon regeneration was assessed close to (2 mm) and farther from (4 and 6 mm) the injury site in cryosections of sciatic nerves immunostained with an antibody against GAP43, which is upregulated in regenerating peripheral nervous system axons (Fig. 13F). At 3 d after injury, a few axons had regenerated to the 2 or 4 mm point in both WT and KIF3C KO sciatic nerves, but very few axons were seen at 6 mm distal to the injury (Fig. 13F,G). However, 5 d after injury, ∼45% fewer axons reached the distance of 6 mm from the crush site in the KIF3C KO nerve compared with axons in control nerves (Fig. 13F,G). This trend of delayed regeneration was also observed at 8 d after injury, where 30% fewer axons regenerated to 6 mm in the KIF3C KO sciatic nerve compared with WT controls (Fig. 13F,G). Thus, KIF3C KO axons in vivo display a significant delay in the time course of axon regeneration after crush injury. These results corroborate our in vitro findings and reinforce the notion that KIF3C is required for axon regeneration after injury.
Discussion
Axon regeneration after injury relies on the reconstruction, reorganization, and stabilization of the microtubule cytoskeleton at the axon tip, where a correct equilibrium between microtubule polymerization and depolymerization is necessary for normal axon growth (Conde and Cáceres, 2009; Lowery and Van Vactor, 2009; Bradke et al., 2012). Strong microtubule stabilization leads to static growth cones, whereas microtubule destabilization leads to retraction bulbs with consequent impaired axonal elongation (Tanaka et al., 1995; Ertürk et al., 2007). An increasing number of microtubule plus-end regulators (Akhmanova and Steinmetz, 2008), including kinesin motor proteins (Wu et al., 2006), have been reported, but there is little information about how they might regulate microtubule dynamics after axonal injury. Here we have shown that KIF3C is an injury-associated kinesin that plays a part in sensory axon regeneration through an effect on the stability of microtubules and their organization. KIF3C's interaction with EB3 through its tail domain is necessary for KIF3C localization at the tips of a subset of growing microtubules in the periphery of the growth cone, where dynamic microtubule behavior is necessary for axon growth. Importantly, we show that loss of KIF3C function adversely affects axon growth in adult DRG neurons and impairs regeneration after injury in both embryonic and adult DRG neurons in vitro and in vivo. Our data show that KIF3C regulates microtubule dynamics by participating as a microtubule-destabilizing factor in growth cones.
Kinesins and microtubule control
Our observations show that the plus-end-directed kinesin KIF3C is involved in the regulation of microtubule dynamics during mammalian axon regeneration. KIF3C has effects during the retraction and growth cone reconstruction phase after axotomy as well as during subsequent axon growth. In growing adult axons, we found that growth cones lacking KIF3C contained higher levels of stable, overgrown, and looped microtubules and that many of these growth cones also had a stunted growth phenotype, suggesting that the impairment in microtubule dynamics led to the abnormal phenotype. In the absence of KIF3C, the enlarged growth cones contained microtubules with a reduced number of dynamic (EB3-positive) tips, similar to those observed in developing hippocampal neurons and glia from mice lacking KIF2A, a depolymerizing plus-end-directed kinesin involved in axon pathfinding and axonal branching (Homma et al., 2003). In embryonic DRG neurons, KIF3C does not localize to the growth cone; therefore, other proteins must regulate growth cone microtubule dynamics at this stage during normal axon growth. However, KIF3C is produced locally after axotomy and is needed for axon regeneration. Further work will determine how developmental changes contribute to microtubule dynamics in axon growth and regeneration. How does KIF3C localize to the dynamic tips of microtubules in the growth cone? Microtubules in adult growth cones are highly tyrosinated (dynamic), whereas embryonic DRG axons and growth cones contain lower levels of tyrosinated microtubules relative to acetylated (stable) microtubules (Bouquet et al., 2004; Jones et al., 2006). Interestingly though, axon injury induces a significant increase in tubulin tyrosination at the injury site in embryonic DRG axons (Cho and Cavalli, 2012). We found that KIF3C preferentially binds to tyrosinated microtubules in the absence of other binding partners or microtubule-associated proteins. Moreover, when microtubule detyrosination is induced by treating the cells with taxol, KIF3C is released from the microtubule tip. The microtubule stabilization by taxol led to large growth cones with looped microtubules in their periphery, very similar to those seen in axons lacking KIF3C. It is probable that the preferential binding of KIF3C to tyrosinated microtubules is responsible for its enrichment at the growth cone where KIF3C will interact with EB3 and localize to the tips of dynamic microtubules in the growth cone periphery. Notably, binding of KIF3C to tyrosinated microtubules could regulate its destabilizing activity, as observed with the kinesin depolymerizers MCAK or KIF2A, which preferentially depolymerize tyrosinated microtubules in mouse embryonic fibroblasts and developing hippocampal neurons (Homma et al., 2003; Peris et al., 2009). Microtubule detyrosination removes bound MCAK or KIF2A from the microtubule plus-ends with a consequent increase in microtubule overgrowth and looping, similar to the stabilization of microtubules we observed when KIF3C is absent. Our data suggest that KIF3C preferential binding to tyrosinated microtubules might be necessary for its enrichment in the growth cone and that further interactions with EB3 are required for localizing KIF3C specifically to growing microtubule tips in the growth cone.
Growth cone reconstruction
After axonal injury, a complex process of restructuring takes place in which the remodeling of the microtubule cytoskeleton accompanies morphological changes in the axon (Gumy et al., 2010). Calcium from the surrounding environment enters the axon, activating calpains and inducing microtubule depolymerization and axon retraction (Ziv and Spira, 1997; Gitler and Spira, 1998; Kamber et al., 2009). After the cut membrane has resealed, microtubules repolymerize, the new growth cone is formed, and the newly polymerized dynamic microtubules invade and explore the growth cone (Bradke et al., 2012). In vitro, when we cut DRG axons, rapid retraction took place. However, in the absence of KIF3C, most axons did not retract, supporting our suggestion that the microtubules are less prone to depolymerization and are more stable. In KIF3C KO-injured adult axons observed in vitro, the growth cone was able to reform, but the axons then failed to elongate during the 1 h that we observed them. Embryonic growth cones lack KIF3C unless the axon is cut and the kinesin is locally synthesized in the newly formed growth cone. Again, lack of KIF3C local protein synthesis prevents normal regeneration. In vivo regeneration is delayed in KIF3C KO animals. Growth cone formation can be achieved in the absence of polymerizing microtubules by the restructuring of the actin cytoskeleton and by the anterograde transport of vesicles that fuse with the plasma membrane (Sahly et al., 2003; Erez et al., 2007; Erez and Spira, 2008), and these processes can presumably still continue in the absence of KIF3C-induced microtubule dynamics. However, only growth cones whose lamellipodia are invaded by dynamic microtubules will achieve axonal elongation (Tanaka et al., 1995). The KIF3C KO mouse is viable and develops normally (Yang et al., 2001); and in agreement with this, we show that knockdown of KIF3C in embryonic DRGs does not affect neurite outgrowth from the cell body. However, overexpression of KIF3C during embryonic stages is detrimental to axon growth as the protein is unable to go into the axon and accumulates proximally, perhaps affecting trafficking of other proteins important for axon growth. Thus, the protein levels of KIF3C must be tightly regulated during development in concert with other processes that are taking place in the cell, such as tubulin modifications. In adult DRGs, KIF3C moves into axons, and overexpression of KIF3C has no effect on axon growth. However, KIF3C knockdown impairs axon growth as well as regeneration. Perhaps during neuronal maturation, as changes in tubulin modifications take place and KIF3C enters the axon, the protein acquires also an important role in the transport of cargos from the cell body into the axon. Therefore, knockdown of KIF3C might also impair axonal transport. Together, our data support the notion that KIF3C is dispensable for the development of sensory neurons but is required for embryonic and adult regeneration after injury, making it a specific injury-associated kinesin.
Mechanism of action of KIF3C
Two main factors appear to underlie a successful axon regeneration program. One is the achievement of the correct balance between microtubule stabilization/destabilization. The other one is the attainment of microtubules growing in the correct direction in the growth cone. Changes in the directionality of microtubule growth could result from excessive microtubule stabilization as seen in static growth cones or from microtubule destabilization as observed in retraction bulbs (Tanaka et al., 1995; Ertürk et al., 2007). Drugs that modulate microtubule dynamics have been used to alter the levels of microtubule stability in the axons and growth cones, and the general consensus is that microtubule stabilization promotes axon regeneration, whereas microtubule destabilization inhibits regrowth (Ertürk et al., 2007). However, uncertainty remains as to which are the parameters that define the appropriate level of microtubule stabilization necessary for successful axon regeneration. We have found that KIF3C is a microtubule-destabilizing factor required for axon regeneration. How to reconcile this finding with those demonstrating that microtubule stabilization is required for regrowth? Analysis of microtubule dynamics from KIF3C knockdown growth cones shows that microtubules experience almost persistent growth with no shrinkage events. KIF3C knockdown does not suppress microtubule dynamics, as the microtubules still do grow, but the balance is tilted toward a steady microtubule growth. How much microtubule stabilization is required for axon regeneration? A better understanding of the parameters that control microtubule dynamics in the growth cone will determine this.
To support axon extension, microtubules in the growth cone periphery or injured axonal tip must have directed and polarized growth. Our data demonstrate that microtubule looping is a result of overgrown microtubules that might bend backwards when they reach the cortex of the growth cone. An additional factor influencing microtubule growth orientation is the interaction of KIF3C with EB3. In Drosophila dendrites, the alignment of microtubules is reached by the interaction of KLP64D, a Kinesin-2 family member, with the EB1 and APC complex on a neighboring growing microtubule plus-end (Mattie et al., 2010). It is also possible that KIF3C contributes to maintaining microtubule growth direction in the growth cone by binding to one microtubule through its MD and by interacting indirectly with a neighboring microtubule through EB3 via its tail domain. In this way, the cytoskeleton can be organized in parallel microtubules to achieve directed microtubule growth. We suggest that, after axonal injury, KIF3C contributes to growth cone motility and axonal regeneration by the following: (1) regulating the dynamic state of microtubules through destabilization and (2) organizing and directing the growth of microtubules. In the future, we will need to understand how KIF3C microtubule-destabilizing activity is regulated in concert with other microtubule dynamics-related processes that are required for axon regeneration (Fig. 14).
Figure 14.
Schematic model showing KIF3C localization and function at the microtubule plus-ends. A, EB3 targets KIF3C to the plus-end of microtubules (MTs) in the growth cone. Here, KIF3C regulates microtubule dynamics and functions as a microtubule-destabilizing factor. B, The microtubule-destabilizing role of KIF3C is necessary to achieve appropriate axonal growth and regeneration after injury (B).
Footnotes
This work was supported by a United Kingdom Medical Research Council Project Grant to L.F.G. and J.W.F., FP7 EU Marie Curie postdoctoral fellowship to L.F.G. and E.T., EU FP7 project Plasticise to D.J.C. and J.F.W., the Christopher and Dana Reeve Foundation to J.W.F., the National Institute for Health Research Cambridge Biomedical Research Centre to J.W.F., the Foundation for Fundamental Research on Matter to E.A.K., and Netherlands Organization for Scientific Research to L.C.K. and C.C.H. We thank Sam van Beuningen for helping with the transfection of cortical neurons.
The authors declare no competing financial interests.
References
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- Bouquet C, Soares S, von Boxberg Y, Ravaille-Veron M, Propst F, Nothias F. Microtubule-associated protein 1B controls directionality of growth cone migration and axonal branching in regeneration of adult dorsal root ganglia neurons. J Neurosci. 2004;24:7204–7213. doi: 10.1523/JNEUROSCI.2254-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradke F, Fawcett JW, Spira ME. Assembly of a new growth cone after axotomy: the precursor to axon regeneration. Nat Rev Neurosci. 2012;13:183–193. doi: 10.1038/nrn3176. [DOI] [PubMed] [Google Scholar]
- Chierzi S, Ratto GM, Verma P, Fawcett JW. The ability of axons to regenerate their growth cones depends on axonal type and age, and is regulated by calcium, cAMP and ERK. Eur J Neurosci. 2005;21:2051–2062. doi: 10.1111/j.1460-9568.2005.04066.x. [DOI] [PubMed] [Google Scholar]
- Cho Y, Cavalli V. HDAC5 is a novel injury-regulated deacetylase controlling axon regeneration. EMBO J. 2012;31:3063–3078. doi: 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Daire V, Poüs C. Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch Biochem Biophys. 2011;510:83–92. doi: 10.1016/j.abb.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Davidovic L, Jaglin XH, Lepagnol-Bestel AM, Tremblay S, Simonneau M, Bardoni B, Khandjian EW. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum Mol Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- Drummond DR. Regulation of microtubule dynamics by kinesins. Semin Cell Dev Biol. 2011;22:927–934. doi: 10.1016/j.semcdb.2011.09.021. [DOI] [PubMed] [Google Scholar]
- Erez H, Spira ME. Local self-assembly mechanisms underlie the differential transformation of the proximal and distal cut axonal. J Comp Neurol. 2008;1030:1019–1030. doi: 10.1002/cne.21522. [DOI] [PubMed] [Google Scholar]
- Erez H, Malkinson G, Prager-Khoutorsky M, De Zeeuw CI, Hoogenraad CC, Spira ME. Formation of microtubule-based traps controls the sorting and concentration of vesicles to restricted sites of regenerating neurons after axotomy. J Cell Biol. 2007;176:497–507. doi: 10.1083/jcb.200607098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler D, Spira ME. Real time imaging of calcium-induced localized proteolytic activity after axotomy and its relation to growth cone formation. Neuron. 1998;20:1123–1135. doi: 10.1016/S0896-6273(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Gordon-Weeks PR, Lang RD. The alpha-tubulin of the growth cone is predominantly in the tyrosinated form. Brain Res. 1988;470:156–160. doi: 10.1016/0165-3806(88)90213-1. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Bampton ET, Tolkovsky AM. Hyperglycaemia inhibits Schwann cell proliferation and migration and restricts regeneration of axons and Schwann cells from adult murine DRG. Mol Cell Neurosci. 2008;37:298–311. doi: 10.1016/j.mcn.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Tan CL, Fawcett JW. The role of local protein synthesis and degradation in axon regeneration. Exp Neurol. 2010;223:28–37. doi: 10.1016/j.expneurol.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Huang CF, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Mol Biol Cell. 2010;21:572–583. doi: 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114:229–239. doi: 10.1016/S0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, Jelesarov I, Winkler FK, Wüthrich K, Akhmanova A, Steinmetz MO. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Jones SL, Selzer ME, Gallo G. Developmental regulation of sensory axon regeneration in the absence of growth cones. J Neurobiol. 2006;66:1630–1645. doi: 10.1002/neu.20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kamber D, Erez H, Spira ME. Local calcium-dependent mechanisms determine whether a cut axonal end assembles a retarded endbulb or competent growth cone. Exp Neurol. 2009;219:112–125. doi: 10.1016/j.expneurol.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol. 2010;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, Borisy GG, Steinmetz MO, Akhmanova A. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Laketa V, Simpson JC, Bechtel S, Wiemann S, Pepperkok R. High-content microscopy identifies new neurite outgrowth regulators. Mol Biol Cell. 2007;18:242–252. doi: 10.1091/mbc.E06-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Nadar VC, Kozielski F, Kozlowska M, Yu W, Baas PW. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J Neurosci. 2010;30:14896–14906. doi: 10.1523/JNEUROSCI.3739-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM. Directed microtubule growth, +TIPs, and kinesin-2 are required for uniform microtubule polarity in dendrites. Curr Biol. 2010;20:2169–2177. doi: 10.1016/j.cub.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin NL, Schnapp BJ. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol Biol Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navone F, Consalez GG, Sardella M, Caspani E, Pozzoli O, Frassoni C, Morlacchi E, Sitia R, Sprocati T, Cabibbo A. Expression of KIF3C kinesin during neural development and in vitro neuronal differentiation. J Neurochem. 2001;77:741–753. doi: 10.1046/j.1471-4159.2001.00277.x. [DOI] [PubMed] [Google Scholar]
- Pazyra-Murphy MF, Segal RA. Preparation and maintenance of dorsal root ganglia neurons in compartmented cultures. J Vis Exp. 2008;20:951. doi: 10.3791/951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purro SA, Ciani L, Hoyos-Flight M, Stamatakou E, Siomou E, Salinas PC. Wnt regulates axon behavior through changes in microtubule growth directionality: a new role for adenomatous polyposis coli. J Neurosci. 2008;28:8644–8654. doi: 10.1523/JNEUROSCI.2320-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson SJ, Burgoyne RD. Differential localisation of tyrosinated, detyrosinated, and acetylated α-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell Motil Cytoskeleton. 1989;12:273–282. doi: 10.1002/cm.970120408. [DOI] [PubMed] [Google Scholar]
- Sahly I, Erez H, Khoutorsky A, Shapira E, Spira ME. Effective expression of the green fluorescent fusion proteins in cultured Aplysia neurons. J Neurosci Methods. 2003;126:111–117. doi: 10.1016/S0165-0270(03)00072-4. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ohi R, Pellman D. Move in for the kill: motile microtubule regulators. Trends Cell Biol. 2012;11:567–575. doi: 10.1016/j.tcb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Ho T, Kirschner MW. The role of microtubule dynamics in growth cone motility and axonal growth. J Cell Biol. 1995;128:139–155. doi: 10.1083/jcb.128.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JR. An introduction to error analysis. Ed 2. Sausalito, CA: University Science; 1997. [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiang X, Hammer JA., 3rd Motor proteins at the microtubule plus-end. Trends Cell Biol. 2006;16:135–143. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Yang Z, Goldstein LS. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol Biol Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Roberts EA, Lawrence SB, Goldstein LS. Functional analysis of mouse kinesin motor Kif3C. Mol Cell Biol. 2001;21:5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Spira ME. Localized and transient elevations of intracellular Ca2+ induce the dedifferentiation of axonal segments into growth cones. J Neurosci. 1997;17:3568–3579. doi: 10.1523/JNEUROSCI.17-10-03568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]