Abstract
Distinct anatomical regions of the neocortex subserve different sensory modalities and neuronal integration functions, but mechanisms for these regional specializations remain elusive. Involvement of epigenetic mechanisms for such specialization through the spatiotemporal regulation of gene expression is an intriguing possibility. Here we examined whether epigenetic mechanisms might play a role in the selective gene expression in the association areas (AAs) and the primary visual cortex (V1) in macaque neocortex. By analyzing the two types of area-selective gene promoters that we previously identified, we found a striking difference of DNA methylation between these promoters, i.e., hypermethylation in AA-selective gene promoters and hypomethylation in V1-selective ones. Methylation levels of promoters of each area-selective gene showed no areal difference, but a specific methyl-binding protein (MBD4) was enriched in the AAs, in correspondence with expression patterns of AA-selective genes. MBD4 expression was mainly observed in neurons. MBD4 specifically bound to and activated the AA-selective genes both in vitro and in vivo. Our results demonstrate that methylation in the promoters and specific methyl-binding proteins play an important role in the area-selective gene expression profiles in the primate neocortex.
Introduction
The neocortex is a structure that is present only in mammals and is enlarged in primates. It consists of anatomically and functionally distinct areas that subserve different sensory modalities and neuronal integration functions (Felleman and Van Essen, 1991; Kaas, 2005). In the last decades, studies have addressed the underlying mechanisms that control the formation of cortical layer and primary sensory areas; but the molecular mechanisms that form and maintain these areas remain to be elucidated (Lukaszewicz et al., 2006; O'Leary and Sahara, 2008). Presumably, the different regional specializations relate to the different gene expression profiles and resultant distinctive histochemical phenotypes seen in different cortical regions. Recent analyses of gene expression patterns in rodents reveal that there are four patterning centers that control graded transcription in the neocortex (O'Leary and Sahara, 2008). It can be expected that primates, with their much greater arealization, will have other molecular specification in addition to that found in rodents.
Our approach has been therefore to isolate genes that are specifically expressed in the neocortical areas in primates. Notably, we have reported two groups of genes that are differentially expressed either in the macaque primary visual cortex in the occipital lobe (Tochitani et al., 2001; Takahata et al., 2009; Watakabe et al., 2009), or in the association areas (AAs) of cortex including the frontal lobes (Komatsu et al., 2005; Takaji et al., 2009; Sasaki et al., 2010). The pronounced area selectivity and activity dependency of these genes occur in macaques and marmosets, but not in the rodents, lagomorphs, or carnivores examined so far. We thus suggest that the mechanisms underlying this gene expression may be an important clue to the evolution of the primate cerebral cortex (Yamamori, 2011; Takahata et al., 2012). Our previous studies clarified the functions of two of the V1-selective genes, HTR1B and HTR2A, in the macaque primary visual cortex (Watakabe et al., 2009), and functional studies are currently underway for other molecules. Identifying such mechanisms underlying these unique area-selective genes is important for understanding the development, evolution, and function of the primate neocortex.
Here, we demonstrate striking differences of DNA methylation between the promoter regions of the genes selectively expressed in the AAs and the primary visual cortex (V1) in macaque monkeys. Although the methylation levels of promoters of each area-selective gene showed no regional difference, MBD4, among five known methyl-binding proteins, was enriched in the AAs. MBD4 was mainly observed in NeuN-positive cells, specifically bound to and activated by the AA-selective genes both in culture and in vivo macaque monkey neocortex. Thus, our results provide evidence for a critical role of DNA methylation and methyl-binding proteins in the differential gene expression profiles in the primate neocortex.
Materials and Methods
Animals.
Eight macaque monkeys (a male and a female Rhesus monkey, Macaca mulatta, and three male and three female Japanese monkeys, M. fuscata) of late juvenile to young adult (from 2 years 9 months to 4 years 5 months) were used in this study. Note that we have been using this stage of macaques throughout our publications of area-selective gene expression and have found no significant individual and sexual differences of normal macaque monkeys in the expression patterns of the genes studied in this paper. In the text we therefore group all these monkeys as adults for simplicity. All the protocols used in this study were approved by the Animal Research Committee of the National Institute for Basic Biology, National Institute for Physiological Sciences, Japan; they were in accordance with the animal care guidelines of the U.S. National Institutes of Health (NIH).
In situ hybridization.
Digoxigenin (DIG)-labeled antisense and sense riboprobes were prepared using a DIG-dUTP labeling kit (Roche Diagnostics). Riboprobes were newly prepared from the cDNA library of each animal by reverse transcription (RT)-PCR and conventional TA cloning techniques. Detailed information on the probes used in this study was described previously (Tochitani et al., 2001; Komatsu et al., 2005; Takaji et al., 2009; Watakabe et al., 2009; Sasaki et al., 2010; Takahata et al., 2010). The sense probes did not detect any signals stronger than the background signal (data not shown). In situ hybridization (ISH) was performed as follows: free-floating sections were soaked in 4% paraformaldehyde (PFA)/0.1 m PB, pH 7.4, overnight at 4°C and then treated with 10 μg/ml proteinase K for 30 min at 37°C. After acetylation, the sections were incubated in the hybridization buffer (5× standard saline citrate [SSC; 1× means 150 mm NaCl, 15 mm Na-citrate, pH 7.0], 50% formamide, 2% blocking reagent, 0.1% N-lauroylsarcosine [NLS], 0.1% SDS, 20 mm maleic acid buffer; pH 7.5) containing 0.5 μg/ml DIG-labeled riboprobe at 60°C overnight. Hybridized sections were washed by successively immersing in the washing buffer (2× SSC, 50% formamide, 0.1% NLS; 60°C, 20 min, twice), RNase A buffer (10 mm Tris-HCl, 10 mm ethylenediaminetetraacetic acid, 0.5 m NaCl, pH 8.0) containing 20 μg/ml RNase A (37°C, 15 min), 2× SSC/0.1% NLS (37°C, 20 min, twice), and 0.2× SSC/0.1% NLS (37°C, 15 min, twice). Hybridization signals were visualized by alkaline phosphatase (AP) immunohistochemical staining using a DIG detection kit (Roche Diagnostics). Because ISH signals were strong for the V1-selective genes (HTR1B, HTR2A, and FSTL1/OCC1), we shortened their reaction time (to 4–6 h) with nitro blue tetrazolium/5-Bromo-4-chloro-3-indolyl phosphate from the original reaction time (18–24 h) to prevent the saturation of ISH signals. Sections were mounted onto glass slides and dehydrated through a graded series of increasing ethanol concentrations followed by xylene and then coverslipped with Entellan New (Merck, No. UN1866).
Fluorescence double-labeling ISH was performed as described previously (Takahata et al., 2009). Fluorescein isothiocyanate (FITC)-labeled riboprobes for green fluorescent protein (GFP; Komine et al., 2006) were prepared. Brain sections were hybridized with both the DIG-labeled and the FITC-labeled probes. The hybridization protocol was the same as that of colorimetric single ISH. DIG was detected by staining with AP-conjugated anti-DIG antibodies and subsequently using an HNPP Fluorescence Detection kit (Roche Diagnostics). FITC was detected by staining with horseradish peroxidase (HRP)-conjugated anti-FITC antibodies (Roche Diagnostics); the staining was enhanced using a TSA-Plus DNP system (PerkinElmer Life Sciences) followed by staining with Alexa 488-conjugated anti-DNP antibodies (Invitrogen). The sections were then counterstained with Hoechst 33342 (Dojindo) diluted in PBS to 1:1000. After mounting onto glass slides, sections were air-dried and coverslipped with CC/Mount (Diagnostic BioSystems, No. K002).
Sodium bisulfite modification and DNA sequencing.
The CpG islands in the promoter of area-selective genes were based on the algorithm described for MethPrimer software (http://www.urogene.org/methprimer/index1.html; Li and Dahiya, 2002). For sodium bisulfite modification, a solution of 1 μg of digested genomic DNA was heated at 98°C for 5 min and incubated with 0.3 N NaOH in a final volume of 27.5 μl for 20 min at 37°C. Two hundred seventy microliters of a solution containing 3.6 m sodium bisulfite (freshly prepared; Sigma) and 0.6 mm hydroquinone (freshly prepared; Sigma) was gently mixed and incubated for 5 h at 55°C in the dark. The sample was precipitated with 600 μl water, 90 μl 3 m NaOAc, 2.5 μl glycogen, and 900 μl isopropanol, then desulfonated by incubation with 0.3 N NaOH for 15 min at 37°C. The DNA was ethanol precipitated and resuspended in 25 μl of TE buffer. Using 2 μl of the solution, each region was amplified by AmpliTaq-Gold DNA polymerase (Applied Biosystems). The information about the primers is described in Table 1). The PCR product was cloned into pBluescript II KS+ (Stratagene), and used as a template for cycle sequencing. The proportion of methylated cytosines was determined after sequencing 10 clones.
Table 1.
Primers and oligonucleotides used in this study
| Gene | Species | Forward (5′ > 3′) | Reverse (5′ >3′) | Length | Application | Accession |
|---|---|---|---|---|---|---|
| RBP4 | Macaque | ATGAGGGTAAGGACATAAATGTCTTGC | CCTTAACAGCATCAAACAAGTCACCAACTC | 413 | Bis-seq, ChIP | AC194680 |
| Macaque | ATGAGGGTAAGGATATAAATGTTTTGT | CCTTAACAACATCAAACAAATCACCAACTC | 413 | Bis-Seq | ||
| Macaque | TCAAAGTGGTTTCAGGGAAGTGCC | GCCCTGGCTCAGTTACCCTGCACT | 467 | Bis-Seq | AC194680 | |
| Macaque | TTAAAGTGGTTTTAGGGAAGTGTT | ACCCTAACTCAATTACCCTACACT | 467 | Bis-Seq | ||
| PNMA5 | Macaque | TGGGAAGTAGGAGAGTTTATGTCACC | GTATGATCTCAGCTCACTGCAACCTC | 626 | Bis-seq, ChIP | AC193042 |
| Macaque | TGGGAAGTAGGAGAGTTTATGTTATT | ATATAATCTCAACTCACTACAACCTC | 626 | Bis-Seq | ||
| Human | TGCAGGAAGTAGGAGGGTTTATGTCACC | GCCAGGTCTCCCTGAAGTTCTGCCTG | 397 | Bis-seq, ChIP | U82671 | |
| Human | TGTAGGAAGTAGGAGGGTTTATGTTATT | ACCAAATCTCCCTAAAATTCTACCTA | 397 | Bis-Seq | ||
| SLIT1 | Macaque | ACAAGGCATCATCCATGGACTATGAGAC | CATTGGCTTTCTTGTATCTTGTTCACAG | 474 | Bis-seq, ChIP | AC215327 |
| Macaque | ATAAGGTATTATTTATGGATTATGAGAT | CATTAACTTTCTTATATCTTATTCACAA | 474 | Bis-Seq | ||
| Macaque | CCACCTGTCAGAAATGGAGGAAC | GGCCTCTAGGCCCCACCTGACT | 409 | Bis-Seq | AC215327 | |
| Macaque | TTATTTGTTAGAAATGGAGGAAT | GACCTCTAAACCCCACCTAACT | 409 | Bis-Seq | ||
| HTR1B | Macaque | AGGCATGGCTGGAGAAAGCCACAGGTC | TTCCCATCCTCCACCCCATCCACCTA | 411 | Bis-Seq, ChIP | AC195613 |
| Macaque | AGGTATGGTTGGAGAAAGTTATAGGTT | TCCCATCCTCCCACCCCATCCACCTAA | 411 | Bis-Seq | ||
| Macaque | GGAGCTTCCTTGGCCAGGAAGGGAA | CTCTTGGATCTGCAGACTTTGCCCACAG | 628 | Bis-Seq | AC195613 | |
| Macaque | GGAGTTTTTTTGGTTAGGAAGGGAA | CACTTAAATCTACAAACTTTACCCACAA | 628 | Bis-Seq | ||
| HTR2A | Macaque | GTAGGAGGAGTGACAGAGCAGGAAC | CAAGCAGGCAGCCCAGGCCTCAGTC | 265 | Bis-Seq, ChIP | AL160397 |
| Macaque | GTAGGAGGAGTGATAGAGTAGGAAT | CAAACAAACAACCCAAACCTCAATC | 265 | Bis-Seq | ||
| FSTL1 | Macaque | ATGCTGGACTCAGGATCTTCCCAGCAGAGC | GGCTGCGGCTGTGGCCAGGTGGGCGGTGC | 319 | Bis-Seq, ChIP | AC199131 |
| Macaque | ATGTTGGATTTAGGATTTTTTTAGTAGAGT | AACTACYACTATAACCAAATAAACYTTAC | 319 | Bis-Seq | ||
| MBD4 | Hu, mcq | ATGGGCACGACTGGGCTGGAGAGTCTGAGT | AGATAGACTTAATTTTTCATGATTTTCCCA | 1743 | OE,RT-PCR | NM003925 |
| Hu, mcq | GGGAAAGAGTTGTGAAGCAAA | sh#1, position: 444–465 | 21 | Knockdown | NM003925 | |
| Hu, mcq | GCAGGAAGATTTGATGTGTAC | sh#2, position: 483–504 | 21 | Knockdown | NM003925 | |
| Hu, mcq | GAGAAGTATGAGGATACCTTT | sh#3, position: 1212–1233 | 21 | Knockdown | NM003925 | |
| Hu, mcq | GAGCTTCATGGGATTGGTAAA | sh#4, position: 1774–1795 | 21 | Knockdown | NM003925 | |
| Hu, mcq | GGTAAATATGGCAACGACTCT | sh#5, position: 1787–1808 | 21 | Knockdown | NM003925 | |
| MBD1 | Hu, mcq | ATGGCTGAGGACTGGCTGGACTGCCCG | TCCCGGTTGAAGGCAATTCTCTGTGCT | 500 | ISH | NM015846 |
| MBD2 | Hu, mcq | ATGCGCGCGCACCCGGGGGGAGGCCGCTGC | TTTCGGATCACTTCCTCCTTCTTCCATCCG | 500 | ISH | NM003927 |
| MBD3 | Hu, mcq | ATGGAGCGGAAGAGGTGGGAGTGC | CCCTGCAGGCCCTTGGGGAGGTCCATGGT | 500 | ISH | NM003926 |
| MBD4 | Hu, mcq | ATGGGCACGACTGGGCTGGAGAGTCTGAGT | AGATGGGATGTCAGGGCTGCCATGCTG | 500 | ISH | NM003925 |
| MeCP2 | Hu, mcq | ATGGTAGCTGGGATGTTAGGGCTCAGGGAA | CGGGAGGGGCTCCCTCTCCCAGTTACCGTG | 500 | ISH | NM015846 |
| GAPDH | Hu, mcq | CTCATGACCACAGTCCATGCC | AGGTCCACCACCCTGTTGCTG | 461 | RT-PCR | NM002046 |
| MBD1 | Hu, mcq | CAAAGACTGTCGAGCACAGAG | ATCATGGGGCAGCTGCAGGAG | 142 | Real-time PCR | NM015846 |
| MBD2 | Hu, mcq | GCTGGCAAGAGCGATGTCTAC | TGTAATTTACTAGGCATCATC | 141 | Real-time PCR | NM003927 |
| MBD3 | Hu, mcq | TCGGCCGGCCACAGGGATGTC | ATCTTGCTCATCAGCATCTTG | 143 | Real-time PCR | NM003926 |
| MBD4 | Hu, mcq | ACCTCTAGTGAGCGCCTAGTC | GCGATGGGTTCTTGTAGCAAG | 149 | Real-time PCR | NM003925 |
| MeCP2 | Hu, mcq | ATGGTAGCTGGGATGTTAG | GCTGCACTGGCTCATGCTTGC | 144 | Real-time PCR | NM015846 |
| GAPDH | Hu, mcq | AGGTGAAGGTCGGAGTCA | GGTCATTGATGGCAACAA | 98 | Real-time PCR | NM002046 |
OE, Overexpression.
Real-time RT-PCR.
To quantify the difference in mRNA expression level between the areas, real-time RT-PCR was performed as previously described (Takahata et al., 2009). Total RNA was isolated from the prefrontal cortex (PFC) and primary visual cortex (V1) of adult macaque monkeys using Isogen (Nippongene, No. 311–02501). Two micrograms of total RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription kits (Applied Biosystems, No. 4368816) and used as the template for real-time RT-PCR. The prepared cDNA was reacted with specific primers for five methyl-binding domain proteins (Table 1) using a KAPA SYBR FAST qPCR kit (KAPA Biosystems, No. KR0389) under the control of the Applied Biosystems 7500 Real-Time PCR System. To amplify only cDNA from mRNA gene transcripts, each primer set was designed to span the first intron. Only a single band was obtained when each reacted sample was electrophoresed, and the sequence of the band was confirmed to be derived from the targeted sequence of each gene (data not shown). A series of diluted solutions of brain samples was examined to determine the amplification efficiency of each primer set. The amount of each mRNA in each area was calculated on the basis of the cycle number, fluorescence intensity, and amplification efficiency. The amount of each mRNA was represented as the relative ratio to that of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, which was used as an internal standard.
Semiquantitative RT-RCR.
To analyze the mRNA expression level of PNMA5, the cDNA from SH-SY5Y cells was amplified using human PNMA5-specific primers for the indicated cycles described in Figure 5H. Human GAPDH-specific primers were used in control reactions. The amplification products were analyzed by agarose gel electrophoresis. The primers used in these experiments are listed in Table 1.
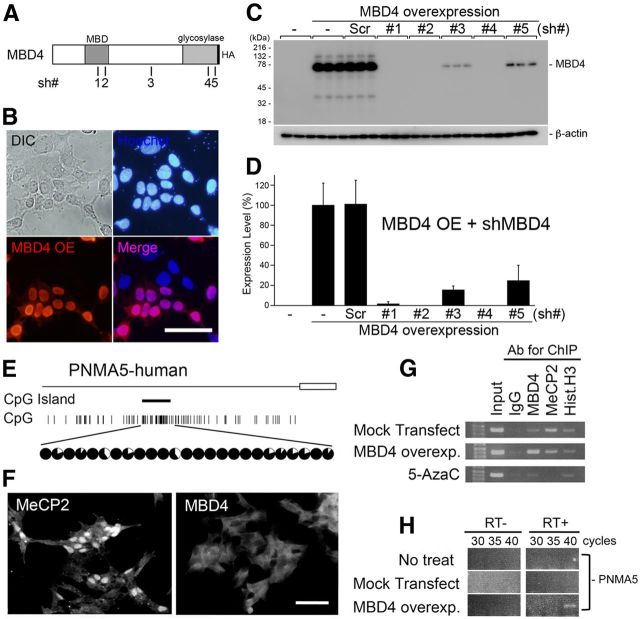
Figure 5.
Overexpression of MBD4 induces the expression of PNMA5 mRNA in human neuroblastoma cell culture. A, Schematic illustration of the full MBD4 protein with an HA-tag in the C terminus, and the positions of the 21 bases of the sh #1–5 vectors used for MBD4 knockdown. B, Overexpression (OE) of MBD4-HA in SH-SY5Y cells. The overexpressed proteins were stained with the anti-HA antibody (red). The localization of MBD4-HA in the nuclei was determined by costaining with Hoechst (blue). C, D, Quantification of the knockdown effects of cotransfection of the U6-shMBD4 (#1–5) vectors with MBD4-HA in SH-SY5Y. Sh-MBD4#4 (with the highest efficiency, 99% knockdown effect) was used in subsequent in vitro and in vivo experiments. Error bars indicate SD. E, The human PNMA5 promoter has a CpG island at a position similar to that in the macaque monkey (Fig. 2A, PNMA5), and this island is also hypermethylated in SH-SY5Y human neuroblastoma cells. F, SH-SY5Y cells express MeCP2 and not MBD4. G, MeCP2 is bound to the CpG islands in the human PNMA5 promoter in control conditions (Mock transfect/MeCP2:IP), whereas blocking methylation by 5-AzaC diminishes this binding (5-AzaC/MeCP2:IP). Overexpressed MBD4 may bind at the same site because MeCP2 binding appears to have been reduced (MBD4 overexp/MBD4 and MeCP2:IP), but this result remains to be confirmed, as the transfection efficacy to the cells was not 100%. H, The overexpression of MBD4 induced PNMA5 expression in SH-SY5Y cells.
Western blotting.
To analyze the vectors for overexpression of MBD4 for determining the efficiency of the knockdown of these proteins, SH-SY5Y cells were grown to 90% confluence in DMEM + 10% fetal bovine serum (FBS) in 24-well plates in a 5% CO2 incubator, then transfected with the indicated plasmid (total 0.8 μg/ml) with Lipofectamine 2000 reagent (2 μl/ml; Invitrogen) following the manufacturer's recommended method. After 24 h, cells were lysed by SDS-sample buffer, and the proteins were separated by 10% SDS-PAGE. The proteins were transferred onto PVDF membranes, and then were immunostained with specific antibodies for MBD4 (Abcam, ab84754, 1:200), or β-actin (Abcam, ab16039–500, 1:1000), followed by the appropriate HRP-conjugated secondary antibody (The Jackson Laboratory; 1:10,000); immunostaining was visualized with a chemiluminescence kit (SuperSignal West Dura; Pierce, #34075). The images were acquired using a Luminescent Image Analyzer LAS-3000 (FUJIFILM), and densitometric analysis for the quantification of the knockdown effect was performed using ImageJ software (NIH).
Immunohistochemistry and immunocytochemistry.
Free-floating sections or cultured SH-SY5Y cells were fixed with 4% PFA in PB and immersed into the blocking buffer: 5% bovine serum albumin, 4% normal goat serum, 0.1% Triton X-100 in Tris-buffered saline (TBS) for 1 h at room temperature. The sections were then reacted with the primary antibodies, anti-HA (Bethyl Laboratory, rabbit polyclonal, A190–108A; 1:1000), anti-NeuN (Millipore, mouse monoclonal; 1:200), anti-MBD1 (Abcam, rabbit polyclonal, ab108510; 1:100), anti-MBD2 (Abcam, rabbit polyclonal, ab109260; 1:250), anti-MBD3 (Abcam, rabbit polyclonal, ab71777; 1:50), anti-MBD4 (Abcam, rabbit polyclonal, ab84754; 1:100), and anti-MeCP2 (Lifespan Biosciences, rabbit polyclonal; 1:400) in the same blocking buffer overnight at 4°C. The sections were then washed in TBST three times and reacted with the secondary antibodies conjugated with anti-mouse IgG-Alexa488 (Invitrogen; 1:200), anti-rabbit-IgG-Cy3 (The Jackson Laboratory; 1:200) or anti-rabbit-IgG-biotin (The Jackson Laboratory; 1:2000) for 2 h at room temperature. The sections were counterstained with the chromosomal dye Hoechst 33342 (Dojindo) and coverslipped with CC/Mount (Diagnostic BioSystems, No. K002). For diaminobenzidine (DAB) detection, immunoreactivity was detected using an avidin/biotin/peroxidase detection kit (Vectastain) and DAB as a substrate, with nickel enhancement. The sections were dehydrated through a graded series of increasing ethanol concentrations followed by xylene, and coverslipped with Entellan New (Merck, No. UN1866). Sections developed without the primary antibodies showed no detectable signals above the background level (data not shown).
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays (ChIP) and double-ChIP were performed as previously described (Nandiwada et al., 2006; Nelson et al., 2006; Onishi et al., 2009). For each reaction, subconfluent SH-SY5Y cells in 10 cm dishes, or a dissected macaque cerebral cortex (PFC and V1, ∼100 mg each), were treated with 1% formaldehyde for 15 min at room temperature to cross-link chromatin binding proteins. After cell lysis, the chromatin was fragmented to an average length of ∼600 bp by sonication, and subjected to immunoprecipitation with 5 μg of the first IP antibody (MBD4, sc10753; Santa Cruz Biotechnology; Histone H3, ab1791, Abcam), or a control IgG in the presence of Protein A or G beads. The immunoprecipitated protein-DNA complexes were washed following to the method of Onishi et al. (2009), and eluted from the beads with 300 μl of an Elution Buffer (50 mm NaHCO3, 1% SDS). One hundred microliters of the supernatant was reserved as a single ChIP sample for PCR analysis later, while the rest (200 μl) was used for the second immunoprecipitation. After being diluted to 600 μl with IP dilution buffer (167 mm NaCl, 16.7 mm Tris-HCl, pH 8.1, 1.2 mm EDTA, 1.1% Triton X-100, 0.01% SDS), each sample was divided into two equal aliquots and immunoprecipitated using 5 μl of the second IP antibody (CBP, sc-1211; Santa Cruz Biotechnology; p300, C20, Santa Cruz Biotechnology) and control IgG, respectively, according to published methods (Nandiwada et al., 2006), in the presence of Protein A or G beads. After washing and elution steps, all the samples were heated at 67°C for 4–5 h to reverse the cross-links. The DNA in the complexes was then purified and analyzed, together with the single ChIP samples, by PCR with candidate gene-specific primers. The positions of the primer in this experiment were set on the same positions as the ones in the methylation analysis in Figure 2, and the detailed primer information is described in Table 1.
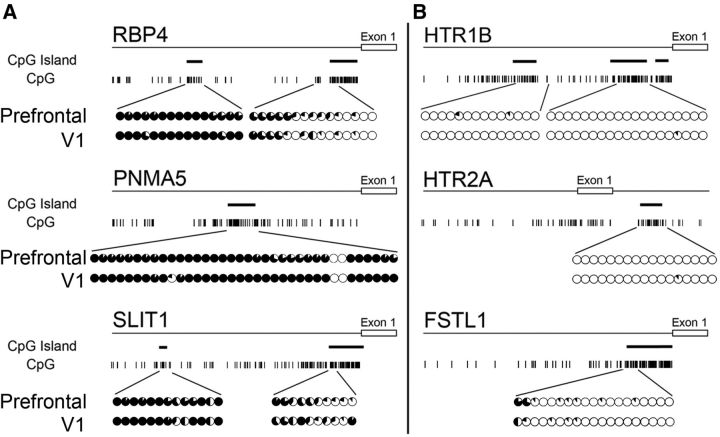
Figure 2.
Methylation of AA- and V1-selective gene promoters. Map of CpG base motifs in each AA-selective (A) and V1-selective gene (B) and their methylation status in the vicinity of the promoter region for tissues from the PFC of area 46 (top rows) and V1 (bottom rows). The sites of CpG islands are represented by thick bars. The fraction of methylated DNA at each CpG site is shown by the proportion of the black area in each circle, with a full black circle indicating complete methylation. Left and right sides show the AA- and V1-selective gene promoters, respectively. The CpG island in the promoter region of the AA-selective genes was hypermethylated, whereas that of the V1-selective genes was almost completely unmethylated. The same pattern was observed for genomic DNA obtained from both adult macaque PFC and V1 tissues.
Construction of the plasmid vectors.
The adeno-associated virus (AAV) vector was based on AAV Helper-Free system (Agilent Technologies). For the overexpression of MBD4 and MeCP2, the pAAV-CMV-T2A-EGFP was constructed as follows: at first, a T2A-EGFP fragment was generated by PCR using a 5′ primer with XbaI and XhoI sites and a 3′ primer with a SalI site. This product was digested by XbaI and SalI and inserted into the XbaI and XhoI sites of the pAAV-MCS plasmid. Next, the full-length fragment of human MBD4 or MeCP2 was inserted into the XhoI site of this vector. The direction and sequence of the inserted fragment were verified by DNA sequencing.
For the MBD4 and MeCP2 knockdown experiments, the pAAV-U6-CMV-GFP was constructed as follows: to generate plasmids encoding small hairpin-RNA (sh-RNA) driven by the U6 promoter, a 0.7-kb PvuII fragment from pSilencer 2.1-U6 neo (Ambion Life Technologies) was inserted into the blunted MluI site of pAAV-hrGFP (Agilent Technologies). The direction and sequence of the inserted fragment were verified by DNA sequencing. Switching of the sh-RNA cassette was performed based on the manufacturer's instructions (Ambion). Briefly, two 63-mer oligonucleotides that corresponded to the forward and reverse sequences were synthesized (The position is described in Fig. 5A). These oligos were annealed in 1 × DNA annealing solution (Ambion) at 90°C for 3 min, then cooled down to room temperature over 1 h. The resulting fragment was ligated to the target plasmid digested by BamHI and HindIII.
Culture, transfection, and stimulation of SH-SY5Y cells.
SH-SY5Y human neuroblastoma cells were grown in DMEM/F12 supplemented with 10% FBS at 37°C in 5% CO2 incubator. For the experiments, 2 × 105 cells were transferred on 13 mm round-glass coverslips precoated with poly-d-lysine in a 24-well culture plate and 500 μl of growth media, then transfected with the MBD4 plasmid (0.8 μg/well) via Lipofectamine 2000 reagent (2 μl/well; Invitrogen, 52887) in 100 μl Opti-MEM I (Gibco, 31985–070) following the manufacturer's recommended method. To reduce basal expression of immediate early gene molecules, the cells were serum starved for 24 h in DMEM/F12 containing 0.5% FBS as described earlier. Experimental treatments were performed in the same medium. After 2 d, cells were fixed by 4% PFA in PBS and enclosed by CC/Mount (Diagnostic BioSystems, No. K002).
Preparation and infection of AAV into monkey neocortex.
The method for AAV preparation was followed as described previously (Yagi et al., 2011). AAV1 viral vectors were injected into the monkey neocortex as follows: 1 μl of AAV1-U6-shMBD4#4-CMV-eGFP-SV40polyA (2.6 × 109 TU/μl) and AAV1-CMV-hMBD4-T2A-EGFP-SV40polyA (4.1 × 109 TU/μl) were injected into the PFC (1 mm in depth) and V1 (0.8 mm in depth), respectively, of two Japanese macaque monkeys (M. fuscata, 2 years 9 months, 3.9 kg, female; 3 years, 6 months, 4.0 kg, female) using a glass pipette (70–100 μm in diameter). Basic techniques for AAV injection were described previously (Kinoshita et al., 2012). Four weeks later, the monkeys were deeply anesthetized and perfused, and the brain was stored frozen for subsequent histological experiments.
Image acquisition and data analysis.
An Olympus fluorescence microscope (BX50) was used and images were acquired with a CCD camera (DP70) driven by a DP Controller and DP manager (Olympus). Quantification of data was performed using ImageJ software. Montages of the images were created using Adobe Photoshop CS4.
Results
Difference in the methylation status between AA- and V1-selective gene promoters
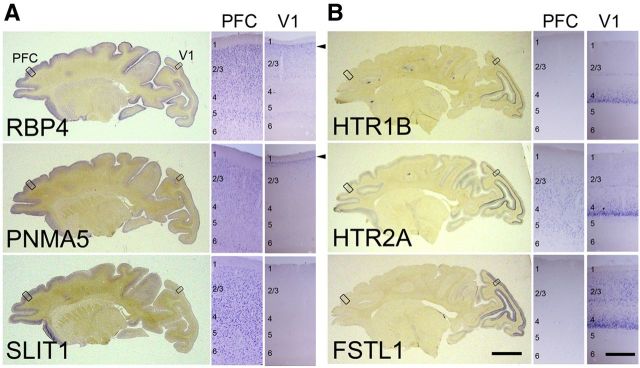
To elucidate the molecular mechanisms underlying area-selective gene expression in the primate neocortex, we used the known selectively AA-expressed genes (AA-selective genes) RBP4, PNMA5, and SLIT1, and the selectively V1-expressed genes (V1-selective genes) HTR1B, HTR2A, and FSTL1/OCC1. We first prepared parasagittal sections that were ∼5 mm apart from the central sulcus of the adult macaque cortex, and performed ISH to confirm the expression pattern of these area-selective genes (Fig. 1). As previously reported (Yamamori, 2011), ISH signals of RBP4, PNMA5, and SLIT1 mRNA are enriched in AAs including the PFC (Komatsu et al., 2005; Takaji et al., 2009; Sasaki et al., 2010; Fig. 1A), but are not generally observed in V1, except in a restricted region at the upper end of layer II (Fig. 1A, arrowheads, V1 columns). In contrast, mRNAs of HTR1B, HTR2A, and FSTL1 are preferentially expressed in V1 where they are subjected to strong activity-dependent gene expression (Tochitani et al., 2001; Watakabe et al., 2009). These ISH signals are strongest in layer IV of V1, especially in layer 4Cβ (Fig. 1B, V1 columns). Over 95% of RBP4-expressing cells express PNMA5 in the macaque AAs (Takaji et al., 2009). Similarly, HTR1B and HTR2A expressing cells also highly overlap in macaque V1 (Watakabe et al., 2009). This suggests that common mechanisms regulate the expression of each of these two groups of genes, and hence provide a good model for investigating the regulatory mechanisms for the region-specific gene expression in the macaque neocortex.
Figure 1.
ISH patterns of cortical area-selective genes in adult macaque cortex. Parasagittal sections (5 mm away from the central sulcus) of ISH for AA-selective genes, RBP4, SLIT1, and PMNA5 (A) and primary visual area (V1)-selective genes, HTR1B, HTR2A, and FSTL1 (B). Scale bar: 10 mm. A, B, Right two sides show enlargements of regions boxed in the left photo. The arrowheads indicate the expression of the AA-selective genes of RBP4 and PNMA5 in upper layer 2 in V1. Scale bar, 0.5 mm.
DNA methylation occurring at CpG dinucleotides is found throughout the vertebrate genome where it is typically associated with repression of DNA transcription (Ballestar and Wolffe, 2001). Hence, different methylation patterns in the AA- and V1-selective genes may potentially mediate the selective expression. We therefore performed bisulfite sequencing of genomic DNAs obtained from adult macaque PFC and V1 to examine the methylation status near the coding region of these area-selective genes. As shown in Figure 2, we found a striking difference in methylation levels within the promoter regions between the two sets of genes. In the AA-selective genes (RBP4, PNMA5, and SLIT1), the bases within the region preceding exon 1 are frequently enriched with CpGs that were partially methylated (∼50%) as compared with the next closest CpG island within the promoter region (>90%; Fig. 2A). In contrast, the CpGs in the vicinity of the promoter regions of the V1-selective genes (HTR1B, HTR2A, and FSTL1) were almost completely unmethylated (Fig. 2B).
There were no significant differences in the methylation state of these regions for all the genes between PFC and V1 (Fig. 2A,B, upper and the lower circles), despite the striking difference in methylation patterns of the AA- and V1-selective genes. This difference hints that methylation may still be important for control of area-selective expression, for example, if there are regional differences in the expression of methyl-CpG binding proteins that contribute to the coupling between methylation and gene expression. We therefore examined the cortical expression of methyl-CpG binding proteins.
The expression pattern of methyl-binding proteins in macaque neocortex
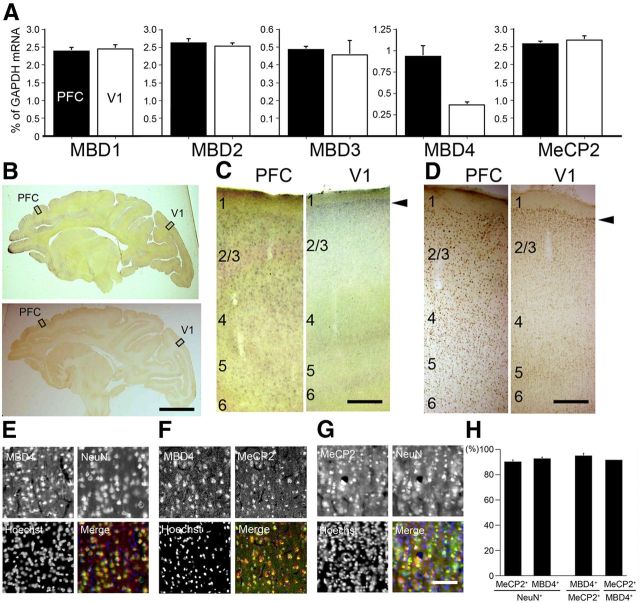
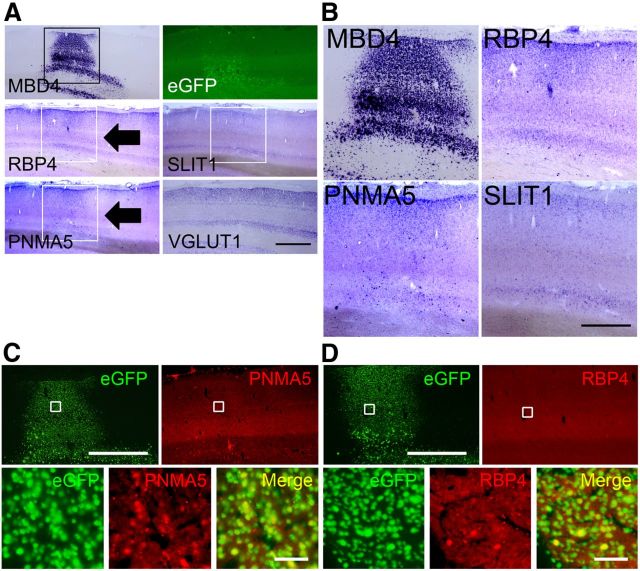
MBD1, 2, 3, and 4 and MeCP2 comprise a family of vertebrate nuclear proteins, each of which contains a methyl-CpG binding domain (MBD). The MBD domain binds regions of heavy methylated DNA, most likely CpG islands associated with promoter regions of transcriptionally silenced genes. All these MBDs, with the exception of MBD4, are regarded to form complexes with histone deacetylases (HDAcs) and contribute to HDAc-dependent gene repression (Ballestar and Wolffe, 2001). Note, however, that the direct binding of MBD4 with Sin3A, a transcriptional regulatory protein that contains paired amphipathic helix domains, and HDAc1 has been reported in cell culture (Kondo et al., 2005). Since there has been little previous investigation of the expression of these proteins in monkey neocortex, we performed real-time RT-PCR (Fig. 3A). Among these MBDs, only MBD4 showed region-selective expression, being highly expressed in PFC and very low in V1 (Fig. 3A). In addition to the full-length, alternatively spliced form (Owen et al., 2007) was also detected in PFC in semiquantitative RT-PCR (data now shown). We confirmed these expression patterns by ISH and immunohistochemistry (IHC) using the specific probes and antibodies for these five MBDs in the adult macaque neocortex (Fig. 3B–D, and others are not shown). Higher ISH signal intensities of MBD4 were observed in PFC than in V1 (Fig. 3C). IHC confirmed the high relative expression level of MBD4 in the AA cortex and the very low level in V1 (Fig. 3D). It is noteworthy that the expression of MBD4 in V1 was restricted to only layer II, for both the mRNA and protein (Fig. 3C,D, arrowheads, right columns). This pattern of expression mostly in layer II in V1 was similar to those for RBP4 and PNMA5 (Fig. 1A, arrowheads). In contrast, MBD1, MBD2, and MBD3 were expressed in both PFC and V1 in macaques (Fig. 3A). The expression patterns of AA-selective RBP and PNMA5 are very similar (Komatsu et al., 2005; Takaji et al., 2009) and show several distinct features, as described previously (Komatsu et al., 2005). The expression levels of AA-selective genes are not only high in PFC but also in the parietal association cortex and ventral visual higher AAs. However, in this study we only focused on the expression in PFC in comparison with that in V1 because our main purpose in this study was to elucidate the methylation and the control mechanism for expression of AA- and V1-selective genes. Next, to determine the nature of the cells in PFC that express MBD4, we conducted double-labeling IHC with the neuronal-specific marker, NeuN. As previously reported, the AA-selective genes we studied here are coexpressed (>90%) with the excitatory neuron marker VGLUT1 (Komatsu et al., 2005; Takaji et al., 2009), and the distribution of RBP4 and PNMA5 in the adult macaque neocortex is very similar (>95%) (Takaji et al., 2009). We found that most cells that expressed NeuN also coexpressed MBD4 (Fig. 3E). Quantification by cell counting revealed that almost 90% of the NeuN-positive neurons coexpressed MBD4 (Fig. 3H). MeCP2, one of the classic MBD family proteins, was also expressed in most neurons (Fig. 3G), consistent with the previous report (Akbarian et al., 2001). The vast majority (>90%) of the cells in PFC that expressed MBD4 also expressed MeCP2 (Fig. 3F,H). In conclusion, among the five MBD family proteins, only MBD4 protein showed a regional cortical distribution pattern similar to the AA-selective genes, and that most of the cells expressing MBD4 were neurons that also express MeCP2.
Figure 3.
Expression patterns of methyl-binding proteins in macaque neocortex. A, The expression levels of MBD1, MBD2, MBD3, MBD4, and MeCP2 in PFC (black bars) and V1 (white bars) were examined by real-time RT-PCR. The amount of each mRNA was normalized as the relative ratio to that of the internal standard, GAPDH mRNA. MBD4 expression level in PFC was higher than that in V1, whereas all four other genes showed no significant difference in expression level between PFC and V1. Error bars indicate SD. B, Parasagittal sections of the adult macaque brains were stained for MBD4 by both ISH (top) and IHC (bottom). Right, Higher magnification photomicrographs of the boxed regions of PFC and V1 (C, ISH; D, IHC). Scale bars: B, 10 mm; C, D, 400 μm. E, MBD4-positive cells express the neuronal marker NeuN. Hoechst dye was used for nuclear staining. All three markers are merged in the right bottom part. F, MBD4 and MeCP2-positive cells highly overlapped. G, MeCP2-positive cells also express the neuronal marker NeuN. Scale bar, 50 μm. H, Quantification of MeCP2- and MBD4-positive cells in NeuN-positive cells. The total proportion of MBD4-positive cells among MeCP2-positive cells, and vice versa, is shown. Three 200 × 200 μm2 windows within layer II in PFC (from three monkeys) were used for counting. Error bars indicate SD.
Relationship between the MBD4 and AA-selective gene expression
The sequence similarity among these five methyl-binding proteins is largely limited to the MBD regions. Sequence alignment of this region in murine MBDs and MeCP2 proteins shows that the MBD-like domain of MBD4 is most similar to that of MeCP2, while those of MBD1, 2, and 3 are more similar to each other than to either MBD4 or MeCP2 (Ballestar and Wolffe, 2001). We aligned the MBD-like regions of the macaque MBDs and MeCP2 proteins, and found that MBD4 and MeCP2 are also more similar to each other than to the other MBD proteins (52.3% identical and 83.1% similarity to MeCP2 vs MBD4; 38.5 ± 2.9% identical and 63.8 ± 4.0% similarity to MBD1, 2, and 3 vs MBD4). It has also been demonstrated previously that both MeCP2 and MBD4 exhibit a significant binding preference for nucleosomes containing methylated DNA (Ishibashi et al., 2008).
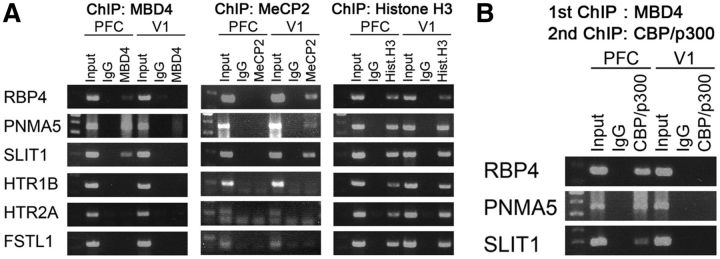
We therefore hypothesized that MBD4 binds to methylated CpGs of the promoter of the AA-selective genes (RBP4, PNMA5, and SLIT1) in PFC. To test this hypothesis, we performed ChIP. MBD4 was indeed bound to the nearest CpG islands in these AA-selective gene promoters in PFC, but bound to neither V1-selective genes nor AA-selective genes in V1 (Fig. 4A, left). This suggests that MBD4 preferably binds to CpGs of the AA-selective gene promoters in PFC. MeCP2 bound to the AA-selective gene promoters of RBP4, PNMA5, and SLIT1 in V1 (Fig. 4A, middle). MeCP2 may act together with other factors to repress the expression of the AA-selective genes in V1. To further identify the molecules that involve interaction with MBD4, we performed double ChIP and found that the CBP/p300 histone acetyltransferases coprecipitated with MBD4 bound to the AA-selective gene promoters (Fig. 4B). In summary, binding of MBD4 to the AA-selective gene promoters recruits CBP/p300 to modify the chromatin structure and likely induces the selective expression of these genes.
Figure 4.
MBD4 binds and recruits CBP/p300 at AA-selective gene promoters in macaque PFC. A, ChIP assay for the binding of MeCP2 and MBD4 to the promoter regions of six target area-selective genes in tissues obtained from macaque PFC and V1. ChIP with an antibody against MBD4 shows that MBD4 is bound to the promoter regions of the AA-selective genes of RBP4, PNMA5, and SLIT1 in PFC and not V1 tissues. MeCP2 was bound to the same genes in V1 tissues. B, Double ChIP using antibodies against MBD4 and CBP/p300 suggests the presence of specific coprecipitation of the histone acetyltransferase CBP/p300 with MBD4 in PFC, and not in V1. An IgG control did not show such coprecipitation.
To further examine the effects of MBD4 on the expression of AA-selective genes, we constructed vectors for gain of function (by overexpression) and for loss of function (knocking down endogenous MBD4 by sh-RNA expression; Fig. 5). We first transfected the MBD4 vectors in SH-SY5Y human neuroblastoma cells (and in HEK293T cells as well, data not shown), and confirmed by Western blot analysis that overexpressed MBD4 protein of correct size was formed (Fig. 5C, MBD4 overexpression lane), and by immunostaining that the protein was localized in the nuclei with immunostainings (Fig. 5B). Next, we selected and validated the vectors for sh-RNA expression to knockdown MBD4 genes. We designed five vectors that included the 21 base sequences that are conserved in human and macaque coding region to use them in human neuroblastoma (Fig. 5E–H) and the macaque neocortex (Figs. 6, 7) shRNA expression was driven by the U6 promoter (Fig. 5A, the sh numbers, #, in illustration bars). We tested these vectors for their capability to knockdown the transfected protein in SH-SY5Y (and HEK293T) cells and selected the most efficacious for the subsequent in vitro and in vivo experiments. The sh #4 and vectors knocked down >95% of the MBD4 protein in both cell lines (MBD4: sh #4, 99%; Fig. 5C,D).
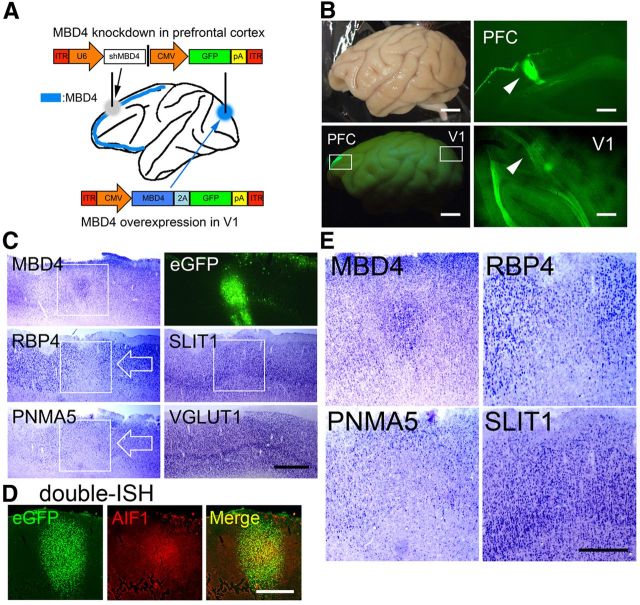
Figure 6.
Expressions of the AA-selective RBP4, PNMA5, and SLIT1 in sections including PFC in AAV-shMBD4-injected monkey. A, An illustration of the AAV1-mediated gene transfer procedures in the macaque brain. AAV1–U6-shMBD4 was injected into PFC to examine the effects of loss of function of MBD4, whereas AAV1-CMV-MBD4 was injected into V1 to examine the gain of function of MBD4. B, Four weeks later, eGFP signals were observed in the injected sites in PFC and V1. Scale bar, 10 mm. C, The ISH signals of MBD4, RBP4, PNMA5, SLIT1, and VGLUT1 (an excitatory neuron-specific marker gene) are shown. The white boxes of MBD4, RBP4, and PNMA5 indicate areas around the injection site, demonstrating decreased expression, but note that MBD4 ISH signals were enhanced at the injected site, possibly due to the damage-induced proliferation of glial cells or the migration of glial cells from uninjected regions. RBP4 and PNMA5 expression were restricted to neurons, and these signals were decreased around the injected site (white arrows). The MBD4 signals were only enhanced in the center region, with decrease observed in the signals just outside this focus, forming a donut-like appearance of MBD4 expression. Scale bar, 2 mm. D, Double-ISH of eGFP (green, left) and AlF1/Iba1 (red, middle), an activated microglial marker. A merged image (right) is also shown. Scale bar, 2 mm. E, Higher magnification photomicrographs of the white boxes shown for MBD4, RBP4, PNMA5, and SLIT1 in the top, delineating clear boundaries of the decreased areas of expressions (except for SLIT1). Scale bar, 1 mm.
Figure 7.
Expressions of AA-selective RBP4, PNMA5, and SLIT1 in the AAV-MBD4-injected monkey V1. A, Expressions of AA-selective RBP4, PNMA5, and SLIT1 in sections of V1 in AAV-MBD4-injected monkey. ISH signals of the ectopic expression of both PNMA5 and RBP4 observed in the MBD4-injected sites in V1 (black arrows). The white boxes for MBD4, RBP4, and PNMA5 indicate the injection sites within these areas showing ectopic expressions of these genes. The photomicrographs of eGFP, SLIT1 (indicated by white boxes), and VGLUT1 expressions around the injection site are also shown. Scale bar, 2 mm. B, Higher magnifications of these boxed areas, demonstrating the ectopic expressions of MBD4, RBP4, and PNMA5, but not that of SLIT1. Scale bar, 1 mm. C, D, Double-labeling fluorescence ISH of AA-selective genes (red) and transgened marker eGFP (green). Although the expression levels of PNMA5 and RBP4 varied, we were able to confirm that the ectopic expressions of PNMA5 (C) and RBP4 (D) in V1 colocalized well with eGFP expression, which demonstrated that the expressions of these genes were affected in the same cells as those with the genetic manipulation. Note that, in the loss of function experiments in PFC, the MBD4 and PNMA5 signal intensities were too low to be detected by our double-ISH protocol (data not shown), by which a signal is more difficult to be detected than by single-labeling ISH (Fig. 6C,D). Scale bars: (for C, D), top, 2 mm; bottom, 50 μm.
For our in vitro analysis of the effects of MBD4 on gene activation, we examined the AA-selective gene PNMA5 (Takaji et al., 2009) in SH-SY5Y human neuroblastoma cells. This human PNMA5 gene promoter has a CpG Island at a similar position to that of the macaque PNMA5 promoter (compare Fig. 5E, Fig. 2A). We first studied the methylation of this CpG island, which turned out to be hypermethylated in SH-SY5Y cells as in macaques (Fig. 5E, circles). MeCP2 was highly expressed in the nuclei of untransfected SH-SY5Y cells, in contrast with only low expression of MBD4 (Fig. 5F), as similar to that of adult macaque V1 (Fig. 3A–D). Overexpression of MBD4 resulted in binding of MBD4 to the PNMA5 promoter as demonstrated by ChIP (Fig. 5G, MBD4 lane). Overexpression of MBD4 also seemed to decrease MeCP2 binding, suggesting competition between MBD4 and MeCP2 for the promoter site (Fig. 5G, MeCP2 lane). The demethylation agent, 5-azacytidine, decreased binding of MeCP2 at the PNMA5 promoter (Fig. 5G, 5-AzaC row), indicating binding of MeCP2 under normal conditions at the CpG sites. Consistent with the low MBD4 expression in SH-SY5Y cells, PNMA5 was also not usually expressed in these cells (Fig. 5H, No treat). Overexpression of MBD4 could now induce PNMA5 expression (Fig. 5H, MBD4 overexp.). These results suggest that in SH-SY5Y cells, MeCP2 binds to the CpGs in the promoter region of PNMA5 to suppress transcription, and overexpressed MBD4 can bind to the same site to in turn induce the expression of this gene.
Regulation of AA-selective gene expression by MBD4 in macaque neocortex
We next examined whether this effect of MBD4 in vitro can also be demonstrated in vivo in the regulation of expression of area-selective genes. For this purpose, we used the AAV vector to manipulate MBD4 gene in the macaque neocortex. The overall scheme of this experiment was described in Figure 6A. We used the sh #4 sequence for MBD4 knockdown that was highly effective in our in vitro model (Fig. 5A–D) in PFC, and MBD4 overexpression vector in V1. And we used two adult macaque (M. fuscata) monkeys aged 2 years and 9 months and 3 years and 9 months, respectively. As shown in Figure 1A, AA-selective genes are expressed in PFC but not in V1 in these ages.
Four weeks after injection of either the AAV1-shMBD4#4 vector (for MBD4 knockdown) into macaque PFC or the AAV1-MBD4 vector (for overexpression) into V1 (Fig. 6A), the control marker eGFP was observed in PFC (Fig. 6B, PFC) and V1 (Fig. 6B, V1) at both injection sites. Then, we examined the expression of AA-selective genes close to the injection sites by ISH (Fig. 6C,E). The expression level of MBD4 mRNA decreased in response to AAV1-shMBD4#4 injection in PFC (Fig. 6C,E, MBD4). It is possible that the residual MBD4 ISH signal at the injection site may be derived from these microglia, possibly migrating from outside to the injection site (Theodore et al., 2008), or proliferating to the extent that they overwhelm the shRNA. Indeed, in a separate experiment, the expression of AIF1/Iba1, an activated microglial marker, was found to be induced at the center of the injection site (Fig. 6D), which shows the merging of this site with the eGFP expression site, whereas only the expression of eGFP was observed in the surrounding region (Fig. 6D, eGFP and Merge). The expressions of mRNAs of the AA-selective genes PNMA5 and RBP4 were suppressed by MBD4 knockdown around the injection site, whereas the expression of the AA-selective gene SLIT1 appeared to be only slightly suppressed and was not markedly affected (Fig. 6C, white arrows; E, higher magnification photos).
On the other hand, in V1 where the AAV1-MBD4 vector was injected, MBD4 mRNA expression was clearly detected (Fig. 7A,B, MBD4). The AA-selective genes RBP4 and PNMA5 were ectopically induced at this site, but not SLIT1 (Fig. 7A, black arrows; B, higher magnification photos). The fluorescence double-labeling ISH of eGFP and PNMA5 (Fig. 7C) or RBP4 (Fig. 7D) showed that the cells that were positive for PNMA5 and RBP4 were exclusively positive for eGFP, indicating that the ectopic expression of PNMA5 and RBP4 in V1 was affected in the same cells as those with the genetic manipulation of MBD4.
These results confirm that MBD4 regulates the AA-selective transcription of the RBP4 and PNMA5 in vivo, as we also demonstrated for PNMA5 in vitro. However, different control mechanisms from those for RBP4 and PNMA5 are likely involved in SLIT1 expression.
Discussion
The major findings of this study are as follows. (1) There are differences in methylation state; the high and low methylation states of the promoters for the AA- and V1-selectively expressed genes, respectively. The methylation levels were not different between PFC and V1 regions. (2) A regional difference was observed in the expression of the methyl-binding protein, MBD4. (3) MBD4 specifically bound to and activated the AA-selective genes in vivo. These regulatory mechanisms, each of which may occur at different developmental stages, activate the expression of a group of genes that play certain critical roles in the development, regionalization, and function of the primate neocortex.
Mechanisms of AA-selective gene expression
We consider that in the mitotic or postmitotic stage of corticogenesis, one or more DNA-methyltransferases (DNMTs) differentially methylate specific groups of preferred genes (Fig. 8). In mammalian embryos, there are two major cycles of epigenetic reprogramming of the genome: preimplantation development and germ cell development (Reik, 2007; Reik et al., 2001). This DNA modification event including demethylation–remethylation has been conventionally viewed as a highly stable epigenetic mark in postmitotic cells. However, recent studies have provided direct evidence that extensive active DNA modifications occur in mature neurons in vivo, including DNA demethylation and de novo methylation (Guo et al., 2011b). Elucidating the mechanisms that explain what epigenetic events occur in the area-selective gene promoters and what types of DNMT are involved in this process remains for future study. In addition, emerging evidence suggests that the hydroxylation of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) is involved in the epigenetic regulatory mechanism in the mammalian brain. That is, a recent study has indicated that the 5-hmC level markedly increases from the early postnatal stage to adulthood (Szulwach et al., 2011). It is noteworthy that the AA-selective genes RBP4 and PNMA5 change their expression patterns in the AAs after birth (Komatsu et al., 2005). It remains to be examined whether area-selective gene promoters are subjected to 5-hmC-mediated epigenetic modifications, because the bisulfite sequencing we used in this study cannot distinguish 5-mC and 5-hmC (Guo et al., 2011a).
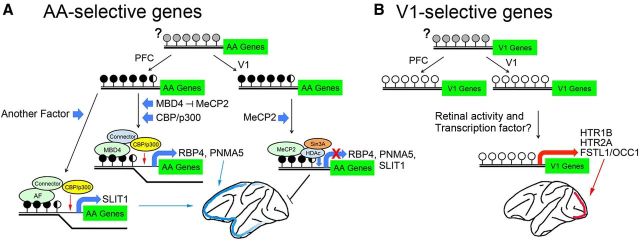
Figure 8.
Summary model of the activity- and region-specific expression of area-selective genes in the macaque monkey. A, The CpGs in the vicinity of the coding region of the adult AA-selective genes are hypermethylated (black circles). Gray circles indicate the unknown methylation status in early stages. The transcriptional corepressor of HDAc/Sin3A is possibly bound with MeCP2 at the methyl-CpGs in the promoters of the V1 neurons, whereas the expression of the AA-selective genes occurs when MBD4 binds in the same CpG position with CBP/p300 histone acetyltransferase. The candidates for the connector that mediates the binding between CBP/p300 and MBD4 include either TRADD/STAT1/FADD (Wang et al., 2000) or ROCK2/MLH1 (Tanaka et al., 2006). B, In the V1-selective genes (right schematic), the CpGs close to these genes are unmethylated (open circles). Gene expression occurs when the appropriate TFs and coactivators induced by activity are bound to the activity-dependent cis-element in the vicinity of the V1-selective genes (orange bar). Histone acetylation by CBP/p300 may also regulate transcriptional activation.
Second, MBD4 expression in a specific region (the AAs in the cortex) switches on the expression of these methylated genes. Regarding the question of when the area-selective expression occurs, Bernard et al. (2012) reported area- and layer-selective gene expressions using microarray on the macaque neocortex, and Kang et al. (2011) on human neocortex. We compared our results with those of others who conducted human (http://humanbraintranscriptome.org) and macaque neocortical expression analyses (http://www.blueprintnhpatlas.org). In their microarray data, five of the area-selective genes we examined in this study, PNMA5, SLIT1, HTR1B, HTR2A, and FSTL1 (RBP4 is not presented in there), also show quite similar area selectivity in macaques. Moreover, five of the area-selective genes, RBP4, PNMA5, SLIT1, HTR1B, and HTR2A (FSTL1 is not presented in these references), also show a quite similar area selectivity in humans. In addition, the MBD4 expression level is also higher in the medial frontal cortex than in the occipital cortex in macaques, which is consistent with our observations shown in Figure 3. In their data, the overall expression level of MBD4 is highest at 0 months (newborn), and gradually decreases with age, i.e., at 3, 12, and 48 months. In regard to the question about how the area-selective expression of MBD4 occurs, there are several possibilities. One possibility is that, in the embryonic stage, its promoters with hypermethylated and the binding of MBD4, regulate the expression by itself; however, the methylation status of its promoters is not known. Another possibility is that the expression of MBD4 is regulated by other proteins, such as the transcription factors Pax6 and Sp8, which show graded expression in cortical progenitors in mice (O'Leary and Sahara, 2008). However, these are not mutually exclusive possibilities.
A number of proteins likely bind to MBD4 to regulate this region-specific transcription. Although these key intermediates also remain to be identified, the candidates that are known to bind MBD4 are TRADD/STAT1/FADD (Wang et al., 2000) and MLH1/ROCK2 (Tanaka et al., 2006). Either complex is reported to bind to both MBD4 and CBP/p300. Histone acetylation by CBP/p300 may increase chromatin accessibility and activate transcription of AA-selective genes. MeCP2 is reported to be expressed near histone octamer levels and is bound genome wide in neurons (Skene et al., 2010). It is bound to the AA-selective gene promoters (RBP4, PNMA5, and SLIT1) in V1 where MBD4 does not exist (except for layer II), which may suppress the expression of these genes, as shown in Figure 5G.
We speculate that there may be other AA-selective genes not examined here that would show similar interactions with MBD4. Recent systematic microarray analyses described above (Bernard et al., 2012) revealed that a large group of genes displays rostrocaudal gradient expressions, which were not detectable by our RLCS method using macrodissected tissues (Yamamori, 2011). Among them, it is possible that some of these genes are AA selective similarly to RBP4 and PNMA5, i.e., their expression levels are high in AA but low in V1 (except for layer II), the expression alters after birth to show AA-selective expression, and they have a highly methylated CpG island in their promoters, although we do not know their characteristics nor the selective binding of MBD4 to their promoters.
Mechanisms of V1-selective gene expression
The expression of each V1-selective gene (HTR1B, HTR2A, and FSTL1) induced by activity in V1 is differently altered among prosimians (galagos), New World monkeys (owl monkeys, marmosets), and Old World monkeys (macaque monkeys; Takahata et al., 2012). This strongly suggests that transactivating factors including transcriptional factors (TFs), specific cis-elements, and methylation status in the promoter are varied even within the monkey phylogeny. Interestingly, in owl monkeys, the mRNA expressions of HTR1B and FSTL1 were observed in layer IV in V1 as in macaques, but monocular inactivation did not apparently influence their expression (Takahata et al., 2012). On the other hand, HTR1B is affected by activity in marmosets (Nakagami et al., 2013). None of these V1-selective genes show V1-selective expression in mice, rabbits, or ferrets (Yamamori, 2011). Although the cis-elements, TFs, and coactivators that are involved in V1-selective, activity-regulated gene expressions in a primate species-specific manner remain to be studied in the future, it is possible that epigenetic mechanisms, including methylation of V1-selective gene promoters, have suppressive effects in the nonprimate neocortex. We are currently working to identify species-specific mechanisms and their relationship with the area-selective gene expression in the primate neocortex in comparison with those in the rodent neocortex. In summary, the data presented here indicate that both the methylation level of promoter regions of the area-selective genes, and their region-specific cis-elements together, play a critical role in controlling gene expression in specific regions of the primate neocortex and the response to environmental stimuli.
Possible roles of methylation of area-selective genes in brain function
Postmitotic cortical expansion during human development occurs at different developmental periods in different cortical regions (Casey et al., 2005). Controlling the expression of area-selective genes by methylation and methyl-binding proteins may be a key underlying molecular mechanism for this regional cortical maturation. In this case, AA-selective genes may play roles in dendritic branching and spine formation in pyramidal neurons in the AAs (Cohen et al., 2011). These features have been reported as characteristic in the primate AAs (Elston et al., 1999, 2009). For example, SLIT1, one of the AA-selective markers we used in this study (Sasaki et al., 2010), has been implicated in the dendritic development of cortical neurons in rodents (Whitford et al., 2002) and in the proliferation of CNS progenitor cells (Borrell et al., 2012).
Our findings may also shed light on the underlying basis of developmental mental disorders caused by an alteration of methylation status and/or methylated DNA binding proteins. MeCP2 mutations cause Rett syndrome, a progressive neurodevelopmental disorder that is one of the most common causes of mental retardation in human females, with symptoms starting at 6–19 months of age (Amir et al., 1999). However, the precise mechanisms by which MeCP2 mutations cause Rett syndrome are unknown. MeCP2 regulates the expression of a wide range of genes, i.e., it can function as both an activator and a repressor of transcription (Chahrour et al., 2008). Although the mechanisms by which MeCP2 shares this dual function with a set of genes have not yet been clarified, it is possible that similar mechanisms underlie MBD4 function, i.e., it can function both as a transcriptional activator (Fig. 8) and repressor (Kondo et al., 2005) in the primate neocortex. Our results suggest a mechanism by which different cortical regions can develop distinct molecular and histochemical phenotypes that will contribute to the development of functional specialization of cortical regions in primates including humans.
Footnotes
This research is supported by the project “Highly Creative Animal Model Development for Brain Sciences” performed under the Strategic Research Program for Brain Sciences by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Part of this study is the result of a Grant-in-Aid for Scientific Research on Innovative Areas (Neocortical Organization) from the Ministry of Education, Culture, Sports, Science, and Technology, of Japan. We thank Drs. Andrew Moorhouse; the University of South Wales, Sydney; and Kathleen Rockland, Boston University School of Medicine, Boston, for their critical comments.
The authors declare no competing financing interests.
References
- Akbarian S, Chen RZ, Gribnau J, Rasmussen TP, Fong H, Jaenisch R, Jones EG. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol Dis. 2001;8:784–791. doi: 10.1006/nbdi.2001.0420. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MM, Serikawa K, Lemon T, Morgan R, Copeland C, Smith K, Cullen V, Davis-Turak J, Lee CK, Sunkin SM, Loboda AP, Levine DM, Stone DJ, Hawrylycz MJ, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Cárdenas A, Ciceri G, Galcerán J, Flames N, Pla R, Nóbrega-Pereira S, García-Frigola C, Peregrín S, Zhao Z, Ma L, Tessier-Lavigne M, Marín O. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Tottenham N, Liston C, Durston S. Imaging the developing brain: what have we learned about cognitive development? Trends Cogn Sci. 2005;9:104–110. doi: 10.1016/j.tics.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Tweedale R, Rosa MG. Cortical integration in the visual system of the macaque monkey: large-scale morphological differences in the pyramidal neurons in the occipital, parietal and temporal lobes. Proc Biol Sci. 1999;266:1367–1374. doi: 10.1098/rspb.1999.0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Oga T, Fujita I. Spinogenesis and pruning scales across functional hierarchies. J Neurosci. 2009;29:3271–3275. doi: 10.1523/JNEUROSCI.5216-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011a;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011b;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, So K, Cupples CG, Ausió J. MBD4-mediated glycosylase activity on a chromatin template is enhanced by acetylation. Mol Cell Biol. 2008;28:4734–4744. doi: 10.1128/MCB.00588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. From mice to men: the evolution of the large, complex human brain. J Biosci. 2005;30:155–165. doi: 10.1007/BF02703695. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Matsui R, Kato S, Hasegawa T, Kasahara H, Isa K, Watakabe A, Yamamori T, Nishimura Y, Alstermark B, Watanabe D, Kobayashi K, Isa T. Genetic dissection of the circuit for hand dexterity in primates. Nature. 2012;487:235–238. doi: 10.1038/nature11206. [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Retinol-binding protein gene is highly expressed in higher-order association areas of the primate neocortex. Cereb Cortex. 2005;15:96–108. doi: 10.1093/cercor/bhh112. [DOI] [PubMed] [Google Scholar]
- Komine Y, Nakamura K, Katsuki M, Yamamori T. Novel transcription factor zfh-5 is negatively regulated by its own antisense RNA in mouse brain. Mol Cell Neurosci. 2006;31:273–283. doi: 10.1016/j.mcn.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Kondo E, Gu Z, Horii A, Fukushige S. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol Cell Biol. 2005;25:4388–4396. doi: 10.1128/MCB.25.11.4388-4396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Cortay V, Giroud P, Berland M, Smart I, Kennedy H, Dehay C. The concerted modulation of proliferation and migration contributes to the specification of the cytoarchitecture and dimensions of cortical areas. Cereb Cortex. 2006;16(Suppl 1):i26–i34. doi: 10.1093/cercor/bhk011. [DOI] [PubMed] [Google Scholar]
- Nakagami Y, Watakabe A, Yamamori T. Monocular inhibition reveals temporal and spatial changes in gene expression in the primary visual cortex of marmoset. Front Neural Circuits. 2013;7:43. doi: 10.3389/fncir.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandiwada SL, Li W, Zhang R, Mueller DL. p300/Cyclic AMP-responsive element binding-binding protein mediates transcriptional coactivation by the CD28 T cell costimulatory receptor. J Immunol. 2006;177:401–413. doi: 10.4049/jimmunol.177.1.401. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A, Peng GH, Hsu C, Alexis U, Chen S, Blackshaw S. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron. 2009;61:234–246. doi: 10.1016/j.neuron.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RM, Baker RD, Bader S, Dunlop MG, Nicholl ID. The identification of a novel alternatively spliced form of the MBD4 DNA glycosylase. Oncol Rep. 2007;17:111–116. [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Komatsu Y, Watakabe A, Sawada K, Yamamori T. Prefrontal-enriched SLIT1 expression in Old World monkey cortex established during the postnatal development. Cereb Cortex. 2010;20:2496–2510. doi: 10.1093/cercor/bhp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Differential expression patterns of occ1-related genes in adult monkey visual cortex. Cereb Cortex. 2009;19:1937–1951. doi: 10.1093/cercor/bhn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata T, Hashikawa T, Tochitani S, Yamamori T. Differential expression patterns of OCC1-related, extracellular matrix proteins in the lateral geniculate nucleus of macaque monkeys. J Chem Neuroanat. 2010;40:112–122. doi: 10.1016/j.jchemneu.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Takahata T, Shukla R, Yamamori T, Kaas JH. Differential expression patterns of striate cortex-enriched genes among old world, new world, and prosimian primates. Cereb Cortex. 2012;22:2313–2321. doi: 10.1093/cercor/bhr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaji M, Komatsu Y, Watakabe A, Hashikawa T, Yamamori T. Paraneoplastic antigen-like 5 gene (PNMA5) is preferentially expressed in the association areas in a primate specific manner. Cereb Cortex. 2009;19:2865–2879. doi: 10.1093/cercor/bhp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nishimura D, Wu RC, Amano M, Iso T, Kedes L, Nishida H, Kaibuchi K, Hamamori Y. Nuclear Rho kinase, ROCK2, targets p300 acetyltransferase. J Biol Chem. 2006;281:15320–15329. doi: 10.1074/jbc.M510954200. [DOI] [PubMed] [Google Scholar]
- Theodore S, Cao S, McLean PJ, Standaert DG. Targeted overexpression of human alpha-synuclein triggers microglial activation and an adaptive immune response in a mouse model of Parkinson disease. J Neuropathol Exp Neurol. 2008;67:1149–1158. doi: 10.1097/NEN.0b013e31818e5e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Liang F, Watakabe A, Hashikawa T, Yamamori T. The occ1 gene is preferentially expressed in the primary visual cortex in an activity-dependent manner: a pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex. Eur J Neurosci. 2001;13:297–307. doi: 10.1046/j.0953-816X.2000.01390.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu TR, Cai S, Welte T, Chin YE. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–4512. doi: 10.1128/MCB.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Sadakane O, Shimegi S, Takahata T, Higo N, Tochitani S, Hashikawa T, Naito T, Osaki H, Sakamoto H, Okamoto M, Ishikawa A, Hara S, Akasaki T, Sato H, Yamamori T. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb Cortex. 2009;19:1915–1928. doi: 10.1093/cercor/bhn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chédotal A, Ghosh A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/S0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Yagi H, Ogura T, Mizukami H, Urabe M, Hamada H, Yoshikawa H, Ozawa K, Kume A. Complete restoration of phenylalanine oxidation in phenylketonuria mouse by a self-complementary adeno-associated virus vector. J Gene Med. 2011;13:114–122. doi: 10.1002/jgm.1543. [DOI] [PubMed] [Google Scholar]
- Yamamori T. Selective gene expression in regions of primate neocortex: Implications for cortical specialization. Prog Neurobiol. 2011;94:201–222. doi: 10.1016/j.pneurobio.2011.04.008. [DOI] [PubMed] [Google Scholar]