Abstract
The mammalian target of rapamycin (mTOR) regulates cell growth in response to various intracellular and extracellular signals. It assembles into two multiprotein complexes: the rapamycin-sensitive mTOR complex 1 (mTORC1) and the rapamycin-insensitive mTORC2. In this study, we inactivated mTORC1 in mice by deleting the gene encoding raptor in the progenitors of the developing CNS. Mice are born but never feed and die within a few hours. The brains deficient for raptor show a microcephaly starting at E17.5 that is the consequence of a reduced cell number and cell size. Changes in cell cycle length during late cortical development and increased cell death both contribute to the reduction in cell number. Neurospheres derived from raptor-deficient brains are smaller, and differentiation of neural progenitors into glia but not into neurons is inhibited. The differentiation defect is paralleled by decreased Stat3 signaling, which is a target of mTORC1 and has been implicated in gliogenesis. Together, our results show that postnatal survival, overall brain growth, and specific aspects of brain development critically depend on mTORC1 function.
Introduction
In eukaryotes, cell growth is regulated by the target of rapamycin (TOR) pathway (Wullschleger et al., 2006; Laplante and Sabatini, 2012). The mammalian ortholog mTOR (mTOR) assembles into two distinct multiprotein complexes mTORC1 and mTORC2 (Jacinto et al., 2004; Sarbassov et al., 2004). Function of mTORC1 requires raptor (gene abbreviated as rptor) and is inhibited by the immunosuppressant rapamycin (Wullschleger et al., 2006; Laplante and Sabatini, 2012). mTORC1 controls metabolism, protein translation, ribosome biogenesis, autophagy, and transcription. However, the function of mTORC1 in the developing brain has so far not been analyzed because constitutive mTOR and raptor-deficient embryos die shortly after implantation resulting from proliferation defects (Gangloff et al., 2004; Murakami et al., 2004; Guertin et al., 2006).
The role of mTOR in the brain has mainly been studied using cultured cells. A few studies addressed differentiation of neural stem cells and came to the seemingly conflicting conclusions that mTORC1 is either crucial for commitment into the neuronal (Han et al., 2008) or into the glial lineage (Rajan et al., 2003; Wang et al., 2008). Different conditions may explain these alterations, thus posing the question to which extent the chosen conditions reflect the physiological situation. In cultured neurons, mTOR is important for polarization, dendrite development, and axonal growth and guidance (Swiech et al., 2008). In particular, the complexity of the dendritic tree is reduced upon addition of rapamycin (Jaworski et al., 2005; Kumar et al., 2005), and local translation in growth cones, which is important for axon guidance, is rapamycin-sensitive (Campbell and Holt, 2001; Verma et al., 2005; Park et al., 2008). Other studies have shown that axon specification and elongation are mTORC1-dependent (Li et al., 2008). In summary, in vitro data have resulted in a variety of potential functions of mTORC1.
The role of mTOR in brain development has only been analyzed in studies of the flat-top mutant (Hentges et al., 1999) caused by a mutation in Mtor that results in a splicing defect (Hentges et al., 2001). Mutant embryos lack a telencephalon and die at midgestation. Interestingly, this effect is phenocopied by prolonged rapamycin application (Hentges et al., 2001). However, interpretation of these results is difficult because embryos with a germline deletion of Mtor die much earlier (Gangloff et al., 2004; Murakami et al., 2004). The generation of floxed alleles for rptor (Bentzinger et al., 2008; Polak et al., 2008) now allowed us to study the specific role of mTORC1 in the developing brain. We find that raptor-deficient brains are uniformly smaller because of decreased cell size and reduced cell number. Moreover, we show that differentiation of glia is impaired with little effect on neuronal differentiation.
Materials and Methods
Generation of rptor knock-out mice in the developing brain.
rptor floxed mice were generated as described previously (Bentzinger et al., 2008). They were crossed with mice that express Cre under the control of the CNS-specific nestin promoter and enhancer (Zimmerman et al., 1994; Tronche et al., 1999; Graus-Porta et al., 2001). Recombination of the floxed rptor alleles induced a frame shift and precocious stop of translation (see Fig. 1A). Genotyping was performed on tail or brain lysates using primers shown in Figure 1A: P1, 5′-ATG GTA GCA GGC ACA CTC TTC ATG; P2, 5′-GCT AAA CAT TCA GTC CCT AAT C; P3, 5′-CAG ATT CAA GCA TGT CCT AAG C; Cre-forward, 5′-TGT GGC TGA TGA TCC GAA TA; Cre-backward, 5′-GCT TGC ATG ATC TCC GGT AT. Mice that were heterozygous or homozygous for the floxed rptor allele and were negative for Cre were used as control. Mice of either sex were used in all the experiments.
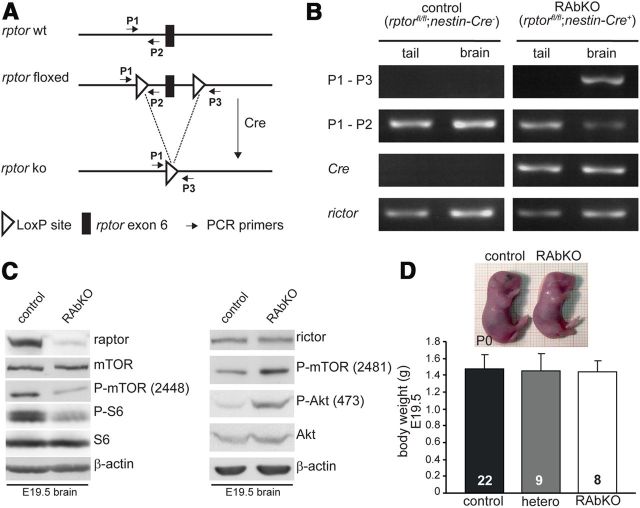
Figure 1.
Rptor knock-out in the CNS causes perinatal death. A, Schematic representation of the wild-type (rptor wt) and the targeted rptor allele before (rptor floxed) and after (rptor ko) recombination. Positions of PCR primers P1, P2, and P3 used in B are indicated. B, PCR analysis of genomic DNA isolated from brains or the tails of newborn control (rptorfl/fl; nestin-Cre−) and RAbKO (rptorfl/fl−; nestin-Cre+) mice. Tissue-specific LoxP recombination using primers P1 and P3 results in an amplicon of 204 bp for the recombined allele and no product from nonrecombined allele. PCR using primers within the rictor allele served as control. C, Western blot analysis for the indicated proteins using brain lysates of E19.5 control and RAbKO mice. The presence of some raptor protein is probably the result of meninges and blood vessels, which both do not express Cre (Graus-Porta et al., 2001). D, Photographs of newborn control and RAbKO mice (top) and body weight of E19.5 mice (bottom). Error bar indicates mean ± SD. N is indicated in the bars. Black bars represent control; gray bars, heterozygous (rptorfl/+; nestin-Cre+); open bars, RAbKO mice.
Antibodies.
Rabbit polyclonal antibodies are as follows: mTOR, S6 ribosomal protein, P-S6 ribosomal protein (Ser235/236), P-mTOR (Ser2448), P-mTOR (2481), Akt, P-Stat3 (Ser727), and cleaved Caspase 3 from Cell Signaling Technology; brain lipid-binding protein and Map2 from Millipore Bioscience Research Reagents; GAD65/67 and neurofilament from Sigma; Ki67 from Novocastra or NeoMarkers; and Tbr2 and Tbr1 from Abcam. Rabbit monoclonal antibodies are as follows: raptor, rictor, P-Akt (Ser473), P-Stat3 (Tyr705), and β-actin from Cell Signaling Technology. Mouse monoclonal antibodies are as follows: Stat3 from Cell Signaling Technology, GFAP from Millipore Bioscience Research Reagents, β-tubulin III (also called Tuj1) from Sigma, and Satb2 from Abcam. Goat polyclonal antibodies to Sox2 were from Santa Cruz Biotechnology. Rat monoclonal antibodies are as follows: Ctip2 from Abcam and BrdU from AbD Serotec. EdU detection was performed using the Click-iT imaging Kit from Invitrogen. Sox5 antibodies raised in guinea pig were a kind gift from Prof. M. Wegner (University of Erlangen).
Brain lysates for Western blots.
RAbKO and control brains were dissected at E19.5 or P0 and lysed in the presence of protease and phosphatase inhibitors. Western blots were conducted using standard procedures.
Brain sections, histochemistry and immunohistochemistry.
For immunohistochemistry, the heads of the embryos were fixed in 4% PFA overnight and then cryoprotected in 30% sucrose. Brains were embedded in OCT reagent and frozen on dry ice. For fresh frozen sections (size comparison, cresyl violet), heads were directly frozen in OCT on dry ice. The 10- or 12-μm-thick sections were prepared with a cryostat. Fresh frozen sections were postfixed for 20 min with 4% PFA. For paraffin sections, brains were fixed at least 2 d in 4% PFA and paraffin-embedded using a Shandon Pathcenter Tissue Processor. The 4-μm-thick sections, cut on a microtome, were deparaffinized and directly subjected to cresyl violet staining. For immunohistochemistry, an antigen retrieval protocol was applied by boiling the sections for 20 min in 0.05% Tween in 10 mm sodium citrate buffer, pH 6.0. Immunostainings were performed using standard procedures. For BrdU stainings, brain sections were incubated in 2 m HCl for 7 min at 37°C and then incubated for 10 min in borate buffer (0.1 m, pH 8.5) before immunostaining. EdU detection was done according to the manufacturer's instructions (Click-iT; Invitrogen).
Quantifications.
For all experiments, at least three mice per genotype were examined. For BrdU pulse-chase experiments, BrdU (50 mg/kg) was administered to a pregnant mouse by a single intraperitoneal injection and the brains of the embryos were harvested 3, 12, or 24 h later. Quantification was done by determining the number of BrdU-positive cells that were localized in all or in specific layers of the cortex above a particular length of the ventricular zone. Thus, the normalized numbers given in the figures represent cells in a specific column of the developing cortex. In contrast to the counting of BrdU-positive cells per total cell number, this paradigm minimizes the introduction of artifacts because of the size difference between the cells in controls and RAbKO mice. Similar quantification was applied for PH3 or Caspase 3-positive cells. Borders between cortical regions R1, R2, and R3 were defined in stainings for BrdU, Sox2, and Tbr2 as described in Figure 4A. All quantifications were carried out in anatomically matched regions. The total number of cells was determined by counting the number of Hoechst-positive nuclei in at least 3 sections per animal in a defined area that spanned from the ventricle to the pial surface. The percentage of cells stained with a layer-specific marker was determined by counting the number of positive cells/total number of DAPI-positive cells in the respective layer. For analysis of the cell cycle length, pregnant dams were injected first with EdU and BrdU 1.5 h apart and killed 30 min later. Analysis of 4 sections per animal of 5 control and 5 RAbKO was done as described previously (Martynoga et al., 2005). The total number of proliferating cells was determined on adjacent sections using antibodies to Ki67 and costaining with DAPI was used to assure that all cells at E16.5 were Ki67-positive. The paired t test was used for statistical analysis. The amount of DNA in tissue homogenates was measured using the method described previously (Labarca and Paigen, 1980).
Figure 4.
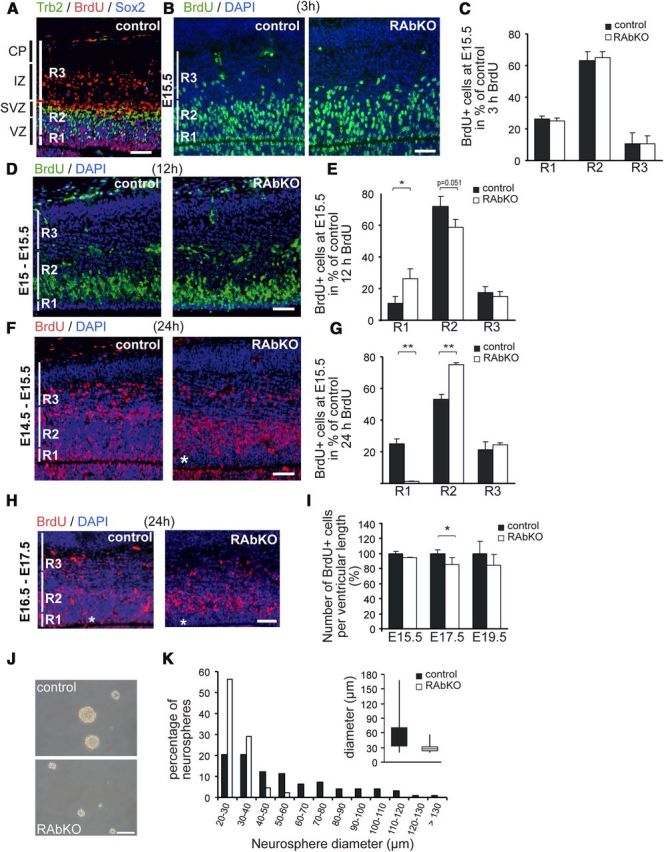
Depletion of raptor alters localization and number of proliferating precursor cells. A, Regions used for counting proliferating cells (BrdU-positive; red) are indicated as R1–R3 and are defined by staining E15.5 cortical sections from control mice with antibodies to Tbr2 (green) and Sox2 (blue). Region R1 is defined by the exclusive presence of Sox2-positive and Tbr2-negative cells. Region R2 contains all Tbr2-positive cells (which are also Sox2-positive). Region R3 is defined by the absence of both Sox2- and Tbr2-positive cells. B, D, F, H, Staining for BrdU (green in B, D; red in F, H) and DAPI (blue) at indicated time points. Immunostaining and quantification of BrdU-positive nuclei: B, C, 3 h after BrdU injection; D, E, 12 hours after BrdU injection; F, G, 24 h after BrdU injection. There is a shift in the localization of proliferating nuclei with progression through the cell cycle. H, Injection of BrdU at E16.5 and analysis at E17.5. *RAbKO mice have only few BrdU-positive cells in R1. I, Quantification of BrdU-positive cells per ventricular length in the entire cortical column. J, Micrographs of secondary neurospheres derived from E19.5 brains. There is a pronounced difference in size. K, Distribution of the diameters of secondary neurospheres. Size distribution of RAbKO neurospheres is shifted to the left. Inset, Box plot of the median size of the neurospheres. Data are from 100 control and 91 mutant neurospheres. C, E, G, I, Data are mean ± SD. N ≥ 3 mice per genotype. Not significant: p > 0.05. *p < 0.05 (two-tailed t test). **p < 0.01 (two-tailed t test). Scale bars, 100 μm. Black bars represent control; open bars, RAbKO mice. VZ, Ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate.
Neurospheres.
Neurospheres were prepared from P0 telencephali as described previously (Giachino et al., 2009). To determine the capacity of secondary neurosphere formation and growth, cells were plated at a clonal density of 3000 cells/ml (Sirko et al., 2007). Six representative images were taken for each well of a 6-well plate to determine neurosphere number and diameter. For differentiation, neurospheres were split after 5–6 d and plated onto coverslips coated with 15 μg/ml poly-L-ornithine and 40 μg/ml laminin (150,000 cells/cm2). The dispersed cultures were differentiated in neurosphere medium lacking FGF2, EGF, and heparin and fixed after 5 d with 4% PFA (Giachino et al., 2009).
Hippocampal cultures.
Hippocampi were dissected from E16.5 mice in HBSS (Sigma) and incubated for 10 min at 37°C in HBSS containing 0.05% trypsin and 0.02% EDTA. Hippocampi were washed twice with HBSS, triturated in plating medium (DMEM, 0.2 mg/ml l-glutamine, 1% penicillin/streptomycin, and 10% FCS), and plated at a density of 100,000 cells/cm2 on glass coverslips coated with 10 μg/ml poly-d-lysine. After 3 h, the medium was replaced by growth medium (Neurobasal medium; Invitrogen), 0.2 mg/ml l-glutamine, 1% penicillin/streptomycin, and 2% B27 (Invitrogen). After 4 d, cells were cotransfected with a plasmid expressing GFP from the synapsin promoter and either a Cre- or a control plasmid, which both contained the CMV promoter. Cells were fixed after 14 d in culture and imaged by confocal microscopy.
Results
mTORC1 controls embryonic brain size and is required for postnatal survival
To address the role of mTORC1 in brain development, we conditionally deleted rptor (gene encoding raptor) in neural progenitors using rptor floxed mice (Bentzinger et al., 2008) and the well-described nestin-Cre mice (Tronche et al., 1999; Graus-Porta et al., 2001). In those rptorfl/fl; nestin-Cre+ (RAbKO) mice (Fig. 1,A), the floxed exon 6 in rptor was successfully deleted in brain but not in tails as verified using genomic DNA (Fig. 1B). Consequently, the level of raptor was very low in brain lysates from RAbKO mice (Fig. 1C). The low levels of raptor remaining in the brain are most likely derived from meninges and blood vessels, which are not targeted by nestin-Cre. Phosphorylation of mTOR at serine 2448, indicative of mTORC1 activity (Copp et al., 2009), and of the mTORC1 target S6 was strongly decreased in the brains of RAbKO mice (see Fig. 1C; Table 1 for quantification). In contrast, abrogation of mTORC1 seemed not to affect the function of mTORC2 as the protein level of rictor and phosphorylation of mTOR at the mTORC2 site serine 2481 (Copp et al., 2009) and of Akt/PKB at serine 473 (Sarbassov et al., 2005) was not abrogated (Fig. 1C, right). To the contrary, phosphorylation at both sites was rather increased in RAbKO mice, consistent with previous results from mice in which rptor was conditionally deleted in other tissues (Bentzinger et al., 2008; Polak et al., 2008; Shende et al., 2011). These data are strong evidence that mTORC1 activity was eliminated and that mTORC2 activity was not reduced. RAbKO mice were born at the expected Mendelian ratio (52.3% rptorfl/+;nestin-Cre− and rptorfl/fl;nestin-Cre−, 25.2% rptorfl/+;nestin-Cre+ and 22.4% rptorfl/fl;nestin-Cre+) and their body weight was normal (Fig. 1D). However, RAbKO mice became cyanotic and died within a few hours after birth (data not shown). Their stomachs never contained milk, and their breathing was irregular (gasping), suggesting that the mice died of respiratory failure. Innervation and the positioning of the postsynaptic acetylcholine receptors at the neuromuscular junctions of RAbKO mice were indistinguishable from control mice (data not shown), indicating that the respiratory difficulties might be based on the malfunctioning of higher brain centers involved in respiratory control.
Table 1.
Quantification of Western blot bands from whole brain lysate of E19.5 control and RAbKO micea
| Protein | % of control, mean | % of control, SD | p | N |
|---|---|---|---|---|
| raptor | 22 | 4 | 2.4E-5 | 3 |
| P-S6 | 30 | 12 | 1.6E-4 | 4 |
| GFAP | 48 | 22 | 0.013 | 4 |
| Stat3 | 69 | 10 | 1.1E-4 | 4 |
| P-Stat3-727 | 49 | 19 | 0.026 | 4 |
| P-Stat3-705 | 41 | 6 | 4.1E-4 | 4 |
a The values are normalized to the intensity of the bands observed in control mice.
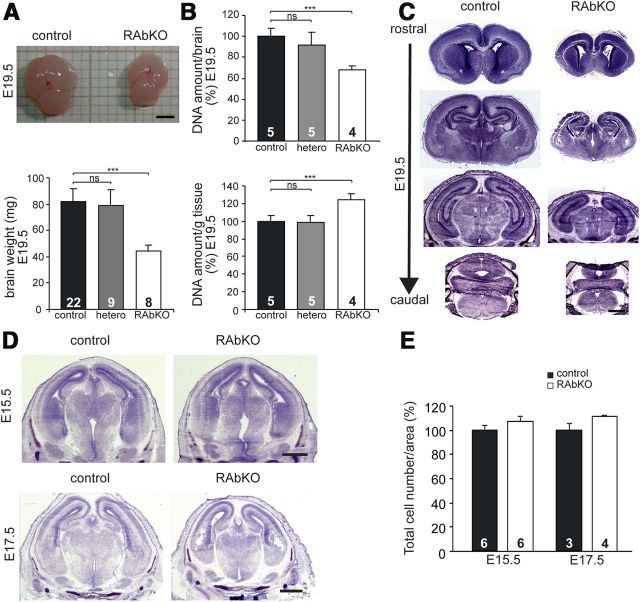
The brains of RAbKO mice at E19.5 appeared smaller and their weight was approximately only half compared with controls (Fig. 2A), even though the body weight was unchanged (Fig. 1D). The difference in brain size could be caused by a decrease in cell number or cell size, both processes that have been reported to be influenced by mTOR (Wullschleger et al., 2006; Laplante and Sabatini, 2012). The total amount of DNA per brain was >30% lower in RAbKO brains compared with controls, indicating that the number of cells was reduced (Fig. 2B, top). However, the relative amount of DNA per gram wet weight was increased in RAbKO mice to >120% compared with controls, indicating that cell density was increased (Fig. 2B, bottom). DNA amount was also increased in RAbKO mice when normalized to the amount of protein (control: 100 ± 8.4%, N = 5 mice; RAbKO: 142 ± 20.7%, N = 3 mice; data represent mean ± SD). These results indicate that the brain of the RAbKO mice contains fewer and smaller cells. Despite the difference in size, the overall structure of the brain in RAbKO mice was normal at birth (Fig. 2C). The difference in brain size was detectable at E17.5 (Fig. 2D, bottom) but not at E15.5 (Fig. 2D, top). Although there was a trend in E17.5 RAbKO mice for an increased cell density compared with control mice, this difference did not reach significance (Fig. 2E), indicating that the smaller size of RAbKO brains at this developmental period was mainly the result of changes in cell number. Together, these data indicate that the microcephaly of RAbKO mice at birth is the result of a decrease in cell number and cell size. The size difference becomes apparent between E15.5 and E17.5 and increases progressively until birth.
Figure 2.
Nestin-Cre-mediated deletion of rptor causes microcephaly. A, The brain from E19.5 RAbKO mice is considerably smaller (top) and weighs less (bottom). B, Relative DNA amount per brain (top) and relative DNA amount per gram wet weight (bottom). C, Cresyl violet-stained brain sections of E19.5 control and RAbKO mice at different levels in the rostrocaudal axis. D, Cresyl violet-stained sections at E15.5 (top) and E17.5 (bottom). The brain of RAbKO mice is smaller at E17.5. E, Quantification of cortical cell number as measured by Hoechst staining. A, B, Error bars indicate mean ± SD. E, Error bar indicates mean ± SEM. N is indicated in the bars. ***p < 0.001 (two-tailed t test). ns, Not significant (p > 0.05). Scale bars: A, 2 mm; C, D, 1 mm.
Raptor deficiency increases apoptosis and affects cell cycle duration
To address the mechanism involved in the reduced cell number in RAbKO mice, the presence of apoptotic cells was assessed by staining brain sections with antibodies to cleaved Caspase 3. Only a few apoptotic cells were found in E15.5 and E17.5 RAbKO mice (Fig. 3A,B). However, at E19.5, the number of apoptotic cells among the β-tubulin III (Tuj1)-positive neurons was ∼8 times higher in RAbKO mice than in control mice (Fig. 3A,B). Staining with the neuronal marker Satb2 underlined that the vast majority of the caspase 3-positive nuclei were neurons (Fig. 3C). Apoptotic cells were not unique to cortical regions but could also be found at E19.5 in other brain regions, such as the hippocampus (see Fig. 7C) or the thalamus (data not shown). This suggests that cell death of neurons contributes to the reduced cell number in the brain of RAbKO mice at late developmental stages but does not contribute to the size reduction at E17.5.
Figure 3.
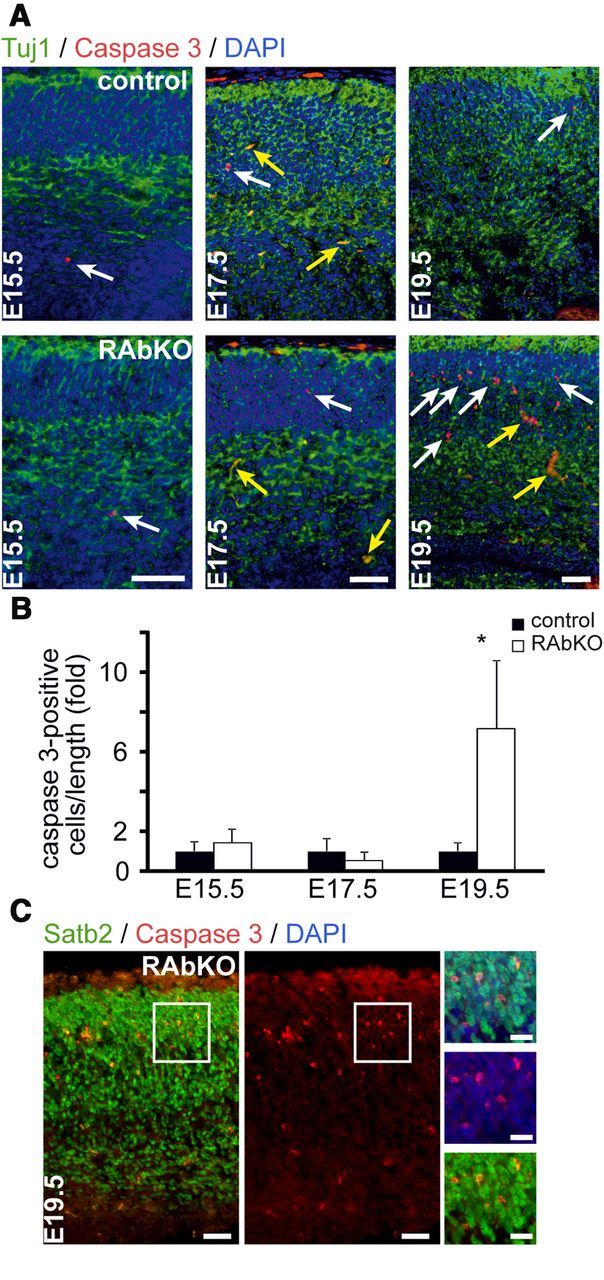
Loss of raptor leads to increased cell death during late brain development. A, Examination of apoptosis by staining for cleaved Caspase 3, β-tubulin III (Tuj1), and DAPI. White arrows indicate apoptotic nuclei; yellow arrows indicate blood vessels. B, Quantification of the number of caspase 3-positive cells per ventricular length in the cortex. The number of apoptotic cells was set to 1 in control mice. C, Staining for cleaved Caspase 3 and Satb2. Apoptotic cells are positive for the neuronal marker Satb2 (inset). Data are mean ± SD; N ≥3 animals per genotype. *p < 0.05 (two-tailed t test). Scale bars: A, C, 50 μm; C insets, 25 μm.
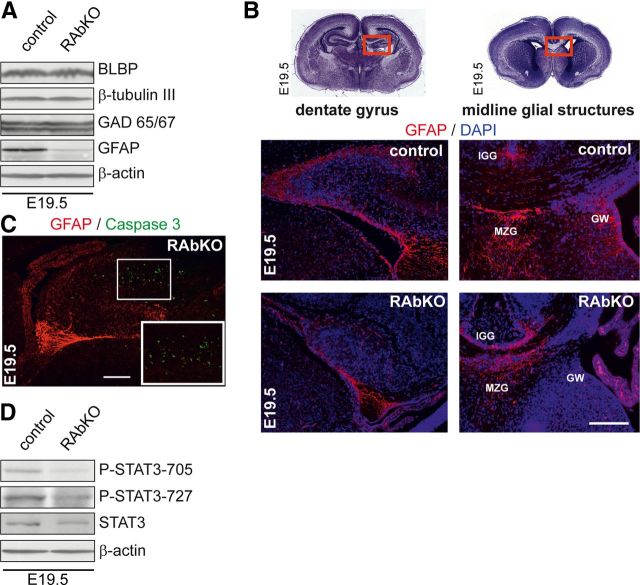
Figure 7.
Astrocytic differentiation is impaired in RAbKO brains in vivo. A, Western blot analysis of E19.5 brain lysates probed for indicated markers of neural development. B, Top, Cresyl violet stainings of brain sections from E19.5 mice. Red boxes represent regions shown below: sections of dentate gyrus and midline glial structure regions stained with antibody to GFAP (red) and DAPI (blue). IGG, Indusium griseum glia; MZG, midline zipper glia; GW, glial wedge. C, Analysis of apoptotic cells in E19.5 hippocampus of RAbKO mice stained for GFAP and Caspase 3. Apoptotic cells are not GFAP-positive. The white frame represents the magnified inset. D, Western blot analysis of E19.5 brain lysates probed for indicated proteins. Scale bars: B, C, 200 μm.
In search for a mechanism responsible for the size difference at the early stages, we assessed cell cycle kinetics and cell differentiation. As mTORC1 has been implicated in controlling G2/M phase transition during the cell cycle (Ramirez-Valle et al., 2010), we analyzed proliferation in embryonic RAbKO mice by BrdU labeling in utero. Radial glial cells in the ventricular zone of the developing cortex display phase-related interkinetic nuclear migration during cell cycle progression (Gotz and Huttner, 2005). DNA replication takes place in the basal region, whereas mitosis occurs in the apical region of the ventricular zone. To determine nuclear position, the developing cortex was divided into three regions R1 to R3, based on Sox2 and Tbr2 staining. R1 covered the apical VZ, R2 the basal VZ and the subventricular zone, and R3 the intermediate zone and the cortical plate (Fig. 4A). E15.5 RAbKO embryos labeled with BrdU 3 h before death did not show differences in the distribution of BrdU-labeled cells in the developing cortex compared with controls (Fig. 4B,C). When RAbKO mice were pulsed with BrdU at E15 and analyzed 12 h later, a significantly larger proportion of the BrdU-labeled cells was detected in R1 at the expense of labeled cells in R2 (Fig. 4D,E). After one pulse of BrdU at E14.5 and analysis 24 h later, approximately one-fourth of the BrdU-positive nuclei in control mice were located in R1, whereas in RAbKO mice, only very few BrdU-positive cells were found in R1 (Fig. 4F,G). In contrast, significantly more BrdU-positive cells were present in R2 in the RAbKO mutants (Fig. 4F,G). This nuclear position phenotype was also observed when BrdU was injected at E16.5 and mice were analyzed at E17.5 (Fig. 4H). Moreover, the total number of BrdU-positive cells in a cortical column of a given length was reduced in RAbKO mice at E17.5 but was not changed at E15.5 and E19.5 (Fig. 4I). Thus, the labeling of cells in S-phase by one pulse of BrdU at E16.5 results in fewer BrdU-positive cells along the entire cortical column 24 h later but not at E15.5 or E19.5.
Loss of raptor affects neurosphere size
Neurosphere formation and maintenance are a surrogate assay for neural stem cells (Reynolds and Weiss, 1996). Thus, we isolated and cultured spherogenic neural stem/progenitor cells from the telencephalon of P0 mice. The number of secondary neurospheres generated from RAbKO and control animals was similar (control, 10.9 ± 3.04; RAbKO, 12.7 ± 5.29; N ≥15; p > 0.05), whereas the size of RAbKO-derived neurospheres was strongly reduced (Fig. 4J,K). Compared with control cultures, the size distribution in neurospheres from RAbKO mice was shifted toward smaller diameters (p < 1.384 × 10−12; Wilcoxon two-sample test). Thus, spherogenic stem cells seemed not to be affected by loss of raptor, but the proliferative capacity of cells within the neurospheres was decreased. Interestingly, a similar proliferation defect was reported for embryonic stem cells deficient for raptor or mTOR (Gangloff et al., 2004; Murakami et al., 2004; Guertin et al., 2006).
Cell cycle exit and proliferation are reduced in raptor-deficient neural progenitor cells
The changes in temporal–spatial distribution of progenitor cells and the reduced number of BrdU-positive cells at E17.5 after a 24 h pulse indicated potential defects in specific periods of the cell cycle. To test whether loss of raptor affected cell cycle exit of progenitors, BrdU was injected into pregnant mice at day 14.5, 16.5, or 18.5 of gestation and cell cycle exit of the progenitors cells of the embryos was examined by costaining for BrdU and Ki67 24 h after injection (Fig. 5A,B). The proportion of BrdU-positive progenitors that exited the cell cycle (i.e., became Ki67-negative) was not affected at E15.5 but was reduced by ∼50% in E17.5 and E19.5 RAbKO cortices (Fig. 5A,B). Hence, precursor cells in the cortex of RAbKO mice in late embryonic development may remain longer in the cell cycle. To address this directly, cell cycle length of control and mutant progenitor cells was tested at E16.5 by EdU/BrdU double-labeling experiments (Martynoga et al., 2005). Cortical progenitors of RAbKO embryos showed a significant increase in length of the S-phase (Ts) and of the entire cell cycle (Tc) compared with control (Fig. 5C). Further, there were significantly fewer Sox2-positive apical progenitor cells (Fig. 5D,F) and a slightly, although not significantly, lower number of Tbr2-positive basal progenitor cells (Fig. 5E,F) in E17.5 mutant embryos. In addition, the number of PH3-positive apical progenitor cells (marker for cells in M-phase) that lined the ventricular surface was lower in the mutant at E17.5 (Fig. 5G,H), but not at E15.5 (Fig. 5H). Thus, all these data point to changes in cell cycle characteristics upon loss of raptor in cortical progenitors.
Figure 5.
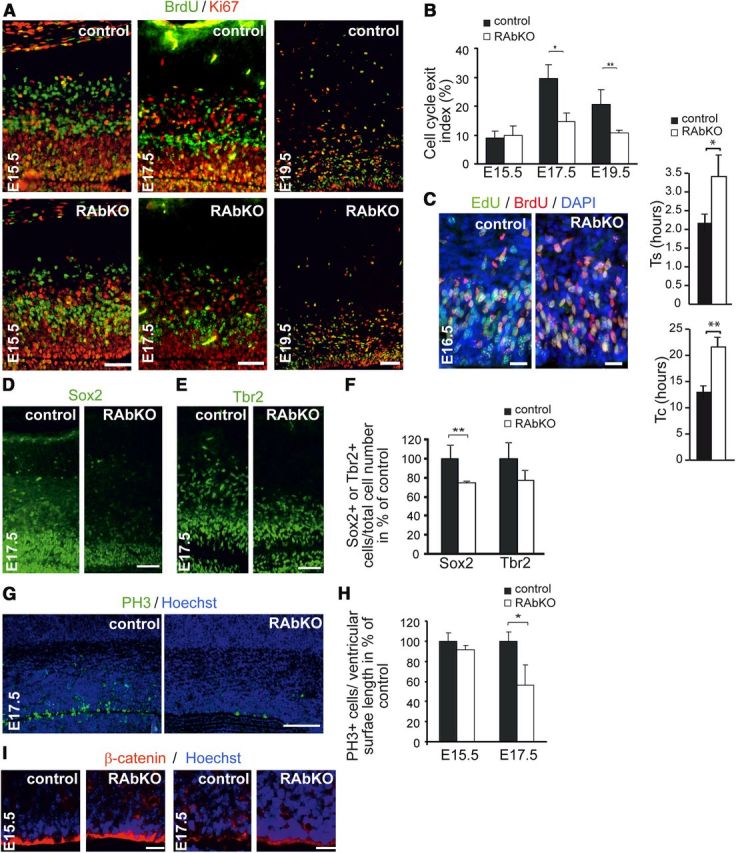
Loss of raptor causes aberrant cell cycle exit and affects cell cycle length. A, Staining of brain sections for BrdU (green) and Ki67 (red) at indicated time points. B, Quantification of BrdU+/Ki67− as percentage of all BrdU+ cells (cell cycle exit index). At E17.5, fewer cells leave the cell cycle in RAbKO mice. C, Cell cycle length analysis at E16.5. S-phase and total cell cycle length are increased in RAbKO mice. Staining of E17.5 cortical sections with antibodies to Sox2 (D) or Tbr2 (E). F, Quantification of the number of Sox2+ and Tbr2+ cells per total cell number. G, Analysis of mitosis at E17.5 by staining for PH3. H, Quantification of mitotic nuclei at E15.5 and E17.5. I, Staining for β-catenin in the ventricular zone of cortical sections from E15.5 and E17.5 control and RAbKO mice. B, F, H, Error bars indicate mean ± SD. C, Error bar indicates mean ± SEM. N ≥ 3 mice per genotype. N = 5 control and 5 RAbKO mice. *p < 0.05 (two-tailed t test). **p < 0.01 (two-tailed t test). Scale bars: A, D, G, 100 μm; C, I, 20 μm.
As loss of cell polarity or detachment from the apical membrane after mitosis could also result in proliferation defects (Loulier et al., 2009), we also examined the structure of radial glial cells in the cortex of RAbKO embryos. Like in control embryos, radial glia in the ventricular zone of RAbKO mutants showed a continuous and apical staining for the adhesion complex protein β-catenin (Fig. 5I). Thus, a disturbed polarity of radial glia cannot explain the observed changes in progenitor proliferation and differentiation in the cortex of RAbKO brains. In summary, these data suggest that changes in cell cycle characteristics are the main contributor to the change in brain size in RAbKO mice at E17.5. Lengthening of the cell cycle is likely to reduce expansion of the progenitor pool. In addition, the delay in cell cycle exit points to potential defects in the differentiation process of mutant progenitor cells at later stages of development, when gliogenesis starts in the mouse cortex.
Effect of raptor deficiency on cortical layering
Cortical precursor proliferation, differentiation, and migration of neurons are intimately linked (Gotz and Huttner, 2005; Baranek et al., 2012). Therefore, we examined the production of differentiated cells in the cortical plate from E12.5 and E14.5 progenitors by BrdU pulse-chase experiments. When progenitors were labeled at E12.5, no differences in BrdU-labeled progeny in the cortical plate at E19.5 were observed and BrdU-labeled cells were distributed throughout the cortical layers in both genotypes (Fig. 6A). However, when labeled at E14.5, cortical progenitor-derived cells showed a different cellular distribution in RAbKO mice compared with controls at E19.5. Whereas in control mice most BrdU-positive cells populated the upper cortical plate (Fig. 6B), the labeled cells in RAbKO mice remained concentrated in the ventricular zone/subventricular zone/intermediate zone and fewer cells born at E14.5 reached the cortical plate (Fig. 6B). As a consequence, the ratio between BrdU-positive cells in the cortical plate and in the ventricular zone/subventricular zone/intermediate zone was significantly reduced in RAbKO mice (Fig. 6C). Despite these initial changes, the overall organization within the mutant cortex seemed normal at E19.5 (Fig. 6D). To detect potentially misguided or misspecified neurons, the distribution of layer-specific markers was assessed by immunohistochemistry at E19.5 (Fig. 6E). The expression pattern of the examined markers, including Satb2 (for layers 2–4), Sox5 (for subplate and layers 5 and 6), Tbr1 (for layer 6), and Ctip2 (for layer 5) was comparable between control and mutant, and no obvious layering defects were detected (Fig. 6E). However, in RAbKO cortices, the respective layers were narrower compared with controls, and the cells appeared more densely packed (Fig. 6E). Thus, these findings suggest that loss of raptor might affect the timing of the differentiation/migration process of neurons (Fig. 6B,C). However, their specification and correct positioning within the cortical layers appear to be unaffected (Fig. 6E).
Figure 6.
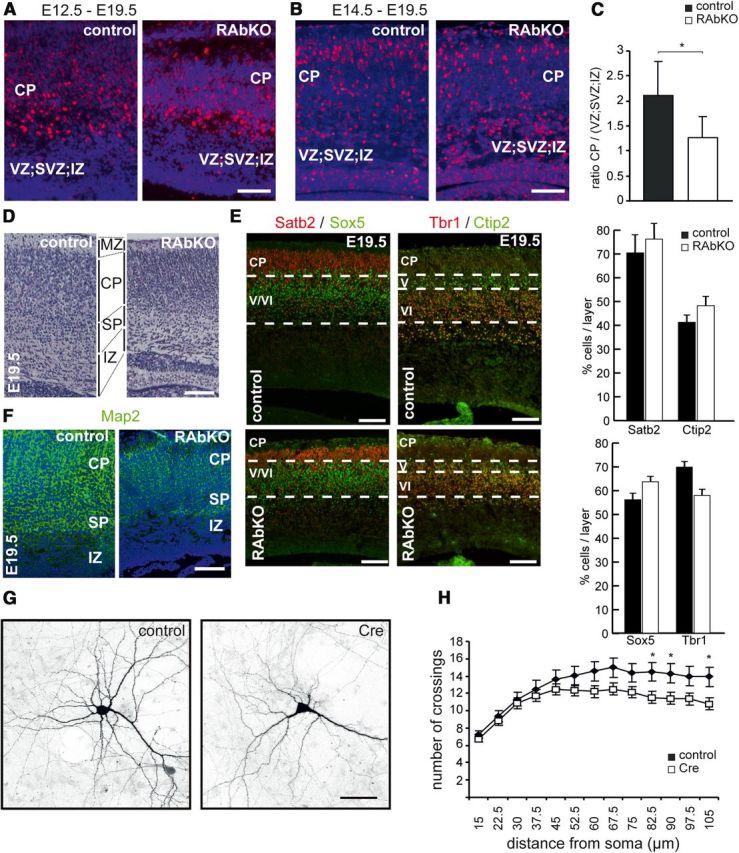
RAbKO mice show changes in the localization of late-generated neurons without affecting overall cortical layering. A, B, BrdU (red) and DAPI (blue) staining in E19.5 cortical sections of mice that were injected with BrdU at E12.5 (A) or E14.5 (B). Fewer BrdU-positive cells reach the CP when labeled at E14.5. C, Quantification of the distribution of BrdU-labeled cells from E14.5-injected mice. D, E19.5 cortical sections stained with cresyl violet. MZ, Marginal zone; CP, cortical plate; SP, subplate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. E, E19.5 cortical sections stained with antibodies to Satb2 (red) and Sox5 (green), Tbr1 (red), and Ctip2 (green). The percentage of cells positive for indicated markers was determined by counting the number of positive cells/total number of cells (DAPI-positive). Data are mean ± SEM; N = 3 mice. F, E19.5 cortical sections stained with antibodies to Map2 (green) and Hoechst (blue). G, Confocal picture of cultured hippocampal neurons isolated from rptorfl/fl mice that were transfected with control plasmid (control) or a Cre-expressing construct (Cre) at 4 DIV and analyzed at 14 DIV. Cells were cotransfected with a GFP-expression plasmid. H, Sholl analysis of Cre-expressing and control neurons. C, N = 5 mice per genotype. H, N = 5 control and 4 RAbKO mice. *p < 0.05 (two-tailed t test). C, Data are mean ± SD. H, Data are confidence intervals. Scale bars: A, B, D–F, 100 μm; G, 50 μm.
We also observed that Map2 staining (a dendritic marker) was weak in the RAbKO mice compared with controls, indicative of a reduction of neuropil in the cortical plate of raptor-deficient mice (Fig. 6F). To test whether the low Map2 signal and the higher cell density in RAbKO cortex were the consequence of a reduced dendritic complexity, we also examined neurite outgrowth in raptor-deficient neurons in vitro. We deleted rptor from E16.5 primary hippocampal neurons of floxed mice after 4 d in vitro by transfection with a Cre-expression vector. Transfected neurons were visualized by cotransfection with a GFP-expressing vector (Fig. 6G). Sholl analysis 10 d after transfection showed a significant decrease in the complexity of the neurites compared with control-transfected neurons (Fig. 6G,H). These results are also in agreement with previous work using rapamycin or RNAi against mTOR (Jaworski et al., 2005; Kumar et al., 2005). Thus, the decreased Map2 staining in cortical sections of RAbKO mice could be the result of a reduction in the neuronal complexity.
Glial differentiation is impaired in raptor-deficient brains
mTORC1 is important for neural stem/progenitor cell differentiation into neurons and glia (Rajan et al., 2003; Han et al., 2008; Wang et al., 2008). Thus, we analyzed the expression of neuronal and glial proteins in the brains of E19.5 control and RAbKO mice by Western blot analysis. We did not detect significant changes in the expression of the neuron-specific isoform β-tubulin III or GAD65/67, a marker for a subpopulation of inhibitory interneurons (Zhu et al., 1999), or brain lipid-binding protein, a radial glia marker reflecting the number of neuronal precursors (Hartfuss et al., 2001) (Fig. 7A). In contrast, the astrocytic marker GFAP was strongly reduced in RAbKO brain lysates (Fig. 7A; Table 1). In newborn mice, when GFAP+ cells start to populate the brain (Sancho-Tello et al., 1995; Shu et al., 2003), GFAP immunostaining in RAbKO brains was weaker compared with controls in the granule cell layer of the dentate gyrus and in the midline glial structures: the indusium griseum glia, the midline zipper glia, and the glial wedge (Fig. 7B). Although apoptosis was increased in the hippocampus of RAbKO mice at E19.5, the cells positive for Caspase 3 were not GFAP-positive (Fig. 7C). In summary, these results suggest that mTORC1 is important for the generation of glial cells.
mTOR signaling has been linked to glial differentiation via its ability to phosphorylate Stat3 (Yokogami et al., 2000; Rajan et al., 2003), which regulates astrocyte development (Johe et al., 1996; Bonni et al., 1997). Phosphorylation of Stat3 was indeed decreased in brain lysates of newborn RAbKO compared with control mice (Fig. 7D; Table 1). We also found a concomitant, although less pronounced, loss of Stat3 protein, which might be the result of instability of the nonphosphorylated Stat3 or the loss of Stat3-expressing cells in the brain lysate. This result suggests that failure of mTORC1-mediated activation of Stat3 may be the cause for the decrease in GFAP-positive cells.
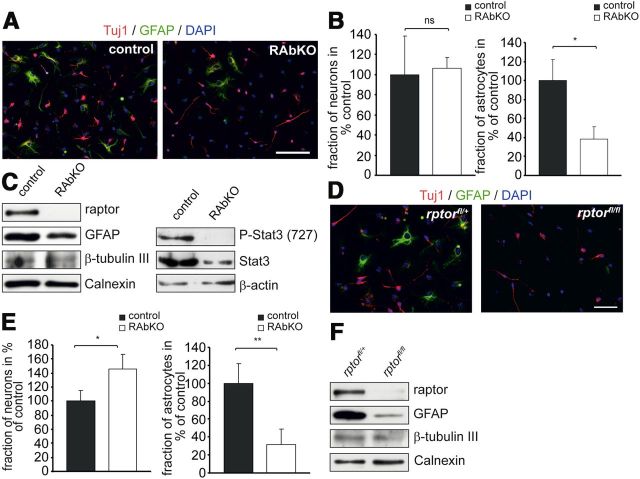
To address whether the reduction in GFAP in RAbKO mice was the result of a reduced ability of RAbKO progenitors to differentiate into the astroglial lineage, differentiation was examined in dissociated neurosphere cultures from RAbKO and control mice. Whereas the number of β-tubulin III (Tuj1)-positive neurons was not changed, the number of GFAP-positive cells was much lower in 5-day-old cultures from RAbKO mice (Fig. 8A,B). In addition, Western blot analysis confirmed that levels of GFAP, but not of β-tubulin III, were lower in RAbKO-derived cell cultures and those changes correlated with a low level of Stat3 phosphorylation (Fig. 8C). To examine whether the changes in glial differentiation were the result of inherent differences in the stem cell pool of RAbKO mice, we ablated rptor acutely from neural stem cells by culturing cells from heterozygously or homozygously floxed rptor mice followed by infection with a Cre-expressing adenovirus. The acute removal of raptor also resulted in fewer GFAP-positive cells (Fig. 8D,E) and a reduction in GFAP (Fig. 8F). In addition, an increase in the number of β-tubulin III-positive immature neurons was observed (Fig. 8D,E). In conclusion, these results indicate that mTORC1 is important for the proper differentiation of neural progenitor cells into the glial lineage.
Figure 8.
Raptor regulates astrocytic differentiation in vitro. A, Cells from dissociated neurospheres after 5 d of differentiation. Cells were stained with antibodies against β-tubulin III (Tuj1; red), GFAP (green), and DAPI (blue). B, Relative number of neurons (left) and astrocytes (right) derived from dissociated neurospheres. C, Western blot analysis of lysates from dissociated neurosphere cultures grown under the same conditions as in A probed with antibodies against the proteins indicated. D–F, Analysis of differentiation in E19.5 rptorfl/+ and rptorfl/fl neurospheres transduced with Cre-expressing adenoviruses. D, Micrographs of neurosphere-derived cells after differentiation. After 5 d, cultures were stained with Tuj1 antibody (red), anti-GFAP antibody (green), and DAPI (blue). E, Quantification of the fraction of neurons (left) and astrocytes (right) in percentage of control. F, Western blot analysis of lysates from cultures grown under the same conditions as in D probed for the indicated marker proteins. Sample number in E is 4 mice per genotype. Error bars indicate mean ± SD from three independent experiments. *p < 0.05 (two-tailed t test). **p < 0.01 (two-tailed t test). ns, Not significant (p > 0.05). Scale bars: A, D, 100 μm.
Discussion
Here we addressed the role of raptor in brain development by conditional inactivation of rptor in nestin-expressing progenitor cells ∼E10.5 (Graus-Porta et al., 2001). Consistent with the concept that mTORC1 controls organismal size, as shown in other species (Oldham et al., 2000; Long et al., 2002), RAbKO mice show microcephaly. Moreover, RAbKO mice die shortly after birth with signs of cyanosis. In contrast, rapamycin treatment of developing embryos results in aberrant development of the telencephalon and death of the embryos already after midgestation (Hentges et al., 2001), suggesting that rapamycin-treated embryos may die because of mTORC1 inhibition in non-CNS cells and/or to loss of activity of both mTOR complexes.
Raptor regulates progenitor cell proliferation in the telencephalon and affects survival
Deletion of rptor in neural progenitors caused changes in cell size and cell number, which together resulted in a reduced brain size that became discernible from E17.5 onwards. It is well established that cell cycle progression and cell growth are linked because cells need to increase their size in the G phase to assure that cell size is maintained after mitosis (Fingar and Blenis, 2004). We here provide direct evidence that deletion of rptor affects cell division in neural progenitors of the developing cortex. At E16.5, cell cycle length in the RAbKO mice was >1.5 times longer than in controls and the number of progenitor cells in the ventricular zone that were in M-phase was lower at E17.5. These data were also corroborated by the strong size difference between secondary neurospheres isolated from RAbKO and control mice. In conclusion, our data provide strong evidence that loss of raptor affects cell proliferation in the developing brain, consistent with the role of mTORC1 in cell cycle progression (Fingar and Blenis, 2004). Moreover, recent evidence indicates that cell cycle regulation by mTORC1 involves phosphorylation of mTOR and raptor (Ramirez-Valle et al., 2010; Ekim et al., 2011) and that cell proliferation depends on mTORC1-mediated phosphorylation of 4E-BP proteins (Dowling et al., 2010).
Our work also provides evidence that mTORC1 promotes survival of cells at late stages of embryonic development as we observed substantial apoptosis at E19.5. These apoptotic events are likely to also contribute to the microcephaly of RAbKO mice at birth. The apoptotic cells were positive for Satb2 but not for GFAP, indicating that this process affects mostly neuronal cells and not glia. However, we do not know whether prevention of apoptosis by mTORC1 is a cell autonomous function or a consequence of a failure of neurons to connect to their targets as cultured neurons that lack raptor showed some deficits in neurite outgrowth.
mTORC1 can be activated by Akt3/PKBγ. Like RAbKO mice, mice deficient for Akt3/PKBγ have small brains and fewer cells (Easton et al., 2005; Tschopp et al., 2005), indicating that at least some of those effects of Akt3/PKBγ go through mTORC1. In addition, S6K, which is one of the main targets of mTORC1, has been shown to affect cell size when ablated in vivo (Shima et al., 1998; Montagne et al., 1999). Thus, the lack of S6K phosphorylation may account for the difference in cell size observed in E19.5 RAbKO mice.
mTORC1 is involved in gliogenesis
Evidence from previous reports led to the hypothesis that mTORC1 has an important role for neural stem/progenitor cell differentiation into neurons and glia (Rajan et al., 2003; Han et al., 2008; Wang et al., 2008). Our work now provides substantial in vivo evidence that loss of raptor mainly affects gliogenesis. Cortical progenitor cells in the mouse brain are programmed to sequentially produce cohorts of neurons and glial cells during embryonic development. Gliogenesis starts at ∼E17.5 in the cortex and is triggered by intrinsic signals. In addition, extrinsic feedback signaling from neurons in the cortical plate is needed for correct cell-fate specification (Seuntjens et al., 2009). The impact of rptor deletion on proliferation of neural precursors was only detected after E15.5. These developmental stages correlate with the beginning of gliogenesis (Franco and Muller, 2013), suggesting that mTORC1 may rather affect gliogenesis than neurogenesis. In agreement with this notion, there was no striking difference in the positioning of layer-specific neurons at E19.5. Importantly, RAbKO mice showed a strong decrease in the amount of GFAP but not of any neuronal markers at E19.5, and dissociated cells derived from E19.5 neurospheres showed a substantial reduction in glial, but not neuronal, differentiation in vitro. Finally, in newborn mice, when GFAP+ cells start to populate the brain (Sancho-Tello et al., 1995; Shu et al., 2003), GFAP immunostaining in RAbKO brains was weaker compared with controls. Together, our results provide strong evidence that mTORC1 signaling is important for gliogenesis during late cortical development but has rather subtle effects on neurogenesis at earlier time points of corticogenesis.
The effect of rptor deletion on gliogenesis correlated with a strong effect on the Jak-Stat pathway, which controls astrocytic differentiation (Johe et al., 1996; Bonni et al., 1997), and where mTORC1 has been implicated in this Stat3-dependent function (Rajan et al., 2003; Wang et al., 2008). Phosphorylation of Stat3 at Ser727 is rapamycin-sensitive, and phosphorylation of Stat3 at Tyr705 and Ser727 is essential for maximal activation (Wen et al., 1995; Yokogami et al., 2000). Stat3 phosphorylation was strongly reduced in the brains of RAbKO mice, and levels of Stat3 protein were lower, suggesting that phosphorylation of Stat3 might be important for the stabilization of the protein. Alternatively, the preponderance of neuronal cells could prevent activation of the positive autoregulatory loop of Jak-Stat signaling important for gliogenesis (He et al., 2005). Interestingly, deletion of the mTORC1 inhibitor Tsc2, a model for the genetic disease tuberous sclerosis in humans, increased Stat3 phosphorylation and GFAP expression (Onda et al., 2002).
The effect of mTORC1 on glial differentiation is also compatible with the observation that a brain-specific knock-out for the upstream activator Rheb1 (using the same nestin-Cre driver that we used to delete rptor) also shows deficits in the glial lineage (Zou et al., 2011). Interestingly, though, those mice did not die perinatally but survived for 5–6 weeks, and their brains were not smaller at birth (Zou et al., 2011). Hence, deletion of rptor causes a significantly more severe phenotype than deletion of Rheb1, suggesting the existence of either functional redundancy in the GTPases that activate mTORC1 during brain development or the presence of alternative pathways that can activate mTORC1 in neural progenitors in the absence of Rheb1.
In conclusion, our data show that mTORC1 is crucial for normal brain growth and postnatal survival. Several aspects of brain development are affected by deletion of rptor and thus loss of mTORC1 function, which may together be responsible for the death of the newborn mice. Regulation of cell size and division, which are both affected in RAbKO mice, are processes that are well known to be associated with mTORC1 signaling. Our data also strongly indicate that Stat3 is an important downstream target of mTORC1 that is required for normal differentiation of precursor cells into GFAP-positive glia. In summary, the multiple phenotypes of RAbKO mice described here show that mTORC1 has a central role in the integration of the inputs from several signaling pathways.
Footnotes
This work was supported by the Cantons of Basel-Stadt and Basel-Landschaft and the Swiss National Science Foundation (M.A.R.). The work in the laboratory of S.A. was supported by the Swiss National Science Foundation, the Novartis Foundation, the Botnar Foundation, and Swiss Life. We thank Drs. J. S. Tchorz and F. Schatzmann for providing a Cre-expressing adenovirus; Prof. M. Wegner (University of Erlangen) for the anti-Sox5 antibodies; Ludmilla Butenko for excellent technical assistance; and Prof. V. Taylor, Drs. M. Gassmann, and J. R. Reinhard for comments on the manuscript.
The authors declare no competing financial interests.
References
- Baranek C, Dittrich M, Parthasarathy S, Bonnon CG, Britanova O, Lanshakov D, Boukhtouche F, Sommer JE, Colmenares C, Tarabykin V, Atanasoski S. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc Natl Acad Sci U S A. 2012;109:3546–3551. doi: 10.1073/pnas.1108718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Rüegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/S0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009;69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VM, Szabolcs M, de Jong R, Oltersdorf T, Ludwig T, Efstratiadis A, Birnbaum MJ. Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol Cell Biol. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekim B, Magnuson B, Acosta-Jaquez HA, Keller JA, Feener EP, Fingar DC. mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol Cell Biol. 2011;31:2787–2801. doi: 10.1128/MCB.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Müller U. Shaping our minds: stem and progenitor cell diversity in the Mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C, Basak O, Taylor V. Isolation and manipulation of mammalian neural stem cells in vitro. Methods Mol Biol. 2009;482:143–158. doi: 10.1007/978-1-59745-060-7_9. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Müller U. β1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/S0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Han J, Wang B, Xiao Z, Gao Y, Zhao Y, Zhang J, Chen B, Wang X, Dai J. Mammalian target of rapamycin (mTOR) is involved in the neuronal differentiation of neural progenitors induced by insulin. Mol Cell Neurosci. 2008;39:118–124. doi: 10.1016/j.mcn.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Götz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, de Vellis J, Sun YE. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges KE, Sirry B, Gingeras AC, Sarbassov D, Sonenberg N, Sabatini D, Peterson AS. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc Natl Acad Sci U S A. 2001;98:13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges K, Thompson K, Peterson A. The flat-top gene is required for the expansion and regionalization of the telencephalic primordium. Development. 1999;126:1601–1609. doi: 10.1242/dev.126.8.1601. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Werner H, Püschel AW. Rheb and mTOR regulate neuronal polarity through Rap1B. J Biol Chem. 2008;283:33784–33792. doi: 10.1074/jbc.M802431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Müller F, Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol. 2002;12:1448–1461. doi: 10.1016/S0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Loulier K, Lathia JD, Marthiens V, Relucio J, Mughal MR, Tang SC, Coksaygan T, Hall PE, Chigurupati S, Patton B, Colognato H, Rao MS, Mattson MP, Haydar TF, Ffrench-Constant C. β1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Crino PB, Zhang H, Murphey RD, Rastelli L, Gould Rothberg BE, Kwiatkowski DJ. Tsc2 null murine neuroepithelial cells are a model for human tuber giant cells, and show activation of an mTOR pathway. Mol Cell Neurosci. 2002;21:561–574. doi: 10.1006/mcne.2002.1184. [DOI] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Rajan P, Panchision DM, Newell LF, McKay RD. BMPs signal alternately through a SMAD or FRAP-STAT pathway to regulate fate choice in CNS stem cells. J Cell Biol. 2003;161:911–921. doi: 10.1083/jcb.200211021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Valle F, Badura ML, Braunstein S, Narasimhan M, Schneider RJ. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol Cell Biol. 2010;30:3151–3164. doi: 10.1128/MCB.00322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Sancho-Tello M, Vallés S, Montoliu C, Renau-Piqueras J, Guerri C. Developmental pattern of GFAP and vimentin gene expression in rat brain and in radial glial cultures. Glia. 1995;15:157–166. doi: 10.1002/glia.440150208. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Shende P, Plaisance I, Morandi C, Pellieux C, Berthonneche C, Zorzato F, Krishnan J, Lerch R, Hall MN, Rüegg MA, Pedrazzini T, Brink M. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–1082. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Puche AC, Richards LJ. Development of midline glial populations at the corticoseptal boundary. J Neurobiol. 2003;57:81–94. doi: 10.1002/neu.10252. [DOI] [PubMed] [Google Scholar]
- Sirko S, von Holst A, Wizenmann A, Götz M, Faissner A. Chondroitin sulfate glycosaminoglycans control proliferation, radial glia cell differentiation and neurogenesis in neural stem/progenitor cells. Development. 2007;134:2727–2738. doi: 10.1242/dev.02871. [DOI] [PubMed] [Google Scholar]
- Swiech L, Perycz M, Malik A, Jaworski J. Role of mTOR in physiology and pathology of the nervous system. Biochim Biophys Acta. 2008;1784:116–132. doi: 10.1016/j.bbapap.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA. Essential role of protein kinase B γ (PKB γ/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132:2943–2954. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xiao Z, Chen B, Han J, Gao Y, Zhang J, Zhao W, Wang X, Dai J. Nogo-66 promotes the differentiation of neural progenitors into astroglial lineage cells through mTOR-STAT3 pathway. PLoS One. 2008;3:e1856. doi: 10.1371/journal.pone.0001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/S0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Zhou L, Wu JY, Rao Y. Cellular and molecular guidance of GABAergic neuronal migration from an extracortical origin to the neocortex. Neuron. 1999;23:473–485. doi: 10.1016/S0896-6273(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Zimmerman L, Parr B, Lendahl U, Cunningham M, McKay R, Gavin B, Mann J, Vassileva G, McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhou L, Du XX, Ji Y, Xu J, Tian J, Jiang W, Zou Y, Yu S, Gan L, Luo M, Yang Q, Cui Y, Yang W, Xia X, Chen M, Zhao X, Shen Y, Chen PY, Worley PF, et al. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20:97–108. doi: 10.1016/j.devcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]