Abstract
The maintained expression of transcription factors throughout the development of mesodiencephalic dopaminergic (mDA) neurons suggests multiple roles at various stages in development. Two members of the forkhead/winged helix transcription factor family, Foxa1 and Foxa2, have been recently shown to have an important influence in the early development of mDA neurons. Here we present data demonstrating that these genes are also involved in the later maintenance of the mDA system. We conditionally removed both genes in postmitotic mDA neurons using the dopamine transporter-cre mouse. Deletion of both Foxa1 and Foxa2 resulted in a significant reduction in the number of tyrosine hydroxylase (TH)-positive mDA neurons. The decrease was predominantly observed in the substantia nigra region of the mDA system, which led to a loss of TH+ fibers innervating the striatum. Further analysis demonstrated that the reduction in the number of TH+ cells in the mutant mice was not due to apoptosis or cell-fate change. Using reporter mouse lines, we found that the mDA neurons were still present in the ventral midbrain, but that they had lost much of their dopaminergic phenotype. The majority of these neurons remained in the ventral mesencephalon until at least 18 months of age. Chromatin immunoprecipitation suggested that the loss of the mDA phenotype is due to a reduction in the binding of the nuclear orphan receptor, Nurr-1 to the promoter region of TH. These results extend previous findings and demonstrate a later role for Foxa genes in regulating the maintenance of dopaminergic phenotype in mDA neurons.
Introduction
The production of mesodiencephalic dopaminergic (mDA) neurons requires a precise transcriptional cascade at a specific time point in development (Ang, 2006). These neurons arise from the floor plate of the caudal diencephalon and ventral mesencephalon (VM) regions of the developing mouse embryo between E10.5 and E13.5 (Marin et al., 2005; Ono et al., 2007). A great deal is now known about the early specification of mDA progenitors, but very little is understood about the maturation of mDA neurons.
One intriguing aspect of this latter stage of mDA neuronal development is the maintained expression of genes that are required for the initial specification. This sustained expression suggests that these genes may have additional roles in the maintenance of the mDA population. The homeodomain transcription factor, orthodenticle homeobox 2 (Otx2), for example, has multiple functions across various stages of the early development of this population. It is required for the early patterning of the mesencephalic region (Acampora et al., 1995; Matsuo et al., 1995; Ang et al., 1996; Broccoli et al., 1999; Millet et al., 1999), and for the specification of the mDA progenitors (Vernay et al., 2005; Omodei et al., 2008). Complementary gain-of-function experiments involving the overexpression of Otx2 result in an increased numbers of mDA progenitors (Ono et al., 2007; Omodei et al., 2008). The expression of Otx2 is maintained into adulthood in a specific subset of the mDA population, called the ventral tegmental area (VTA) (Chung et al., 2005, 2010; Di Salvio et al., 2010a). This expression was recently shown to have a functional role in maintaining specific mDA subpopulation identity and conferring resistance to toxins (Di Salvio et al., 2010b). Thus, it appears that the sustained expression of early mDA developmental genes has additional roles in the later maturation/maintenance of mDA neurons.
The forkhead/winged helix genes, Foxa1 and Foxa2 (Foxa), are also expressed throughout the life span of the mDA neurons (Thuret et al., 2004; Wijchers et al., 2006; Ferri et al., 2007; Cai et al., 2009; Lin et al., 2009). Both genes have been shown to play a fundamental role in the specification and early development of mDA neurons (Ferri et al., 2007; Ang, 2009; Lin et al., 2009). The expression of the Foxa genes in the adult mDA neurons was previously reported (Kittappa et al., 2007; Cai et al., 2009), but little is known about what role this maintained expression serves. Here, we looked at what functions Foxa genes may have in the mDA neuronal population, by conditionally deleting the genes using the Cre-loxp system. We found that in the absence of sustained Foxa gene expression, mDA neurons lose much of their “dopaminergic” program. The cells remain in the mDA domain of the VM and are still present in aged mice (≥18 months old), but they fail to express most of the mDA markers examined. These results suggest that Foxa genes are critical for the maintenance of the mDA phenotype.
Materials and Methods
Animals.
Foxa1flox/flox;Foxa2flox/flox mice were generated as previously described (Ferri et al., 2007). They were crossed with DATcre/+ mice (Zhuang et al., 2005) and conditional Rosa26-YFP reporter mice referred to as R26RYFP/YFP (Srinivas et al., 2001). Both male and female DATcre/+;Foxa1flox/flox;Foxa2flox/flox (referred to as DATcre/+;Foxa1/2 cko) mice from mixed backgrounds were used in this study. At all times, animals were handled according to the Society of Neuroscience Policy on the Use of Animals in Neuroscience Research, as well as the European Communities Council Directive.
Gait analysis.
A 50 cm runway was placed between a bright light and the home cage in a darkened room. Mice were trained 3 consecutive days to walk along the runway (5× trials each day). On the day of testing, two trial runs were performed before black paint was applied to the feet of the mice for the third run. A 40 cm length of paper was placed on the runway and the mice were timed from the moment that they were placed on the runway until they reached the home cage (Jolicoeur et al., 1979; Patel and Hillard, 2004).
Immunohistochemistry of brain sections.
Embryonic brains were immersion fixed overnight in 4% paraformaldehyde in 0.1 m PBS at 4°C, whereas postnatal brains were perfused with 4% paraformaldehyde and left overnight in 4% paraformaldehyde at 4°C. All tissues were then cryoprotected with 30% sucrose in PBS, embedded in optimum cutting temperature compound (VWR International) and sectioned on a cryostat (CM3050S, Leica). For embryonic tissues, 12 μm sections were collected on Superfrost glass slides, whereas for adult tissue 35 μm sections were collected with a brush, kept in PBS at 4°C, and stained as floating sections. For immunohistochemistry, sections were incubated overnight at room temperature with the appropriate primary antibody diluted in 1% BSA in PBS. Sections were then extensively washed in PBS and incubated 1 h at room temperature with a secondary antibody conjugated with a fluorochrome (The Jackson Laboratory). Sections were then washed and mounted in Vectashield H-1000 (Vector Laboratories). The following primary antibodies were used: rabbit anti-AADC (1:200, AB1569, Millipore), rabbit anti-ALDH1A1 (1:200, ab24343, Abcam), mouse anti-Brn3a (1:5, MAB1585, Millipore), rabbit anti-calbindin D-28k (1:1000, CB38a, Swant), rabbit anti-calretinin (1:1000, AB5054, Millipore), rabbit anti-activated caspase 3 (1:500, R&D Systems), rat anti-DAT (1:200, MAB369, Millipore), mouse anti-En1 (1:40, 4G11, DSHB), guinea pig anti-Foxa1 (1:500) (Wan et al., 2005), rabbit anti-Foxa2 (1:1000) (Filosa et al., 1997), goat anti-Foxa2 (1:500, Ab5074, Abcam), rabbit anti-GATA2 (1:100, SC-9008, Santa Cruz Biotechnology), rabbit anti-GFAP (1:500, Z0334, Dako), sheep anti-GFP (1:200, 4745–1051, AbD Serotec), rabbit anti-Girk2 (1:100, APC-006, Alomone Labs), rabbit anti-Islet-1/2 (1:500) (Tsuchida et al., 1994), rabbit anti-Lmx1a (1:1000, gift from M. German, University of California San Francisco Diabetes Center, San Francisco, California), guinea pig anti-Lmx1b (1:2000, generous gift from Drs. T. Mueller and C. Birchmeier, Max Delbruck Center of Molecular Medicine, Berlin, Germany), rabbit anti-Nurr1 (1:200, sc-990, Santa Cruz Biotechnology), goat anti-OTX2 (1:500, AF1979, R&D Systems), rabbit anti-Pitx3 (1:200, 38–2850, Zymed), rabbit anti-s100β (1:1000, s2644, Sigma-Aldrich), rabbit anti-serotonin (1:5000, S5545, Sigma-Aldrich), mouse anti-TH (1:1000, 22941, Immunostar), rabbit anti-TH (1:200, AB152, Millipore), sheep anti-TH (1:500, AB1542, Millipore), rabbit anti-TH (1:1000, P40101-0, Pel-Freez), and rabbit anti-VMAT2 (1:500, AB1767, Millipore).
All images were collected on a Leica TCS SP2 confocal microscope and processed with Adobe Photoshop 7.0 software (Adobe Systems). Quantitative immunohistochemical data are presented as mean ±SE. For embryonic and adult cell counts, 1:8 and 1:6 series were used throughout the entire midbrain, respectively. Quantifications were made from single confocal plane images.
In situ hybridization.
The procedures for whole-mount and section in situ hybridization were described previously (Schaeren-Wiemers and Gerfin-Moser, 1993). Mouse antisense probes for AADC, Foxa1, Foxa2, GAD63, Nurr1, and TH were generated from cDNA templates using RT-PCR as described previously (Krawchuk and Kania, 2008). cDNA template was generated from E18.5 ventral midbrain RNA. Primer sequences are available upon request.
Fiber density analysis.
To determine the fiber density in the striatum and nucleus accumbens, the mean optical intensity was measured from the TH-positive stained sections (Carlsson et al., 2007). Images were captured by a digital camera (AxioCam HRc, Zeiss) and analyzed using the ImageJ software platform, version 1.42q for Mac OSX (National Institutes of Health; http://rsb.info.nih.gov/ij/). The measurements were conducted from 12 coronal levels for the striatum and six levels for the nucleus accumbens (see Fig. 3A, as delineated). Nonspecific background was determined by readings made from the TH-corpus callosum and normalized to the white light background surrounding the section on the glass slide.
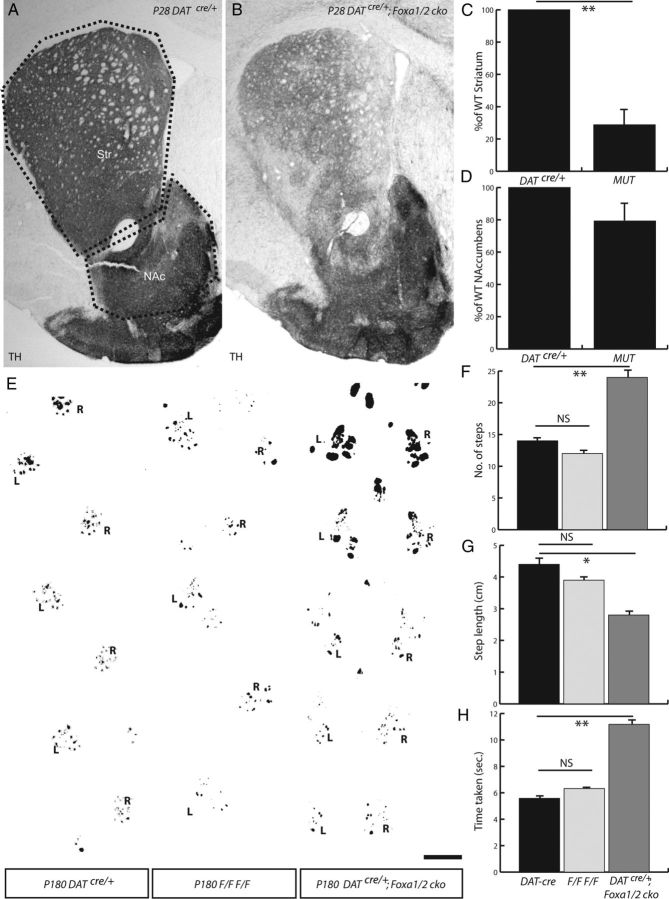
Figure 3.
Striatal fiber density and gait analysis of the DATcre/+;Foxa1/2 cko mice. A–D, Fiber density analysis of the striatum and nucleus accumbens was conducted on DATCre/+ (A) and DATcre/+;Foxa1/2 cko mice (B) using the areas delineated in A. DATcre/+;Foxa1/2 cko (MUT) mice demonstrated a 71.2% reduction in the number of TH+ fibers innervating the striatum (C), but only a 20.7% decrease in the nucleus accumbens (D) compared with DATCre/+ littermates. E, Foot gait analysis was also performed at 6 months of age on DATCre/+ mice, Foxa1flox/flox; Foxa2flox/flox (F/F F/F) mice, and DATCre/+;Foxa1/2 cko mice to demonstrate the hopping phenotype observed in the DATcre/+;Foxa1/2 cko mice. Left (L) and right (R) paw prints were recorded with black paint. F–H, The gait analysis demonstrated an increase in the number of steps made (F), and a reduction in step length (G), and time taken (H) by DATcre/+;Foxa1/2 cko mice while crossing the runway used. NAc, Nucleus accumbens; Str, striatum. *p < 0.05; **p < 0.01. Scale bar: (in E) A, B, 100 μm; E, 1 cm.
TUNEL.
Cryostat sections were washed once for 5 min in PBS containing 0.1% Triton X-100, permeabilized in ice-cold 0.01 m citrate buffer and 0.1% Triton X-100 for 2 min, and washed again in PBS containing 0.1% Triton X-100. The enzymatic reaction was then performed at 37°C according to the manufacturer's protocol (Roche Diagnostics).
Cell migration analysis.
Coronal sections of E18.5 midbrain were divided into bins (see Fig. 5A, represented along the bottom), taking into account the full spread of mDA cell migration from the midline to the most lateral point on the A–P axis. The total number of YFP+ cells were counted from seven sections of both DATcre/+;R26RYFP/+ and DATcre/+;Foxa1/2;R26RYFP/+ cko embryos (N = 3).
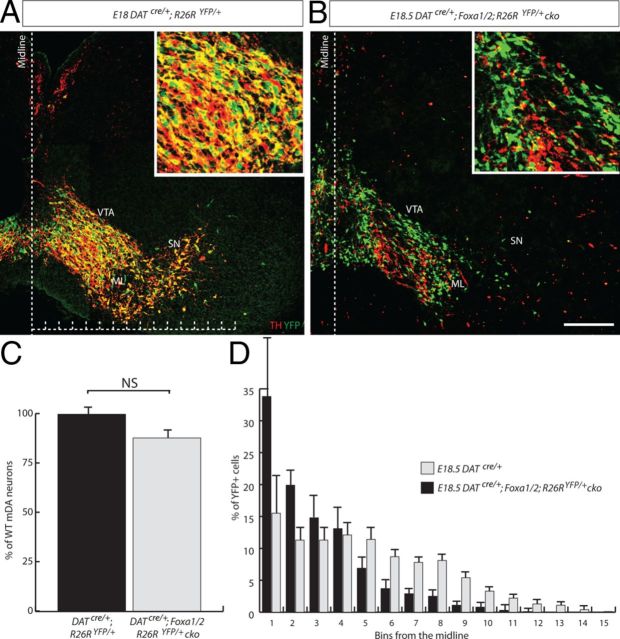
Figure 5.
TH−/YFP+ cells are present in the midbrain of DATcre/+;Foxa1/2;R26RYFP/+ cko mice. A, B, YFP+/TH+ cells were observed in the DATcre/+;R26RYFP/+ (A, inset), but very few double labeled cells were found in the DATcre/+;Foxa1/2;R26RYFP/+ cko E18 midbrain (B, inset). C, When the total number of YFP+/TH− and TH+/YFP− cells are counted in the DATcre/+;Foxa1/2;R26RYFP/+ cko mouse and compared with the DATcre/+;R26RYFP/+, there was only a small reduction in the number of mDA neurons. A greater percentage of YFP+ cells were located near the ventral midline in the midbrain of a DATcre/+;Foxa1/2;R26RYFP/+ cko embryo compared with the midbrain of a DATcre/+;R26RYFP/+ midbrain. The data were plotted as the percentage of cells versus distance from the midline. The medial-lateral axis was divided into bins that are equally spaced from each other (as indicated in A), with bin 1 being closest to the midline. *p < 0.05. Scale bar: (in B) A, B, 50 μm. NS, Not significant.
Cell size analysis.
Single plane, 40× magnification confocal images were taken of TH+/DAPI+ or YFP+/DAPI+ cells from the VM of DATcre;Foxa1/2 cko mice. The TH+ or YFP+ channel was changed to grayscale so that the cell body appeared white against a black background. The DAPI channel was changed to grayscale so that the nucleus of the cell was black and the background was made transparent. The DAPI image was then overlaid on the TH+ or YFP+ image and the circumference of the DAPI+ cell bodies was calculated using the ImageJ software platform. The diameter of each cell was calculated from this number.
ChIP binding assay.
Chromatin immunoprecipitation was performed on freshly dissected VM from E18.5 DATcre; Foxa1/2 cko embryos and their Cre-negative littermates (Foxa1 flox/flox;Foxa2 flox/flox) according to the procedure used previously (Lin et al., 2009). qPCR was performed using the following primers: Aadc, F: TGTGCCCCATGTACACAAGT, R: CATGCAAAGAGAGCAGACACA; Vmat2, F: GTGTCTACTGTCTATCACAG, R: GTCAAAGTGTCCATGAAGCC; Th and Foxa2 open reading frame primers are provided in Lin et al. (2009).
Statistical analysis.
Statistical analysis was performed using statistical software (Microsoft Excel and GraphPad Prism v4.0, GraphPad Software). All comparisons between mutant samples and controls were performed using two-tailed Student's t test or ANOVA. Differences were considered significant when p values were <0.05. In the text and figures, percentages are presented with the SE.
Results
Foxa1/2 are expressed in mDA progenitors and postmitotic neurons
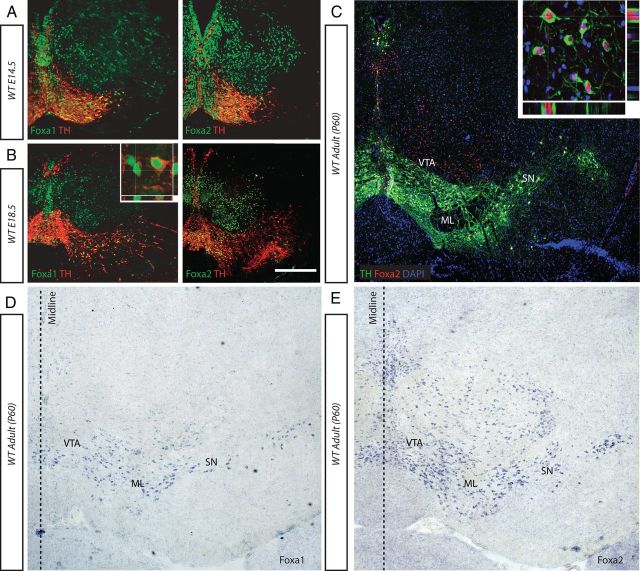
The expression pattern of Foxa1/2 during early mDA progenitor development has been described in previous publications (Ferri et al., 2007). We determined to see whether this expression was maintained throughout later embryonic stages, the early postnatal period, and into adulthood by in situ hybridization and immunohistochemistry on midbrain sections. Between E14.5 and 18.5, Foxa1 and Foxa2 were expressed in the VM (Fig. 1A,B, respectively). Both genes were present in more nuclei than just the mDA neurons (Fig. 1A,B). Postnatally, Foxa1 and Foxa2 were expressed by mDA neurons at all time-points analyzed through to P60 (Fig. 1C–E). Interestingly, Foxa1 expression in the adult VM was more specific to the mDA neurons, whereas Foxa2 was maintained in additional VM nuclei (Fig. 1C–E). At all time points examined, Foxa1 and Foxa2 were present in all of the mDA neurons analyzed. Their expression was located specifically in the nucleus and was not seen in the cytoplasm (Fig. 1B,C, insets). The maintenance of Foxa2 gene expression in the adult mDA neurons demonstrated here agreed with previous observations (Kittappa et al., 2007; Cai et al., 2009). This persistent expression of Foxa1/2 throughout the maturation of the mDA system suggested potential roles of these genes in the differentiation and/or maintenance of mDA neurons.
Figure 1.
The expression of the Foxa genes in the mDA neurons is maintained into adulthood. A, B, Immunological staining with Foxa1 and Foxa2 antibodies demonstrates their expression patterns on coronal sections of VM at E14.5 and E18.5 (A, B, respectively). C, E, Immunological and in situ stainings demonstrate that both Foxa2 are expressed in the ventral midbrain of 60-d-old adult mice (C, E, respectively). D, Foxa1 expression was detected in the mDA region only by in situ hybridization, as the Foxa1-specific antibody did not work in adult sections. Foxa1 and Foxa2 expression was localized to the nucleus both embryonically (B, inset) and postnatally (C, inset), respectively. ML, Medial lemniscus. Scale bar: (in B) A, 80 μm; B, 100 μm; C, D, 50 μm.
Deletion of Foxa1/2 in mDA neurons
To conditionally delete Foxa1 and Foxa2 in postmitotic mDA neurons, we used the dopamine transporter-Cre (DATcre/+) mouse (Zhuang et al., 2005). We crossed DATcre/+ male animals with the Rosa26LacZflox/flox female mice to determine precisely when recombination was occurring in mDA neurons. No βgal expression was seen when the VM of these mice were analyzed at E12.5 (data not shown). The first βgal+ cells appeared at E13.5 in only a small number of TH+ cells in the VM, but their number increased markedly by E14.5 (Fig. 2A). Earlier studies have shown that the expression of βgal was strong in mDA neurons, and only weak staining was observed in dopaminergic neurons of the olfactory bulb and hypothalamus in adult DATcre/+;Rosa26LacZflox/+ mice (Zhuang et al., 2005). Given that 95% of mDA neurons are postmitotic by E13.5 (Bayer et al., 1995), the DATcre/+ mouse strain is useful for inactivating genes specifically in mDA neurons.
Figure 2.
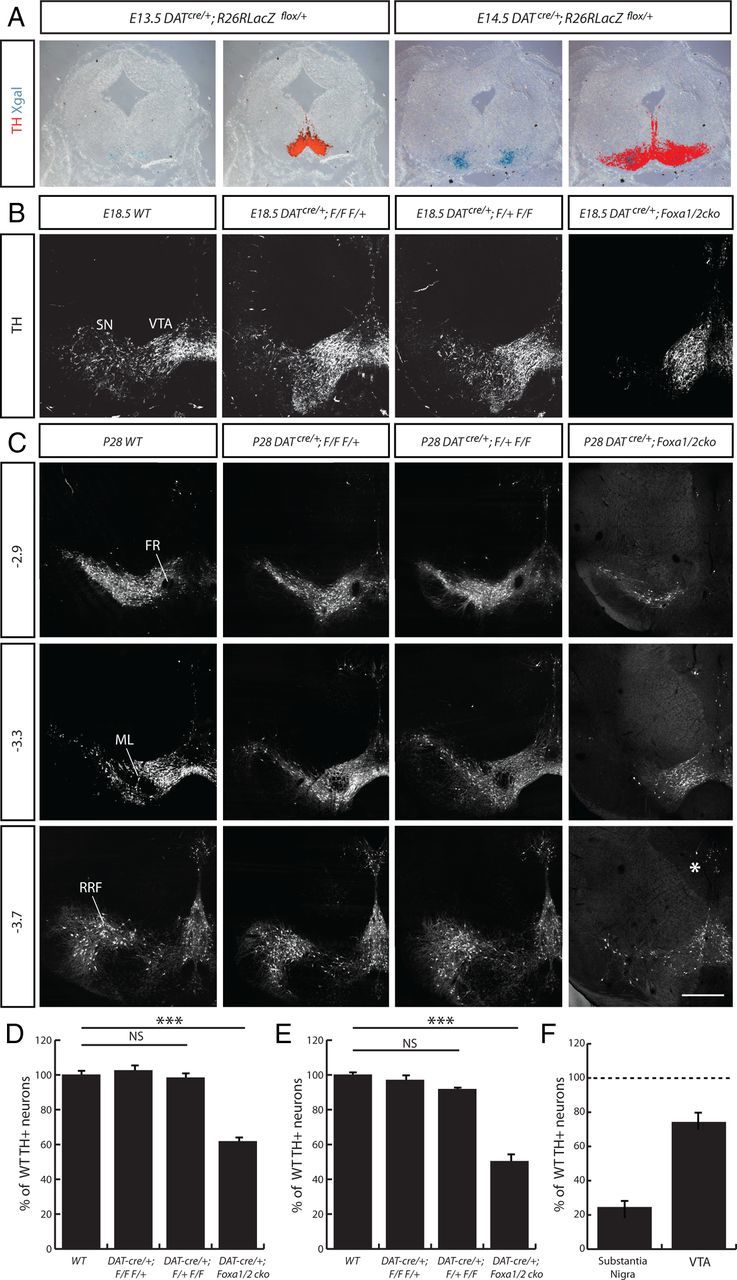
Deletion of Foxa genes in postmitotic mDA neurons using the DATcre/+ mouse. A, Activation of LacZ expression in DATcre/+;Rosa26LacZflox/+ embryos occurred between E13.5 and E14.5, as determined by Xgal staining on coronal sections of embryonic midbrain. B, C, TH+ mDA neurons were observed in the transgenic mice at both E18.5 (B), and P28 (C). D, E, Coronal sections of P28 midbrain are presented as relative to bregma. The percentage of TH+ cells at E18.5 (D) and P28 (E) in the VM of WT, DATCre/+;Foxa1flox/flox; Foxa2flox/+ (DATcre/+; F/F F/+), DATCre/+;Foxa1flox/+; Foxa2flox/flox (DATcre/+; F/+ F/F), and DATCre/+;Foxa1/2 cko mice were counted and significant differences were observed. ***p < 0.001. NS, Not significant. The percentage of TH+ cells in the mDA subregions was compared, highlighting a more dramatic loss in the SN than the VTA (F). The loss of mDA neurons in the SN region is in part due to a defect in migration of these mDA neurons from medial to lateral positions in the midbrain (Fig. 5D). FR, Fasciculus retroflexus; RRF, retrorubral field. Scale bars: (in C) A, B, 50 μm; C, 100 μm.
DATcre/+;Foxa1/2 cko mice were born at a normal frequency, developed into adulthood and were fertile. Upon dissection, we found no observable differences in size, weight, or morphology when comparing the brains of DATcre/+;Foxa1/2 cko mice with their wild-type (WT) littermates (data not shown). When assessing the mDA neurons in the VM of the DATcre/+;Foxa1/2 cko mice, we observed little if any change at E14.5 when Cre is first beginning to be expressed. By E18.5, however, there was a 40% reduction in the number of TH+ neurons in the VM of the DATcre/+;Foxa1/2 cko mice, compared with WT mice and mice with one functional copy of Foxa1 or Foxa2 gene in mDA neurons (n = 3; WT = 995.7 ± 8.7, DATcre/+;Foxa1flox/flox;Foxa2flox/+ = 1003.3 ± 11.2, DATcre/+;Foxa1flox/+;Foxa2flox/flox = 1020.3 ± 19; DATcre/+;Foxa1/2 cko = 619.3 ± 13.9 TH+ cells; p = 0.00009) (Fig. 2B,D). This loss of TH+ cells increased with time to 50.3% in adult mice at P28 (n = 3; WT = 2079.3 ± 32.2, DATcre/+;Foxa1flox/flox;Foxa2flox/+ = 2016.0 ± 39.3, DATcre/+;Foxa1flox/+;Foxa2flox/flox = 1849.0 ± 60.6; DATcre/+;Foxa1/2 cko = 1045.9 ± 32.9 TH+ cells; p = 0.000006) (Fig. 2C,E). The mDA neurons in the substantia nigra (SN) of the DATcre/+;Foxa1/2 cko mice were more affected with a 75.6% reduction in the number of TH+ cells (n = 3; WT = 918 ± 10.7 TH+ cells, DATcre/+;Foxa1/2 cko = 224.3 ± 37.6), whereas the VTA subset lost 25.9% of the WT number of TH+ neurons (n = 3; WT = 929 ± 12.2 TH+ cells, DATcre/+;Foxa1/2 cko = 688.4 ± 20.0) (Fig. 2F). Other mDA subregions (such as the mDA neurons near the ventricular zone) were less dramatically affected (Fig. 2C, asterisk; data not shown).
Given that previous work from this laboratory has demonstrated that Foxa1 and Foxa2 regulate early mDA development in a dose-dependent manner (Ferri et al., 2007), we next analyzed multiple combinations of allele deletion using the DATcre/+ mouse. At both E18.5 and P28, we found no change in the number of TH+ cells in the VM of single Foxa gene deletions (DATcre/+;Foxa1flox/flox and DATcre/+;Foxa2flox/flox) compared with WT (data not shown). The DATcre/+;Foxa1flox/+;Foxa2floxflox animals showed a large reduction in the number of TH+ cells, compared with WT but this was not significant (p = 0.057) (Fig. 2C,E). These results demonstrate that the Foxa1 and Foxa2 can largely compensate for each other, but are both required for the correct maintenance of the mDA system at the adult stage.
Behavioral phenotype in the DATcre/+;Foxa1/2 cko mice
In addition to the loss of TH+ cells, the DATcre/+;Foxa1/2 cko mice exhibited a 71.2 ± 12.2% loss of TH+ fiber density in the striatum (n = 4; p=0.0035) (Fig. 3A–C), whereas there was only a 20.7 ± 13.7% reduction of TH+ fiber density in the nucleus accumbens (n = 4; p = 0.21) (Fig. 3A,B,D). The preferential loss of TH+ fibers to medial and lateral regions of the striatum is interesting, but this phenotype may be because of a specific role of Foxa1/2 on the projection pattern or on the medial-lateral positioning of mDA neurons in the striatum and midbrain respectively (see below), which could indirectly influence the targeting of these neurons in mutant mice. Despite the absence of TH+ fiber innervation in the striatum, we observed no altered posture or tremor in the DATcre/+;Foxa1/2 cko mice based on gross observation. In addition, these mice did not demonstrate any difficulties in initiating or maintaining movement, which suggested that motor control itself was not affected in these mice, similar to what has been observed in Pitx3−/− mice (Smidt et al., 2004). We noted, however, that from an early postnatal age the mice displayed a “hopping” style of walking (Fig. 3E) compared with WT mice. Although they were capable of walking with the alternating step pattern of a WT mouse, the DATcre/+;Foxa1/2 cko mice predominantly moved both their rear legs at the same time (Fig. 3E).
To quantify this hopping behavior, we conducted a gait analysis on 6-month-old DATcre/+;Foxa1/2 cko and WT mice. The DATcre/+;Foxa1/2 cko mice took significantly more steps (24 ± 2.0 steps) to cross the 40 cm runway used in the gait analysis compared with DATcre/+ mice (14 ± 0.58 steps) and Foxa1flox/flox;Foxa2flox/flox mice (12 ± 1.0 steps; p = 0.0086) (Fig. 3F). This was primarily because of a shortening of the average step length (4.4 ± 0.51 cm for DATcre/+ mice, compared with 3.9 ± 0.14 cm for the Foxa1flox/flox; Foxa2flox/flox mice, and 2.8 ± 0.15 cm for DATcre/+;Foxa1/2 cko mice; p = 0.036) (Fig. 3G), and resulted in the mutant mice taking significantly longer (11.18 ± 0.52 s) to traverse the length of the runway than the DATcre/+ mice (5.58 ± 0.59 s) and Foxa1flox/flox;Foxa2flox/flox mice (6.32 ± 0.08 s; p = 0.002) (Fig. 3H). These findings indicated that the DATcre/+;Foxa1/2 cko mice have a significant reduction in the level of mDA fibers innervating the striatum, and display an alternative means of locomotion.
Loss of TH+ neurons is not due to cell death or fate change
Given that we observed a 40% reduction in the number of TH+ neurons in the DATcre/+;Foxa1/2 cko mice between E14.5 and 18.5, we wanted to determine whether cell death was occurring at any of the time points in between to explain the loss of cell number. Both activated caspase 3 staining and TUNEL expression revealed little or no increase in cell death compared with the WT VM (Fig. 4A,B; data not shown). In contrast, control tissues such as the E15.5 thymus showed a number of activated Caspace3+ apoptotic cells (Fig. 4A). This experiment suggested that the loss of TH+ cells in the VM of DATcre/+;Foxa1/2 cko mice was not due to cell death.
Figure 4.
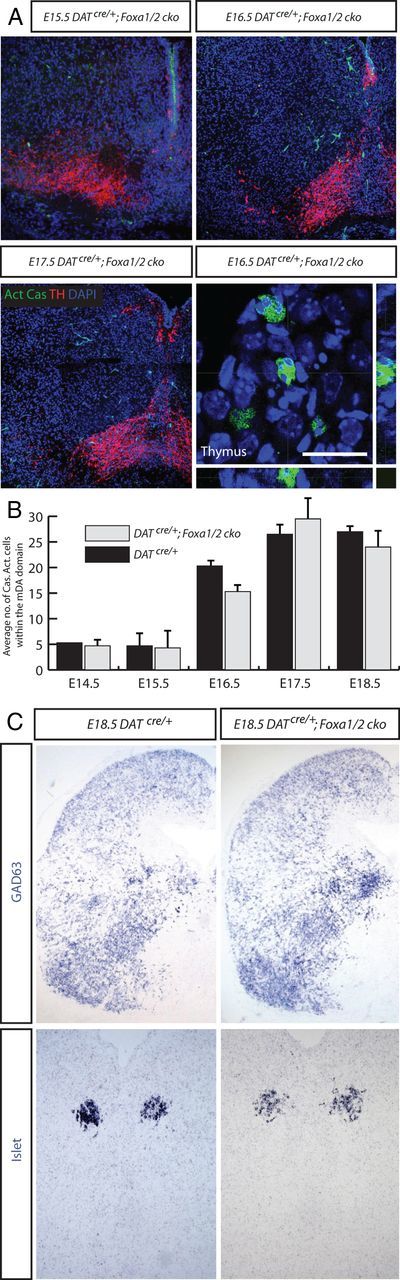
No apoptotic cells or change of fate in the mDA region of DATCre/+;Foxa1/2 cko mice. A, Coronal sections of the VM from E15.5, 16.5, and 17.5 DATCre/+;Foxa1/2 cko mice presented little to no activated phosphorylated caspase3 (Act Cas) staining, although the thymus from the same section of the E16.5 embryo had many positive cells. B, There was no difference compared with DATCre/+ sections at the same time points. C, Staining coronal sections of DATCre/+ and DATCre/+;Foxa1/2 cko midbrain with alternative cell type markers, like GAD63 and Islet suggested that there was no obvious change of fate in the Foxa1-Foxa2- mDA neurons in the DATCre/+;Foxa1/2 cko mice. Scale bars: (in A) A, 10 μm for the thymus image; A (for the other images), C, 50 μm.
We next examined the hypothesis of whether the loss of Foxa genes in mDA neurons resulted in changes of cell fate. We ran a battery of alternative midbrain neuronal markers (such as GABA, GAD, Brn3a, Islet, serotonin, GATA2, and glutamate) and glial markers (GFAP and s100β) (Fig. 4C; data not shown), but witnessed no obvious increase in any of the expression of these genes that could account for the reduction in TH+ neuronal number in the VM of the DATcre/+;Foxa1/2 cko mice at E18.5. Together, these stainings demonstrated that the loss of TH+ cell in the VM of the DATcre/+;Foxa1/2 cko mice between E14.5 and 18.5 was not due to cell death or fate change.
mDA neurons lose their phenotype in the DATcre/+;Foxa1/2 cko mouse
Lack of evidence for cell death and cell fate changes suggested the possibility that mDA neurons were still present but not expressing appropriate dopaminergic markers such as tyrosine hydroxylase. To follow cells that have undergone Cre-mediated excision of Foxa1/2, we crossed the DATcre/+;Foxa1/2 cko mouse with a R26RYFP/YFP reporter strain of mice (Srinivas et al., 2001). At E14.5, few YFP+ cells were observed in the VM of either the DATcre/+;R26RYFP/+ or DATcre/+;Foxa1/2;R26RYFP/+ cko mice, consistent with results using Rosa26LacZflox/flox reporter mouse strain (Fig. 2A; data not shown). By E18.5, however, almost all of the YFP+ cells in the ventral midbrain of the DATcre/+;R26RYFP/+ mouse VM were TH+ (Fig. 5A, inset). In contrast, double antibody labeling using anti-TH and anti-GFP specific antibodies showed a large number of YFP+TH− neurons present both in the VTA and SN regions of the VM of DATcre/+;Foxa1/2;R26RYFP/+ cko mouse (Fig. 5B, inset). This result indicates that expression of TH was not maintained in the mDA neurons in which Foxa1 and Foxa2 had been deleted. When we determined the total number of mDA neurons in DATcre/+;Foxa1/2;R26RYFP/+ cko mutant embryos by combining the numbers of TH+/YFP− and TH−/YFP+ cells, we found that there was only a 12.02% reduction in total cell number compared with the total number of TH+/YFP+ mDA neurons in DATcre/+;R26RYFP/+ mouse embryos at E18.5 (WT = 2157.3 data not shown and ± 96.9 TH+ cells vs DATcre/+;Foxa1/2;R26RYFP/+ cko = 1898.0 ± 66.7 YFP+/TH− and YFP−/TH+ cells; p = 0.092) (Fig. 5C). We also noted that an unusually high proportion of these cells were restricted to the medial portion of the VM in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice, suggesting a possible role in the regulation of mDA cell migration for Foxa genes (Fig. 5D).
This phenotype was maintained into adulthood in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice. At the P28 time point, the YFP+/TH− cells in the VM of the DATcre/+;Foxa1/2;R26RYFP/+ cko mice were still present and largely restricted to the mDA domain (Fig. 6A,D compare with B,E). Although the TH+ cells all had normal mDA cell morphology, we noticed that the TH−/YFP+ cells were smaller than the TH+/YFP− mDA neurons. The YFP+/TH− cells in the SN were on average 34.4% smaller in cell diameter than their TH+/YFP− counterparts, and in the VTA this reduction was 31.6% (n = 478 cells for the VTA and 184 cells for the SN from 3 brains; 941.7 ± 107.3 μm2 for TH+ SN cells and 617.8 ± 58.4 μm2 for YFP+ SN cells; p = 0.057; 700.1 ± 70.2 μm2 for TH+ VTA cells and 478.6 ± 37.2 μm2 for YFP+ VTA cells; p = 0.049) (Fig. 6G–I).
Figure 6.
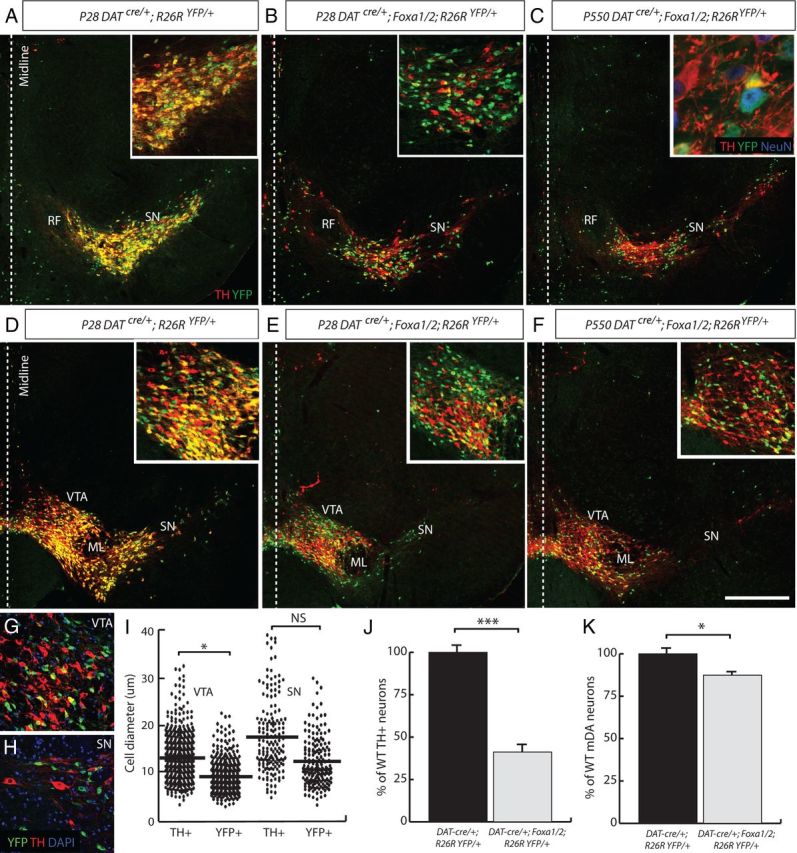
TH−/YFP+ mutant mDA neurons survive until late adulthood, but are smaller in size than their WT counterparts. A–F, Images taken from rostral (A–C) and medial (D–F) sections of the midbrain are presented for comparison. YFP+/TH+ cells were found in the midbrain of DATcre/+;R26RYFP/+ P28 adult mice (A, D, insets), but very few double-labeled cells were observed in the midbrain of a DATcre/+;Foxa1/2;R26RYFP/+ cko mouse at P28 (B, E, insets) and P550 (C, F, inset). The YFP+/TH− cells were all neurons as determined by the neuronal marker, NeuN (C,inset). G–I, When the diameter of YFP+ and TH+ cells was measured in the VTA (G) and SN (H), a significant difference was found in size of the YFP+ cells (I). J, Although the total number of TH+ cells in the DATcre/+;Foxa1/2;R26RYFP/+ cko animals was severely reduced, the total number of mDA neurons (YFP+/TH− and TH+/YFP−) was only slightly reduced in these mice compared with DATcre/+;R26RYFP/+ mice. K, The quantification of cell numbers are represented as a ratio of WT cell numbers and in percentages. *p < 0.05; ***p < 0.001. Scale bar: (in D) A–F, 150 μm; G, H, 60 μm.
To see how long these cells remained in the VM of the DATcre/+;Foxa1/2;R26RYFP/+ cko mice, we analyzed these mice all the way to 18 months of age (P550). In the DATcre/+;R26RYFP/+ mice, the YFP expression was well maintained in the TH+ cells in the VM (data not shown). In all of the mutant mice examined at P550 (n = 4), TH−/YFP+ cells were found in the mDA region of the VM (Fig. 6C,F). The total number of TH+ cells in the VM of the DATcre/+;Foxa1/2;R26RYFP/+ cko mice represented only 41.2% of the WT population (WT = 2014.3 ± 77.2 TH+ cells vs 830.3 ± 49.1 TH+ cells; p = 0.0002) (Fig. 6J). However, when we counted the total number of YFP+/TH− and TH+/YFP− cells in the VM of DATcre/+;R26RYFP/+ mice and DATcre/+;Foxa1/2;R26RYFP/+ cko mice at P550, we found only a 10.6% reduction in the total number of cells (WT = 2014.3 ± 77.2 TH+ cells vs 1759.7 ± 77.0 TH+ cells; p = 0.044) (Fig. 6K). All of the YFP+/TH− cells that were analyzed at P550 were positive for the neuronal marker, NeuN demonstrating that although the cells had lost their mDA phenotype, they were still neurons (Fig. 6C, inset). Together these stainings and quantifications suggested that the reduction in the number of TH+ cells in the VM of the DATcre/+;Foxa1/2 cko mouse was not due to cell death or change of cell fate, but rather due to loss of TH expression in mDA neurons.
Maintenance of certain mDA genes in the absence of Foxa1 and Foxa2
We next performed double immunolabeling analysis on these YFP+/TH− cells at E18.5 to determine whether other mDA differentiation genes were lost in addition to TH when Foxa1 and Foxa2 were deleted in these neurons (Fig. 7A). As expected, the YPF+/TH− neurons in the VM of DATcre/+;Foxa1/2;R26RYFP/+ cko mice did not express Foxa1 (Fig. 7B) and Foxa2 (Fig. 7C), indicating successful deletion of the floxed alleles. The expression of both LIM homeobox transcription factor, Lmx1a, and Lmx1b, which are involved in mDA development were maintained (Yan et al., 2011) (Fig. 7D,E, respectively). In addition, nuclear receptor related 1 (Nurr1, also known as Nr4a2) was also present in YFP+/TH− cells (Fig. 7F). In contrast, the paired-like homeodomain 3 factor (Pitx3), aromatic l-amino acid decarboxylase (AADC) and the vesicular monoamine transporter (Vmat2) were all not expressed in YFP+ mDA cells (Fig. 7G–I).
Figure 7.
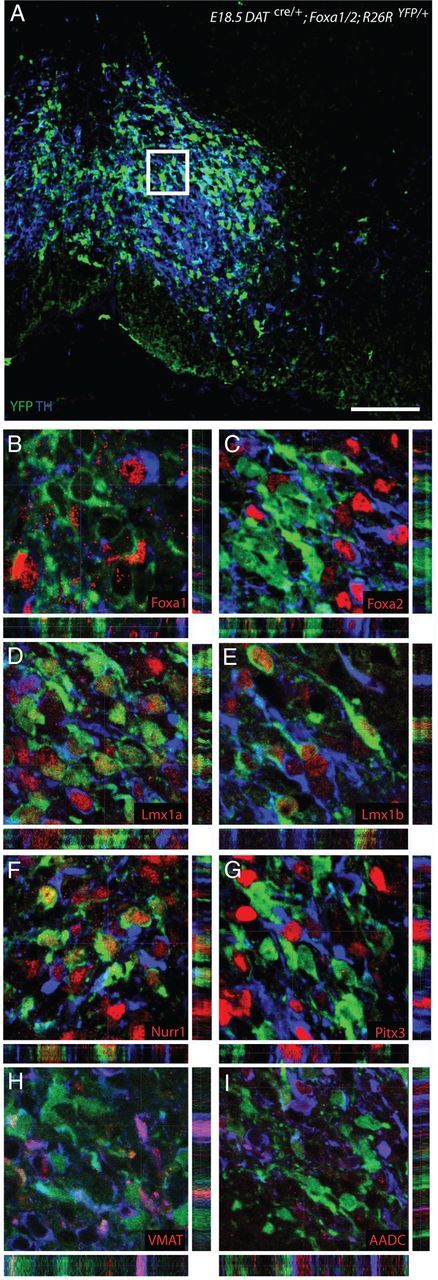
Certain mDA-related genes maintain their expression in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice. A–I, High-magnification coronal images of the YFP+/TH− mDA cells in the VTA region of E18.5 DATcre/+;Foxa1/2;R26RYFP/+ cko embryos (A) were taken from sections triple labeled with YFP (green), TH (blue), and various mDA-related markers, such as Foxa1 (B), Foxa2 (C), Lmx1a (D), Lmx1b (E), Nurr1 (F), Pitx3 (G), VMAT2 (H), and AADC (I). Scale bar: (in A) A, 40 μm; B–I, 15 μm.
Because the expression of Nurr1 was maintained in the TH−/YFP+ cells, we also determined the total number of Nurr1+ cells in the VM of both control DATcre/+;R26RYFP/+ and DATcre/+;Foxa1/2;R26RYFP/+ cko mice and found that there was only a 5% reduction in the total number of Nurr1+ cells at E18.5 (DATcre/+;R26RYFP/+ = 3085.0 ± 95 Nurr1+ cells vs DATcre/+;Foxa1/2;R26RYFP/+ cko = 2888 ± 42 TH+ cells; p = 0.13). Nurr1 was also maintained in all of the YFP+/TH− neurons analyzed at the P28 and P550 time points (data not shown).
Mature mDA neurons can be molecularly divided into predominantly two major subsets by the expression of the G-protein regulated inward-rectifier potassium channel 2 (Girk2) (Mendez et al., 2005; Thompson et al., 2005) and Otx2 (Chung et al., 2005, 2010; Di Salvio et al., 2010a). Girk2 is preferentially expressed in SN mDA neurons (Fig. 8A), whereas Otx2 specifically marks VTA neurons (Fig. 8E). Compared with the WT situation, we found that the total number of Girk2+ neurons in the mutant was 92% reduced in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice (WT = 1358.3 ± 16.6 vs MUT = 108.3 ± 8 Girk2+ cells; p = 0.000,028). Of the remaining Girk2+ cells, only 9.2% were also YFP+ (Fig. 8A,B). We found a similar reduction in the expression of another SN marker, aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), in the YFP+/TH− population of mDA neurons in DATcre/+;Foxa1/2;R26RYFP/+ cko mice (data not shown). We also observed a severe loss of DAT expression (Fig. 8C,D), which is more highly expressed in the SN than the VTA (Gonzalez-Hernandez et al., 2004).
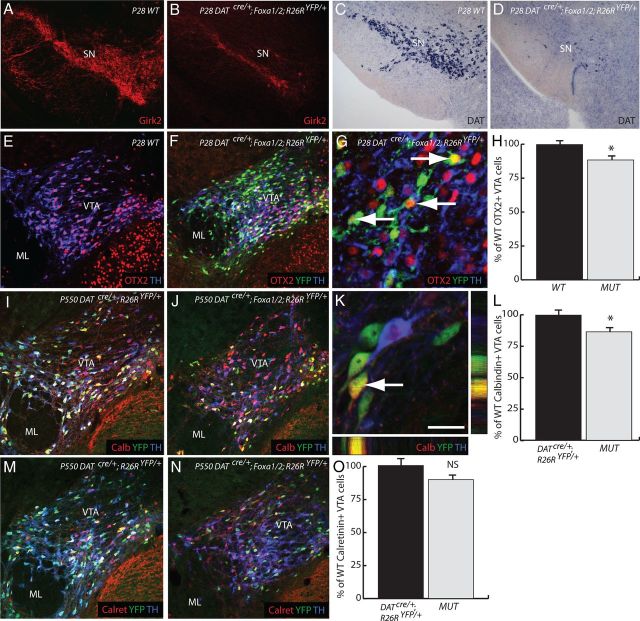
Figure 8.
SN mDA neurons are more affected than the VTA in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice. A, B, Markers specific to the SN, such as Girk2 were dramatically reduced in the mDA neurons of the DATcre/+;Foxa1/2;R26RYFP/+ cko mice. C, D, Even the dopamine transporter (DAT) was almost lost in the absence of Foxa1/2. E–O, Markers of the VTA subregion, such as OTX2 (E–H), calbindin (I–L), and calretinin (M–O) exhibited only a small reduction in their expression in the DATcre/+;Foxa1/2;R26RYFP/+ cko mice (MUT). Arrows in G and K highlight YFP+/TH− cells still expressing OTX2 and calbindin, respectively. *p < 0.05. Scale bar: (in K) A–D,100 μm; E, F, I, J, M, N, 80 μm; G, 20 μm; K, 10 μm.
In contrast, there was only a reduction of 11.4% of Otx2+ neurons in mutant VTA compared with WT mice (WT = 686.3 ± 14.5 vs MUT = 608 ± 16.7 Otx2+ cells; p = 0.024) (Fig. 9E–H). Many of the YFP+/TH− neurons in the VM of DATcre/+;Foxa1/2;R26RYFP/+ cko mice were observed with robust Otx2 expression (Fig. 9G). In contrast, there was only a reduction of 11.4% of Otx2+ neurons in mutant VTA compared with WT mice (WT = 686.3 ± 14.5 vs MUT = 608 ± 16.7 Otx2+ cells; p = 0.024) (Fig. 9E–H). Many of the YFP+/TH− neurons in the VM of DATcre/+;Foxa1/2;R26RYFP/+ cko mice were observed with robust Otx2 expression (Fig. 9G). Previous studies have shown that Otx2 is expressed in VTA but not SN mDA neurons and that the majority (93%) of Otx2+ neurons in the VTA express TH (Di Salvio et al., 2010a). We therefore reasoned that the percentage of YFP+Otx2+/Otx2+ neurons in the mDA region would give a reasonable estimate of the efficiency of recombination in the VTA and this was determined to be ∼43% in adult mutant mice at P28. The recombination efficiency in the total mDA neuronal population was determined to be 47% and 49% respectively, based on the percentage of YFP+Foxa1−/total number of mDA neurons and YFP+/Foxa2−/total number of mDA neurons in E18.5 DATcre/+;Foxa1/2;R26RYFP/+ cko embryos (Fig. 7B,C). These numbers suggest that the deletion of Foxa1/2 occurs with comparable efficiencies in both SN and VTA and therefore support the conclusion that Foxa1/2 are not required for the maintenance of Otx2 in adult mDA neurons. We next determined whether the expression of Calbindin and Calretinin (McRitchie et al., 1996) are modified in DatCre/+;Foxa1/2 cko mice. These additional quantifications were conducted at the P550 time point. We found that compared with WT mice, there was 13.3% reduction in the number of Calbindin+ cells in the VTA (WT = 549 ± 7 vs MUT = 476 ± 10; p = 0.027), and only a 10.5% reduction in the number of Calretinin+ cells (WT = 491 ± 25 vs MUT = 439 ± 0; p = 0.17) (Fig. 8I–O). Altogether, these results indicate that Foxa1/2 is not required for the maintenance of expression of Otx2, calbindin, and calretinin in adult VTA neurons.
Figure 9.
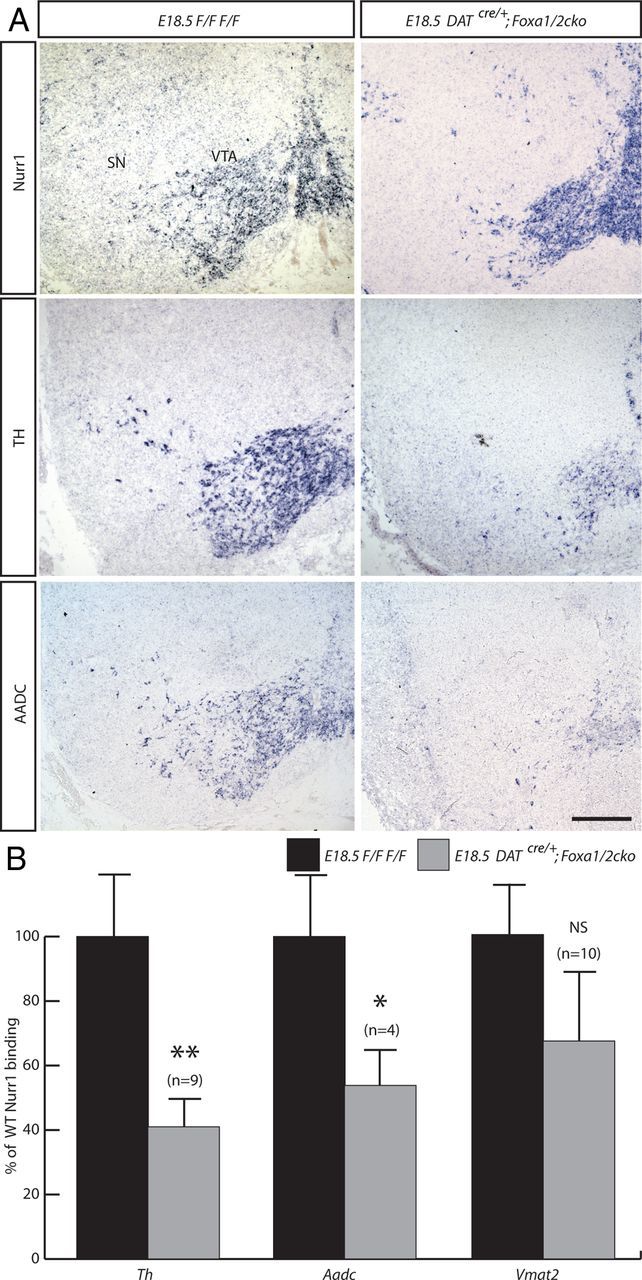
A, In situ hybridization staining of Nurr1, TH, and AADC on coronal sections from E18.5 Foxa1flox/flox;Foxa2flox/flox (F/F F/F) and DATcre/+;Foxa1/2 cko mice midbrain, demonstrate the maintenance of Nurr1 expression, and the reduction in TH and AADC. ChIP analysis suggests that the reduction of TH and AADC expression may result from a reduction of Nurr1 binding to the Th and Aadc promoters. No significant change was observed in Nurr1 binding to the Vmat2 promoter. B, C, Independent ChIP-qPCR experiments were performed with chromatin prepared from E18.5 ventral midbrain tissue from WT or DATcre/+;Foxa1/2 cko, where enrichment is shown as percentage of WT percentage input. *p < 0.05; **p < 0.01. Scale bar, 100 μm.
Reduced binding of Nurr1 to TH promoter region in the absence of the Foxa1 and Foxa2 genes
The nuclear orphan receptor transcription factor, Nurr1, plays an important role in the generation and maintenance of mDA neurons (Kadkhodaei et al., 2009). Deletion of Nurr1 in DATCre/+;Nurr1flox/flox embryos also leads to a severe reduction in the expression of dopaminergic markers, including DAT, TH, AADC, and to a lesser extent the expression of Pitx3 and Lmx1b, in mDA neurons starting at E15.5 (Kadkhodaei et al., 2009). This loss of dopaminergic properties persisted and was accompanied by loss of these neurons in adult mutant mice that was shown using FluoroGold retrograde labeling experiments (Kadkhodaei et al., 2009). Together these findings indicate that Nurr1 is initially required for the maintenance of TH expression and then cell survival. Combined with published data showing that Nurr1 can directly bind and activate the TH promoter in cell lines by chromatin immunoprecipitation (Kim et al., 2003), these results suggest that Nurr1 regulates expression of TH by direct binding. We found that Nurr1 was maintained in the VM of DATcre/+;Foxa1/2 cko embryos, whereas TH and AADC were severely reduced (Fig. 9A). Because we have previously shown that Foxa2 also binds to the TH promoter (Lin et al., 2009), we determined whether Foxa2 and Nurr1 cooperate to bind this DNA fragment by chromatin immunoprecipitation using chromatin isolated from the midbrain of E18.5 control or DATcre/+;Foxa1/2 cko embryos. Interestingly, our results show that Nurr1 binding to Th promoter is significantly reduced in mutant chromatin compared with WT chromatin (p = 0.0058) (Fig. 9B). Because the neuronal promoter of Aadc is bound by both Foxa2 and Nurr1 (Raynal et al., 1998; Ye et al., 1999; Jacobs et al., 2009), we also determined whether the ability of Nurr1 to bind this promoter element depends on Foxa1/2 proteins. We also found a significant reduction in Nurr1 binding to the Aadc neuronal promoter in the DATcre/+;Foxa1/2cko embryos (p = 0.041) (Fig. 9B). In contrast, no significant change was detected in Nurr1 binding to the Vmat2 promoter previously described by Jacobs et al. (2009) with or without Foxa1/2 (p = 0.128) (Fig. 9B). These results strongly suggest a role for Foxa1/2 in recruiting Nurr1 to the Th and Aadc promoters but not to the Vmat2 promoter.
Discussion
The expression of Foxa genes in mDA neurons
There have been some conflicting reports surrounding the expression pattern of the Foxa genes in the mDA neurons. For example, Cai et al. (2009) observed a decrease in Foxa2 expression in mDA neurons during development, resulting in no detectable expression of this gene at E15.5. The authors reported a reappearance of expression of Foxa2 at E18.5 and robust expression in the mDA neurons in the adult brain. In addition, although Wijchers et al. (2006) failed to detect Foxa1 in the VM using RT-PCR, they did observe Foxa1 expression in the adult mDA neurons using in situ hybridization. They also failed to detect Foxa2 with both RT-PCR and in situ hybridization (Wijchers et al., 2006). Thus, there have been some differences in opinion about the expression pattern of the Foxa genes in the mDA neurons at various time points. We found no variation over time in Foxa gene expression in mDA neurons using both antibody staining and in situ hybridization. We analyzed numerous embryonic, early postnatal and adult time points and found both genes to be robustly expressed by the mDA neurons, in agreement with other publications (Thuret et al., 2004; Kittappa et al., 2007). Our conclusion is that Foxa1 and Foxa2 are expressed in mDA progenitors, immature and mature neurons during embryogenesis. In adults, Foxa1 expression becomes restricted to mDA neurons as development progresses, whereas Foxa2 exhibits a broader pattern of expression, being present in both mDA neurons and other surrounding nuclei in the VM.
Foxa1 and Foxa2 are required to maintain dopaminergic properties in mDA neurons
Our data showed that when Foxa1 and Foxa2 are deleted in mDA neurons from E13.5 onward, these cells failed to maintain their dopaminergic properties, such as expression of TH, VMAT2, AADC, and DAT. However, these neurons continue to express other mDA neuron markers such as Lmx1a, Lmx1b, and Nurr1. Despite the absence of characteristic dopaminergic proteins, mDA neurons remained in the midbrain at least until 18 months of age, and continued to express the neuronal marker, NeuN. The mDA neurons in the Nurr1 cko mice exhibit a similar phenotype, i.e., failure to maintain their dopamine phenotype. However, these mDA neurons progressively disappear in adult mice, suggesting a role for Nurr1 in regulating survival of these neurons (Kadkhodaei et al., 2009). Because Nurr1 is still expressed in Foxa1−/Foxa2− mDA neurons in DATcre/+;Foxa1/2 cko animals, expression of Nurr1 may contribute to the survival of these neurons. Alternatively, Foxa1/2 may contribute to the regulation of genes essential for cell death thereby leading to survival of the mutant mDA neurons in DATcre/+;Foxa1/2 cko adult mice. Determining downstream target genes in mDA neurons by expression profiling studies will help to distinguish these two possibilities.
The remaining mDA neurons in DATcre/+;Foxa1/2 cko adult mice appear to maintain the expression of VTA specific markers, such as Otx2 and calbindin, although losing the expression of SN-related proteins, for example, Girk2 and ALDH1A1. This result suggests that SN mDA neurons are more severely affected in DATcre/+;Foxa1/2 cko adult mice. As Foxa1 and Foxa2 are similarly deleted in Otx2+ VTA-specific mDA neurons as in the total mDA population, the specific requirements for maintenance of genes expressed in SN mDA neurons is interesting and is not a consequence of region-specific deletion of Foxa1/2. In contrast to the sparing of Otx2, calbindin, and calretinin expression, the expression of dopamine-specific genes such as TH, AADC, and Vmat are affected cell-autonomously in almost all mDA neurons in DATcre/+;Foxa1/2 animals. Further studies are needed to determine how Foxa1/2 contributes to the regulation of Girk2 and Aldh1a1 expression in SN mDA neurons.
Foxa1/2 and Nurr1 cooperate to regulate TH expression
As Nurr1 and Foxa1/2 genes are both required for TH expression and they synergistically cooperate to activate TH expression in forebrain neural progenitors (Lee et al., 2010), we examined potential mechanism of transcriptional activation by both factors. ChIP assays using chromatin prepared from DATcre/+;Foxa1/2 cko versus chromatin from WT embryos suggest that Foxa2 facilitate the interaction of Nurr1 with DNA (both the Th and Aadc promoters in this study). Reciprocally, Nurr1 has been suggested to facilitate binding of Foxa to its target sequences (Lee et al., 2010). Our results did not show a synergistic interaction for binding to the Vmat2 promoter by Foxa2 and Nurr1. Either this interaction is weaker and may require more cases to be tested for the result to acquire statistical significance, or Foxa2 and Nurr1 regulate Vmat2 by a different mechanism that does not involve cooperative binding to the same regulatory sequences.
Previous studies show that Foxa1/2 are required for the expression of Nurr1 (Ferri et al., 2007) and here we show that Foxa1/2 subsequently cooperate with Nurr1 in a feedforward manner to regulate common downstream target genes, such as Th and Aadc in mDA neurons. We suggest that Foxa1/2 are required for Nurr1 to bind to its target sequence on the Th and Aadc promoter. Interestingly, this function of Foxa1/2 is similar to established roles for Foxa1 in facilitating the binding of other nuclear hormone receptors to their cognate DNA sites, such as estrogen receptor in breast cancer cells (Carroll et al., 2005; Laganiere et al., 2005) and androgen receptor in prostate cancer cells (Wang et al., 2007), respectively. Given the common roles of Foxa1/2 and Nurr1 in regulating the maintenance of the mDA phenotype, it will be interesting in the future to determine what are common target genes of Foxa1/2 and Nurr1 in mDA neurons.
Abnormal hopping gait in DATcre/+;Foxa1/2 cko mice
Hopping behavior in mice has previously been associated with spinal cord issues (such as the Ephrin-B3−/− mouse) (Kullander et al., 2001). The main dopaminergic projections down the spinal cord originate in the diencephalic A11 dopamine population (Bjorklund and Skagerberg, 1979). In the DATcre/+;Foxa1/2 cko mice, we found no effect on this population of cells compared with the WT A11 population and their projections down the spine appeared normal (data not shown). It is interesting to note that a previous publication of the Foxa2+/− mouse reported that the mice develop late-onset gait problems with incomplete penetrance (Kittappa et al., 2007). In contrast to the severe reduction in the number of mDA neurons reported in some Foxa2+/− mice with abnormal gait (Kittappa et al., 2007), most of the Foxa1/2-deleted mDA neurons survive in adult DATcre/+;Foxa1/2 cko mice. These differing phenotypes may be due to the different genetic modifications in the two strains of mutant mice, i.e., loss of one copy of Foxa2 at conception in Foxa2+/− mice or a mosaic deletion of Foxa1 and Foxa2 in approximately one-half of the mDA neurons from E13.5 onward in DATcre/+;Foxa1/2 cko mice. Future work examining the role of Foxa1 and Foxa2 in adult mDA neurons will provide insights into understanding these distinct phenotypes.
Footnotes
This work was supported by Medical Research Council United Kingdom, and by a research Grant from Parkinson's United Kingdom (S.-L.A.). We thank Suzanne Claxton for technical support and members of the laboratory for critical reading of this paper.
The authors declare no competing financial interests.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brûlet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Ang SL. Foxa1 and Foxa2 transcription factors regulate differentiation of midbrain dopaminergic neurons. Adv Exp Med Biol. 2009;651:58–65. doi: 10.1007/978-1-4419-0322-8_5. [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Wills KV, Triarhou LC, Ghetti B. Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp Brain Res. 1995;105:191–199. doi: 10.1007/BF00240955. [DOI] [PubMed] [Google Scholar]
- Björklund A, Skagerberg G. Evidence for a major spinal cord projection from the diencephalic A11 dopamine cell group in the rat using transmitter-specific fluorescent retrograde tracing. Brain Res. 1979;177:170–175. doi: 10.1016/0006-8993(79)90927-2. [DOI] [PubMed] [Google Scholar]
- Broccoli V, Boncinelli E, Wurst W. The caudal limit of Otx2 expression positions the isthmic organizer. Nature. 1999;401:164–168. doi: 10.1038/43670. [DOI] [PubMed] [Google Scholar]
- Cai J, Donaldson A, Yang M, German MS, Enikolopov G, Iacovitti L. The role of Lmx1a in the differentiation of human embryonic stem cells into midbrain dopamine neurons in culture and after transplantation into a Parkinson's disease model. Stem Cells. 2009;27:220–229. doi: 10.1634/stemcells.2008-0734. [DOI] [PubMed] [Google Scholar]
- Carlsson T, Carta M, Winkler C, Bjorklund A, Kirik D. Serotonin neuron transplants exacerbate l-DOPA-induced dyskinesias in a rat model of Parkinson's disease. J Neurosci. 2007;27:8011–8022. doi: 10.1523/JNEUROSCI.2079-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Licznerski P, Alavian KN, Simeone A, Lin Z, Martin E, Vance J, Isacson O. The transcription factor orthodenticle homeobox 2 influences axonal projections and vulnerability of midbrain dopaminergic neurons. Brain. 2010;133:2022–2031. doi: 10.1093/brain/awq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Omodei D, Acampora D, Simeone A. Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int J Dev Biol. 2010a;54:939–945. doi: 10.1387/ijdb.092974ms. [DOI] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci. 2010b;13:1481–1488. doi: 10.1038/nn.2661. [DOI] [PubMed] [Google Scholar]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Filosa S, Rivera-Pérez JA, Gómez AP, Gansmuller A, Sasaki H, Behringer RR, Ang SL. Goosecoid and HNF-3beta genetically interact to regulate neural tube patterning during mouse embryogenesis. Development. 1997;124:2843–2854. doi: 10.1242/dev.124.14.2843. [DOI] [PubMed] [Google Scholar]
- González-Hernández T, Barroso-Chinea P, De La Cruz Muros I, Del Mar Pérez-Delgado M, Rodríguez M. Expression of dopamine and vesicular monoamine transporters and differential vulnerability of mesostriatal dopaminergic neurons. J Comp Neurol. 2004;479:198–215. doi: 10.1002/cne.20323. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- Jolicoeur FB, Rondeau DB, Hamel E, Butterworth RF, Barbeau A. Measurement of ataxia and related neurological signs in the laboratory rat. Can J Neurol Sci. 1979;6:209–215. doi: 10.1017/s0317167100119663. [DOI] [PubMed] [Google Scholar]
- Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S, Sumi-Ichinose C, Nomura T, Metzger D, Chambon P, Lindqvist E, Larsson NG, Olson L, Björklund A, Ichinose H, Perlmann T. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29:15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, Lim JK, Anderson T, Isacson O. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem. 2003;85:622–634. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk D, Kania A. Identification of genes controlled by LMX1B in the developing mouse limb bud. Dev Dyn. 2008;237:1183–1192. doi: 10.1002/dvdy.21514. [DOI] [PubMed] [Google Scholar]
- Kullander K, Croll SD, Zimmer M, Pan L, McClain J, Hughes V, Zabski S, DeChiara TM, Klein R, Yancopoulos GD, Gale NW. Ephrin-B3 is the midline barrier that prevents corticospinal tract axons from recrossing, allowing for unilateral motor control. Genes Dev. 2001;15:877–888. doi: 10.1101/gad.868901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganière J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguère V. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Bae EJ, Yi SH, Shim JW, Jo AY, Kang JS, Yoon EH, Rhee YH, Park CH, Koh HC, Kim HJ, Choi HS, Han JW, Lee YS, Kim J, Li JY, Brundin P, Lee SH. Foxa2 and Nurr1 synergistically yield A9 nigral dopamine neurons exhibiting improved differentiation, function, and cell survival. Stem Cells. 2010;28:501–512. doi: 10.1002/stem.294. [DOI] [PubMed] [Google Scholar]
- Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH, Ang SL. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Marin F, Herrero MT, Vyas S, Puelles L. Ontogeny of tyrosine hydroxylase mRNA expression in mid- and forebrain: neuromeric pattern and novel positive regions. Dev Dyn. 2005;234:709–717. doi: 10.1002/dvdy.20467. [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- McRitchie DA, Hardman CD, Halliday GM. Cytoarchitectural distribution of calcium binding proteins in midbrain dopaminergic regions of rats and humans. J Comp Neurol. 1996;364:121–150. doi: 10.1002/(SICI)1096-9861(19960101)364:1<121::AID-CNE11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Mendez I, Sanchez-Pernaute R, Cooper O, Viñuela A, Ferrari D, Björklund L, Dagher A, Isacson O. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson's disease. Brain. 2005;128:1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet S, Campbell K, Epstein DJ, Losos K, Harris E, Joyner AL. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature. 1999;401:161–164. doi: 10.1038/43664. [DOI] [PubMed] [Google Scholar]
- Omodei D, Acampora D, Mancuso P, Prakash N, Di Giovannantonio LG, Wurst W, Simeone A. Anterior–posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Patel S, Hillard CJ. Cannabinoid CB1 receptor agonists produce cerebellar dysfunction in mice. J Pharmacol Exp Ther. 2004;297:629–637. [PubMed] [Google Scholar]
- Raynal JF, Dugast C, Le Van Thaï A, Weber MJ. Winged helix hepatocyte nuclear factor 3 and POU-domain protein brn-2/N-oct-3 bind overlapping sites on the neuronal promoter of human aromatic l-amino acid decarboxylase gene. Brain Res Mol Brain Res. 1998;56:227–237. doi: 10.1016/S0169-328X(98)00048-5. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, Graw J, Burbach JP. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L, Barraud P, Andersson E, Kirik D, Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J Neurosci. 2005;25:6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret S, Bhatt L, O'Leary DD, Simon HH. Identification and developmental analysis of genes expressed by dopaminergic neurons of the substantia nigra pars compacta. Mol Cell Neurosci. 2004;25:394–405. doi: 10.1016/j.mcn.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Hoekman MF, Burbach JP, Smidt MP. Identification of forkhead transcription factors in cortical and dopaminergic areas of the adult murine brain. Brain Res. 2006;1068:23–33. doi: 10.1016/j.brainres.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31:12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Masson J, Gingrich JA, Rayport S, Hen R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J Neurosci Methods. 2005;143:27–32. doi: 10.1016/j.jneumeth.2004.09.020. [DOI] [PubMed] [Google Scholar]