Abstract
Mechanisms that regulate oligodendrocyte (OL) precursor migration and differentiation are important in normal development and in demyelinating/remyelinating conditions. We previously found that the tetraspanin CD82 is far more highly expressed in O4+ OL precursors of the adult rat brain than those of the neonatal brain. CD82 has been physically linked to cMet, the hepatocyte growth factor (HGF) receptor, in tumor cells, and this interaction decreases downstream signaling. We show here that CD82 inhibits the HGF activation of cMet in neonatal and adult rat OL precursors. CD82 expression is sufficient to allow precursor differentiation into mature OLs even in the presence of HGF. In contrast, CD82 downregulation in adult O4+/CD82+ cells inhibits their differentiation, decreases their accumulation of myelin proteins, and causes a reversion to less mature stages. CD82 expression in neonatal O4+/CD82− cells also blocks Rac1 activation, suggesting a possible regulatory effect on cytoskeletal organization and mobility. Thus, CD82 is a negative regulator of HGF/cMet during OL development and overcomes HGF inhibitory regulation of OL precursor maturation.
Introduction
One remarkable feature of oligodendrocyte (OL) development is that a significant number of OL precursors do not differentiate into myelinating OLs but rather remain in an immature and cycling state into and during adulthood. Many of them express NG2 and PDGF receptor α (PDGFRα), whereas others are less mature and others appear to have progressed farther along in the lineage. This overall population is thought to provide new OLs in later stages of myelination, during normal myelin turnover and during remyelination subsequent to demyelinating pathologies. What keeps these cells immature in the normal adult brain and how they begin a differentiation program that leads to myelination are not fully understood.
Growth factor levels regulate the numbers of immature OLs in the adult CNS. The best-known factor is PDGF, a powerful mitogen that is known to promote precursor proliferation both in vitro and in vivo (Noble et al., 1988; van Heyningen et al., 2001; Assanah et al., 2006, 2009). PDGF signaling is turned off by the loss of PDGFRα as precursors mature and by the limiting concentrations of PDGF in the adult CNS. However, several observations indicate that the hepatocyte growth factor (HGF)–cMet signaling pathway is active during OL development, keeps precursors immature, and inhibits myelination in vivo (Ohya et al., 2007). HGF is also found in OL precursors, promotes proliferation, and keeps them immature in vitro (Yan and Rivkees, 2002). Because HGF continues to be expressed in the adult CNS (Jung et al., 1994; Achim et al., 1997) and cMet is expressed by both immature and mature OLs (Yan and Rivkees, 2002), the mechanism(s) for inhibiting this pathway must be different from those that inhibit PDGF-driven proliferation.
In this study, we show that CD82 inhibits cMet activation in OL precursors. We initially found CD82, which encodes a transmembrane tetraspanin glycoprotein, to be more highly expressed in O4+ cells isolated from the adult rat white matter than in those from neonatal forebrain (Lin et al., 2009). CD82 had not been identified previously in OLs, although it is well known for its roles in T-cell activation (Lagaudriere-Gesbert et al., 1998; Shibagaki et al., 1998, 1999) and metastasis (Dong et al., 1995; Ono et al., 1999; Yang et al., 2001; Jackson et al., 2005; Liu and Zhang, 2006; Miranti, 2009). We found that CD82 is expressed in OL lineage cells and that its constitutive expression in vivo promotes precursor differentiation and myelination, whereas its downregulation keeps precursors immature (Mela and Goldman, 2009).
Here we report that CD82 expression in neonatal O4+/CD82− precursors inhibits cMet activation by HGF and promotes their differentiation into later-stage OLs, even in the continued presence of HGF. In contrast, downregulation of CD82 in adult O4+/CD82+ cells inhibits their differentiation, reverts them to a less mature stage, and decreases myelin protein levels. This reversion appears to be more complete in the presence of HGF. CD82 also acts as a regulator of HGF–cMet effects on Rac1 activation: CD82 overexpression in neonatal O4+/CD82− cells blocks Rac1 activation, suggesting a possible regulatory effect on cytoskeletal organization and mobility.
Materials and Methods
Isolation of O4+ cells.
Sprague Dawley adult female (3 months old) and neonatal rats (P0–P1) were used. Neonatal forebrains (with striata, hippocampi, and meninges removed) and adult subcortical white matter were mechanically and enzymatically dissected as described previously (Mela and Goldman, 2009). All animal experiments were performed under the guidelines of the Columbia University Institutional Animal Care and Use Committee. After isolation, O4+ cells were cultured in modified N2B3 (promotes differentiation) or O2A medium (promotes growth) as described previously (Mela and Goldman, 2009).
Antibodies.
The following primary antibodies were used: rabbit anti-CD82 C-16 (1:100; Santa Cruz Biotechnology), mouse anti-cMet (1:50; Santa Cruz Biotechnology), mouse anti-cMet (1:200; Cell Signaling Technology), rabbit anti-phospho-cMet (1:50; Invitrogen), mouse IgMs O4 and O1 (1:50, hybridoma supernatants; American Type Culture Collection), mouse anti-GFP (1:200; Invitrogen), rabbit anti-GFAP (1:500; Dako), mouse anti-CC1 (1:50; Calbiochem), mouse anti-MBP (1:500; Covance), rabbit anti-PDGFRα (1:200; Cell Signaling Technology), rabbit anti-olig2 (1:100; Millipore), mouse anti-NG2 (1:500; kind gift from Dr. W. Stallcup, Burnham Institute, La Jolla, CA), and mouse anti-MAG (1:500; kind gift from Dr. M. Filbin, Hunter College, New York, NY).
Cell culture and immunofluorescence.
O4+ progenitors were resuspended in N2B3 medium and plated on poly-l-lysine-coated eight-well glass chamber slides (Nunc) at 2000 cells per well. N2B3 medium was changed every 2–3 d. Immunofluorescence was performed as described previously (Mela and Goldman, 2009) after fixation in 4% paraformaldehyde (PFA).
Immunofluorescence on rat and human brain sections.
Neonatal and adult rat brains were obtained after transcardial perfusion with 4% PFA. Brains were cryoprotected in 30% sucrose and stored in OCT at −80°C. Cryosections (10 μm) were fixed in 4% PFA for 10 min at room temperature, washed in PBS, blocked with 5% horse serum (Sigma) for 30 min, and then labeled with primary antibodies overnight at 4°C. Sections were then washed three times with PBS and incubated in Alexa Fluor-conjugated secondary antibodies (1:1000; Invitrogen) for 1 h at room temperature and counterstained.
Postmortem normal adult human white matter paraffin sections were obtained by the Neuropathology Department at Columbia University Medical Center. Sections were processed for paraffin removal through xylene and ethanol washes, followed by antigen retrieval (boiling sections for 15 min in 10 mm sodium citrate, pH 6.0). Sections were washed in PBS, blocked with 5% horse serum (Sigma) for 30 min, and then labeled with primary antibodies at 4°C for 48 h. Sections were then washed three times with PBS, incubated in Alexa Fluor-conjugated secondary antibodies (1:1000; Invitrogen) for 1 h at room temperature, and counterstained.
Microscopy and cell counting.
Stained fixed cells and tissue sections were examined and photographed using a Carl Zeiss Axiophot 200 fluorescent microscope equipped with an Axiocam (Carl Zeiss) and OpenLab imaging software (Improvision). Micrographs were merged using Adobe Photoshop (Adobe Systems) or NIH ImageJ. Five representative 40× (objective) fields were photographed, and cells staining positive for each marker were manually counted.
Western blot.
O4+ cells were lysed using the CelLyticM Cell Lysis Reagent with Protease Inhibitor Cocktail (both from Sigma) or MLB buffer (for Rho GTPase activation assay; Mg2+ Lysis Wash Buffer, Millipore), and Western blot analysis of proteins was performed as described previously (Mela and Goldman, 2009).
Retroviral production.
The retroviruses that we used were described previously (Mela and Goldman, 2009). The CD82–IRES–EGFP retrovirus expresses the rat CD82 cDNA inserted into a modified pQCXIX–IRES–EGFP vector (Clontech). The modified vector pQCXIX–IRES–EGFP was used for the production of control retrovirus. The shRNA–ZsGreen retrovirus expresses the s1 (GAGCAGCTTCATTTCCGTC) CD82-specific shRNA cloned into the pSIREN–RetroQ–ZsGreen vector (Clontech). Retroviral aliquots (10 μl with titer 106 colony forming units) were stored at −80°C. To infect neonatal and adult O4+ cells in vitro, we added 10 μl of retrovirus in 5 ml of N2B3 medium immediately after isolation. After 2 d, we changed the medium and we were able to observe infected GFP+ cells (70–80% of neonatal cells or 40–50% of adult cells).
Immunoprecipitation.
Total protein extracts (prey proteins) from acutely isolated adult white matter O4+ cells were incubated with antibody-coupled gel according to the guidelines of the Mammalian Immunoprecipitation Kit (Pierce). The following antibodies were used as bait: rabbit anti-CD82 (70 μg; Santa Cruz Biotechnology), rabbit anti-c-Met (70 μg; Santa Cruz Biotechnology), and mouse anti-GAPDH (70 μg; EnCor) as control. The resulting eluted coimmunoprecipitation complex was analyzed by Western blot with the other antibody.
Rho GTPase activation assay.
The activation of Rac1, cdc42, and RhoA was measured using pull-down assay kits (Rac1/Cdc42 and Rho Activation Assay Kits; Millipore). Acutely isolated O4+ cells from the neonatal rat forebrain were infected with retrovirus and were left in culture for 2 d. On the day of the assay, fresh medium was added and the cells were left to settle for 3 h. New medium with HGF (50 μg/ml; Invitrogen) was added, and cells were lysed at 0, 5, 15, 30, and 60 min after HGF addition. Protein extracts were prepared and analyzed according to the kit guidelines, and samples containing active GTPases were analyzed by Western blot.
Statistical analyses.
Cell culture results (process number and marker expression) were analyzed using χ2 test analyses. cMet activation and Rho activation assay results were analyzed using paired, two-tailed Student's t test. Differences were considered significant when p < 0.05.
Results
cMet is expressed in O4+ cells and in adult rat and human white matter OLs
cMet mRNA and protein are present throughout the mouse brain (Judson et al., 2009), particularly in axons, but they have not been localized to specific glial cell types. We performed immunofluorescence in the neonatal (P1–P2), young (P7), and adult (P90) rat brain for cMet and CD82 as well as for markers of OL development (CC1). cMet is found throughout the neonatal brain in individual cells in the white matter that are CC1+ (Fig. 1A). As we showed previously (Mela and Goldman, 2009), CD82 is not present in the neonatal brain. In the P7 brain, cMet colocalizes to some extent with CD82 and CC1 (Fig. 1B). However, not all cMet+ cells were also CD82+ or CC1+ (Fig. 1B). At this time, many of the cMet+/CD82− and CC1− cells are too immature to have either CD82 or CC1. Note that Ohya et al. (2007) found that NG2+ cells were also cMet+. In the adult brain, cMet is localized to cells in the white matter that also express CC1 or CD82 (Fig. 1C), but we also found CC1+/cMet− and CD82+/cMet− cells, suggesting that cMet may eventually be turned off in more mature OLs the adult brain. We also performed immunofluorescence in white matter sections taken from normal adult human brain, in which cMet appears to be present in cells that have the morphology and distribution of OLs. There is no colocalization with GFAP (Fig. 1D).
Figure 1.
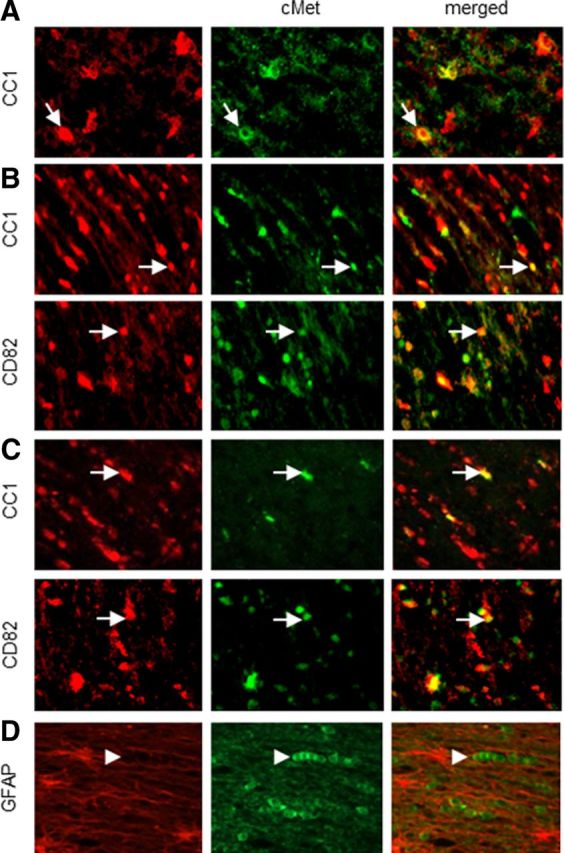
cMet expression in the normal rat and human brain. Immunofluorescence in white matter from neonatal (P0–P1) (A), young (P7) (B), and adult (3 months old) (C) rat brain. cMet is present in CD82+ and CC1+ cells in the white matter. D, Immunofluorescence in normal adult human white matter. cMet is not present in GFAP+ astrocytes. Representative 40× fields are shown. Arrows show double-positive cells, and arrowheads show cMet+ cells. Final magnification, 250×.
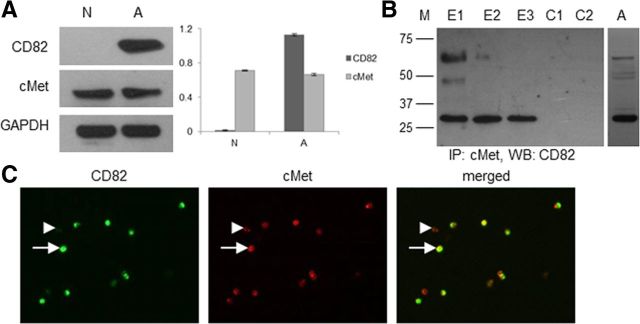
To determine whether cMet is present in O4+ cells, we performed Western blot analysis on acutely isolated O4+ cells from neonatal rat forebrain and adult rat white matter. We found cMet in both adult and neonatal O4+ cells, whereas CD82 is present only in the adult cells (Fig. 2A). Immunofluorescence on acutely isolated adult O4+ cells revealed that all of the O4+/CD82+ cells also express cMet (Fig. 2C). There is a small percentage (11.96%) of adult O4+/cMet+/CD82− cells (in a total of 92 cells, 81 were cMet+/CD82+ and 11 were cMet+/CD82−). These adult O4+/cMet+/CD82− cells may represent an earlier stage in the OL lineage, before the expression of CD82.
Figure 2.
CD82 and cMet expression in acutely isolated O4+ cells. A, Western blot analysis and normalization diagram of total protein extracts from O4+ cells isolated from neonatal rat forebrain and adult rat white matter. GAPDH is used as control (N, neonatal; A, adult). Analysis was performed three times, and a representative blot is shown. B, Total protein extracts from acutely isolated adult O4+ cells were used for immunoprecipitation (IP) for cMet and subsequent Western blot analysis (WB) for CD82 under nonreducing conditions. Glycosylated (∼50 and ∼60 kDa) and nonglycosylated (∼30 kDa) forms of CD82 are present in the precipitate (M, protein marker bands at 75, 50, 37, and 25 kDa; E1–E3, sequential elutions of the precipitate; C1, control quenched gel slurry–activated gel with no bait antibody; C2, control activated gel with GAPDH antibody). Analysis was performed three times, and a representative blot is shown. The glycosylated and nonglycosylated forms are also shown in a representative blot of total protein extract from adult O4+ cells analyzed under native conditions (A). C, Immunofluorescence for CD82 and cMet on acutely isolated O4+ cells from the adult rat white matter. Arrows show CD82+/cMet+ cells, and arrowheads show cMet+/CD82− cells. A representative 40× field is shown. Final magnification, 250×.
In several cancer cell types, cMet and CD82 can associate with each other, providing evidence for a physical link between these two membrane proteins and a possible interaction (Takahashi et al., 2007; Todeschini et al., 2007). Therefore, we wanted to determine whether these two molecules interact, because they are both present in adult O4+ cells. Immunoprecipitation for cMet and subsequent Western blot analysis for CD82 revealed that cMet and CD82 coimmunoprecipitate in acutely isolated adult O4+ cells (Fig. 2B). We used nonreducing conditions that permit isolation with the least possible interference in protein structure. As a result, we were able to isolate CD82 in both native (29 kDa) and glycosylated (46–60 kDa) forms after three sequential elution steps (Fig. 2B, E1–E3). Specificity was assayed by omitting the cMet antibody (Fig. 2B, C1) or by using an antibody against GAPDH (irrelevant to the cMet/CD82 interaction but present in adult O4+ cells; Fig. 2B, C2). Both controls confirmed specificity of the assay.
Effects of HGF and CD82 on neonatal OL precursors
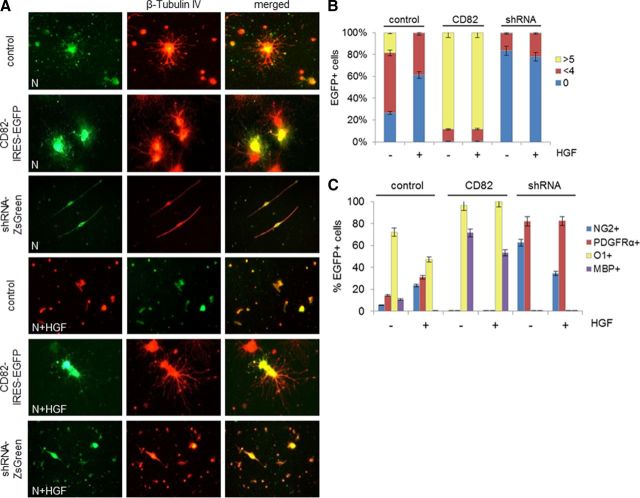
To define the effects of HGF and CD82 on neonatal OLs, we isolated O4+ cells from the neonatal brain and grew them in culture in the presence or absence of HGF. To visualize cell morphology, we performed immunofluorescence for β-tubulin IV, a protein of the cytoskeleton that is expressed in cells of the OL lineage (Terada et al., 2005), and to assess maturity, we immunostained cultures with antibodies to NG2, PDGFRα, and MBP and with the O1 monoclonal antibody. We infected cells directly after isolation with a CD82–IRES–EGFP or an shRNA–ZsGreen retrovirus, which constitutively express CD82 or downregulate CD82, respectively (Mela and Goldman, 2009). We also used a control EGFP-expressing retrovirus (pQCXIX–IRES–EGFP). Cells were cultured in the continued presence of HGF (50 ng/ml) or without HGF for 7 d.
Control virus-infected cells showed some degree of maturation in the absence of HGF: most were O1+, 10% were MBP+, and small proportions were PDGFRα+ and NG2+. In the presence of HGF, greater percentages were PDGFRα+ and NG2+, fewer were O1+, and none was MBP+. Note that, although none of the neonatal O4+ cells initially was CD82+, all of the O1+ cells in control cultures became CD82+, showing the acquisition of the tetraspanin over the culture period.
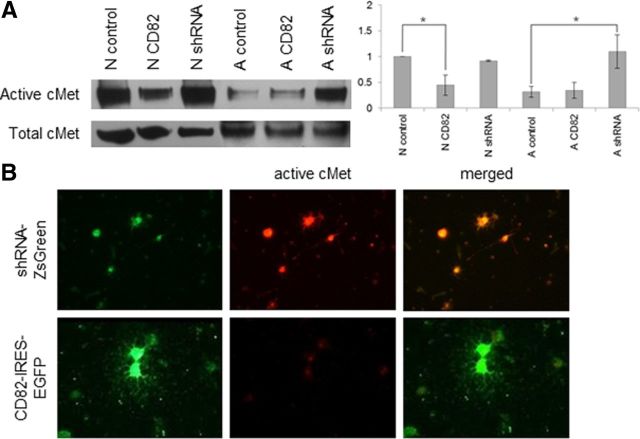
When we infected neonatal cells with the CD82-expressing virus, the cells became more mature. In the presence of HGF, most became positive for O1 and MBP (100 and 53.33% of CD82–EGFP+ cells, respectively), none remained either PDGFRα+ or NG2+, and the vast majority of them had grown more than five processes (88.64%) (Fig. 3, Table 1). In the absence of HGF, we observed the same maturation O1+ (96.67%) or MBP+ (71.43%), and no cells were either PDGFRα+ or NG2+. In the presence of CD82, these cells no longer activated cMet (Fig. 4A,B).
Figure 3.
CD82 and HGF effects on neonatal O4+ cells in vitro. A, Immunofluorescence for β-tubulin IV on neonatal O4+ cells infected with CD82–IRES–EGFP, shRNA–ZsGreen, or control–IRES–EGFP retrovirus after 7 d in vitro in the absence or presence of HGF. Representative 40× fields are shown. Final magnification, 250×. B, Diagram of percentages (as presented in Table 1) of GFP+ cells (neonatal O4+ cells infected with CD82–IRES–EGFP, shRNA–ZsGreen, or control–IRES–EGFP retrovirus) in relation to number of processes: round cells with no processes (0) and cells with less than four (<4) or more than five (>5) processes, after 7 d in culture with (+) or without (−) HGF. C, Diagram of percentages of GFP+ cells (as described in B) in relation to NG2, PDGFRα, O1, and MBP expression after 7 d in culture with (+) or without (−) HGF (N, neonatal; control, control–IRES–EGFP; CD82, CD82–IRES–EGFP; shRNA, shRNA–ZsGreen).
Table 1.
CD82 expression effects in neonatal and adult O4+ cells after 7 d in vitro in the presence or absence of HGF
|
Control–EGFP |
CD82–IRES–EGFP |
shRNA–ZsGreen |
||||
|---|---|---|---|---|---|---|
| −HGF | +HGF | −HGF | +HGF | −HGF | +HGF | |
| Neonatal | ||||||
| Process number | ||||||
| 0 | 23 (26.7%) | 37 (61.7%) | 0 (0%) | 0 (0%) | 42 (84%) | 58 (78.4%) |
| <4 | 47 (54.7%) | 23 (38.3%) | 6 (11.1%) | 5 (11.4%) | 8 (16%) | 16 (21.6%) |
| >5 | 16 (18.6%) | 0 (0%) | 48 (88.9%) | 39 (88.6%) | 0 (0%) | 0 (0%) |
| Marker expression | ||||||
| NG2 | 3 of 56 (5.4%) | 11 of 47 (23.4%) | 0 (0%) | 0 (0%) | 25 of 40 (62.5%) | 20 of 58 (34.5%) |
| PDGFRα | 7 of 49 (14.3%) | 13 of 42 (30.9%) | 0 (0%) | 0 (0%) | 41 of 50 (82%) | 42 of 51 (82.4%) |
| O1 | 31 of 43 (72.1%) | 26 of 55 (47.3%) | 29 of 30 (96.7%) | 35 of 35 (100%) | 0 (0%) | 0 (0%) |
| MBP | 5 of 47 (10.6%) | 0 (0%) | 40 of 56 (71.4%) | 32 of 60 (53.3%) | 0 (0%) | 0 (0%) |
| Adult | ||||||
| Process number | ||||||
| 0 | 16 (11.8%) | 14 (18.4%) | 0 (0%) | 0 (0%) | 5 (12.2%) | 23 (56.1%) |
| <4 | 44 (32.3%) | 23 (30.3%) | 14 (29.2%) | 12 (28.6%) | 28 (68.3%) | 18 (43.9%) |
| >5 | 76 (55.9%) | 39 (51.3%) | 34 (70.8%) | 30 (71.4%) | 8 (19.5%) | 0 (0%) |
| Marker expression | ||||||
| NG2 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 14 of 40 (35%) |
| PDGFRα | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 44 of 54 (81.5%) | 26 of 44 (59.1%) |
| O1 | 52 of 52 (100%) | 49 of 49 (100%) | 44 of 44 (100%) | 37 of 37 (100%) | 24 of 64 (37.5%) | 0 (0%) |
| MBP | 20 of 42 (47.6%) | 14 of 55 (25.5%) | 25 of 32 (78.1%) | 28 of 58 (48.3%) | 0 (0%) | 0 (0%) |
Five representative 40× fields from two separate experiments were photographed per marker. Green and double-positive cells and their number of processes were manually counted. Statistical analysis was performed using χ2 test. There are statistically significant differences in process numbers between all populations in the presence and absence of HGF, with the exception of control–EGFP+ adult cells. There are statistically significant differences in marker expression between all populations in the presence and absence of HGF, with the exception of NG2, PDGFRα (0% expected and observed cells), and O1 expression between adult CD82–IRES–EGFP+ and control–EGFP+ cells. Number of processes: neonatal cells, x2CD82–IRES–EGFP+/control–EGFP+ (−HGF) = 9.098e−72, x2shRNA–ZsGreen+/control–EGFP+ (−HGF) = 2.056e−37, x2CD82–IRES–EGFP+/control–EGFP+ (+HGF) = 2.773e−19, x2shRNA–ZsGreen+/control–EGFP+(+HGF) = 0.000592, x2control–EGFP+ (±HGF) = 8.523e−16; adult cells, x2CD82–IRES–EGFP+/control–EGFP+ (−HGF) = 0.000324, x2shRNA–ZsGreen+/control–EGFP+ (−HGF) = 1.372e−14, x2CD82–IRES–EGFP+/control–EGFP+ (+HGF) = 1.878e−06, x2shRNA–ZsGreen+/control–EGFP+ (+HGF) = 5.774e−30, x2control–EGFP+ (±HGF) = 0.12283 (x20.05 = 0.103, df = 2). Marker expression: neonatal cells, CD82–IRES–EGFP+/control–EGFP+, x2NG2+ = 8.025e−08, x2PDGFRα+ = 1.779e−11, x2O1+ = 2.568e−16, x2MBP+ = 1.177e−24; shRNA–ZsGreen+/control–EGFP+, x2NG2+ = 1.805e−134, x2PDGFRα+ = 2.292e−90, x2O1+ = 8.56e−28, x2MBP+ = 0.00113, x2control–EGFP+ (±HGF) = 3.431e−21; adult cells, CD82–IRES–EGFP+/control–EGFP+, x2O1+ = 1, x2MBP+ = 2.63371e−10; shRNA–ZsGreen+/control–EGFP+, x2NG2+ = 0, x2PDGFRα+ = 1.96793e−32, x2O1+ = 4.26804e−32, x2MBP+ = 1.23245e−17, x2control–EGFP+ (±HGF) = 0.00136 (x20.05 = 0.00393, df = 1).
Figure 4.
CD82 effects on cMet activation in vitro. A, Western blot analysis and normalization diagram for total and active cMet on total protein extracts isolated from neonatal and adult O4+ cells, uninfected or infected with CD82–IRES–EGFP or shRNA–ZsGreen retrovirus, after 2 d in vitro in the presence of HGF. Neonatal cells expressing CD82 showed significantly less cMet activation than control cells (p = 0.0398). Adult cells after downregulating CD82 showed significantly higher activation of cMet than controls (p = 0.0414). The experiment was repeated three times, and a representative Western blot is shown (N, neonatal; A, adult; control, uninfected cells; CD82, CD82–IRES–EGFP; shRNA, shRNA–ZsGreen). B, Immunofluorescence for active cMet in neonatal O4+ cells infected with CD82–IRES–EGFP or shRNA–ZsGreen retrovirus after 7 d in vitro in the presence of HGF (representative 40× fields). *p < 0.05. Final magnification, 250×.
In contrast, shRNA virus-infected neonatal O4+ cells did not acquire O1 or MBP in the continued presence of HGF but rather remained immature, NG2+ (34.48%), or PDGFRα+ (82.35%) round cells with few (21.62%) or no (78.38%) processes. In the absence of HGF, cells also remained immature NG2+ (62.5%) or PDGFRα+ (82%).
Thus, the cells matured with CD82 and failed to mature without CD82 regardless of the presence of HGF. Control virus-infected cells matured to some extent but significantly less so in the presence of HGF.
Effects of HGF and CD82 on adult OL precursors
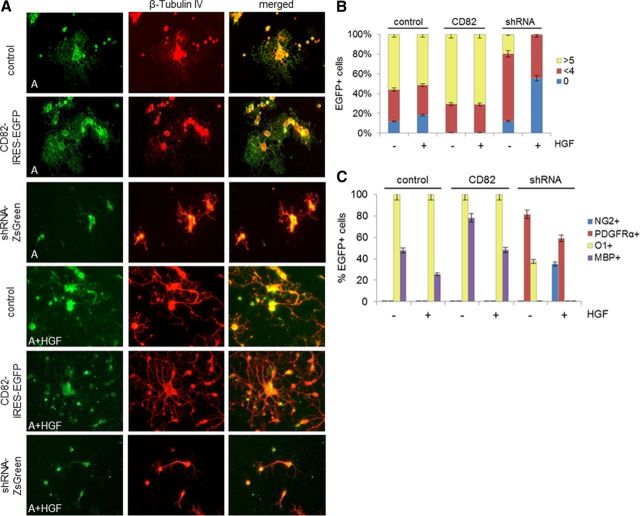
Adult O4+/CD82+ cells infected with the control virus remained O1+ (100%) or MBP+ (48%), although a smaller percentage of precursors were MBP+ in the presence of HGF than in its absence (Fig. 5, Table 1).
Figure 5.
CD82 and HGF effects on adult O4+ cells in vitro. A, Immunofluorescence for β-tubulin IV on adult O4+ cells infected with CD82–IRES–EGFP, shRNA–ZsGreen, or control–IRES–EGFP retrovirus after 7 d in vitro in the absence or presence of HGF. Representative 40× fields are shown. Final magnification, 250×. B, Diagram of percentages (as presented in Table 1) of GFP+ cells (adult O4+ cells infected with CD82–IRES–EGFP, shRNA–ZsGreen, or control–IRES–EGFP retrovirus) in relation to number of processes: round cells with no processes (0) and cells with less than four (<4) or more than five (>5) processes, after 7 d in culture with (+) or without (−) HGF. C, Diagram of percentages of GFP+ cells (as described in B) in relation to NG2, PDGFRα, O1, and MBP expression after 7 d in culture with (+) or without (−) HGF (A, adult; control, control–IRES–EGFP; CD82, CD82–IRES–EGFP; shRNA, shRNA–ZsGreen).
Adult O4+ cells infected with the CD82–IRES–EGFP retrovirus and grown in the presence of HGF became mature: O1+ (100%) or MBP+ (48.28%) with complex morphology (71.43% with more than five processes) and in the absence of HGF O1+ (100%) or MBP+ (78.13%) with lacy, extended processes (70.83% with more five processes). This maturation is not surprising because these cells already express CD82.
In contrast, CD82 downregulation in adult cells grown in the presence of HGF caused many to become PDGFRα+ (59.09%) or NG2+ (35%) and to appear as round cells with few (43.9%) or no (56.1%) processes (Fig. 5). None was O1+ or MBP+. Furthermore, these cells now activated cMet in response to HGF (Fig. 4). When we downregulated CD82 in adult precursors grown without HGF, we observed a different phenotype. Most cells became PDGFRα+ (81.48%), some were O1+ (37.5%), but surprisingly none became NG2+. Most of these cells displayed few (68.29%) or no (12.2%) processes, but a small proportion displayed more than five processes (19.51%). After CD82 was downregulated, the cells then activated cMet in response to HGF (Fig. 4A).
Thus, when CD82 was downregulated, precursors reverted to a less mature stage. In the presence of HGF, there were no O1+ cells and a significant number of NG2+ cells, but in the absence of HGF, 40% of the cells continued to be O1+ and none of the cells became NG2+.
Effects of CD82 on myelin protein levels
The O4+ cells isolated from adult rat white matter contain transcripts for a number of myelin and mature OL genes, such as MAG, MOG, PLP, and MBP (Lin et al., 2009). The encoded myelin proteins are also present in acutely isolated adult cells, with the exception of MBP (Lin et al., 2009). However, the adult cells will accumulate MBP in culture (Fig. 5). We asked whether CD82 expression or downregulation altered the levels of proteins.
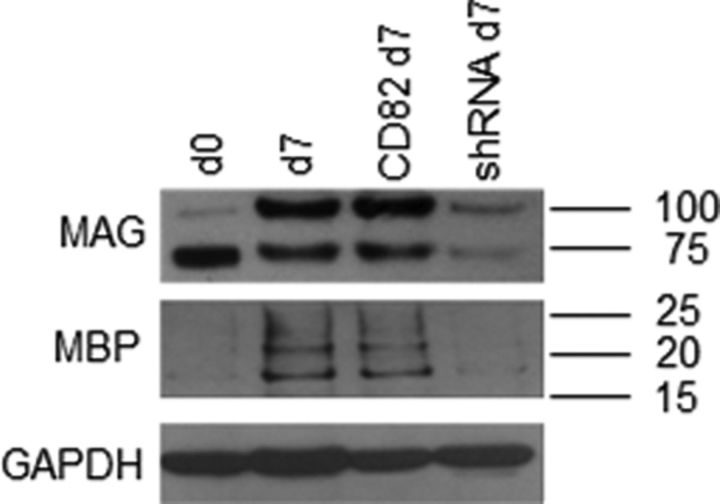
Adult O4+ cells were infected with CD82–IRES–EGFP, shRNA–ZsGreen, or control retroviruses, and, after 7 d in culture, total proteins were analyzed by Western blot. Acutely isolated adult O4+ cells contained MAG—the migration on the gel suggests the nonglycosylated form (Pedraza et al., 1990)—but not MBP, as reported previously (Lin et al., 2009), but over 7 d in culture, they accumulated MAG (both nonglycosylated and glycosylated forms) and MBP. Infecting these cells with the CD82–IRES–EGFP virus appeared to have no effect on protein levels, as might be anticipated. However, downregulating CD82 with the shRNA–ZsGreen virus decreased substantially protein levels of both MAG and MBP (Fig. 6). These observations imply that CD82 affects myelin protein levels. However, we do not know whether this represents a transcriptional or posttranscriptional regulation. Additionally, this effect may be separate from the function of CD82 in inhibiting cMet activation.
Figure 6.
CD82 downregulation prevents accumulation of MAG and MBP proteins. Western blot analysis for MAG and MBP proteins on total protein samples from acutely isolated (d0) adult O4+ cells or after 7 d (d7) in vitro and infected with either CD82–IRES–EGFP (CD82) or shRNA–ZsGreen (shRNA) retrovirus (GAPDH was used as control).
Rac1 RhoGTPase activation is delayed by CD82 expression
As another measure of the effect of CD82 on HGF–cMet activation, we examined the ability of HGF to activate small RhoGTPases in the presence and absence of CD82. The activation of Rac1, cdc42, and RhoA was measured using a time course assay, because small RhoGTPases can become active in a few minutes. We were not able to detect significant activation of RhoA or cdc42, but we found significant differences in the activation of Rac1.
Acutely isolated O4+ cells from the neonatal rat forebrain were infected with the CD82–IRES–EGFP retrovirus and were allowed to grow in culture, without HGF, for 2 d. On the day of the experiment, we changed the medium, added new medium without HGF, and let the cells settle for 3 h. Then, we fixed some cells (time 0), added fresh medium with HGF (50 ng/ml), and lysed the cells at specific time points after the addition of HGF (0, 5, 15, 30, and 60 min). Half of each sample was analyzed for the GTP-bound (active) form, whereas an equal amount from each sample was saved to analyze for total GTPase. As controls, we performed the same set of experiments using neonatal O4+ cells that were uninfected and also using neonatal cells infected with a control retrovirus that derives from the same pQCXIX vector with EGFP but without the CD82 cDNA (Mela and Goldman, 2009).
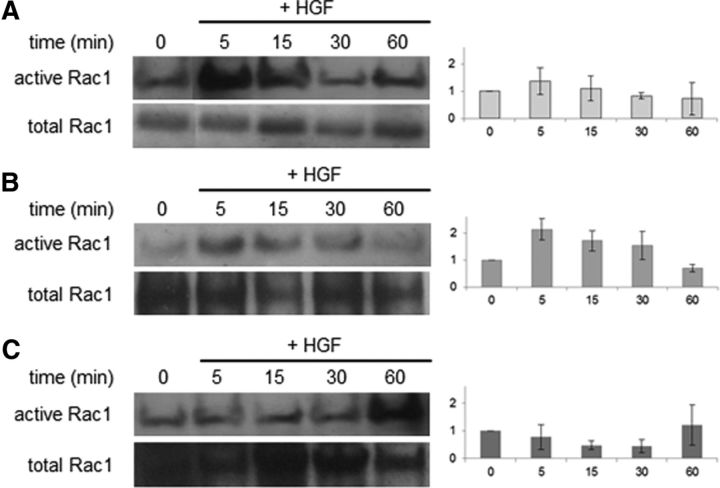
In the control virus-infected and uninfected cells, the addition of HGF induced a rapid activation of Rac1 (5 min) that gradually decreased (15–60 min) (Fig. 7A,B). In contrast, we found that the expression of CD82 significantly affected the kinetics of HGF-induced Rac1 activation. In CD82-expressing cells, Rac1–GTP levels remained approximately the same (5 min) or decreased (15–30 min) after the addition of HGF (Fig. 7C). Low Rac1 activation occurred at 60 min after the addition of HGF. This finding suggests that CD82 significantly blocks HGF-induced Rac1 activation in neonatal O4+/CD82− cells.
Figure 7.
Rac1 activation assay. Western blot analysis and normalization diagrams of active and total Rac1 in uninfected cells (A), infected cells with control–IRES–EGFP (B), or CD82–IRES–EGFP retrovirus, neonatal O4+ cells (C) at 2 d in vitro (0) and at 5, 15, 30, and 60 min after the addition of HGF. Rac1 activation is significantly different between the A and C cell populations at 5 and 30 min (pt test 5 min = 0.0015, pt test 15 min = 0.0663, pt test 30 min = 0.0294, pt test 60 min = 0.0777). Rac1 activation is significantly different between the B and C populations at 30 min (pt test 5 min = 0.1053, pt test 15 min = 0.056, pt test 30 min = 0.0294, pt test 60 min = 0.455). There are no significant differences between the two control (A, B) populations (pt test 5 min = 0.2657, pt test 15 min = 0.3337, pt test 30 min = 0.1904, pt test 60 min = 0.9418). Assay was performed three times, and representative results are shown.
Discussion
In this study, we describe the role of CD82 as a regulator of maturation and of HGF/cMet signaling in O4+ OL progenitors from the rat brain. We show that CD82 associates with cMet, blocking its activation by HGF and blocking HGF-induced Rac1 activation.
HGF and its receptor cMet, a tyrosine kinase encoded by the proto-oncogene cMet, are required for normal development. Mice lacking either HGF or cMet die during embryogenesis with defects in liver, placenta, or muscle development (Schmidt et al., 1995; Uehara et al., 1995). Both molecules are expressed during brain development and persist in the adult (Honda et al., 1995; Achim et al., 1997). cMet is expressed in neurons, OLs and their precursors, and microglia (Yan and Rivkees, 2002; Lalive et al., 2005; Kitamura et al., 2007; Ohya et al., 2007; Judson et al., 2009). Our finding that CD82 is expressed in NG2−/O4+ OL precursors but not in less mature cells suggests a potential role for this molecule during late OL development. The finding that NG2+ cells are cMet+ (Ohya et al., 2007) and CD82− (Mela and Goldman, 2009) further suggests that the HGF/cMet interaction plays a role in keeping these cells in cycle and/or preventing them from differentiating farther along the OL lineage. The transition from an NG2+/CD82− to an NG2−/CD82+ OL precursor is an essential step in normal OL development and in remyelination and indicates that the initial expression of CD82 occurs at a critical stage of OL development.
We found that adult O4+ (PDGFRα−/NG2−) OL precursors were able to revert to a PDGFRα+ phenotype after CD82 had been downregulated in the absence of HGF. Cells had also lost MBP. Interestingly, none of the precursors became NG2+ unless HGF was also added to the medium. This observation suggests that HGF–cMet signaling may play a role in positively regulating NG2 expression or that the loss of HGF–cMet signaling by CD82 during OL development is an important factor in the loss of NG2 in developing OL precursors. Another effect of HGF was to decrease the proportion of MBP+ cells in both control cells and in cells expressing CD82, in both the adult and neonatal precursors (Figs. 3, 5). In control cells from the neonates, HGF kept a greater percentage immature (PDGFRα+, NG2+). Thus, HGF inhibits maturation, to some extent, in these O4+ cells. This may represent less of an effect than HGF has on earlier OL precursors A2B5+, which are kept proliferating and immature by HGF (Yan and Rivkees, 2002). Different stages of OL development may be affected differently by this growth factor, and we know that the O4+ population from neonates is different from that in adults (Mason and Goldman, 2002; Lin et al., 2009).
CD82 downregulation in adult O4+ cells decreased levels of the myelin proteins MBP and MAG. This finding may suggest that CD82 regulates myelin protein gene transcription or stabilizes protein levels. How might CD82 regulate myelin protein levels? A recent study (Laursen et al., 2011) showed that activation of β1 integrins in OLs positively regulates the translation of MBP mRNA by binding an mRNA-binding protein that interacts with the 5′ UTR of the MBP mRNA. Interactions between CD82 and β-1 integrins have been found in other cell types (Mannion et al., 1996). Integrin activation is also required for the maturation of OLs and myelination (Decker et al., 2004; Barros et al., 2009). The possibility that CD82 activates β-1 integrin before or during myelination should be explored in the future.
We found little MBP protein in acutely isolated adult O4+ cells, in keeping with our previous results (Lin et al., 2009), despite the presence of MBP transcripts (Lin et al., 2009) and despite the presence of CD82 (Fig. 2). However, after 7 d in culture, MBP protein accumulated (Fig. 6). A possible explanation for the lack of MBP despite the presence of CD82 is that MBP is translated in cell processes, which are sheared off during the isolation process. After a few days in culture, these adult precursors reconstitute cell processes. However, when CD82 was downregulated, MBP did not accumulate and cells did not grow multiple processes (Fig. 5). Thus, CD82 could function in two ways to promote MBP accumulation: first, to activate β-1 integrins, and second, to promote cell process growth.
We found that, in the adult OL progenitors, CD82 coimmunoprecipitates with cMet; we cannot conclude at this time whether other molecules are part of the protein complex. In other cell types, particularly cancer cells, the CD82/cMet binding can be direct (Takahashi et al., 2007) or through complex formation involving other transmembrane proteins and lipids (Todeschini et al., 2007). Indeed, CD82 forms complexes with other tetraspanins and a wide range of other transmembrane proteins, and it can be a component of lipid rafts (Odintsova et al., 2003; Delaguillaumie et al., 2004; Takahashi et al., 2007; Laursen et al., 2011). OL precursor development is dependent in part on signaling through growth factors and integrins regulated by their associations with lipid rafts (Baron et al., 2003; Decker et al., 2004). Thus, a change in cMet/CD82 binding or in raft composition by the inclusion of CD82 could have important effects on signaling pathways regulating OL development.
Several studies suggest that HGF/cMet signaling is important for OL and neuronal survival in experimental autoimmune encephalomyelitis (EAE) models. Increased HGF levels are detected in the CSF of patients with inflammatory and demyelinating diseases (Tsuboi et al., 2002), whereas a recent study suggests a possible relationship between CSF HGF levels and disease activity in multiple sclerosis (Muller et al., 2012). HGF/cMet interactions are important through the proinflammatory phase in macrophages (Moransard et al., 2010), whereas HGF acts as a chemoattractant for OL progenitors in vitro and is expressed by macrophages in EAE models (Lalive et al., 2005). In addition, a recent study has shown that mesenchymal stem cells secrete HGF, and its interaction with cMet has therapeutic effects on EAE, such as enhanced myelin repair and immunomodulation (Bai et al., 2012). Together with these studies, our observations suggest that HGF/cMet interactions increase the immature OL precursor pool, including cells that express PDGFRα and NG2, and may promote increased migration of these cells to a remyelinating lesion. However, the expression of other molecules, notably CD82, is required for these cells to mature and myelinate. Our observations also imply that the adult O4+/CD82+ OL precursors are not influenced by HGF/cMet signaling and thus cannot be expanded or induced to migrate by HGF. These precursors may be already primed for rapid remyelination, because they already express myelin genes and rapidly remyelinate demyelinated zones (Mason et al., 2004).
Downstream molecules of HGF/cMet signaling also regulate OL development. For example, RhoA GTPase plays a significant role in OL development by promoting progenitor migration when activated, whereas its inactivation is a critical factor to initiate myelination (Liang et al., 2004; Mi et al., 2005; Kippert et al., 2007; Feltri et al., 2008). Conversely, the roles of Rac1 and cdc42 in the CNS have not been clarified. Rac1 has been studied extensively in Schwann cell development and PNS myelination (Feltri et al., 2008). Rac1 and cdc42 activation seems to be important for proper OL differentiation in vitro (Liang et al., 2004), but tissue-specific conditional ablation of cdc42 or Rac1 in OLs does not affect proliferation, migration, or in vitro differentiation; instead it causes enlargement of the inner tongue of OL processes and formation of a different type of myelin sheath (Thurnherr et al., 2006). Thus, Cdc42 and Rac1 appear to play different roles in PNS and CNS myelination; this suggests that the signaling role of a given small Rho GTPase in a specific cell type cannot predict its function in another cell type (Symons and Settleman, 2000; Liang et al., 2004; Wang and Zheng, 2007). Our finding that CD82 significantly blocks HGF-induced Rac1 activation supports the view that Rho GTPase activation effects not only depend on cell type but are also time and stage specific. Because CD82 expression correlates well with the ability of precursors to form long, complex, cell processes, it is possible that the HGF-induced Rac1 activation inhibits process formation or a multiprocessed morphology. Indeed, adult O4+ cells lose their complex multiprocessed phenotype when CD82 is downregulated (Fig. 5) but still maintain several short processes. In contrast, when HGF is added, the cells assume a much simpler morphology with few or no processes, with many of them appearing bipolar or unipolar (Fig. 5).
In conclusion, we show that CD82 is a negative regulator of HGF/cMet activation in rat O4+/CD82+ OL precursors. The timing of CD82 expression in OL development is significant because it promotes a transition from the NG2+ precursor stage to a more mature, premyelinating OL. Additional studies on CD82 and its interactions with other molecules, as well as on its effects on more downstream HGF/cMet signaling pathways, may provide new key insights into late OL precursor development and myelination.
Footnotes
This work was supported by National Multiple Sclerosis Society Postdoctoral Fellowship Award FG 1837-A-1.
The authors declare no competing financial interests.
References
- Achim CL, Katyal S, Wiley CA, Shiratori M, Wang G, Oshika E, Petersen BE, Li JM, Michalopoulos GK. Expression of HGF and cMet in the developing and adult brain. Brain Res Dev Brain Res. 1997;102:299–303. doi: 10.1016/S0165-3806(97)00108-9. [DOI] [PubMed] [Google Scholar]
- Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. 2009;57:1835–1847. doi: 10.1002/glia.20895. [DOI] [PubMed] [Google Scholar]
- Bai L, Lennon DP, Caplan AI, DeChant A, Hecker J, Kranso J, Zaremba A, Miller RH. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat Neurosci. 2012;15:862–870. doi: 10.1038/nn.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron W, Decker L, Colognato H, ffrench-Constant C. Regulation of integrin growth factor interactions in oligodendrocytes by lipid raft microdomains. Curr Biol. 2003;13:151–155. doi: 10.1016/S0960-9822(02)01437-9. [DOI] [PubMed] [Google Scholar]
- Barros CS, Nguyen T, Spencer KS, Nishiyama A, Colognato H, Müller U. Beta1 integrins are required for normal CNS myelination and promote AKT-dependent myelin outgrowth. Development. 2009;136:2717–2724. doi: 10.1242/dev.038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker L, ffrench-Constant C. Lipid rafts and integrin activation regulate oligodendrocyte survival. J Neurosci. 2004;24:3816–3825. doi: 10.1523/JNEUROSCI.5725-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker L, Baron W, ffrench-Constant C. Lipid rafts: microenvironments for integrin-growth factor interactions in neural development. Biochem Soc Trans. 2004;32:426–430. doi: 10.1042/BST0320426. [DOI] [PubMed] [Google Scholar]
- Delaguillaumie A, Harriague J, Kohanna S, Bismuth G, Rubinstein E, Seigneuret M, Conjeaud H. Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J Cell Sci. 2004;117:5269–5282. doi: 10.1242/jcs.01380. [DOI] [PubMed] [Google Scholar]
- Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barrett JC. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- Feltri ML, Suter U, Relvas JB. The function of RhoGTPases in axon ensheathment and myelination. Glia. 2008;56:1508–1517. doi: 10.1002/glia.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Kagoshima M, Wanaka A, Tohyama M, Matsumoto K, Nakamura T. Localization and functional coupling of HGF and c-Met/HGF receptor in rat brain: implication as neurotrophic factor. Brain Res Mol Brain Res. 1995;32:197–210. doi: 10.1016/0169-328X(95)00075-4. [DOI] [PubMed] [Google Scholar]
- Jackson P, Marreiros A, Russell PJ. KAI1 tetraspanin and metastasis suppressor. Int J Biochem Cell Biol. 2005;37:530–534. doi: 10.1016/j.biocel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Judson MC, Bergman MY, Campbell DB, Eagleson KL, Levitt P. Dynamic gene and protein expression patterns of the autism-associated met receptor tyrosine kinase in the developing mouse forebrain. J Comp Neurol. 2009;513:511–531. doi: 10.1002/cne.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Castren E, Odenthal M, Vande Woude GF, Ishii T, Dienes HP, Lindholm D, Schirmacher P. Expression and functional interaction of hepatocyte growth factor-scatter factor and its receptor c-met in mammalian brain. J Cell Biol. 1994;126:485–494. doi: 10.1083/jcb.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert A, Trajkovic K, Rajendran L, Ries J, Simons M. Rho regulates membrane transport in the endocytic pathway to control plasma membrane specialization in oligodendroglial cells. J Neurosci. 2007;27:3560–3570. doi: 10.1523/JNEUROSCI.4926-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Iwanami A, Nakamura M, Yamane J, Watanabe K, Suzuki Y, Miyazawa D, Shibata S, Funakoshi H, Miyatake S, Coffin RS, Nakamura T, Toyama Y, Okano H. Hepatocyte growth factor promotes endogenous repair and functional recovery after spinal cord injury. J Neurosci Res. 2007;85:2332–2342. doi: 10.1002/jnr.21372. [DOI] [PubMed] [Google Scholar]
- Lagaudrière-Gesbert C, Lebel-Binay S, Hubeau C, Fradelizi D, Conjeaud H. Signaling through the tetraspanin CD82 triggers its association with the cytoskeleton leading to sustained morphological changes and T cell activation. Eur J Immunol. 1998;28:4332–4344. doi: 10.1002/(SICI)1521-4141(199812)28:12<4332::AID-IMMU4332>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Lalive PH, Paglinawan R, Biollaz G, Kappos EA, Leone DP, Malipiero U, Relvas JB, Moransard M, Suter T, Fontana A. TGF-beta-treated microglia induce oligodendrocyte precursor cell chemotaxis through the HGF-c-Met pathway. Eur J Immunol. 2005;35:727–737. doi: 10.1002/eji.200425430. [DOI] [PubMed] [Google Scholar]
- Laursen LS, Chan CW, ffrench-Constant C. Translation of myelin basic protein mRNA in oligodendrocytes is regulated by integrin activation and hnRNP-K. J Cell Biol. 2011;192:797–811. doi: 10.1083/jcb.201007014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Draghi NA, Resh MD. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci. 2004;24:7140–7149. doi: 10.1523/JNEUROSCI.5319-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Mela A, Guilfoyle EM, Goldman JE. Neonatal and adult O4(+) oligodendrocyte lineage cells display different growth factor responses and different gene expression patterns. J Neurosci Res. 2009;87:3390–3402. doi: 10.1002/jnr.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Zhang XA. KAI1/CD82, a tumor metastasis suppressor. Cancer lett. 2006;240:183–194. doi: 10.1016/j.canlet.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Mannion BA, Berditchevski F, Kraeft SK, Chen LB, Hemler ME. Transmembrane-4 superfamily proteins CD81 (TAPA-1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29) J Immunol. 1996;157:2039–2047. [PubMed] [Google Scholar]
- Mason JL, Goldman JE. A2B5+ and O4+ Cycling progenitors in the adult forebrain white matter respond differentially to PDGF-AA, FGF-2, and IGF-1. Mol Cell Neurosci. 2002;20:30–42. doi: 10.1006/mcne.2002.1114. [DOI] [PubMed] [Google Scholar]
- Mason JL, Toews A, Hostettler JD, Morell P, Suzuki K, Goldman JE, Matsushima GK. Oligodendrocytes and progenitors become progressively depleted within chronically demyelinated lesions. Am J Pathol. 2004;164:1673–1682. doi: 10.1016/S0002-9440(10)63726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mela A, Goldman JE. The tetraspanin KAI1/CD82 is expressed by late-lineage oligodendrocyte precursors and may function to restrict precursor migration and promote oligodendrocyte differentiation and myelination. J Neurosci. 2009;29:11172–11181. doi: 10.1523/JNEUROSCI.3075-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Miranti CK. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell Signal. 2009;21:196–211. doi: 10.1016/j.cellsig.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Moransard M, Sawitzky M, Fontana A, Suter T. Expression of the HGF receptor c-met by macrophages in experimental autoimmune encephalomyelitis. Glia. 2010;58:559–571. doi: 10.1002/glia.20945. [DOI] [PubMed] [Google Scholar]
- Müller AM, Jun E, Conlon H, Sadiq SA. Cerebrospinal hepatocyte growth factor levels correlate negatively with disease activity in multiple sclerosis. J Neuroimmunol. 2012;251:80–86. doi: 10.1016/j.jneuroim.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- Odintsova E, Voortman J, Gilbert E, Berditchevski F. Tetraspanin CD82 regulates compartmentalisation and ligand-induced dimerization of EGFR. J Cell Sci. 2003;116:4557–4566. doi: 10.1242/jcs.00793. [DOI] [PubMed] [Google Scholar]
- Ohya W, Funakoshi H, Kurosawa T, Nakamura T. Hepatocyte growth factor (HGF) promotes oligodendrocyte progenitor cell proliferation and inhibits its differentiation during postnatal development in the rat. Brain Res. 2007;1147:51–65. doi: 10.1016/j.brainres.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Ono M, Handa K, Withers DA, Hakomori S. Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res. 1999;59:2335–2339. [PubMed] [Google Scholar]
- Pedraza L, Owens GC, Green LA, Salzer JL. The myelin-associated glycoproteins: membrane disposition, evidence of a novel disulfide linkage between immunoglobulin-like domains, and posttranslational palmitylation. J Cell Biol. 1990;111:2651–2661. doi: 10.1083/jcb.111.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Hanada K, Yamaguchi S, Yamashita H, Shimada S, Hamada H. Functional analysis of CD82 in the early phase of T cell activation: roles in cell adhesion and signal transduction. Eur J Immunol. 1998;28:1125–1133. doi: 10.1002/(SICI)1521-4141(199804)28:04<1125::AID-IMMU1125>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Hanada KI, Yamashita H, Shimada S, Hamada H. Overexpression of CD82 on human T cells enhances LFA-1/ICAM-1-mediated cell-cell adhesion: functional association between CD82 and LFA-1 in T cell activation. Eur J Immunol. 1999;29:4081–4091. doi: 10.1002/(SICI)1521-4141(199912)29:12<4081::AID-IMMU4081>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Symons M, Settleman J. Rho family GTPases: more than simple switches. Trends Cell Biol. 2000;10:415–419. doi: 10.1016/S0962-8924(00)01832-8. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sugiura T, Abe M, Ishii K, Shirasuna K. Regulation of c-Met signaling by the tetraspanin KAI-1/CD82 affects cancer cell migration. Int J Cancer. 2007;121:1919–1929. doi: 10.1002/ijc.22887. [DOI] [PubMed] [Google Scholar]
- Terada N, Kidd GJ, Kinter M, Bjartmar C, Moran-Jones K, Trapp BD. Beta IV tubulin is selectively expressed by oligodendrocytes in the central nervous system. Glia. 2005;50:212–222. doi: 10.1002/glia.20175. [DOI] [PubMed] [Google Scholar]
- Thurnherr T, Benninger Y, Wu X, Chrostek A, Krause SM, Nave KA, Franklin RJ, Brakebusch C, Suter U, Relvas JB. Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci. 2006;26:10110–10119. doi: 10.1523/JNEUROSCI.2158-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2-tetraspanin CD82 complex inhibits met and its cross-talk with integrins, providing a basis for control of cell motility through glycosynapse. J Biol Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- Tsuboi Y, Kakimoto K, Akatsu H, Daikuhara Y, Yamada T. Hepatocyte growth factor in cerebrospinal fluid in neurologic disease. Acta Neurol Scand. 2002;106:99–103. doi: 10.1034/j.1600-0404.2002.01125.x. [DOI] [PubMed] [Google Scholar]
- Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- van Heyningen P, Calver AR, Richardson WD. Control of progenitor cell number by mitogen supply and demand. Curr Biol. 2001;11:232–241. doi: 10.1016/S0960-9822(01)00075-6. [DOI] [PubMed] [Google Scholar]
- Wang L, Zheng Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol. 2007;17:58–64. doi: 10.1016/j.tcb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Yan H, Rivkees SA. Hepatocyte growth factor stimulates the proliferation and migration of oligodendrocyte precursor cells. J Neurosci Res. 2002;69:597–606. doi: 10.1002/jnr.10323. [DOI] [PubMed] [Google Scholar]
- Yang X, Wei LL, Tang C, Slack R, Mueller S, Lippman ME. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:5284–5288. [PubMed] [Google Scholar]