Abstract
Proteolytic shedding of cell surface proteins generates paracrine signals involved in numerous signaling pathways. Neuregulin 1 (NRG1) type III is involved in myelination of the peripheral nervous system, for which it requires proteolytic activation by proteases of the ADAM family and BACE1. These proteases are major therapeutic targets for the prevention of Alzheimer's disease because they are also involved in the proteolytic generation of the neurotoxic amyloid β-peptide. Identification and functional investigation of their physiological substrates is therefore of greatest importance in preventing unwanted side effects. Here we investigated proteolytic processing of NRG1 type III and demonstrate that the ectodomain can be cleaved by three different sheddases, namely ADAM10, ADAM17, and BACE1. Surprisingly, we not only found cleavage by ADAM10, ADAM17, and BACE1 C-terminal to the epidermal growth factor (EGF)-like domain, which is believed to play a pivotal role in signaling, but also additional cleavage sites for ADAM17 and BACE1 N-terminal to that domain. Proteolytic processing at N- and C-terminal sites of the EGF-like domain results in the secretion of this domain from NRG1 type III. The soluble EGF-like domain is functionally active and stimulates ErbB3 signaling in tissue culture assays. Moreover, the soluble EGF-like domain is capable of rescuing hypomyelination in a zebrafish mutant lacking BACE1. Our data suggest that NRG1 type III-dependent myelination is not only controlled by membrane-retained NRG1 type III, but also in a paracrine manner via proteolytic liberation of the EGF-like domain.
Introduction
Protease signaling is an important cellular mechanism in health and disease, and sheddases often liberate membrane-bound substrates for paracrine signaling (Turk et al., 2012). Sheddases are also involved in the generation of the Alzheimer's disease-associated amyloid β-peptide from the amyloid precursor protein (APP) (Lichtenthaler et al., 2011). Production of amyloid β-peptide is initiated by the β-site APP-cleaving enzyme (BACE1 or β-secretase) (Haass, 2004), which is therefore a promising drug target for the treatment of Alzheimer's disease (De Strooper et al., 2010). Physiologically, BACE1 is required to process Neuregulin 1 (NRG1) type III, a key regulator of myelination in the peripheral nervous system (PNS; Hu et al., 2006; Willem et al., 2006; Birchmeier and Nave, 2008; Brinkmann et al., 2008). Therefore, understanding the precise function of BACE1 in NRG1 type III processing and signaling is crucial to avoid side effects upon therapeutic inhibition of BACE1. NRG1 is predominantly expressed in neurons and multiple variants are generated by alternative splicing. All NRG1 isoforms contain an epidermal growth factor (EGF)-like domain, which binds and activates ErbB receptor tyrosine kinases (Falls, 2003; Mei and Xiong, 2008). Although most isoforms are single transmembrane domain (TMD) proteins, NRG1 type III contains two TMDs and forms a hairpin-like protein with the EGF-like domain in its extracellular loop (Fig. 1A; Wang et al., 2001).
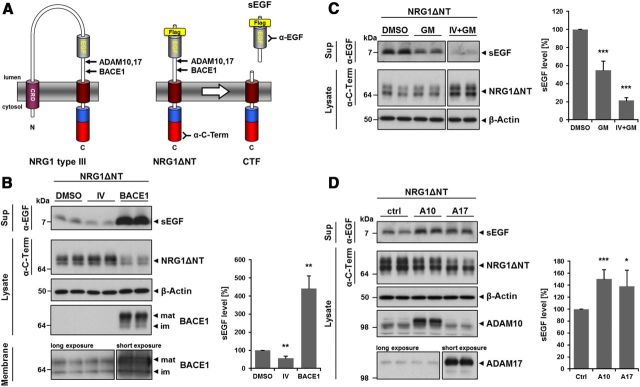
Figure 1.
BACE1, ADAM10, and ADAM17 cleave in the stalk region of NRG1 type III. A, Schematic representation of NRG1ΔNT. NRG1ΔNT comprises the EGF-like domain and the C terminus but lacks the N terminus of NRG1 type III. A Flag-tag was inserted immediately N-terminal of the EGF-like domain to facilitate immunoprecipitation for MS analysis. Shedding of NRG1ΔNT by BACE1, ADAM10, or ADAM17 releases an sEGF domain that was detected directly in the supernatant using an EGF antibody. NRG1ΔNT was detected in the cell lysate with a C-terminal antibody. CRD indicates cysteine-rich domain. B, BACE1 is shedding NRG1 type III in the stalk region. HEK293 cells transfected with NRG1ΔNT were treated with the specific BACE1 inhibitor IV (10 μm) or cotransfected with BACE1. Note that, due to its rapid turnover, the CTF generated by shedding can only be detected upon γ-secretase inhibition (data not shown). Expression of transfected and endogenous BACE1 was confirmed in cell lysates and isolated membranes (bottom). mat indicates mature; im, immature. Bar graph: Quantification of experiments (mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student's t test, inhibitor: n = 5, overexpression: n = 6). C, D, ADAM10 and ADAM17 contribute to the shedding of NRG1 type III in the stalk region. C, Cells expressing NRG1ΔNT were treated with the broad-spectrum ADAM inhibitor GM6001 (GM, 25 μm). Coinhibition of ADAMs and BACE1 was achieved by combined treatment with GM and inhibitor IV (25 and 10 μm, respectively). D, Coexpression of NRG1ΔNT with ADAM10 (A10) or ADAM17 (A17). Bar graphs in C, D: Quantification of experiments (mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student's t test, in C, n = 5; in D, n = 6).
NRG1 type III is essential for Schwann cell development in both mouse and zebrafish and determines myelin sheath thickness (Wolpowitz et al., 2000; Michailov et al., 2004; Taveggia et al., 2005; Monk and Talbot, 2009; Perlin et al., 2011). BACE1 knock-out mice exhibit severe hypomyelination and accumulation of uncleaved NRG1 type III, demonstrating that NRG1 type III is a natural BACE1 substrate (Hu et al., 2006; Willem et al., 2006). NRG1-mediated signaling requires proteolytic processing and shedding in the stalk (juxtamembrane) region of NRG1 type I, and NRG1 type II releases the ectodomain as a paracrine signal. Conversely, shedding of NRG1 type III generates a membrane-tethered N-terminal fragment (NTF), which presents the EGF-like domain as juxtacrine signal (Falls, 2003). Because BACE1 knock-out mice still show some degree of myelination, other proteases may compensate for the loss of BACE1 (Velanac et al., 2012). Indeed, members of the ADAM (a disintegrin and metalloproteinase) family also cleave NRG1 (Montero et al., 2000; Shirakabe et al., 2001; Horiuchi et al., 2005; La Marca et al., 2011; Luo et al., 2011), and the NRG1 type III NTF generated by ADAM10 activates ErbB receptors similar to the BACE1-processed fragment (Luo et al., 2011).
Based on the finding that recombinant soluble NRG1 type III NTFs are sufficient for signaling in vitro (Syed et al., 2010), we searched for a proteolytic pathway that would physiologically liberate a signaling-competent EGF-like domain from NRG1 type III. We found that the EGF-like domain of NRG1 type III is liberated by a dual BACE1 or ADAM17 cleavage. Moreover, the soluble EGF-like (sEGF) domain not only stimulates ErbB3 receptor phosphorylation, but also rescues hypomyelination in a bace1 mutant zebrafish.
Materials and Methods
cDNA constructs, primers, and lentivirus production.
NRG1 type III β1a (GenBank: AF194438.1) cDNA was cloned into the pcDNA4-myc-HisA vector (Invitrogen) using EcoRI and XhoI restriction sites. For the construct V5-IIINRG1, a V5-tag (GKPIPNPLLGLDST) was inserted directly after M1 by fusion PCR. The V5-IIINRG1-HA construct was generated by introduction of an HA-tag (YPYDVPDYA) between V281 and M282. For the truncated construct NRG1ΔNT, the respective NRG1 type III sequence was subcloned into the pSecTag2A (Invitrogen) vector that features an N-terminal secretion signal. A suboptimal Furin cleavage site (RAVRSL) after the secretion signal sequence was optimized (RARRSV) by QuikChange mutagenesis (Stratagene) and a Flag-tag (DYKDDDDK) was inserted N-terminal of I236. The constructs α- and β-sEGF were generated by subcloning the respective sequences from V5-IIINRG1-HA into the modified pSecTag2A vector using the SfiI and XhoI restriction sites. BACE1 and ADAM10 expression constructs have been described previously (Capell et al., 2000; Wild-Bode et al., 2006). The construct expressing ADAM17 was kindly provided by Dr. R.A. Black at Amgen and has been described previously (Black et al., 1997). For expression in primary neurons, V5-IIINRG1-HA was cloned into a lentiviral expression vector AD149FhSynW2 under control of the human synapsin promoter using the NheI and EcoRI restriction sites. Lentiviral particles were produced in HEK293FT cells as described previously (Orozco et al., 2012). Briefly, HEK293FT cells were cotransfected with the lentiviral expression construct psPAX2 and pVSVg for 24 h. After another 24 h, supernatant was collected and virus particles were concentrated by ultracentrifugation and resuspended in neurobasal medium. The sequences of all constructs were verified by sequencing. Construct and oligonucleotide sequences are available upon request.
Stable cell lines, transfection, and inhibitor treatment.
HEK293T cells (Invitrogen) and MCF-7 cells (Karey and Sirbasku, 1988) were cultured in DMEM with Glutamax (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen). CHO ldlD cells deficient in O-linked glycosylation were a kind gift from Monty Krieger and have been described previously (Kingsley et al., 1986). CHO wild-type (wt) and ldlD cells were cultured in DMEM with 10% fetal calf serum and nonessential amino acids. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The following inhibitors were dissolved in DMSO and used at the indicated final concentrations for 12–24 h: BACE1 inhibitor IV (5 or 10 μm; Calbiochem); GM6001, a broad-spectrum inhibitor of matrix metalloproteinases and ADAMs (25 μm; Enzo Life Sciences), the ADAM10-specific inhibitor GI254023X (5 μm; a kind gift from Dr. Schmidt, Technical University of Darmstadt, described previously by Ludwig et al., 2005), and the ADAM17-specific inhibitor GL 506–3 (5 μm; a kind gift from Galderma). O-linked glycosylation was blocked with benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside (4 mm; Merck).
Primary neuronal culture, transduction, and inhibitor treatment.
Hippocampal neurons were prepared from embryonic day 18 Sprague-Dawley rat embryos as described previously (Orozco et al., 2012) and cultured in neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), Pen/Strep, and 2 mm l-glutamine. After 4 d in vitro, cells were incubated with lentiviral particles for 8 h, and after 2 d, transduction inhibitor treatment was performed for 16 h. Supernatants and cells were collected and analyzed by immunoblotting.
Primary Schwann cell culture.
Primary rat Schwann cells were prepared as described previously (Einheber et al., 1997) and proliferation was induced using high-glucose DMEM with Glutamax (Invitrogen), 10% FBS, 2 mm l-glutamine (Invitrogen), 2 μm forskolin (Sigma-Aldrich), and 10 μg/ml pituitary extract (Sigma-Aldrich). One day before the experiment, cells were washed twice with HBSS (Invitrogen) and proliferation was stopped by maintaining the cells in DMEM with Glutamax, 10% FBS, and 2 mm l-glutamine.
Antibodies.
The following monoclonal neoepitope-specific antibodies were generated by immunization with the respective peptides: 4F10: rat, SFYKHLGIEF; 10E8: mouse, MEAEELYQKR; 7E6: mouse, QTAPKLSTS. Hybridoma supernatants were used 1:40 for immunoblotting. The following antibodies were used for immunoblotting: antibody to NRG1 C terminus (pRb, 1:10000, SC348; Santa Cruz Biotechnology), to NRG1 EGF-like domain (HRGβ pRb, 1:2000, Ab-2; Thermo Scientific), to β-actin (1:5000; Sigma-Aldrich), to BACE1 (pRb, 1:1000, AB5940; Millipore), to ADAM17/TACE (pRb, 1:1000, ab39162; Abcam), to ADAM10 (pRb, 1:5000, 422751; Calbiochem), to Calnexin (pRb, 1:10000; Stressgen), to V5-tag (1:5000; Invitrogen), to HA-tag (HRP-conjugated mRat, 1:2000, 3F10; Roche), to ErbB3 (pRb, 1:1000, sc-285, C-17; Santa Cruz Biotechnology), to p-ErbB3 (pRb, 1:1000, sc-135654, Tyr1328; Santa Cruz Biotechnology), to AKT (mRb, 1:4000, C67E7; Cell Signaling Technology), and to p-AKT (mRb, 1:3000, D9E XP Ser473; Cell Signaling Technology). Secondary antibodies were HRP-conjugated anti-mouse and anti-rabbit IgG (pGoat, 1:10000; Promega) or anti-rat IgG (pGoat, 1:4000, sc-2006; Santa Cruz Biotechnology).
Sample preparation and immunoblotting.
Medium was conditioned overnight, immediately cooled upon collection, and supplemented with protease inhibitor mixture (Sigma-Aldrich). Cell debris was removed by centrifugation (5 min, 5500 × g, 4°C) and the supernatant was subjected directly to standard SDS-PAGE. For total cell lysates, cells were washed with ice-cold PBS, scraped off, and pelleted by centrifugation (5 min, 1000 × g, 4°C). Cells were lysed in lysis buffer (20 mm citrate, pH 6.4, 1 mm EDTA, 1% Triton X-100) freshly supplemented with protease inhibitor mixture for 30 min on ice. After clarification (15 min, 10000 × g, 4°C), protein concentration was determined with the BCA protein assay (Pierce) and equal amounts of protein were subjected to SDS-PAGE. Proteins were transferred onto PVDF (Immobilon-P; Millipore) or nitrocellulose (Protran; Whatman) membranes and the indicated antibodies were used for immunodetection. Bound antibodies were detected with HRP-conjugated secondary antibodies using the chemiluminescence detection reagents ECL and ECL Plus (GE Healthcare). For quantification, images were acquired with a Luminescent Image Analyzer LAS-4000 (Fujifilm) and analyzed with the Multi Gauge V3.0 software.
siRNA-mediated knock-down and membrane preparation.
For RNA interference, cells were plated in polylysine-coated dishes and reverse transfected with siGENOME pool targeting ADAM10 (10 nm; Thermo Scientific), ON-TARGETplus SMARTpool targeting ADAM17 (15 nm; Dharmacon) or respective controls using Lipofectamine 2000 (Invitrogen). Fresh medium was added 24 h after transfection and (if applicable) substrate cDNA was transfected 48 h after initial transfection. Medium was conditioned starting 68 h after siRNA transfection and cells were harvested 12–24 h later. For detection of endogenous BACE1, ADAM10 and ADAM17 cell membranes were prepared as described previously (Sastre et al., 2001).
MS analysis after immunoprecipitation.
Conditioned medium was prepared as described above and incubated with anti-Flag M2 or anti-HA (Sigma-Aldrich) agarose beads overnight (rotation, 4°C). Beads where washed three times with IP/MS buffer (0.1% N-octylglucoside, 10 mm Tris-HCl, pH 8.0, 5 mm EDTA, 140 mm NaCl) and two times with water. Immunoprecipitated proteins were eluted with trifluoroacetic acid/acetonitrile/water (0.3%/40%) saturated with α-cyano-4-hydroxycinnamic acid and matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) analysis was performed as described previously (Okochi et al., 2002) using Voyager DE STR (Applied Biosystems). Molecular masses were calibrated using the Sequazyme Peptide Mass Standards Kit (Applied Biosystems).
Preparation of sEGF domains and phosphorylation assays.
sEGF domains were expressed in CHO wt and CHO ldlD cells and supernatants were conditioned for 24 h. After quantification of their initial abundance by Western blotting, medium from control cells was used to adjust the concentrations of the EGF-like domains in the supernatants by dilution. Equal concentrations were controlled by Western blotting again (and further adjustment was done if necessary) and the adjusted supernatants were used for phosphorylation assays. The assay was performed as follows: subconfluent MCF-7 or purified primary Schwann cells were incubated with conditioned media containing equal amounts of EGF-like domains for 30 min. Medium from cells expressing an empty vector was used as negative control and incubation with 0.5 nm recombinant NRG1 EGF-like domain (NRG1-β1, 396-HB/CF; R&D Systems) served as a positive control. Cells were then washed with ice-cold PBS and lysates were prepared as described above using a modified RIPA buffer supplemented with phosphatase inhibitor mixture (PhosSTOP; Roche). Eventually, the phosphoprotein/total protein readout of the assay was normalized to the Western blot intensities of the EGF-like domains in the media used for stimulation.
Zebrafish maintenance and transgenic lines used.
All zebrafish embryos were raised at 28°C in E3 media (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm MgSO4) and were staged as described previously (Kimmel et al., 1995). To suppress the growth of mold, methylene blue (10−5%) was added to E3 media. Animals of either sex were used for this study. All experiments were performed in accordance with animal protection standards of the Ludwig-Maximilians University Munich and have been approved by the government of Upper Bavaria (Regierung von Oberbayern, München, Germany). In addition to the AB wt strain, a transgenic zebrafish line, Tg(claudin k:GFP) in which the claudin k promoter drives expression of a membrane-bound GFP (Münzel et al., 2012), was also used. This transgenic line is hereafter referred to as claudin k:GFP. Furthermore, a newly generated bace1 mutant (van Bebber et al., 2013) was crossed to claudin k:GFP, and bace1 homozygous mutants carrying the claudin k:GFP were used to analyze the activity of NRG1 type III-derived β-sEGF in vivo.
mRNA injections and image acquisition.
β-sEGF mRNA was synthesized in vitro using the mMessage mMACHINE kit (Ambion) according to standard protocols. β-sEGF mRNA was injected at a concentration of 425 ng/μl in fertilized eggs at the one-cell stage. Zebrafish larvae were anesthetized with tricaine (0.016% w/v) at 3 d postfertilization (dpf) and oriented in 3% methylcellulose on coverslips. Fluorescence of claudin k:GFP was imaged with an LSM510 META inverted confocal microscope (Zeiss). Pictures were assembled in Photoshop 8.0 (Adobe Systems). Brightness and contrast were adjusted with ImageJ.
Results
Juxtamembrane cleavage of NRG1 type III by ADAM10, ADAM17, and BACE1
To investigate NRG1 type III processing in living cells, we generated a truncated Flag-tagged NRG1 type III construct comprising the C-terminal region, the transmembrane domain, the juxtamembrane region, and the EGF-like domain (NRG1ΔNT; Fig. 1A). Expression of the truncated protein not only facilitates the analysis of proteolytic processing within living cells, but also allows the determination of potential cleavage sites by MS. We coexpressed three different sheddases suggested to be involved in NRG1 type III processing, BACE1, ADAM10, and ADAM17 (Montero et al., 2000; Hu et al., 2006; Willem et al., 2006; La Marca et al., 2011; Luo et al., 2011), or repressed their endogenous activities with inhibitors. Inhibition of endogenous BACE1 (by the BACE1 inhibitor IV) resulted in reduced liberation of the EGF-like domain into the supernatant (Fig. 1B). Conversely, ectopic BACE1 expression strongly enhanced shedding reflected by significantly increased amounts of the sEGF domain in the supernatant and reduced substrate levels in the cell lysate (Fig. 1B). Due to its rapid turnover by the γ-secretase (Bao et al., 2003, 2004), the C-terminal fragment (CTF) resulting from shedding could only be detected upon γ-secretase inhibition (data not shown).
We then investigated whether both ADAM10 and ADAM17 were also able to cleave NRG1ΔNT. The broad-spectrum ADAM inhibitor GM6001 significantly reduced the amount of sEGF detected in the supernatant. Combined inhibition of BACE1 and ADAMs by treatment with both inhibitor IV and GM6001 almost completely abolished endogenous shedding activity, as reflected by strongly reduced amounts of sEGF in the supernatant and the accumulation of NRG1ΔNT substrate within the cell lysate (Fig. 1C). Consistent with these findings, ectopic expression of either ADAM10 or ADAM17 resulted in enhanced shedding of NRG1ΔNT and subsequent sEGF accumulation in the conditioned media (Fig. 1D). These results suggest that BACE1, ADAM10, and ADAM17 are able to cleave NRG1 type III between the extracellular EGF-like domain and the C-terminal transmembrane domain.
Identification of BACE1, ADAM10, and ADAM17 cleavage sites within the juxtamembrane region of NRG1 type III
To map the cleavage sites of the respective proteases, we immunoprecipitated the sEGF domain from the supernatants of NRG1ΔNT-expressing HEK293 cells using antibodies to the Flag epitope (Fig. 1A). Isolated peptides were analyzed using MALDI-TOF MS. Mass spectra derived from the supernatant of control cells revealed four prominent peptide species corresponding to cleavages after A283, F285, Y286, and F293 (Fig. 2A,G). To identify the specific cleavage sites of the three sheddases shown to be involved in processing of NRG1ΔNT, we inhibited their endogenous activities with selective inhibitors or enhanced their cleavage by overexpression of the corresponding protease. This revealed that the peptide peak at 7846.8 kDa corresponding to a cleavage site after F293 was strongly reduced upon inhibition of endogenous BACE1 (Fig. 2A). Conversely, production of the same peptide was greatly enhanced to the expense of all other cleavage products upon BACE1 overexpression (Fig. 2A), indicating competing shedding activities.
Figure 2.
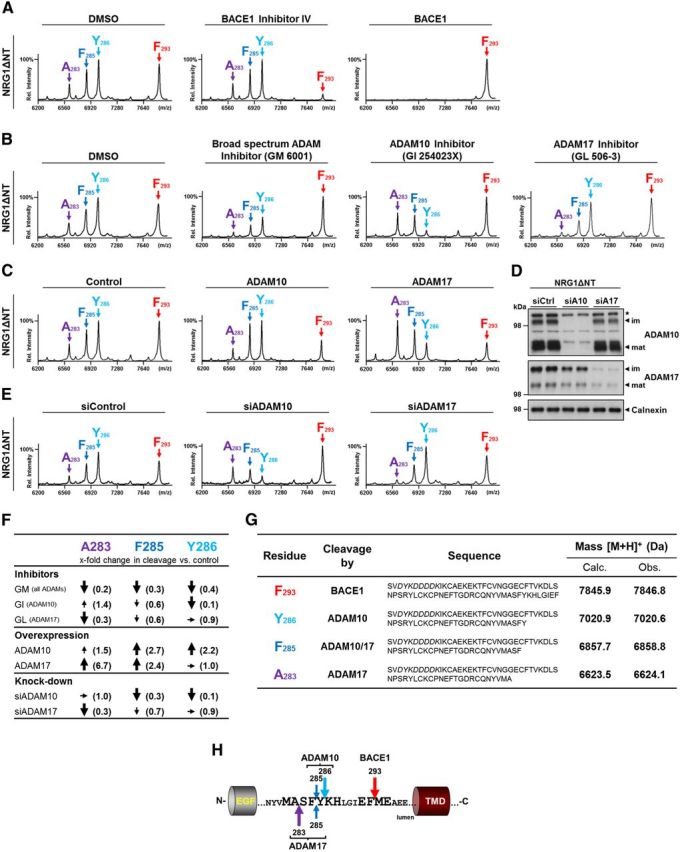
Identification of shedding sites of BACE1, ADAM10, and ADAM17 in the stalk region of NRG1 type III. A, BACE1 cleaves NRG1 type III after F293. Supernatants of Figure 1B were immunoprecipitated with Flag-agarose beads and precipitated sEGF peptides were analyzed by MALDI-TOF MS. The peptide corresponding to a cleavage after F293 is generated by BACE1 (red arrow). Note that with overexpression of BACE1, the other peaks are below detection limit. B, C, ADAM10 and ADAM17 shed NRG1 type III after A283, F285, and Y286. Cells expressing NRG1ΔNT were either treated with indicated inhibitors (GM6001, 25 μm; GI254023X and GL506–3, 5 μm) or cotransfected with indicated proteases and supernatants were analyzed as in A. ADAM-specific peaks were compared with the peak caused by BACE1 in each spectrum. Broad-spectrum ADAM inhibition (GM6001) reduced cleavage at all ADAM cleavage sites. Inhibition and overexpression of ADAM10 revealed its main cleavage sites to be after Y286 and F285 (light blue and blue arrows, respectively). Conversely, ADAM17 cleaved NRG1ΔNT mainly after A283 (purple arrows). D, E, Knock-down of ADAM10 and ADAM17 confirms shedding sites. Cells expressing NRG1ΔNT were transfected with siRNA (10 nm) against ADAM10 (siA10) or ADAM17 (siA17) and a nontargeting siRNA as a control (siCtrl). D, Western blot analysis of membrane preparations confirmed efficient downregulation of both the immature (im) and mature (mat) form of ADAM10 and ADAM17. E, sEGF peptides were isolated from the supernatant and analyzed as in B. F, Summary of MS data for ADAM10 and ADAM17 cleavage after A283, F285, and Y286. Peak intensities (areas) of ADAM-specific peaks were normalized to the signal generated by BACE1 in each spectrum (mean ± SD; n = 3). The normalized peak intensities then were compared with the respective controls. Changes of cleavage (fold) under different conditions (inhibition, overexpression, and knock-down of ADAM10 and ADAM17) are summarized as follows: increased: >1.6× (↑, big arrows), mildly increased: 1.5–1.2× (↑, small arrows) unchanged: 1.1–0.9× (→), mildly decreased: 0.8–0.5× (↓, small arrows), or decreased: <0.4× (↓, big arrows). G, List of peptides identified by MS. Peptide sequences with corresponding protease(s) are given and observed (Obs.) peptide masses are compared with calculated (Calc.) masses. Italic letters indicate Flag-tag; [M+H]+, a singly charged peptide. H, Graphic representation of shedding sites in the stalk region of NRG1 type III. Cleavage sites of BACE1, ADAM10, and ADAM17 are shown and preferred cleavage positions of ADAM10 and ADAM17 are indicated by longer arrows, respectively.
Similarly, we analyzed the secreted EGF-like domains upon overexpression or inhibition of ADAM proteases. Inhibition of ADAMs with the broad-spectrum ADAM inhibitor GM6001 decreased the abundance of the three peptides A283, F285, and Y286 compared with the peptide generated by BACE1, suggesting that these peptides are indeed generated by protease activities associated with members of the ADAM family (Fig. 2B,F). To identify the cleavage sites of ADAM10 and ADAM17, we used inhibitors that preferentially inhibit either ADAM10 (GI254023X) or ADAM17 (GL506–3). Inhibition of ADAM10 reduced the intensity of peaks corresponding to cleavages after F285 and Y286 but increased cleavage after A283. Conversely, blocking ADAM17 resulted in reduced cleavage after A283 (and, to a minor extent, after F285), whereas cleavage after Y286 remained unaffected (Fig. 2B,F). Consistent with these findings, overexpression of either protease caused increased cleavage after the sites affected by their respective inhibitors (Fig. 2C,F). Therefore, ADAM10 expression leads to enhanced cleavage after residues F285 and Y286, whereas expression of ADAM17 increases cleavage after residues A283 and F285 (Fig. 2C).
To verify these cleavage sites under endogenous conditions, we knocked down ADAM10 and ADAM17 in HEK293 cells expressing NRG1ΔNT. The siRNA pools efficiently reduced the protein levels of ADAM10 and ADAM17 (Fig. 2D). As before, sEGF peptides were isolated from the conditioned medium and analyzed by MALDI-TOF MS (Fig. 2E). Knock-down of ADAM10 dramatically reduced cleavage after Y286 and F285, whereas cleavage after A283 was unaffected. Conversely, reduction of ADAM17 strongly impaired cleavage after A283 and reduced cleavage after F285 to some extent, whereas cleavage after Y286 was not affected (Fig. 2E,F). These findings suggest that BACE1 specifically cleaves NRG1 type III after F293, whereas ADAM17 cleaves after A283 and, to some extent, also after F285. ADAM10 processes NRG1 type III after Y286, but also shares the minor cleavage site after F285 with ADAM 17 (Fig. 2H).
Processing of NRG1 type III liberates the EGF-like domain
Shedding is thought to activate NRG1 type III by generating a membrane-tethered NTF comprising the EGF-like domain (Falls, 2003; Fig. 3A). However, we hypothesized that the resulting NTF may even be further processed by sheddases to liberate an sEGF domain (Fig. 3A). To examine this possibility, we expressed full-length NRG1 type III with an N-terminal V5-tag (V5-IIINRG1) in HEK293 cells and coexpressed BACE1, ADAM10, or ADAM17 (Fig. 3B). Expression of either protease was controlled as before (Fig. 1) and comparable amounts were observed (data not shown). Full-length V5-IIINRG1 was detected as multiple bands, suggesting intensive posttranslational modification such as glycosylation (Fig. 3B). Decreased full-length protein was present in the lysate upon coexpression of either protease. This was most apparent when BACE1 was overexpressed (Fig. 3B). The CTF generated upon BACE1 overexpression (β-CTF) migrated slightly faster than the CTFs generated by either ADAM (α-CTF; Fig. 3B). This is consistent with the BACE1 cleavage site being closer to the transmembrane domain than the cleavage sites identified for ADAM10 and ADAM17 (Fig. 2H). We also analyzed the cell lysate for the NTF generated upon shedding of NRG1 type III. Detection with an antibody against the N-terminal V5-tag and reprobing with an anti-EGF antibody confirmed that the observed NTF still contained the EGF-like domain (Fig. 3B, dashed box).
Figure 3.
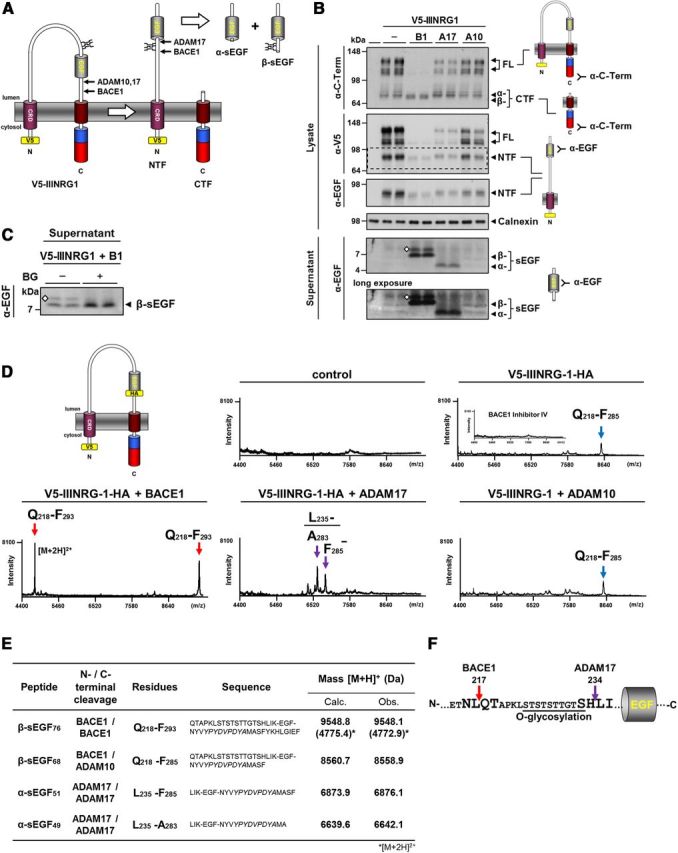
BACE1 and ADAM17 liberate an sEGF domain from NRG1 type III. A, Model depicting NRG1 type III processing and EGF liberation. Shedding of the V5-tagged NRG1 type III (V5-IIINRG1) in the C-terminal stalk region generates an NTF and CTF. Further cleavage of the NTF by ADAM17 and BACE1 liberates the EGF-like domain into the lumen (α- and β-sEGF). Glycosylation N-terminal of the EGF-like domain is indicated. B, Processing of NRG1 type III generates both a membrane-tethered and an sEGF domain. V5-IIINRG1 was coexpressed with BACE1 (B1), ADAM17 (A17), and ADAM10 (A10), and membrane-tethered fragments were detected in the cell lysate by Western blotting. Due to the different shedding sites in the stalk region of NRG1 type III, the CTFs generated by BACE1 and ADAMs differ in size (β-CTF and α-CTF, respectively). The observed NTFs contained the EGF-like domain, as confirmed by reprobing of the membrane with an EGF antibody (dashed box). Analysis of the supernatants revealed that BACE1 and ADAM17 liberate sEGF domains of different sizes (β-sEGF and α-sEGF, respectively). The diamonds denote posttranslational modification. C, The β-sEGF is subject to O-linked glycosylation. Cells expressing V5-IIINRG1 and BACE1 were treated or not with a blocker of O-glycosylation (benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside, BG, 4 mm). Soluble β-sEGF was detected with an antibody against the EGF-like domain. The diamond denotes O-linked glycosylation. D, Cleavage of NRG1 type III NTF before Q218 by BACE1 and before L235 by ADAM17 liberates the EGF-like domain. For immunoprecipitation, an HA-tag was inserted immediately after the EGF-like domain into the construct shown in A. To determine the exact cleavage sites, fragments were isolated from supernatants by IP with HA agarose and analyzed by MALDI-TOF MS. BACE1 liberated a fragment comprising residues Q218-F293, whereas ADAM17 generated smaller fragments containing residues L235-F285 and L235-A283. Processing by endogenous proteases or by coexpressed ADAM10 caused low level secretion of a fragment with a BACE1-cleaved N terminus and an ADAM-cleaved C terminus (Q218-F285). Inhibition of BACE1 (BACE1 inhibitor IV, 10 μm) abolished secretion of the EGF-like domain completely (inset in second panel). E, List of peptides identified by MS. Peptide sequences (without residues of the EGF-like domain) and corresponding proteases are listed. Observed masses (Obs.) are compared with calculated (Calc.) masses. Peptide numbers indicate number of residues comprised by each peptide (excluding the HA-tag). [M+H]+ indicates singly charged peptide; [M+2H]2+, a doubly charged peptide; italic letters, HA-tag. F, Graphic representation of BACE1 and ADAM17 cleavage sites N-terminal of the EGF-like domain. Cleavage sites and the serine/threonine-rich region where O-linked glycosylation occurs are indicated.
Enhanced proteolytic cleavage of the full-length V5-IIINRG1 should lead to the accumulation of the NTF as cleavage product. However, we did not detect higher amounts of NTF in cells coexpressing one of the three sheddases, but rather observed reduced NTF levels especially upon BACE1 or ADAM17 coexpression (Fig. 3B). Because this may be indicative of further NTF processing by these proteases, we analyzed the supernatants for soluble peptides liberated from the membrane-bound NTF. Both BACE1 and ADAM17 were found to liberate sEGF domains of different sizes (β-sEGF and α-sEGF) into the medium (Fig. 3B). Moreover, processing of V5-IIINRG1 by endogenous proteases also resulted in the secretion of a β-sEGF, albeit to a much smaller extent (Fig. 3B, long exposure). β-sEGF, but not α-sEGF, migrated as a double band, suggesting posttranslational modifications (Fig. 3B). A serine/threonine-rich stretch, which may be a site for O-linked glycosylation, is located close to the N terminus of the EGF-like domain (Fig. 3A). We treated cells expressing V5-IIINRG1 and BACE1 with benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside, a specific blocker of O-linked glycosylation. Western blot analysis of supernatants using an antibody against the EGF-like domain revealed that inhibition of O-glycosylation abolished formation of the higher-molecular-weight band and caused β-sEGF to appear as a single peptide (Fig. 3C). This demonstrates that, in contrast to α-sEGF, β-sEGF is subject to O-linked glycosylation.
A second BACE1 and ADAM17 cleavage N-terminal of the EGF-like domain
The results described above suggest that the EGF-like domain is released by cleavages that occur both N-terminal and C-terminal of the EGF-like domain. To identify the putative cleavage site(s), we used a combined immunoprecipitation/MALDI-TOF MS approach. An HA-tag was inserted immediately after the EGF-like domain to enable immunoprecipitation. This construct (V5-IIINRG1-HA; Fig. 3D) was transiently expressed in HEK293 cells with and without BACE1, ADAM10, or ADAM17. Western blot analysis of lysates and supernatants confirmed the generation of membrane-tethered and soluble fragments from this construct in a similar fashion to the untagged construct (data not shown). MS analysis of peptides secreted from cells expressing V5-IIINRG1-HA yielded one major peak at 8558.9 kDa (Fig. 3D). This corresponds to a peptide (β-sEGF68) having Q218 as an N-terminal residue and F285 as a C-terminal residue (Fig. 3E). Although the C terminus is the result of endogenous ADAM-mediated shedding in the stalk region after F285 (Fig. 2H), the N terminus results from a novel cleavage after L217, 16 residues N-terminal of the EGF-like domain. Inhibition of endogenous BACE1 activity with the specific inhibitor IV abolished generation of the peptide, suggesting BACE1-mediated processing after L217 (Fig. 3D, inset). Strikingly, this novel cleavage site (ETNL|QTAP) resembles that of the Swedish mutation of APP (EVNL|DAEF), which strongly increases BACE1-mediated processing (Citron et al., 1992, 1995; Cai et al., 1993). Consistent with this, coexpression of BACE1 strongly enhanced cleavage at this novel site and, in agreement with the shedding data shown above (Fig. 2H), produced a slightly larger peptide (β-sEGF76) ending with the BACE1 shedding site F293 at its C terminus (Fig. 3D,E).
Expression of ADAM17 liberated a shorter sEGF from V5-IIINRG1-HA beginning with L235 and ending after either A283 or F285, respectively (α-sEGF49 and 51; Fig. 3D,E). This is consistent with the data obtained for ADAM17-mediated processing of NRG1-ΔNT (Fig. 2H). The newly identified ADAM17 cleavage site after H234 is located immediately N-terminal of the EGF-like domain and is therefore responsible for the observed size difference of α-sEGF and β-sEGF (Fig. 3F). Cleavage at this site excludes the serine/threonine-rich stretch from α-sEGF and explains why, in contrast to β-sEGF, α-sEGF is not subject to O-linked glycosylation (Fig. 3B,C,F). These findings demonstrate that BACE1 and ADAM17, but not ADAM10, are capable of liberating the EGF-like domain from NRG1 type III by dual cleavage (Fig. 3A).
Membrane-bound and soluble fragments of NRG1 type III are detected by neo-epitope-specific antibodies to BACE1 cleavage sites
To further validate and facilitate detection of NRG1 type III fragments, we generated neo-epitope-specific antibodies against the above identified cleavage sites. Monoclonal antibodies 10E8 and 4F10 were raised against the neo-epitopes generated by BACE1-mediated shedding in the stalk region (epitopes M294EAEELYQKR and SFYKHLGIEF293, respectively). An additional antibody was raised against the novel BACE1 cleavage site N-terminal of the EGF-like domain (7E6, epitope Q218TAPKLSTS; Fig. 4A). We then investigated whether these antibodies are suitable as sensitive tools for the detection of the different processing products generated by BACE1, ADAM10, and ADAM17.
Figure 4.
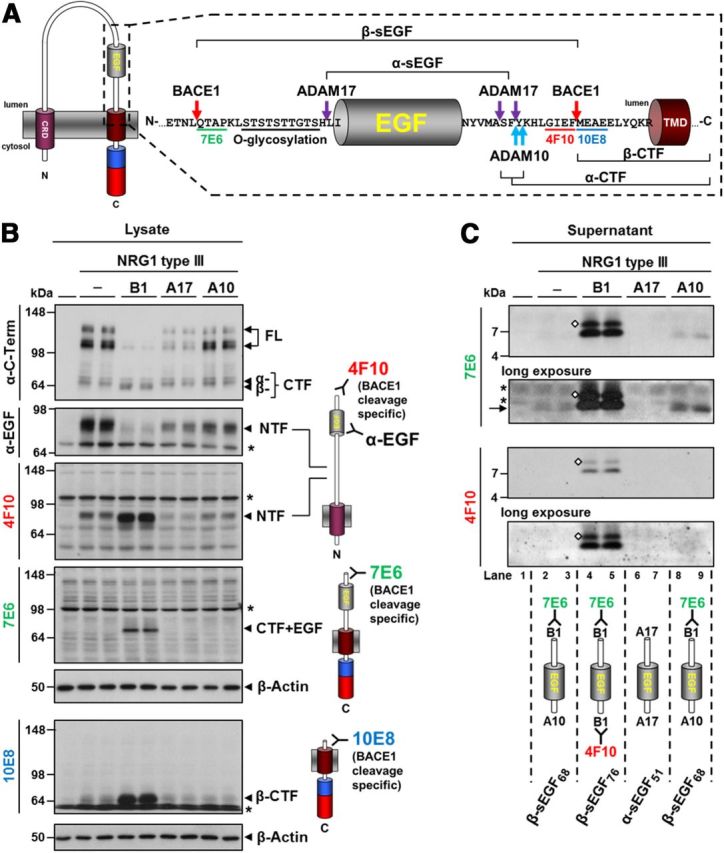
Monoclonal antibodies generated against BACE1 cleavage sites in NRG1 type III detect membrane-bound and soluble fragments. A, Scheme summarizing identified cleavage sites in NRG1 type III. Cleavage sites of respective proteases are marked by arrows and α-sEGF and β-sEGF and α-CTF and β-CTF are indicated. Epitopes of the generated antibodies and the site of glycosylation are shown. B, Membrane-tethered NRG1 type III fragments are recognized by antibodies raised against BACE1 cleavage sites. Total lysates of cells expressing untagged NRG1 type III and BACE1 (B1), ADAM17 (A17), or ADAM10 (A10) were analyzed by Western blotting. BACE1-cleaved fragments were detected with the indicated antibodies. C, Antibodies 7E6 and 4F10 recognize β-sEGF. Media from B were analyzed by Western blot. The N terminus of β-sEGF was detected using 7E6 and the C terminus was detected with 4F10. Note that in the case of endogenous processing or ADAM10 overexpression, the β-sEGF is liberated by N-terminal BACE1 cleavage (7E6 signal in lanes 2 and 3 and lanes 8 and 9, indicated by an arrow) but not C-terminal BACE1 shedding (no 4F10 signal in lanes 2 and 3 and lanes 8 and 9). Overexpression of ADAM17 prevents formation of β-sEGF (no signals for either antibody in lanes 6 and 7). Diamonds indicate glycosylation. Asterisks indicate unspecific background bands that are also present in the untransfected control (lane 1).
In cell lysates of HEK293 cells coexpressing NRG1 type III and either protease, antibody 4F10 selectively detected NTFs generated by BACE1- but not ADAM-mediated shedding (Fig. 4B). Upon coexpression of BACE1, the 7E6 antibody detected the CTF (comprising the EGF-like domain) resulting from BACE1 cleavage after L216 (Fig. 4B). This indicates that, upon elevated BACE1 expression, cleavage may first occur N-terminal of the EGF-like domain. Antibody 10E8 confirmed the generation of the β-CTF upon shedding of NRG1 type III by BACE1, but not by ADAMs (Fig. 4B). Being specific for the BACE1-generated neo-epitopes, the antibodies did not recognize the full-length NRG1 type III.
Western blot analysis of supernatants with antibodies 7E6 and 4F10 (specific to the BACE1 cleaved N and C terminus of the EGF-like domain, respectively) revealed robust amounts of soluble β-sEGF76 upon expression of BACE1 (Fig. 4C, lanes 4 and 5). As determined by MS (Fig. 3D, E), endogenous protease activity in HEK293 cells generates β-sEGF68 through ADAM10-mediated (C-terminal) shedding and BACE1-mediated (N-terminal) cleavage. Consistent with this, we detected low amounts of β-sEGF68 with the 7E6 antibody (Fig. 4C, lanes 2 and 3, arrow), whereas the 4F10 antibody did not recognize this fragment due to the absence of a BACE1-generated C-terminal epitope. Accordingly, enhanced shedding by ectopic expression of ADAM10 further increased the amounts of β-sEGF68 in the supernatant (Fig. 4C, compare lanes 8 and 9 with lanes 2 and 3). ADAM17-mediated cleavage N- and C-terminal of the EGF-like domain releases α-sEGF49 (Fig. 3D,E), which does not contain the epitopes recognized by 7E6 or 4F10 (Fig. 4A). Consistent with that, overexpression of ADAM17 prevented endogenous formation of β-sEGF68, and no 7E6 signal is observed in lanes 6 and 7 in Figure 4C. Therefore, the neo-epitope-specific antibodies are sensitive and selective tools for the investigation of NRG1 type III processing, and their specific immunoreactivity with defined processing products confirms the cleavage sites described above.
Dual cleavage of NRG1 type III in primary neurons releases the EGF-like domain
The neo-epitope-specific antibodies characterized in Figure 4 were used to investigate proteolytic processing of NRG1 type III in primary hippocampal neurons. To allow detection of NRG1 proteolytic fragments in neurons, we expressed V5-IIINRG1-HA using lentiviral transduction and studied its processing by endogenous secretases (Fig. 5). As revealed by Western blot analysis of lysates, shedding of V5-IIINRG1-HA in neurons generated a CTF and an NTF containing the EGF-like domain (Fig. 5A). Inhibition of BACE1 activity (by the BACE inhibitor IV) decreased the overall turnover of the full-length protein, as shown by the accumulation of the full-length precursor and decreased generation of the CTF. This was amplified by concomitant inhibition of ADAM proteases (by IV + GM6001), supporting the idea that BACE1 and ADMAs can compete for shedding in the stalk region of NRG1 type III under physiological conditions. CTFs and NTFs resulting from BACE1 shedding only were specifically detected with antibodies 10E8 and 4F10 and their generation was completely abolished upon inhibition of BACE1. Using antibody 7E6 for immunodetection, we did not observe a CTF containing the EGF-like domain (CTF+EGF; Fig. 4B) that would be generated by a single BACE1 cleavage of NRG1 type III after L216 (data not shown). This is consistent with the concept that, physiologically, the EGF-like domain of NRG1 type III is tethered to the membrane via its NTF (Taveggia et al., 2005). As observed in non-neuronal cells (Fig. 3), additional N-terminal cleavages release the EGF-like domain into the supernatant (β-sEGF and α-sEGF, respectively; Fig. 5A). Blocking BACE1 activity selectively prevented generation of the slightly larger β-sEGF, whereas the shorter α-sEGF was abolished upon ADAM inhibition. Reprobing of the membrane with the 7E6 antibody (Fig. 5A) revealed that the β-sEGF contains a BACE1-generated N terminus. We then performed immunoprecipitations from supernatants of neurons expressing V5-IIINRG1-HA using the neo-epitope-specific antibodies 7E6 and 4F10 (Fig. 5B). With both antibodies, the slightly larger β-sEGF, but not the smaller α-sEGF (lower band in upper panel of Fig. 5B), was detected, further confirming the identified BACE1 cleavage sites.
Figure 5.
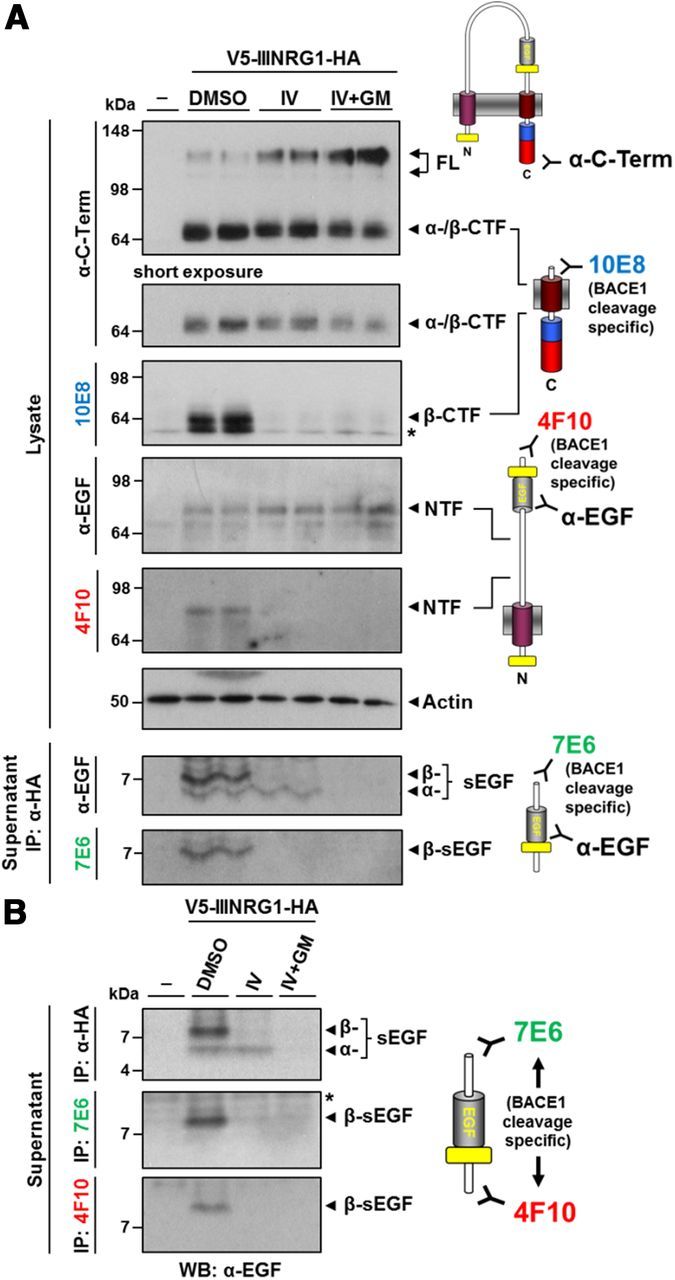
Cleavage of NRG1 type III in primary neurons releases the EGF-like domain. A, Processing of NRG1 type III in primary neurons generates both a membrane-tethered and an sEGF domain. V5-IIINRG1-HA was expressed in primary hippocampal neurons and cells were treated with indicated inhibitors (BACE1 inhibitor IV, 5 μm; ADAM inhibitor GM6001, 25 μm) for 16 h. Cells were lysed, conditioned supernatants were immunoprecipitated with HA-agarose beads, and protein levels were determined by immunoblotting with indicated antibodies. β-CTFs and NTFs generated by BACE1 cleavage were specifically detected by antibodies 10E8 and 4F10, respectively. Additional cleavages N-terminal of the EGF-like domain by BACE1 and ADAMs liberated β-sEGF and α-sEGF into the supernatant, respectively. B, BACE1 cleavage N- and C-terminal of the EGF-like domain liberates β-sEGF from neurons. Supernatants from neurons as described in A were immunoprecipitated with antibodies against the HA-tag or previously identified BACE1 cleavage sites (7E6 and 4F10). Purified peptides were detected using an α-EGF antibody.
These findings demonstrate that processing of NRG1 type III by endogenous BACE1 and ADAM proteases in primary neurons results in the release of the EGF-like domain as β-sEGF and α-sEGF.
α-sEGF and β-sEGF activate ErbB3 receptors in a paracrine fashion
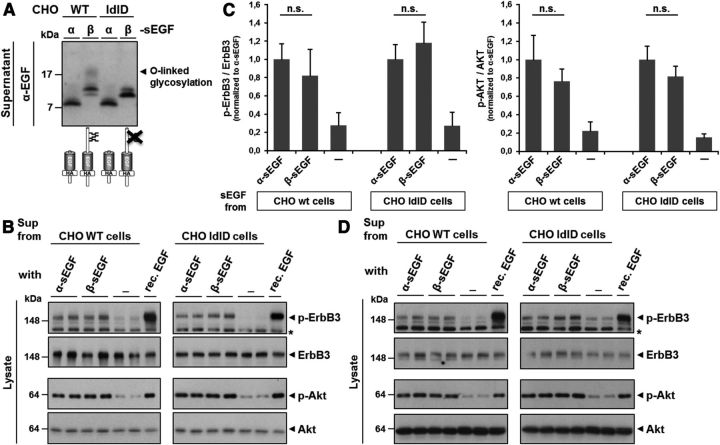
Myelination in the PNS is proposed to be regulated through the juxtacrine activation of ErbB receptors on Schwann cells by the NRG1 type III NTF-tethered EGF-like domain on axonal membranes (Wang et al., 2001; Birchmeier and Nave, 2008). In particular, stimulation of ErbB3 receptor phosphorylation and subsequent activation of the downstream PI3 kinase signaling pathway has been shown to promote PNS myelination (Newbern and Birchmeier, 2010). Because we were able to show that the EGF-like domain of NRG1 type III is liberated by BACE1 and ADAM17, we also investigated whether these small, sEGF domains were functional and if they could signal through ErbB3 receptors in a paracrine fashion. We expressed α-sEGF and β-sEGF in CHO wt cells and collected conditioned media (Fig. 6A). MCF-7 cells that express the ErbB3 receptor and are known to allow monitoring of NRG1 signaling via ErbB3 (Luo et al., 2011) were then incubated with the conditioned media containing equal concentrations of α-sEGF or β-sEGF or with supernatants from cells transfected with an empty vector. The ability of the sEGF domains to activate ErbB3 receptors and initiate PI3 kinase downstream signaling was monitored via phosphorylation of ErbB3 and AKT. Stimulation with 0.5 nm recombinant NRG1 EGF-like domain served as a positive control. Western blot analysis of total and phosphorylated levels of ErbB3 and AKT confirmed robust activation of both receptor and downstream signaling pathway upon stimulation with α-sEGF and β-sEGF (Fig. 6B, left). No significant differences between α-sEGF and β-sEGF in ErbB3 and AKT activation were observed (Fig. 6C).
Figure 6.
sEGF domains activate ErbB3 receptors on MCF-7 and Schwann cells and initiate AKT downstream signaling. A, Preparation of a-sEGF and glycosylated and nonglycosylated β-sEGF using CHO cells. α-sEGF and β-sEGF constructs were expressed in wt and O-glycosylation deficient (ldlD) CHO cells. Conditioned supernatants were analyzed by Western blotting and the concentrations of α-sEGF and β-sEGF were adjusted by dilution with medium from control cells. O-linked glycosylation causes β-sEGF to migrate as an additional and diffuse band of higher molecular weight, which is abolished in CHO ldlD cells. B–D, α-sEGF and β-sEGF activate ErbB3 receptors and AKT signaling in MCF-7 and primary Schwann cells independently of glycosylation. MCF-7 cells (B) and primary Schwann cells (D) were incubated with supernatants from A or from cells expressing an empty vector as a control. A recombinant NRG1 EGF-like domain (0.5 nm) was used as a positive control. After cell lysis, the levels of (phosphorylated) ErbB3 and AKT were determined by Western blotting. Quantification of phosphorylated protein/total protein ratio of experiments with MCF-7 cells is shown in C (mean ± SD; n.s., not significant, *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student's t test, n = 3).
In contrast to the smaller α-sEGF, β-sEGF contains a serine/threonine-rich sequence that can be O-glycosylated (Fig. 3C). To determine whether this difference in glycosylation affects activation of ErbB3 and AKT signaling we expressed α-sEGF and β-sEGF in CHO ldlD cells (Kingsley et al., 1986), which are deficient in O-linked glycosylation (Fig. 6A). Treatment of MCF-7 cells with conditioned media containing α-sEGF or nonglycosylated β-sEGF again stimulated activation of ErbB3 and AKT signaling in a very similar way (Fig. 6B, right; C). This suggests that O-linked glycosylation does not significantly alter the ability of β-sEGF to activate and signal through ErbB3 receptors.
Schwann cells are the recipient cells of NRG1 type III-mediated signaling during PNS myelination. Therefore, we repeated the experiments described above and incubated purified rat primary Schwann cells with α-sEGF and β-sEGF. Western blot analysis of phosphorylated ErbB3 and AKT levels in these cells confirmed our previous results. No difference in the activation of ErbB3 and AKT was observed upon stimulation with either α-sEGF or glycosylated or nonglycosylated β-sEGF (Fig. 6D).
These findings are in conflict with a recent study claiming an inhibitory effect of ADAM17-processed NRG1 type III on PNS myelination (La Marca et al., 2011). In this study, a slightly different ADAM17 cleavage site (G290) was identified. Moreover, although a recombinant NRG1 EGF-like domain cleaved by BACE1 induced AKT signaling in Schwann cells, an ADAM17-cleaved fragment failed to do so. We therefore investigated whether the different cleavage sites found for ADAM17 (G290 and A283) may explain the discrepancy in signaling of ADAM17- and BACE1-generated sEGF. We constructed sEGF domains that only differ in their very C-terminal residues. β-sEGF-F was designed to end with F293, simulating BACE1 processing, whereas β-sEGF-G terminates at G290, mimicking the ADAM17 cleavage site proposed by La Marca et al. (2011). Conditioned media containing similar amounts of the peptides (Fig. 7A) were then used to stimulate MCF-7 (Fig. 7B,C) and rat primary Schwann cells (Fig. 7D) as before. No difference in ErbB3 activation or induction of downstream AKT signaling in these cells was observed upon stimulation with β-sEGF-F or β-sEGF-G.
Figure 7.
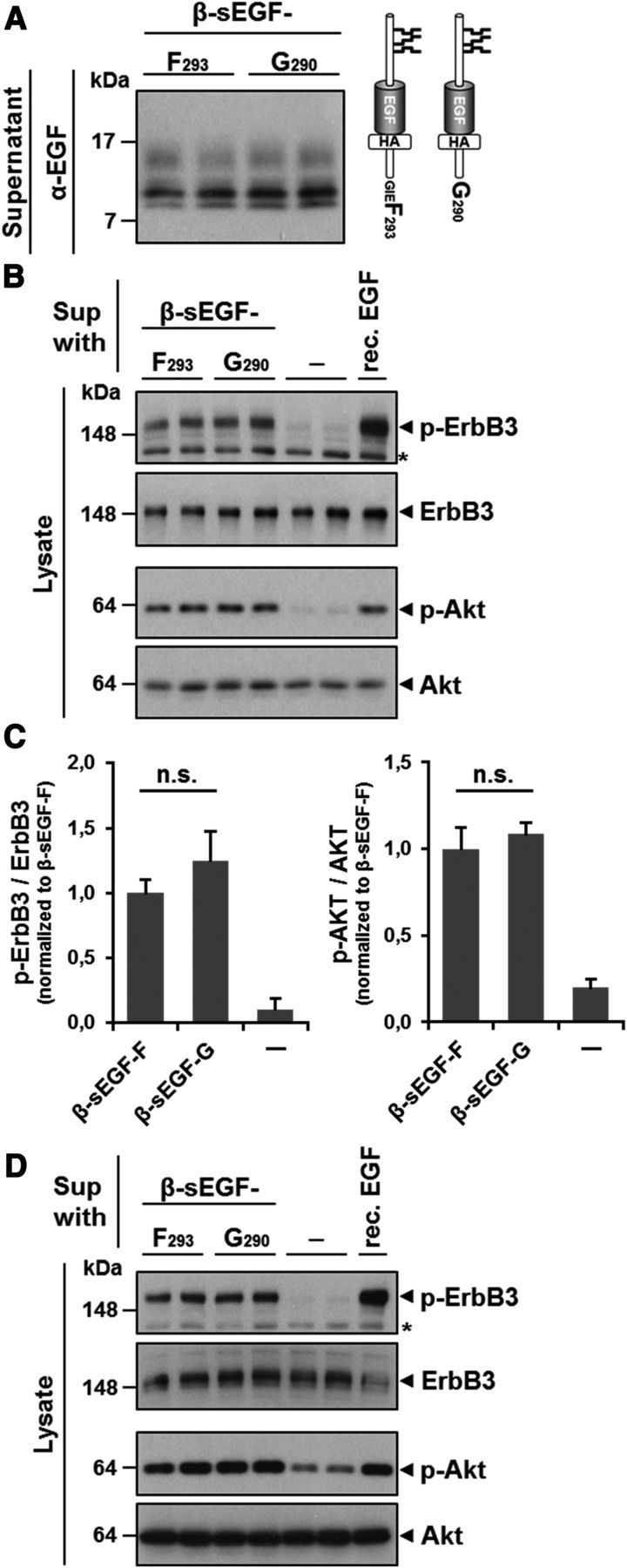
β-sEGF generated by BACE1- or ADAM17-mediated shedding similarly activates ErbB3 receptors and AKT downstream signaling A, β-sEGF-F and β-sEGF-G mimic shedding of NRG1 type III by BACE1 and ADAM17. The constructs β-sEGF-F and β-sEGF-G have identical N termini but comprise C termini generated by either BACE1- or ADAM17-mediated shedding: F293, BACE1 shedding; G290, ADAM17 shedding (see Results for details). Conditioned supernatant was collected from CHO wt cells and analyzed by Western blot. B–D, β-sEGF-F and β-sEGF-G activate ErbB3 receptors and AKT signaling in MCF-7 and primary Schwann cells. MCF-7 cells (B) and primary Schwann cells (D) were incubated with supernatants from A or from cells expressing an empty vector as a control. Recombinant NRG1 EGF-like domain (0.5 nm) was used as a positive control. Cells were lysed and levels of (phosphorylated) ErbB3 and AKT were determined by Western blotting. Quantification of phosphorylated protein/total protein ratio of experiments with MCF-7 cells is shown in C (mean ± SD; n.s., not significant, *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student's t test, n = 3).
These findings demonstrate that sEGF domains generated from NRG1 type III by BACE1 and ADAM17 are biologically active and signal through ErbB3 receptors on Schwann cells in a paracrine manner.
β-sEGF promotes PNS myelination in vivo
We also investigated whether the sEGF domain possesses signaling activity in vivo. We expressed the BACE1-derived β-sEGF in homozygous mutant bace1 (bace1−/−) zebrafish (van Bebber et al., 2013) and investigated whether the β-sEGF could compensate for the lack of BACE1-mediated NRG1 type III processing. To visualize myelination in vivo, we generated bace1−/− mutants expressing GFP under the control of the claudin k promoter, which labels Schwann cells and oligodendrocytes (Fig. 8A,B; Münzel et al., 2012). Phenotypically, bace1 homozygous zebrafish mutants become distinguishable from their wt siblings at 3 dpf due to severely reduced Schwann cell myelination (Fig. 8A,B). Myelination of lateral line axons by Schwann cells in the PNS is severely impaired, whereas myelination of Mauthner axons by oligodendrocytes in the CNS is unaffected (Fig. 8A,B). The observed selective hypomyelination in the PNS of bace1−/− zebrafish is consistent with the reduced myelination of BACE1 knock-out mice (Willem et al., 2006). To determine whether paracrine signaling could stimulate myelination, bace1−/− zebrafish carrying the claudin k:GFP transgene were injected with β-sEGF mRNA and analyzed at 3 dpf for rescued myelination. In 24 of 63 bace1−/− zebrafish, hypomyelination in the PNS was partially rescued upon expression of β-sEGF (Fig. 8B,C).
Figure 8.
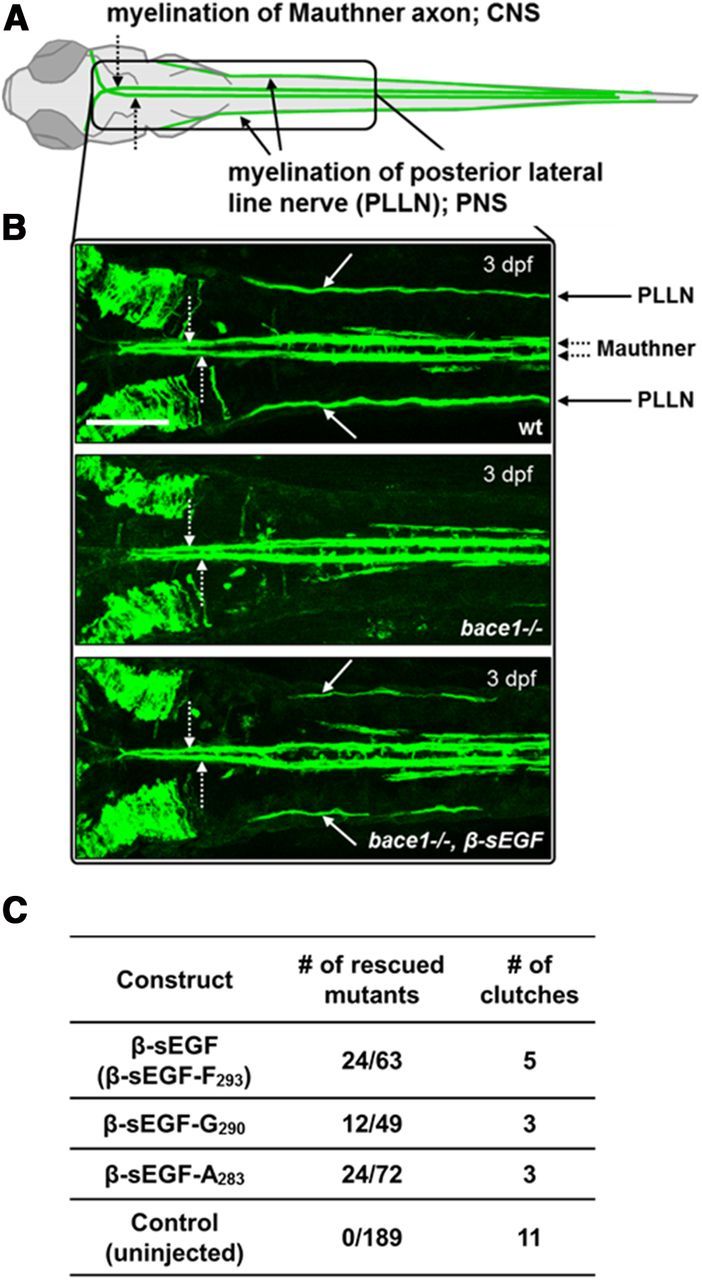
The soluble NRG1 type III EGF-like domain liberated by BACE1 promotes PNS myelination in vivo. A, The transgenic claudin k:GFP line allows visualization of myelin sheaths in zebrafish larvae. Schematic view of claudin k:GFP labeled myelin around Mauthner axons (CNS, dotted arrows) and lateral line axons (PNS, arrows) at 3 dpf (dorsal view). B, The sEGF domain rescues PNS myelination defect in bace1−/− zebrafish carrying the claudin k:GFP transgene. Dorsal views of wt, uninjected bace1 mutants (bace1−/−) and bace1 mutants injected with β-sEGF mRNA (bace1−/−, β-sEGF). Middle: In bace1−/− mutants myelination of the Mauthner axons (dotted arrows) is not affected, whereas myelination of the lateral line axons is severely reduced to absent (arrows in other panels). Bottom: Upon injection of β-sEGF mRNA, bace1−/− mutants display a partial rescue of hypomyelination in the PNS. Scale bar, 100 μm. C, sEGF domains generated by ADAM17-mediated shedding of NRG1 type III also promote PNS myelination. In addition to β-sEGF (B), β-sEGF constructs with C termini mimicking ADAM17 mediated shedding (β-sEGF-G290 and β-sEGF-A283; see Results for details) were injected into bace1−/− mutant zebrafish. Regardless of their very C-terminal residues, all constructs partially rescued the hypomyelination phenotype.
Finally, we used our zebrafish model to further test in vivo whether the C terminus of the EGF-like domain generated by ADAM17 cleavage abolishes its signaling capacity, as observed by La Marca et al. (2011). We injected mRNA encoding a NRG1 type III EGF-like domain terminating at either G290 (β-sEGF-G; La Marca et al., 2011) or A283 (β-sEGF-A; this study) into bace1−/− zebrafish. Both mRNAs partially rescued the PNS hypomyelination, similar to the rescue observed with β-sEGF (Fig. 8C). Although it is not possible to compare quantitatively the extent of the rescue effects, the results nevertheless provide strong evidence that the ADAM17-cleaved C terminus of the EGF-like domain does not abolish its signaling ability, as was claimed previously (La Marca et al., 2011).
In conclusion, our findings demonstrate that the sEGF domain of NRG1 type III liberated by N-terminal BACE1 cleavage promotes PNS myelination in vivo.
Discussion
Release of growth factors by proteolytic processing has emerged as an important regulator of many signaling pathways. For example, shedding particularly regulates signaling of ErbB receptor ligands (Sanderson et al., 2006). Sheddases either generate signaling-competent, membrane-retained proteins or release ectodomains that subsequently signal in a paracrine manner. At the same time, ectodomain shortening also triggers intramembrane cleavage of the remaining membrane fragments by intramembrane-cleaving proteases, releasing the substrate's intracellular domains into the cytosol, which may then act as transcription factors (Blobel et al., 2009; Lal and Caplan, 2011).
NRG1 type III is subject to such processing, which is also called regulated intramembrane proteolysis (Brown et al., 2000). Shedding in its stalk region activates NRG1 type III by generating a membrane-anchored NTF that presents the EGF-like domain to the luminal space and signals to ErbB receptors in a juxtacrine/contact-dependent manner (Falls, 2003; Taveggia et al., 2005). In addition, the resulting NRG1 CTF is cleaved by the γ-secretase and regulates transcription in neurons (Bao et al., 2003, 2004; Chen et al., 2010). Analysis of knock-out mouse models established BACE1 and ADAM17 as important NRG1 type III sheddases that control myelination in the PNS. Although BACE1-mediated shedding of NRG1 type III promotes myelination (Hu et al., 2006; Willem et al., 2006), shedding by ADAM17 was shown to inhibit this process (La Marca et al., 2011). This differential regulation was suggested to be due to small differences in the C termini of the NRG1 type III NTFs generated after shedding by BACE1 or ADAM17 (La Marca et al., 2011).
Despite the proposed importance, most NRG1 type III cleavage sites have only been investigated in vitro using short recombinant peptides in cleavage assays. Moreover, although the CTF resulting from shedding is eventually turned over by the γ-secretase, little is known about further processing of the corresponding NTF. It has been suggested that cleavage close to its N-terminal cysteine-rich TMD could release a large, soluble fragment containing the EGF-like domain (Wang et al., 2001). However, it remained unclear which proteases could mediate such a second cleavage event and whether the resulting soluble protein would possess signaling activity. We have now analyzed proteolytic processing of NRG1 type III in living cells to investigate whether, indeed, an sEGF domain was generated. To confirm the biological activity of such an sEGF domain, we generated a zebrafish mutant that lacks BACE1 and allows in vivo rescuing assays.
We first investigated the cleavages occurring between the EGF-like domain and the C-terminal TMD. Consistent with Montero et al. (2000), who suggested that the region from methionine 282 to tyrosine 286 may be a site of ADAM-mediated cleavage, we now assigned the ADAM17 cleavage to alanine 283 and, to a minor extent, to phenylalanine 285. Cleavage between alanine 283 and serine 284 fits well with the substrate preference of ADAM 17, which is known to favor alanine residues at the P1 position and to cleave several substrates with serine at P1′ (Caescu et al., 2009). For BACE1 and ADAM10, we report cleavage sites after phenylalanine 293 and phenylalanine 285, respectively, thereby confirming previous data from assays with recombinant proteins (Hu et al., 2008, Luo et al., 2011). In the case of ADAM10, we also observed cleavage after tyrosine 286, which has not been reported previously. These findings demonstrate homogenous shedding of NRG1 type III by BACE1 10 residues N-terminal of the TMD and heterogeneous shedding by ADAM10 and ADAM17 at close but distinct sites 7–10 residues N-terminal of the BACE1 cleavage site. We note that the shedding sites reported here for ADAM10 (Y286) and ADAM17 (A283 and F285) differ from the sites reported by others (La Marca et al., 2011; Luo et al., 2011). For ADAM10, the difference is marginal and may be due to the different experimental setups, namely in vitro digest of recombinant peptides (Luo et al., 2011) and cellular expression system (this study). For ADAM17, it is currently unclear why Luo et al. (2011) and we could not observe cleavage after G290 as reported by La Marca et al. (2011). However, the fact that the cleavage site motifs of ADAM10 and ADAM17 are fairly similar to each other and that both enzymes can cleave peptides in vitro at the same peptide bonds (Caescu et al., 2009) support our finding of close cleavage sites for these proteases within the stalk region of NRG1 type III.
BACE1- and ADAM-mediated shedding of NRG1 type III was also detected under endogenous protease levels in primary neurons. Although we cannot compare the contribution of individual proteases quantitatively, we observed additive effects of BACE1 and ADAM inhibitors, supporting the idea that these enzymes compete for shedding in the stalk region of NRG1 type III in neurons.
In addition to the shedding events taking place C-terminal of the EGF-like domain, we observed proteolytic processing at novel sites located N-terminal of the EGF-like domain, which indicated that the EGF-like domain may be secreted. We observed liberation of the EGF-like domain from NRG1 type III by additional N-terminal cleavages in HEK cells and primary neurons generating α-sEGF and β-sEGF. Using MS and site-specific antibodies, we were able to demonstrate that BACE1 is responsible for the N-terminal cleavage generating β-sEGF in both cell types. Interestingly, the novel BACE1 cleavage site resembles the BACE1 cleavage site in APP with the Swedish mutation. This mutation dramatically increases the affinity of BACE1 to its substrate (Citron et al., 1992, 1995), strongly indicating that this site may be used efficiently in vivo. In addition to BACE1, ADAM17 (but not ADAM10) was also found to cleave at another novel site close to the N terminus of the EGF-like domain, thereby generating α-sEGF. The detection of a similar fragment in the supernatant of primary neurons expressing NRG1 type III suggests that α-sEGF is generated by ADAM17-mediated cleavage in these cells as well. However, due to the lack of antibodies against the novel ADAM17 cleavage site, we currently cannot exclude an additional contribution of other ADAMs.
Both α-sEGF and β-sEGF are functionally active and induce ErbB3 receptor phosphorylation and AKT downstream signaling in MCF-7 and Schwann cells. Moreover, we demonstrated the in vivo signaling potential of β-sEGF as an instructive factor in the process of peripheral myelination, because β-sEGF was able to rescue the peripheral hypomyelination in a bace1 mutant zebrafish.
Our data are in agreement with a recent study reporting paracrine stimulation of Schwann cells and myelination by recombinant NRG1 type III (Syed et al., 2010). However, whereas this study used a mixture of recombinant peptides comprising the entire NRG1 type III N terminus, we now provide evidence for the generation of signaling-competent, soluble NRG1 type III fragments by BACE1- and ADAM17-mediated proteolysis. Unfortunately, it is not possible to generate an N-terminally uncleavable NRG1 type III NTF, so we cannot investigate whether NRG1 type III signaling in the context of myelination occurs exclusively via the sEGF domain. However, our results, together with the data of others (Syed et al., 2010), suggest that this novel paracrine signaling pathway may at least partially contribute to NRG1 type III signaling.
We observed a similar activation of ErbB3 and AKT in Schwann cells after stimulation with ADAM17-generated α-sEGF (C terminus A283) and BACE1 generated β-sEGF (C terminus F293). In contrast, others did not detect such activation upon stimulation with an EGF-like domain cleaved by ADAM17 (C terminus G290, La Marca et al., 2011). Because this was attributed to the very C-terminal residues of the EGF-like domain, we sought to reconcile these controversial findings by investigating the impact of these residues on ErbB activation. However, we could not detect any difference regarding ErbB signaling between sEGF domains terminating at the identified BACE1 (F293) or the ADAM17 cleavage sites (A283 and G290). Likewise, neither C termini attributed to ADAM17 cleavage prevented rescue of the hypomyelination phenotype in a bace1−/− zebrafish model. Moreover, in support of our findings, another study recently found no difference in ErbB activation by the membrane-anchored EGF-like domain after BACE1 (C terminus F293) or ADAM10 (C terminus F285, which excludes G290) processing (Luo et al., 2011). We currently have no definite explanation for the observed discrepancies; however, in vitro digests with recombinant peptides and enzymes imply the risk of additional cleavages that would not occur in a cellular environment. Such an additional cleavage within the EGF-like domain might abolish its signaling capacity and could partially account for the observed differences.
In summary, we have shown here that cleavage of NRG1 type III by BACE1 and ADAM17 at as-yet-unknown sites releases the EGF-like domain from its membrane anchor and allows for paracrine signaling of NRG1 type III via ErbB receptors. The proteases involved in NRG1 type III processing are major drug targets in the prevention or therapy of Alzheimer's disease and cancer (Duffy et al., 2011; Vassar and Kandalepas, 2011). The fact that these proteases have multiple—and until now unappreciated—roles in NRG1 signaling calls for caution when manipulating their activities in the course of therapy.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft Collaborative Research Center (Grant #SFB596 “Molecular Mechanisms of Neurodegeneration” A9 to M.W. and C.H., A16 to B.S., and Z2 to E.K.), the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013)/ ERC Grant Agreement No. 321366-Amyloid (advanced grant to C.H.), the Helmholtz Young Investigator program (to D.E.), the Hans and Ilse Breuer Foundation (fellowship to D.F.), the Federal Ministry of Education and Research (“IG Alzheimer” to M.W. and C.H., Competence Network for Neurodegenerative Diseases (KNDD) to S.F.L. and C.H.). We thank Dr. Boris Schmidt (Technical University of Darmstadt) for kindly providing the inhibitor GI254023X, Dr. Paul Saftig (University Kiel) and Galderma for kindly providing the inhibitor GL 506-3, and Drs. Ignasi Forné, Lars Israel, and Axel Imhof (Center for Protein Analysis, Ludwig-Maximilians-University, Munich) for support with the MS analysis.
The authors declare no competing financial interests.
References
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Lin H, Ouyang Y, Lei D, Osman A, Kim TW, Mei L, Dai P, Ohlemiller KK, Ambron RT. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004;7:1250–1258. doi: 10.1038/nn1342. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Blobel CP, Carpenter G, Freeman M. The role of protease activity in ErbB biology. Exp Cell Res. 2009;315:671–682. doi: 10.1016/j.yexcr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BG, Agarwal A, Sereda MW, Garratt AN, Müller T, Wende H, Stassart RM, Nawaz S, Humml C, Velanac V, Radyushkin K, Goebbels S, Fischer TM, Franklin RJ, Lai C, Ehrenreich H, Birchmeier C, Schwab MH, Nave KA. Neuregulin-1/ErbB signaling serves distinct functions in myelination of the peripheral and central nervous system. Neuron. 2008;59:581–595. doi: 10.1016/j.neuron.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/S0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XD, Golde TE, Younkin SG. Release of excess amyloid beta protein from a mutant amyloid beta protein precursor. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- Capell A, Steiner H, Willem M, Kaiser H, Meyer C, Walter J, Lammich S, Multhaup G, Haass C. Maturation and propeptide cleavage of beta-secretase. J Biol Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. J Neurosci. 2010;30:9199–9208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ. Mutation of the beta-amyloid precursor protein in familial Alzheimer's disease increases beta-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- Citron M, Teplow DB, Selkoe DJ. Generation of amyloid beta protein from its precursor is sequence specific. Neuron. 1995;14:661–670. doi: 10.1016/0896-6273(95)90323-2. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Mullooly M, O'Donovan N, Sukor S, Crown J, Pierce A, McGowan PM. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin Proteomics. 2011;8:9. doi: 10.1186/1559-0275-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einheber S, Zanazzi G, Ching W, Scherer S, Milner TA, Peles E, Salzer JL. The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J Cell Biol. 1997;139:1495–1506. doi: 10.1083/jcb.139.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls DL. Neuregulins and the neuromuscular system: 10 years of answers and questions. J Neurocytol. 2003;32:619–647. doi: 10.1023/B:NEUR.0000020614.83883.be. [DOI] [PubMed] [Google Scholar]
- Haass C. Take five–BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Dev Biol. 2005;283:459–471. doi: 10.1016/j.ydbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- Hu X, He W, Diaconu C, Tang X, Kidd GJ, Macklin WB, Trapp BD, Yan R. Genetic deletion of BACE1 in mice affects remyelination of sciatic nerves. FASEB J. 2008;22:2970–2980. doi: 10.1096/fj.08-106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karey KP, Sirbasku DA. Differential responsiveness of human breast cancer cell lines MCF-7 and T47D to growth factors and 17 beta-estradiol. Cancer Res. 1988;48:4083–4092. [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Kozarsky KF, Hobbie L, Krieger M. Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase deficient mutant. Cell. 1986;44:749–759. doi: 10.1016/0092-8674(86)90841-X. [DOI] [PubMed] [Google Scholar]
- Lal M, Caplan M. Regulated intramembrane proteolysis: signaling pathways and biological functions. Physiology (Bethesda) 2011;26:34–44. doi: 10.1152/physiol.00028.2010. [DOI] [PubMed] [Google Scholar]
- La Marca R, Cerri F, Horiuchi K, Bachi A, Feltri ML, Wrabetz L, Blobel CP, Quattrini A, Salzer JL, Taveggia C. TACE (ADAM17) inhibits Schwann cell myelination. Nat Neurosci. 2011;14:857–865. doi: 10.1038/nn.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Haass C, Steiner H. Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J Neurochem. 2011;117:779–796. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen. 2005;8:161–171. doi: 10.2174/1386207053258488. [DOI] [PubMed] [Google Scholar]
- Luo X, Prior M, He W, Hu X, Tang X, Shen W, Yadav S, Kiryu-Seo S, Miller R, Trapp BD, Yan R. Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J Biol Chem. 2011;286:23967–23974. doi: 10.1074/jbc.M111.251538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Monk KR, Talbot WS. Genetic dissection of myelinated axons in zebrafish. Curr Opin Neurobiol. 2009;19:486–490. doi: 10.1016/j.conb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JC, Yuste L, Díaz-Rodríguez E, Esparís-Ogando A, Pandiella A. Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-alpha-converting enzyme. Mol Cell Neurosci. 2000;16:631–648. doi: 10.1006/mcne.2000.0896. [DOI] [PubMed] [Google Scholar]
- Münzel EJ, Schaefer K, Obirei B, Kremmer E, Burton EA, Kuscha V, Becker CG, Brösamle C, Williams A, Becker T. Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia. 2012;60:253–270. doi: 10.1002/glia.21260. [DOI] [PubMed] [Google Scholar]
- Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. doi: 10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C. Presenilins mediate a dual intramembranous γ-secretase cleavage of Notch-1. EMBO J. 2002;21:5408–5416. doi: 10.1093/emboj/cdf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep. 2012;13:759–764. doi: 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin JR, Lush ME, Stephens WZ, Piotrowski T, Talbot WS. Neuronal Neuregulin 1 type III directs Schwann cell migration. Development. 2011;138:4639–4648. doi: 10.1242/dev.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MP, Dempsey PJ, Dunbar AJ. Control of ErbB signaling through metalloprotease mediated ectodomain shedding of EGF-like factors. Growth Factors. 2006;24:121–136. doi: 10.1080/08977190600634373. [DOI] [PubMed] [Google Scholar]
- Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C. Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa-Sehara A. Roles of Meltrin beta /ADAM19 in the processing of neuregulin. J Biol Chem. 2001;276:9352–9358. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- Syed N, Reddy K, Yang DP, Taveggia C, Salzer JL, Maurel P, Kim HA. Soluble neuregulin-1 has bifunctional, concentration-dependent effects on Schwann cell myelination. J Neurosci. 2010;30:6122–6131. doi: 10.1523/JNEUROSCI.1681-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, Chao MV, Falls DL, Role L, Salzer JL. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, Turk du SA, Turk V. Protease signalling: the cutting edge. EMBO J. 2012;31:1630–1643. doi: 10.1038/emboj.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bebber F, Hruscha A, Willem M, Schmid B, Haass C. Loss of Bace2 in zebrafish affects melanocyte migration and is distinct from Bace1 knock out phenotypes. J Neurochem. 2013 doi: 10.1111/jnc.12198. doi: 10.1111/jnc.12198. Advance online publication. Retrieved February 14, 2013. [DOI] [PubMed] [Google Scholar]
- Vassar R, Kandalepas PC. The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer's disease. Alzheimers Res Ther. 2011;3:20. doi: 10.1186/alzrt82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velanac V, Unterbarnscheidt T, Hinrichs W, Gummert MN, Fischer TM, Rossner MJ, Trimarco A, Brivio V, Taveggia C, Willem M, Haass C, Möbius W, Nave KA, Schwab MH. Bace1 processing of NRG1 type III produces a myelin-inducing signal but is not essential for the stimulation of myelination. Glia. 2012;60:203–217. doi: 10.1002/glia.21255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Miller SJ, Falls DL. The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem. 2001;276:2841–2851. doi: 10.1074/jbc.M005700200. [DOI] [PubMed] [Google Scholar]
- Wild-Bode C, Fellerer K, Kugler J, Haass C, Capell A. A basolateral sorting signal directs ADAM10 to adherens junctions and is required for its function in cell migration. J Biol Chem. 2006;281:23824–23829. doi: 10.1074/jbc.M601542200. [DOI] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- Wolpowitz D, Mason TB, Dietrich P, Mendelsohn M, Talmage DA, Role LW. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron. 2000;25:79–91. doi: 10.1016/S0896-6273(00)80873-9. [DOI] [PubMed] [Google Scholar]