Abstract
Noncoding RNA plays an important role in all aspects of the cellular life cycle, from the very basic process of protein synthesis to specialized roles in cell development and differentiation. However, many noncoding RNAs remain uncharacterized and the function of most of them remains unknown. Mid‐size noncoding RNAs (mncRNAs), which range in length from 50 to 400 nucleotides, have diverse regulatory functions but share many fundamental characteristics. Most mncRNAs are produced from independent promoters although others are produced from the introns of other genes. Many are found in multiple copies in genomes. mncRNAs are highly structured and carry many posttranscriptional modifications. Both of these facets dictate their RNA‐binding protein partners and ultimately their function. mncRNAs have already been implicated in translation, catalysis, as guides for RNA modification, as spliceosome components and regulatory RNA. However, recent studies are adding new mncRNA functions including regulation of gene expression and alternative splicing. In this review, we describe the different classes, characteristics and emerging functions of mncRNAs and their relative expression patterns. Finally, we provide a portrait of the challenges facing their detection and annotation in databases.
This article is categorized under:
Regulatory RNAs/RNAi/Riboswitches > Regulatory RNAs
RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry
RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems
RNA Evolution and Genomics > RNA and Ribonucleoprotein Evolution
Keywords: mid‐size noncoding RNA, noncoding RNA, snoRNA, snRNA, tRNA
1. INTRODUCTION
Noncoding RNAs are often subdivided into long and small RNAs around a cut‐off of 200 nucleotides. The long noncoding RNAs over 200 nucleotides are mostly generated from intergenic regions (Brosnan & Voinnet, 2009; Dhanoa, Sethi, Verma, Arora, & Mukhopadhyay, 2018; Zampetaki, Albrecht, & Steinhofel, 2018). However, the small noncoding RNAs are a disparate and heterogeneous group of distinct RNA families. The most extensively studied small noncoding RNAs are very short (<50 nucleotides) unstructured and generally expressed at low abundance, and include microRNA (miRNA) and piwi‐interacting RNA (Cheng, Dong, Zhang, Zhao, & Li, 2018; Le Thomas, Toth, & Aravin, 2014). In contrast, the 50–200 nucleotides size range includes several highly structured and highly expressed RNA families including transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs; Boivin, Deschamps‐Francoeur, & Scott, 2018; Brosnan & Voinnet, 2009; Haeusler & Engelke, 2006; Iben & Maraia, 2014). These abundant highly structured families of “small” noncoding RNA share fundamental characteristics in terms of structure, maturation, and function that hinder their detection by RNA‐sequencing (Boivin et al., 2018). Some members of these small noncoding RNA families and other additional RNA groups (including some snoRNAs and snRNAs, as well as the 7SL, 7SK, and RNaseP RNAs) are longer than 200 nucleotides and are often categorized as long noncoding RNAs. However, we propose here that all RNAs between 50–400 nucleotides are a distinct class, the mid‐size noncoding RNAs (Figure 1). Many RNA of this type have been the subject of separate reviews that discuss specific functional characteristics (e.g., Decatur, Liang, Piekna‐Przybylska, & Fournier, 2007; Fang & Guo, 2017; Lui & Lowe, 2013; Orioli, 2017; Reynolds, Vargas‐Rodriguez, Soll, & Crnkovic, 2017; Tuorto & Lyko, 2016; Woolford & Baserga, 2013). In this review, we deviate from earlier work by treating ncRNA that vary from 50–400 as a single class and describe their structural and functional features with a focus on human mncRNAs.
Figure 1.
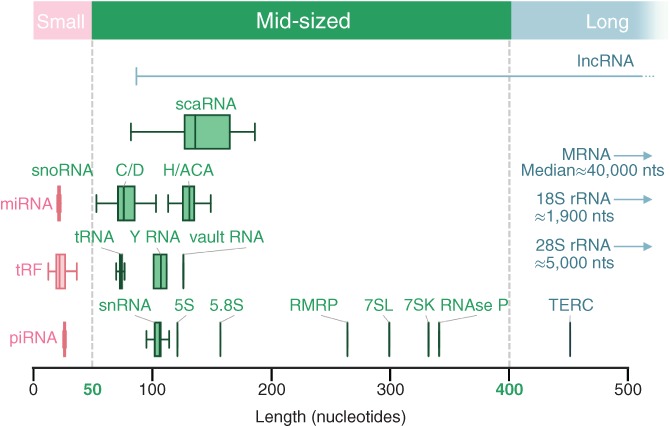
Types and size distribution of human RNA. The different types of human RNAs are illustrated as a function of their length. The average size of small, mid‐size and long RNA is indicated in red, green and blue boxes, respectively. Dotted lines indicate the size delimitation between these RNA families. The median size of mRNAs and the size of rRNAs that are too large to be visible on the figure are indicated on the right
While human RNA is the focus of this review it is important to note, however, that mncRNAs exist in diverse forms across all domains of life and the origin of some stretch as far back as the Last Universal Common Ancestor (LUCA) (Figure 2). Notable examples and differences between species will be highlighted when appropriate. mncRNAs can be divided into 5 main classes: (a) translation‐related RNAs like tRNA, ribosomal RNA (rRNA) and the signal recognition particle (SRP) RNA 7SL, (b) catalytic RNAs like RNase P and MRP, (c) RNA modification guides, like snoRNAs and scaRNAs, (d) spliceosomal RNA which include snRNA, and (e) regulatory RNAs including vault RNAs, 7SK and Y RNAs.
Figure 2.
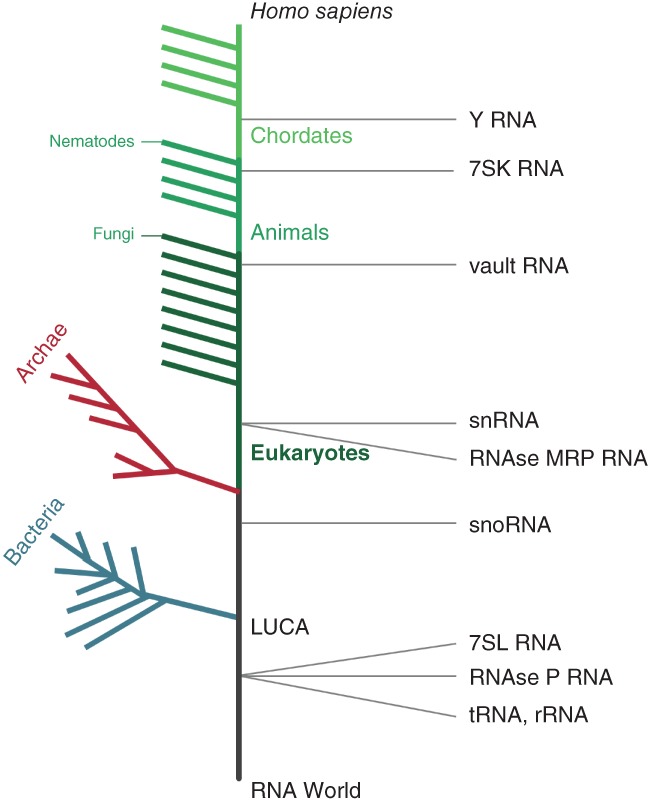
Schematics of the evolutionary history of human mncRNA. The vertical time‐scale is a descriptive representation (not to scale) of the appearance time of different RNA types with the bottom representing the RNA world and the top representing human‐specific RNA. Eukaryotic branches close to the human lineage are indicated in light green while those further away are indicated in dark green. The branching events relevant to the appearance of the different types of mncRNA are indicated on the right
1.1. Translation‐related RNAs
The most conserved mncRNAs are the translation‐related RNAs (Figure 2). In all domains of life, protein translation from a messenger RNA template is carried out both by rRNAs and tRNAs. These RNAs were the first noncoding RNAs to be characterized, as their function is so fundamental for every living organism (McQuillen, Roberts, & Britten, 1959; Rich & RajBhandary, 1976). The discovery of tRNA and the standard genetic code added a whole new dimension to sequence selection in evolution (Itzkovitz & Alon, 2007; Koonin & Novozhilov, 2009; Pechmann & Frydman, 2013). The advantages of this protein‐coding system in the genomes of every known living organism probably explains why no known modern cell has reverted to an RNA‐only form or has developed an alternative mechanism for protein translation, although small variations of the standard code do exist (Crick, 1968; Hinegardner & Engelberg, 1963; Koonin & Novozhilov, 2009). For example, tRNA fragments and derivatives have started to dominate the space of unannotated transcriptome being discovered through sequencing (Anderson & Ivanov, 2014; Diebel, Zhou, Clarke, & Bemis, 2016; Keam & Hutvagner, 2015; Shen et al., 2018; Sun et al., 2018). Like other translation‐associated mncRNAs, 7SL RNA (SRP RNA) is highly conserved and required for protein localization even in bacteria. Indeed, 7SL RNA is the single most abundant RNA after rRNA in the human transcriptome (Boivin, Deschamps‐Francoeur, Couture, et al., 2018). The sequence and structure of this RNA is well conserved. In all domains of life, the 7SL RNA has a two stem‐loop S domain and an Alu domain, although in some bacteria species, 7SL is reported to have only a single stem‐loop S domain. This conservation suggests early evolution of the protein trafficking system, presumably to send proteins into the membranes of primitive cells. This is supported by recent evidence that the LUCA proteome included proteins that are localized to membranes (Weiss et al., 2016).
1.2. Catalytic RNAs
The catalytic RNA class of mncRNAs includes the RNA processing enzymes RNase P and the RNase mitochondrial RNA processing (MRP). The RNAse P RNA is also conserved in all domains of life (Figure 2). This ribozyme is essential for the cleavage of the 5′‐leader elements of precursor tRNAs as well as for the proper transcription of other pol III transcripts, including 5S rRNA and 7SL (Reiner, Ben‐Asouli, Krilovetzky, & Jarrous, 2006). In bacteria, the RNase P RNA has only one associated protein, C5, which is not necessary for its catalytic activity. In archaea and eukaryotes, the protein content of the RNase P is much higher (up to 10 proteins in higher eukaryotes) and the RNA component has a weaker catalytic activity when isolated, as if it had progressively evolved to become dependent on and regulatable by its associated proteins (Guerrier‐Takada, Gardiner, Marsh, Pace, & Altman, 1983; Kikovska, Svard, & Kirsebom, 2007; Marquez, Chen, Evans, & Pace, 2006). More strikingly, it was shown that functional RNase P exists without an RNA component in human mitochondria, bacteria, archaea and in spinach chloroplast (Gossringer et al., 2017; Holzmann et al., 2008; Nickel et al., 2017). This suggests that, even though an RNA is functionally relevant and well conserved in all domains of life, it is not impossible that functional redundancy by proteins or other RNAs can lead to its progressive substitution. In early eukaryotes, the RNase P RNA gene duplicated and one copy became the RNase MRP, which is essential for precursor rRNA processing and mitochondrial DNA replication (Esakova & Krasilnikov, 2010; Jarrous & Gopalan, 2010; Sbisa, Pesole, Tullo, & Saccone, 1996; Schmitt & Clayton, 1993).
1.3. RNA modification guides
The RNA modification class of mncRNAs includes scaRNAs and the C/D and H/ACA snoRNAs. The small Cajal body‐specific RNAs (scaRNA) are specialized in the biogenesis and modification of small nuclear ribonucleoprotein complexes (snRNAs) including those required for splicing (Bohnsack & Sloan, 2018). The C/D box snoRNAs, required for RNA 2′‐O‐ribose methylation, evolved after branching from bacteria (Lafontaine & Tollervey, 1998; Figure 2). In contrast, bacterial RNA methylation relies solely on site‐specific RNA‐binding proteins to modify their rRNA (Sergeeva, Bogdanov, & Sergiev, 2015). H/ACA and C/D snoRNAs probably both evolved from single ancestors and have spread throughout genomes by hitchhiking on retro‐transposable elements (Weber, 2006). This is especially evident in supraprimates where many H/ACA snoRNAs arose from mutations and intronic insertion of the H/ACA‐containing Alu repeated elements (Jady, Ketele, & Kiss, 2012; Ketele, Kiss, & Jady, 2016; Kriegs, Churakov, Jurka, Brosius, & Schmitz, 2007). Since Alu repeated elements are known to be derived from the Alu domain of 7SL, it is tempting to speculate that H/ACA snoRNAs first evolved and spread from a similar event involving the retro‐transposition of the Alu domain of 7SL at the beginning of the archaea/eukarya phylogenic branch. Sequence homology suggests that the snoRNA‐dependent modification system evolved from a closely related system that uses tRNA‐introns as guides for tRNA modifications. This mechanism involves a protein that recognizes a guide RNA (snoRNA) that in turn recognizes rRNA. This system allows greater modification site specificity and faster evolution rate of the modification site than protein‐alone systems (Terns & Terns, 2002). Indeed, snoRNAs can duplicate using retro‐transposable elements and only a few nucleotide substitutions can result in the recognition of novel target sites. For an RNA‐binding protein, a whole mRNA duplication is necessary, and many mutations may be required to change the RNA binding motif. This is probably one of the reasons why archaea and eukaryote rRNAs have many more modifications than bacterial rRNAs (Lafontaine & Tollervey, 1998).
1.4. Spliceosomal RNA
Spliceosomal RNAs are among the most highly expressed mammalian mncRNAs and are often encoded by multiple genes (Cordin, Hahn, & Beggs, 2012; Irimia & Roy, 2014; Villa, Pleiss, & Guthrie, 2002). These RNAs were acquired during evolution to respond to the modification of the mechanism of protein production in eukaryotes, which changed from simple transcription coupled translation to highly regulated separate processes. The evolution of introns and the requirement of splicing required the development of an intron removal machinery capable of detecting exon–intron boundaries (Rogozin, Carmel, Csuros, & Koonin, 2012). The U1, U2, U4, U5, and U6 snRNA of the major spliceosome clearly appeared early in eukaryotic evolution (Figure 2) as they are found in higher eukaryotes as well as deep‐branching organisms like Giardia lamblia (Brow & Guthrie, 1988; Hudson et al., 2012; Marz, Kirsten, & Stadler, 2008; Vanacova, Yan, Carlton, & Johnson, 2005). The U11, U12, U4atac, and U6atac of the minor spliceosome have only been identified in higher eukaryotes Metazoa, Fungi, Viridiplantaea, and Heterokonta (Lorkovic, Lehner, Forstner, & Barta, 2005; A. G. Russell, Charette, Spencer, & Gray, 2006; Turunen, Niemela, Verma, & Frilander, 2013). The snRNA class of mncRNA, which is required for pre‐mRNA splicing, also contributes to transcriptome and proteome diversity through the process of alternative splicing. In addition, snRNAs play an important role in the regulation of RNA expression and stability through splicing‐associated processes like nonsense‐mediated decay (Bush, Chen, Tovar‐Corona, & Urrutia, 2017; Grutzmann et al., 2014; Kelemen et al., 2013).
1.5. Regulatory RNA
The last class of mncRNAs is an ever‐expanding eclectic group of regulatory RNAs involved in different aspects of gene expression. This class of mncRNA appeared late during evolution (Figure 2) and plays an important role in the biology of vertebrate cells. Examples of this class of RNA include the vault RNAs of the enigmatic vault particles which have only been found in animals and a few fungi (Kedersha, Miquel, Bittner, & Rome, 1990; Stadler et al., 2009). The vault complex consists of one major vault protein (MVP), two minor vault proteins (VPARP and TEP1), and a variety of associated untranslated small RNA molecules called vault RNAs. Given the association of the complex with the nuclear membrane it was proposed that vault particles participate in intracellular and nucleocytoplasmic transport (Stadler et al., 2009; van Zon, Mossink, Scheper, Sonneveld, & Wiemer, 2003). Another example of regulatory mncRNA are the 7SK RNAs, which are slightly more recent than vault RNAs as they have only been found in chordates and nematodes (Marz et al., 2009). 7SK RNAs play a role in transcription regulation through their interaction and inhibition of the positive transcription elongation factor b (Peterlin, Brogie, & Price, 2012). The telomerase RNA component (TERC), essential for telomere repair, is yet another example of this class of mncRNAs. Although its function has been thoroughly studied in vertebrates, ciliates and yeast, its evolutionary origin is not clear due to the heterogeneous origin of telomeric repeats (Y. Brown et al., 2007; Podlevsky & Chen, 2016; Qi et al., 2013; Romero & Blackburn, 1991). Finally, this class also includes Y RNAs, which are believed to promote DNA replication and interact with the autoimmune protein Ro60 (Hall, Turnbull, & Dalmay, 2013). This RNA class encompasses the most recently evolved members of the mncRNA family (Mosig, Guofeng, Stadler, & Stadler, 2007). Below we will discuss the structural and functional characteristics of the different classes of mncRNA and their possible association with human disease.
2. mncRNA GENOMIC ARCHITECTURE AND EXPRESSION STRATEGIES
mncRNA use diverse strategies for expression. These expression strategies differ on multiple levels including both the genomic architecture of their expression module and the polymerase employed for transcription. The genomic architecture determines the relationship between the mncRNA and its neighboring genes. For example, a mncRNA gene located in an intron depends on the expression level of its host gene and on the efficiency of its splicing. However, at the same time, the processing of a mncRNA and its proximity to the splice site may influence the expression of its host gene (Hirose & Steitz, 2001; Vincenti, De Chiara, Bozzoni, & Presutti, 2007). On the other hand, the number of genes encoding mncRNAs is greatly affected by their association with transposable elements (Davis et al., 2017; Klimenko, 2017). Controlling the overall abundance and thus the function of a single RNA species produced from multiple genes, like for example U1, which is encoded by hundreds of copies, requires intergene regulation. However, the coordination mechanisms remain largely unclear. Below, we describe the different architectures and expression routes of mncRNAs.
2.1. Independent mncRNA genes
Many families of mncRNAs have their own promoters and are independently transcribed. Most independently expressed mncRNAs are transcribed by the RNA polymerase III (Dieci, Fiorino, Castelnuovo, Teichmann, & Pagano, 2007). These include the eukaryotic tRNA, U6 snRNA, RNase P, 7SL, vault, RNase MRP, Y and 7SK RNA genes (Canella, Praz, Reina, Cousin, & Hernandez, 2010; Dieci et al., 2007). This group of abundant and highly transcribed RNAs represents the bulk of the RNA pol III transcriptome and more than 50% of the non‐ribosomal RNA in human cells (Boivin, Deschamps‐Francoeur, Couture, et al., 2018). The RNA pol III machinery recognizes specific upstream and internal promoter elements for these genes, including a distal sequence element, a proximal sequence element, and a TATA‐box as well as A and B boxes (Dieci et al., 2007). mncRNAs transcribed by the RNA pol III do not accumulate in capped or polyadenylated form, although in many cases certain transcripts acquire 5′ cap structures as they mature (Abdelhamid et al., 2014; Massenet, Bertrand, & Verheggen, 2017; Perumal & Reddy, 2002). In addition, although many pol III transcribed mncRNAs come from intergenic regions, others are located along with their own promoter, in either sense or antisense orientation, in the introns of other genes (Dieci et al., 2007). While most mncRNA families are thus transcribed by the RNA pol III, all the snRNA genes except U6 are independently transcribed by the RNA polymerase II (Guiro & Murphy, 2017; Jawdekar & Henry, 2008). The pol II promoters of these snRNA genes are more basic than most RNA pol II promoters, consisting of a simple distal sequence element and a proximal sequence element (Guiro & Murphy, 2017). The snRNA transcripts are co‐transcriptionally capped but are not polyadenylated (Guiro & Murphy, 2017). Intriguingly, the U6atac snRNA genes have been reported to be transcribed by RNA pol I, II and III (Grummt, 2003; J. Russell & Zomerdijk, 2005; Younis et al., 2013). In addition, a small number of snoRNA have been identified in intergenic regions and are independently transcribed either by RNA pol II or RNA pol III (Dieci et al., 2007; Jawdekar & Henry, 2008). It remains possible that many or even most independently transcribed snoRNAs are hidden in the genome awaiting discovery. Annotating the full repertoire of noncoding RNAs, with few structural clues, in regions without potential host genes, will require independent predictors that rely on sequencing read accumulation.
2.2. Gene‐embedded mncRNAs
Unlike other mid‐size RNAs, most vertebrate snoRNAs and scaRNAs are embedded in introns of longer RNA pol II transcribed genes referred to as host genes, and thus require the expression of their host gene to be generated (Dieci, Preti, & Montanini, 2009). The production of these embedded mid‐size RNAs not only requires host gene transcription by the RNA polymerase II, but also splicing of the intron containing the embedded gene and removal of the excess intronic sequences by exonuclease trimming (Boivin, Deschamps‐Francoeur, & Scott, 2018; Kiss, Fayet, Jady, Richard, & Weber, 2006; Massenet et al., 2017). These intronic mncRNAs are not capped or polyadenylated in their mature form, which differentiates them from other independently transcribed mncRNA. While still in the intron, the embedded gene transcript is bound by RNA‐binding proteins that initiate assembly of the mature ribonucleoprotein (RNP) particle and protect the nascent RNA from degradation during the intron trimming (Hirose & Steitz, 2001; Ooi, Samarsky, Fournier, & Boeke, 1998; Petfalski, Dandekar, Henry, & Tollervey, 1998). In flies, there are often more than one snoRNA per intron, whereas in mammals there is usually just one snoRNA per intron, although host genes often contain several snoRNA‐encoding introns (Boivin, Deschamps‐Francoeur, & Scott, 2018; Dieci et al., 2009). Intronic mncRNAs are not restricted to the introns of protein coding genes but also occur in the introns of other noncoding genes (J. W. Brown, Marshall, & Echeverria, 2008; Hesselberth, 2013). Protein coding snoRNA host genes are often members of the family of terminal oligopyrimidine (TOP) genes, many of which encode proteins involved in translation and gene expression. TOP gene expression modules thus encode several products, protein and snoRNAs, impacting protein production (Dieci et al., 2009). However, the snoRNA class of mncRNA may behave as mobile genetic elements and a given snoRNA can be found in different host genes across organisms (Weber, 2006). The level and temporality of host gene expression have been proposed as other important determinants of the position of a snoRNA in the genome. Indeed, it was suggested that snoRNAs could move to any host gene with appropriate expression levels, regardless of its function. Supporting this, many snoRNA are encoded in host genes with functions unrelated to translation or protein synthesis (Hoeppner & Poole, 2012). Consistently, in mammals and flies mncRNAs and particularly snoRNAs were found in genes that do not code for proteins at all as is the case of lncRNA. Such host genes consist of short exons separated by short introns, most of which contain a snoRNA (Dieci et al., 2009). Many lncRNA snoRNA host genes display striking conservation patterns, the intronic snoRNAs being much more highly conserved than the exons, suggesting that the expression of snoRNAs is the main cellular function of these lncRNAs (Ono et al., 2010).
2.3. Expression of multicopied mncRNAs
Many mncRNAs exist in multiple copies in genomes presumably due to retrotransposon L1 (long interspersed nuclear element 1) or other mobile element retroposition events, resulting in the amplification of clusters of mncRNA. Transposition‐dependent cluster creation and amplification is most evident in the translation‐related mncRNAs, like tRNAs (Doucet, Droc, Siol, Audoux, & Gilbert, 2015). Studies of the snRNA copies in the human genome indicate the existence of truncated copies, with preferential positions for the truncation in regions that are single‐stranded and resemble the L1 endonuclease cleavage site, implying that these copies may be functional (Doucet et al., 2015). Many mncRNAs, whether truncated or not, are annotated as pseudogenes, although many are expressed (Boivin, Deschamps‐Francoeur, Couture, et al., 2018). mncRNA copy number ranges from a handful of copies, for some snoRNAs, to more than a hundred copies for certain tRNAs, and for 5S rRNAs, in various organisms including cows, rats and plants (Bermudez‐Santana et al., 2010; Haeusler & Engelke, 2006; Michaud, Cognat, Duchene, & Marechal‐Drouard, 2011; Wong, Abrahamson, & Nazar, 1984).
Although many of the mncRNA copies might not be functional, a large number of these copies generate detectable RNA and in some cases, they are strongly expressed (Boivin, Deschamps‐Francoeur, Couture, et al., 2018; Hoeppner, Denisenko, Gardner, Schmeier, & Poole, 2018).
For example, of the 147 annotated U1 copies in human, 10 copies are each expressed at above 1,000 transcripts per million in the ovarian cancer cell line SKOV3ip1, accounting for the vast majority of the detected U1 transcripts (Boivin, Deschamps‐Francoeur, Couture, et al., 2018). In addition, snRNA variants have been found to be differentially expressed in different cell types and stages, with switching between copies during development seen in Drosophila, Xenopus, mouse and human (Lu & Matera, 2014; O'Reilly et al., 2013). Importantly, variants of U1 snRNA were found to regulate different target transcripts and thus play different biological functions (O'Reilly et al., 2013). Similarly, copies of tRNAs are also differentially expressed across Caenorhabditis elegans tissues (Sagi et al., 2016) and the expression of tRNAs varies in disease (Goodarzi et al., 2016). This suggests that expression of individual mncRNA copies is strongly regulated and it will be important to characterize the regulation of all mncRNAs, under different conditions in health and in disease.
3. mncRNA STRUCTURE AND MATURATION
3.1. Structure
Different mncRNAs classes form specific secondary structures that are essential for their stability, biogenesis, and function. Simple mutations that disrupt the structure are sufficient to suppress the accumulation of mncRNA (Filipowicz, Pelczar, Pogacic, & Dragon, 1999; Watkins et al., 1998). The energetic stability of mncRNA structures also often impairs their detectability by standard sequencing and polymerase chain reaction techniques (Boivin, Deschamps‐Francoeur, Couture, et al., 2018; Deschamps‐Francoeur et al., 2014). The tRNAs have the most distinctive secondary structure, a three‐leafed clover composed of three hairpin loops that folds into a 3D L‐shape (Giege et al., 2012). tRNA structure is intimately linked to its function permitting its insertion into the P and A sites of the ribosome, and its anticodon loop is essential for recognizing the mRNA code (Giege et al., 2012). Similarly, the 7SL RNA is characterized by a tight secondary fold including the long central fifth helix that is universally conserved (Rosenblad, Larsen, Samuelsson, & Zwieb, 2009). 7SL is thus recognized by core SRP proteins, enabling the assembly of a functional SRP complex, and forming an integral part of both the small and large domains of the SRP (Rosenblad et al., 2009). snoRNAs also display distinctive structures. H/ACA box snoRNAs consist of two hairpin loops separated by an H box and terminated by an ACA box. The hairpin loops have internal bulges with guide sequences that recognize the targets. C/D box snoRNA are comparatively less structured than other mncRNAs, consisting generally of a loose hairpin. Their distinctive feature is their terminal kink‐turn motif, which consists of the non‐Watson–Crick base pairing of residues of the C and D boxes, representing a binding site for core interactors. The structure of both the H/ACA and C/D box snoRNAs influences the binding of the proteins forming the core of the stable RNP particle, and ultimately also their functions (Bachellerie, Cavaille, & Huttenhofer, 2002; Dupuis‐Sandoval, Poirier, & Scott, 2015).
There are two classes of spliceosomal snRNAs, the Sm and “like Sm” (Lsm) class. The Sm particles include the U1, U2, U4, U4atac, U5, U7, U11, and U12 RNAs, which are widely studied and constitute the bulk of spliceosomal RNA. The sm‐snRNA has a hyper‐methylated trimethylguanosine 5′ cap and forms a distinct 3′ stem‐loop structure that is necessary for recognition by the survival of motor neuron (SMN) protein and thus for the stability and function of this class of snRNAs (Ares & Weiser, 1995; Kolb, Battle, & Dreyfuss, 2007; Moreno Diaz de la Espina, Alverca, Cuadrado, & Franca, 2005; Piekna‐Przybylska, Liu, & Fournier, 2007; Schumperli & Pillai, 2004). In contrast, Lsm snRNAs, which include U6 and U6atac, have a 5′‐γ‐monomethylphosphate cap and their 3′ stem loops terminate in a stretch of uridines that form the binding site for the distinct heteroheptameric Lsm protein ring (Beggs, 2005; Brow & Guthrie, 1988, 1990; Decker, Teixeira, & Parker, 2007; Frendewey, Barta, Gillespie, & Potashkin, 1990; Spiller, Boon, Reijns, & Beggs, 2007; Verdone, Galardi, Page, & Beggs, 2004). In summary, all mncRNAs contain distinctive structures consisting of conserved stem loops that function as flags for assembly of structural protein components. Disruption of these structures disassembles the RNP particle and leads to degradation of the RNA due to the resulting exposure to cellular ribonucleases.
3.2. mncRNA biogenesis and assembly into RNPs
Whether transcribed from their own promoter or excised from host gene transcripts, all mncRNAs require several maturation steps, including binding to protein components for stability and function, and transport to specific cellular compartments where they function or where they are further processed. The sequential protein binding events and processing steps, and ultimately assembly into RNPs have been extensively characterized for specific mncRNAs. For example, while still in their host transcript intron, snoRNAs are bound by core proteins in the nucleoplasm. The binding of these co‐factors defines the boundary of the mature RNA and protects it from nuclear ribonucleases, while the absence of these co‐factors flags defects in the RNA structure leading to the activation of the quality control or nuclear surveillance machinery (J. W. Brown et al., 2008; Didychuk, Butcher, & Brow, 2018; Dupuis‐Sandoval et al., 2015; Massenet et al., 2017; Ogami, Chen, & Manley, 2018; Quinn & Chang, 2016; Schmidt & Butler, 2013; Szymanski, Barciszewska, Zywicki, & Barciszewski, 2003; Xing & Chen, 2018). H/ACA box snoRNAs are bound by a preformed protein complex, consisting of the pseudouridylase dyskerin and assembly factors, which are replaced by the GAR1, NHP2 and NOP10 proteins (Li, Duan, Li, Ma, & Ye, 2011). Similarly, the terminal k‐turn of C/D box snoRNAs is bound by the SNU13 (also known as 15.5K) and NOP58 proteins. The C/D snoRNA is then further bound by two additional core proteins, NOP56 and fibrillarin, which provides the enzymatic activity and completes the core complex (Massenet et al., 2017; Rothe et al., 2014). The final processing of both C/D and H/ACA snoRNAs occurs in Cajal bodies. The mature snoRNPs are then targeted to the nucleolus where they function (Massenet et al., 2017). 7SL assembly involves quality control after transcription and this includes binding to the La protein and remodeling of its 3′ termini through its 3′ oligo (U) trimming and monoadenylation (E. Leung & Brown, 2010). The nucleolar 7SL RNA protein complex then recruits the core SRP proteins, SRP9, SRP14, SRP68, and SRP72, after they are imported into the nucleus (E. Leung & Brown, 2010). Subsequent binding of SRP19 to this complex makes it competent for export to the cytoplasm, where additional protein factors are recruited to form a mature functional SRP (E. Leung & Brown, 2010).
Like the 7SL RNA, tRNAs are transcribed by RNA polymerase III in the nucleus or in organelles such as mitochondria, chloroplasts, or plastids. Regardless of their location, nascent tRNAs are processed as local structures formed during transcription. As the D‐arm, anticodon arm, and finally the TΨC arm and the acceptor stem form, the maturation process terminates with 5′ and 3′ end trimming, the removal of introns, if present, and the addition of the characteristic CCA tails. At any point in this maturation pathway, posttranscriptional modifications can be added to influence folding (Belostotsky, Frishberg, & Entelis, 2012; Chatterjee, Nostramo, Wan, & Hopper, 2018; Fang & Guo, 2017; Shen et al., 2018; Wichtowska, Turowski, & Boguta, 2013). In contrast spliceosomal RNAs mature posttranscriptionally. After pol II transcription and capping, U1, U2, U4, and U5 pre‐snRNAs are assembled into an export complex, which crosses the nuclear pore and disassembles in the cytoplasm. There, the SMN proteins recruit the pre‐snRNAs to form the core snRNP complex and enable modification of the cap and exonucleolytic trimming of the pre‐snRNAs (Kiss, 2004). The complex is then imported into the nucleus where it transits through the Cajal bodies for chemical modification (pseudouridylation and 2′‐O‐methylation) by scaRNAs, and binding to additional proteins (Kiss, 2004). The mature snRNPs then accumulate in nucleoplasmic splicing speckles close to actively transcribed genes (Kiss, 2004). Unlike the pol II transcribed snRNAs, the biogenesis of U6 occurs entirely in the nucleus. Newly synthesized U6 RNA is bound and stabilized by the La protein which helps snRNP assembly, leading to its replacement by the heteroheptameric doughnut‐shaped Lsm (Sm like) complex (Kiss, 2004; Mroczek & Dziembowski, 2013). The maturing U6 snRNP visits the nucleolus to be chemically modified by snoRNAs to render the U6 catalytically active (Mroczek & Dziembowski, 2013). The U6 snRNP then localizes to the Cajal body where it is assembled into the U4/U6 di‐snRNP complex (Mroczek & Dziembowski, 2013).
Although the core RNP complexes involving the main mncRNAs have been extensively characterized in terms of structure and biogenesis, recent evidence indicates these pictures may be incomplete or over‐simplified. Indeed, some members of these families bind noncanonical proteins and localize to different cellular compartments, and likely have different functions. For example, specific snoRNAs do not depend on core snoRNP interactors for their biogenesis and bind to other proteins including splicing factors (Chu et al., 2012; Deschamps‐Francoeur et al., 2014; Kishore et al., 2010; Soeno et al., 2010). Variations in the canonical processing pathways as well as alternative derivatives of mncRNA precursors and mature sequences were reported to give rise to multiple subspecies. For example, stable fragments of tRNA and snoRNA were found in several species (Cole et al., 2009; M. S. Scott et al., 2012; Taft et al., 2009). tRNA‐derived RNA fragments are generated through endonucleolytic cleavages under certain conditions, for example, stress and hypoxia. These fragments may signal stress responses or regulate gene expression (Shen et al., 2018). Similarly, small fragments generated from snoRNA were implicated in gene regulation, including by RNAi interference (M. S. Scott & Ono, 2011). Together, these observations suggest that mncRNA processing variations are an evolutionary nursery for new RNA species.
3.3. Posttranscriptional modification
Many families of mncRNAs are heavily posttranscriptionally modified, affecting their structure, interaction partners and function. For example, snRNA modifications, like base methylations, 2′‐O‐methylation, and pseudouridylation, play an important role in spliceosome assembly and regulate splicing efficiency (Bohnsack & Sloan, 2018; Kiss, 2004). tRNAs are the most modified RNAs, often by deamination to inosine, various forms of methylation and acetylation (Gogakos et al., 2017; Lorenz, Lunse, & Morl, 2017). Modification of tRNA affects folding, stability and function including the decoding of the wobble position and frameshifting (El Yacoubi, Bailly, & de Crecy‐Lagard, 2012; Lorenz et al., 2017; Nachtergaele & He, 2017). Many other mncRNA families are also modified. Indeed, it was proposed that snoRNAs modify each other to explain the crosslinking of one snoRNA to another (Kishore et al., 2013). Recently, differential posttranscriptional modification of mncRNAs were linked to growth conditions and cell type, suggesting a dynamic regulation that can fine‐tune the functionality of the main cellular RNPs (Bohnsack & Sloan, 2018; El Yacoubi et al., 2012; Nachtergaele & He, 2017). Furthermore, variation in posttranscriptional modifications of mncRNAs are being linked to human pathologies (Torres, Batlle, & Ribas de Pouplana, 2014). The large variety and dynamic nature of mncRNA modifications increase their functional diversity and provide additional levels of regulation of biogenesis and function.
4. EMERGING FUNCTIONS OF mncRNA
In recent years, many new functions of mncRNAs have been discovered suggesting a much larger implication in cell biology than previously envisioned (Diebel et al., 2016; Dupuis‐Sandoval et al., 2015; Keam & Hutvagner, 2015; Nachtergaele & He, 2017; Orioli, 2017; Stadler et al., 2009; van Zon et al., 2003; Yang, Wen, & Zhu, 2015). This is particularly evident in the case of snoRNAs, which are the mncRNA family with the largest literature describing emerging functions. In addition to their canonical role as guides for the site‐specific modification of rRNA and snRNAs, snoRNAs have been found to modulate coding gene expression in multiple ways including through the regulation of alternative splicing, of mRNA 3′ processing and as serving as mediators of metabolic stress (Dupuis‐Sandoval et al., 2015; Falaleeva, Welden, Duncan, & Stamm, 2017; Shi, Huang, Huang, & Yao, 2018). For example, SNORD115, which shares an 18‐nt sequence complementarity with the alternative exon Vb of the serotonin receptor 2C, was shown to promote the inclusion of this exon (Kishore & Stamm, 2006). Interestingly, the functions of snoRNAs in splicing and modification are not mutually exclusive. Indeed, SNORD27 has been known to both methylate rRNA and modify the splice site selection. Mutational analysis indicated that SNORD27 binds to the 5′ splice sites of the E2F7 transcripts, presumably to block U1 snRNP access, leading to exon skipping (Falaleeva et al., 2016). Depletion of SNORD27 represses the splicing of several other exons suggesting a general function of the snoRNA in the repression of splicing (Falaleeva et al., 2016). In addition to splicing, a subset of snoRNAs was found associated with the mRNA 3′ processing complex in mammals. One such snoRNA, SNORD50A, blocks binding of the cleavage and polyadenylation specificity factor to poly(A) sites, affecting alternative polyadenylation profiles and transcript levels (Huang et al., 2017). On the other hand, several reports have found snoRNAs to play a role in lipotoxicity and metabolic stress (Caputa & Schaffer, 2016). For example, the rRNA modification snoRNAs located in the intron of the Rpl13a locus accumulate in the cytoplasm and play a critical role in response to metabolic stresses, suggesting once again a dual cellular role (Michel et al., 2011). snoRNAs were also found to be the predominant class of RNAs bound to the double stranded RNA‐dependent protein kinase PKR during stress response. Overexpression of specific snoRNAs activates PKR and leads to phosphorylation of eIF2 alpha resulting in modification of translation profiles (Youssef et al., 2015).
Noncanonical functions were also reported for snRNAs in processes other than splicing, including the regulation of transcription, polyadenylation, and RNA stability (Valadkhan & Gunawardane, 2013). For example, U1 is reported to be involved in a process called telescripting in which the U1 snRNP prevents premature cleavage and polyadenylation of pre‐mRNA, promoting the synthesis of long transcripts (Oh et al., 2017). The U1 snRNP was also implicated in the degradation of specific pre‐mRNA by binding to sequences resembling 5′ splice sites in their 3′ untranslated region, leading to inhibition of polyadenylation (Buratti & Baralle, 2010). tRNAs have also been found to play many diverse noncanonical roles, in particular in bacteria. Such roles include functions in the biosynthesis of diverse cellular macromolecules, and regulatory functions in which tRNAs are used as messengers or sensors for the metabolic state of the cell (Katz, Elgamal, Rajkovic, & Ibba, 2016). In addition to the emerging noncanonical roles of full‐length mncRNAs, fragments of mncRNAs, which are typically shorter and less highly structured than their full‐length precursors are increasingly reported to play cellular roles (Falaleeva & Stamm, 2013; Soares & Santos, 2017).
4.1. mncRNA and disease
Many mncRNAs including snRNAs, TERC and snoRNAs are linked to human diseases. Mutations in mncRNAs contributing to housekeeping functions, like the spliceosomal RNAs, are found in several diseases including cancer (Cazzola, Rossi, Malcovati,, & Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators, 2013; Harbour, 2013; Lee & Rio, 2015; L. M. Scott & Rebel, 2013; Visconte, Makishima, Maciejewski, & Tiu, 2012; Wan & Wu, 2013; Zent & Burack, 2014). Mutations in TERC, which possesses structural features of H/ACA snoRNAs, were linked to dyskeratosis congenita (Chen & Greider, 2004; Li et al., 2011; Rashid et al., 2006). More recently, mncRNAs with less established functions including canonical and noncanonical snoRNAs are being increasingly associated with human disease. For example, several snoRNAs were either upregulated or downregulated in different types of cancer including breast (Langhendries, Nicolas, Doumont, Goldman, & Lafontaine, 2016), lung (Langhendries et al., 2016; Liao et al., 2010; Mannoor, Shen, Liao, Liu, & Jiang, 2014; Mei et al., 2012; Su et al., 2016), brain (Dong et al., 2009), pancreatic (Cui et al., 2017; Kitagawa et al., 2019), and prostate (Martens‐Uzunova et al., 2015). These cancer‐associated snoRNAs have been shown to both induce and suppress tumor development (Mannoor et al., 2014). For example, SNORA42 is overexpressed in non‐small cell lung cancer, and knocking it down in this cancer caused cell growth arrest and apoptosis (Mei et al., 2012). SNORD50A, on the other hand, acts as a tumor suppressor in prostate cancer cells (Dong et al., 2009). The snoRNAs likely alter tumor cell function through their effect on ribosome production, but equally may affect RNA stability and splicing. snoRNAs have roles in other diseases like Prader–Willi syndrome, which is a genetic disorder linked to mutations in chromosome 15. Two large families of C/D box snoRNAs, SNORD115 and SNORD116, are located on this chromosome at the locus IC‐SNURF‐SNRPN (Bortolin‐Cavaille & Cavaille, 2012; Cavaille et al., 2000; Dong et al., 2009). As mentioned above, the SNORD115 snoRNAs do not target the modification of rRNA and have rather been shown to affect the splicing of the serotonin receptor 5‐HT(2C)R through specific binding of its pre‐mRNA. In Prader–Willi patients, SNORD115 can be absent, affecting the production of functional serotonin receptors 5‐HT(2C)R (Kishore & Stamm, 2006).
5. ANNOTATION AND CURATION OF mncRNA
The annotation of human noncoding RNAs is ongoing. Currently, there are databases focusing on individual subclasses, but no dedicated databases exist for mncRNA as a whole. There are also databases that are more inclusive. Some meta‐databases regroup information on one or more subclass from other databases. Databases also differ in the way they identify noncoding RNAs. Most of these databases use predictive algorithms based on the structural and sequence features of each group of RNAs, and some consider direct experimental evidence. Here, we discuss the different types of databases that house different classes of mncRNAs and present their main features to help users identify appropriate databases for their subfield of interest. The main databases housing mncRNAs and their family scope are listed in Figure 3.
Figure 3.
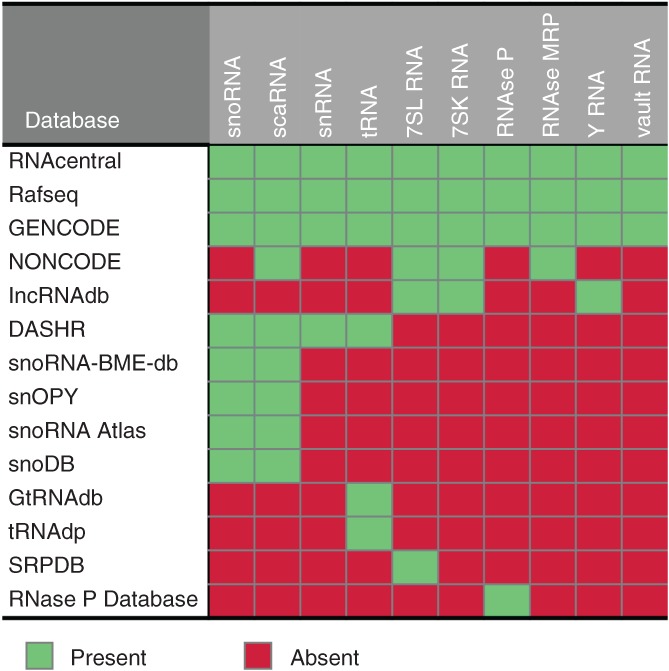
Summary of the RNA contents in mncRNA databases. The RNA content found in major genome annotation databases is indicated
5.1. Types of databases housing mncRNA subclasses
General databases bring together the annotations of different RNAs including mncRNA, and tend to rely on experimental evidence of expression, and predictions drawn from publicly available literature. Examples of this type of database are GENCODE (Harrow et al., 2012) and RefSeqGene (O'Leary et al., 2016). Currently, these databases house variable numbers of mncRNAs including tRNA, snoRNA, snRNA, Y RNA and vault RNA, and it is not clear if any one database is near complete. The main focus of this type of database is to give the genomic location and functional information for each gene. These annotations are especially helpful for research involving several types of RNA, or that may be interested in the relationship between different types of RNAs. Specialized databases like snoRNA‐LBME‐db (Lestrade & Weber, 2006) or GtRNAdb (Chan & Lowe, 2016) provide information about a subclass of mncRNA. For example, snoRNA databases like snoRNA‐LBME‐db, snOpy (Yoshihama, Nakao, & Kenmochi, 2013) and snoRNA Atlas (Jorjani et al., 2016) give information about the position and nature of each snoRNA target. The tRNA databases like GtRNAdb and tRNAdb show information about sequence alignment of tRNAs, secondary structure and give the anticodon type for each tRNA gene (Juhling et al., 2009). In addition, some mncRNA databases have a genomic focus on single or just a small number of genes, like the SRPDB (Andersen et al., 2006; Gorodkin, Knudsen, Zwieb, & Samuelsson, 2001; Rosenblad, Gorodkin, Knudsen, Zwieb, & Samuelsson, 2003) and RNAse P databases (J. W. Brown, 1999). Their central purpose is to focus on orthology by showing the sequence and/or structure in a wide range of species. snOpy, GtRNAdb, and tRNAdb also give extensive orthological information, allowing the users to compare the genomic organization of snoRNAs or differences in codon usage between species. The information held in these specialized databases is well suited for researchers with an interest in specific RNA types.
Hybrid databases mix features of both general and specialized types, which increase their scope, but they fall short of covering all RNA types. For example, NONCODE (Zhao et al., 2016) and lncRNAdb (Quek et al., 2015) both mainly focus on long noncoding RNAs, but both have extended their purpose by including shorter noncoding RNAs. NONCODE, for example, includes scaRNAs, 7SL, 7SK. and RNAse MRP while lncRNAdb includes annotation of 7SL, 7SK, and Y RNAs. The reason for the inclusion and exclusion of specific noncoding RNA types in these databases is unclear, but may be related to the length of these RNA species. NONCODE and lncRNAdb both manually curate annotation from the literature but NONCODE also fetches extra information from other databases, including RefSeqGene, GENCODE, and lncRNAdb. On the other hand, the DASHR database (Y. Y. Leung et al., 2016) provides annotation for only the “major classes of small noncoding RNAs,” which includes miRNAs (primary and mature), snoRNAs, snRNAs, scRNAs, tRNAs, rRNAs, and tRNA‐fragments. It fetches its information from miRbase, GENCODE and tRNAdb and complements this with the computational analysis of 802 small‐RNA‐Seq datasets from various tissues and studies. In contrast to these databases that aggregate and filter information from experiments and/or predictions gathered in the literature, the RNAcentral noncoding RNA database indiscriminately collects information from other databases (RNAcentral, 2017). By filtering and integrating the data of 27 databases, RNAcentral provides a more comprehensive view of the transcriptome across many species. Therefore, this general database may be very helpful for researchers considering many different genes and gene types. The user will inevitably accept a certain amount of false‐positive data from this gathering exercise. However, the database is great to find if any given sequence is annotated in any database.
5.2. Features and benefits of different noncoding RNA databases
The architecture and features of these different databases influence their suitability for different research questions. For example, RNA‐Seq analysis often requires a file holding gene annotation information, usually in general transfer format (gtf) or general feature format (gff). Generalized databases like RNAcentral, GENCODE, RefSeqGene, and DASHR as well as the specialized database snoRNA Atlas provide downloads for these formats, making them directly useful for RNA‐Seq analysis. Other databases provide other forms of search and download features in various formats like csv or fasta, which can easily be converted to gtf or gff format by providing gene coordinates and features. For automated queries, RNAcentral, GENCODE and NCBI offer an application programming interface for fetching relevant data. The availability of a direct connection to GENCODE, NCBI, and DASHR facilitates the fetching and filtering of data for specific interests. In most cases, these databases provide users with gene names, sequence, identifiers and links pointing to other databases and tools, but they vary in their presentation of the structure, expression and/or conservation data. Structural information is extremely important to mncRNA researchers, but only a few RNAs have experimentally validated structures. Expression values are also of great interest, but most of these data come from RNA‐Seq analyses, which varies in quality between experiments and RNA type. Expression of structured RNAs like tRNAs and snoRNAs is particularly underestimated by standard RNA‐Seq methods, while pre‐sequencing selection of a specific RNA type may introduce biases in the relative presentation of snoRNA (Boivin, Deschamps‐Francoeur, Couture, et al., 2018). The methods of data analysis may also affect the estimation of RNA abundance. For example, using the RNAcentral database gff3 annotation file would result in the elimination of overlapping reads generated by different databases. In contrast using GENCODE data may result in read loss due to overlap between host genes and the nested noncoding RNAs (Frankish et al., 2015). Choosing the GENCODE “basic” annotation set or filtering transcripts by support level may help reduce the read loss problem. General annotation sets are more suitable for routine analysis of standard RNA while more inclusive databases should be privileged for exploratory purpose. For RNA type‐specific analyses, more specialized databases would be more appropriate as they provide comprehensive information for the targeted RNA group. In the end, no database is perfect and periodic evaluation of the data provenance and update dates should be considered before a database is selected. That said, RefSeqGene, GENCODE, and RNAcentral databases are currently reliable options as they include the largest numbers of mncRNAs (Figure 4).
Figure 4.
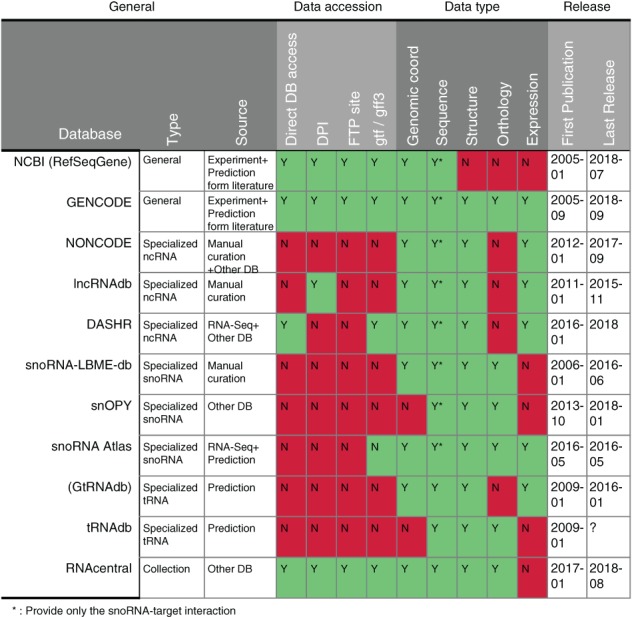
Key features of major human mncRNA annotation databases. General information for each database, as of December 2018, including methods for data accession, data type and release dates, are indicated
6. CONCLUSION
The mncRNAs are nonpolyadenylated RNAs between 50 and 400 nucleotides that have stable structures and form stable RNP particles. This class of RNA influences the different levels of gene expression: RNA maturation, processing, modification, and translation. In addition, mncRNA are associated with several human diseases including cancer. In terms of mass, mncRNA exceed that of mRNA, and some mncRNAs have the highest copy number of any transcripts in the cell (Boivin, Deschamps‐Francoeur, Couture, et al., 2018; Palazzo & Lee, 2015). As such, miscalculating the expression level of this class of noncoding RNA undoubtedly affects the analysis of the entire transcriptome. Indeed, the small size, lack of polyadenylation and strong structure of mncRNA complicates their detection using standard techniques, and as a consequence, their abundance is often underestimated. Recent developments in the sequencing of noncoding RNA opens the door to new avenues for the analysis of these classes of mncRNA but challenges remain in ascertaining their absolute expression levels and assigning function to these emerging classes of regulatory RNA.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
Single cell transcriptomics of noncoding RNAs and their cell‐specificity
ACKNOWLEDGMENTS
This work was supported by Canadian Institutes of Health Research (CIHR) grant PJT 153171 (S.A. and M.S.S.) and Research Chair in RNA Biology and Cancer Genomics (S.A.). V.B. was supported by a scholarship from the Fonds de Recherche du Québec ‐ Santé (FRQS). M.S.S. holds a FRQS Research Scholar Junior 2 Career Award.
Boivin V, Faucher‐Giguère L, Scott M, Abou‐Elela S. The cellular landscape of mid‐size noncoding RNA. WIREs RNA. 2019;10:e1530. 10.1002/wrna.1530
Funding information Canada Research Chairs; Institute of Genetics, Grant/Award Number: PJT153171; Fonds de Recherche du Québec ‐ Santé
REFERENCES
- Abdelhamid, R. F. , Plessy, C. , Yamauchi, Y. , Taoka, M. , de Hoon, M. , Gingeras, T. R. , … Carninci, P. (2014). Multiplicity of 5′ cap structures present on short RNAs. PLoS One, 9(7), e102895 10.1371/journal.pone.0102895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, E. S. , Rosenblad, M. A. , Larsen, N. , Westergaard, J. C. , Burks, J. , Wower, I. K. , … Zwieb, C. (2006). The tmRDB and SRPDB resources. Nucleic Acids Research, 34, D163–D168. 10.1093/nar/gkj142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. , & Ivanov, P. (2014). tRNA fragments in human health and disease. FEBS Letters, 588(23), 4297–4304. 10.1016/j.febslet.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares, M., Jr. , & Weiser, B. (1995). Rearrangement of snRNA structure during assembly and function of the spliceosome. Progress in Nucleic Acid Research and Molecular Biology, 50, 131–159. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J. P. , Cavaille, J. , & Huttenhofer, A. (2002). The expanding snoRNA world. Biochimie, 84(8), 775–790. [DOI] [PubMed] [Google Scholar]
- Beggs, J. D. (2005). Lsm proteins and RNA processing. Biochemical Society Transactions, 33(Pt. 3), 433–438. 10.1042/BST0330433 [DOI] [PubMed] [Google Scholar]
- Belostotsky, R. , Frishberg, Y. , & Entelis, N. (2012). Human mitochondrial tRNA quality control in health and disease: A channelling mechanism? RNA Biology, 9(1), 33–39. 10.4161/rna.9.1.18009 [DOI] [PubMed] [Google Scholar]
- Bermudez‐Santana, C. , Attolini, C. S. , Kirsten, T. , Engelhardt, J. , Prohaska, S. J. , Steigele, S. , & Stadler, P. F. (2010). Genomic organization of eukaryotic tRNAs. BMC Genomics, 11, 270 10.1186/1471-2164-11-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack, M. T. , & Sloan, K. E. (2018). Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biological Chemistry, 399(11), 1265–1276. 10.1515/hsz-2018-0205 [DOI] [PubMed] [Google Scholar]
- Boivin, V. , Deschamps‐Francoeur, G. , Couture, S. , Nottingham, R. M. , Bouchard‐Bourelle, P. , Lambowitz, A. M. , … Abou‐Elela, S. (2018). Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. RNA, 24(7), 950–965. 10.1261/rna.064493.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin, V. , Deschamps‐Francoeur, G. , & Scott, M. S. (2018). Protein coding genes as hosts for noncoding RNA expression. Seminars in Cell & Developmental Biology, 75, 3–12. 10.1016/j.semcdb.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Bortolin‐Cavaille, M. L. , & Cavaille, J. (2012). The SNORD115 (H/MBII‐52) and SNORD116 (H/MBII‐85) gene clusters at the imprinted Prader‐Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Research, 40(14), 6800–6807. 10.1093/nar/gks321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan, C. A. , & Voinnet, O. (2009). The long and the short of noncoding RNAs. Current Opinion in Cell Biology, 21(3), 416–425. 10.1016/j.ceb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Brow, D. A. , & Guthrie, C. (1988). Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature, 334(6179), 213–218. 10.1038/334213a0 [DOI] [PubMed] [Google Scholar]
- Brow, D. A. , & Guthrie, C. (1990). Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes and Development, 4(8), 1345–1356. [DOI] [PubMed] [Google Scholar]
- Brown, J. W. (1999). The ribonuclease P database. Nucleic Acids Research, 27(1), 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. W. , Marshall, D. F. , & Echeverria, M. (2008). Intronic noncoding RNAs and splicing. Trends in Plant Science, 13(7), 335–342. 10.1016/j.tplants.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Brown, Y. , Abraham, M. , Pearl, S. , Kabaha, M. M. , Elboher, E. , & Tzfati, Y. (2007). A critical three‐way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Research, 35(18), 6280–6289. 10.1093/nar/gkm713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti, E. , & Baralle, D. (2010). Novel roles of U1 snRNP in alternative splicing regulation. RNA Biology, 7(4), 412–419. [DOI] [PubMed] [Google Scholar]
- Bush, S. J. , Chen, L. , Tovar‐Corona, J. M. , & Urrutia, A. O. (2017). Alternative splicing and the evolution of phenotypic novelty. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 372(1713), 20150474. 10.1098/rstb.2015.0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella, D. , Praz, V. , Reina, J. H. , Cousin, P. , & Hernandez, N. (2010). Defining the RNA polymerase III transcriptome: Genome‐wide localization of the RNA polymerase III transcription machinery in human cells. Genome Research, 20(6), 710–721. 10.1101/gr.101337.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputa, G. , & Schaffer, J. E. (2016). RNA regulation of lipotoxicity and metabolic stress. Diabetes, 65(7), 1816–1823. 10.2337/db16-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J. , Buiting, K. , Kiefmann, M. , Lalande, M. , Brannan, C. I. , Horsthemke, B. , … Huttenhofer, A. (2000). Identification of brain‐specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proceedings of the National Academy of Sciences of the United States of America, 97(26), 14311–14316. 10.1073/pnas.250426397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola, M. , Rossi, M. , Malcovati, L. , & Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators . (2013). Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood, 121(2), 260–269. 10.1182/blood-2012-09-399725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, P. P. , & Lowe, T. M. (2016). GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Research, 44(D1), D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, K. , Nostramo, R. T. , Wan, Y. , & Hopper, A. K. (2018). tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochimica et Biophysica Acta, Gene Regulatory Mechanisms, 1861(4), 373–386. 10.1016/j.bbagrm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. L. , & Greider, C. W. (2004). Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends in Biochemical Sciences, 29(4), 183–192. 10.1016/j.tibs.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Dong, L. , Zhang, J. , Zhao, Y. , & Li, Z. (2018). Recent advances in microRNA detection. Analyst, 143(8), 1758–1774. 10.1039/C7AN02001E [DOI] [PubMed] [Google Scholar]
- Chu, L. , Su, M. Y. , Maggi, L. B., Jr. , Lu, L. , Mullins, C. , Crosby, S. , … Tomasson, M. H. (2012). Multiple myeloma‐associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. Journal of Clinical Investigation, 122(8), 2793–2806. 10.1172/JCI63051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, C. , Sobala, A. , Lu, C. , Thatcher, S. R. , Bowman, A. , Brown, J. W. , … Hutvagner, G. (2009). Filtering of deep sequencing data reveals the existence of abundant dicer‐dependent small RNAs derived from tRNAs. RNA, 15(12), 2147–2160. 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin, O. , Hahn, D. , & Beggs, J. D. (2012). Structure, function and regulation of spliceosomal RNA helicases. Current Opinion in Cell Biology, 24(3), 431–438. 10.1016/j.ceb.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Crick, F. H. (1968). The origin of the genetic code. Journal of Molecular Biology, 38(3), 367–379. [DOI] [PubMed] [Google Scholar]
- Cui, L. , Nakano, K. , Obchoei, S. , Setoguchi, K. , Matsumoto, M. , Yamamoto, T. , … Hiraoka, N. (2017). Small nucleolar noncoding RNA SNORA23, up‐regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat‐containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology, 153(1), 292–306.e2. 10.1053/j.gastro.2017.03.050 [DOI] [PubMed] [Google Scholar]
- Davis, M. P. , Carrieri, C. , Saini, H. K. , van Dongen, S. , Leonardi, T. , Bussotti, G. , … Enright, A. J. (2017). Transposon‐driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Reports, 18(7), 1231–1247. 10.15252/embr.201744059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W. A. , Liang, X. H. , Piekna‐Przybylska, D. , & Fournier, M. J. (2007). Identifying effects of snoRNA‐guided modifications on the synthesis and function of the yeast ribosome. Methods in Enzymology, 425, 283–316. 10.1016/S0076-6879(07)25013-X [DOI] [PubMed] [Google Scholar]
- Decker, C. J. , Teixeira, D. , & Parker, R. (2007). Edc3p and a glutamine/asparagine‐rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae . Journal of Cell Biology, 179(3), 437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps‐Francoeur, G. , Garneau, D. , Dupuis‐Sandoval, F. , Roy, A. , Frappier, M. , Catala, M. , … Scott, M. S. (2014). Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Research, 42(15), 10073–10085. 10.1093/nar/gku664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanoa, J. K. , Sethi, R. S. , Verma, R. , Arora, J. S. , & Mukhopadhyay, C. S. (2018). Long non‐coding RNA: Its evolutionary relics and biological implications in mammals: A review. Journal of Animal Science and Technology, 60, 25 10.1186/s40781-018-0183-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk, A. L. , Butcher, S. E. , & Brow, D. A. (2018). The life of U6 small nuclear RNA, from cradle to grave. RNA, 24(4), 437–460. 10.1261/rna.065136.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebel, K. W. , Zhou, K. , Clarke, A. B. , & Bemis, L. T. (2016). Beyond the ribosome: Extra‐translational functions of tRNA fragments. Biomarker Insights, 11(Suppl. 1), 1–8. 10.4137/BMI.S35904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci, G. , Fiorino, G. , Castelnuovo, M. , Teichmann, M. , & Pagano, A. (2007). The expanding RNA polymerase III transcriptome. Trends in Genetics, 23(12), 614–622. 10.1016/j.tig.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Dieci, G. , Preti, M. , & Montanini, B. (2009). Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics, 94(2), 83–88. 10.1016/j.ygeno.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Dong, X. Y. , Guo, P. , Boyd, J. , Sun, X. , Li, Q. , Zhou, W. , & Dong, J. T. (2009). Implication of snoRNA U50 in human breast cancer. Journal of Genetics and Genomics, 36(8), 447–454. 10.1016/S1673-8527(08)60134-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet, A. J. , Droc, G. , Siol, O. , Audoux, J. , & Gilbert, N. (2015). U6 snRNA pseudogenes: Markers of retrotransposition dynamics in mammals. Molecular Biology and Evolution, 32(7), 1815–1832. 10.1093/molbev/msv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis‐Sandoval, F. , Poirier, M. , & Scott, M. S. (2015). The emerging landscape of small nucleolar RNAs in cell biology. WIREs RNA, 6(4), 381–397. 10.1002/wrna.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi, B. , Bailly, M. , & de Crecy‐Lagard, V. (2012). Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annual Review of Genetics, 46, 69–95. 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- Esakova, O. , & Krasilnikov, A. S. (2010). Of proteins and RNA: The RNase P/MRP family. RNA, 16(9), 1725–1747. 10.1261/rna.2214510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva, M. , Pages, A. , Matuszek, Z. , Hidmi, S. , Agranat‐Tamir, L. , Korotkov, K. , … Stamm, S. (2016). Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre‐mRNA splicing. Proceedings of the National Academy of Sciences of the United States of America, 113(12), E1625–E1634. 10.1073/pnas.1519292113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva, M. , & Stamm, S. (2013). Processing of snoRNAs as a new source of regulatory non‐coding RNAs: snoRNA fragments form a new class of functional RNAs. BioEssays, 35(1), 46–54. 10.1002/bies.201200117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva, M. , Welden, J. R. , Duncan, M. J. , & Stamm, S. (2017). C/D‐box snoRNAs form methylating and non‐methylating ribonucleoprotein complexes: Old dogs show new tricks. BioEssays, 39(6), 1–28. 10.1002/bies.201600264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, P. , & Guo, M. (2017). Structural characterization of human aminoacyl‐tRNA synthetases for translational and nontranslational functions. Methods, 113, 83–90. 10.1016/j.ymeth.2016.11.014 [DOI] [PubMed] [Google Scholar]
- Filipowicz, W. , Pelczar, P. , Pogacic, V. , & Dragon, F. (1999). Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochimica Polonica, 46(2), 377–389. [PubMed] [Google Scholar]
- Frankish, A. , Uszczynska, B. , Ritchie, G. R. , Gonzalez, J. M. , Pervouchine, D. , Petryszak, R. , … Harrow, J. (2015). Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics, 16(Suppl. 8), S2 10.1186/1471-2164-16-S8-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey, D. , Barta, I. , Gillespie, M. , & Potashkin, J. (1990). Schizosaccharomyces U6 genes have a sequence within their introns that matches the B box consensus of tRNA internal promoters. Nucleic Acids Research, 18(8), 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege, R. , Juhling, F. , Putz, J. , Stadler, P. , Sauter, C. , & Florentz, C. (2012). Structure of transfer RNAs: Similarity and variability. WIREs RNA, 3(1), 37–61. 10.1002/wrna.103 [DOI] [PubMed] [Google Scholar]
- Gogakos, T. , Brown, M. , Garzia, A. , Meyer, C. , Hafner, M. , & Tuschl, T. (2017). Characterizing expression and processing of precursor and mature human tRNAs by hydro‐tRNAseq and PAR‐CLIP. Cell Reports, 20(6), 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi, H. , Nguyen, H. C. B. , Zhang, S. , Dill, B. D. , Molina, H. , & Tavazoie, S. F. (2016). Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell, 165(6), 1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin, J. , Knudsen, B. , Zwieb, C. , & Samuelsson, T. (2001). SRPDB (signal recognition particle database). Nucleic Acids Research, 29(1), 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossringer, M. , Lechner, M. , Brillante, N. , Weber, C. , Rossmanith, W. , & Hartmann, R. K. (2017). Protein‐only RNase P function in Escherichia coli: Viability, processing defects and differences between PRORP isoenzymes. Nucleic Acids Research, 45(12), 7441–7454. 10.1093/nar/gkx405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, I. (2003). Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Development, 17(14), 1691–1702. [DOI] [PubMed] [Google Scholar]
- Grutzmann, K. , Szafranski, K. , Pohl, M. , Voigt, K. , Petzold, A. , & Schuster, S. (2014). Fungal alternative splicing is associated with multicellular complexity and virulence: A genome‐wide multi‐species study. DNA Research, 21(1), 27–39. 10.1093/dnares/dst038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier‐Takada, C. , Gardiner, K. , Marsh, T. , Pace, N. , & Altman, S. (1983). The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35(3, Pt. 2), 849–857. [DOI] [PubMed] [Google Scholar]
- Guiro, J. , & Murphy, S. (2017). Regulation of expression of human RNA polymerase II‐transcribed snRNA genes. Open Biology, 7(6), 1–9. 10.1098/rsob.170073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler, R. A. , & Engelke, D. R. (2006). Spatial organization of transcription by RNA polymerase III. Nucleic Acids Research, 34(17), 4826–4836. 10.1093/nar/gkl656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. E. , Turnbull, C. , & Dalmay, T. (2013). Y RNAs: Recent developments. Biomolecular Concepts, 4(2), 103–110. 10.1515/bmc-2012-0050 [DOI] [PubMed] [Google Scholar]
- Harbour, J. W. (2013). Genomic, prognostic, and cell‐signaling advances in uveal melanoma. American Society of Clinical Oncology Educational Book, 388–391. 10.1200/EdBook_AM.2013.33.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow, J. , Frankish, A. , Gonzalez, J. M. , Tapanari, E. , Diekhans, M. , Kokocinski, F. , & Hubbard, T. J. (2012). GENCODE: The reference human genome annotation for the ENCODE project. Genome Research, 22(9), 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth, J. R. (2013). Lives that introns lead after splicing. WIREs RNA, 4(6), 677–691. 10.1002/wrna.1187 [DOI] [PubMed] [Google Scholar]
- Hinegardner, R. T. , & Engelberg, J. (1963). Rationale for a universal genetic code. Science, 142(3595), 1083–1085. [DOI] [PubMed] [Google Scholar]
- Hirose, T. , & Steitz, J. A. (2001). Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 98(23), 12914–12919. 10.1073/pnas.231490998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner, M. P. , Denisenko, E. , Gardner, P. P. , Schmeier, S. , & Poole, A. M. (2018). An evaluation of function of multicopy noncoding RNAs in mammals using ENCODE/FANTOM data and comparative genomics. Molecular Biology and Evolution, 35(6), 1451–1462. 10.1093/molbev/msy046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner, M. P. , & Poole, A. M. (2012). Comparative genomics of eukaryotic small nucleolar RNAs reveals deep evolutionary ancestry amidst ongoing intragenomic mobility. BMC Evolutionary Biology, 12, 183 10.1186/1471-2148-12-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann, J. , Frank, P. , Loffler, E. , Bennett, K. L. , Gerner, C. , & Rossmanith, W. (2008). RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell, 135(3), 462–474. 10.1016/j.cell.2008.09.013 [DOI] [PubMed] [Google Scholar]
- Huang, C. , Shi, J. , Guo, Y. , Huang, W. , Huang, S. , Ming, S. , … Yao, C. (2017). A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Research, 45(15), 8647–8660. 10.1093/nar/gkx651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. J. , Moore, A. N. , Elniski, D. , Joseph, J. , Yee, J. , & Russell, A. G. (2012). Evolutionarily divergent spliceosomal snRNAs and a conserved non‐coding RNA processing motif in Giardia lamblia . Nucleic Acids Research, 40(21), 10995–11008. 10.1093/nar/gks887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben, J. R. , & Maraia, R. J. (2014). tRNA gene copy number variation in humans. Gene, 536(2), 376–384. 10.1016/j.gene.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia, M. , & Roy, S. W. (2014). Origin of spliceosomal introns and alternative splicing. Cold Spring Harbor Perspectives in Biology, 6(6), a016071. 10.1101/cshperspect.a016071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz, S. , & Alon, U. (2007). The genetic code is nearly optimal for allowing additional information within protein‐coding sequences. Genome Research, 17(4), 405–412. 10.1101/gr.5987307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady, B. E. , Ketele, A. , & Kiss, T. (2012). Human intron‐encoded Alu RNAs are processed and packaged into Wdr79‐associated nucleoplasmic box H/ACA RNPs. Genes & Development, 26(17), 1897–1910. 10.1101/gad.197467.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous, N. , & Gopalan, V. (2010). Archaeal/eukaryal RNase P: Subunits, functions and RNA diversification. Nucleic Acids Research, 38(22), 7885–7894. 10.1093/nar/gkq701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawdekar, G. W. , & Henry, R. W. (2008). Transcriptional regulation of human small nuclear RNA genes. Biochimica et Biophysica Acta, 1779(5), 295–305. 10.1016/j.bbagrm.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani, H. , Kehr, S. , Jedlinski, D. J. , Gumienny, R. , Hertel, J. , Stadler, P. F. , … Gruber, A. R. (2016). An updated human snoRNAome. Nucleic Acids Research, 44(11), 5068–5082. 10.1093/nar/gkw386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhling, F. , Morl, M. , Hartmann, R. K. , Sprinzl, M. , Stadler, P. F. , & Putz, J. (2009). tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Research, 37, D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, A. , Elgamal, S. , Rajkovic, A. , & Ibba, M. (2016). Non‐canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Molecular Microbiology, 101(4), 545–558. 10.1111/mmi.13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam, S. P. , & Hutvagner, G. (2015). tRNA‐derived fragments (tRFs): Emerging new roles for an ancient RNA in the regulation of gene expression. Life, 5(4), 1638–1651. 10.3390/life5041638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. L. , Miquel, M. C. , Bittner, D. , & Rome, L. H. (1990). Vaults. II. Ribonucleoprotein structures are highly conserved among higher and lower eukaryotes. Journal of Cell Biology, 110(4), 895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen, O. , Convertini, P. , Zhang, Z. , Wen, Y. , Shen, M. , Falaleeva, M. , & Stamm, S. (2013). Function of alternative splicing. Gene, 514(1), 1–30. 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketele, A. , Kiss, T. , & Jady, B. E. (2016). Human intron‐encoded AluACA RNAs and telomerase RNA share a common element promoting RNA accumulation. RNA Biology, 13(12), 1274–1285. 10.1080/15476286.2016.1239689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska, E. , Svard, S. G. , & Kirsebom, L. A. (2007). Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proceedings of the National Academy of Sciences of the United States of America, 104(7), 2062–2067. 10.1073/pnas.0607326104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, S. , Gruber, A. R. , Jedlinski, D. J. , Syed, A. P. , Jorjani, H. , & Zavolan, M. (2013). Insights into snoRNA biogenesis and processing from PAR‐CLIP of snoRNA core proteins and small RNA sequencing. Genome Biology, 14(5), R45 10.1186/gb-2013-14-5-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, S. , Khanna, A. , Zhang, Z. , Hui, J. , Balwierz, P. J. , Stefan, M. , … Stamm, S. (2010). The snoRNA MBII‐52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Human Molecular Genetics, 19(7), 1153–1164. 10.1093/hmg/ddp585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, S. , & Stamm, S. (2006). The snoRNA HBII‐52 regulates alternative splicing of the serotonin receptor 2C. Science, 311(5758), 230–232. [DOI] [PubMed] [Google Scholar]
- Kiss, T. (2004). Biogenesis of small nuclear RNPs. Journal of Cell Science, 117(Pt. 25), 5949–5951. 10.1242/jcs.01487 [DOI] [PubMed] [Google Scholar]
- Kiss, T. , Fayet, E. , Jady, B. E. , Richard, P. , & Weber, M. (2006). Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harbor Symposia on Quantitative Biology, 71, 407–417. [DOI] [PubMed] [Google Scholar]
- Kitagawa, T. , Taniuchi, K. , Tsuboi, M. , Sakaguchi, M. , Kohsaki, T. , Okabayashi, T. , & Saibara, T. (2019). Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Molecular Oncology, 13(2), 212–227. 10.1002/1878-0261.12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimenko, O. V. (2017). Small non‐coding RNAs as regulators of structural evolution and carcinogenesis. Noncoding RNA Research, 2(2), 88–92. 10.1016/j.ncrna.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, S. J. , Battle, D. J. , & Dreyfuss, G. (2007). Molecular functions of the SMN complex. Journal of Child Neurology, 22(8), 990–994. 10.1177/0883073807305666 [DOI] [PubMed] [Google Scholar]
- Koonin, E. V. , & Novozhilov, A. S. (2009). Origin and evolution of the genetic code: The universal enigma. IUBMB Life, 61(2), 99–111. 10.1002/iub.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs, J. O. , Churakov, G. , Jurka, J. , Brosius, J. , & Schmitz, J. (2007). Evolutionary history of 7SL RNA‐derived SINEs in supraprimates. Trends in Genetics, 23(4), 158–161. 10.1016/j.tig.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Lafontaine, D. L. , & Tollervey, D. (1998). Birth of the snoRNPs: The evolution of the modification‐guide snoRNAs. Trends in Biochemical Sciences, 23(10), 383–388. [DOI] [PubMed] [Google Scholar]
- Langhendries, J. L. , Nicolas, E. , Doumont, G. , Goldman, S. , & Lafontaine, D. L. (2016). The human box C/D snoRNAs U3 and U8 are required for pre‐rRNA processing and tumorigenesis. Oncotarget, 7(37), 59519–59534. 10.18632/oncotarget.11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas, A. , Toth, K. F. , & Aravin, A. A. (2014). To be or not to be a piRNA: Genomic origin and processing of piRNAs. Genome Biology, 15(1), 204 10.1186/gb4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. , & Rio, D. C. (2015). Mechanisms and regulation of alternative pre‐mRNA splicing. Annual Review of Biochemistry, 84, 291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade, L. , & Weber, M. J. (2006). snoRNA‐LBME‐db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Research, 34, D158–D162. 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, E. , & Brown, J. D. (2010). Biogenesis of the signal recognition particle. Biochemical Society Transactions, 38(4), 1093–1098. 10.1042/BST0381093 [DOI] [PubMed] [Google Scholar]
- Leung, Y. Y. , Kuksa, P. P. , Amlie‐Wolf, A. , Valladares, O. , Ungar, L. H. , Kannan, S. , … Wang, L. S. (2016). DASHR: Database of small human noncoding RNAs. Nucleic Acids Research, 44(D1), D216–D222. 10.1093/nar/gkv1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Duan, J. , Li, D. , Ma, S. , & Ye, K. (2011). Structure of the Shq1‐Cbf5‐Nop10‐Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO Journal, 30(24), 5010–5020. 10.1038/emboj.2011.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J. , Yu, L. , Mei, Y. , Guarnera, M. , Shen, J. , Li, R. , … Jiang, F. (2010). Small nucleolar RNA signatures as biomarkers for non‐small‐cell lung cancer. Molecular Cancer, 9, 198 10.1186/1476-4598-9-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, C. , Lunse, C. E. , & Morl, M. (2017). tRNA modifications: Impact on structure and thermal adaptation. Biomolecules, 7(2), E35. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z. J. , Lehner, R. , Forstner, C. , & Barta, A. (2005). Evolutionary conservation of minor U12‐type spliceosome between plants and humans. RNA, 11(7), 1095–1107. 10.1261/rna.2440305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z. , & Matera, A. G. (2014). Developmental analysis of spliceosomal snRNA isoform expression. G3 (Bethesda), 5(1), 103–110. 10.1534/g3.114.015735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui, L. , & Lowe, T. (2013). Small nucleolar RNAs and RNA‐guided post‐transcriptional modification. Essays in Biochemistry, 54, 53–77. 10.1042/bse0540053 [DOI] [PubMed] [Google Scholar]
- Mannoor, K. , Shen, J. , Liao, J. , Liu, Z. , & Jiang, F. (2014). Small nucleolar RNA signatures of lung tumor‐initiating cells. Molecular Cancer, 13, 104 10.1186/1476-4598-13-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez, S. M. , Chen, J. L. , Evans, D. , & Pace, N. R. (2006). Structure and function of eukaryotic ribonuclease P RNA. Molecular Cell, 24(3), 445–456. 10.1016/j.molcel.2006.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens‐Uzunova, E. S. , Hoogstrate, Y. , Kalsbeek, A. , Pigmans, B. , Vredenbregt‐van den Berg, M. , Dits, N. , … Jenster, G. (2015). C/D‐box snoRNA‐derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget, 6(19), 17430–17444. 10.18632/oncotarget.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz, M. , Donath, A. , Verstraete, N. , Nguyen, V. T. , Stadler, P. F. , & Bensaude, O. (2009). Evolution of 7SK RNA and its protein partners in metazoa. Molecular Biology and Evolution, 26(12), 2821–2830. 10.1093/molbev/msp198 [DOI] [PubMed] [Google Scholar]
- Marz, M. , Kirsten, T. , & Stadler, P. F. (2008). Evolution of spliceosomal snRNA genes in metazoan animals. Journal of Molecular Evolution, 67(6), 594–607. 10.1007/s00239-008-9149-6 [DOI] [PubMed] [Google Scholar]
- Massenet, S. , Bertrand, E. , & Verheggen, C. (2017). Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biology, 14(6), 680–692. 10.1080/15476286.2016.1243646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillen, K. , Roberts, R. B. , & Britten, R. J. (1959). Synthesis of nascent protein by ribosomes in Escherichia coli . Proceedings of the National Academy of Sciences of the United States of America, 45(9), 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, Y. P. , Liao, J. P. , Shen, J. , Yu, L. , Liu, B. L. , Liu, L. , … Jiang, F. (2012). Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene, 31(22), 2794–2804. 10.1038/onc.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, M. , Cognat, V. , Duchene, A. M. , & Marechal‐Drouard, L. (2011). A global picture of tRNA genes in plant genomes. The Plant Journal, 66(1), 80–93. 10.1111/j.1365-313X.2011.04490.x [DOI] [PubMed] [Google Scholar]
- Michel, C. I. , Holley, C. L. , Scruggs, B. S. , Sidhu, R. , Brookheart, R. T. , Listenberger, L. L. , … Schaffer, J. E. (2011). Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metabolism, 14(1), 33–44. 10.1016/j.cmet.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Diaz de la Espina, S. , Alverca, E. , Cuadrado, A. , & Franca, S. (2005). Organization of the genome and gene expression in a nuclear environment lacking histones and nucleosomes: The amazing dinoflagellates. European Journal of Cell Biology, 84(2–3), 137–149. [DOI] [PubMed] [Google Scholar]
- Mosig, A. , Guofeng, M. , Stadler, B. M. , & Stadler, P. F. (2007). Evolution of the vertebrate Y RNA cluster. Theory in Biosciences, 126(1), 9–14. 10.1007/s12064-007-0003-y [DOI] [PubMed] [Google Scholar]
- Mroczek, S. , & Dziembowski, A. (2013). U6 RNA biogenesis and disease association. WIREs RNA, 4(5), 581–592. 10.1002/wrna.1181 [DOI] [PubMed] [Google Scholar]
- Nachtergaele, S. , & He, C. (2017). The emerging biology of RNA post‐transcriptional modifications. RNA Biology, 14(2), 156–163. 10.1080/15476286.2016.1267096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel, A. I. , Waber, N. B. , Gossringer, M. , Lechner, M. , Linne, U. , Toth, U. , … Hartmann, R. K. (2017). Minimal and RNA‐free RNase P in Aquifex aeolicus . Proceedings of the National Academy of Sciences of the United States of America, 114(42), 11121–11126. 10.1073/pnas.1707862114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami, K. , Chen, Y. , & Manley, J. L. (2018). RNA surveillance by the nuclear RNA exosome: Mechanisms and significance. Noncoding RNA, 4(1), 1–21. 10.3390/ncrna4010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J. M. , Di, C. , Venters, C. C. , Guo, J. , Arai, C. , So, B. R. , … Dreyfuss, G. (2017). U1 snRNP telescripting regulates a size‐function‐stratified human genome. Nature Structural & Molecular Biology, 24(11), 993–999. 10.1038/nsmb.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary, N. A. , Wright, M. W. , Brister, J. R. , Ciufo, S. , Haddad, D. , McVeigh, R. , … Pruitt, K. D. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Research, 44(D1), D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, M. , Yamada, K. , Avolio, F. , Scott, M. S. , van Koningsbruggen, S. , Barton, G. J. , & Lamond, A. I. (2010). Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Molecular Biology of the Cell, 21(9), 1569–1584. 10.1091/mbc.E10-01-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi, S. L. , Samarsky, D. A. , Fournier, M. J. , & Boeke, J. D. (1998). Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat‐debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA, 4(9), 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly, D. , Dienstbier, M. , Cowley, S. A. , Vazquez, P. , Drozdz, M. , Taylor, S. , … Murphy, S. (2013). Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Research, 23(2), 281–291. 10.1101/gr.142968.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli, A. (2017). tRNA biology in the omics era: Stress signalling dynamics and cancer progression. BioEssays, 39(3). 10.1002/bies.201600158 [DOI] [PubMed] [Google Scholar]
- Palazzo, A. F. , & Lee, E. S. (2015). Non‐coding RNA: What is functional and what is junk? Frontiers in Genetics, 6, 2 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann, S. , & Frydman, J. (2013). Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature Structural & Molecular Biology, 20(2), 237–243. 10.1038/nsmb.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal, K. , & Reddy, R. (2002). The 3′ end formation in small RNAs. Gene Expression, 10(1–2), 59–78. [PMC free article] [PubMed] [Google Scholar]
- Peterlin, B. M. , Brogie, J. E. , & Price, D. H. (2012). 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. WIREs RNA, 3(1), 92–103. 10.1002/wrna.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski, E. , Dandekar, T. , Henry, Y. , & Tollervey, D. (1998). Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Molecular and Cellular Biology, 18(3), 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna‐Przybylska, D. , Liu, B. , & Fournier, M. J. (2007). The U1 snRNA hairpin II as a RNA affinity tag for selecting snoRNP complexes. Methods in Enzymology, 425, 317–353. 10.1016/S0076-6879(07)25014-1 [DOI] [PubMed] [Google Scholar]
- Podlevsky, J. D. , & Chen, J. J. (2016). Evolutionary perspectives of telomerase RNA structure and function. RNA Biology, 13(8), 720–732. 10.1080/15476286.2016.1205768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Li, Y. , Honda, S. , Hoffmann, S. , Marz, M. , Mosig, A. , … Chen, J. J. (2013). The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Research, 41(1), 450–462. 10.1093/nar/gks980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek, X. C. , Thomson, D. W. , Maag, J. L. , Bartonicek, N. , Signal, B. , Clark, M. B. , … Dinger, M. E. (2015). lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Research, 43, D168–D173. 10.1093/nar/gku988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, J. J. , & Chang, H. Y. (2016). Unique features of long non‐coding RNA biogenesis and function. Nature Reviews. Genetics, 17(1), 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Rashid, R. , Liang, B. , Baker, D. L. , Youssef, O. A. , He, Y. , Phipps, K. , … Li, H. (2006). Crystal structure of a Cbf5‐Nop10‐Gar1 complex and implications in RNA‐guided pseudouridylation and dyskeratosis congenita. Molecular Cell, 21(2), 249–260. 10.1016/j.molcel.2005.11.017 [DOI] [PubMed] [Google Scholar]
- Reiner, R. , Ben‐Asouli, Y. , Krilovetzky, I. , & Jarrous, N. (2006). A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes & Development, 20(12), 1621–1635. 10.1101/gad.386706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, N. M. , Vargas‐Rodriguez, O. , Soll, D. , & Crnkovic, A. (2017). The central role of tRNA in genetic code expansion. Biochimica et Biophysica Acta ‐ General Subjects, 1861(11, Pt. B), 3001–3008. 10.1016/j.bbagen.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich, A. , & RajBhandary, U. L. (1976). Transfer RNA: Molecular structure, sequence, and properties. Annual Review of Biochemistry, 45, 805–860. 10.1146/annurev.bi.45.070176.004105 [DOI] [PubMed] [Google Scholar]
- RNAcentral . (2017). RNAcentral: A comprehensive database of non‐coding RNA sequences. Nucleic Acids Research, 45(D1), D128–D134. 10.1093/nar/gkw1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I. B. , Carmel, L. , Csuros, M. , & Koonin, E. V. (2012). Origin and evolution of spliceosomal introns. Biology Direct, 7, 11 10.1186/1745-6150-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, D. P. , & Blackburn, E. H. (1991). A conserved secondary structure for telomerase RNA. Cell, 67(2), 343–353. [DOI] [PubMed] [Google Scholar]
- Rosenblad, M. A. , Gorodkin, J. , Knudsen, B. , Zwieb, C. , & Samuelsson, T. (2003). SRPDB: Signal recognition particle database. Nucleic Acids Research, 31(1), 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad, M. A. , Larsen, N. , Samuelsson, T. , & Zwieb, C. (2009). Kinship in the SRP RNA family. RNA Biology, 6(5), 508–516. [DOI] [PubMed] [Google Scholar]
- Rothe, B. , Back, R. , Quinternet, M. , Bizarro, J. , Robert, M. C. , Blaud, M. , … Branlant, C. (2014). Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Research, 42(3), 2015–2036. 10.1093/nar/gkt1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. G. , Charette, J. M. , Spencer, D. F. , & Gray, M. W. (2006). An early evolutionary origin for the minor spliceosome. Nature, 443(7113), 863–866. 10.1038/nature05228 [DOI] [PubMed] [Google Scholar]
- Russell, J. , & Zomerdijk, J. C. (2005). RNA‐polymerase‐I‐directed rDNA transcription, life and works. Trends in Biochemical Sciences, 30(2), 87–96. 10.1016/j.tibs.2004.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, D. , Rak, R. , Gingold, H. , Adir, I. , Maayan, G. , Dahan, O. , … Rechavi, O. (2016). Tissue‐ and time‐specific expression of otherwise identical tRNA genes. PLoS Genetics, 12(8), e1006264 10.1371/journal.pgen.1006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbisa, E. , Pesole, G. , Tullo, A. , & Saccone, C. (1996). The evolution of the RNase P‐ and RNase MRP‐associated RNAs: Phylogenetic analysis and nucleotide substitution rate. Journal of Molecular Evolution, 43(1), 46–57. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. , & Butler, J. S. (2013). Nuclear RNA surveillance: Role of TRAMP in controlling exosome specificity. WIREs RNA, 4(2), 217–231. 10.1002/wrna.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M. E. , & Clayton, D. A. (1993). Nuclear RNase MRP is required for correct processing of pre‐5.8S rRNA in Saccharomyces cerevisiae . Molecular and Cellular Biology, 13(12), 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli, D. , & Pillai, R. S. (2004). The special Sm core structure of the U7 snRNP: Far‐reaching significance of a small nuclear ribonucleoprotein. Cellular and Molecular Life Sciences, 61(19–20), 2560–2570. 10.1007/s00018-004-4190-0 [DOI] [PubMed] [Google Scholar]
- Scott, L. M. , & Rebel, V. I. (2013). Acquired mutations that affect pre‐mRNA splicing in hematologic malignancies and solid tumors. Journal of the National Cancer Institute, 105(20), 1540–1549. 10.1093/jnci/djt257 [DOI] [PubMed] [Google Scholar]
- Scott, M. S. , & Ono, M. (2011). From snoRNA to miRNA: Dual function regulatory non‐coding RNAs. Biochimie, 93(11), 1987–1992. 10.1016/j.biochi.2011.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, M. S. , Ono, M. , Yamada, K. , Endo, A. , Barton, G. J. , & Lamond, A. I. (2012). Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Research, 40(8), 3676–3688. 10.1093/nar/gkr1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, O. V. , Bogdanov, A. A. , & Sergiev, P. V. (2015). What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie, 117, 110–118. 10.1016/j.biochi.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Yu, X. , Zhu, L. , Li, T. , Yan, Z. , & Guo, J. (2018). Transfer RNA‐derived fragments and tRNA halves: Biogenesis, biological functions and their roles in diseases. Journal of Molecular Medicine, 96(11), 1167–1176. 10.1007/s00109-018-1693-y [DOI] [PubMed] [Google Scholar]
- Shi, J. , Huang, C. , Huang, S. , & Yao, C. (2018). snoRNAs associate with mRNA 3′ processing complex: New wine in old bottles. RNA Biology, 15(2), 194–197. 10.1080/15476286.2017.1416278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, A. R. , & Santos, M. (2017). Discovery and function of transfer RNA‐derived fragments and their role in disease. WIREs RNA, 8(5). 10.1002/wrna.1423 [DOI] [PubMed] [Google Scholar]
- Soeno, Y. , Taya, Y. , Stasyk, T. , Huber, L. A. , Aoba, T. , & Huttenhofer, A. (2010). Identification of novel ribonucleo‐protein complexes from the brain‐specific snoRNA MBII‐52. RNA, 16(7), 1293–1300. 10.1261/rna.2109710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller, M. P. , Boon, K. L. , Reijns, M. A. , & Beggs, J. D. (2007). The Lsm2‐8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Research, 35(3), 923–929. 10.1093/nar/gkl1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, P. F. , Chen, J. J. , Hackermuller, J. , Hoffmann, S. , Horn, F. , Khaitovich, P. , … Ullmann, K. (2009). Evolution of vault RNAs. Molecular Biology and Evolution, 26(9), 1975–1991. 10.1093/molbev/msp112 [DOI] [PubMed] [Google Scholar]
- Su, J. , Liao, J. , Gao, L. , Shen, J. , Guarnera, M. A. , Zhan, M. , … Jiang, F. (2016). Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget, 7(5), 5131–5142. 10.18632/oncotarget.4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C. , Fu, Z. , Wang, S. , Li, J. , Li, Y. , Zhang, Y. , … Yin, Y. (2018). Roles of tRNA‐derived fragments in human cancers. Cancer Letters, 414, 16–25. 10.1016/j.canlet.2017.10.031 [DOI] [PubMed] [Google Scholar]
- Szymanski, M. , Barciszewska, M. Z. , Zywicki, M. , & Barciszewski, J. (2003). Noncoding RNA transcripts. Journal of Applied Genetics, 44(1), 1–19. [PubMed] [Google Scholar]
- Taft, R. J. , Glazov, E. A. , Lassmann, T. , Hayashizaki, Y. , Carninci, P. , & Mattick, J. S. (2009). Small RNAs derived from snoRNAs. RNA, 15(7), 1233–1240. 10.1261/rna.1528909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M. P. , & Terns, R. M. (2002). Small nucleolar RNAs: Versatile trans‐acting molecules of ancient evolutionary origin. Gene Expression, 10(1–2), 17–39. [PMC free article] [PubMed] [Google Scholar]
- Torres, A. G. , Batlle, E. , & Ribas de Pouplana, L. (2014). Role of tRNA modifications in human diseases. Trends in Molecular Medicine, 20(6), 306–314. 10.1016/j.molmed.2014.01.008 [DOI] [PubMed] [Google Scholar]
- Tuorto, F. , & Lyko, F. (2016). Genome recoding by tRNA modifications. Open Biology, 6(12). 10.1098/rsob.160287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen, J. J. , Niemela, E. H. , Verma, B. , & Frilander, M. J. (2013). The significant other: Splicing by the minor spliceosome. WIREs RNA, 4(1), 61–76. 10.1002/wrna.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S. , & Gunawardane, L. S. (2013). Role of small nuclear RNAs in eukaryotic gene expression. Essays in Biochemistry, 54, 79–90. 10.1042/bse0540079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon, A. , Mossink, M. H. , Scheper, R. J. , Sonneveld, P. , & Wiemer, E. A. (2003). The vault complex. Cellular and Molecular Life Sciences, 60(9), 1828–1837. 10.1007/s00018-003-3030-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S. , Yan, W. , Carlton, J. M. , & Johnson, P. J. (2005). Spliceosomal introns in the deep‐branching eukaryote Trichomonas vaginalis . Proceedings of the National Academy of Sciences of the United States of America, 102(12), 4430–4435. 10.1073/pnas.0407500102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone, L. , Galardi, S. , Page, D. , & Beggs, J. D. (2004). Lsm proteins promote regeneration of pre‐mRNA splicing activity. Current Biology, 14(16), 1487–1491. 10.1016/j.cub.2004.08.032 [DOI] [PubMed] [Google Scholar]
- Villa, T. , Pleiss, J. A. , & Guthrie, C. (2002). Spliceosomal snRNAs: Mg(2+)‐dependent chemistry at the catalytic core? Cell, 109(2), 149–152. [DOI] [PubMed] [Google Scholar]
- Vincenti, S. , De Chiara, V. , Bozzoni, I. , & Presutti, C. (2007). The position of yeast snoRNA‐coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre‐mRNA. RNA, 13(1), 138–150. 10.1261/rna.251907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconte, V. , Makishima, H. , Maciejewski, J. P. , & Tiu, R. V. (2012). Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia, 26(12), 2447–2454. 10.1038/leu.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , & Wu, C. J. (2013). SF3B1 mutations in chronic lymphocytic leukemia. Blood, 121(23), 4627–4634. 10.1182/blood-2013-02-427641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N. J. , Gottschalk, A. , Neubauer, G. , Kastner, B. , Fabrizio, P. , Mann, M. , & Luhrmann, R. (1998). Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA‐binding protein, are present together with Gar1p in all H BOX/ACA‐motif snoRNPs and constitute a common bipartite structure. RNA, 4(12), 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. J. (2006). Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genetics, 2(12), e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, M. C. , Sousa, F. L. , Mrnjavac, N. , Neukirchen, S. , Roettger, M. , Nelson‐Sathi, S. , & Martin, W. F. (2016). The physiology and habitat of the last universal common ancestor. Nature Microbiology, 1(9), 16116 10.1038/nmicrobiol.2016.116 [DOI] [PubMed] [Google Scholar]
- Wichtowska, D. , Turowski, T. W. , & Boguta, M. (2013). An interplay between transcription, processing, and degradation determines tRNA levels in yeast. WIREs RNA, 4(6), 709–722. 10.1002/wrna.1190 [DOI] [PubMed] [Google Scholar]
- Wong, W. M. , Abrahamson, J. L. , & Nazar, R. N. (1984). Are DNA spacers relics of gene amplification events? Proceedings of the National Academy of Sciences of the United States of America, 81(6), 1768–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford, J. L., Jr. , & Baserga, S. J. (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics, 195(3), 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y. H. , & Chen, L. L. (2018). Processing and roles of snoRNA‐ended long noncoding RNAs. Critical Reviews in Biochemistry and Molecular Biology, 53, 1–11. 10.1080/10409238.2018.1508411 [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Wen, L. , & Zhu, H. (2015). Unveiling the hidden function of long non‐coding RNA by identifying its major partner‐protein. Cell & Bioscience, 5, 59 10.1186/s13578-015-0050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama, M. , Nakao, A. , & Kenmochi, N. (2013). snOPY: A small nucleolar RNA orthological gene database. BMC Research Notes, 6, 426 10.1186/1756-0500-6-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis, I. , Dittmar, K. , Wang, W. , Foley, S. W. , Berg, M. G. , Hu, K. Y. , … Dreyfuss, G. (2013). Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife, 2, e00780 10.7554/eLife.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef, O. A. , Safran, S. A. , Nakamura, T. , Nix, D. A. , Hotamisligil, G. S. , & Bass, B. L. (2015). Potential role for snoRNAs in PKR activation during metabolic stress. Proceedings of the National Academy of Sciences of the United States of America, 112(16), 5023–5028. 10.1073/pnas.1424044112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki, A. , Albrecht, A. , & Steinhofel, K. (2018). Long non‐coding RNA structure and function: Is there a link? Frontiers in Physiology, 9, 1201 10.3389/fphys.2018.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zent, C. S. , & Burack, W. R. (2014). Mutations in chronic lymphocytic leukemia and how they affect therapy choice: Focus on NOTCH1, SF3B1, and TP53. Hematology/The Education Program of the American Society of Hematology, 2014(1), 119–124. 10.1182/asheducation-2014.1.119 [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Li, H. , Fang, S. , Kang, Y. , Wu, W. , Hao, Y. , … Chen, R. (2016). NONCODE 2016: An informative and valuable data source of long non‐coding RNAs. Nucleic Acids Research, 44(D1), D203–D208. 10.1093/nar/gkv1252 [DOI] [PMC free article] [PubMed] [Google Scholar]


