Abstract
The ideal sampling method and benefit of qualitative versus quantitative culture, for Carbapenemase-resistant Enterobacteriaceae recovery in hospitalized patients’ rooms/bathrooms is unknown. Although nylon-flocked swab improved overall Gram-negative organism recovery compared with cellulose sponge, both were similar for Carbapenemase-resistant Enterobacteriaceae. Quantitative culture was inferior and unrevealing beyond the qualitative results.
Background:
Carbapenem-resistant Enterobacteriaceae (CRE) are important healthcare-associated pathogens with high mortality rates.1,2 CRE recovery from the patient environment may be informative for evaluation of efficiency of cleaning and disinfection in routine and outbreak setting and infection prevention research studies3. Although the rayon-tipped swab has better sensitivity than cellulose sponge (CS) methods for detection of Acinetobacter in the near patient environment, the ideal sampling method for CRE detection from high touch surfaces (HTS) in the patient room is unknown.4 We compared two sampling methods (nylon-flocked swab [NFS] and CS) and two culturing methods (qualitative and quantitative) for detection of CRE, non-CRE carbapenem-resistant organisms and all other Gram-negative organisms in rooms where the occupant harbored CRE.
Methods:
We prospectively identified patients at the Johns Hopkins Hospital, a 1,145 patient bed tertiary academic center in Baltimore, Maryland, with recent (past 3 days) clinical or surveillance culture(s) growing CRE who had occupied the same hospital room for the most recent 2 days. High Touch Surfaces (HTS) from patient room were sampled. One half of each HTS was sampled using NFS (eSwab™; Copan Diagnostics, Murrieta, CA) dipped in neutralizer buffer (Hardy Diagnostics; Santa Maria, CA) using a previously published method.4,5 An individual NFS was used per half HTS. The other half of the HTS was sampled using CS with neutralizer (3M; Maplewood, MN), where up to 5 HTS were sampled with the specific sides of the CS (e.g. a composite).4,6 Due to right hand dominance, bacteria may have been more likely to be removed from that side of the HTS during cleaning. To avoid introducing a systematic bias, alternating sides of the HTS were sampled by each method.
Qualitative (PBS with tween broths were held for up to 3 days and subcultured if turbid) and quantitative cultures (including positive and negative controls) were performed following CDC protocols using MacConkey agar for selection of Gram-negative organisms, incubated overnight at 35°C.5,6 Identification and antimicrobial susceptibility testing of recovered organisms were performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker Daltonics Inc.) and via the BD Phoenix™ Automated Microbiology System, respectively.4 Enterobacteriaceae resistant to at least ertapenem were identified as CRE. If a CS was culture positive, then all HTS of the composite were deemed positive. Limit of Detection was determined by preparing a 0.5 McFarland standard, plating 100 μl aliquots of 10-fold dilution series of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae ATCC BAA-1705 onto sterile Formica slabs and sampled by CS and NFS in a similar manner to HTS sampling.
The frequency of Gram-negative organism recovery for each method was compared to a gold standard, defined as recovery using either NFS or CS method. Each HTS was categorized according to frequency of Gram-negative organism recovery by either method: 0–10%, 10–20%, and >20%. Descriptive statistical analysis was conducted using SAS 9.4.
This study was acknowledged by the Johns Hopkins University institutional review board as non-human subjects research.
Results:
There were 229 HTS sampled in 17 unique patient rooms from May to December 2016. Eight of 17 (47%) patient rooms had Gram-negative bacteria detected from at least 1 HTS by either method; 2/17 (12%) had CRE recovered, 2/17 (12%) had a non-CRE carbapenem-resistant organism recovered; 7/17 (41%) had other or additional Gram-negative organisms (Table 1). For the two rooms where CRE was detected, one was by NFS and one by CS (see Table 1, Patient 3, Patient 5). Due to low overall recovery of CRE and non-CRE carbapenem-resistant organisms, we grouped these with all other Gram-negative organism recovery to define the gold standard. The sensitivity for detection of any Gram-negative organisms in the environment was 100% for NFS and 21% for CS. Seven of the eight (88%) positive rooms were identified using qualitative culture, and four of eight (50%) by quantitative culture. The Limit of Detection for NFS and CS methods was ~2 × 107 CFU/ml. Figure 1 shows frequency of Gram-negative organism recovery from HTS in patient room and bathroom.
Table 1:
Patient Carbapenemase-resistant Enterobacteriacea (CRE) culture and Environmental Culture Results for Nylon flocked swab and Cellulose Sponge Methods
| Patienta | Patient CRE Culture | # days patient had occupied room on day of sampling |
Environmental Culture Results | ||||
|---|---|---|---|---|---|---|---|
| Source | Organism | Nylon flocked swab: Qualitative |
Nylon flocked swab: Quantitative |
Cellulose Sponge: Qualitative |
Cellulose Sponge: Quantitative |
||
| 1 | Urine | New Delhi metallo-β lactamse-producing Escherichia coli | 37 | ||||
| 2 | Sputum | Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae | 80 | ||||
| 3a | Sputum | KPC-producing K. pneumoniae | 66 | IV pump: KPC-producing K. pneumoniae Bathroom sink: Pseudomonas putida, Acinetobacter species |
|||
| 4 | Blood | KPC-producing K. pneumoniae |
28 | Bathroom light switch, side bed rail, foot bed rail, over bed table & toilet seat: Stenotrophomonas maltophilia Bathroom sink: Pseudomonas aeruginosa & Enterobacter cloacae |
Grab bar, vital sign monitor, bathroom sink, bathroom light switch, bathroom door knob, bedroom inside knob, side bed rail, foot bed rail, over bed table IV pump/pole: 1.6 - >3 × 102 CFU/mL S. maltophilia |
Composite 4 -Room and bathroom sink and faucet: E. cloacae |
Composite 1 - Bed rails/vital signs monitor/ call bell >9× 103 CFU/Sponge S. maltophilia |
| 5 b | Sputum | Non-Carbapenemase producing-CRE (Non-CP-CRE) E. cloacae | 50 | Composite 1 - Bed rails/vital signs monitor/ call bell & Composite 4 - Room and bathroom sink and faucet: E. cloacae |
|||
| 6 | Sputum | KPC-producing Citrobacter freundii complex & KPC-producing Citrobacter amalonaticus |
31 | Composite 2 – Bedside table/ IV pole/ IV pump/ bathroom outside door handle 1.5 CFU/sponge Enterobacter aerogenes | |||
| 7c | Blood | KPC-producing K. pneumoniae |
18 | Patient room sink: Non-Carbapenemase resistant K. pneumoniae Bathroom sink: P. aeruginosa |
Patient room sink: 70 CFU/mL P. aeruginosa, 10 CFU/mL P. putida, 64 CFU/mL Klebsiella oxytoca Bathroom sink: 75 CFU/mL P. aeruginosa, 2.5 × 102 CFU/mL Pseudomonas fluorescence |
||
| 8 | Rectal Swab | New Delhi metallo-β lactamse-producing K. pneumoniae | 5 | Patient bathroom sink: P. aeruginosa | Patient bathroom sink: 29 CFU/mL P. aeruginosa, 8 CFU/mL C. freundii | ||
| 9 | Blood | Non-CP-CRE E. cloacae complex |
7 | ||||
| 10 | Sputum | Non-CP-CRE E. cloacae complex |
17 | ||||
| 11 | Urine | Non-CP-CRE E. cloacae complex |
8 | Patient bathroom sink: S. maltophilia | |||
| 12 | Urine | KPC-producing K. pneumoniae | 10 | ||||
| 13 | Rectal Swab | Non-CP-CRE E. aerogenes |
21 | ||||
| 14 | Blood | OXA-48-like and New Delhi metallo-β lactamse -producing K. pneumoniae | 30 | ||||
| 15 | Tissue | Non-CP-CRE K. pneumoniae | 22 | ||||
| 16 | Tissue | KPC-producing C. freundii complex | 94 | Bedside inside rail/grip, beside outside rail/grip, vital sign monitor and bedside tray: E. coli |
|||
| 17 | Urine | Non-CP-CRE E. cloacae |
14 | ||||
Patient 3: The clinical and environmental KPC-producing K. pneumoniae were identical strains confirmed by pulsed-field gel electrophoresis.
Patient 5: The clinical carbapenem resistant E. cloacae and environmental carbapenem susceptible E. cloacae were identical strains confirmed by pulsed-field gel electrophoresis.
Patient 7: The clinical and environmental K. pneumoniae were different strains confirmed by pulsed-field gel electrophoresis.
Figure 1.
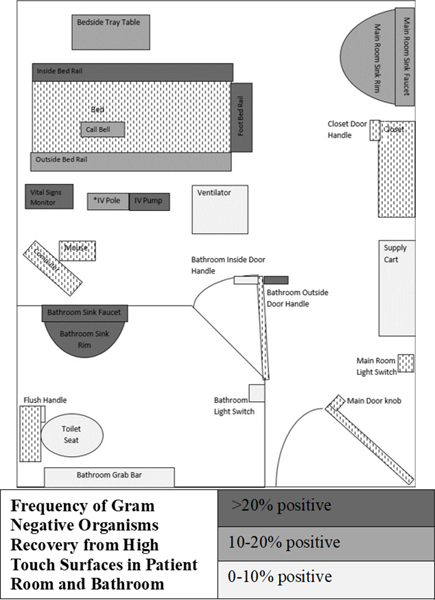
Frequency of Gram-Negative Organism Recovery from High Touch Surfaces in Patient Room and Bathroom
Discussion
We infrequently recovered CRE in the room and bathroom of in-patients known to be infected or colonized with CRE. Although we were unable to ascertain if NFS method or CS method was superior at CRE recovery from environmental surfaces we did find some practical advantages of NFS method. With NFS the specific positive HTS is known, rather than the CS composite where detected organisms could have been recovered from up to 5 HTS. In addition, NFS method requires less microbiologist time and expertise, and less specialized equipment (stomacher or large centrifuge) than CS, making is less costly. Although, it took less time to process a single NFS (~ 8 mins) compared to the sponge (~ 35 mins), the additional NFS’s per patient room (up to 23) were more time intensive than the CS approach (up to 5 CS per room) due to the higher number of samples collected. We found qualitative cultures had a higher sensitivity for Gram-negative organism recovery than quantitative. NFS are likely readily available in many healthcare facilities where they are used for patient multidrug-resistant organism surveillance programs, making it a feasible option when sampling the environment in a CRE outbreak situation, or in research studies when assessing cleaning practices. Some studies have favored different sampling methods over NFS for CRE recovery. An Israeli study compared recovery of Klebsiella pneumoniae carbapenemase -Carbapenemase-producing Enterobacteriaceae in the hospital setting using contact plates and NFS with either direct plating to Klebsiella pneumoniae carbapenemase selective agar or broth enrichment, and found enhanced recovery with contact plates (contact plate 32% vs NFS with direct plating 24% vs 16% NFS with broth enrichment).7 However, these studies did not use neutralizer prior to sampling with NFS. Use of NFS with neutralizer rather than phosphate-buffered saline has been found to be superior at recovery of Staphylococcus aureus, and neutralizer with NFS in this study may have helped with bacteria recovery.8 Another potential strength of our study design was accounting for the important confounding variable of right hand dominance during cleaning whereby we alternated which half of HTS was sampled by each method.
We found the environment of patients known to harbor CRE frequently contaminated with other Gram-negative organisms. We are not aware of any other studies looking at all Gram-negative organism recovery, however MDR Gram-negative organism recovery can range from 1.8 to 30% of surfaces.9,10 Shams et al found 34% of high touch surfaces contaminated with MDROs post-daily cleaning, although mostly with Gram-positive organisms.6
Our study has some limitations. The Limit of Detection for CRE was ~2 × 107 CFU/mL, therefore it is possible that HTS with lower Gram-negative organism burden may have given negative results by sampling methods. Although we sampled a small number of patient rooms, this study supports the use of NFS when recovering Gram-negative organisms in the patient environment. The NFS method is more feasible, due to decreased cost, increased availability, and less lab expertise necessary, and may be advantageous during outbreak investigations as the specific contaminated HTS is identified.
Acknowledgements
Financial Support. This work was supported by funding from The Sherrilyn and Ken Fisher Center for Environmental Diseases and the National Institutes of Health (R21-AI130608) awarded to P.J.S., Centers for Disease Control and Prevention (CDC) Epicenters Program (C.R., V.S., A.M.M., L.L.M., P.J.S.)
Past Presentation Statement: This study was previously presented in oral format at the Society for Healthcare Epidemiology of America Spring Conference, Atlanta, GA in May 2016.
Footnotes
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article.
References:
- 1.Vital Signs: Carbapenem-Resistant Enterobacteriaceae. Morbidity and Mortality Weekly Report (MMWR). Centers for Disease Control and Prevention website. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6209a3.htm. Published 2013. Accessed June 25, 2018. [PMC free article] [PubMed]
- 2.Rock C, Curless MS, Cantara M, et al. Resolution of Carbapenemase-Producing Klebsiella pneumoniae Outbreak in a Tertiary Cancer Center; the Role of Active Surveillance. Infect Control Hosp Epidemiol. 2017;38:1117–1119. [DOI] [PubMed] [Google Scholar]
- 3.Galvin S, Dolan A, Cahill O, Daniels S, Humphreys H. Microbial monitoring of the hospital environment: why and how? J Hosp Infect. 2012;82:143–151. [DOI] [PubMed] [Google Scholar]
- 4.Thom KA, Howard T, Sembajwe S, et al. Comparison of swab and sponge methodologies for identification of Acinetobacter baumannii from the hospital environment. J Clin Microbiol. 2012;50:2140–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges LR, Rose LJ, O’Connell H, Arduino MJ. National validation study of a swab protocol for the recovery of Bacillus anthracis spores from surfaces. J Microbiol Methods. 2010;81:141–146. [DOI] [PubMed] [Google Scholar]
- 6.Shams AM, Rose LJ, Edwards JR, et al. Assessment of the Overall and Multidrug-Resistant Organism Bioburden on Environmental Surfaces in Healthcare Facilities. Infect Control Hosp Epidemiol. 2016;37:1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerner A, Adler A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental Contamination by Carbapenem-Resistant Enterobacteriaceae. J Clin Microbiol. 2013;51:177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedin G, Rynbäck J, Loré B. New technique to take samples from environmental surfaces using flocked nylon swabs. J Hosp Infect. 2010;75:314–317. [DOI] [PubMed] [Google Scholar]
- 9.Sbarra AN, Harris AD, Johnson JK, et al. Guidance on Frequency and Location of Environmental Sampling for Acinetobacter baumannii. Infect Control Amp Hosp Epidemiol. 2018;39:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torkar KG, Ivić S . Surveillance of bacterial colonisation on contact surfaces in different medical wards. Arh Hig Rada Toksikol. 2017;68:116–126. [DOI] [PubMed] [Google Scholar]


