Abstract
Communication between and within cells is essential for multicellular life. While intracellular signal transduction pathways are often specified in molecular terms, the information content they transmit remains poorly defined. Here, we review research efforts to merge biological experimentation with concepts of communication that emerge from the engineering disciplines of signal processing and control theory. We discuss the challenges of performing experiments that quantify information transfer at the molecular level, and we highlight recent studies that have advanced toward a clearer definition of the information content carried by signaling molecules. Across these studies, we emphasize a theme of increasingly well-matched experimental and theoretical approaches to decode the data streams directing cellular behavior.
Introduction: Parallels between systems for information transfer
The study of cellular signal transduction –the transfer of non-genetic information within and between cells - has been a key interface point between experimental biology and systems biology. For biomedical researchers and experimental biologists, signal transduction pathways are of interest because of their central role in coordinating organismal development and physiological homeostasis. The etiology of most chronic human diseases can be traced to abnormal function of a regulatory network, such as mutations that alter signaling protein activity. For engineers and scientists trained in quantitative methods, these inter- and intra-cellular communication networks have characteristics that parallel well-studied problems in communication, presenting an attractive challenge for the application of well-established theoretical tools with the hope of overcoming some of the limitations of purely experimental research. The relationship between these fields has been renewed and revisited many times over the past 30 years (1–3). The most important biological insights have emerged when experimental and quantitative tools are carefully and thoughtfully conjoined. In this essay we focus on a particular area of renewed collaboration, in which advances in the ability to detect signaling events with a high level of detail in individual cells have enabled connections to the engineering discipline of signal processing. Our discussion is intended as a guide to further reading, rather than a comprehensive review, with the goal of drawing interest to emerging questions in this area that will likely be rewarding over the next few years.
In traditional signal transduction experiments, the significance of a biochemical event is often evaluated (sometimes subconsciously) by its apparent magnitude. For example, following a stimulus, a band on western blot representing the phosphorylation of a protein may be 10-fold more intense than the corresponding band from unstimulated cells. This increase may be judged more significant than one in which a 2-fold change is induced. However, such judgements are often made in the absence of knowledge of whether these differences have functional importance within the cells of interest. A 2-fold increase may be sufficient to saturate the process being studied, while a 10-fold increase evokes no further response. Conversely, both activity levels might fall below the threshold to induce a cellular response of interest. The true significance of the result (for the cell) depends on the strength of the signal relative to the responsiveness of the next step in the process. Establishing this quantitative relationship between signal and response is often challenging and leads to a great deal of ambiguity in both conceptual and formal models of signaling processes. In this essay, we explore both the experimental challenges inherent in addressing such questions within signaling pathways and the broader biological concepts that have emerged from research in this area.
The scenario described above is an example of a problem inherent in any multi-stage communication system, whether natural or human-engineered. Communication systems - including signaling pathways, neural networks, or electronic circuits - consist of multiple elements in sequence, each of which receives an input signal and produces an output signal (Fig. 1A). The basic function of each element, whether it be a kinase within a signaling cascade, a neuron within a neural pathway, or a transistor within a radio, is to produce an output signal that is variable and dependent on the input signal. The simplest types of element simply relay the input signal without changing it, while more complex elements can transform the input signal in a number of ways to create the output signal. In engineering, the relationship between the input and the output for each element is known as its “transfer function”. Importantly, transfer functions can be used to characterize the behavior of individual elements within a system (e.g., the response of a single protein kinase within a cascade; Fig. 1B) or larger sets of connected components (e.g. an entire kinase cascade; Fig. 1C). This concept has played a central role in the development of electronic communication systems, from radio broadcasting to cellular telephones, and is also essential to our understanding of information processing in neural and sensory systems. Yet, the application of such ideas to intracellular signaling pathways has been limited, especially in mammalian systems.
Figure 1.
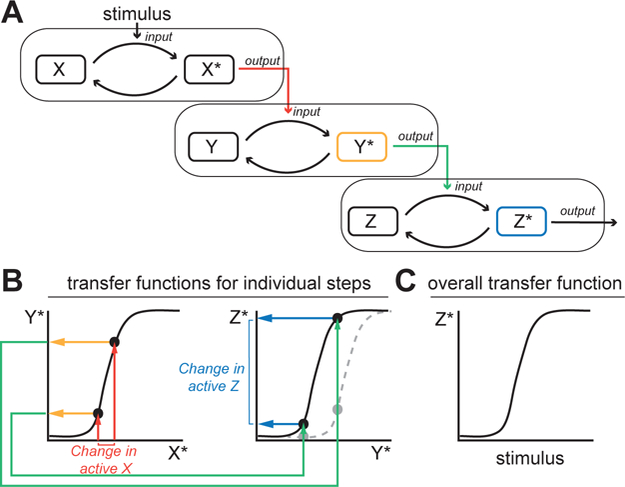
Alignment of transfer functions in signal transduction cascades. A. Hypothetical signaling cascade function. Many known signaling pathways share a similar structure, in which proteins A,B, and C exist in both inactive (X,Y,and Z) and active states (X*,Y*, and Z*). For each step, the “input” is the concentration of the active upstream stimulating molecule, and the “output” is the steady-state concentration of active molecules produced. B. Curves relating the input to the output for two of the stages shown in A. For each signaling step, these curves indicate the concentration of active output molecules as a function of the input. If a change in concentration of active X occurs (red arrows in A and B), the concentration of active Y changes accordingly (yellow oval in A and yellow arrows in B). This change in active Y becomes the input to the next step (green arrows in A and B), resulting in a change in active Z (blue ovals in A and blue arrows in B). Note that if the Y*→Z* transfer function were misaligned (gray dotted line), the induced change in Z* would be much smaller (gray circles), reducing the ability of the pathway to transmit changes in the activity of X. C. A potential transfer curve for the overall cascade, relating the stimulus strength to the final output of the pathway, Z*. The shape of this curve depends on the alignment of the curves for the individual steps as shown in (B).
A fundamental concept has emerged from these other communication-related fields that has strong implications for signal transduction pathways: for a pathway to transmit information - such as the concentration of an extracellular ligand - the transfer functions of every element in the pathway must be well aligned. If they are not, the ability of the system to act as a conduit for information is severely compromised (4). If elements are connected without attention to alignment, it is more likely than not that their input and output ranges will be mismatched, and the output of one element will either over- or under-stimulate the next element, leading to either saturation of the downstream element or failure to stimulate a response. A simple illustration of such a situation can be obtained visually, by walking from a dark room into bright sunlight; the initial discomfort and visual difficulty of this experience results, in part, from saturation of the rhodopsin-coupled G-protein signaling pathway in the photoreceptor cells of the eye. Fortunately, the visual system has adaptive properties that quickly adjust the transfer function of the system, enabling more intense input signals to be effectively processed (5). Within both engineering and neurobiology, such adaptation is termed “gain control”. Mechanisms facilitating gain control have been well-studied in neuronal and sensory systems (6), where they are essential for ensuring that the outputs and inputs of successive neurons are appropriately matched.
The application of concepts from the signal processing field (see Box 1), such as transfer functions and gain control, to intracellular signaling pathways has remained limited (7). Much of this conceptual gap can be attributed to a lack of appropriate experimental data with which to accurately measure transfer functions. To fully employ concepts from the signal processing field, the ideal data collection method would quantify specific signaling protein activities within individual cells to avoid artifacts from averaging across heterogeneous cells. It would also provide high temporal resolution, to determine when signaling reaches steady state or whether frequency-modulated responses occur, and would permit monitoring multiple molecular signals to allow for repeated stimulation of the same cell. While fully realizing this ideal remains difficult, studies based on live-cell imaging now enable many of these criteria to be achieved with reasonable effort. These experiments can be conducted with relatively inexpensive widefield epifluorescence microscopes, and detailed protocols for all stages of setup and analysis are available (8–10). When combined with appropriate quantitative methods, such studies have uncovered noteworthy characteristics that are common among mammalian signal transduction pathways (11). We focus on three topics in cellular communication that stand out with recent works bringing together theoretical and experimental aspects: dynamic range, signal processing, and information transfer.
Box 1. Planning a multidisciplinary signal transduction study.
For experimental biologists, incorporating quantitative methods from other fields into their area of study can seem intimidating. However, what is frequently perceived as the main obstacle – an inherent difficulty in mathematical approaches – is more often in reality a lack of familiarity with the existence and capabilities of quantitative methods. Thus, a collaborator broadly experienced in quantitative analysis is often an essential resource. Of course, the most important ingredient in any successful collaboration is a shared interest in the topic of study. But a second main question is where to look for such a collaborator, and what areas of expertise are most germane for a potential project. We provide a short summary below of quantitative approaches that integrate well with modern signal transduction studies. On a university campus, individuals skilled in these areas can be found in a wide range of departments, including the usual suspects of systems biology, statistics, physics, and various engineering fields. However, there are now many similarly trained researchers in less obvious disciplines, including plant biology, epidemiology, and sociology – especially as these fields incorporate larger datasets and quantitative methods.
Dynamical systems.
Methods to simulate how systems evolve over time, using differential equations or similar types of models to represent the changes in interlinked parameters. Important for the majority of modeling studies in signal transduction.
Commonly used in:
Most engineering disciplines, systems biology, economics, ecology.
Biological example:
Will the activity of a kinase cascade remain elevated following stimulation, return to baseline levels, or oscillate? (48)
System identification.
Methods to determine which model best represents the relationship between measured variables. Important for characterizing the transfer functions between signaling molecules and pathways. Many applications include machine learning techniques.
Commonly used in:
Electrical, mechanical, and aeronautical engineering, systems biology, computer science and bioinformatics.
Biological example:
Can a mathematical model accurately predict which genes are activated under a given stimulus? (49)
Control theory.
Methods to predict the behavior of systems with feedback loops, and to control devices using feedback (for example, thermostats). Important for systems with heavy or complex regulation, and for designing strategies to modulate systems (e.g. therapeutics).
Commonly used in:
Electrical, mechanical, and aeronautical engineering.
Biological example:
How does the cell maintain constant levels of metabolites such as ATP? (50).
Information theory.
Methods to quantify information – how much a given measurement tells you about a system. Important for assessing the reliability of a signaling system, especially in the presence of confounding noise.
Commonly used in:
Electrical engineering, communications, applied physics.
Biological example:
How different must two concentrations of ligand be for a signaling pathway to distinguish between them? (20)
Accurately measuring signaling events across their dynamic range
The challenge of quantifying the informational content of a signaling event is intimately linked to the problem of measuring that event within the cell. This connection is fundamental: in such experiments, the experimentalist is attempting to perform, in essence, the same task that the signaling pathway itself performs within the cell – that of distinguishing different levels of the input signal, with sufficient accuracy to control a cellular process (or to understand the regulation of that process, in the case of the researcher). Both the experimentalist and the signaling pathway face limits on the accuracy with which this signal can be quantified. These include: 1) measurement noise, 2) limits on sensitivity that determine the lower limit of input signal that is reliably detectable, and 3) saturation when the detection process reaches its maximum value at a sub-maximal input strength, preventing it from distinguishing further increases in the input. In any experiment, it must be remembered that the measurement process itself is an element with its own transfer function, and taking this transformation into account is essential for correctly interpreting the data.
An instructive example of the relationship between cellular signals and measurements can be found in recent experiments in which two fluorescent protein-based reporters for the same kinase were monitored in the same cell (12, 13). One reporter, EKAR3, was a FRET-based construct, in which phosphorylation by the kinase ERK induces a conformational change leading to a shift in the emission properties of a CFP/YFP FRET pair. In the other reporter, ERK-TR, phosphorylation of the reporter by ERK disrupts a nuclear localization sequence fused to RFP, causing the fluorescent protein to be exported from the nucleus (Fig. 2A). Both reporters are reversible, respond rapidly to changes in ERK activity induced by upstream growth factors or pharmacological inhibitors, and typically show strong qualitative agreement in their responses (12). However, when reporter responses were compared on a cell-by-cell for different stimulus strengths, a clear differential relationship emerged: EKAR3 was capable of detecting smaller impulses of ERK, but reached saturation at lower levels than ERK-TR (Fig. 2B, C) (13). Because these measurements used different detection modalities but were made simultaneously within same cell, technical artifacts could be ruled out and the differences attributed to different dynamic ranges of response for the different reporters, providing a clear demonstration that such differences exist at the level of an individual cell.
Figure 2.
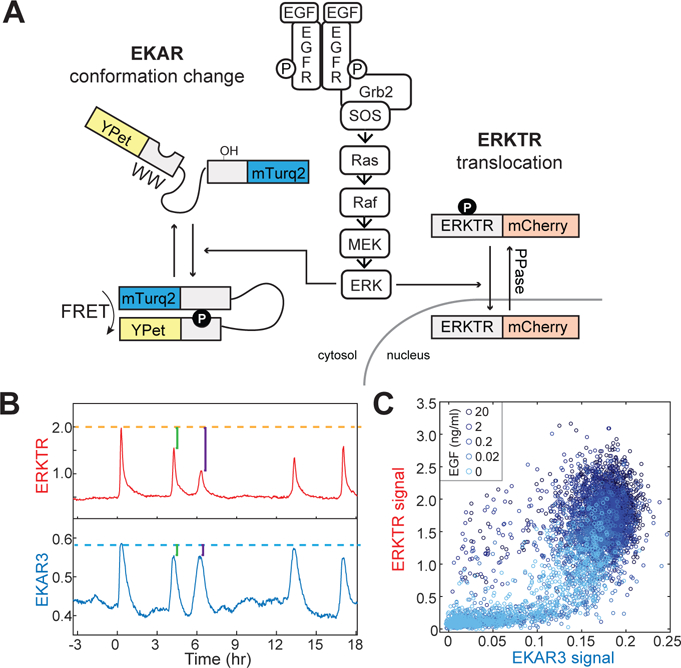
Parallel signaling processes with differing transfer parameters. A. Experimental setup for measuring two synthetic ERK substrates. EKAR is a FRET-based reporter that undergoes a conformational change upon phosphorylation by ERK, leading to changes in fluorescence emission characteristics, while ERKTR is a translocation-based reporter that shifts its predominant localization from the nucleus to the cytosol upon phosphorylation by ERK. B. Measurements of ERK activity pulses in a single cell expressing both EKAR and ERKTR. C. Scatter plot of peak heights as measured by both EKAR and ERKTR. Data are re-printed with permission from (13).
From this result, it becomes clear that each of the reporter signals represents a different view of cellular ERK activity, as filtered through their respective transfer functions. Considering this difference a bit more deeply, it is noteworthy that while the reporters are synthetic proteins, they operate on principles similar to many endogenous kinase substrates, in which phosphorylation induces changes in conformation or cellular localization. Thus, it can be inferred that among the many natural protein substrates of ERK, there may be significant variation in the dynamic range of input to which they respond. Such differences have in fact been documented among endogenous substrates, and may function to create unique responses to different strengths of signals. One example of such ordering can be found in the substrates of cyclin dependent kinases, whose sensitivities to phosphorylation vary such that they respond in a staged manner to progressively higher CDK activity over the course of the cell cycle (14). Similarly, differences in promoter sensitivity underlie ordered responses to morphogen gradients (15), and may enable time-dependent protein synthesis such that large protein complexes can be assembled more efficiently (16). Thus, in considering carefully the transfer properties involved in accurately measuring even one signaling node, we not only learn about the quality and limitations of the data, but also gain insight into organizing principles for how cells process the information contained within the pathway’s activity.
Another transformation inherent in many experimental approaches is the averaging of a measured quantity across a large number of cells (17). Determining an average behavior is often an essential step in analysis and interpretation of data, even when data are collected with single-cell resolution. However, relying on averages alone can severely distort the apparent transfer function for a process. For example, individual cells within a population might display a 3-fold increase in signal intensity between their unstimulated and stimulated responses, but vary by 10-fold in the concentration of stimulus to which they respond (Fig. 3). Population-average measurements of these cells would indicate that the overall response spans a more than 10-fold range, with an EC50 defined by the median individual response, while individual cell measurements would indicate only a 3-fold range. Thus, a population measurement of dynamic range will in fact represent a convolution of the individual cell behaviors and the range of response sensitivities among cells, rather than the range of the response in any individual cell. Both quantities may contain important information for the system and may be useful in characterizing the response, but need to be distinguished if the ultimate goal is a quantitative model of typical cell behavior. Fitting a model to the 10-fold range of output would impose unrealistic constraints on the model and would likely lead to errors as other parts of the model are adjusted to accommodate this inaccuracy.
Figure 3.
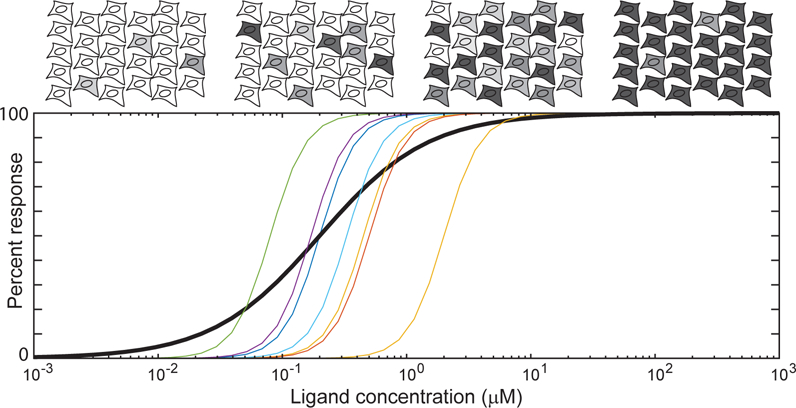
Relationship between single-cell and population-level responses. A hypothetical population of cells is depicted, which vary in their responsiveness to ligand (colored curves). The bold curve represents the aggregate response for a population of cells with a distribution of individual response curves. At increasing concentrations of ligand, an increasing number of cells (top, gray) are progressively recruited to the response.
Signal processing by pathways: lessons from single-node measurements
Signaling events are now routinely measured with single-cell resolution. The most accessible approach is to use activity-specific antibodies or other fluorescent probes, to capture a “snapshot” measurement of the signaling activity status of many cells at a fixed point in time (18, 19). Because of the need to chemically fix cells, such datasets are limited in their time resolution, but they can nonetheless establish the distribution of responses possible for a given cell type. When coupled with information theory, a mathematical framework for quantifying the relationship between the stimulus and intracellular responses, such datasets can be used to estimate the ability of the pathway to resolve different strengths of input stimuli (20, 21). Such studies have revealed the variability inherent in signal processes, but are limited in their ability to quantify time-dependent responses, potentially failing to account for some information.
To capture information carried in the time domain, live-cell measurements with fluorescent protein-based reporters allow for the tracking of cells for minutes, hours, or days following a stimulus (22). A growing number of studies have revealed that many signaling pathways respond to a simple step-up stimulus with diverse kinetic outputs (23–25)(for a comprehensive review, see (11)). One way in which kinetics may carry information is termed “dose-to-duration” encoding, in which different input strengths induce transient pulses of response with varying durations (26). Downstream responses that integrate the amount of time that the responding signal remains active are thus not limited in their response by the range of amplitudes of the upstream signal. Another advantage of live-cell data is the ability to measure each cell in multiple states, such as pre- and post-stimulus. Such within-cell comparisons can be essential, because some pathways employ fold-change detection in which the response is proportional to the relative change, as opposed to the absolute magnitude of the input signal (27, 28).
Live cell measurements also allow for a more sophisticated form of analysis, in which complex time-dependent input signals are used as stimuli, and measurements are recorded at a downstream node (29). Stimulus patterns can be generated using microfluidics to make rapid changes in extracellular ligand concentration (30, 31), or optogenetic tools such as engineered light responsive signaling elements (32). This approach comes much closer to methods used to characterize transfer functions in electronic circuits, because it allows the experimenter to determine the response to both the magnitude and the frequency of the stimulus. Moreover, the same cell can be repeatedly interrogated, avoiding the assumptions inherent in combining data from different cells’ responses.
One insight that is consistent among all of these studies is that signaling pathways operating in individual cells have fairly limited dynamic ranges. Moreover, cells within a population often diverge substantially in their threshold of response to a stimulus (13, 33–35). Thus, the situation depicted in Figure 3 is relatively common and has implications for many commonly held models of cellular functions. Perhaps most strikingly, it means that any individual cell cannot mount reliably distinct responses to more than a small range of input concentration. While studies examining this issue have debated different approaches to quantifying the output function, collectively they suggest that the upper limit of the “channel capacity” for many mammalian signaling pathways is approximately 2 bits. This means that the output magnitude of a stimulated pathway, at any given time, can distinguish up to about four different concentrations of stimulus.
A solution to this seemingly limited view of cellular information processing, is that, because of the high degree of variability between cells, “decision making” functions may instead be performed at the population level rather than at the single-cell level. For example, at a sub-saturating dose of a ligand, only a subset of cells will respond due to the heterogeneity in responsiveness. Further increases in ligand concentration will push a number of the non-responders over their activation threshold, leading to a response to both ligands that is easily distinguished from activation by a single ligand. Such population-level information processing is supported by theoretical models, which point out that such heterogeneity may be essential when regulating inherently binary responses such as cell death (36). For example, if all cells shared the same sensitivity to a death-inducing ligand like TNF, the only regulatory possibilities would be to either retain or eliminate the entire population. With a broadly distributed range of sensitivities, it becomes possible to reduce the population by a desired fraction depending on the dose of the ligand. Thus, broad distributions of signal pathway responsiveness can provide an organism with a higher degree of control over the behavior of a tissue (36, 37).
An elegant experimental example of such population-level control can be found in the Drosophila embryo. Nuclei respond to the morhphogen Bicoid – which is distributed in an anterior-posterior gradient - by activating certain genes wherever the Bicoid concentration exceeds a certain level. This response has a high degree of precision, enabling each nucleus to determine unambiguously whether it lies on the anterior or posterior side of the boundary (38). Detailed studies of the kinetics of this process in individual nuclei reveal that their individual ranges of response are too small to explain the full dynamic range. Instead, the organism-level response depends on the fact that each nucleus is probabilistic in its capacity to respond and thus is independent from the strength or duration of response (39). Because the mRNAs produced by each nuclei are exported and shared within the local area of the syncytium, the collective mRNA production in each region averages the local nuclei, such that the stochastic nature of induction enhances the overall dynamic range.
Stepwise transfer of information in signaling pathways
Population-level information processing is a reasonable strategy for functions that are collective properties of multiple cells, such as the total amount of a molecule synthesized, or the fraction of cells taking on a given fate. However, other cellular functions appear to demand careful cell-autonomous regulation (36). For example, maintenance of cellular ATP levels is essential for cell viability, and deviations from the normative concentration must be quickly and accurately corrected to prevent cell death. This need for accuracy implies that some regulatory functions may require tighter regulatory control than has been measured for signaling pathways thus far. Accordingly, cells have developed strategies for gain control, also termed Dose Response Alignment (or DoRA) to reduce the tendency toward mismatches in the dynamic range in signaling cascades. Despite the fact that it is relatively difficult to ensure that the quantitative parameters of a pathway maintain DoRA, such regulation appears to be pervasive across many systems, implying that DoRA is a trait selected for in the evolution of regulatory systems (4).
A key component for DoRA is negative feedback from a lower tier of the cascade to a higher tier. The capacity of negative feedback to shift multi-tiered responses toward alignment is well-known from many human-designed systems in which it is used to linearize the system response (4). Such a strategy has also been confirmed to operate and produce DoRA within the yeast MAPK cascade, and may potentially explain other instances of DoRA (40). Importantly, however, negative feedback alone is insufficient to produce DoRA; also required is a mechanism for comparing the input to the output and adjusting the output to match the input (41). Such a mechanism is analogous to “proportional control” in engineered systems, where the strength of feedback is adjusted to be proportional to the difference between output and input. Further exploration of the mechanisms for DoRA in biochemical networks is ongoing, and differences in the results of theoretical studies to date underscore the complexity of this problem (41, 42), which remains surprisingly understudied despite its fundamental importance to intracellular signaling functions.
Ultimately, to measure the transfer function of a signaling process as it occurs in the cell, the researcher must make accurate measurements of both the input and output signal in such a way that the dynamic range of each measurement matches or exceeds the range of the input and output values as they occur in the cell. Given the caveats of population measurements relative to single-cell ones (discussed above), such measurements would best be made using time-lapse single-cell techniques. However, despite advances in live-cell signaling reporters (43), there remain few cases in which multiple reporters are available for sequential steps within a pathway. One area where such sequential measurements are both achievable and of high interest is in understanding information transfer from signaling pathways to their target genes. Studies using the yeast transcription factor Msn2, have pioneered concepts in this area (24, 44). Msn2 responds to multiple stresses, including glucose limitation, high salt, and oxidative stress, raising the question of how subsets of genes specific to each stress are activated by the same transcription factor. Live-cell measurements have made it possible to link the dynamics of Msn2 localization, which varies according to the initiating stress, to the expression of different Msn2 client genes. This work has revealed that different dynamic patterns and magnitudes of Msn2, when coupled to differential response parameters for target gene promoters, can encode specific gene expression profiles. In mammalian cells, similar concepts have begun to be explored in the p53 and Ras/MAPK systems (13, 45, 46), where gene expression profiles are more complex.
Echoing the theme of the single-node measurements discussed above, from these studies it is clear that there is a high degree of cell-to-cell variability that limits the amount of information that can be transmitted by any one pathway. What remains to be determined is whether this variability results primarily from stochastic events, or whether it reflects differences in the activity of pathways not measured. Ultimately, answering this question will be of great importance in interpreting genome-wide single-cell expression profiling, which are now possible.
Conclusion
For many, the ultimate goal of signal transduction research is to understand the cellular communication systems that regulate human physiology, in sufficient detail to know which components are required for healthy functioning, and to identify means to restore function when these systems break down. For much of its history, work in this field has focused on enumeration of the proteins, small molecules, and nucleic acids involved in these processes and elucidation of their interactions. It has been noted many times that while this cataloging is an essential first step, models that capture systems-level behavior will be needed to integrate this knowledge. While a wave of “systems biology” research over the past 15 years has endeavored to build such models, it remains unclear to many whether this goal has been achieved, given the failure of broadly useful and predictive quantitative models to emerge. If this goal has not yet been achieved, is it because we still lack key molecular details, or because we have yet to develop the correct modeling approaches? It can be argued that, while both are likely to be true, there is an important third obstacle: many of the experiments reported thus far have failed to quantify the key operational properties of signaling networks that enable predictive modeling (7, 47). Many of the studies discussed here have appreciated this gap and have contributed to an emerging conceptual framework that stresses the importance of these properties, while others have developed new tools aimed at making such measurements accessible. With these technologies now in place, we anticipate that the next few years will yield substantial insights into the properties of signal transduction pathways that enable them to transmit information, an appreciation of the limitations on reliability, and ultimately quantitative models of how cell fates and identities are specified through signaling pathways.
Summary Points.
Concepts used to engineer communication systems can augment cell signaling studies.
The dynamic range of signaling processes is an essential factor determining how they transfer information about stimuli.
Single cells are often limited in their information processing capacity, but variability between cells can improve signal discrimination at the tissue level.
Operational properties of signaling pathways are essential parameters for predictive models.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences (1R01GM115650). The authors declare that they have no competing interests.
References
- 1.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol 2010;11(6):414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiley HS, Shvartsman SY, Lauffenburger DA. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol 2003;13(1):43–50. [DOI] [PubMed] [Google Scholar]
- 3.Schoeberl B, Kudla A, Masson K, Kalra A, Curley M, Finn G, et al. Systems biology driving drug development: from design to the clinical testing of the anti-ErbB3 antibody seribantumab (MM-121). NPJ Syst Biol Appl 2017;3:16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brent R Cell signaling: what is the signal and what information does it carry? FEBS Lett 2009;583(24):4019–24. [DOI] [PubMed] [Google Scholar]
- 5.Sagoo MS, Lagnado L. G-protein deactivation is rate-limiting for shut-off of the phototransduction cascade. Nature 1997;389(6649):392–5. [DOI] [PubMed] [Google Scholar]
- 6.Lizbinski KM, Dacks AM. Intrinsic and Extrinsic Neuromodulation of Olfactory Processing. Front Cell Neurosci 2017;11:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antebi YE, Nandagopal N, Elowitz MB. An operational view of intercellular signaling pathways. Curr Opin Syst Biol 2017;1:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pargett M, Gillies TE, Teragawa CK, Sparta B, Albeck JG. Single-Cell Imaging of ERK Signaling Using Fluorescent Biosensors. Methods Mol Biol 2017;1636:35–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum Y, Fritz RD, Ryu H, Pertz O. Measuring ERK Activity Dynamics in Single Living Cells Using FRET Biosensors. Methods Mol Biol 2017;1487:203–21. [DOI] [PubMed] [Google Scholar]
- 10.Hansen AS, Hao N, O’Shea EK. High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat Protoc 2015;10(8):1181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science 2013;342(6163):1193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sparta B, Pargett M, Minguet M, Distor K, Bell G, Albeck JG. Receptor Level Mechanisms Are Required for Epidermal Growth Factor (EGF)-stimulated Extracellular Signal-regulated Kinase (ERK) Activity Pulses. J Biol Chem 2015;290(41):24784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies TE, Pargett M, Minguet M, Davies AE, Albeck JG. Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation. Cell Syst 2017. [DOI] [PMC free article] [PubMed]
- 14.Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell 2016;167(7):1750–61 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papatsenko D, Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc Natl Acad Sci U S A 2005;102(14):4966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, et al. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 2001;292(5524):2080–3. [DOI] [PubMed] [Google Scholar]
- 17.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell 2013;152(5):945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito TH, Uda S, Tsuchiya T, Ozaki Y, Kuroda S. Temporal decoding of MAP kinase and CREB phosphorylation by selective immediate early gene expression. PLoS One 2013;8(3):e57037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JY, Lin JR, Cimprich KA, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol Cell 2012;45(2):196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A. Information transduction capacity of noisy biochemical signaling networks. Science 2011;334(6054):354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uda S, Saito TH, Kudo T, Kokaji T, Tsuchiya T, Kubota H, et al. Robustness and compensation of information transmission of signaling pathways. Science 2013;341(6145):558–61. [DOI] [PubMed] [Google Scholar]
- 22.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, et al. Systems biology. Accurate information transmission through dynamic biochemical signaling networks. Science 2014;346(6215):1370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 2004;36(2):147–50. [DOI] [PubMed] [Google Scholar]
- 24.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 2008;455(7212):485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu H, Chung M, Dobrzynski M, Fey D, Blum Y, Lee SS, et al. Frequency modulation of ERK activation dynamics rewires cell fate. Mol Syst Biol 2015;11(11):838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behar M, Hao N, Dohlman HG, Elston TC. Dose-to-duration encoding and signaling beyond saturation in intracellular signaling networks. PLoS Comput Biol 2008;4(10):e1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol Cell 2009;36(5):872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frick CL, Yarka C, Nunns H, Goentoro L. Sensing relative signal in the Tgf-beta/Smad pathway. Proc Natl Acad Sci U S A 2017;114(14):E2975–E82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci U S A 2008;105(20):7165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen AS, O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol 2013;9:704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellogg RA, Tay S. Noise facilitates transcriptional control under dynamic inputs. Cell 2015;160(3):381–92. [DOI] [PubMed] [Google Scholar]
- 32.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 2013;155(6):1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell 2008;30(1):11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strasen J, Sarma U, Jentsch M, Bohn S, Sheng C, Horbelt D, et al. Cell-specific responses to the cytokine TGFbeta are determined by variability in protein levels. Mol Syst Biol 2018;14(1):e7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 2010;466(7303):267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suderman R, Bachman JA, Smith A, Sorger PK, Deeds EJ. Fundamental trade-offs between information flow in single cells and cellular populations. Proc Natl Acad Sci U S A 2017;114(22):5755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovary KM, Taylor B, Zhao ML, Teruel MN. Expression variation and covariation impair analog and enable binary signaling control. Mol Syst Biol 2018;14(5):e7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 2013;154(4):789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol 2013;23(21):2140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu RC, Pesce CG, Colman-Lerner A, Lok L, Pincus D, Serra E, et al. Negative feedback that improves information transmission in yeast signalling. Nature 2008;456(7223):755–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews SS, Peria WJ, Yu RC, Colman-Lerner A, Brent R. Push-Pull and Feedback Mechanisms Can Align Signaling System Outputs with Inputs. Cell Syst 2016;3(5):444–55 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan L, Ouyang Q, Wang H. Dose-response aligned circuits in signaling systems. PLoS One 2012;7(4):e34727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 2014;157(7):1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao N, O’Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol 2011;19(1):31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter JR, Fisher BE, Batchelor E. p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks. Cell Syst 2016;2(4):272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE. Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell 2017;67(5):757–69 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munsky B, Li G, Fox ZR, Shepherd DP, Neuert G. Distribution shapes govern the discovery of predictive models for gene regulation. Proc Natl Acad Sci U S A 2018;115(29):7533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol 2004;164(3):353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neuert G, Munsky B, Tan RZ, Teytelman L, Khammash M, van Oudenaarden A. Systematic identification of signal-activated stochastic gene regulation. Science 2013;339(6119):584–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra FA, Buzi G, Doyle JC. Glycolytic oscillations and limits on robust efficiency. Science 2011;333(6039):187–92. [DOI] [PubMed] [Google Scholar]


