SUMMARY
Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine to HIV or other difficult pathogens due to the immunological hurdles posed, including B cell immunodominance and germinal center (GC) quantity and quality. We found that two independent methods of slow delivery immunization of rhesus monkeys (RM) resulted in more robust T follicular helper (TFH) cell responses and GC B cells with improved Env-binding, tracked by longitudinal fine needle aspirates. Improved GCs correlated with the development of > 20-fold higher titers of autologous nAbs. Using a new RM genomic immunoglobulin loci reference, we identified differential IgV gene usage between immunization modalities. Ab mapping demonstrated targeting of immunodominant non-neutralizing epitopes by conventional bolus immunized animals, while slow delivery immunized animals targeted a more diverse set of epitopes. Thus, alternative immunization strategies can enhance nAb development by altering GCs and modulating immunodominance of non-neutralizing epitopes.
Graphical Abstract
eTOC blurb
A study in rhesus macaques shows that a slow delivery immunization modality improves HIV neutralization antibody responses by improving germinal center size, immune cell responses, driving differential IgV usage and improved Env binding.
INTRODUCTION
A majority of licensed vaccines provide protection through the induction of protective antibodies (Plotkin, 2010). The isolation of HIV-1 broadly neutralizing antibodies (bnAbs) from HIV-infected individuals and the finding that passive transfer of bnAbs can protect non-human primates (NHPs) from SHIV infection support the feasibility of an antibody-based HIV vaccine (Burton and Hangartner, 2016; Nishimura and Martin, 2017). Elicitation of neutralizing antibodies (nAbs) against clinically relevant HIV strains (i.e., tier 2 and tier 3 strains) by immunization has been difficult (Montefiori et al., 2018). Much of that challenge centers on structural features of HIV envelope (Env), which have complex and incompletely understood immunological implications. Env consists of gp120 and gp41 components that form a trimeric spike, which is the only viral protein on HIV virions and the only target for nAbs (Burton and Hangartner, 2016). Human immunizations with monomeric gp120 have failed to elicit tier 2 nAbs in clinical trials (Haynes et al., 2012; Mascola et al., 1996; Rerks-Ngarm et al., 2009). The reasons for this are not obvious, as nAb epitopes are present on gp120. Key developments in protein design have been made towards the expression of soluble native-like HIV Env trimers (Julien et al., 2013; Kulp et al., 2017; Lyumkis et al., 2013; Sanders et al., 2013). Immunization with these Env trimers elicited substantial strain-specific tier 2 nAbs in rabbits and guinea pigs, but failed to elicit nAbs in mice (Feng et al., 2016; Hu et al., 2015; Sanders et al., 2015). Trimer immunizations of NHPs have been sporadically successful (Havenar-Daughton et al., 2016a; Pauthner et al., 2017; Sanders et al., 2015; Zhou et al., 2017). For some regimes in NHPs, autologous tier 2 nAbs have been elicited within 10 weeks, which is comparable to the speed of nAb development in HIV-infected individuals (Pauthner et al., 2017; Richman et al., 2003; Wei et al., 2003). Thus, while nAb epitopes are presented on native-like trimers, the immunological parameters controlling the development of nAbs to Env remain to be elucidated. These parameters are also likely important for nAbs to other pathogens.
Germinal centers (GCs) are essential for HIV nAb development, which requires antibody (Ab) somatic hypermutation (SHM) (Klein et al., 2013; West et al., 2014). GCs are sites where B cells compete for antigen and undergo repeated rounds of SHM of their BCRs and selection by GC T follicular helper cells (GC-TFH) to evolve high affinity Abs (Crotty, 2014; Mesin et al., 2016). B cells with higher affinity to Ag present more peptide:MHC complexes to GC-TFH cells and receive more help (Crotty, 2014; Gitlin et al., 2014; Victora et al., 2010). GC-TFH help signals to GC B cells (BGC) result in proliferation and further SHM (Gitlin et al., 2015). TFH help quality was associated with HIV nAb development in trimer immunized rhesus macaques (RM) (Havenar-Daughton et al., 2016a). Frequencies of highly functional memory TFH cells in blood were associated with bnAb development in HIV-infected humans (Locci et al., 2013; Moody et al., 2016). GC-TFH cells were also positively correlated with nAb development in SIV+ RMs and SHIV+ RMs (Chowdhury et al., 2015; Petrovas et al., 2012; Yamamoto et al., 2015).
B cell responses to protein Ags are polyclonal, targeting several epitopes across an Ag. The composition of the Ag-specific B cell repertoire can be complex. B cells initially engage in interclonal competition, and then engage in interclonal and intraclonal competition, resulting in complex outcomes (Kuraoka et al., 2016; Tas et al., 2016). Theoretically, the surface of a protein represents a continuum of epitopes. In reality, the Ab response to a protein predominantly targets a limited number of epitopic sites. Immunodominance is the phenomenon in which B cells that recognize an epitopic site dominate a response at the expense of B cells that recognize other sites. Immunodominance is well described for influenza HA, in which the epitopes are recognized in a hierarchical manner (Angeletti and Yewdell, 2018; Angeletti et al., 2017). Immunodominance appears to be a key immunological process limiting the development of broad nAb responses to influenza (Andrews et al., 2018; Angeletti and Yewdell, 2018; Angeletti et al., 2017; Victora and Wilson, 2015) and may also be important for nAb development against other refractory pathogens, including HIV (Havenar-Daughton et al., 2017). Soluble Env trimer can be subject to in vivo degradation, resulting in breakdown products that are not exposed on the virion surface. These non-native epitopes are likely ‘dark antigen’ and can be immunodominant (Kuraoka et al., 2016). Evidence of immunodominance impairing HIV nAb development includes: the lack of tier 2 nAb responses by gp120 immunized humans, the lack of autologous tier 2 nAb responses in non-native Env trimer immunized RMs, the sporadic nature of tier 2 nAb development in Env trimer immunized RMs, and the role of immunodominance in the response of rare and low affinity HIV CD4-binding-site specific B cells in a mouse model (Abbott et al., 2018; Havenar-Daughton et al., 2017).
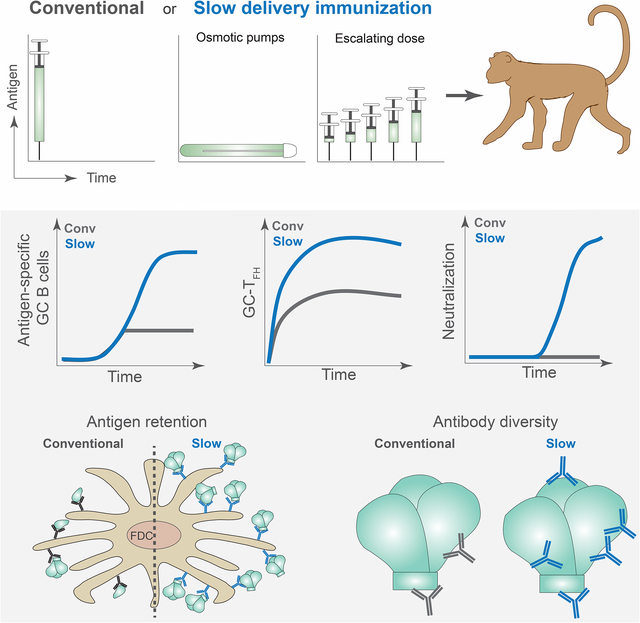
Much of the focus in HIV vaccine development is on the choice of antigen and adjuvant, but another parameter is the kinetics of antigen availability. Slow delivery immunization is an attractive vaccine strategy because it more closely mimics a natural, acute infection (Cirelli and Crotty, 2017). While the adjuvanticity of alum has been believed to be in part due to a ‘depot’ effect of antigen, many antigens rapidly elute from alum in vivo (Hogenesch, 2002; Shi et al., 2001; Weissburg et al., 1995) and several studies reported that this depot did not affect Ab responses (Hogenesch, 2012; Hutchison et al., 2012; Noe et al., 2010), suggesting that alum adjuvanticity does not primarily function via a slow release mechanism. In contrast, two-week slow release immunization using nonmechanical osmotic pumps (OP) and a soluble adjuvant resulted in enhanced BGC and TFH cell responses in mouse models (Hu et al., 2015; Tam et al., 2016). Two-week escalating dose (ED) immunization resulted in similar outcomes and enhanced deposition of immune complexes onto follicular dendritic cells (FDC) (Tam et al., 2016).
Understanding the importance of different aspects of B and T cell biology in the development of HIV nAbs has been limited by the fact that wildtype mice do not develop tier 2 nAbs in response to trimer immunization. While NHPs are important animal models for HIV vaccine design because of their close evolutionary relationship to humans, it has been difficult to study the early response to Env immunization due to the inaccessible nature of lymph nodes (LN). As a first proof-of-concept in NHPs, RMs were immunized with soluble native-like Env trimers in a soluble ISCOMs-class saponin adjuvant delivered via OPs (Pauthner et al., 2017). OP-immunized RMs responded with the most robust autologous tier 2 nAb responses in the study, which developed by week 10 in all animals. The rapidity and magnitude of the nAb response suggested that improved affinity maturation, B cell lineage recruitment, Env-specific TFH cell responses, or other factors may be responsible for the improved nAb response. Antigen-specific B cell and TFH cells were not examined. Here we have examined the early B and T cell responses to Env trimers in RMs using new tools and comparative immunology between conventional and slow release concepts to gain insights into the development of HIV nAbs, which may also be applicable to other refractory pathogens.
RESULTS
Env-specific GC responses are more robust upon slow release immunization
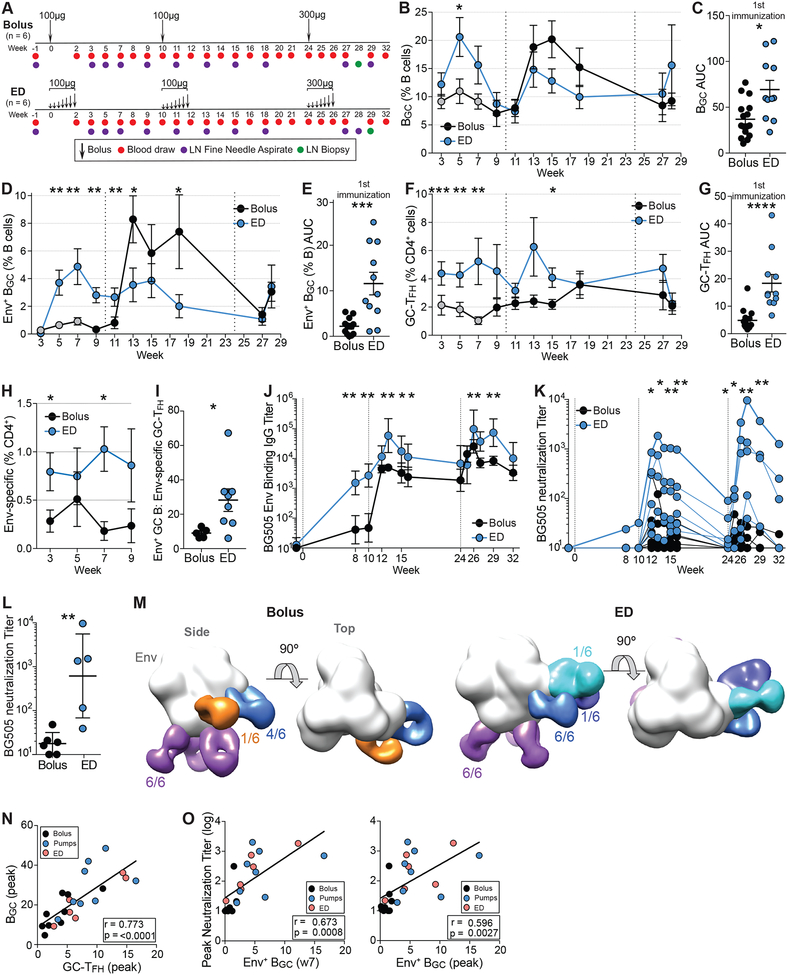
Three groups of RMs were immunized with soluble native-like Env trimer BG505 Olio6CD4ko protein (Kulp et al., 2017) in a soluble ISCOMs-class saponin adjuvant. Three delivery strategies were tested: conventional bolus immunization via subcutaneous (SubQ) injection (n = 9), two-week SubQ nonmechanical osmotic pumps (‘OP’, n = 4) and four-week SubQ OP (n = 4) (Fig 1A). All immunizations were given bilaterally in left and right thighs. To determine the kinetics of the GC response to primary immunization, longitudinal LN fine needle aspirates (FNA) were employed to sample the draining inguinal LNs weekly in each hind limb. This study is the first longitudinal (i.e., same individuals sampled) weekly kinetic analysis of a GC response in any species. Previous work demonstrated that LN FNAs well represented the cellular composition of the whole LN and were well tolerated (Havenar-Daughton et al., 2016a).
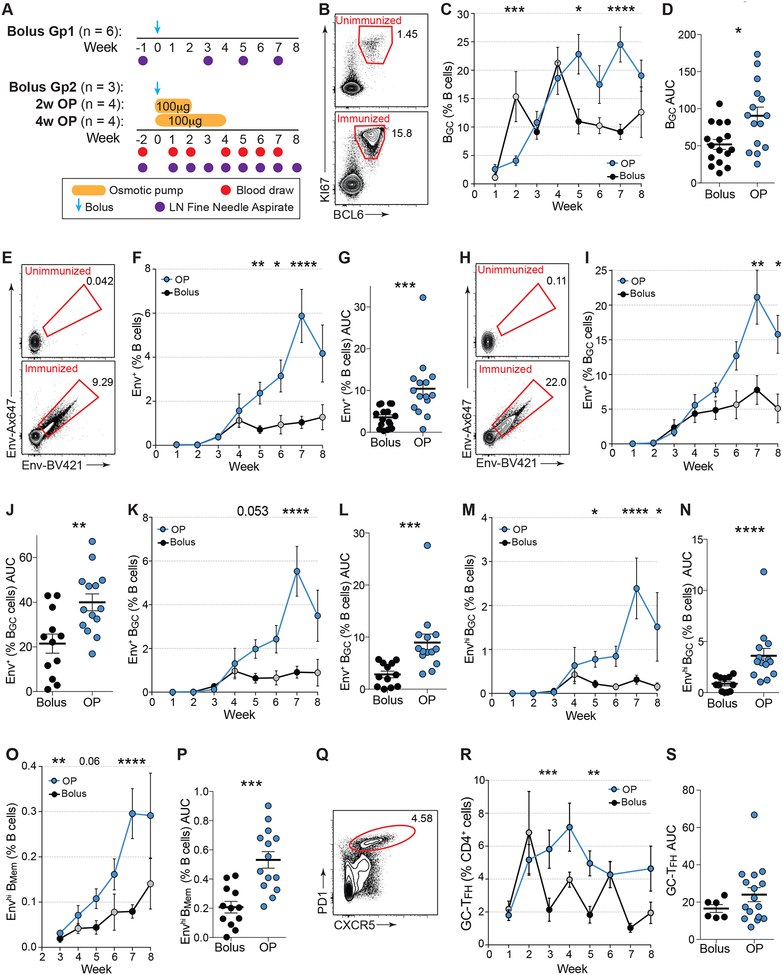
Figure 1. Sustained delivery immunization enhances BGC cell responses.
(A) Immunization and sampling schedule of first immunization. Bolus Group 2 (Gp2), 2w osmotic pump (OP), and 4w OP RMs were immunized and sampled at the same time. Bolus Gp1 were immunized and sampled separately. Bolus Gp1&2 data have been pooled.
(B) Representative BGC cell flow cytometry, gated on viable CD20+ B cells pre- and post-immunization. See Fig S1 for full gating strategy.
(C) BGC cell frequencies over time. Black circles, pooled bolus Gp1&2; grey circles, bolus Gp2.
(D) Cumulative BGC cell responses (AUC of C) to immunization within individual LNs at weeks 1, 3–7 [AUC].
(E) Representative flow cytometry of Env trimer-specific B cells pre- and post-immunization.
(F) Env-specific B cell frequencies over time.
(G) Cumulative Env trimer-specific B cells (AUC of F) within each LN.
(H) Representative flow cytometry Env trimer-specific BGC cells pre- and post-immunization.
(I) Env trimer-specific BGC cell frequencies over time.
(J) Cumulative Env trimer-specific BGC cells (AUC of I) within each LN.
(K) Env trimer-specific BGC cells over time.
(L) Cumulative Env trimer-specific BGC cells (AUC of K) within each LN.
(M) Frequencies of high-affinity Env trimer-specific BGC cells over time.
(N) Cumulative high-affinity Env trimer-specific BGC cells (AUC of M) within each LN.
(O) High-affinity Env trimer-specific BMem cells over time.
(P) Cumulative high-affinity Env trimer-specific BMem cell responses within individual LNs.
(Q) Representative flow cytometry of GC-TFH cells, gated on CD4+ T cells. See Fig S2 for full gating strategy.
(R) GC-TFH cells over time.
(S) Cumulative GC-TFH cell response between w1 plus w3-6 [AUC].
Mean ± SEM are graphed. Statistical significance tested using unpaired, two-tailed Mann-Whitney U tests. *p≤0.05, **p≤0.01. ***p≤0.001, ****p≤0.0001
See also Figures S1–2 and Data S1.
GCs developed slower than expected, based on comparison to mouse data of LN GC kinetics after protein immunization, with almost no BGC cells (BCL6+KI67+ or CD38−CD71+ of CD20+CD3−) detectable at week 1 (w1) postimmunization (Fig 1B–C, S1A–B). Substantially greater BGC frequencies were present at w2 in both groups (w1 v w2, p = 0.0015). No differences were observed in GC kinetics between the OP groups, so data from these groups were pooled in subsequent analyses (n = 8 animals; n = ~16 LN FNAs per time point. Fig S1C–D). Total BGC in the LNs peaked at w7 in OP-immunized RMs after a single immunization, substantially later than after bolus immunization (Fig 1C). OP animals had significantly more BGC cells throughout the first immunization (p = 0.017 [Area under the curve (A∪C)]. Fig 1D, S1D).
Given that RMs are not kept in a sterile environment, interpretation of BGC kinetics, in the absence of antigen-specific probes, is confounded by uncertainty regarding the antigenic targets of the GCs. In previous studies, total BGC cell responses were measured, but antigen-specificity was not determined (Havenar-Daughton et al., 2016a; Pauthner et al., 2017). Detection of antigen-specific BGC cells is a challenge, as BGC cells express less BCR than non-BGC B cells (Fig S1E). Using Olio6 Env trimers conjugated to two fluorochromes as probes (EnvAx647 and EnvBV421), we measured the kinetics and magnitude of the Env trimer-specific B and BGC cell response (EnvAx647+ EnvBV421+ BCL6+KI67+ or CD38−CD71+ of CD20+CD3−, Fig 1E–N, S1F–O). This method was specific, with little experimental ‘noise’, as naive B cells and BGC cells from unimmunized animals did not bind these probes (Fig 1E, H, S1H, S1M, Data S1). Despite observing considerable GCs at w2–3, Env-specific BGC cells with detectable affinity to the probes were rare at w2–3, but consistently detectable at w4 in both groups (Fig 1I–K, S1H–O). Env-specific BGC frequencies were relatively stable between w4–8 in bolus RMs. In OP animals, frequencies of Env-specific BGC increased over time (p = 0.006 compared to bolus as Env+ % of BGC over time [AUC], and p = 0.0001 compared to bolus as Env+ BGC % of total B cells over time [AUC]. Fig 1H–L). Enhanced BGC cell binding of Env by OP animals was not due to an increase in BCR expression (Fig S1I). High binding Env-specific BGC cells became more abundant in OP animals over time (p < 0.0001 compared to bolus [AUC]. Fig 1M–N, S1J–O, R), suggesting that OP administration resulted in more affinity maturation compared to conventional bolus immunization.
Env trimer-specific memory B cells (BMem. BCL6− KI67− or CD38+CD71− EnvAx647+/hi EnvBV421+/hi CD20+ cells) developed in draining LNs in each group (Fig 1O–P, S1P–Q). OP animals developed higher frequencies of high affinity Env trimer-specific BMem. Overall, these data demonstrate that slow immunization delivery resulted in more robust GCs and suggested substantially greater affinity maturation to Env after a single immunization than occurred upon conventional bolus immunization.
Slow release immunization enhanced Env-specific GC-TFH cell responses
While OP RMs had higher total GC-TFH cell (CXCR5+ PD1hi of CD4+CD8−) frequencies than bolus RMs at several points during the 1st immunization, overall GC-TFH cells did not differ between groups (Fig 1Q–S, S2A). The specificity of GC-TFH cells at these time points could not be measured due to limited cell numbers and experimental prioritization of the B cell assays.
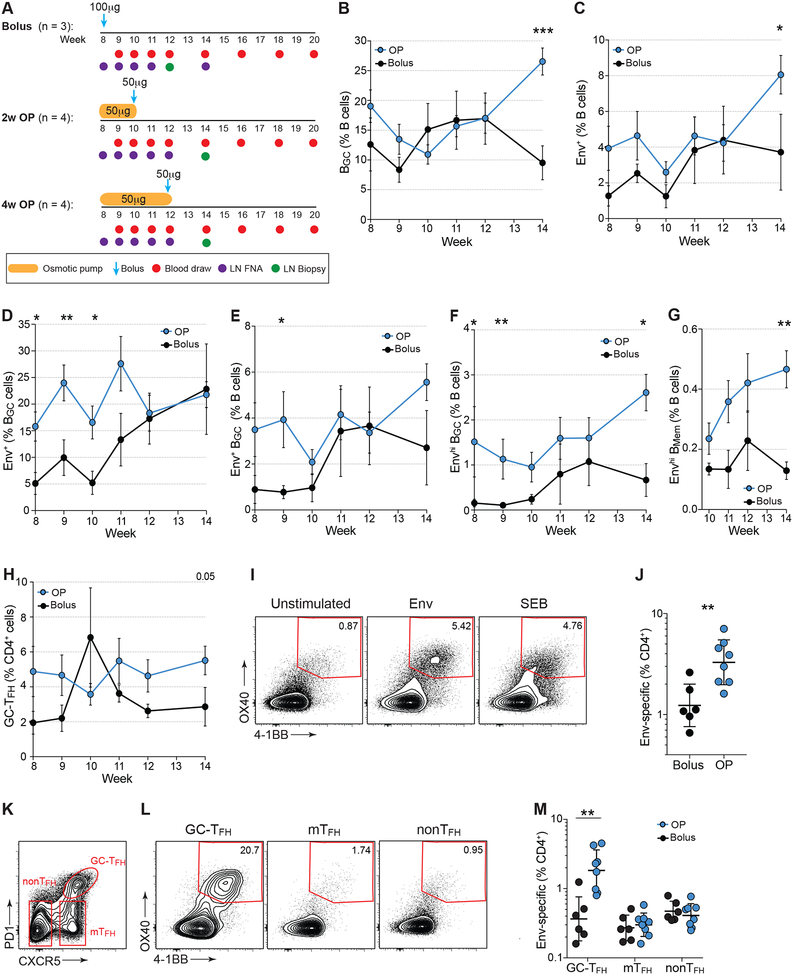
Based on previous immunization regimens (Pauthner et al., 2017), we administered a 2nd immunization at w8 (Fig 2A). For OP groups, the immunization was split evenly between pumps and a bolus administered at the end of OP delivery to simulate an escalating dose immunization. We hypothesized that a bolus immunization at the end of the slow release delivery may enhance plasma cell (BPC) differentiation and Ab titers. The total dose of Env trimer was matched between groups (100μg, Fig 2A). Draining LN GC responses observed after the 2nd immunization were relatively flat (Fig 2B), perhaps due to ongoing GC reactions immediately prior to the 2nd immunization (Fig 1C, 2B). OP RMs had significantly larger BGC responses at w14 (Fig 2B). Env-specific B and BGC frequencies in bolus RMs increased after the 2nd immunization (Fig 2C–F). High affinity Env-specific BGC recall responses were largely comparable between OP and bolus immunized animals (Fig 2G). Overall, kinetics of the secondary GC responses differed from those in the primary GC responses.
Figure 2. Germinal center responses following 2nd Env trimer immunization.
(A) Immunization and sampling schedule of 2nd immunization. Groups were immunized and sampled contemporaneously.
(B) Frequencies of total BGC cells over time.
(C) Env trimer-specific B cell frequencies over time.
(D-E) Env trimer-specific BGC cells over time.
(F) High-affinity Env trimer-specific BGC cells over time.
(G) High-affinity Env trimer-specific BMem cells over time.
(H) GC-TFH cell frequencies after 2nd immunization.
(I) Representative flow cytometry of Env-specific CD4+ T cells from LNs. Cells were left unstimulated or stimulated with a peptide pool spanning Olio6CD4ko.
(J) Env-specific CD4+ T cells at w12 (bolus) or w14 (OP).
(K) Representative flow cytometry of GC-TFH, mantle (m)TFH and non-TFH cell subsets.
(L) Flow cytometry of AIMOB assay (OX40+4-1BB+), gated on GC-TFH, mTFH or non-TFH cells.
(M) Quantification of Env-specific CD4+ T cells by subset.
Mean ± SEM are graphed. Statistical significance tested using unpaired, two-tailed Mann-Whitney U test. *p≤ 0.05, **p≤0.01, ***p≤0.00. See also Figure S2.
Total GC-TFH cell frequencies were similar in response to the 2nd Env trimer immunization (Fig 2H). To identify Env-specific GC-TFH cells, we performed cytokine-agnostic activation induced marker (AIM) flow cytometry assays with biopsied LN cells. Higher frequencies of Env-specific CD4+ T cells were present in OP animals compared to bolus RMs (Fig 2I–J, S2B). The Env-specific CD4+ T cell response enhancement was selective to Env-specific GC-TFH cells (Fig 2K–M). Thus, OP immunization elicited an immune response that generated substantially more Env-specific GC-TFH cells, commensurate with the development of significantly higher frequencies of high affinity Env-specific BGC cells.
Slow release immunization enhanced humoral responses
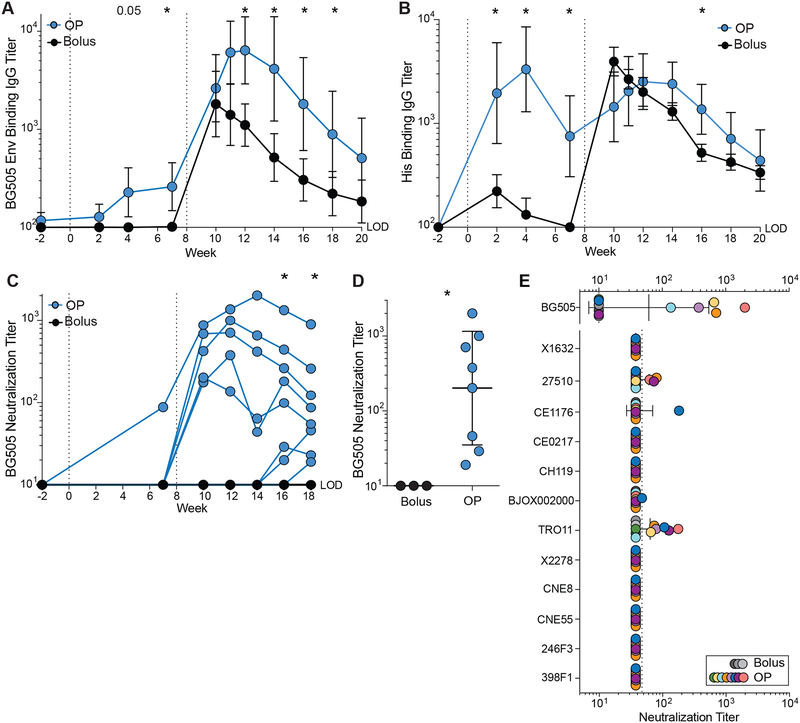
Antibody responses to the immunization approaches were examined, in light of the differences in GC responses. A single bolus immunization failed to elicit detectable BG505 Env trimer-specific plasma IgG titers (Fig 3A, S2C–D). In contrast, a single OP immunization elicited modest but significant Env-specific plasma IgG titers (w7, p = 0.048). The Olio6CD4ko Env trimer design included a His tag, which elicited a strong Ab response after one OP immunization (Fig 3B, S2E–F), while bolus RMs made anti-His IgG after the booster immunization. A fraction of the Env-specific B and BGC cells likely recognized the His tag epitope. The 2nd Env trimer immunization induced anamnestic Env IgG responses in both groups, with OP outperforming bolus immunization (Fig 3A). Env-specific IgG titers increased in response to the 2nd OP immunization prior to the end-of-regimen bolus injection, demonstrating that OP immunization alone was sufficient for substantial anamnestic BPC development (w7 vs w10, p = 0.008). Env-binding IgG titers between bolus groups and between OP groups were similar to the previous RM study (Fig S2G).
Figure 3. Sustained delivery immunization induces higher nAb titers.
(A) BG505 Env trimer IgG binding endpoint titers over time.
(B) His IgG binding endpoint titers over time.
(C) BG505 (autologous) nAb titers over time.
(D) Peak BG505 nAb titers after two immunizations.
(E) Neutralization breadth on a 12-virus panel.
GMT and ± geometric SD. Statistical significance was tested using unpaired, two-tailed Mann-Whitney U test. *p≤0.05. See also Figure S2.
To assess the development of tier 2 nAb titers over time, sera were tested for autologous neutralization of BG505 N332 pseudovirus using TZM-bl neutralization assays (Fig 3C–D, S2H–J). By w10, 5/8 OP animals developed nAbs in contrast to 0/3 bolus animals (1:99 vs < 1:10 geometric mean titer [GMT]). All OP RMs developed nAbs by w18. Peak neutralization titers of OP RMs were ~20-fold higher than bolus animals (1:202 vs 1:10 GMT, p = 0.01). Neutralization breadth was assessed using a 12Avirus panel of tier 2 isolates. 6/8 OP RMs demonstrated partial neutralization breadth, neutralizing one to three heterologous isolates (Fig 3E, S2K). No heterologous nAbs were detected in bolus animals. Thus, slow release immunization enhanced the magnitude and quality of the Ab response to Env immunization, which was associated with the enhanced Env-specific BGC and GC-TFH cell responses.
Slow delivery immunization alters the antigen-specific B cell repertoire
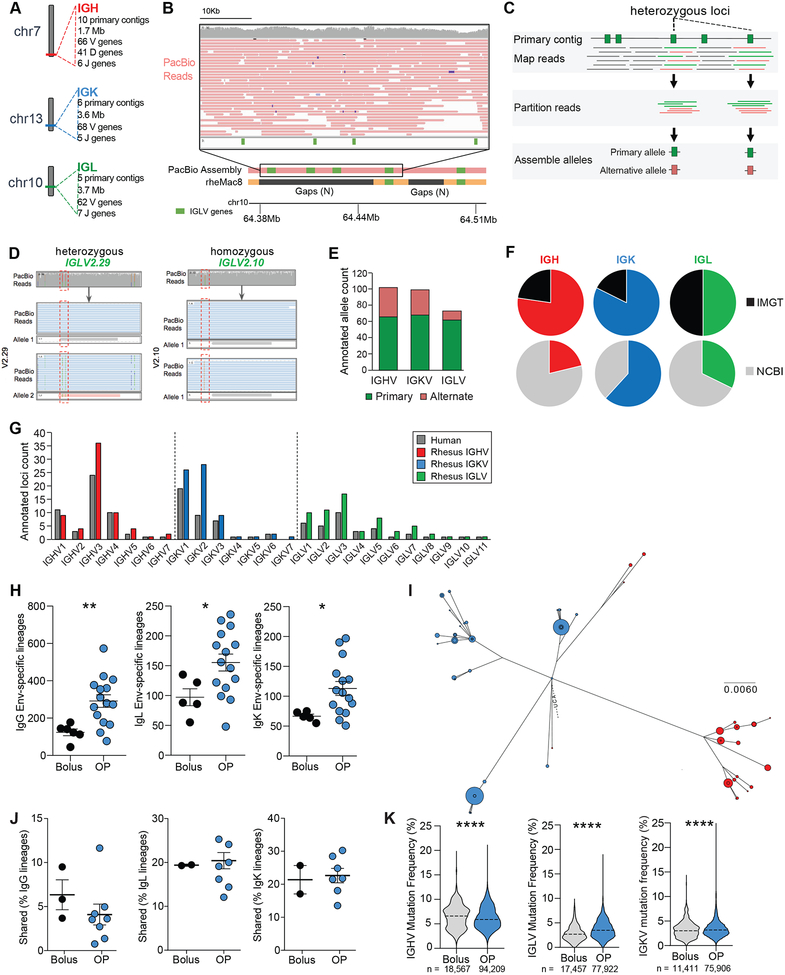
Because of the higher frequencies of high-affinity B cells and nAb titers observed in the OP animals, we hypothesized that slow release immunization delivery may affect several aspects of B cell responses. Firstly, slow delivery immunization may activate (direct effect) or recruit (via T cell help) more diverse B cell lineages. Inclusion of more independent B cell lineages would increase the likelihood that B cells with rarer and/or lower-affinity BCRs capable of developing into nAbs will be expanded. Secondly, slow delivery may increase immune complex formation, impacting GC development and maintenance. Thirdly, slow delivery may result in the generation of more BMem capable of re-circulating and reseeding new GCs among multiple LNs upon booster immunization. Finally, slow delivery immunization may drive higher rates of SHM. A major technical challenge for testing several of these hypotheses was the lack of a complete reference sequence of the RM immunoglobulin (Ig) gene loci, which is required for proper B cell lineage assignment and identification of authentic SHM. While a RM genome sequence was available (Gibbs et al., 2007), the Ig genes were largely unmapped. Ig genes reside within highly complex genomic regions that are characterized by high levels of repetitive sequences and inter-individual haplotype variation, which make genomic characterization and Ig gene annotation challenging (Watson and Breden, 2012; Watson et al., 2017). Most next generation sequencing techniques use short read technologies (~150bp), which can be insufficient for resolving large (>15kb) repetitive segments (Alkan et al., 2011). Therefore, we sequenced the genome of a RM using Pacific Biosciences long-read sequencing technology to 60-fold coverage. Overall, reads had a median length of 16.6kb and a maximum individual read length of 69.4kb. Genome assembly was conducted using FALCON/FALCON-Unzip, resulting in a total of 1,633 primary contigs with a median length of 8.4mb (2.83gb total bases).
Contigs containing the Ig loci were identified, and V, D, and J genes were annotated via a combination of bioinformatics and manual curation (Fig 4A). 66 IGHV, 41 IGHD, 6 IGHJ, 68 IGKV, 5 IGKJ, 62 IGLV, and 7 IGLJ genes were identified by focusing on gene segments with open reading frames (ORFs; Fig 4A). Notably, the long reads allowed for the characterization of regions that were unresolved in previous assemblies, including the current RM reference genome (rheMac8), facilitating identification of novel gene loci (Fig 4B). It was also possible to identify heterozygous allelic variants at loci identified in primary contigs by using a combination of raw read data and alternate contigs from FALCON-Unzip (Fig 4C–E). We determined that 37/66 IGHV, 31/68 IGKV, and 12/65 IGLV genes were heterozygous, amounting to a germline database of 103, 99, and 77 V alleles for each respective locus (Fig 4E, Table S1). Sequencing BCRs of mature B cells from the same animal and close relatives supported the presence of these annotated ORF sequences (data not shown). A significant fraction of alleles identified in the assemblies were not represented by sequences in either the IMGT database or NCBI Ig gene repositories, highlighting the utility of this approach for improving upon existing genomic databases (Fig 4F). Ig gene family sizes are comparable between human and RM (Fig 4G).
Figure 4. Immunoglobulin gene germline annotations using long-read genomic DNA sequencing.
(A) Locus and assembly summaries for the RM Ig loci.
(B) A representative region where PacBio primary contigs resolved gaps in the current RM reference genome. PacBio reads span these gaps (inset).
(C) Overview of V gene allelic variant discovery process. Reads overlapping annotations on primary contigs were assessed for the presence of SNPs, which were used to partition reads for allele-specific assemblies.
(D) SNPs (e.g., green and red) within or near genes (red boxes) were used to partition reads to each respective haplotype, allowing for the identification of heterozygous (pink) and homozygous (grey) gene segments.
(E) Primary and alternate contig alleles.
(F) Variable (V) genes from PacBio assembly that were present in IMGT or NCBI V gene repositories.
(G) V gene counts from PacBio primary contig assemblies, compared to human gene loci.
(H) Quantification of Env-specific B cells lineages from individual LNs.
(I) Phylogenetic analysis of a lineage found in both LNs in one animal. Blue, left LN; Red, right LN. Dot size represents number of reads with that sequence.
(J) Lineages shared between R and L LNs within an animal.
(K) Mutation frequencies in IGHV, IGLV, or IGKV. Violin plots; Dash = mean.
Mean ± SEM; statistical significance in H and J tested using unpaired, two-tailed Mann-Whitney U test. Significance in K tested using Student’s t-test. *p≤ 0.05, **p≤0.01, ***p≤0.001
To assess how slow delivery immunization affected the Env-specific B cell repertoire, we isolated and sequenced BCRs from Env-specific B cells in the biopsied draining LNs after the 2nd immunization (Fig 2A, S3A). The majority of the sequenced Env-specific B cells were BGC (77%), providing a window into this difficult-to-study cell type (Table S2). More Env-specific B cells were isolated from OP animals compared to bolus RMs (303,644 v 52,302 cells [total]; 20,242 v 8,717 cells [mean], p = 0.029. Data S1), consistent with the higher frequencies identified by flow cytometry (Fig 1–2). Utilizing the new RM Ig genomic reference, we assigned each unique Env-specific BCR sequence to the V and J genes with most similarity, performed lineage analysis, and determined SHMs. Larger numbers of Env-specific BCR sequences were isolated from OP animals than bolus animals, both for heavy chain (IgG: 94,209 v 18,567 [total]; 5,691 v 2,846 [mean]) and light chains (IgL: 77,922 v 17,457 [total]; 5,240 v 3,643 [mean]. IgK: 75,906 v 11,411 [total]; 4,827 v 2,261 [mean]). Furthermore, significantly more unique IgG, IgL and IgK B cell lineages were identified in LNs of OP animals compared to bolus-immunized animals (Fig 4H, S3B). Clonal abundance and Shannon H index analyses confirmed increased clonal diversity in pump animals (Fig S3C–D). While most BCR lineages were found in only one LN (Fig S3E), 0.7 – 30.2% were found in both R and L LNs (Fig 4I–J). SHM rates in V genes and across the BCR were largely similar between groups, as were CDR3 lengths (Fig 4K, S3F–G). Thus, substantially more Env-specific B cell lineages were recruited and sustained in animals receiving a slow release immunization, while SHM rates were comparable.
Slow delivery immunization results in greater Ab diversity
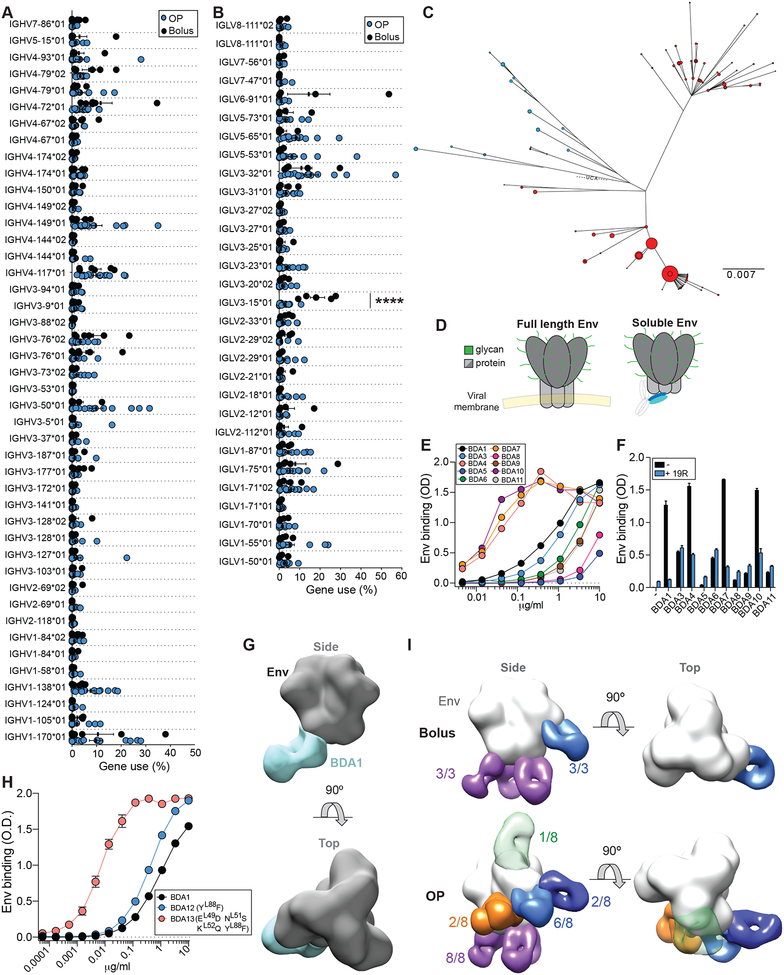
Given that slow delivery immunization resulted in more Env-specific B cell lineage diversity, we sought to determine if differential IgV gene usage occurred, which may suggest differences in the epitopes targeted on the Env trimer. Bolus RMs utilized IGLV3–15*01 (IGLV3-15) significantly more frequently than OP RMs (q = 0.00003. Fig 5A–B, S4A). 18.75% and 2.6% of Env-specific B cells from LNs of bolus and OP animals, respectively, utilized IGLV3-15. Using IMGT for similar analysis, a difference in IGLV3-15 (aka IGLV3-10*01) was also identified between groups (Fig S4B–D). Env-specific B cells that used IGLV3-15 were phylogenetically diverse (bolus, 11.4 mean per LN; OP, 6.4) and could be found in both draining LNs within a single animal (Fig 5C).
Figure 5. Slow delivery immunization shifts immunodominance.
(A) IGHV or (B) IGLV use by antigen-specific B cells within a LN. Each data point is single LN. Statistical significance tested using multiple t-tests with FDR = 5%; ****q<0.0001.
(C) Phylogenetic tree of an IGLV3-15+ lineage. Blue, left LN; Red, right LN. Dot size represents number of reads with that sequence.
(D) The base of Env is hidden on the virion surface. Soluble trimer allows access of the base to B cells. Glycans restrict access to the main Env trimer surface.
(E) Binding curves of mAbs isolated from w7 to BG505 Env trimer.
(F) Cross-competition ELISA assay. Data is representative of two experiments, each performed in duplicate.
(G) 3D EM reconstruction of BDA1 Fab (blue) in complex with BG505 Env trimer.
(H) Binding curves of BDA1 and related mutants to BG505 Env trimer. BDA12 has a single w12 mutation in L-CDR3. BDA13 contains this mutation and three additional w12 mutations in L-CDR2. Data is representative of two experiments, each performed in duplicate.
(I) Composite 3D reconstruction of Env trimer bound to Fabs isolated from all animals as determined by polyclonal EM analysis. Numbers of individuals with Fab that binds region are listed. Base (purple), glycan hole-I (light blue), C3/V5 (dark blue), fusion peptide (orange), V1/V3 apex (green). Apex specific Fab is transparent to represent rarity.
Mean ± SEM. See also Figures S4–6 and Table S3.
The differential use of IGLV3-15 suggested that the Env-specific B cells elicited by bolus immunization preferentially targeted epitopes distinct from the Env-specific B cells elicited by OP immunization. Taken together with the lack of nAbs in the bolus animals, we hypothesized that B cells that utilized IGLV3-15 recognized the base of the trimer. This region is normally hidden on full length Env expressed on HIV virions. In contrast, the base is the largest proteinaceous region exposed on soluble Env trimer, due to the unusually dense glycans covering most of the remainder of the surface of Env (Stewart-Jones et al., 2016) (Fig 5D). The base is a major non-neutralizing Ab target in mice and RMs immunized with soluble Env trimer, and base-specific B cells are proposed to be immunodominant to nAb-epitope-specific B cells (Havenar-Daughton et al., 2017; Hu et al., 2015; Kulp et al., 2017). To test this hypothesis, we sequenced Env-specific single B cells from the draining LNs of two bolus immunized animals at w7 to obtain paired BCR sequences utilizing IGLV3-15. We synthesized a panel of IGLV3-15+ mAbs, termed BDA1-11, representing 11 unique B cell lineages (Table S3). Almost all of the IGLV3-15+ mAbs bound BG505 Env trimer, but not monomeric BG505 gp120 or His peptide (Figure 5E, S5A–C). Binding of several BDA mAbs to Env was selectively blocked by 19R, a high affinity Env base-binding mAb, demonstrating that IGLV3-15+ Abs recognize the Env trimer base (Figure 5F). EM analysis of a BDA mAb Fab complex with Env trimer confirmed binding of BDA1 to the trimer base (Figure 5G). We next sought to determine how BDA1 was related to the Env-specific B cells isolated from the same LN after booster immunization with Env trimer (w12). Alignment and phylogenetic analysis of the BDA1 lineage consisted of BDA1, three related w12 sequences, and the inferred germline sequence (Fig S5D–E). The BDA1 lineage heavy and light chains accumulated more mutations at w12, indicating recall GC responses of IGLV3-15+ cells and ongoing SHM. Introduction of wk12 IgL mutations into BDA1 resulted in an increase in Env binding, suggesting that the BDA1 light chain contacts the Env trimer base (Fig 5H, S5D–F).
We utilized polyclonal EM serological analysis as an independent approach to assess the Ab responses to Env trimer between the immunization strategies. This technique allows for simultaneous visualization of Abs targeting distinct epitopes from polyclonal serum (Bianchi et al., 2018). Ab responses in bolus animals targeted two sites on Env: the trimer base (3/3 animals), and the glycan hole I region (GH-1, 3/3) (Fig 5I, S6). In contrast, the Ab responses in OP animals were more diverse. In addition to the base and GH-1 regions, three potential nAb epitopes, the fusion peptide, V1/V3-glycan, and C3/V5 regions (Klasse et al., 2018; Kong et al., 2016), were targeted by OP animals. In sum, slow release immunization resulted in a substantial shift in the BGC cell and Ab response towards Env epitopes that are more diverse than those targeted by bolus animals. The shifted response was towards nAb epitopes, which are likely immunorecessive relative to the Env trimer base, indicating that slow delivery immunization modulates immunodominance or changes the immunodominance hierarchy.
Escalating dose immunization enhances germinal center and nAb responses
Escalating dose (ED) is an immunization strategy to achieve extended antigen exposure that is an approach distinct from OP administration (Tam et al., 2016). ED immunization has the added advantage of mimicking the antigen dose dynamics of an acute infection. Therefore, an RM ED study was performed with Env trimer as an independent assessment of the immunological implications of extended antigen delivery in a vaccine setting. The control group was given conventional bolus immunizations at w0, w10, and w24, totaling 100μg, 100μg, and 300μg of Olio6 native-like Env trimer protein, respectively, mixed with an ISCOMs-class adjuvant. ED immunizations were administered as 7 injections over 2 weeks (Fig 6A), with a total antigen dose equivalent to that of the conventional bolus immunization group. Significantly higher frequencies of BGC cells in draining LNs were observed at w5 in the ED group compared to the conventional bolus immunization group (Fig 6B–C, S7A). ED immunization resulted in significantly more Env-specific B and BGC cell after the 1st immunization (p = 0.0002 [AUC]. Fig 6D–E, S7B–F, Data S2). Bolus immunized animals had significantly higher frequencies of Env-specific BGC than ED animals after the 2nd immunization. These frequencies were comparable after the 3rd immunization. ED immunization elicited improved affinity maturation, as indicated by the enhanced development of Envhi BGC cells compared to conventional immunization after the 1st immunization (Fig S7G–L). Additionally, ED immunization resulted in significantly more Env-specific BMem cells compared to conventional immunization after the 1st immunization (Fig S7M–P). Total BGC frequencies, and Env-specific BGC and BMem cell frequencies increased upon the 2nd and 3rd ED immunizations, though not above the peak frequencies observed in response to the 1st ED regimen. Analysis of CD4+ T cells in the draining LNs revealed that ED resulted in significantly higher total GC-TFH and Env-specific GC-TFH cells after the 1st immunization (Fig 6F–H, S7Q–R). ED immunized animals showed a higher ratio of Env+ BGC: Env-specific GC-TFH cells, suggesting that ED immunization resulted in greater antigen-specific help to B cells than conventional immunization (Fig 6I). The magnitude of the improved primary Env-specific BGC cell response, the increased GC-TFH cell response, and the enhanced Envhi BGC cells upon ED immunization were comparable to those observed after OP immunization.
Figure 6. Dose escalating immunization strategy results in higher nAb titers.
(A) Immunization and sampling schedule. Groups were immunized and sampled contemporaneously.
(B) Total BGC frequencies over time. Data from bolus Gp2 (Fig 1) are included in these analyses (grey circles).
(C) Cumulative BGC cell response to the first immunization, calculated between w3-7 [AUC].
(D) Env trimer-specific BGC cells frequencies over time.
(E) Cumulative Env trimer-specific BGC cell responses to one immunization.
(F) Total GC-TFH cell frequencies over time.
(G) Cumulative GC-TFH responses to one immunization (AUC of F).
(H) Env-specific CD4+ responses after one immunization.
(I) Ratio of Env+ BGC to Env-specific GC-TFH cells at w5, calculated as Env+ BGC (% B cells)/ Env-specific GC-TFH (% CD4+).
(J) BG505 Env trimer binding IgG titers over time.
(K) Autologous BG505 nAb titers over time.
(L) Peak BG505 nAb titers after three immunizations.
(M) Composite 3D reconstruction of Env trimer bound to Fabs isolated from all animals after two immunizations. 3D EM reconstructions from individual animals can be seen in Figure S8A.
(N) Correlation between peak GC-TFH and BGC cell % during 1st immunization from both studies.
(O) Correlation between Env+ BGC cells (% B cells) and peak neutralization titers. Env+ BGC cell values are from w7 or peak frequencies during 1st immunization. Peak nAb titers are after 2nd immunization.
Serological data represent GMT ± geometric SD. Cell-frequency data represent mean ± SEM. Statistical significance was tested using unpaired, two-tailed Mann-Whitney U tests. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See also Figure S7–8 and Data S2.
A single ED immunization regimen was sufficient to elicit a BG505 Env-specific IgG response (Fig 6J). Anamnestic Env-binding plasma Ab responses were observed after the 2nd and 3rd ED and conventional immunizations. All ED immunized animals developed autologous tier 2 nAbs after the 2nd immunization, while only 3/6 conventionally immunized animals developed nAbs (Fig 6K). Peak autologous nAb titers after the 3rd immunization were ~30-fold higher in ED RMs, significantly greater than bolus animals (1:615 vs 1:18 GMT, p = 0.009. Fig 6L; heterologous nAb breadth in Fig S7S).
Bolus animals targeted three regions on Env (base, GH-I and fusion peptide), while ED immunized animals targeted four (base, GH-I, C3/V5-I and II) (Fig 6M). 6/6 ED animals had Ab responses against multiple regions of Env, while 2/6 bolus animals targeted a single site. (Fig 6M, S8A). Thus, ED immunization resulted in greater Ab diversity.
Total GC-TFH cell frequencies correlated with total BGC frequencies during the 1st immunization (r = 0.773, p = < 0.0001 [peak of 1st immunization], Fig 6N). In a previous study, total BGC frequencies correlated with nAb development (Pauthner et al., 2017). A primary hypothesis of this study was that the magnitude of Env-specific BGC cells to the 1st immunization might predict autologous nAb development. Peak Env-specific BGC frequencies to the 1st immunization correlated with peak autologous nAb titers in response to the 2nd immunization (r = 0.673, p = 0.0008 [w7]; r = 0.596, p = 0.0027 [peak of 1st immunization]. Fig 6O), indicating that Env-specific BGC and GC-TFH cell responses can predict subsequent nAb development.
Taken together, the data show that the ED immunization modality generated greater GC and humoral responses than dose-matched conventional immunization, recapitulating the immune responses elicited to OP immunization, indicating that modulation of BGC and TFH cell responses is a general property of slow delivery immunization strategies, which can result in dramatically different B cell specificities and nAb development.
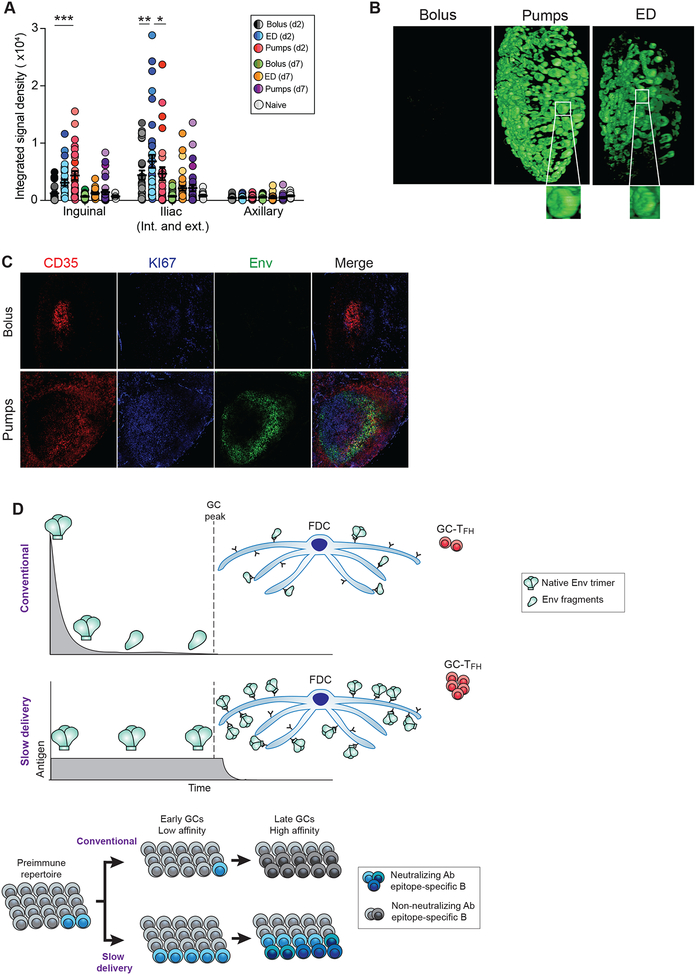
An ED regimen resulted in enhanced FDC deposition of antigen in mice (Tam et al., 2016). We hypothesized that the enhanced GC responses observed in Env trimer immunized RMs with both slow delivery immunization modalities were, at least in part, due to increased availability of antigen to BGC and GC-TFH cells (Cirelli and Crotty, 2017). An in vivo antigen tracking study was performed with fluorescently labeled Env trimer and ISCOMs-class adjuvant administered via a conventional bolus, 2w OP, or an ED regimen. LNs of OP- and ED-immunized animals contained significantly more Env trimer at d2 as measured by laser scanning imaging (Fig 7A, S8B). Light sheet microscopy of whole LNs revealed substantially more Env-containing B cell follicles in animals immunized with either slow delivery regimen (Fig 7B, Movie S1). Histological analyses of LNs confirmed Env colocalized with FDCs and GCA adjacent cells after OP or ED immunization (Fig 7C, S8C). Thus, slow delivery immunization leads to enhanced antigen retention within LNs in NHPs.
Figure 7. Slow delivery immunization results in enhanced antigen retention in LNs.
(A) Quantitation of fluorescent Env in draining inguinal and iliac and non-draining axillary LNs. Mean ± SEM; statistical significance was tested using two-way ANOVA. *adjusted p <0.05, **p<0.01, ***p<0.001
(B) Light sheet microscopy visualizing Env in intact draining LNs at d2. 360° views available in video S1.
(C) Histology of draining LNs at d7. Green, Env; red, CD35; blue, KI67. Scale bars, 100μm.
(D) Model of GC response in conventional immunization vs. slow delivery. Slow delivery immunization likely alters early (~d1-d7) activation and differentiation of TFH cells and activation and recruitment of a diverse set of B cells. Greater GC-TFH help supports a wider repertoire of B cells, which is more likely to contain nAb precursors, later in the response (w3–7). Antigen delivered via conventional bolus immunization can be subject to degradative processes and nonnative forms of antigen can be presented by FDCs late in the response, while OPs protect the antigen prior to release. Immune complex (IC) formation is enhanced by slow delivery immunization. See also Figure S8 and Movie S1.
DISCUSSION
Understanding the underlying immunological challenges to nAb development against difficult pathogens may be important for understanding why protective immunity to such pathogens is elusive. Direct examination of primary immune responses in lymphoid tissue is required to develop such an understanding. Strategies to enhance humoral and GC responses to immunization are likely needed for the development of vaccines against some complex pathogens, particularly HIV. Using two independent methods, we have demonstrated that slow delivery immunization resulted in enhanced autologous tier 2 nAb development in NHPs. We found several aspects of GC biology were affected by slow delivery immunization. OP and ED immunization induced higher frequencies of total and Env-specific GC-TFH cells. Greater availability of GC-TFH cell help and antigen to B cells was accompanied by larger and more enduring BGC responses. Both slow delivery strategies resulted in substantially more Env-specific BGC cells. The BGC cells were more diverse, as defined by unique Env-specific B cell lineages, which may be a consequence of broader initial activation of antigen-specific B cells and/or of sustaining larger GCs over time. The biological relevance of those processes is reinforced by the observation of more diverse nAb Env-binding specificities generated in slow delivery immunized RMs compared with conventional bolus immunization.
To examine the immune responses directly in the draining LNs, we employed weekly LN FNAs. From this, we found that bolus immunization elicited a robust GC response, but that slow delivery immunization altered the kinetics and overall magnitude of the GC response. Larger Env-specific B cell responses during the primary immunization were positively correlated with the larger nAb response that subsequently developed in OP and ED immunized animals, suggesting that much of the failure of a bolus immunization to a difficult antigen is intrinsic to early B cell events associated with immunodominance features of multi-epitope complex antigens. Strikingly, slow delivery modulated the immunodominance of the B cell response to non-neutralizing epitopes on Env trimer. OP and bolus RMs had comparable Env-binding ELISA titers at w10. Nevertheless, OP animals had considerably higher autologous nAb titers at that same timepoint. This was also observed between ED and bolus RMs at w11 and w25. These data strongly suggested that the composition of the Ab response was altered by slow delivery, such that nAbs were a significantly greater fraction of the responses, The data show that slow delivery immunization does not simply elicit a larger total Ab response.
Nearly 20% of Env trimer-specific B cells in bolus immunized animals were IGLV3-15+. Several IGLV3-15+ Abs targeted the non-neutralizing base of the Env trimer. The base is a major site recognized by Abs of soluble trimer immunized animals. The trimer base appears to be immunodominant because it is a large exposed protein surface with many potential epitopes and acceptable BCR angles of approach when compared to the other surfaces of Env trimer, which are predominantly shielded by large glycans (Havenar-Daughton et al., 2017). Taken together, the differences in the Env-specific B cell repertoire, nAb titers, and the polyclonal Ab EM mapping demonstrate substantial immunodominance of non-neutralizing B cells that outcompete B cells specific for neutralizing epitopes after a conventional bolus injection.
Slow delivery immunization altered the repertoire of the responding Env-specific B cells and the range of Ab specificities and nAb specificities. The simplest explanation for this outcome is that slow delivery increases the likelihood that rare and/or lower affinity immunorecessive nAb precursors are recruited into the B cell response, resulting in more diversity in the epitopes targeted among the BGC cells. We reiterate that the antigen dose was equal between the bolus and OP or ED animals, thus total dose is not the driver of these differential outcomes. It has been reported that naive antigen-specific B cells normally have a narrow window of time of a few days to be recruited into a GC response (Turner et al., 2017). A narrow window of time for B cell recruitment disproportionately affects B cells with rare precursor frequencies. Slow delivery immunization may substantially expand the pool of recruited B cells by extending that window, thereby increasing the breadth of the B cell repertoire sampled by the draining LN. Additionally, TFH selection of B cells based on affinity may be most stringent prior to the GC response (Schwickert et al., 2011; Yeh et al., 2018); slow antigen delivery may reduce that stringency by substantially broadening the time window for TFH interactions with Env-specific B cells of differing epitope specificities at the border of the follicle. The diversity of the B cell response would then likely be greater at the end of the immunization.
Despite a considerable apparent difference in affinity maturation (Envhi BGC and BMem cells), SHM rates were largely equivalent between OP and bolus groups at the times measured. The data suggest that differential rates of SHM were not the cause of the improved affinity maturation and improved nAb responses. A study examining SHM in BMem after RM immunizations with non-native Env trimers and a range of adjuvants (which did not elicit tier 2 nAbs) did not observe differences in SHM between groups (Francica et al., 2015). The SHM data herein are consistent with the model where the primary cause of the difference in the nAb outcomes was the altered immunodominance profile of the B cell response.
A small fraction of Env-specific B cells from OP RMs utilized IGLV3.15, but Abs isolated from OP animals still targeted the base, consistent with diverse epitopes accessible on the base allowing BCRs targeting this site utilizing diverse IGHV and IGLV genes. While the Env trimer base was exposed in both contexts, differences in epitope accessibility on the Env trimer may exist between immunization strategies (Fig 7D). Immune complexes (ICs), composed of Env trimer and Abs, are bound by FDCs for presentation to B cells. Binding of an Ab to its cognate epitope, however, may block access to that epitope by BGC cells undergoing selection. We speculate that a large fraction of the early Ab response targets the base. During a slow delivery immunization, early base-specific non-neutralizing Abs may form ICs with newly available Env trimers, enhancing presentation on FDCs and possibly increasing the likelihood that nAb epitope-specific BGC cells will be selected for survival due to increased antigen availability in the GC and the orientation of the Env trimer on FDCs occluding the base.
We predicted that slow release immunization would reduce the B cell response to protein breakdown products and fragments that occur in vivo, such as the internal face of gp120 and V3, by protecting the antigen in its native form, thus having a greater percentage of intact Env trimer antigen on FDCs at 2A10 weeks postimmunization (Cirelli and Crotty, 2017; Tam et al., 2016). BGC cell responses to breakdown products are surely present in each immunization group. These cells likely make up a substantial fraction of the ‘dark antigen’ GC response (Kuraoka et al., 2016), and may have immunodominant specificities, as a majority of BGC cells did not bind intact native Env trimer with measurable affinity (Fig 1I, S7E). Adjuvant alone does not induce a BGC cell response, consistent with the conclusion that the BGC cells elicited in these immunizations are predominantly specific for Env (Havenar-Daughton et al., 2016a). While those specificities are of interest, experiments here focused on the Env trimer-binding B cells, due to limited cell numbers per sample.
Despite their enhanced and more diverse responses to immunization, slow delivery immunized animals still targeted non-neutralizing epitopes. Immunogens should therefore be optimized to focus the response towards neutralizing epitopes and minimize responses against non-neutralizing epitopes. OPs have been used in humans for drug delivery and are feasible for early human vaccine trials. However, Ops are impractical for large-scale vaccination efforts, as immunization requires a simple surgery. Nevertheless, ED is technically available immediately as a GC enhancing alternative to conventional bolus immunization. Less cumbersome slow delivery immunization technologies are worthy of further development, including degradable encapsulating biomaterials and depot forming adjuvants that make antigen available over time (i.e., not rendered inert in the depot) in ways that sustain GCs (DeMuth et al., 2013; 2014). Such technologies may be able to rescue protective immune responses to antigens that have previously failed by conventional bolus immunization, if immunodominance of non-neutralizing epitopes was a factor in their failure.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shane Crotty (shane@lji.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rhesus Macaques
Outbred Indian RMs (Macaca mulatta) were sourced and housed at the Yerkes National Primate Research Center and maintained in accordance with NIH guidelines. This study was approved by the Emory University Institutional Animal Care and Use Committee (IACUC). When osmotic pumps were implanted, animals were kept in single, protected contact housing. At all other times, animals were kept in paired housing. Animals were treated with anesthesia and analgesics for procedures as per veterinarian recommendations and IACUC approved protocols. In all studies, animals were grouped to divide age, weight and gender as evenly as possible.
OP study: Animals were between 2.5 – 3 years of age at time of 1st immunization. Bolus group 2: 2 males (M), 1 female (F); 2w OP group: 3M, 1F; 4w OP group: 3M, 1F.
ED study: Animals were between 3 – 6.5 years of age at time of 1st immunization. Bolus group 1: 3M, 3F; ED group: 2M, 4F.
Antigen tracking study: animals were between 3 – 6 years of age at time of immunization. Bolus group: 2M, 1F; OP group: 3M; ED group: 3M.
METHOD DETAILS
Immunizations
Osmotic pump study: Animals were immunized at 2 time points: week 0 and week 8. All immunizations were administered subcutaneously (SubQ) divided between the left and right mid-thighs. Bolus animals were given two SubQ injections of 50μg of Olio6CD4ko + 187.5 units (U) of saponin adjuvant in PBS, for a total of 100μg Olio6CD4ko trimer protein + 375U of saponin adjuvant. At week 0, osmotic pumps (Alzet, models −2002 and −2004) were loaded with 50μg Olio6CD4ko + 187.5U saponin adjuvant, for a total of 100μg Olio6CD4ko trimer + 375U of saponin adjuvant. Pumps were implanted SubQ in the same location as bolus immunizations. At week 8, osmotic pump animals were immunized with osmotic pumps loaded each with 25μg Olio6CD4ko + 93.75U saponin adjuvant. At the end of the osmotic pump delivery, a SubQ bolus immunization of 25μg Olio6CD4ko + 93.75U was given in each leg, totaling 50μg Olio6CD4ko + 187.5U saponin adjuvant at weeks 12 and 14 for 2 week and 4 week osmotic pump groups, respectively. Each pump was loaded with Olio6CD4ko and saponin adjuvant in a total volume of 200ul. During the 1st immunization, each 2w and 4w OP released 3.57ug Olio6CD4ko + 13.4U saponin adjuvant and 1.78ug Olio6CD4ko + 6.7U saponin adjuvant per day, respectively. During the 2nd immunization, each 2w and 4w OP released 1.78ug Olio6CD4ko + 6.7U saponin and 0.89ug Olio6CD4ko + 3.35U saponin adjuvant per day, respectively.
Dose escalation study: Animals were immunized at 3 time points: weeks 0, 10, and 24. All immunizations were administered SubQ in the left and right mid-thighs. Bolus animals were given two injections of 50μg of Olio6 + 187.5U of saponin adjuvant in PBS, for total of 100μg immunogen and 375U saponin adjuvant at weeks 0 and 8. At week 24, two injections of 150μg of Olio6 + 187.5U saponin adjuvant were administered for a total of 300μg Olio6 + 375U saponin adjuvant. For each immunization, escalating dose animals were given seven injections of Olio6 and saponin adjuvant in each thigh over 12 days (on days 0, 2, 4, 6, 8, 10, 12 for each immunization). The total doses of Olio6 at each injection during the first two immunizations were: 0.2, 0.43, 1.16, 3.15, 8.56, 23.3, 63.2μg (the doses per immunization site were 0.1, 0.215, 0.58, 1.575, 4.28, 11.65, 31.6μg). The total doses of Olio6 at each injection during the third immunization were: 0.6, 1.29, 3.48, 9.45, 25.68, 69.9, 189.6μg (the doses per immunization site were 0.3, 0.645, 1.74, 4.725, 12.84, 34.95, 94.8μg). The total doses of saponin adjuvant at each injection during all immunizations were: 0.75, 1.61, 4.35, 11.81, 32.1, 87.38, 237.0U (the doses per immunization site were 0.375, 0.805, 2.175, 5.905, 16.05, 43.69, 118.5U).
Antigen tracking study: Animals were immunized at week 0 with a total dose of 100ug untagged MD39 conjugated to Alexa Fluor 647. All immunizations were administered SubQ in the left and right mid-thighs. Bolus animals were given 2 injections of 50μg MD39 + 187.5U of saponin adjuvant in PBS. Osmotic pumps (Alzet, models 2002) were loaded with 50μg MD39 + 187.5U saponin adjuvant. Escalating dose animals were given a series of 7 injections over 12 days (on days 0, 2, 4, 6, 8, 10, 12 for each immunization). The total dose of MD39 at each injection were: 0.2, 0.43, 1.16, 3.15, 8.56, 23.3, 63.2μg (the doses per immunization site were 0.1, 0.215, 0.58, 1.575, 4.28, 11.65, 31.6μg). The total doses of saponin adjuvant at each injection were: 0.75, 1.61, 4.35, 11.81, 32.1, 87.38, 237.0U (the doses per immunization site were 0.375, 0.805, 2.175, 5.905, 16.05, 43.69, 118.5U). Animals were sacrificed at 2 or 7 days (3 animals per group per day) after immunization (bolus, d2 and d7; pumps, d16 and d21; escalating dose, d14 and d19). All inguinal LNs were harvested and fixed in PLP buffer (pH7.4 50mM PBS + 100mM lysine, 1% paraformaldehyde, 2mg/mL sodium periodate) for 1 week at 4°C and then washed and stored in PBS with 0.05% sodium azide at 4°C until used for imaging.
Lymph node fine needle aspirates, whole LN biopsy tissue, blood collection and processing
LN FNAs were used to sample at both right and left inguinal LNs. FNAs were performed by a veterinarian. Draining lymph nodes were identified by palpitation. Cells were collected by passing a 22A gauge needle attached to a 3mL syringe into the lymph node 4 times. Samples were expelled into RPMI containing 10% fetal bovine serum, 1X penicillin/streptomycin. Samples were centrifuged and Ammonium-Chloride-Potassium (ACK) lysing buffer was used if sample was contaminated with red blood cells. Excisional LNs were conducted at weeks 12 (bolus) or 14 (osmotic pump groups). LNs were dissociated through 70μM strainers and washed with PBS. Blood was collected at various time points into CPT tubes for PBMC and plasma isolation. Serum was isolated using serum collection tubes and frozen.
ISCOMs-class saponin adjuvant
The adjuvant used for all the described studies was a ISCOM-like saponin nanoparticle comprised of self-assembled cholesterol phospholipid, and quillaja saponin prepared as previously described (Plotkin, 2010). Briefly, 10 mg each of cholesterol (Avanti Polar Lipids) and DPPC (Avanti Polar Lipids) were dissolved separately in 20% MEGA-10 (Sigma-Aldrich) detergent at a final concentration of 20 mg/mL and 50 mg Quil-A saponin (InvivoGen) was dissolved in MilliQ H2O at a final concentration of 100 mg/mL. Next, DPPC solution was added to cholesterol followed by addition of Quil-A saponin in rapid succession and the volume was brought up with PBS for a final concentration of 1 mg/mL cholesterol and 2% MEGA-10. The solution was allowed to equilibrate at 25°C overnight, followed by 5 days of dialysis against PBS using a 10k MWCO membrane. The adjuvant solution was filter sterilized using a 0.2 μm Supor syringe filter, concentrated using 50k MWCO Centricon filters, and further purified by FPLC using a Sephacryl S-500 HR size exclusion column. Each adjuvant batch was finally characterized by negative stain transmission electron microscopy (TEM) and dynamic light scattering (DLS) to confirm uniform morphology and size and validated for low endotoxin content by Limulus Amebocyte Lysate assay (Lonza). Final adjuvant concentration was determined by cholesterol quantification (Sigma-Aldrich).
Immunogen and probe generation
Olio6, Olio6CD4ko, and MD39 Env trimers were generated as previously described. Avi-tagged Olio6, Olio6CD4ko, and MD39 DNA constructs were synthesized, protein was produced and purified, and the proteins were then biotinylated using BirA-500 (Avidity) and assessed for biotin conjugation efficiency using SDS-PAGE. All Env immunogens and probes contained a six histidine tag (His tag) for purification. Immunogens were tested for endotoxin contamination with Endosafe PTS (Charles River). Proteins with an endotoxin level <10 EU/mg were used in immunizations. Immunogens and probes were aliquoted and kept frozen at -80°C until immediately before use.
Flow cytometry and cellular analyses
Biotinylated protein were individually premixed with fluorochrome-conjugated streptavidin (SA-Alexa Fluor 647 [Ax647] or SA-Brilliant Violet 421 [BV421]) at RT for 20 minutes. Olio6CD4ko probes were used in figures 1, 3, 6, and S1 (osmotic pump study) from weeks -1 to 8. Olio6 probes were used from weeks 9 to 14. Olio6 and Olio6CD4ko differ by a single amino acid . MD39 probes were used in figures 7 and S7 (dose escalation study). MD39 is closely related to Olio6.
For the full LN GC panel, cells were incubated with probes for 30 minutes at 4°C, washed twice and then incubated with surface antibodies for 30 minutes at 4°C. Cells were fixed and permeabilized for 30 minutes using FoxP3/Transcription Factor Staining Buffer Set (Thermo Scientific) according to manufacturer’s protocols. Cells were stained with intranuclear antibodies in 1X permeabilization buffer for 30 minutes, 4°C. Cells were washed twice with 1x permeabilization buffer and acquired on an LSR I (BD Biosciences). For Ag-specific B cell sort panels, cells were incubated with probes for 30 minutes at 4°C, washed twice and then incubated with surface antibodies for 30 minutes at 4°C. Cells were sorted on a FACSAria II.
For the osmotic pump study, full LN GC panel was used on fresh cells at weeks -2, 1-7, 9-12, 14. At weeks 7,12, and 14, cells were sorted using the Ag-specific B cell sort panel. Cells were stained fresh at week 7 and single cell sorted. At weeks 12 (bolus) and week 14 (osmotic pump animals), biopsied LNs were thawed, stained and bulk sorted for BR sequencing. Sorted cells were defined as Viability dye− CD4− CD8a− CD16− CD20+ (IgM+ IgG+)− Olio6-Alexa647+ Olio6-BV421+. For the dose escalation study, the full LN GC panel was used at every time point. Data reported are raw flow cytometry values at each time point.
Validation of CD38 and CD71 as surface markers of BGC cells: frozen, biopsied mesenteric LNs were used. Cells were stained as described above.
B cell analysis: LN FNA samples 3% of the LN on average. Because of the nature of the technique, some samples do not have enough cells to be included in the analyses. Generally, for GC and Env-specific B cell gating, a threshold of 1,000 and 10,000 B cells, respectively, is used. For Env-specific BGC cell gating, a threshold of 1,000 BGC cells is used.
BMem cells: BMem cells (% Env+ or Envhi) were calculated as the percentage of Env-specific or high-affinity Env-specific B cells that were not BCL6+ KI67+ or CD38− CD71+. BMem Env+ and Envhi (% B) cells were calculated as % Env+ (% B cells) − % Env+ BGC (% B) and % Envhi (% B cells) − % Envhi BGC (% B), respectively.
Area under the curve [AUC]: AUC was calculated for individual LNs. For figures 1 and S1, AUC was calculated from weeks 1, 3 to 7. Bolus gr1 did not have FNA data at week 1. For these samples, the median of the week 1 values from bolus gr2 was used. Raw values were used at other time points. For figure 2, AUC was calculated from weeks 1, 3 to 6. GC-TFH frequencies were not collected for bolus grp2, 2w pumps or 4w pump animals at week 7. For figure 3, AUC was calculated between weeks 9 and 12 because of poor cell recovery at weeks 8 and 14. For figures 7 and S7, AUC was calculated between weeks 3-7 (1st immunization) and between weeks 11-15 (2nd immunization) using raw values. Parameters used: baseline = 0; peaks less than 10% of distance from minimum to maximum y were ignored.
Antigen-specific CD4+ T cell assay
AIM assays were conducted as previously described (Dan et al., 2016; Havenar-Daughton et al., 2016b; Reiss et al., 2017).
Osmotic pump study: Frozen macaque lymph nodes from week 12 (bolus animals) or week 14 (osmotic pump animals) were thawed. Cells were treated with DNAse (Stemcell Technologies) for 15 minutes, 37°C washed and then rested for 3 hours. Cells were cultured under the following conditions: media only (RPMI containing 10% fetal bovine serum, 1X penicillin/streptomycin, 2mM L-glutamine), 5ug/mL Olio6CD4ko peptide megapool, or 1ng/mL SEB (positive control, Toxin Technology, Inc.). After 18 hours, cells were stained and acquired on FACSCelesta (BD Biosciences).
Dose escalation study: About 50% of lymphocytes are lost during the freeze-thaw process. To maximize the number of viable cells to identify Env-specific CD4+ cells, cells were shipped overnight at 4°C to LJI. Cells were centrifuged and treated with DNAse for 15 minutes, 37°C. Cells were washed, cultured for 18 hours under the conditions described above. All values reported are background subtracted ((% OX40+ 4-1BB+ CD4+ (Env-stimulated condition) – % OX40+ 4–1BB+ CD4+ (unstimulated condition)).
Whole genome sequencing and genome assembly
High molecular weight (>50kb) genomic DNA was isolated from the kidney of a perfused, female rhesus macaque. A full genome 30kb library was prepared according to manufacturer’s protocols. Sequencing was performed on a PacBio RS II (Pacific Biosciences). Genome assembly was performed using FALCON and FALCON-Unzip (Pacific Biosciences) (Chin et al., 2016). The final assembly contained 1633 contigs made up of 2.83 Gbp. The N50 contig length is 8.4Mbp, with a maximum contig length of 28.8Mbp.
Immunoglobulin loci annotation
Primary contigs from FALCON/FALCON-Unzip assemblies containing IG sequences were identified by aligning V, D, and J sequences from multiple sources, including sequences for RM and the crab-eating macaque (Macaca fascicularis) from the IMGT reference directory (http://www.imgt.org/vquest/refseqh.html) and using BLAT (Corcoran et al., 2016; Kent, 2002). Gene annotation of primary contigs was carried out in two stages: (1) rough coordinates in each contig harboring putative V, D, and J segments were identified by mapping existing sequences (i.e., those noted above for contig identification, as well as human IG D and J gene sequences from IMGT); followed by (2) manual curation, during which precise 5’ and 3’ gene segment boundaries were determined for each annotation, based on alignments to previously reported sequences, as well as the identification of flanking recombination signal sequence (RSS) heptamers within the contig assembly. Each gene annotation was assigned to a given subfamily based on the closest matching published sequence. Only ORF annotations lacking premature stop codons and/or insertion-deletions resulting in drastic frameshifts were considered.
Additional V gene allelic variants in the IGH, IGK, and IGL loci were identified by mapping PacBio raw reads back to IG-associated primary and alternate contigs from the FALCON/FALCON-Unzip assemblies using BLASR (Chaisson and Tesler, 2012). Putative heterozygous ORF genes were identified based on variants present in PacBio reads mapping to a given ORF locus (Fig 6C). To characterize putative alternate alleles, raw reads were partitioned and assembled locally at heterozygous ORFs using MsPAC (Rodriguez et al., in prep; https://bitbucket.org/oscarlr/mspac). Raw reads and assembled allelic variants were visually inspected in the context of primary and alternate FALCON/FALCON-Unzip contigs and confirmed using the Integrated Genomics Viewer (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). To classify genes/alleles annotated from PacBio assembly data as “known” or “novel”, sequences were cross-referenced with the RM IMGT reference database and publicly available sequences annoted as Ig sequences in the NCBI nucleotide collection using BLAT and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), respectively.
Bulk BCR sequencing
The protocol for rhesus macaque repertoire sequencing was obtained by courtesy of Dr. Daniel Douek, NIAID/VRC (Huang et al., 2016). Bulk Env-specific B cells were sorted into 350uL Qiagen RLT buffer. RNA was extracted using the RNeasy Micro-DNase Digest protocol (QIAGEN) on QIAcube automation platforms (Valencia, CA). Reverse transcription (RT) was performed using Clontech SMARTer cDNA template switching: 5’ CDS oligo(dT) (12 μM) was added to RNA and incubated at 72°C for 3 minutes and 4°C for at least 1 minute. The RT mastermix (5x RT Buffer (250 mM Tris-HCl (pH 8.3), 375 mM KCl, 30 mM MgCl2), Dithiothreitol, DTT (20 mM), dNTP Mix (10 mM), RNAse Out (40U/μL), SMARTer II A Oligo (12 μM), Superscript II RT (200U/μL)) was added to the reaction and incubated at 42°C for 90 minutes and 70°C for 10 minutes. First-strand cDNA was purified using AMPure XP beads (Beckman Coulter). Following RT, two PCR rounds were carried out to generate immunoglobulin amplicon libraries compatible with Illumina sequencing. All oligos were ordered from Integrated DNA Technologies. The first PCR amplification was carried out using KAPA Real-Time Library Amplification Kit (Kapa Biosciences). cDNA was combined with master mix (2X KAPA PCR Master Mix, 12 μM μL 5PIIA and 5 μL IgG/IgK/IgL Constant Primer (2 μM)). The amplification was monitored using real-time PCR and was stopped during the exponential phase. The amplified products were again purified using AMPure XP beads. A second round of PCR amplification was carried out for addition of barcodes and Illumina adapter sequences: master mix (2X KAPA PCR Master Mix 2x, SYBR Green 1:10K, Nuclease-free water), 10 μM of P5_Seq BC_XX 5PIIA, 10 μM of P7_ i7_XX IgG/IgK/IgL and were combined with amplified Immunoglobulin from the first round PCR and amplified using real-time PCR monitoring. The P5_Seq BC_XX 5PIIA primers contain a randomized stretch of four to eight random nucleotides. This was followed by purification with AMPure XP beads. A final PCR step was performed for addition of remaining Illumina adaptors by mixing master mix (2X KAPA PCR Master Mix, 10 μM P5_Graft P5_seq, Nuclease-free water), 10 μM of P7_ i7_XX IgG/IgK/IgL oligo and amplified products from the previous PCR step followed by purification with AMPure XP beads. The quality of library was assessed using Agilent Bioanalyzer. The amplicon libraries were pooled and sequenced on an Illumina MiSeq as a 309 paired-end run.
Single cell RNA-seq
Single cells were sorted by flow cytometry into 10 uL of QIAGEN RLT buffer. RNA was purified using RNACleanXP Solid Phase Reversible Immobilization (SPRI) beads (Beckman Coulter). Full-length cDNA amplification of single-cells was performed using a modified version of the SMART-Seq II protocol (Picelli et al., 2014), as described previously (Upadhyay et al., 2018). Amplified cDNA was fragmented using Illumina Nextera XT DNA Library Preparation kits and dual-indexed barcodes were added to each sample. Libraries were validated using an Agilent 4200 Tapestation, pooled, and sequenced at 101 SR on an Illumina HiSeq 3000 to an average depth of 1M reads in the Yerkes NHP Genomics Core (http://www.yerkes.emory.edu/nhp_genomics_core/).
V gene and somatic hypermutation analyses
Illumina bcl files from IgG, IgK and IgL amplicons were converted to fastq files usng the bcl2fastq tool. FastQC v0.11.5 (Andrew, 2010) was used to check the quality of fastq files. The repertoire sequence analysis was carried out using the pRESTO 0.5.6, Change-O 0.3.12, Alakazam 0.2.10.999 and SHazaM 0.1.9 packages from the Immcantation pipeline (Gupta et al., 2015; Vander Heiden et al., 2014). Pre-processing was performed using tools in the pRESTO package. Paired-end reads were first assembled with AssemblePairs tool. Reads with a mean quality score of less than 20 were filtered out using FilterSeq. The MaskPrimers tool was used to remove the forward primers and the random nucleotides from the assembled sequences. Data from each of two technical replicates were combined. Duplicates were removed and the duplicate counts were obtained for each unique sequence using CollapseSeq. SplitSeq was used to select sequences that had duplicate counts of at least two to eliminate singletons that may arise due to sequencing errors. The pre-processed sequences were then annotated using IgBLAST v1.6.1 (Ye et al., 2013).
Since the IMGT database (Lefranc and Lefranc, 2001) is lacking several V genes, a custom IgBLAST database was created for V genes using sequences from the genomic assembly in this study. The sequences were aligned using MUSCLE v3.8.1551 (Edgar, 2004) and only the V genes with complete sequence and no unknown amino acid (X) were selected. The corresponding nucleotide sequences of these V genes were clustered using CD-HIT v4.7(Fu et al., 2012) to remove 100% redundant sequences. The protein sequences for this non-redundant set were submitted to the IMGT DomainGapAlign tool (Ehrenmann and Lefranc, 2011; Ehrenmann et al., 2010) to obtain gapped V sequences. Corresponding gaps were introduced in the nucleotide sequences and the positions for framework (FR) and complementarity-determining regions (CDR) regions determined using custom scripts. These sequences were used to create the IgBLAST database for V genes. The databases for J and D genes was obtained from the IgBLAST ftp site (ftp://ftp.ncbi.nih.gov/blast/executables/igblast/release/internal_data/rhesus_monkey/). The annotations from IgBLAST were saved into a Change-O database and functional sequences were selected using Change-O. The gene usage and clonal frequencies were obtained from the Alakazam package and SHM estimations were obtained from the SHazaM package.
To obtain paired heavy and light chain sequences from single cell RNA-Seq data, we used the BALDR pipeline, as previously described (Upadhyay et al., 2018), with the Unfiltered method for rhesus macaques. Parallel instances of BALDR were run using the gnu parallel utility (Tange, 2011). The reads were trimmed using Trimmomatic v0.36 (Bolger et al., 2014). The trimmed reads were assembled using Trinity v2.6.5 (Grabherr et al., 2011). The assembled transcripts were annotated with the sequenced V(D)J genes in this study using IgBLAST v1.6.1. Reads greater than 50 bp were aligned to the assembled transcripts using bowtie2-2.3.0 (Langmead and Salzberg, 2012) to rank assembled transcripts based on the number of mapped reads. The assembled transcripts were filtered to remove non-productive sequences and those with the same V(D)J and CDR3 sequence as a higher ranked transcript. Out of the remaining sequences, the top ranking transcript sequence was chosen for the heavy and light chains.
Lineage analysis
For the quantification of B cell lineages, two independent analyses were performed with largely equivalent results.
Lineage analyses in figures 5 and S4 utilized only the sequences from the genomic assembly generated in this study. The annotations from IgBLAST were saved into a Change-O database and functional sequences were selected using Change-O. The functional sequences were assigned to a clone using a custom script based on the following criteria: (i) same V gene, (ii) same J gene, (iii) same CDR3 length and (iv) percentage identity of CDR3 nucleotide sequence > 85%. The analysis was also performed with the larger IgBLAST database with comparable results.
Phylogenetic trees were generated using a larger IgBLAST database created for V genes using sequences from the genomic assembly in this study or by combining sequences from previously published studies (Corcoran et al., 2016; Lefranc and Lefranc, 2001; Ramesh et al., 2017; Sundling et al., 2012) and the sequences from the assembly in this study. Lineage assignment was performed using a clustering procedure that exploited both germline inference and sequence similarity. Two sequences were deemed to potentially belong to the same lineage when: (i) their inferred UCA sequences (ignoring the junction and D region) are within 1% of each other (using a kmer-based distance approximation from (Kumar et al., 2018) for computational efficiency), tolerating calls to closely related V and J genes; and (ii) when the length-normalized Levenshtein distance between their junction+D sequences is within 10%. The clustering algorithm itself maintains a set of candidate lineages, storing all sequences for each lineage, and each new sequence in turn is added to the lineage where the largest proportion of sequences match the above two criteria. If no existing candidate cluster has >50% of its reads match the new sequence, then that sequence is used to seed a new candidate cluster containing this sequence as its sole member. Where members of a lineage had different inferred UCA sequences, the modal UCA was chosen as the UCA for the entire lineage. This lineage clustering algorithm was implemented in the Julia language for scientific computing (v0.6.2). Each lineage was aligned with MAFFT (Katoh and Standley, 2013) , and phylogenetic trees were inferred using FastTree2 (Price et al., 2010). Phylogenies were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/), using automated coloring and annotation scripts implemented in Julia.
ELISAs
BG505 trimer, gp120 and His ELISAs: Half-area 96-well high binding plates (Corning) were coated with streptavidin at 2.5μg/mL (Thermo Fisher Scientific) overnight at 4°C. Plates were washed with PBS + 0.05% Tween (PBS-T) three times. Biotinylated BG505, biotinylated His peptide conjugated to mouse CD1d or biotinylated gp120 was diluted to 1.0μg/mL in PBS + 1% BSA were captured for 2 hours, 37°C. Plates were washed three times and then blocked with PBS+ 3% BSA for 1 hour, RT. Plasma samples or monoclonal antibodies were serially diluted in PBS + 1% BSA and incubated for 1 hour, RT. Plates were washed three times and horseradish peroxidase goat anti-rhesus IgG (H+L) secondary (Southern Biotech) was added at 1:3000 dilution in PBS + 1% PBS for 1 hour, RT. Plates were washed three times with PBS-T and absorption was measured at 450nm following addition of TMB substrate (Thermo Scientific). Endpoint titers were calculated as dilution at which O.D. signal was 0.1 above background using GraphPad Prism v7.0 or 8.0. Antibody data panels show geometric mean titers with geometric SD.
Lectin-capture BG505 trimer ELISA: To maximize access to the base of the trimer, we utilized a lectin-capture assay. Env trimer is heavily glycosylated, except at the base. Capture with a lectin, which binds glycans, increases the likelihood that the base will be exposed more than in a streptavidin-capture ELISA. Half-area 96- well high binding plates (Corning) were coated with 5μg/mL lectin from Galanthus nivalis (snowdrop) (Sigma) in PBS overnight at 4°C. Plates were washed with 0.05% PBS-Tween (PBS-T) three times. 1ug/ml BG505 trimer in PBS + 1% BSA was bound to plates for 2 hours at 37°C and then washed three times. Plates were blocked with PBS + 3% BSA for 1 hour, RT. Monoclonal antibodies were serially diluted in PBS + 1% BSA and incubated for 1.5 hours at RT. Plates were washed three times with PBS-T before incubation with horseradish peroxidase goat anti-human IgG, Fcγ fragment specific (Jackson ImmunoResearch) at 1:5000 in PBS + 1% BSA for 1hr, RT. Plates were washed five times with PBS-T and absorption was measured at 450nm following addition of TMB substrate (Thermo Fisher Scientific). O.D. values presented are background subtracted.
Cross-competition trimer ELISA: We used a modified lectin capture ELISA for this assay. Plates were coated with GNL and BG505 and blocked as previously described. Plates were incubated with 0 or 10μg/mL 19R (fab) in PBS + 1% BSA for 1.5 hours at RT. Plates were washed three times with PBS-T. 2.5μg/mL of whole monoclonal antibody was added for 1 hour at RT and then washed three times with PBS-T before incubation with horseradish peroxidase goat anti-human IgG, Fcγ fragment specific (Jackson ImmunoResearch) at 1:5000 in PBS + 1% BSA for 1hr, RT. Plates were washed five times with PBS-T and absorption was measured at 450nm following addition of TMB substrate (Thermo Fisher Scientific). Data presented are background (no fab or mAb) subtracted. As an additional background control, 19R fab was incubated without mAb.
Pseudovirus neutralization assay
Autologous neutralization assays were performed as previously described (Pauthner et al., 2017). BG505 pseudovirus neutralization was tested using the BG505.W6M.ENV.C2 isolate (AIDS Reagents Program), carrying the T332N mutation to restore the N332 glycosylation site.
Heterologous neutralization breadth was tested on a panel of 12 cross-clade isolates, representative of larger virus panels composed of isolated from diverse geography and clades (deCamp et al., 2014). All viruses in this panel are Tier 2. Week 10 (bolus), w12 (2w OPs), and w14 (4w OPs) were tested in osmotic pump study. Limit of detection (LOD) for heterologous viruses is 1:50 (dotted line). Titers below LOD are set at 1:40. Week 26 (bolus) and w27 (ED) were tested in ED study.
Neutralization titers are reported as ID50 titers. All neutralization Ab data panels show geometric mean titers with geometric SD.
19R
The genes encoding the 19R rhesus macaque IgG1 heavy chain and kappa light chain were synthesized and separately cloned into the pcDNA3.4 plasmid by Thermo Fisher Scientific. The 19R IgG was expressed in Expi293 cells and purified using Protein A by Thermo Fisher Scientific. 19R Fab was generated by digesting 19R IgG using the Pierce Fab Preparation Kit (Thermo Fisher Scientific).
Monoclonal EM analysis
The heavy and light chains of the BDA monoclonal antibodies were codon-optimized, synthesized and cloned into pFUSE2ss-CHIg-hG1 and pFUSE2ss-CLIg-hl2, respectively, by GenScript. Antibodies were expressed and purified by GenScript. Antibody sequences are available in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fixable Viability Dye eFluor506 | Thermo Fisher Scientific | Cat # 65–0866-18 |
| Fixable Viability Dye eFluor780 | Thermo Fisher Scientific | Cat # 65–0865-18 |
| Mouse anti-human CD20 PE-Texas Red (clone: 2H7) | Beckman Coulter | Cat # IM3607U |
| Mouse anti-human CD4 BV650 (clone: OKT-4) | BioLegend | Cat # 317436 |
| Mouse anti-human CD8a Qdot705 (clone: 3B5) | Thermo Fisher Scientific | Cat # Q10059 |
| Mouse anti-human IgG PE-Cy7 (clone: G18–145) | BD Biosciences | Cat # 561298 |
| Mouse anti-human CXCR5 PE (clone: MU5UBEE) | Thermo Fisher Scientific | Cat # 12–9185-42 |
| Mouse anti-human PD1 BV605 (clone: EH12.2H7) | BioLegend | Cat # 329924 |
| Mouse anti-human CD3 BV786 (clone: SP34–2) | BD Biosciences | Cat # 563918 |
| Mouse anti-human IgM PerCP-Cy5.5 (clone:G20–127) | BD Biosciences | Cat # 561285 |
| Mouse anti-human Ki67 Alexa Fluor 700 (clone: B56) | BD Biosciences | Cat # 561277 |
| Mouse anti-human Ki67 Alexa Fluor 488 (clone: B56) | BD Biosciences | Cat # 558616 |
| Mouse anti-human Bcl6 Alexa Fluor 488 (clone: K112–91) | BD Biosciences | Cat # 561524 |
| Mouse anti-human CD4 APC eFluor 780 (clone: SK3) | Thermo Fisher Scientific | Cat # 47–0047-42 |
| Mouse anti-human CD8 APC eFluor 780 (clone: RPA-T8) | Thermo Fisher Scientific | Cat # 47–0088-42 |
| Mouse anti-human CD16 APC eFluor 780 (clone: ebioCB16) | Thermo Fisher Scientific | Cat # 47–0168-42 |
| Mouse anti-human CD20 Alexa Fluor 488 (clone: 2H7) | BioLegend | Cat # 302316 |
| Mouse anti-human CD38 PE (Clone: OKT) | NHP Reagents | Cat # PR-3802 |
| Mouse anti-human CD71 PE-CF594 (Clone: L01.1) | BD Biosciences | Custom conjugate |
| Mouse anti-human CD4 BV650 (Clone: OKT4) | BioLegend | Cat # 317436 |
| Mouse anti-human CD20 BV570 (Clone: 2H7) | BioLegend | Cat # 302332 |
| Mouse anti-human PD1 BV785 (Clone: EH12.2H7) | BioLegend | Cat # 329930 |
| Mouse anti-human CXCR5 PE-Cy7 (Clone: MU5UBEE) | Thermo Fisher Scientific | Cat # 25–9185-42 |
| Mouse anti-human CD25 FITC (Clone: BC96) | BioLegend | Cat # 302604 |
| Mouse anti-human OX40 PE (Clone: L106) | BD Biosciences | Cat # 340420 |
| Mouse anti-human 4–1BB APC (Clone:4B4–1) | BioLegend | Cat # 309810 |
| Mouse anti-human CD14 APC/Cy7 (Clone: M5E2) | BioLegend | Cat # 301820 |
| Mouse anti-human CD16 APC/Cy7 (Clone: 3G8) | BioLegend | Cat # 302018 |
| Mouse anti-human CD20 BV650 (Clone: 2H7) | BioLegend | Cat # 302336 |
| Mouse anti-human CD4 APC (Clone: OKT4) | BioLegend | Cat # 317416 |
| Mouse anti-human Bcl6 BV421 (Clone: K112–91) | BD Biosciences | Cat # 563363 |
| Mouse anti-human Bcl6 Alexa Fluor 647 (Clone: K112–91) | BD Biosciences | Cat # 561525 |
| Mouse anti-human IgM BV421 (Clone: G20–127) | BD Biosciences | Cat # 562618 |
| Mouse anti-human IgG PE (Clone: G18–145) | BD Biosciences | Cat # 560951 |
| Mouse anti-human IgD Alexa Fluor 488 | Southern Biotech | Cat # 2030–30 |
| Mouse anti-human Lambda biotin (Clone: IS7–24C7) | Miltenyi | Cat # 130–093-025 |
| Mouse anti-human CD35 (Clone: E11) | BD Biosciences | Cat # 565327 |
| Streptavidin-Alexa Fluor 647 | Thermo Fisher Scientific | Cat # S32357 |
| Streptavidin-BV421 | BioLegend | Cat # 405225 |
| Streptavidin-BV711 | BioLegend | Cat # 405241 |
| Goat anti-rhesus IgG (H+L) - HRP | Southern Biotech | Cat # 6200–05 |
| Goat anti-Human IgG, Fcγ fragment specific-HRP | Jackson ImmunoResearch | Cat # 109–035-098 |
| 19R | Thermo Fisher Scientific, unpublished | Custom |
| BDA1 HC:QVQLQESGPGLVKPSETLSLTCAVSGASISIYWWGWIRQPPGKGLEWIGEIIGSSGSTNSNPSFKSRVTISKDASKNQFSLNLNSVTAADTAVYYCVRVGAAISLPFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCYSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA2 HC:QVQLQESGPGLLKPSETLSLTCAVSGGSFSSYWWSWIRQPPGKGLEWIGEINGNSGSTHYNPSLKSRVTISKDASKNQFSLKLNSVTAADTAVYYCARWGPTGVTQGEPDFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITYSGDALPKRYAYWFQQKPGQSPVLIIYEDSKRPSGIPERFSGSSSGTVATLTISGAQVEDEADYYCYSTDSSGNHFFGAGTRLTVL |
Genscript | Custom |
| BDA3 HC:QVQLQESGPGLVKPSETLSLTCAVSGHSVSSGYGWGWIRQPPGKGLEWIGQIYGYSGSTSYNPSLKSRVTVSTDTSKNQFSLRLSSLTAADTAVYYCARWHHGSFDIWGPGTPITISS LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTINGAQVEDEGDYYCYSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA4 HC:QVQLQESGPGLVKPSETLSLTCAVSGASIRIYWWGWIRQPPGKGLEWIGEIIGSSGSTNSNPSFKSRVTISKDASKNQFSLNLNSVTAADTAVYYCVRVGAAISFPFDYWGQGVLVTVSS LC:SSELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEGDFYCYSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA5 HC:QVQLQESGPGLLKPSETLSLTCAVSGGSFSSYWWSWIRQPPGKGLEWIGEINGNSGNTHYNPSLKSRVTISKDASKNHFSLKLSSVTAADTAVYYCARWGPTGVTQGEPDFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITYSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEADYYCYSTDSSGNHFFGAGTRLTVL |
Genscript | Custom |
| BDA6 HC:QVQLQESGPGLLKPSETLSLTCAVSGGSFSSYWWSWIRQPPGKGLEWIGEINGNSGSTHYNPSLKSRVTISKDASKNQFSLKLNSVTAADTAVYYCARWGPTGVTQGEPDFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYVYWFQQKPGQSPVLIIYEDSKRPSGIPERFSGSSSGTVATLTISGAQVEDEADYYCYSTISSGNDRIFGAGTRLTVL |
Genscript | Custom |
| BDA7 HC:QVQLQESGPGLVKPSETLSLTCAVSGASISIYWWSWIRQPPGKGLEWIGEIIGNSGSTNSNPSFKSRVTISKDASKNQFSLKLSSVTAADTAVYYCVRVGAAISLPYDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCYSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA8 HC:QVQLQESGPGLLKPSETLSLTCAVSGGSFSSYWWSWIRQPPGKGLEWIGEINGNSGNTHYNPSLKSRVTISKDASKNQFSLKLSSVTAADTAVYYCARWGPTGVTQGEPEFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITYSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEADYYCYSTDSSGNHFFGAGTRLTVL |
Genscript | Custom |
| BDA9 HC:QVQLQESGPGLLKPSETLSLTCAVSGGSFSNYWWSWIRQPPGKGLEWIGEINGNSGSTHYNPSLKSRVTISKDASKNQFSLKLNSVTAADTAVYYCARWGPTGVTQGEPDFDFWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITYSGDALPKRYAYWFQQKPGQSPVLIIYEDSKRPSGIPERFSGSSSGTVATLTISGAQVEDEADYYCYSTDSSGNHFFGVGTRLTVL |
Genscript | Custom |
| BDA10 HC:QVQLQESGPGLVKPSETLSLTCAVSGVSISIYWWSWIRQPPGKGLEWIGEIIGNSGNTNSSPSFKSRVTISKDASKNQFSLKLSSVTAADTAVYYCVRVGAAISLPFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITCSGDALPEKYAYWFQQKPGQSPVLIIYDDNIRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCYSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA11 HC:QVQLQESGPGLVKPSETLSLTCAVSGGSFSSYWWSWIRQPPGKGLEWIGEINGNSGSTNYNPSLKSRVTISKDASKNQFSLKLSSVTAADTAVYYCARVRVGAAISLPFDYWGQGVLVTVSS LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCYSRHISGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA12 HC: BDA1HC LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYEDNKRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCFSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| BDA13 HC: BDA1 HC LC:SYELTQPPSVSVSPGQTARITCSGDALPKKYAYWFQQKPGQSPVLIIYDDSQRPSGIPERFSGSSSGTVATLTISGAQVEDEGDYYCFSRHSSGNHGLFGGGTRLTVL |
Genscript | Custom |
| Bacterial and Virus Strains | ||
| BG505.W6M.ENV.C2 | Wu et al., 2006 / NIH AIDS Reagent Program | Cat # 11518 |
| 12 virus panel – global isolates | (deCamp et al., 2014) | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BG505 Olio6 | Produced inhouse (Kulp et al., 2017) | |
| BG505 Olio6CD4-KO | Produced inhouse (Kulp et al., 2017) | |
| BG505 MD39 | Produced inhouse (Kulp et al., 2017) | |
| BG505 Olio6CD4-KO – Biotin | Produced inhouse | |
| BG505 Olio6 – Biotin | Produced inhouse | |
| BG505 MD39 – Biotin | Produced inhouse | |
| BG505 SOSIP – Biotin | Produced inhouse | |
| BG505 gp120 - Biotin | Produced inhouse | |
| BG505 SOSIP.664 v5.2 | Produced inhouse (Torrents de la Peña et al., 2017) | |
| His-Tag biotin peptide biotin - ALDGGGGSHHHHHHHH | A&A Labs LLC | N/A |
| Recombinant Protein A Sepharose | GE Healthcare | Cat # 17127902 |
| Lectin from Galanthus nivalis (snowdrop), lyophilized powder | Sigma-Aldrich | Cat # L9275–5mg |
| Olio6-CD4ko peptide megapool | A&A Labs LLC | N/A |
| Staphylococcal enterotoxin B (SEB) | Toxin Technology Inc. | Cat # BT202 |
| DNAse I Solution | Stemcell Technologies | Cat # 07900 |
| Streptavidin | Jackson ImmunoResearch Laboratories, Inc | Cat # 016–000-084 |
| TMB Substrate | Thermo Fisher Scientific | Cat # 34021 |
| Cholesterol | Avanti Polar Lipids | Cat # 700000 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (16:0 PC (DPPC)) | Avanti Polar Lipids | Cat # 850355 |
| N-Decanoyl-N-methylglucamine (MEGA-10) | Sigma-Aldrich | Cat # D6277 |
| Quil-A® saponin | InvioGen | Cat # vac-quil |
| Critical Commercial Assays | ||
| Pierce™ Fab Preparation Kit | Thermo Fisher Scientific | Cat # 44985 |
| eBioscience FoxP3/ Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat # 00–5523-00 |
| QCL-1000™ Endpoint Chromogenic LAL Assay | Lonza | Cat # 50–647U |
| BirA biotin-protein ligase standard reaction kit | Avidity Inc | Cat # BirA500 |
| Cholesterol Quantitation Kit | Sigma- Aldrich | Cat # MAK043–1KT |
| AMPure XP beads | Beckman Coulter | Cat # A63882 |
| KAPA HiFi HotStart Real-time PCR Master Mix (2X) | Kapa Biosystems, Inc. | Cat # KK2702 |
| Deposited Data | ||
| Rhesus macaque genomic raw sequences | NCBI SRA | PRJNA509445 |
| Rhesus macaque genome assembly | NCBI Assembly | SBKD01000000 |
| Week 7 single-cell RNA-seq sequences | NCBI SRA | PRJNA520929 |
| IgG BCR sequence reads from 224–13 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCVI00000000 |
| IgG BCR sequence reads from 224–13 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVJ00000000 |
| IgK BCR sequence reads from 224–13 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVK00000000 |
| IgL BCR sequence reads from 224–13 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVL00000000 |
| IgG BCR sequence reads from RCn16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCVM00000000 |
| IgK BCR sequence reads from RCn16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCVN00000000 |
| IgL BCR sequence reads from RCn16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCVO00000000 |
| IgG BCR sequence reads from RCn16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVP00000000 |
| IgK BCR sequence reads from RCn16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVQ00000000 |
| IgL BCR sequence reads from RCn16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCVR00000000 |
| IgG BCR sequence reads from RRk16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCWO00000000 |
| IgK BCR sequence reads from RRk16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCWP00000000 |
| IgL BCR sequence reads from RRk16 (bolus) right LN | NCBI SRA, GenBank | PRJNA520929, KCWQ00000000 |
| IgG BCR sequence reads from RRk16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCWR00000000 |
| IgK BCR sequence reads from RRk16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCWS00000000 |
| IgL BCR sequence reads from RRk16 (bolus) left LN | NCBI SRA, GenBank | PRJNA520929, KCWT00000000 |
| IgG BCR sequence reads from RVh16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCVS00000000 |
| IgK BCR sequence reads from RVh16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCVT00000000 |
| IgL BCR sequence reads from RVh16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCVU00000000 |
| IgG BCR sequence reads from RVh16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCVV00000000 |
| IgK BCR sequence reads from RVh16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCVW00000000 |
| IgL BCR sequence reads from RVh16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCVX00000000 |
| IgG BCR sequence reads from RYm16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXA00000000 |
| IgK BCR sequence reads from RYm16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXB00000000 |
| IgL BCR sequence reads from RYm16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXC00000000 |
| IgG BCR sequence reads from RYm16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXD00000000 |
| IgK BCR sequence reads from RYm16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXE00000000 |
| IgL BCR sequence reads from RYm16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXF00000000 |
| IgG BCR sequence reads from RWr16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWU00000000 |
| IgK BCR sequence reads from RWr16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWV00000000 |
| IgL BCR sequence reads from RWr16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWW00000000 |
| IgG BCR sequence reads from RWr16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWX00000000 |
| IgK BCR sequence reads from RWr16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWY00000000 |
| IgL BCR sequence reads from RWr16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWZ00000000 |
| IgG BCR sequence reads from RQq16 (2w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCVY00000000 |
| IgG BCR sequence reads from RQq16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCVZ00000000 |
| IgK BCR sequence reads from RQq16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWA00000000 |
| IgL BCR sequence reads from RQq16 (2w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWB00000000 |
| IgG BCR sequence reads from RWh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXG00000000 |
| IgK BCR sequence reads from RWh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXH00000000 |
| IgL BCR sequence reads from RWh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXI00000000 |
| IgG BCR sequence reads from RWh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXJ00000000 |
| IgK BCR sequence reads from RWh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXK00000000 |
| IgL BCR sequence reads from RWh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXL00000000 |
| IgG BCR sequence reads from ROw16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXM00000000 |
| IgK BCR sequence reads from ROw16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXN00000000 |
| IgL BCR sequence reads from ROw16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCXO00000000 |
| IgG BCR sequence reads from ROw16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXP00000000 |
| IgK BCR sequence reads from ROw16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXQ00000000 |
| IgL BCR sequence reads from ROw16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCXR00000000 |
| IgG BCR sequence reads from RTh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWI00000000 |
| IgK BCR sequence reads from RTh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWJ00000000 |
| IgL BCR sequence reads from RTh16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWK00000000 |
| IgG BCR sequence reads from RTh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWL00000000 |
| IgK BCR sequence reads from RTh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWM00000000 |
| IgL BCR sequence reads from RTh16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWN00000000 |
| IgG BCR sequence reads from RFr16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWC00000000 |
| IgK BCR sequence reads from RFr16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWD00000000 |
| IgL BCR sequence reads from RFr16 (4w pumps) right LN | NCBI SRA, GenBank | PRJNA520929, KCWE00000000 |
| IgG BCR sequence reads from RFr16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWF00000000 |
| IgK BCR sequence reads from RFr16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWG00000000 |
| IgL BCR sequence reads from RFr16 (4w pumps) left LN | NCBI SRA, GenBank | PRJNA520929, KCWH00000000 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM 224–13 (bolus) | EMDataBank | EMD-9175 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RCn16 (bolus) | EMDataBank | EMD-9176 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RRk16 (bolus) | EMDataBank | EMD-9180 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RVh16 (2w pumps) | EMDataBank | EMD-9181 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RYm16 (2w pumps) | EMDataBank | EMD-9185 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RWr16 (2w pumps) | EMDataBank | EMD-9184 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RQq16 (2w pumps) | EMDataBank | EMD-9179 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RWh16 (4w pumps) | EMDataBank | EMD-9183 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM ROw16 (4w pumps) | EMDataBank | EMD-9178, EMD-9186 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RTh16 (4w pumps) | EMDataBank | EMD-9182 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RFr16 (4w pumps) | EMDataBank | EMD-9177 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM 145–11 (bolus) | EMDataBank | EMD-0571 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RHw16(bolus) | EMDataBank | EMD-0580 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RAa14 (bolus) | EMDataBank | EMD-0575 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM 5N6 (bolus) | EMDataBank | EMD-05694e |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM REj15 (bolus) | EMDataBank | EMD-0578 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM REv16 (bolus) | EMDataBank | EMD-0579 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RAv16 (ED) | EMDataBank | EMD-0576, EMD-0577 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM B077(ED) | EMDataBank | EMD-0573, EMD-0574 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM BM57 (ED) | EMDataBank | EMD-0572 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM 99–13(ED) | EMDataBank | EMD-0570 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RMI15 (ED) | EMDataBank | EMD-0581 |
| Negative stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from RM RWo15 (ED) | EMDataBank | EMD-0582 |
| Negative stain EM map of BDA1 in complex with BG505 SOSIP.664 | EMDataBank | EMD-9138 |
| Experimental Models: Cell Lines | ||
| TZM-bl cells | NIH AIDS Reagent Program | Cat #8129 |
| Experimental Models: Organisms/Strains | ||
| Indian-origin rhesus macaques (outbred) | Yerkes National Primate Research Center | |
| Olionucleotides | ||
| CDS Oligo (dT): TTTTTTTTTTTTTTTTTTTTTTTTTVN | Integrated DNA Technologies, Inc. | |
| SMARTer II A Oligo: AAGCAGTGGTATCAACGCAGAGTACATrGrGrG | Integrated DNA Technologies, Inc. | |
| IgG Constant Primer: GCCAGGGGGAAGACCGATGGGCCCTTGGTGGA | Integrated DNA Technologies, Inc. | |
| IgK Constant Primer: GCGGGAAGATGAAGACAGATGGTGCAGCCACAG | Integrated DNA Technologies, Inc. | |
| IgL Constant Primer: GGCCTTGTTGGCTTGAAGCTCCTCAGAGGAGGG | Integrated DNA Technologies, Inc. | |
| P5_Seq BC_XX 5PIIA: CACGACGCTCTTCCGATCT 4–8xN AACCACTA AAGCAGTGGTATCAACGCAGAGT | Integrated DNA Technologies, Inc. | |
| P7_ i7_XX IgG: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GCCAGGGGGAAGACCGATGGGCCCTTGGTGGA | Integrated DNA Technologies, Inc. | |
| P7_ i7_XX IgK: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GCGGGAAGATGAAGACAGATGGTGCAGCCACAG | Integrated DNA Technologies, Inc. | |
| P7_ i7_XX IgL: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GGCCTTGTTGGCTTGAAGCTCCTCAGAGGAGGG | Integrated DNA Technologies, Inc. | |
| P5_Graft P5_seq: AATGATACGGCGACCACCGAGATCTACAC TCTTTCCCTACACGACGCTCTTCCGATCT | Integrated DNA Technologies, Inc. | N/A |
| Software and Algorithms | ||
| Prism v7.0/v8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v10.4 | FlowJo LLC | https://www.flowjo.com |
| Appion database | (Lander et al., 2009) | |
| Leginon | (Suloway et al., 2005) | |
| DoG Picker | (Voss et al., 2009) | |
| Relion | (Scheres, 2012) | |
| FALCON-Unzip | (Chin et al., 2016) | |
| IMGT | http://www.imgt.org | |
| BLAT | (Kent, 2002) | |
| BLASR | (Chaisson and Tesler, 2012) | |
| BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | |
| MsPAC | https://bitbucket.org/oscarlr/mspac | |
| Integrated Genomics Viewer | (Robinson et al., 2011; Thorvaldsdóttir et al., 2013) | https://software.broadinstitute.org/software/igv/ |
| BALDR | (Upadhyay et al., 2018) | |
| FastQC v0.11.5 | (Andrew, 2010) | |
| Immcantation | (Gupta et al., 2015; Vander Heiden et al., 2014) | |
| IgBLAST v1.6.1 | (Ye et al., 2013) | |
| MUSCLE v3.8.1551 | (Edgar, 2004) | |
| CD-HIT v4.7 | (Fu et al., 2012) | |
| IMGT DomainGAPAlign | (Ehrenmann and Lefranc, 2011; Ehrenmann et al., 2010) | |
| MAFFT | (Katoh and Standley, 2013) | |
| FastTree2 | (Price et al., 2010) | |
| FigTree | http://tree.bio.ed.ac.uk/software/figtree/ | |
| ImageJ | https://imagej.nih.gov/ij/ | |
| Other | ||
| Mini-osmotic pumps (0.5uL/hr, 14 day release) | ALZET | Model 2002 |
| Mini-osmotic pumps (0.25uL/hr, 28 day release) | ALZET | Model 2004 |
| Costar Assay Plate, 96 well flat-bottom, half area, high binding | Corning | Cat # 3690 |
Fab was generated using Pierce Fab preparation kit (Thermo Fisher Scientific). 15 μg of BG505 SOSIPv5.2 Env trimer (untagged) was complexed with 41μg BDa1 Fab at room temperature overnight in a total reaction volume of 50 μL. The complex was diluted 1:20 with TBS and 3 μL was applied to a glow-discharged, carbon-coated 400-mesh copper grid and blotted off after 15 seconds. 3 μL of 2% (w/v) uranyl formate stain was applied and immediately blotted off, followed by another application of 3 μL of stain for 45 seconds, blotted once more, and allowed to air-dry. Images were collected via Leginon (Potter et al., 1999) using an FEI Talos microscope (1.98A°/pixel; 72,000× magnification; 25 e−/ A°2). Particles were picked from the raw images using DoG Picker(Voss et al., 2009). 2D classification, 3D sorting and refinement of the complex was conducted using RELION 3.0b0 (Nakane et al., 2018).
Polyclonal EM analysis
Plasma (Pump study: week 10 (bolus), week 12 (2 week pumps) or week 14 (4 week pumps); Dose escalation study: weeks 13 and 15 pooled) was diluted 4X with PBS and incubated with protein A sepharose beads (GE Healthcare) overnight at 4C. Resin was washed 3X with PBS and eluted with 0.1M glycine pH2.5 and immediately neutralized with 1M Tris-HCL pH 8. Fabs were purified using Pierce Fab preparation kit (Thermo). Fab was generated using Pierce Fab Preparation Kit (Thermo Scientific). Reaction was incubated with protein A sepharose resin for 1 hour, RT. Fabs were buffer exchanged using Amicon ultra 0.5ml centrifugal filters (Millipore Sigma).
Upon buffer exchange into TBS, 0.5 to 0.8 mg of total Fab was incubated overnight with 10 μg BG505 trimers at RT in ~36 μL total volume. The formed complexes were then separated from unbound Fab via size exclusion chromatography (SEC) using Superose 6 Increase 10/300 column (GE Healthcare) equilibrated with TBS. The flow-through fractions containing the complexes were pooled and concentrated using 100 kDa cutoff centrifugal filters (EMD Millipore). The final trimer concentration was adjusted to approximately 0.04 mg/mL prior to application onto carbon-coated copper grids.
Complexes were applied to glow-discharged, carbon-coated 400-mesh copper grids, followed by applying 3 μL of 2% (w/v) uranyl formate stain that was immediately blotted off, and followed by application of another 3 μL of stain for 45–60 s, and blotted once more. Stained grids were allowed to air-dry and stored under ambient conditions until imaging. Images were collected via Leginon using a Tecnai T12 electron microscopes operated at 120 kV; ×52,000 magnification; 2.05 A°/pixel. In all cases, the electron dose was 25 e−/ A°2. Particles were picked from the raw images using DoG and placed into stacks using Appion software (Lander et al., 2009). 2D reference-free alignment was performed using iterative MSA/MRA) (Sorzano et al., 2010). Finally, the particle stacks were then converted from IMAGIC to RELION-formatted MRC stacks and subjected to RELION 2.1 2D and 3D classification (Scheres, 2012).
Epitopes are pseudocolored as: base (purple), glycan hole-I (light blue), C3/V5 (dark blue), fusion peptide (orange), V1/V3 apex (green).
Whole LN Imaging
All NHP LNs were harvested and immediately placed in PLP buffer (pH 7.4 50 mM PBS + 100 mM lysine, 1% paraformaldehyde, 2 mg/mL sodium periodate) for fixation. After 4-5 days at 4°C, the tissues were washed and stored in PBS with 0.05% sodium azide at 4°C until taken for imaging.
Total antigen signal within LNs from the antigen tracking study was measured by placing the tissues directly on the glass scanning surface of a Typhoon FLA 9500 biomolecular imager (GE Healthcare Life Sciences) and using a 635 nm excitation laser and a ≥665 nm long-pass filter. The integrated signal density corresponding to Alexa Fluor 647-labeled MD39 in each LN was calculated using ImageJ and plotted using GraphPad Prism 8.
Selected LNs were clarified via a combination / modification of the iDISCO (Renier et al., 2016) and CUBIC (Kubota et al., 2017) organ-clearing methods. The LNs were first delipidated based on the iDISCO methanol incubation protocol. First, the tissues were washed in water for 1 hour, followed by 20% methanol in water for 2 hours. A series of step increases in methanol percentage (40%, 60%, 100%, 100%) followed, each step for 2 hours. The LNs were then placed into 2:1 MeOH:DCM overnight, and the next day were rehydrated with the following series of methanol solutions for 2 hours each: 100%, 100%, 80%, 60%, 40%, 20%, 0%, 0%. Next, the LNs were placed into 10-20 mL of a 1:1 mixture of CUBIC-R solution for 1 day, followed by at least 20 mL of undiluted CUBIC-R for 2 days or as long as needed for adequate clarification. Larger organs were moved into a fresh 20 mL of CUBIC-R solution to ensure that the refractive index of the solution would not be significantly lowered by residual water in the tissue.
Clarified LNs were imaged in CUBIC-R using the LaVision Ultramicroscope II focused beam ligh tsheet using Olympus MVPLAPO 2x Dry lens, magnification: 2.0x, NA: 0.50, WD: 10 mm with short dipping cap. The Alexa Fluor 647-labeled SOSIP was imaged using the 640 nm laser at 100 ms exposure time on an Andor Neo camera with focus magnification: 1.25x. Snapshots and movies were generated using the 3D viewer in the FIJI package of ImageJ.
Histology
Selected LNs were embedded in 3% low melting temperature agarose (Sigma-Aldrich), and then sliced into 350 μm-thick sections using a vibratome. The slices were blocked and permeabilized for 2 days in PBS with 10% goat serum and 0.2% Triton-X-100, followed by staining for 3 days with BV421-labeled mouse anti-human CD35 (E11,BD Biosciences) and Alexa Fluor 488-labeled mouse anti-Ki67 (B56, BD Biosciences) in the blocking buffer. Stained slices were then washed for 3 days with PBS containing 0.2% Tween-20, and then mounted onto glass slides with coverslips. Images were captured using an automated spinning disc confocal slide scanner (TissueFAXS Confocal SL, TissueGnostics USA) utilizing a Zeiss Axio Imager Z2 equipped with a Zeiss 20x Plan-Apochromat 0.8NA objective, Lumencor Spectra X light engine, Maerzhauser motorized stage and 120 slide loader, and a Crest Optics X Light V2 confocal imager, along with TissueFAXS slide scanning software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Graphpad Prism v7.0 or 8.0 was used for all statistical analyses. Significance of differences in neutralization, BG505 binding titers, cellular frequencies and mean fluorescent intensities were calculated using unpaired, two-tailed Mann-Whitney U tests. Differences in mutation frequencies and CDR3 length between groups were calculated using unpaired student’s t tests. Significance of differences in V gene use between groups and between Env MFIs (q value) were calculated using multiple t tests, corrected for multiple comparisons with a false discovery rate (FDR) of 5% (Benjamini, Krieger, and Yekutieli). Differences in BCR expression of GC vs non-BGC cells were calculated using paired, Wilcoxon test. Correlations between neutralization and cell frequencies were calculated using log transformed Ab titer values in two-tailed Pearson correlation tests. Differences in fluorescence intensity in LNs were calculated using two-way ANOVA.
DATA AND SOFTWARE AVAILABILITY
Env-specific B cell BCR and whole genome raw reads used in this paper are available at NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession SBKD00000000. The version described in this paper is version SBKD01000000. The raw genomic reads and genome assembly are both under BioProject ID PRJNA509445. Env-specific BCR sequences are available under BioProject ID PRJNA520929 and are deposited at DDBJ/ENA/GenBank under the accession: KCVI00000000, KCVJ00000000, KCVK00000000, KCVL00000000, KCVM00000000, KCVN00000000, KCVO00000000, KCVP00000000, KCVQ00000000, KCVR00000000, KCVS00000000, KCVT00000000, KCVU00000000, KCVV00000000, KCVW00000000, KCVX00000000, KCVY00000000, KCVZ00000000, KCWA00000000, KCWB00000000, KCWC00000000, KCWD00000000, KCWE00000000, KCWF00000000, KCWG00000000, KCWH00000000, KCWI00000000, KCWJ00000000, KCWK00000000, KCWL00000000, KCWM00000000, KCWN00000000, KCWO00000000, KCWP00000000, KCWQ00000000, KCWR00000000, KCWS00000000, KCWT00000000, KCWU00000000, KCWV00000000, KCWW00000000, KCWX00000000, KCWY00000000, KCWZ00000000, KCXA00000000, KCXB00000000, KCXC00000000, KCXD00000000, KCXE00000000, KCXF00000000, KCXG00000000, KCXH00000000, KCXI00000000, KCXJ00000000, KCXK00000000, KCXL00000000, KCXM00000000, KCXN00000000, KCXO00000000, KCXP00000000, KCXQ00000000, KCXR00000000. 3D EM reconstructions have been deposited in the Electron Microscopy Databank (http://www.emdatabank.org/) under the accession numbers listed in the Key Resources Table.
| Lymph node GC Panel | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Env probe-biotin | Alexa Fluor 647 | Invitrogen | |
| Env probe-biotin | Brilliant Violet 421 | BioLegend | |
| Viability | efluor506 | Thermo Fisher | |
| CD20 | PE-Texas Red | Beckman Coulter | 2H7 |
| CD4 | Brilliant Violet 650 | Biolegend | OKT-4 |
| CD8a | Qdot 705 | Thermo Fisher | 3B5 |
| IgG | PE-Cy7 | BD Biosciences | G18-145 |
| CXCR5 | PE | Thermo Fisher | MU5UBEE |
| PD1 | Brilliant Violet 605 | Biolegend | EH12.2H7 |
| CD3 | Brilliant Violet 786 | BD Biosciences | SP34-2 |
| IgM | PerCP-Cy5.5 | BD Biosciences | G20-127 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| BCL6 | Alexa Fluor 488 | BD Biosciences | K112-91 |
| Antigen-specific B cell sorts | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Env probe-biotin | Alexa Fluor 647 | Invitrogen | |
| Env probe-biotin | Brilliant Violet 421 | BioLegend | |
| Viability | efluor780 | Thermo Fisher | |
| CD4 | APC efluor780 | Thermo Fisher | SK3 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD16 | APC efluor780 | Thermo Fisher | ebioCD16 |
| CD20 | Alexa Fluor 488 | BioLegend | 2H7 |
| IgG | PE-Cy7 | BD Biosciences | G18-145 |
| IgM | PerCP-Cy5.5 | BD Biosciences | G20-127 |
| CD38 | PE | NHP Reagents | OKT |
| CD71 | PE-CF594 | BD Biosciences (custom) | L01.1 |
| AIM Assay Panel | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| CD4 | Brilliant Violet 650 | BioLegend | OKT4 |
| CD20 | Brilliant Violet 570 | BioLegend | 2H7 |
| PD1 | Brilliant Violet 785 | BioLegend | EH12.2H7 |
| CXCR5 | PE-Cy7 | Thermo Fisher | MU5UBEE |
| CD25 | FITC | BioLegend | BC96 |
| 0X40 | PE | BD Biosciences | L106 |
| 4-1BB | APC | BioLegend | 4B4-1 |
| Viability | efluor780 | Thermo Fisher | |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD14 | APC/Cy7 | BioLegend | M5E2 |
| CD16 | APC/Cy7 | BioLegend | 3G8 |
| BGC cell surface marker validation | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Viability | efluor780 | Thermo Fisher | |
| CD20 | Brilliant Violet 650 | BioLegend | 2H7 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD4 | APC | BioLegend | 0KT4 |
| CD38 | PE | NHP Reagents | OKT |
| CD71 | PE-CF594 | BD Biosciences (custom) | L01.1 |
| BCL6 | BV421 | BD Biosciences | K112-91 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| Macaque BCR expression | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Viability | efluor780 | Thermo Fisher | |
| CD4 | APC efluor780 | Thermo Fisher | SK3 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD16 | APC/Cy7 | BioLegend | 3G8 |
| CD20 | Brilliant Violet 650 | BioLegend | 2H7 |
| BCL6 | Alexa Fluor 647 | BD Biosciences | K112-91 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| IgM | BV421 | BD Biosciences | G20-127 |
| IgG | PE | BD Biosciences | G18-145 |
| IgD | Alexa Fluor 488 | Southern Biotech | |
| Lambda | Biotin | Miltenyi | IS7-24C7 |
| Streptavidin | Brilliant Violet 711 | BioLegend | |
Supplementary Material
(A) Full gating strategy of BGC cells. KI67+ BCL6+ expression was used at weeks -1-6, 8-14. CD38−CD71+ expression used at week 7.
(B) Validation of CD38 and CD71 as surface markers for bona fide BGC cells in mesenteric LNs of RM.
(C) BGC frequencies of bolus and individual OP groups. ** p<0.01. Blue asterisks, 2w OP vs. bolus. Pink asterisks, 4w OP vs. bolus.
(D) Cumulative BGC cell responses to first immunization of individual groups between w1 plus w3-7.
(E) Lambda expression in BGC and non-BGC cells. Top panel, gating strategy for BGC and non-BGC cells in LNs. Lower panel, histogram and quantification of lambda expression in lambda+ BGC and non-BGC cells.
(F) Gating strategy of Env trimer-specific B cells, gated on CD20+ cells as per Figure S1A.
(G) Olio6CD40KO differs from Olio6 by a single amino acid, to prevent binding to human CD4. Neither trimer binds RM CD4.
(H) Gating strategy and flow cytometry analysis of Env trimer-specific BGC cells over time within an individual LN, gated on CD20+ cells as per Figure S1A.
(I) Quantification of IgG gMFI of Env+ BGC cells. Differences in gMFI between weeks is due to use of different panels or flow cytometers for acquisition.
(J) Representative flow cytometry plot of High-affinity Env trimer-specific B cells, gated on CD20+ cells.
(K) Quantification of high affinity Env trimer-specific B cells over time.
(L) Cumulative High-affinity Env trimer-specific B cell response between w1 plus w3-7 [AUC].
(M) Flow cytometry analysis of High-affinity Env-specific BGC cells in a bolus immunized animal between w-1 and 8.
(N) Quantification of High-affinity Env trimer-specific BGC cells over time.
(O) Cumulative High-affinity Env-specific BGC cell responses within individual LNs between w2 and w7.
(P) Quantification of Env trimer-specific BMem cells. BMem were defined as the non-GC (BCL6− KI67− or CD38+ CD71−) B cells within the Env-specific B cell gate as per figure S1E.
(Q) Cumulative Env trimer-specific BMem cell response between weeks 3 and 7 [AUC]. All data represent mean ± SEM.*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
(R) Mean fluorescent intensities (MFIs) of Env trimer-specific B cells over time. Value calculated as: average of EnvAx647 and EnvBV421 MFIs (Env+ B cells) – average of EnvAx647 and EnvBV421 MFIs (Env− B cells).
As bolus Gp1 was not stained at same time as other groups, Gp1 was not included in these analyses. Number of LNs graphed are: bolus, 6 (w4), 4 (w5), 5 (w6), 5 (w7), 4 (w8); OP, 15 (w4), 15 (w5), 12 (w6), 15 (w7), 15 (w8). Statistical significance was tested using multiple t tests with 5% FDR.
Env-specific and High-affinity Env-specific B and germinal center (GC) B cell counts in osmotic pump study, Related to Figure 1. *data was not included in analyses due to low cell number
Env-specific and High-affinity Env-specific B and germinal center (GC) B cell counts in dose escalation study, Related to Figure 6. *data was not included in analyses due to low cell number.
Sequences of annotated IGH, IGK and IGL ORFs, Related to Figure 4.
Table S2. Quantification of Env-specific BGC cells used for BCR sequence analyses, Related to Figures 4 and 5.
Table S3. Sequences of BDA monoclonal antibodies, Related to Figure 5.
(A) Full gating strategy of GC-TFH cells. GC-TFH cells were measured at all time points, except for w7.
(B) Bolus and OP immunized animals have comparable frequencies of 4-1BB+ OX40+ CD4+ T cells under SEB stimulation of AIMOB assay. SEB stimulation serves as a positive control for the overall health of the cells.
(C) Representative BG505 Env IgG binding IgG curves at w11. Unimmunized control is at w-2.
(D) BG505 Env trimer endpoint binding IgG titers over time.
(E) Representative anti-His binding IgG curves at w11. Unimmunized control is at w-2.
(F) Quantification of anti-His endpoint binding IgG titers over time.
(G) Direct comparison of BG505 binding IgG titers of animals in this study vs animals in Pauthner et al. For Pauthner et al. groups, plasma from w8 and w12 was used. For bolus and 2w OP groups, plasma from w7 and w12 was used.
(H) Representative BG505 pseudovirus TZM-bl neutralization curves at w14.
(I) Quantification of BG505 neutralization titers between groups over time.
(J) Peak autologous neutralization titers of each animal after two immunizations.
(K) Representative TRO11 pseudovirus TZM-bl neutralization curves at w10 (bolus), w12 (2w OP group, and w14 (4w OP group).
ELISA endpoint titers calculated as dilution at which the O.D. is 0.1 above background. Neutralization titers are reported as ID50 titers. All BG505 binding and neutralization data represent geometric mean titers ± geometric SD using BG505 N332.
A) Frequencies of Env+ and Envhi B cells at sorted time points (bolus, w12 and OP, w14). Env+ cells were used for BCR sequencing.
(B) Quantification of IgG, IgL, and IgK lineages normalized by number of cells sorted. Each data point is an individual LN.
(C) Representative phylogenetic analysis of a single Env-specific lineage found in only one LN.
(D) IgG, IgL, and IgK clonal abundance in individual LNs and within each group.
(E) Shannon H indices of individual LNs. **p<0.01
(F) Comparison of mutation frequencies across BCR. Statistical significance tested using student’s t-test, ****p<0.0001
(G) Comparison of length of CDR3 in heavy and light chains.
(A) Frequency of IGKV gene use, assessed using genomic RM reference in this study.
(B) Frequency of IGHV genes used by BCRs sequenced in Figure 6, assessed using IMGT database.
(C) Frequency of IGKV gene use, assessed using IMGT database.
(D) Frequency of IGLV gene use, assessed using IMGT database. ***q<0.001, FDR = 5%. Mean ± SEM are graphed.
(A) Binding curves of BDA2, and control mAb, 19R to Env trimer. 19R is a High-affinity base-binding mAb. BDA2 showed no binding to Env trimer.
(B) Binding curves of isolated mAbs and control VRC01 and 19R mAbs to gp120. VRC01 is an HIV-1 bnAb that recognizes the CD4 binding site and gp120 monomer.
(C) Binding curves of isolated mAbs and control anti-his mAb to his peptide.
(D) Nucleotide alignment of week 7 BDA1 heavy and light chains, its inferred germline sequences and three closest w12 Env-specific BCR sequences.
(E) Phylogenetic analyses of sequences in B.
(F) Protein alignment of BDA1 and related mAbs.
3D reconstructions of Fab-binding in individual animals. Fabs targeting regions in some animals were rare and were not included in individual 3D reconstructions, but are presented in the 2D images.
(A) Cumulative BGC cell responses to the 2nd immunizations [AUC]. AUC was calculated between w11-15.
(B) Quantification of Env trimer-specific B cells over time.
(C) Cumulative Env trimer-specific B cell responses to one and two immunizations [AUC].
(D) Cumulative Env trimer-specific BGC cell responses to the 2nd immunization [AUC].
(E) Frequency of Env trimer-specific BGC cells over time.
(F) Cumulative Env trimer-specific BGC cell responses to one and two immunizations [AUC].
(G) Mean fluorescent intensities (MFIs) of Env trimer-specific B cells over time. Value calculated as: average of EnvAx647 and EnvBV421 MFIs (Env+ B cells) – average of EnvAx647 and EnvBV421 MFIs (Env− B cells). Number of LNs graphed are: bolus, 12 (w5), 11 (w7), 11 (w9), 10 (w11), 11 (w13), 11 (w15), 7 (w18), 9 (w27), 12 (w28); ED, 10 (w5), 12 (w7), 12 (w9), 12 (w11), 12 (w13), 12 (w15), 10 (w18), 8 (w27), 6 (w28). Statistical significance was tested using multiple t tests with 5% FDR. *q<0.05, **q<0.01, ***q<0.001, ****q<0.0001.
(H) Frequency of High-affinity Env trimer-specific B cells over time.
(I) Cumulative High-affinity B cell responses to one and two immunizations [AUC].
(J) Flow cytometry analysis of High-affinity Env trimer-specific BGC cells in a bolus immunized animal between w-1 and w8.
(K) Frequency of High-affinity Env trimer-specific BGC cells over time.
(L) Cumulative High-affinity BGC cell response to one and two immunizations [AUC].
(M) Quantification of Env-specific BMem cells over time.
(N) Cumulative Env trimer-specific BMem cells to one and two immunizations [AUC].
(O) Quantification of High-affinity Env trimer-specific BMem cells over time.
(P) Cumulative High-affinity Env trimer-specific BMem cells responses to one and two immunizations [AUC].
(Q) Cumulative GC-TFH cell responses to the 2nd immunization [AUC].
(R) Quantification of Env trimer-specific GC-TFH cells after 1 immunization.
(S) Neutralization breadth using 12-virus panel at w26 (bolus) and w27 (ED).
Cell-frequency data represent mean ± SEM. Statistical significance was tested using unpaired, two-tailed Mann-Whitney U tests, unless otherwise noted. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
(A) 3D reconstructions of Fab-binding in individual animals.
(B) Quantitation of EnvAX647 (Alexa647-labelled MD39) in all harvested LNs from RMs immunized via conventional bolus , 2w OP, or an ED regimen. Mean ± SEM are graphed. Statistical significance was tested using two-way ANOVA. *adjusted p <0.05, **p<0.01, ***p<0.001
(C) Histology of draining LNs at d2 and d7 of all three immunization groups. Green, Env; red, CD35; blue, Ki67. Scale bars, 100μm. D7 bolus image panel is same as that in figure 8C.
360° view of iliac LNs of immunized animals, Related to Figure 7.
Highlights.
Slow delivery immunization enhances HIV neutralizing antibody development in monkeys
Slow delivery immunization alters immunodominance of the responding B cells
Weekly longitudinal germinal center (GC) B and Tfh analyses provided new GC insights
High resolution rhesus immunoglobulin loci genomic reference sequence
ACKNOWLEDGEMENTS
We thank A. Fungtammasan and B. Hannigan for genome assembly, S. Gumber of Yerkes National Primate Research Center (YNPRC) for tissue isolation assistance, T. Diefenbach of the Ragon Institute Imagine Core for assistance, the Harvard Neurobiology Imaging Facility for consultation (NS072030), and J. Crotty for graphical assistance. This study was funded by the National Institutes of Health grants AI125068 (D.J.I., S.C.), Al100663 (Scripps CHAVI-ID: S.C., G.S., D.J.I., W.S., A.B.W., D.R.B.), RR00165/OD011132 (YNPRC), AI124436 (S.C., G.S., S.B.), AI136621 (A.B.W.), F31Al131873 (C.A.C.), and by the Ragon Institute and Howard Hughes Medical Institute (D.J.I).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Sajjadian S, and Eichler EE (2011). Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew S (2010). FastQC: A quality control tool for high throughput sequence data. 2010. [Google Scholar]
- Andrews SF, Graham BS, Mascola JR, and McDermott AB (2018). Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Immunogenetic Considerations Underlying B-Cell Biology in he Development of a Pan-Subtype Influenza A Vaccine Targeting the Hemagglutinin Stem. Cold Spring Harb Perspect Biol 10, a029413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, and Yewdell JW (2018). Understanding and Manipulating Viral Immunity: Antibody Immunodominance Enters Center Stage. Trends Immunol 39, 549–561. [DOI] [PubMed] [Google Scholar]
- Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat. Immunol 18, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, et al. (2018). Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49, 288–300.e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, and Hangartner L (2016). Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol 34, 635–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, and Tesler G (2012). Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Del Rio PME, Tharp GK, Trible RP, Amara RR, Chahroudi A, Reyes-Teran G, Bosinger SE, and Silvestri G (2015). Decreased T Follicular Regulatory Cell/T Follicular Helper Cell (TFH) in Simian Immunodeficiency Virus-Infected Rhesus Macaques May Contribute to Accumulation of TFH in Chronic Infection. J. Immunol 195, 3237–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, and Crotty S (2017). Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol 47, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran MM, Phad GE, Vázquez Bernat N, Stahl-Hennig C, Sumida N, Persson MAA, Martin M, and Karlsson Hedestam GB (2016). Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7, 13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, and Crotty S (2016). A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J. Immunol 197, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, et al. (2014). Global Panel of HIV-1 Env Reference Strains for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J. Virol 88, 2489–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMuth PC, Garcia-Beltran WF, Ai-Ling ML, Hammond PT, and Irvine DJ (2013). Composite dissolving microneedles for coordinated control of antigen and adjuvant delivery kinetics in transcutaneous vaccination. Adv Funct Mater 23, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMuth PC, Min Y, Irvine DJ, and Hammond PT (2014). Implantable silk composite microneedles for programmable vaccine release kinetics and enhanced immunogenicity in transcutaneous immunization. Adv Healthc Mater 3, 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenmann F, and Lefranc M-P (2011). IMGT/DomainGapAlign: IMGT standardized analysis of mino acid sequences of variable, constant, and groove domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb Protoc 2011, 737–749. [DOI] [PubMed] [Google Scholar]
- Ehrenmann F, Kaas Q, and Lefranc M-P (2010). IMGT/3DstructureADB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res 38, D301–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, et al. (2016). Thermostability of Well-Ordered HIV Spikes Correlates with the Elicitation of Autologous Tier 2 Neutralizing Antibodies. PLoS Pathog 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francica JR, Sheng Z, Zhang Z, Nishimura Y, Shingai M, Ramesh A, Keele BF, Schmidt SD, Flynn BJ, Darko S, et al. (2015). Analysis of immunoglobulin transcripts and hypermutation following SHIV(AD8) infection and protein-plus-adjuvant immunization. Nat Commun 6, 6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CDAHIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. (2007). Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234. [DOI] [PubMed] [Google Scholar]
- Gitlin AD, Mayer CT, Oliveira TY, Shulman Z, Jones MJK, Koren A, and Nussenzweig MC (2015). HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science 349, 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, and Nussenzweig MC (2014). Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, and Kleinstein SH (2015). Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Torrents de la Peña A, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL, et al. (2016a). Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep 17, 2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Lee JH, and Crotty S (2017). Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunological Reviews 275, 49–61. [DOI] [PubMed] [Google Scholar]
- Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents de la Peña A, Kasturi SP, Dan JM, Bothwell M, Sanders RW, et al. (2016b). Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. J. Immunol 197, 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. (2012). Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med 366, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch H (2002). Mechanisms of stimulation of the immune response by aluminum adjuvants. Vaccine 20, S34–S39. [DOI] [PubMed] [Google Scholar]
- Hogenesch H (2012). Mechanism of immunopotentiation and safety of aluminum adjuvants. Front Immunol 3, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, et al. (2015). Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J. Virol 89, 10383–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison S, Benson RA, Gibson VB, Pollock AH, Garside P, and Brewer JM (2012). Antigen depot is not required for alum adjuvanticity. Faseb J 26, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ (2002). BLAT--the BLAST-like alignment tool. Genome Res 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, Ketas TJ, Cottrell CA, Ozorowski G, Debnath G, Camara D, Francomano E, Pugach P, Ringe RP, LaBranche CC, et al. (2018). Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog 14, e1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, and Nussenzweig MC (2013). Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota SI, Takahashi K, Nishida J, Morishita Y, Ehata S, Tainaka K, Miyazono K, and Ueda HR (2017). Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution. Cell Rep 20, 236–250. [DOI] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. (2017). Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun 8, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Vollbrecht T, Chernyshev M, Mohan S, Hanst B, Bavafa N, Lorenzo A, Ketteringham R, Eren K, Golden M, et al. (2018). Long-read amplicon denoising. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, and Kelsoe G (2016). Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity 44, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, and Lefranc G (2001). The immunoglobulin factsbook.
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. (2013). Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39, 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, et al. (1996). Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis 173, 340–348. [DOI] [PubMed] [Google Scholar]
- Mesin L, Ersching J, and Victora GD (2016). Germinal Center B Cell Dynamics. Immunity 45, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Roederer M, Morris L, and Seaman MS (2018). Neutralization tiers of HIV-1. Curr Opin HIV AIDS 13, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Pedroza-Pacheco I, Vandergrift NA, Chui C, Lloyd KE, Parks R, Soderberg KA, Ogbe AT, Cohen MS, Liao H-X, et al. (2016). Immune perturbations in HIV-1-infected individuals who make broadly neutralizing antibodies. Science Immunology 1, aag0851–aag0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kimanius D, Lindahl E, and Scheres SH (2018). Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife 7, 1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, and Martin MA (2017). Of Mice, Macaques, and Men: Broadly Neutralizing Antibody Immunotherapy for HIV-1. Cell Host Microbe 22, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe SM, Green MA, Hogenesch H, and Hem SL (2010). Mechanism of immunopotentiation by aluminum-containing adjuvants elucidated by the relationship between antigen retention at the inoculation site and the immune response. Vaccine 28, 3588–3594. [DOI] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088.e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. (2012). CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest 122, 3281–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Plotkin SA (2010). Correlates of protection induced by vaccination. Clin. Vaccine Immunol 17, 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. (1999). Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 77, 153–161. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Darko S, Hua A, Overman G, Ransier A, Francica JR, Trama A, Tomaras GD, Haynes BF, Douek DC, et al. (2017). Structure and Diversity of the Rhesus Macaque Immunoglobulin Loci through Multiple De Novo Genome Assemblies. Front Immunol 8, 220–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Baxter AE, Cirelli KM, Dan JM, Morou A, Daigneault A, Brassard N, Silvestri G, Routy J-P, Havenar-Daughton C, et al. (2017). Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE 12, e0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med 361, 2209–2220. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, and Petropoulos CJ (2003). Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proceedings of the National Academy of Sciences 100, 4144–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, la Peña, de AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. (2015). HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223–aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, and Nussenzweig MC (2011). A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J. Exp. Med 208, 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hogenesch H, and Hem SL (2001). Change in the degree of adsorption of proteins by aluminum-containing adjuvants following exposure to interstitial fluid: freshly prepared and aged model vaccines. Vaccine 20, 80–85. [DOI] [PubMed] [Google Scholar]
- Sorzano COS, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernández G, Li M, Xu G, Marabini R, and Carazo JM (2010). A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol 171, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GBE, Soto C, Lemmin T, Chuang G-Y, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens A-J, et al. (2016). Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 165, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, and Karlsson Hedestam GB (2012). High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med 4, 142ra96–142ra96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam HH, Melo MB, Kang M, Pelet JM, Ruda VM, Foley MH, Hu JK, Kumari S, Crampton J, Baldeon AD, et al. (2016). Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. U.S.a 113, 201606050–E201606648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O (2011). Gnu parallel-the command-line power tool. [Google Scholar]
- Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill J-C, Reynaud C-A, Browne EP, et al. (2016). Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JS, Benet ZL, and Grigorova IL (2017). Antigen Acquisition Enables Newly Arriving B Cells To Enter Ongoing Immunization-Induced Germinal Centers. J. Immunol 199, 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay AA, Kauffman RC, Wolabaugh AN, Cho A, Patel NB, Reiss SM, Havenar-Daughton C, Dawoud RA, Tharp GK, Sanz I, et al. (2018). BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O’Connor KC, Hafler DA, Vigneault F, and Kleinstein SH (2014). pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30, 1930–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, and Wilson PC (2015). Germinal center selection and the antibody response to influenza. Cell 163, 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B (2009). DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, and Breden F (2012). The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 13, 363–373. [DOI] [PubMed] [Google Scholar]
- Watson CT, Glanville J, and Marasco WA (2017). The Individual and Population Genetics of Antibody Immunity. Trends Immunol 38, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422, 307–312. [DOI] [PubMed] [Google Scholar]
- Weissburg RP, Berman PW, Cleland JL, Eastman D, Farina F, Frie S, Lim A, Mordenti J, Peterson MR, Yim K, et al. (1995). Characterization of the MN gp120 HIV-1 Vaccine: Antigen Binding to Alum. Pharmaceutical Research 12, 1439–1446. [DOI] [PubMed] [Google Scholar]
- West AP, Scharf L, Scheid JF, Klein F, Bjorkman PJ, and Nussenzweig MC (2014). Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Lynch RM, Gautam R, Matus-Nicodemos R, Schmidt SD, Boswell KL, Darko S, Wong P, Sheng Z, Petrovas C, et al. (2015). Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Sci Transl Med 7, 298ra120–298ra120. [DOI] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41, W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C-H, Nojima T, Kuraoka M, and Kelsoe G (2018). Germinal center entry not selection of B cells is controlled by peptide-MHCII complex density. Nat Commun 9, 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang G-Y, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep 19, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Full gating strategy of BGC cells. KI67+ BCL6+ expression was used at weeks -1-6, 8-14. CD38−CD71+ expression used at week 7.
(B) Validation of CD38 and CD71 as surface markers for bona fide BGC cells in mesenteric LNs of RM.
(C) BGC frequencies of bolus and individual OP groups. ** p<0.01. Blue asterisks, 2w OP vs. bolus. Pink asterisks, 4w OP vs. bolus.
(D) Cumulative BGC cell responses to first immunization of individual groups between w1 plus w3-7.
(E) Lambda expression in BGC and non-BGC cells. Top panel, gating strategy for BGC and non-BGC cells in LNs. Lower panel, histogram and quantification of lambda expression in lambda+ BGC and non-BGC cells.
(F) Gating strategy of Env trimer-specific B cells, gated on CD20+ cells as per Figure S1A.
(G) Olio6CD40KO differs from Olio6 by a single amino acid, to prevent binding to human CD4. Neither trimer binds RM CD4.
(H) Gating strategy and flow cytometry analysis of Env trimer-specific BGC cells over time within an individual LN, gated on CD20+ cells as per Figure S1A.
(I) Quantification of IgG gMFI of Env+ BGC cells. Differences in gMFI between weeks is due to use of different panels or flow cytometers for acquisition.
(J) Representative flow cytometry plot of High-affinity Env trimer-specific B cells, gated on CD20+ cells.
(K) Quantification of high affinity Env trimer-specific B cells over time.
(L) Cumulative High-affinity Env trimer-specific B cell response between w1 plus w3-7 [AUC].
(M) Flow cytometry analysis of High-affinity Env-specific BGC cells in a bolus immunized animal between w-1 and 8.
(N) Quantification of High-affinity Env trimer-specific BGC cells over time.
(O) Cumulative High-affinity Env-specific BGC cell responses within individual LNs between w2 and w7.
(P) Quantification of Env trimer-specific BMem cells. BMem were defined as the non-GC (BCL6− KI67− or CD38+ CD71−) B cells within the Env-specific B cell gate as per figure S1E.
(Q) Cumulative Env trimer-specific BMem cell response between weeks 3 and 7 [AUC]. All data represent mean ± SEM.*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
(R) Mean fluorescent intensities (MFIs) of Env trimer-specific B cells over time. Value calculated as: average of EnvAx647 and EnvBV421 MFIs (Env+ B cells) – average of EnvAx647 and EnvBV421 MFIs (Env− B cells).
As bolus Gp1 was not stained at same time as other groups, Gp1 was not included in these analyses. Number of LNs graphed are: bolus, 6 (w4), 4 (w5), 5 (w6), 5 (w7), 4 (w8); OP, 15 (w4), 15 (w5), 12 (w6), 15 (w7), 15 (w8). Statistical significance was tested using multiple t tests with 5% FDR.
Env-specific and High-affinity Env-specific B and germinal center (GC) B cell counts in osmotic pump study, Related to Figure 1. *data was not included in analyses due to low cell number
Env-specific and High-affinity Env-specific B and germinal center (GC) B cell counts in dose escalation study, Related to Figure 6. *data was not included in analyses due to low cell number.
Sequences of annotated IGH, IGK and IGL ORFs, Related to Figure 4.
Table S2. Quantification of Env-specific BGC cells used for BCR sequence analyses, Related to Figures 4 and 5.
Table S3. Sequences of BDA monoclonal antibodies, Related to Figure 5.
(A) Full gating strategy of GC-TFH cells. GC-TFH cells were measured at all time points, except for w7.
(B) Bolus and OP immunized animals have comparable frequencies of 4-1BB+ OX40+ CD4+ T cells under SEB stimulation of AIMOB assay. SEB stimulation serves as a positive control for the overall health of the cells.
(C) Representative BG505 Env IgG binding IgG curves at w11. Unimmunized control is at w-2.
(D) BG505 Env trimer endpoint binding IgG titers over time.
(E) Representative anti-His binding IgG curves at w11. Unimmunized control is at w-2.
(F) Quantification of anti-His endpoint binding IgG titers over time.
(G) Direct comparison of BG505 binding IgG titers of animals in this study vs animals in Pauthner et al. For Pauthner et al. groups, plasma from w8 and w12 was used. For bolus and 2w OP groups, plasma from w7 and w12 was used.
(H) Representative BG505 pseudovirus TZM-bl neutralization curves at w14.
(I) Quantification of BG505 neutralization titers between groups over time.
(J) Peak autologous neutralization titers of each animal after two immunizations.
(K) Representative TRO11 pseudovirus TZM-bl neutralization curves at w10 (bolus), w12 (2w OP group, and w14 (4w OP group).
ELISA endpoint titers calculated as dilution at which the O.D. is 0.1 above background. Neutralization titers are reported as ID50 titers. All BG505 binding and neutralization data represent geometric mean titers ± geometric SD using BG505 N332.
A) Frequencies of Env+ and Envhi B cells at sorted time points (bolus, w12 and OP, w14). Env+ cells were used for BCR sequencing.
(B) Quantification of IgG, IgL, and IgK lineages normalized by number of cells sorted. Each data point is an individual LN.
(C) Representative phylogenetic analysis of a single Env-specific lineage found in only one LN.
(D) IgG, IgL, and IgK clonal abundance in individual LNs and within each group.
(E) Shannon H indices of individual LNs. **p<0.01
(F) Comparison of mutation frequencies across BCR. Statistical significance tested using student’s t-test, ****p<0.0001
(G) Comparison of length of CDR3 in heavy and light chains.
(A) Frequency of IGKV gene use, assessed using genomic RM reference in this study.
(B) Frequency of IGHV genes used by BCRs sequenced in Figure 6, assessed using IMGT database.
(C) Frequency of IGKV gene use, assessed using IMGT database.
(D) Frequency of IGLV gene use, assessed using IMGT database. ***q<0.001, FDR = 5%. Mean ± SEM are graphed.
(A) Binding curves of BDA2, and control mAb, 19R to Env trimer. 19R is a High-affinity base-binding mAb. BDA2 showed no binding to Env trimer.
(B) Binding curves of isolated mAbs and control VRC01 and 19R mAbs to gp120. VRC01 is an HIV-1 bnAb that recognizes the CD4 binding site and gp120 monomer.
(C) Binding curves of isolated mAbs and control anti-his mAb to his peptide.
(D) Nucleotide alignment of week 7 BDA1 heavy and light chains, its inferred germline sequences and three closest w12 Env-specific BCR sequences.
(E) Phylogenetic analyses of sequences in B.
(F) Protein alignment of BDA1 and related mAbs.
3D reconstructions of Fab-binding in individual animals. Fabs targeting regions in some animals were rare and were not included in individual 3D reconstructions, but are presented in the 2D images.
(A) Cumulative BGC cell responses to the 2nd immunizations [AUC]. AUC was calculated between w11-15.
(B) Quantification of Env trimer-specific B cells over time.
(C) Cumulative Env trimer-specific B cell responses to one and two immunizations [AUC].
(D) Cumulative Env trimer-specific BGC cell responses to the 2nd immunization [AUC].
(E) Frequency of Env trimer-specific BGC cells over time.
(F) Cumulative Env trimer-specific BGC cell responses to one and two immunizations [AUC].
(G) Mean fluorescent intensities (MFIs) of Env trimer-specific B cells over time. Value calculated as: average of EnvAx647 and EnvBV421 MFIs (Env+ B cells) – average of EnvAx647 and EnvBV421 MFIs (Env− B cells). Number of LNs graphed are: bolus, 12 (w5), 11 (w7), 11 (w9), 10 (w11), 11 (w13), 11 (w15), 7 (w18), 9 (w27), 12 (w28); ED, 10 (w5), 12 (w7), 12 (w9), 12 (w11), 12 (w13), 12 (w15), 10 (w18), 8 (w27), 6 (w28). Statistical significance was tested using multiple t tests with 5% FDR. *q<0.05, **q<0.01, ***q<0.001, ****q<0.0001.
(H) Frequency of High-affinity Env trimer-specific B cells over time.
(I) Cumulative High-affinity B cell responses to one and two immunizations [AUC].
(J) Flow cytometry analysis of High-affinity Env trimer-specific BGC cells in a bolus immunized animal between w-1 and w8.
(K) Frequency of High-affinity Env trimer-specific BGC cells over time.
(L) Cumulative High-affinity BGC cell response to one and two immunizations [AUC].
(M) Quantification of Env-specific BMem cells over time.
(N) Cumulative Env trimer-specific BMem cells to one and two immunizations [AUC].
(O) Quantification of High-affinity Env trimer-specific BMem cells over time.
(P) Cumulative High-affinity Env trimer-specific BMem cells responses to one and two immunizations [AUC].
(Q) Cumulative GC-TFH cell responses to the 2nd immunization [AUC].
(R) Quantification of Env trimer-specific GC-TFH cells after 1 immunization.
(S) Neutralization breadth using 12-virus panel at w26 (bolus) and w27 (ED).
Cell-frequency data represent mean ± SEM. Statistical significance was tested using unpaired, two-tailed Mann-Whitney U tests, unless otherwise noted. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
(A) 3D reconstructions of Fab-binding in individual animals.
(B) Quantitation of EnvAX647 (Alexa647-labelled MD39) in all harvested LNs from RMs immunized via conventional bolus , 2w OP, or an ED regimen. Mean ± SEM are graphed. Statistical significance was tested using two-way ANOVA. *adjusted p <0.05, **p<0.01, ***p<0.001
(C) Histology of draining LNs at d2 and d7 of all three immunization groups. Green, Env; red, CD35; blue, Ki67. Scale bars, 100μm. D7 bolus image panel is same as that in figure 8C.
360° view of iliac LNs of immunized animals, Related to Figure 7.
Data Availability Statement
Env-specific B cell BCR and whole genome raw reads used in this paper are available at NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession SBKD00000000. The version described in this paper is version SBKD01000000. The raw genomic reads and genome assembly are both under BioProject ID PRJNA509445. Env-specific BCR sequences are available under BioProject ID PRJNA520929 and are deposited at DDBJ/ENA/GenBank under the accession: KCVI00000000, KCVJ00000000, KCVK00000000, KCVL00000000, KCVM00000000, KCVN00000000, KCVO00000000, KCVP00000000, KCVQ00000000, KCVR00000000, KCVS00000000, KCVT00000000, KCVU00000000, KCVV00000000, KCVW00000000, KCVX00000000, KCVY00000000, KCVZ00000000, KCWA00000000, KCWB00000000, KCWC00000000, KCWD00000000, KCWE00000000, KCWF00000000, KCWG00000000, KCWH00000000, KCWI00000000, KCWJ00000000, KCWK00000000, KCWL00000000, KCWM00000000, KCWN00000000, KCWO00000000, KCWP00000000, KCWQ00000000, KCWR00000000, KCWS00000000, KCWT00000000, KCWU00000000, KCWV00000000, KCWW00000000, KCWX00000000, KCWY00000000, KCWZ00000000, KCXA00000000, KCXB00000000, KCXC00000000, KCXD00000000, KCXE00000000, KCXF00000000, KCXG00000000, KCXH00000000, KCXI00000000, KCXJ00000000, KCXK00000000, KCXL00000000, KCXM00000000, KCXN00000000, KCXO00000000, KCXP00000000, KCXQ00000000, KCXR00000000. 3D EM reconstructions have been deposited in the Electron Microscopy Databank (http://www.emdatabank.org/) under the accession numbers listed in the Key Resources Table.
| Lymph node GC Panel | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Env probe-biotin | Alexa Fluor 647 | Invitrogen | |
| Env probe-biotin | Brilliant Violet 421 | BioLegend | |
| Viability | efluor506 | Thermo Fisher | |
| CD20 | PE-Texas Red | Beckman Coulter | 2H7 |
| CD4 | Brilliant Violet 650 | Biolegend | OKT-4 |
| CD8a | Qdot 705 | Thermo Fisher | 3B5 |
| IgG | PE-Cy7 | BD Biosciences | G18-145 |
| CXCR5 | PE | Thermo Fisher | MU5UBEE |
| PD1 | Brilliant Violet 605 | Biolegend | EH12.2H7 |
| CD3 | Brilliant Violet 786 | BD Biosciences | SP34-2 |
| IgM | PerCP-Cy5.5 | BD Biosciences | G20-127 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| BCL6 | Alexa Fluor 488 | BD Biosciences | K112-91 |
| Antigen-specific B cell sorts | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Env probe-biotin | Alexa Fluor 647 | Invitrogen | |
| Env probe-biotin | Brilliant Violet 421 | BioLegend | |
| Viability | efluor780 | Thermo Fisher | |
| CD4 | APC efluor780 | Thermo Fisher | SK3 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD16 | APC efluor780 | Thermo Fisher | ebioCD16 |
| CD20 | Alexa Fluor 488 | BioLegend | 2H7 |
| IgG | PE-Cy7 | BD Biosciences | G18-145 |
| IgM | PerCP-Cy5.5 | BD Biosciences | G20-127 |
| CD38 | PE | NHP Reagents | OKT |
| CD71 | PE-CF594 | BD Biosciences (custom) | L01.1 |
| AIM Assay Panel | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| CD4 | Brilliant Violet 650 | BioLegend | OKT4 |
| CD20 | Brilliant Violet 570 | BioLegend | 2H7 |
| PD1 | Brilliant Violet 785 | BioLegend | EH12.2H7 |
| CXCR5 | PE-Cy7 | Thermo Fisher | MU5UBEE |
| CD25 | FITC | BioLegend | BC96 |
| 0X40 | PE | BD Biosciences | L106 |
| 4-1BB | APC | BioLegend | 4B4-1 |
| Viability | efluor780 | Thermo Fisher | |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD14 | APC/Cy7 | BioLegend | M5E2 |
| CD16 | APC/Cy7 | BioLegend | 3G8 |
| BGC cell surface marker validation | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Viability | efluor780 | Thermo Fisher | |
| CD20 | Brilliant Violet 650 | BioLegend | 2H7 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD4 | APC | BioLegend | 0KT4 |
| CD38 | PE | NHP Reagents | OKT |
| CD71 | PE-CF594 | BD Biosciences (custom) | L01.1 |
| BCL6 | BV421 | BD Biosciences | K112-91 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| Macaque BCR expression | |||
|---|---|---|---|
| Marker | Fluorochrome | Company | Clone |
| Viability | efluor780 | Thermo Fisher | |
| CD4 | APC efluor780 | Thermo Fisher | SK3 |
| CD8a | APC efluor780 | Thermo Fisher | RPA-T8 |
| CD16 | APC/Cy7 | BioLegend | 3G8 |
| CD20 | Brilliant Violet 650 | BioLegend | 2H7 |
| BCL6 | Alexa Fluor 647 | BD Biosciences | K112-91 |
| KI67 | Alexa Fluor 700 | BD Biosciences | B56 |
| IgM | BV421 | BD Biosciences | G20-127 |
| IgG | PE | BD Biosciences | G18-145 |
| IgD | Alexa Fluor 488 | Southern Biotech | |
| Lambda | Biotin | Miltenyi | IS7-24C7 |
| Streptavidin | Brilliant Violet 711 | BioLegend | |