Abstract
Objective:
The high mortality associated with cervical cancer is due to low uptake of Pap smear test, lack of early diagnostic biomarkers and less-invasive approach, and late presentation of the disease. This study evaluated the expression of hsa-miR-let-7b, hsa-miR-21, hsa-miR-125b, hsa-miR-143, hsa-miR-145, hsa-miR-146a, hsa-miR-155, hsa-miR-182, hsa-miR-200c, and p53 in serum and cervix in relation to classes of Pap smear, in a bid to identify a serum panel for early diagnosis of cervical lesions.
Methodology:
This study included 329 women; 159 healthy women (HW), 46 cervicitis, 46 atypical squamous cells of undetermined significance (ASCUS), 40 low-grade squamous cell intraepithelial lesion (LSIL), 28 high-grade squamous cell intraepithelial lesion (HSIL), and 10 squamous cell carcinoma (SCC). Expression of microRNAs (miRNAs) and p53 was assessed using reverse transcriptase polymerase chain reaction.
Results:
Except for miR-143 and miR-146a, significant correlations were observed between serum and cervix expression of miRNAs and p53 in relation to levels and classes of Pap smear (P < 0.05). Relatively, higher expression of miR-21, miR-146a, miR-155, miR-182, and miR-200c and lower expression of let-7b and miR-145 were observed in sera associated with cervical abnormalities than in sera associated with normal cervix (P < 0.0001, P = 0.001, P < 0.0001, P = 0.003, P = 0.007, P = 0.036, and P = 0.046, respectively). Higher and lower expression of p53 was observed in women diagnosed of LSIL and SCC, respectively, than in HW (P < 0.0001).
Conclusion:
This study suggests that serum expression of miR-21, miR-182, let-7b, miR-145, and p53 is comparable to cervical cell expression and could be useful in differentiating abnormal cervix from the healthy cervix.
Keywords: Cancer diagnosis, cervix, microRNAs, p53, serum
Introduction
About 500,000 new cases of cervical cancer are reported each year while approximately 49% of these cases result in death worldwide.[1] Approximately 85% of these deaths occur in developing countries, majorly due to the absence of discomforting signs and symptoms, inadequate access to screening facilities, fear of the test procedure,[2,3] and ineffective implementation of government policies regarding laboratory testing of high-risk women.[4-7] Current innovative trends in cancer diagnosis are associated with microRNA (miRNA) expression. miRNAs, a group of short non-coding RNAs, are estimable in various body fluids including serum.[8] In the past decade, dysregulated miRNAs were associated with histologically confirmed cervical cancer.[9] The variation in the expression of miRNAs allows for risk stratification, staging and prognosis of cervical diseases, especially in high-risk Human papillomavirus (HPV) infection.[10,11] Due to the stability against RNase degradation and external influences, serum miRNAs qualify as non-invasive biomarkers for investigating diseases.[12,13] However, there is a paucity of data on the dysregulated pattern of miRNA in serum in relation to cervical carcinogenesis.
Bumrungtha et al. observed a 2-fold increase of tissue miR-21 among women with cervicitis than their healthy counterparts. They also observed higher expression of the miRNA in HPV negative (HPV-) cervicitis than HPV positive (HPV+) cervicitis or HPV- normal cells.[14] Their study suggests that cervicitis could be a risk factor for cervical cancer, irrespective of HPV status. However, they did not assess miRNA expression in serum among their participants. Over the years, a potentially less invasive diagnostic panel for the early identification and differentiation of women with precancerous lesions and cervical cancer from healthy women has proved elusive. Xin et al. in their study observed that serum miR-9, miR10a, miR-20a, and miR-196a were able to distinguish between cervical intraepithelial neoplasm (CIN) and healthy controls.[15] However, the expression of miRNAs in serum and cervical cells associated with cervicitis, precancerous lesions, and squamous cell carcinoma (SCC) was not compared. Serum HPV antibody was also not investigated. Identifying a reliable miRNA panel with a comparable expression both in serum and cervical cells in this study will allow for efficient monitoring of high-risk groups. It will also reduce the need for invasive approaches, especially in areas with low uptake of Pap smear test.
Methodology
Study participants and design
This study included 329 consenting women (mean age = 39.45 ± 11.16 years), living in Abeokuta metropolis, Ogun State. It was carried out between the months of December 2016 and May 2018.
Sample collection, handling, and assays
Consequent on receiving ethical clearances (SHA/RES/VOL.2/177 and BUHREC549/18), written informed consents were obtained from participants who opted for cervical screening by Papanicolaou (Pap) smear investigation. Samples were collected at the family planning unit in State Hospital Ijaiye. Cervical cells were collected into liquid-based cytology using cytobrush and preserved (according to SurePath protocol) while whole blood samples were collected into plain bottles and allowed to stand for not more than 2 h. Blood samples were centrifuged for 30 min at 1500 rpm. Sera were separated, aliquoted in Eppendorf tubes and stored at −20°C until analyzed. Commercial ELISA kits (from Qingdao Hightop Biotech Co. Ltd, China) were used in testing the separated sera for antibodies (Immunoglobulin G and Immunoglobulin M) against HPV.
RNA isolation
Cervical cells and serum from each woman were homogenized in Eppendorf tubes containing 50 µl Trizol reagent by pipetting up and down and later vortex using vortex mixer (BioCote; Cat #: SA8; Serial №: R800007537) and centrifuged at 2500 rpm for 15 min. 100 µl gradient separation medium (chloroform) was added to the homogenates which were subsequently vortexed and centrifuged for 30 min at 1500 rpm. The supernatants containing the RNA were aspirated into new labeled tubes. 100 µl of precipitating medium (iso-amyl alcohol) was added to the supernatant with subsequent centrifugation for 30 min at 1500 rpm. The RNA was recovered in pellet form after decanting the supernatant. 50 µl of 70% ethanol was added and centrifuged for 5 min at 1500 rpm, the supernatant was decanted and all tubes were uncapped to allow the ethanol to dry. 50 µl of nuclease-free water (NFW) was added to the total RNA isolated across the board to form the RNA solution. The concentration of total RNA (48 µl of deionized distilled water and 2 µl of RNA solution) was determined by ultraviolet (UV) absorbance spectrophotometry (JENWAY 6305) at 260 nm and 280 nm. The acceptable RNA absorbance reading was set at ≥ 0.05–1.0 while acceptable RNA quality was set at 1.8–2.2 (based on OD260/OD280 calculation).
The cDNA synthesis was carried out by adding miRNA-Universal Stem Loop Primer (USLP) cocktail which contain: Water, sterile nuclease-free (VWR Life Science Biotechnology, Code: E476-500ML); the reverse transcriptase buffer; the reverse transcriptase, miRNA-USLP specific oligos, the oligo dNTPs. 2 µl of the cocktail was aliquoted into 20 µl (containing 1.0 µg) of total RNA across the samples (to both cell and serum homogenates) for the conversion of RNA to complimentary DNA (cDNA). The samples were then incubated at room temperature overnight. The concentration of cDNA was determined spectrophotometrically for cDNA quantification at 260 nm. To establish homogeneity of cDNA (HcDNA) concentration across all samples, the formula HcDNA= VB–VA was applied; where VB (dilution volume) = CA × VA/CB; CA= absorbance reading of RNA × 40 × 25, VA= needed volume of cDNA (25) and CB= least absorbance reading. All samples were diluted to the same concentration.
Reverse transcriptase polymerase chain reaction (PCR)
The primers for miRNA quantification included the following: hsa-miR-Let-7b forward (5’-GTTTCGGGGTGAGGTAGTA-3’), hsa-miR-16 forward (5’-GTTGTCAGCAGTGCCTTAG-3’), hsa-miR-21 forward (5’-GGTGTCGGGTAGCTTATCA-3’), hsa-miR-125b forward (5’-GTTTTGCGCTC CTCTCAGT-3’), hsa-miR-143 forward (5’-TTTTTGCGCAGCGCCCTG-3’), hsa-miR-145 forward (5’-GTTTCACCTTGTCCTCACG-3’), hsa-miR-155 forward (5’-GTTTCT GTTAATGCTAATCGTGATA-3’), hsa-miR-182 forward (5’-GTTTTAGAACTCACACGTGT GA-3’), hsa-miR-200cforward (5’-GTTTCCCTCGTCTTACCCA-3’), universal reverse primer (5’-GTGCAGGGTCCGAGGT-3’), p53 gene forward (5’-GCTCAAGACTGGCGCTAA AA-3’) p53 gene reverse (5’-GTGACTCAGAGAGGACTC AT-3’); β-actin forward (5’-ACACTTTCTA CAATGAGCTGCG-3’) β-actin reverse (5’-ACCAGAGGCATACAGGACAAC-3’) and miRNA-USLP (Universal stem-loop primer; 5’-AGTGCAGGGTCCGAGGTATTCGCACCAG AGCCAACATGTCACG-3’). The amplification was performed following optimization. A mixture of template cDNA (2 µl), forward primers and reverse primers (0.5 µl each; 100 µm stock solution; Inqaba Biotechnical industries Ltd, South Africa), master mix (12.5 µl; Biolabs South Africa, concentration= 2xMMw; 100 µm; lot: 0241706), and NFW (9.5 µl) were added to each sample for complete enzymatic reaction making a final reaction volume of 25 µl, according to manufacturer’s instruction. The PCR was carried out using Multigene Optimax PCR machine (Serial No.: 1405010). Amplification conditions were: 94°C pre-denaturation for 5 min, 94°C for 30 s, annealing 55°C for 30 s, and extension 72°C for 30 s and then 5 min at 72°C by 45 cycles.
Gel electrophoresis
PCR products (amplicons) were electrophoresed in 0.5% (0.5 g) of agarose gel using 50 ml of 0.5× TBE buffer (9.8 g of Tris base, 5.0 g of Tris-boric acid, 2 ml of 0.5 M EDTA, and 48 ml of distilled water and adjusted to pH 8.3 using sodium hydroxide solution) with 0.2 µl ethidium as fluorescent tag. The expression products were visualized as bands by UV-transilluminator (foto/photophoresis; Serial No: FPG1-0204-6277). Snapshots were taken from the trans-illuminator for densitometric analysis using ImageJ software (1.49V). Snapshots of the amplicons (bands) were imported into the software and the contrast was adjusted such that the bands were vivid. The areas around each band were selected and background intensity was subtracted from the amplicons (bands). The intensities (data) of the selected bands displayed in excel format were exported for further statistical analyses. Data generated from miRNA and mRNA were quantified following endogenous normalization using miR-16 for other miRNAs and β-actin for p53 gene.[16,17] The relative expression of the miRNAs and p53 gene was calculated against the expression of miR-16 and β-actin (their respective normalizing genes) using the formula: Density of the target gene divided by the density of normalizing gene, multiplied by hundred.[18,19]
Statistical analysis
Data generated were subjected to analysis of variance, Pearson’s (bivariate), and Partial correlations using SPSS (version 21), and GraphPad Prism (version 6). Results were presented as the mean ± standard error of the mean. Significance was set at P ≤ 0.05.
Results
Following cytological evaluation of Pap smears by two cytopathologists, the age-matched participants were further grouped into normal cervix (n = 159 [48.3%]), cervicitis (n = 46 [14%]), atypical squamous cell of undetermined significance (ASCUS; n = 46 [14%]), low-grade squamous cell intraepithelial lesion (LSIL; n = 40 [12.2%]), high-grade squamous cell intraepithelial lesion (HSIL; n = 28 [8.5%]) and SCC; n = 10 (3.0%), and serum HPV positivity (n = 114/329 [34.7%]): Normal cervix 40/159 (25.2%), cervicitis 14/46 (30.4%), ASCUS 16/46 (34.8%), LSIL 23/40 (57.5%), HSIL 14/28 (50.0%), and SCC 7/10 (70%). All SCCs were confirmed histologically.
Figure 1 shows higher expression of all oncomirs (miR-21, miR-146a, miR-155, and miR-200c) in SCC both in serum and cervical cells when compared with other lesions (P < 0.05) while in serum, higher expression of miR-182 was observed in HSIL when compared other lesions and SCC (P < 0.05). Statistics showed a progressive upregulation of miR-21, miR-146a, and miR-182 with increasing severity of cervical abnormality (P < 0.05), especially in cervical cells. In cervical cells, higher expression of miR-21, miR-182, and miR-200c was observed in HSIL when compared with other precancerous lesions (P < 0.05). Both in serum and cervical cells, relatively higher expression of the miRNAs was observed in cervicitis when compared with the normal cervix (negative for lesion and cervicitis) at P > 0.05. In serum, higher expression of miR-146a and miR-200c was observed in LSIL and cervicitis, respectively, when compared other precancerous lesions (P < 0.05).
Figure 1.
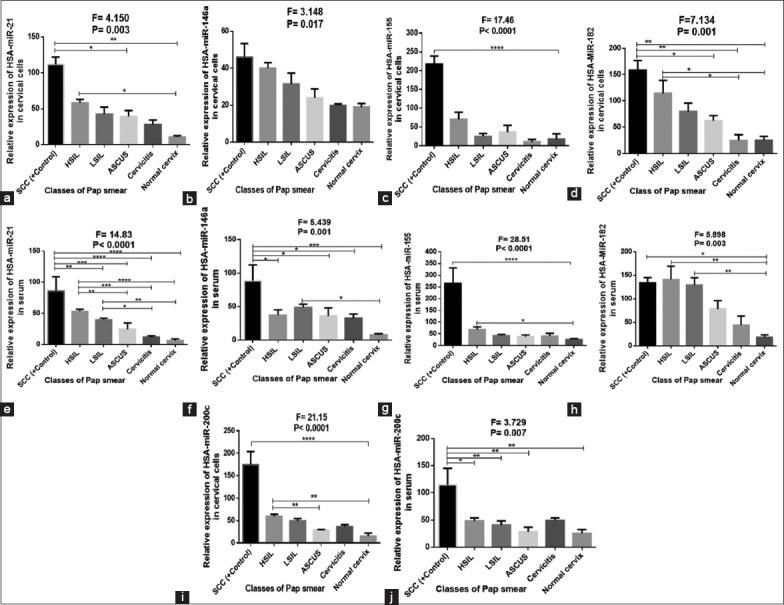
(a-j) Expression of miR-21, miR-146a, miR-155, miR-182, and miR-200c in serum and cervical cells
Figure 2 shows lower expression of let-7b, miR-125b, and p53 in SCC when compared with other lesions (P < 0.05, P > 0.05, and P < 0.05, respectively) while higher expression of miR-143 was observed in SCC when compared Negative for intraepithelial lesions (NILM) (P > 0.05). Both in serum and cervical cells, downregulation of miR-145 was observed in cervical abnormalities when compared with NILM (P > 0.05 in cervical cells and P < 0.05 in serum). Cervicitis was associated with the lowest expression of miR-145, both in serum and cervical cells. In cervical cells, higher expression of let-7b, miR-125b, miR-143, miR-145, and p53 gene was observed in cervicitis, ASCUS, HSIL, NILM, and LSIL, respectively. The gel electrophoretic amplicons are shown in Figure 3.
Figure 2.
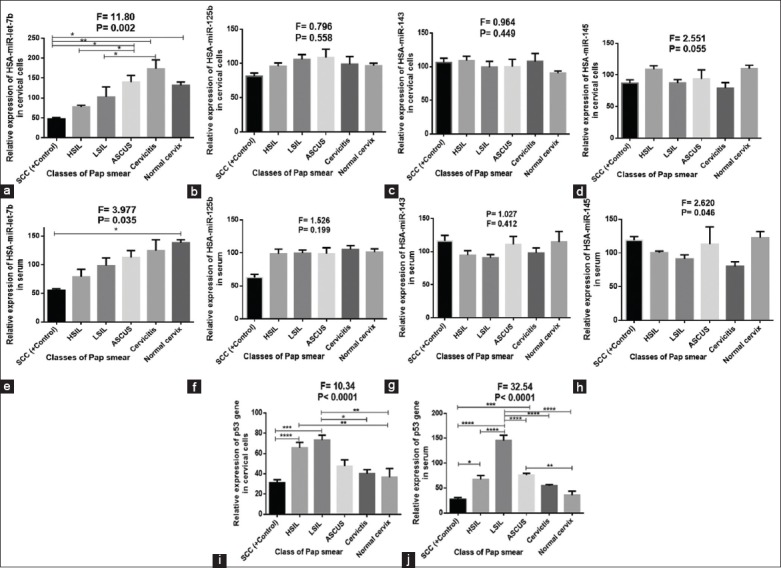
(a-j) Expression of miR-let-7b, miR-125b, miR-143, miR-145, and p53 in serum and cervical cells
Figure 3.
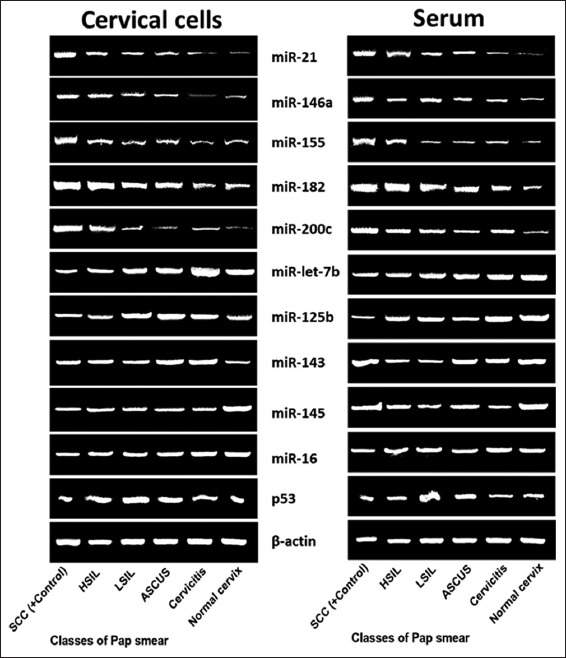
Gel electrophoretic the amplicons of miR-let-7b, miR-21, miR-125b, miR-143, miR-145, miR-146a, miR-155, miR-182, miR-200c, and p53 gene expressions associated with classes of Pap smear both in cervical cells and serum
In Figure 4, higher expression of miR-146a and p53 gene was observed in serum compared with cervical cells at P = 0.010 and P < 0.0001, respectively.
Figure 4.
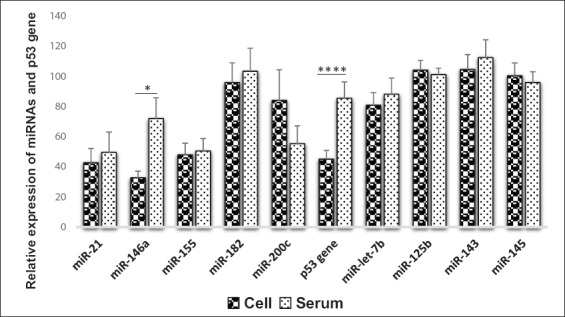
Mean comparison of biomarker expression in serum and cervical cells (Pair sample t-test)
In Figure 5, HPV+ sera had higher expression of miR-21 (2.6; P = 0.080), miR-146a (1.7; P = 0.056), miR-155 (1.1; P = 0.636), miR-182 (1.8; P = 0.092), miR-200c (3.0; P = 0.001), p53 (1.6; P = 0.030), and miR-143 (1.4; P = 0.064) than HPV- sera while HPV- sera had higher expression of let-7b (0.9; P = 0.232), miR-125b (0.9; P = 183), and miR-145 (1.4; P = 0.064) than HPV+ sera.
Figure 5.
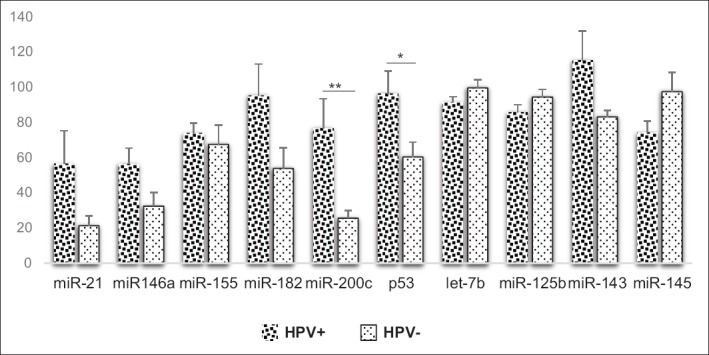
Mean comparison of biomarker expression in HPV+ and HPV- sera (independent t-test)
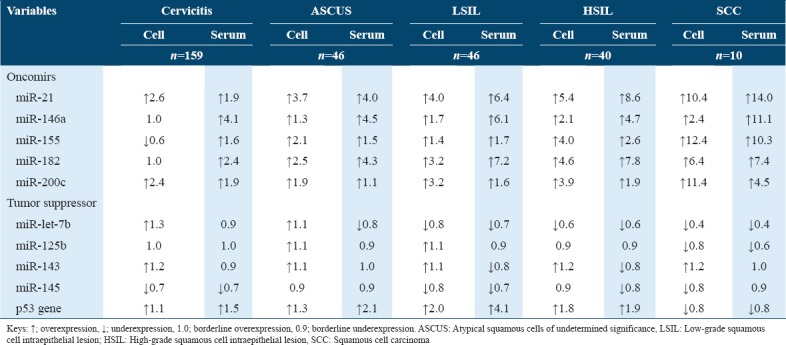
Table 1 shows overexpressed oncomirs in precancerous lesions, both in serum and cervical cells than in the normal cervix. It also showed that the sera of women positive for cervicitis had overexpressed oncomirs while their cervical cells had relatively overexpressed oncomirs. However, miR-21 and miR-200c showed a progressive increase from cervicitis to cervical cancer. Several differences were observed in relation to the expression of tumor suppressors in serum and cervical cells when individuals diagnosed with precancerous and cancers were compared with those with normal Pap smears. Table 1 shows progressive underexpression of let-7b with increasing severity of the cervical abnormality. Its expression in serum and cervical cells become uniform as the disease progresses. While miR-145 was underexpressed in cervicitis, p53 gene was overexpressed both in serum and cervical cells of individuals whose diagnosis was consistent with cervicitis. However, the miRNA was relatively decreased across the cervical abnormalities. Interestingly, miR-125b and p53 gene were relatively overexpression in precancerous lesions but underexpressed in SCC.
Table 1.
Comparison of miRNA expression between the normal cervix and compromised cervix
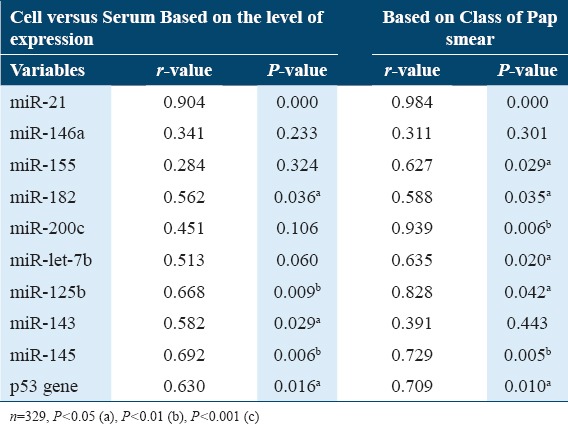
Concerning the expression of biomarkers based of levels and class of Pap smear, statistics [Table 2] showed a significant correlation between serum and cervical cells in relation to miR-21, miR-182, miR-125b, miR-145, and p53 gene (P < 0.05). In terms of expression levels of miR-182 and miR-143, a significant correlation was observed between serum and cervical cells (P < 0.05). Based on classes of Pap smear, a significant correlation was observed between serum and cervical cells in relation to miR-200c and let-7b expression (P < 0.05). However, in terms of the level of expression and classes of Pap smear, there was an insignificant correlation between serum and cervical cells expression of miR-146a (P > 0.05).
Table 2.
Correlation between cell and serum expression of miRNAs in relation to classes of Pap smear and status of viral infection (Partial correlation)
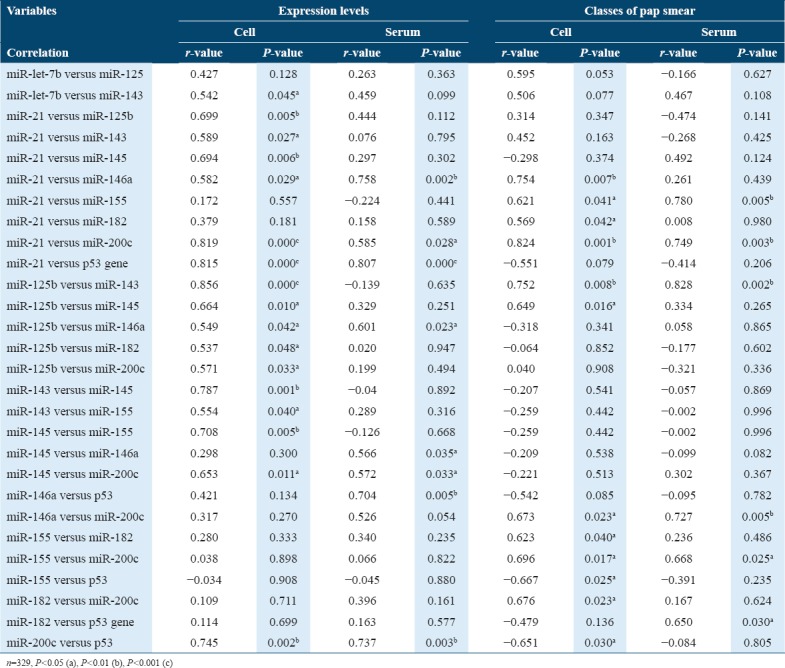
Based on expression levels and classes of Pap smears, Table 3 shows a significant correlation between miR-21 and miR-200c both in serum and cervical cells (P < 0.05). Based on expression levels, a significant correlation was observed between miR-21 and miR-146a, miR-21 and p53 gene, miR-125b and miR-146a, miR-145 and miR-200c, and miR-200c and p53, irrespective of nature of the sample (P < 0.05). Based on expression levels, in cervical cells, a significant correlation was observed between let-7b and miR-143, miR-21 and some miRNAs (125b, 143, and 146a), miR-125b and some miRNAs (143, 145, 182, and 200c), miR-143 and some miRNAs (145 and 155), and miR-145 and miR-155 (P < 0.05) while in serum a significant correlation was observed miR-145 and 146a, miR-146a and p53 (P < 0.05). Based on classes of Pap smear, a significant correlation was observed between miR-21 and miR-155, miR-125b and miR-143, miR-146a and miR-200c, and miR-155 and miR-200c both in serum and cervical cells (P < 0.05). Based on classes of Pap smear, in relation to cervical cells, a significant positive correlation was observed between miR-21 and miR-146a, miR-21 and miR-182, miR-125b and miR-145, and miR-155 and miR-182 (P< 0.05), while a significant negative correlation was observed between miR-155 and p53 gene, and miR-200c and p53 (P< 0.05) but in serum, a significant positive correlation was observed between miR-182 and p53 gene (P< 0.05).
Table 3.
Correlation between biomarkers in relation to expression levels (Pearson’s correlation), classes of pap smear (Partial correlation), and nature of samples
Discussion
This study evaluated the expression of some oncomirs (miR-21, miR-146a, miR-155, miR-182, and miR-200c) and tumor suppressor (let-7, miR-125b, miR-143, miR-145, and p53) in cervical cells and serum. The expression of miR-200c, miR-125b, and miR-145 is higher in cervical cells than in serum, while the reverse was the case for miR-146a, miR-155, miR-182, p53, let-7b, and miR-143. However, significant differences were only observed between cervical cells and serum in relation to miR-146a and p53 gene expression. The expressions of these miRNAs based on the class of Pap smear, except for that of miR-146a and miR-143 suggests that they could be used to monitor cervical carcinogenesis. The correlation between cervical cells and serum in relation to the expression levels of these miRNAs further underscores this assertion.
Although studies have shown that HPV upregulates miR-21,[20,21] previous studies show that HPV status does not significantly impact on miR-21 expression in cervical abnormalities.[14,22] The latter reports are similar to the findings of this study. Furthermore, the findings of Park et al. show that miR-21 has a predictive value of 7 and 2 folds for HPV- and HPV+ cervical cancer, respectively.[22] This might be the reason for the progressive increase in miR-21 expression both in serum and cervical cells, from cervicitis through SCC. This also may explain why miR-21 expression remains uncontroversial in cervical abnormalities.[23] This supports earlier reports which suggest that serum miRNA can be used for predicting and diagnosing cervical cancer.[24-27] The approximate two-fold change in miR-21 expression observed between cervicitis and normal cervix in this study is in consonance with the findings of Bumrungtha et al.[14] This supports the unpopular opinion that cervicitis, when observed among cervical cancer high-risk group, should be taken seriously except when it is associated with underexpressed oncomir. More so, the 1.5 fold change observed between LSIL and HSIL is in consonance with the finding of Gocze et al. between CIN and CIN 2–3.[20] Taken together, these findings suggest that serum miR-21 could be used in monitoring cervical transformation, irrespective of HPV status.
In this study, the higher expression of miR-146a observed in cervical abnormalities when compared with the normal cervix is in line with the findings of Banno et al. who did not only find upregulated miR-21 in cervical cancer cells but also upregulated miR-146a.[24] The early upregulation of miR-146a and miR-21 observed in cervicitis and precancerous lesions in this study agree with the findings of Wilting et al.[29] who recorded a similar result in precancerous cervical cells. The difference in miR-146a expression between serum and cervical cells may be due to the effect of HPV status, which may limit its application as an adjunct serum biomarker to Pap smear. However, the correlation between miR-146a and miR-21 in serum and cervical cells suggests that miR-146a could sufficiently be used to investigate cervical transformation at the cellular level.
The high expression of miR-155 in SCC suggests that the miRNA could be used to differentiate between malignant and non-malignant cervix. The low fold change observed between the normal cervix and cervicitis through precancerous lesions when compared with the high fold change observed in SCC is in concordance with the findings of other studies which posit that miR-155 is upregulated late in CIN 1–3 and cervical cancer.[29,30] The reason for the delayed upregulation of miR-155 is still unknown. Evidence suggests that the late upregulation may be due to the regulatory effects of miR-143, miR-145, and p53 in premalignant cervical lesions, which were all found to be negatively correlated with miR-155.
In vitro, studies have shown that miR-182 is overexpressed in HPV positive human cervical cancer cell lines such as CaSki and SiHa cell lines.[31-33] Similar findings were observed in this study which showed higher expression of the miRNA in HPV+ serum. More so, the findings of Li et al. show late expression of miR-182 in cervical cancer,[30] while other studies show early expression of the oncomir in precancerous lesions and higher expression in cervical cancer.[34-36] The latter is similar to the findings of this study, which showed overexpressed miR-182 with increasing severity of cervical abnormality both in serum and cervical cells. The overexpression of the miRNA in serum among individuals positive for cervicitis and precancerous lesions suggest that it is a potential less-invasive biomarker for early detection of the disease. At the cellular level, a significant correlation was observed between miR-182 and miR-200c.
The upregulation of miR-200c among women positive for cervical abnormalities align with earlier studies which demonstrated overexpressed miR-200c in invasive and metastasizing cervical cancer.[29,32,37] The correlation between its expression in serum and cervical cells and its correlation with miR-21, irrespective of nature of sample and class of Pap smear, suggests that serum miR-200c could be used as a potential biomarker for investigating cervical abnormalities. Again, its 3 fold increase in HPV+ when compared with HPV- individuals also supports its use in investigating high-risk groups.
Wild type p53 (wtp53) is a tumor suppressor which regulates differentiation, cell cycle, and apoptosis especially in the advent of cellular stress evoked by oncoviruses.[38] Some viruses thrive during active wtp53 activity while others successfully replicate in restrained or dysregulated wtp53 activity with resultant malignant transformation.[39,40] The findings of this study suggest that the p53 increases with increasing upregulation of oncomirs up to the point of LSIL, a stage of cervical abnormality that precedes viral integration. It also shows that sustained upregulation of oncomirs was associated with the expression of p53 diminishes. This explains why there was a negative correlation between p53 and the oncomirs. The decreased expression of p53 in serum and cervical cells of women diagnosed with HSIL and SCC could be due to the integration of viral DNA in the host genome. The shift in the expression of p53 before, during, and after cervical transformation, both in serum and cervical cells suggest that is a potential biomarker for early diagnosis of cervical cancer and monitoring high-risk group.
The downregulation of let-7b in precancerous lesions and cervical cancer, when compared with normal cervical cells, is in line with an earlier study which shows that underexpressed let-7b in the cervix correlate well with malignant transformation.[10,41] The cellular upregulation of let-7b among individuals whose Pap smear were consistent with cervicitis and ASCUS appear to align with the findings of Li et al. who reported overexpressed let-7b in precancerous lesions.[30] This suggests that let-7b is upregulated during the early stages of cervical carcinogenesis, perhaps following immune response to high-risk HPV and downregulated soon after significant cervical transformation.[10] However, the transient upregulation observed in cervical cells associated with cervicitis and ASCUS was not observed in serum.
miRNA 125b is abundantly expressed in the cervical transformation zone. It naturally decreases cell proliferation and induces cancer-cell apoptosis.[42,43] The relative upregulation of miR-125b both in serum and cervical cells associated with precancerous lesions could be an attempt to forestall malignant transformation. This finding is similar to the findings of earlier studies in which miR-125b was transiently overexpressed shortly after HPV infection,[24,44-46] and underexpressed following malignant transformation.[10,29,46] The latter is associated with HPV-L2 induced miR-125b inactivation and concomitant formation of koilocytes.[47]
Despite the fact that both miR-143 and miR-145 are found on the locus of chromosome 5 (q32), there are controversies regarding their expressions and specific functions in cervical carcinogenesis.[8,48] Series of in vitro (involving CaSki and SiHa cells) and in vivo (among individuals diagnosed with cervical cancer) investigations show that miR-143 and miR-145 are underexpressed in HPV16/18/31 positive cells.[10,49,50] Interestingly, an array of data also show that HPV- cancer cell line (C-33A) had underexpressed miR-143 and miR-145.[32] In 2011, Li et al. reported that miR-145 is overexpressed in CIN but underexpressed in cervical cancer.[30] Subsequently, Wilting et al., following in vitro and in vivo analysis, observed that miR-143 and miR-145 are transiently overexpressed in HPV+ premalignant lesions (1.38 and 1.05 fold changes, respectively). They also observed 1.92 and 0.85 fold changes of miR-143 and miR-145, respectively, in malignant cells when compared with the healthy cervix. The reason for the discordance is yet to be understood.[29] However, in this study, miR-143 was relatively and progressively upregulated in abnormal cervical cells while the expression varied in serum. The observed variation in miR-143 expression may be influenced by HPV type, hence more studies on this are warranted. Contrastingly, in this study, miR-145, which was relatively downregulated across the classes of cervical abnormalities, is in consonance with the discoveries in some earlier studies.[24,26,28]
Limitations
This study is limited due to the inability to conduct a molecular or immunohistochemical testing and genotyping for HPV in cervical cells. Again, the use of endpoint PCR analysis, a semi-quantitative method, in this study, could be a limitation. Employing quantitative real-time PCR may offer better results. However, this may serve as a baseline for future studies.
Conclusion
This study showed a comparable result of miRNA expression in serum and cervical cells. Since there were upregulated oncomirs and downregulated tumor suppressor miRNAs in cervicitis, all cases of cervicitis should be considered to have the potential to progress and should be closely monitored, especially when it occurs among high-risk individuals. This study suggests that underexpressed miR-21 and miR-182, overexpressed miR-let-7b and miR-145 and moderately expressed p53 could be indicative of healthy cervix. Moderately overexpressed miR-21, miR-182, and p53, and moderately underexpressed let-7b and miR-145 and highly expressed p53 may be indicative of premalignant lesions (LSIL/HSIL). In addition, overexpression miR-21 and miR-182, underexpressed let-7b, miR-145 and p53 may be associated with SCC.
Acknowledgments
Special thanks are to staff of Centre for Biocomputing and Drug Development, Adekunle Ajasin University, and HCT and Family planning clinics, State Hospital Abeokuta for their technical assistance.
References
- 1.Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–86. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Frida KM, Atieno WM, Habtu M. Socio-demographic factors associated with advanced stage of cervical cancer at diagnosis in kenyatta national hospital, Kenya:A cross sectional study. J Cancer Sci Ther. 2017;9:554–61. [Google Scholar]
- 3.Shiferaw S, Addissie A, Gizaw M, Hirpa S, Ayele W, Getachew S, et al. Knowledge about cervical cancer and barriers toward cervical cancer screening among HIV-positive women attending public health centers in Addis Ababa city, Ethiopia. Cancer Med. 2018;7:903–12. doi: 10.1002/cam4.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moscicki AB, Ellenberg JH, Farhat S, Xu J. Persistence of human papillomavirus infection in HIV-infected and -uninfected adolescent girls:Risk factors and differences, by phylogenetic type. J Infect Dis. 2004;190:37–45. doi: 10.1086/421467. [DOI] [PubMed] [Google Scholar]
- 5.Hawes SE, Critchlow CW, Sow PS, Touré P, N'Doye I, Diop A, et al. Incident high-grade squamous intraepithelial lesions in senegalese women with and without human immunodeficiency virus Type 1 (HIV-1) and HIV-2. J Natl Cancer Inst. 2006;98:100–9. doi: 10.1093/jnci/djj010. [DOI] [PubMed] [Google Scholar]
- 6.Agaba PA, Thacher TD, Ekwempu CC, Idoko JA. Cervical dysplasia in Nigerian women infected with HIV. Int J Gynaecol Obstet. 2009;107:99–102. doi: 10.1016/j.ijgo.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Peedicayil A, Thiyagarajan K, Gnanamony M, Pulimood SA, Jeyaseelan V, Kannangai R, et al. Prevalence and risk factors for human papillomavirus and cervical intraepithelial neoplasia among HIV-positive women at a tertiary level hospital in India. J Low Genit Tract Dis. 2009;13:159–64. doi: 10.1097/LGT.0b013e31818fb40d. [DOI] [PubMed] [Google Scholar]
- 8.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, et al. Altered microRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–42. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 10.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–43. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich D, Meller S, Uhl B, Ralla B, Stephan C, Jung K, et al. Nucleic acid-based tissue biomarkers of urologic malignancies. Crit Rev Clin Lab Sci. 2014;51:173–99. doi: 10.3109/10408363.2014.906130. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum:A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 14.Bumrungthai S, Ekalaksananan T, Evans MF, Chopjitt P, Tangsiriwatthana T, Patarapadungkit N, et al. Up-regulation of miR-21 is associated with cervicitis and human papillomavirus infection in cervical tissues. PLoS One. 2015;10:e0127109. doi: 10.1371/journal.pone.0127109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin F, Liu P, Ma CF. A circulating serum miRNA panel as early detection biomarkers of cervical intraepithelial neoplasia. Eur Rev Med Pharmacol Sci. 2016;20:4846–51. [PubMed] [Google Scholar]
- 16.Solayman MH, Langaee T, Patel A, El-Wakeel L, El-Hamamsy M, Badary O, et al. Identification of suitable endogenous normalizers for qRT-PCR analysis of plasma microRNA expression in essential hypertension. Mol Biotechnol. 2016;58:179–87. doi: 10.1007/s12033-015-9912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 18.Royo F, Zuñiga-Garcia P, Torrano V, Loizaga A, Sanchez-Mosquera P, Ugalde-Olano A, et al. Transcriptomic profiling of urine extracellular vesicles reveals alterations of CDH3 in prostate cancer. Oncotarget. 2016;7:6835–46. doi: 10.18632/oncotarget.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolanowska M, Wójcicka A, Kubiak A, Świerniak M, Kotlarek M, Maciąg M, et al. Functional analysis of a novel, thyroglobulin-embedded microRNA gene deregulated in papillary thyroid carcinoma. Sci Rep. 2017;7:9942. doi: 10.1038/s41598-017-10318-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gocze K, Gombos K, Kovacs K, Juhasz K, Gocze P, Kiss I, et al. MicroRNA expressions in HPV-induced cervical dysplasia and cancer. Anticancer Res. 2015;35:523–30. [PubMed] [Google Scholar]
- 21.Ben W, Yang Y, Yuan J, Sun J, Huang M, Zhang D, et al. Human papillomavirus 16 E6 modulates the expression of host microRNAs in cervical cancer. Taiwan J Obstet Gynecol. 2015;54:364–70. doi: 10.1016/j.tjog.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Eom K, Kim J, Bang H, Wang HY, Ahn S, et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17:658. doi: 10.1186/s12885-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okoye JO. Discordant reports of miRNA expression in cervical cancer:An upshot of overlapping factors. Res Cancer Tumor. 2015;4:15–23. [Google Scholar]
- 24.Banno K, Iida M, Yanokura M, Kisu I, Iwata T, Tominaga E, et al. MicroRNA in cervical cancer:OncomiRs and tumor suppressor miRs in diagnosis and treatment. ScientificWorldJournal. 2014;2014:178075. doi: 10.1155/2014/178075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilting SM, Snijders PJ, Verlaat W, Jaspers A, van de Wiel MA, van Wieringen WN, et al. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2013;32:106–16. doi: 10.1038/onc.2012.20. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J, et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224:484–95. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, Ji J, Huo G, Song Q, Zhang X. MiR-182 induces cervical cancer cell apoptosis through inhibiting the expression of DNMT3a. Int J Clin Exp Pathol. 2015;8:4755–63. [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA, et al. Human papillomavirus Type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–82. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javadi H, Lotfi AS, Hosseinkhani S, Mehrani H, Amani J, Soheili ZS, et al. The combinational effect of E6/E7 siRNA and anti-miR-182 on apoptosis induction in HPV16-positive cervical cells. Artif Cells Nanomed Biotechnol. 2018;46:727–36. doi: 10.1080/21691401.2018.1468770. [DOI] [PubMed] [Google Scholar]
- 34.Zheng ZM, Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim Biophys Acta. 2011;1809:668–77. doi: 10.1016/j.bbagrm.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang J, Tang Y, Yang J, Huang S. Chemoradiation and adjuvant chemotherapy in advanced cervical adenocarcinoma. Gynecol Oncol. 2012;125:297–302. doi: 10.1016/j.ygyno.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 36.Villegas-Ruiz V, Juárez-Méndez S, Pérez-González OA, Arreola H, Paniagua-García L, Parra-Melquiadez M, et al. Heterogeneity of microRNAs expression in cervical cancer cells:Over-expression of miR-196a. Int J Clin Exp Pathol. 2014;7:1389–401. [PMC free article] [PubMed] [Google Scholar]
- 37.Rao Q, Shen Q, Zhou H, Peng Y, Li J, Lin Z, et al. Aberrant microRNA expression in human cervical carcinomas. Med Oncol. 2012;29:1242–8. doi: 10.1007/s12032-011-9830-2. [DOI] [PubMed] [Google Scholar]
- 38.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 39.Koromilas AE, Li S, Matlashewski G. Control of interferon signaling in human papillomavirus infection. Cytokine Growth Factor Rev. 2001;12:157–70. doi: 10.1016/s1359-6101(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 40.Aloni-Grinstein R, Charni-Natan M, Solomon H, Rotter V. P53 and the viral connection:Back into the future‡. Cancers (Basel) 2018;10:E178. doi: 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–73. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 42.Cui F, Li X, Zhu X, Huang L, Huang Y, Mao C, et al. MiR-125b inhibits tumor growth and promotes apoptosis of cervical cancer cells by targeting phosphoinositide 3-kinase catalytic subunit delta. Cell Physiol Biochem. 2012;30:1310–8. doi: 10.1159/000343320. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Cao X, Zheng Y, Tang J, Cai W, Wang H, et al. Relationship between cervical disease and infection with human papillomavirus Types 16 and 18, and herpes simplex virus 1 and 2. J Med Virol. 2012;84:1920–7. doi: 10.1002/jmv.23353. [DOI] [PubMed] [Google Scholar]
- 44.Nicolete LD, Nicolete R, Haddad R, Azevedo R, Castro FA, Tanaka Y, et al. Upregulation of hsa-miR-125b in HTLV-1 asymptomatic carriers and HTLV-1-associated myelopathy/tropical spastic paraparesis patients. Mem Inst Oswaldo Cruz. 2012;107:824–7. doi: 10.1590/s0074-02762012000600020. [DOI] [PubMed] [Google Scholar]
- 45.Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. doi: 10.1186/1742-4690-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro J, Marinho-Dias J, Monteiro P, Loureiro J, Baldaque I, Medeiros R, et al. MiR-34a and miR-125b expression in HPV infection and cervical cancer development. Biomed Res Int. 2015;2015:304584. doi: 10.1155/2015/304584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuovo GJ, Wu X, Volinia S, Yan F, di Leva G, Chin N, et al. Strong inverse correlation between microRNA-125b and human papillomavirus DNA in productive infection. Diagn Mol Pathol. 2010;19:135–43. doi: 10.1097/PDM.0b013e3181c4daaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Yu X, Guo X, Tian Z, Su M, Long Y, et al. MiR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting bcl-2. Mol Med Rep. 2012;5:753–60. doi: 10.3892/mmr.2011.696. [DOI] [PubMed] [Google Scholar]
- 49.Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA. MicroRNA expression variability in human cervical tissues. PLoS One. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunasekharan V, Laimins LA. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J Virol. 2013;87:6037–43. doi: 10.1128/JVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


