Abstract
Several studies have linked mitochondrial genetic variation to phenotypic modifications; albeit the identity of the mitochondrial polymorphisms involved remains elusive. The search for these polymorphisms led to the discovery of small noncoding RNAs, which appear to be transcribed by the mitochondrial DNA (“small mitochondrial RNAs”). This contention is, however, controversial because the nuclear genome of most animals harbors mitochondrial pseudogenes (NUMTs) of identical sequence to regions of mtDNA, which could alternatively represent the source of these RNAs. To discern the likely contributions of the mitochondrial and nuclear genome to transcribing these small mitochondrial RNAs, we leverage data from six vertebrate species exhibiting markedly different levels of NUMT sequence. We explore whether abundances of small mitochondrial RNAs are associated with levels of NUMT sequence across species, or differences in tissue-specific mtDNA content within species. Evidence for the former would support the hypothesis these RNAs are primarily transcribed by NUMT sequence, whereas evidence for the latter would provide strong evidence for the counter hypothesis that these RNAs are transcribed directly by the mtDNA. No association exists between the abundance of small mitochondrial RNAs and NUMT levels across species. Moreover, a sizable proportion of transcripts map exclusively to the mtDNA sequence, even in species with highest NUMT levels. Conversely, tissue-specific abundances of small mitochondrial RNAs are strongly associated with the mtDNA content. These results support the hypothesis that small mitochondrial RNAs are primarily transcribed by the mitochondrial genome and that this capacity is conserved across Amniota and, most likely, across most metazoan lineages.
Keywords: mitochondria, NUMT, miRNA, mitonuclear, genomics
Introduction
For the past four decades, biologists have routinely harnessed the genetic variability found within the mitochondrial DNA (mtDNA) sequence as an evolutionary marker, to help in resolving the population genetic processes shaping patterns of evolutionary divergence across natural populations and divergent taxa (Avise et al. 1987; Bandelt et al. 1995; Avise 2000; van Oven 2010). These inferences were typically made under the assumption that this genetic variation is associated with no phenotypic alterations to the organism, and therefore has evolved under a neutral equilibrium model (Ballard and Kreitman 1995). However, the results of numerous studies to emerge over the past two decades have challenged the assumption of selective neutrality of the mitochondrial genome, by showing that mitochondrial genetic variation is often tied to phenotypic expression, or by revealing adaptive signatures of mutational variation within the mtDNA sequence (Ballard and Whitlock 2004; Bazin et al. 2006; Dowling 2014; Ballard and Pichaud 2014; James et al. 2016; Vaught and Dowling 2018).
For example, several studies on vertebrate and invertebrate study systems have leveraged experimental designs able to partition mitochondrial from nuclear genetic effects on organismal function (Harrison and Burton 2006; Innocenti et al. 2011; Estopinal et al. 2014; Mossman et al. 2016; Camus et al. 2017; Tourmente et al. 2017; Morales et al. 2018). These uncovered a role for mitochondrial genetic variation in the regulation of physiological, life-history, and health-related phenotypes. These studies were further augmented by studies reporting intriguing associations between particular mitochondrial haplogroups and patterns of disease penetrance in humans (Singh and Kulawiec 2009; Hudson et al. 2014; Sloan et al. 2015; Marom et al. 2017; Chalkia et al. 2018). For example, Marom et al. (2017) observed that certain diseases, such as Type 2 diabetes and Parkinson’s disease, are enriched in humans carrying particular mtDNA haplogroups (haplogroups H, J, and K), whereas Hudson et al. (2014) noted associations between mtDNA haplogroups and the risk of developing several complex diseases, including ischemic stroke, Parkinson’s disease, multiple sclerosis, and schizophrenia. Yet, while it is clear that mitochondrial genetic variation regularly affects phenotypic expression, uncovering the candidate mtDNA mutations involved in the regulation of these effects has proven difficult, given that the mutations that lie in the mtDNA sequence generally appear in tight linkage disequilibrium and are thus inherited as a block.
Most studies that have sought to uncover the candidate polymorphisms that underpin the mitochondrial genotype–phenotype linkage, have focused on genetic variants within the protein-coding region of the mtDNA (Harrison and Burton 2006; Singh and Kulawiec 2009; Weihe et al. 2009; Florencia Camus et al. 2015). Some of these studies identified candidate polymorphisms that confer a modification to the protein-coding sequence (Florencia Camus et al. 2015; Dowling et al. 2015; Patel et al. 2016). However, in several other cases, the polymorphisms underpinning the association between particular mtDNA haplogroups and patterns of phenotypic expression remained elusive, but do not appear to involve substitutions to the amino acid sequence (Takasaki 2009; Camus et al. 2017; Hopkins et al. 2017). For example, Hopkins et al. (2017) reported that specific mutations in the mitochondrial origin of replication (ORI) in humans are associated with the progression to aggressive grades of prostate cancer. Other studies have proposed that mitochondrial regulation of the phenotype may proceed via mechanisms other than direct changes to the protein coding sequence. Indeed, recent studies have reported the discovery of previously uncharacterized RNAs, which are putatively transcribed by the mitochondria, and that may serve important functions (Mercer et al. 2011; Ro et al. 2013; Pozzi et al. 2017; Larriba et al. 2018; Riggs et al. 2018).
The importance of mitochondrial RNAs (mtRNAs), such as ribosomal RNAs (mt-rRNA) and transfer RNAs (mt-tRNA), which are necessary for translation of messenger RNA (mt-mRNA), has long been recognized (Storz 2002). However, in recent years, several independent studies have reported the presence of small noncoding RNAs that appear to be encoded by the mtDNA in fish, mammals, and clams, and the authors of these studies have suggested a role for these RNAs in driving phenotypic changes such as improved anoxia tolerance and involvement in sex determination (Sanchez et al. 2011; Pozzi et al. 2017; Riggs et al. 2018; Larriba et al. 2018). The presence of these “small mitochondrial RNAs,” across several species, suggests they might play a role in cell regulation.
This suggestion, however, is controversial, and it remains debated whether these small RNAs are nonfunctional, representing degraded residues of longer, well-known RNAs (Houseley and Tollervey 2009), or functional in their own right, representing a nucleic part of a ribonucleoprotein (Hogan et al. 2008). Ribonucleoproteins usually use small RNA to recognize specific DNA or RNA targets with which to bind, to exert their function (Hogan et al. 2008). A classic example of such a mechanism is RNA interference (RNAi), a process in which microRNAs (miRNA) of ∼22 nucleotides in length, each with a specific sequence, lead the RNA-induced silencing complex (RISC) to the target mRNA where the protein can inhibit the translation of the mRNA (Ambros 2004; Ha and Kim 2014; Cloonan 2015).
In 2017, Pozzi and colleagues proposed that some of these mitochondrial small RNAs could use RNAi to shape patterns of nuclear gene regulation, thus invoking a hitherto unrecognized mechanism by which the mitochondria could broadly shape patterns of phenotypic expression (Pozzi et al. 2017). Furthermore, this prediction coincides with recent studies of Innocenti et al. (2011) and Baris et al. (2017), both of which identified an abundance of nuclear genes that are seemingly involved in the regulation of mitochondrial–nuclear interactions, but which had no previously known involvement in mitochondrial function. The proteins encoded by these nuclear genes are generally not transported into the mitochondrion, and thus it is unclear how their regulation could be affected by genetic polymorphisms within the mtDNA. The discovery of small mitochondrial RNAs may help to address these outstanding questions, if these RNAs are able to target a broad range of nuclear genes that have no previous known association with mitochondrial function. However, currently, even the origin of these small RNAs remains controversial. This is because the nuclear genome of eukaryotes is littered with mtDNA sequence in the form of nuclear mitochondrial pseudogenes (Nuclear Mitochondrial DNA Segment, NUMT), which in theory could represent the source of these small RNAs.
The presence of NUMTs makes it difficult to definitively identify the origin of small mitochondrial RNA, since in many metazoans, much of the mtDNA sequence is duplicated via shorter inserts throughout the nuclear genome, with the NUMT sequence being almost identical to that of the mitochondrial DNA of origin (Lopez et al. 1994; Gaziev and Shaikhaev 2010). However, despite their widespread presence among most eukaryotes, the function of NUMTs remains largely unknown, and for the most part, they are considered nonfunctional elements (Hazkani-Covo et al. 2010). Indeed, because the genetic code differs between mitochondrial and nuclear genomes, the mitochondrial sequences would not make the same protein using the nuclear code, and therefore these NUMTs are widely regarded as pseudogenes (Lopez et al. 1994). Furthermore, these sequences are “tuned” to be transcribed and translated using proteins specific to the mitochondria, suggesting the inability of the NUMTs to produce functional proteins even when entire genes are translocated to the nuclear genome (Smits et al. 2010). Yet, although NUMTs may not express proteins, they may still express small RNAs. Because NUMTs are mitochondrial in origin, any small RNAs they encode would be nearly identical in sequence to mRNAs encoded by mtDNA, rendering it difficult to determine whether small RNAs that have previously been identified are transcribed by the mitochondrial or nuclear genome.
Fortunately, NUMTs have several unique features relative to the mtDNA, and these features can be used to formulate two sets of testable predictions that allow the putative source of these small mitochondrial RNAs to be discerned. One of the features is that the amount of NUMT sequence differs greatly across metazoan species (Hazkani-Covo et al. 2010; Rogers and Griffiths-Jones 2012). For example, the mouse harbors around 25 times more NUMT sequence than the chicken (∼37 kb or 0.15% of the mouse nuclear genome, ∼1.52 kb or 0.01% of the chicken nuclear genome (Bensasson et al. 2003; Calabrese et al. 2012). Another feature is that the NUMT amount is stable across tissues, whereas the mtDNA copy number changes greatly across different tissues. Here, we take advantage of these species differences in NUMT content, and tissue-specific differences mtDNA copy number, to home in on the probable origin of the small mitochondrial RNAs. Our inferences hinge on the assumption that the abundance of the small mitochondrial RNAs is correlated with the abundance of their source molecules, be that the amount of NUMT sequence, or copies of mtDNA. We contend that this assumption is plausible given the large differences (greater than a thousandfold) in both the amount of NUMT sequence present across distinct species, and in the number of mitochondria found across distinct tissues within a species. On the basis of this assumption, we formulate two predictions. Firstly, if the small mitochondrial RNAs are encoded primarily by NUMTs, then we predict that the level of transcription of these RNAs across species will be correlated to the amount of species-specific NUMT sequence. Species with large amounts of NUMT sequence should possess more small mitochondrial RNAs than species with low amounts of NUMTs. On the contrary, if the mitochondrial DNA encodes the small mitochondrial RNAs, then it is predicted that there will be no correlation between the amount of NUMT sequence and the abundance of small mitochondrial RNAs across species. The second prediction leverages the observation that the copy number of NUMT pseudogenes will be stable across tissue types within a species, but the copy number of mtDNA molecules will differ greatly. Thus, if generally transcribed by NUMT sequence, we predict that the small mitochondrial RNAs will not exhibit predictable differences in abundance across tissue-types. In contrast, if transcribed by the mtDNA, then we predict the abundance of these small RNAs will be higher in tissues that are rich in mitochondria relative to tissues that are poor in mitochondrial content. In particular, tissues that exhibit naturally enriched levels of mitochondria, such as the cancerous cells, would be predicted to show increased abundances of small mitochondrial RNAs when compared with their healthy counterparts that have less mitochondria (Williams et al. 2015; Vyas et al. 2016; Zhu et al. 2017).
Here, we tested these predictions, using publicly available data stored in the Short Read Archive (SRA) in the NCBI. Many miRNA-sequencing data sets are available for model organisms, such as chicken and mice, which can be repurposed to examine the profiles of small mitochondrial RNAs across species and tissues. To clarify, these data sets consist of RNA sequencing data in which the transcripts have been selected using a size-selection approach that selects transcripts <50 nt. Because of this method, the small mitochondrial RNAs, the targets of our study, are represented in these libraries. Using these data, we first tested whether the amount of NUMTs present in the nuclear genome of a given species is associated with the amount of small mitochondrial RNA transcribed, across several species of the Amniota clade. We then focused on expression patterns of small mitochondrial RNAs across three tissues of mice and chicken, two species with very different amounts of NUMT sequence. To address the second prediction, we compared abundances of small mitochondrial RNA from a mitochondria-rich tissue and a mitochondria-poor tissue, in human data sets. Then, to further probe this prediction, we compared small mitochondrial RNAs abundance in a tissue naturally enriched in mitochondria (cancerous tissue) to its healthy counterpart with physiological levels of mtDNA copies.
Materials and Methods
Data
The data for the first analysis, which includes samples from the brain, cerebellum, heart, kidney, and testis of several species (Monodelphis domestica, Homo sapiens, Macaca mulatta, Ornithorhynchus anatinus, Mus musculus, and Gallus gallus) are published in (Meunier et al. 2013), and are accessible in the Short Reads Archive in NCBI with the ID PRJNA174234. All individuals analysed were males, and in all mammals the brain samples originated from the prefrontal cortex. Full information and origin of all samples are described in Meunier et al. (2013).
To compare the correlation between the NUMT content and small mitochondrial RNAs abundance, we used the data from over 10 samples for three different tissues in two different species: M.musculus and G.gallus. The data from M.musculus, which includes samples from the brain, liver, and heart, comes from several studies. The 13 brain samples include four from PRJNA232648 (Hu et al. 2014), five from PRJNA283972 (Malmevik et al. 2016), and four from PRJNA326768 (Woldemichael et al. 2016). As described by the articles just cited, the samples originated from hippocampal neurons and neuroblastoma cell cultures. The 14 liver samples include one from PRJNA203543 (Yamtich et al. 2015), one from PRJNA401785, and 12 from PRJNA407374 (Hao and Waxman 2018). The original articles did not specify any specific section of the liver used for the sequencing. The 12 heart samples include three from PRJNA219640 (Crippa et al. 2016), seven from PRJNA314812 (Ooi et al. 2017), and two from PRJNA421014 (Huang et al. 2018). As described in the original articles, all samples were obtained from mouse heart ventricles. The data from G.gallus, which includes samples from brain, liver, and heart, comes from several studies. The 13 brain samples are from PRJNA396511 (Warnefors et al. 2017). The 13 liver samples include 10 from PRJNA396511 (Warnefors et al. 2017) and three from PRJNA434773. The 14 heart samples include 12 from PRJNA396511 (Warnefors et al. 2017), and two from PRJNA204941. Details on the sections from which these samples were extracted are unavailable.
To study the correlation between the mtDNA content and the abundance of small mitochondrial RNAs, we used data from H.sapiens, which includes samples from brain and bladder from several studies. The 36 brain samples comprised 11 from PRJNA272617 (Hoss et al. 2015), and 25 from PRJNA394722 (Pantazatos et al. 2017). All the tissues were samples from the prefrontal cortex. The 10 bladder samples (five controls and five cancer) are from PRJDB2583 (Itesako et al. 2014).
NUMT Sequence Estimation
The amount of NUMT sequence per genome of each species was extracted from a previously published article (Hazkani-Covo et al. 2010). These authors had documented NUMT content by aligning the mitochondrial genome of each species on the respective nuclear genome using BlastN with a cut-off value of 0.0001 (e-score). For our study, we selected only high quality genomes, thus eliminating the chance of mischaracterization of NUMTs due to assembly problems. The amount of NUMTs for each species (absolute and relative to the nuclear genome) is: Chicken (1.52 kb, 0.0001%), mouse (37.67 kb, 0.0015%), platypus (244.198 kb, 0.0081%), monkey (261.622 kb, 0.0087%), human (266.478 kb, 0.0087%), opossum (2093.63 kb, 0.0698%).
The amount of NUMTs in each species is not always stable. The NUMT amount might differ slightly across individuals analyzed within a given species, however this possibility cannot be tested without specific genomic data from the individuals analyzed. Nonetheless, we expect that variability in NUMT amount across individuals within a species will be far lower than levels of variability across species. The differences between chicken and other high-NUMT amount species is between ∼15 (mouse) and ∼700-fold (opossum). Thus, we argue that such variability should not affect our results.
The genome sequences of the individuals included in this study are likely to be slightly different from the reference genomes used. In fact, both NUMTs and mtDNA show slight variation across individuals. However, we estimate that any such variation is likely to have had at most a negligible effect on our study. Indeed, we accounted for these differences by using nonstringent criteria during the alignment: A relatively small seed and allowing for a mismatch of one nucleotide in the sequence. Thus, unless multiple SNPs would arise in the short sequences encoding the small mitochondrial RNAs under study, the effects would likely be negligible.
Library Analysis
To obtain the percentage of reads mapping exclusively to the mtDNA (denoted mtDNA-only reads) versus those that map both to NUMTS and mtDNA (denoted NUMT reads), we applied a custom pipeline for each of the samples. At first, we aligned the RNA library to the reference mitochondrial genome of the species of interest, obtaining the mtDNA reads (these are all reads that map to mtDNA, both mtDNA-only and the NUMT reads). Then, we aligned the mtDNA reads onto the nuclear genome to obtain the reads mapping both to the nuclear and mitochondrial genome; which we defined as the NUMT reads (i.e., these reads mapped to both the mtDNA and to NUMT sequence). The reads of each library were aligned to their NCBI reference genomes using Bowtie2 (Langmead and Salzberg 2012), using the default settings of the –local function and a seed of 18 nucleotides with one mismatch allowed (-n 1). From the output of Bowtie2, we retained the overall percentage of reads mapping to the mitochondrial and nuclear (NUMTs) genomes. All the format conversion of the data sets during the analysis was done using standard approaches with samtools (Li et al. 2009) and bedtools (Quinlan and Hall 2010). The results are plotted using MATLAB and Statistics Toolbox Release 2018b, The MathWorks, Inc., Natick, Massachusetts, United States. To identify hotspots of transcription, the alignment files (mtDNA reads) for each sample were converted to a BedGraph format using bedtools (Quinlan and Hall 2010), and all samples from each tissue were merged using the cat function in the terminal. These data were then plotted using the package Circlize in R (Gu et al. 2014).
Statistical Analysis
The percentages of aligned small mitochondrial RNAs (mtDNA reads) extracted from (Meunier et al. 2013) were grouped first by species, creating six groups in which each data point represented the value of a different tissue from the same species (brain, cerebellum, heart, kidney, and testis). A formal statistical test of these data was not appropriate, because there is no independent replication of data points for each tissue in each species. The mtDNA content changes drastically among tissues, thus pooling the tissues in each species to perform a formal statistical test would result in an unreliable result.
The analyses of the chicken and mouse samples (heart, brain, liver), as well as the human samples (brain and bladder), were performed using the same tools. We conducted the analysis using standard functions in R and Past3 (https://folk.uio.no/ohammer/past/; Last accessed on June 26, 2019). The percentage of mtDNA reads aligned from each sample was clustered by tissue type. The distributions obtained were tested for normality using the Shapiro–Wilk test (Shapiro and Wilk 1965), and in every case, the null hypothesis was rejected. Therefore, to test the difference between each distribution we used a two way Mann–Whitney U test (Mann and Whitney 1947).
Results
The Small Mitochondrial RNAs Are Widely Expressed across Tissues in Amniota
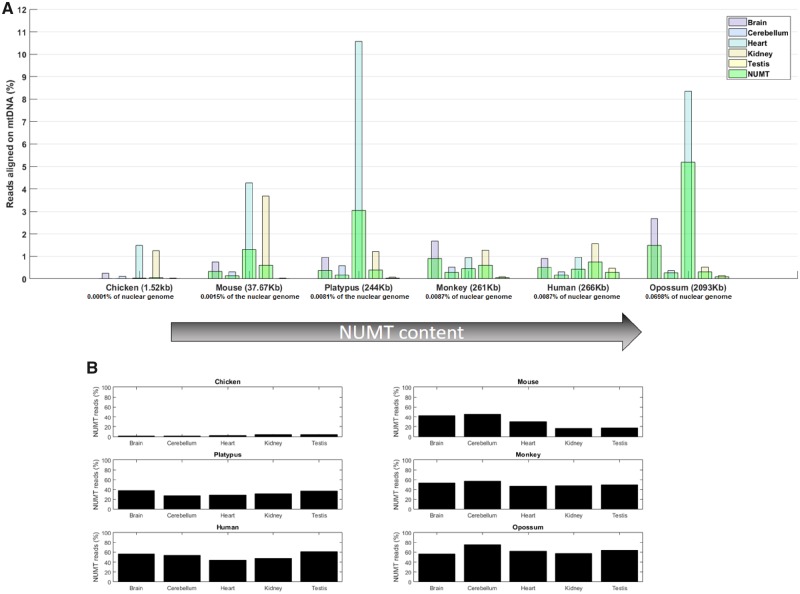
We first tested whether the abundance of small mitochondrial RNAs changes with the amount of NUMT sequence across species. We used data from Meunier et al. (2013), who analyzed tissue-specific RNA libraries from each of six species of the Amniota clade. Each of these species is characterized by differing degrees of NUMT sequence insertions. We quantified the percentage of small RNAs that map to the mtDNA in each of these samples, and compared this to the percentage of reads mapping to NUMT sequence (fig. 1A). Accordingly, we can define three different types of reads: The mtDNA reads, which include the total pool of reads that map to the mtDNA (those that map exclusively to the mtDNA as well as those that map both to mtDNA and NUMT sequence); mtDNA-only reads, which map unambiguously and exclusively to the mtDNA and exclude the sequences present in the NUMTs; and NUMT reads, which represent the reads that map both to the mtDNA and NUMT sequence. Generally, species with high levels of NUMTs have high proportions of reads that map jointly to both mtDNA and NUMTs (fig. 1B). However, even in species that have very high levels of NUMTS, such as the opossum, at least 25% of the mtDNA reads are nonetheless comprised of mtDNA-only reads. Notably, the chicken is characterized by very low levels of NUMT sequence. In this species, the percentage of mtDNA reads that map exclusively to the mtDNA sequence is ∼95% across all sampled tissues.
Fig. 1.
—The abundance of small mitochondrial RNAs across species exhibiting different NUMT contents. In (A), the percentage of small RNAs, relative to the total RNAs in the sample, that align exclusively to the mtDNA in five distinct tissues of six separate species. The NUMT content of each species is denoted in parentheses beside the species name, and the species are ordered by ascending NUMT content. Furthermore, the percentage of NUMTs present in their nuclear genome is denoted below the name of each species. Different tissues are represented by distinct colors: Brain (violet), cerebellum (cyan), heart (light blue), kidney (light brown), and testis (yellow). The percentage of reads mapping to the mtDNA, both mtDNA-only and NUMT reads together, is shown on the Y-axis. On the same axis, the percentage of reads in the RNA libraries that map both to the mtDNA and to the nuclear genome (NUMTs reads) is highlighted in light green. For example, in the case of Ornithorhynchus anatinus (platypus), the heart samples show ∼3% of NUMT reads and ∼7.5% of mtDNA-only reads, for a total of 10.5% mtDNA reads. In (B), the percentage of NUMT reads as a proportion of the overall mtDNA reads, for each sample. While in Gallus gallus, <10% of mtDNA reads map the NUMTs, in most samples of the other species almost half of the mtDNA reads map jointly to the NUMT sequence. Despite this, even in species with the highest level of NUMT sequence, around one-third of the mtDNA reads map uniquely to the mtDNA.
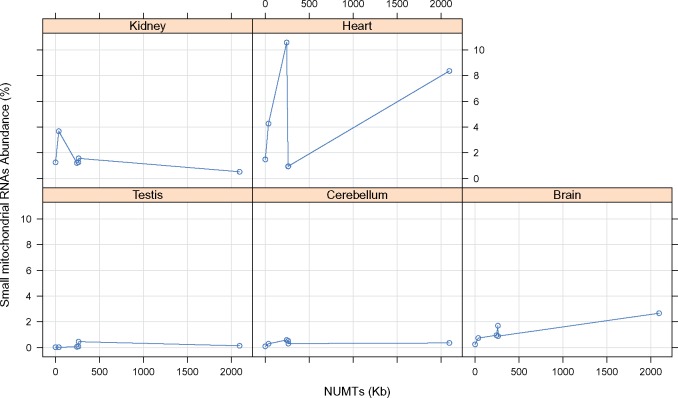
We next investigated whether associations exist between the amount of NUMT sequence and the percentage of mtDNA reads across the different tissues of each species (fig. 2). Although we note there are insufficient samples per tissue and species to warrant formal statistical analysis (n = 1 data point per tissue per species), the analysis highlights an absence of any clear relationship between the NUMT content and total abundance of mtDNA reads across the six species.
Fig. 2.
—The relationship between the percentage of small mitochondrial RNAs in each tissue and NUMT content (kb), across six species. There are six samples for each combination species/tissue, and each data point represents a sample of a single tissue from a single species. For example in the top left plot we can see a data point at the 2,000 mark on the X-axis, which indicates that small mitochondrial RNAs comprised <2% of the RNA library extracted from the kidney of a species with over 2,000 kb NUMTs length (Opossum).
Small Mitochondrial RNA Abundance Is Independent of the NUMT Content
To further probe associations between NUMT content and abundances of mtDNA reads, we conducted a further set of analyses that focused on two of the six species surveyed above (chicken and mouse). These species exhibit very different NUMT contents (chicken 1.52 kb, mouse 37.67 kb), with a large number of supporting small RNA-seq data sets available for each. Our analyses focused on three different tissue types, for which there are numerous samples available in public databases: The brain, heart, and liver (Rooney et al. 2015; Reznik et al. 2016).
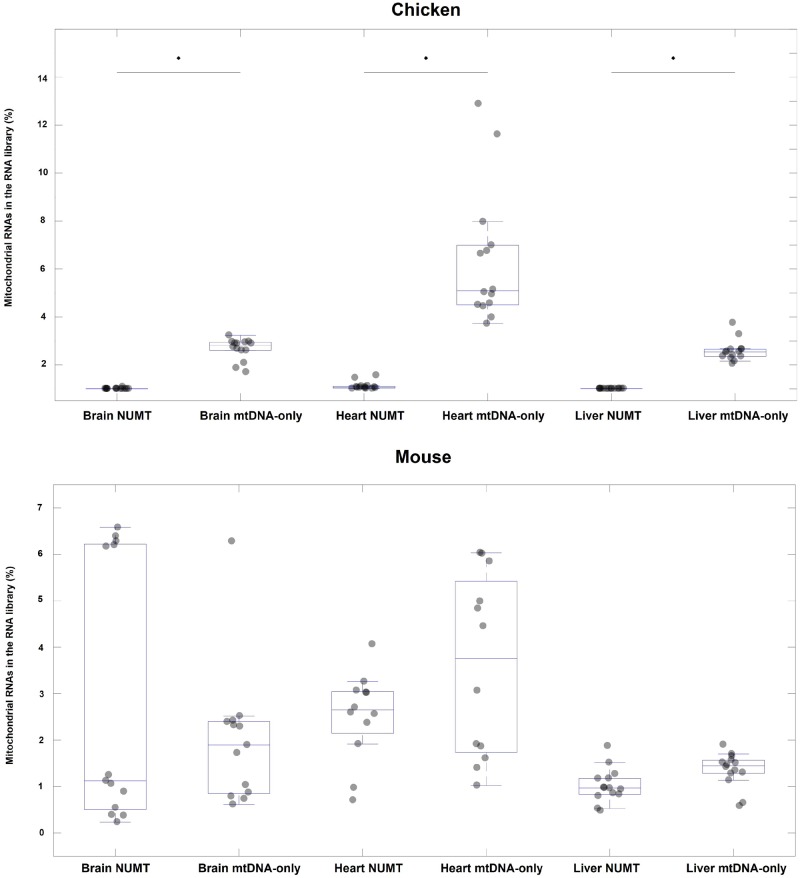
Our analyses of tissue-specific abundances of mtDNA reads reinforce the large difference in levels of NUMT reads between the two species. In the chicken data sets, there are consistently lower levels of NUMT reads per tissue than in the mouse data sets (Mann–Whitney, U = 0, n = 80 P = 0.0001, fig. 3). The mouse data sets show moderate levels of NUMT reads, which nonetheless rarely surpass the amount of mtDNA-only reads. Specifically, while the percentage of NUMT reads in the brain (median 1.18 ± 0.73%) is similar to the percentage of mtDNA-only reads (median 1.87 ± 0.4%), the heart and liver have a greater representation of mtDNA-only reads (medians of 3.75 ± 0.53% and 1.44 ± 0.1% respectively) than NUMT reads (medians of 2.65 ± 0.27% and 1.00 ± 0.1% respectively, supplementary table S2, Supplementary Material online).
Fig. 3.
—Boxplots of percentages of NUMT reads (those mapping both to the mtDNA and the nuclear genome) and mtDNA-only reads in the brain, heart, and liver, across samples taken from chicken and mouse. The percentage of reads in the RNA library mapping to the mtDNA is shown on the vertical axis. The NUMT boxes show the percentage of reads mapping jointly to both mtDNA and NUMT sequence (NUMT reads). The mtDNA-only boxes show reads mapping exclusively to the mtDNA and not to NUMT sequences (mtDNA-only reads). The horizontal line in each box indicates the median of the distribution, and the light gray circles indicate individual data points. The distributions were tested using Mann–Whitney U test and significance (P < 0.05) is indicated with an asterisk.
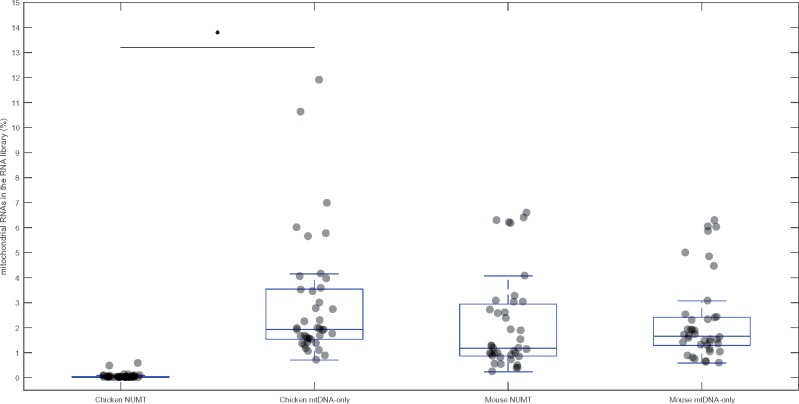
Overall, when comparing mtDNA-only to NUMT reads across all of the chicken samples, by pooling together the data from all tissues, the percentage of NUMT reads usually comprises <0.1% of the small RNA library (fig. 4), confirming observations derived from the data set of Meunier et al. (2013) that we report in figure 1. Furthermore, we observed high abundances of mtDNA-only reads in the chicken samples, represented as a proportion of the total small RNA reads per library, ranging from 0.8 to 12% across the three tissues. Patterns observed across mouse samples similarly reinforced the results of our analysis of the Meunier et al. (2013) data set. We detected a high percentage of NUMT reads in all tissues, which reflects the high amount of NUMT sequence in the mouse genome (∼37 kb).
Fig. 4.
—Boxplots showing the percentages of NUMT reads and mtDNA-only reads across all tissues pooled (heart, brain, and liver) of chicken and mouse. The percentage of reads in the RNA library mapping to the mtDNA is shown on the Y-axis. The NUMT boxes show the percentage of reads mapping to both mtDNA and NUMT sequence (NUMT reads). The mtDNA-only boxes show the reads mapping only the mtDNA and not the NUMTs (mtDNA-only reads). The horizontal line in each box indicates the median of the distribution, and the light gray circles indicate individual data points. The distributions were tested using Mann–Whitney U test and significance (P < 0.05) is indicated with an asterisk.
This pattern suggests that, at least in the chicken, the majority of the mtDNA reads must be transcribed directly from the mtDNA. The source of the mtDNA reads in mice remains open to question, and technically, it is possible that the NUMTs play a secondary role in their transcription. Notwithstanding, it must be noted that all NUMT reads also map to the mtDNA, and thus the mtDNA would remain the most likely candidate site for their transcription. Indeed, if the NUMTs play a sizable role in the transcription of mtDNA reads, we would expect that the total pool of mtDNA reads would be significantly lower in the chicken than in the mouse, given the much lower NUMT content of the chicken genome of NUMT sequence in the chicken. Although there are no differences in the proportion of mtDNA-only reads between the species (U = 621, n = 79, P = 0.12), the chicken samples do exhibit a meaningful reduction in the abundance of mtDNA reads when compared with the mouse samples (U = 493, n = 79, P = 0.003). While at first, this result would suggest a role for the NUMTs in the transcription of small mitochondrial RNA, closer scrutiny of the tissue-specific samples suggests otherwise. Although both the chicken liver and brain samples show statistically significant reductions in the abundance of mtDNA reads relative to the equivalent tissues in mice (liver U = 37, n = 27, P = 0.004; brain U = 38, n = 26, P = 0.009), the heart samples of the two species do not differ (U = 68.5, n = 26, P = 0.43). See supplementary table S1, Supplementary Material online for full statistical analyses.
The Abundance of Small Mitochondrial RNAs Is due to a Few Transcriptional Hotspots
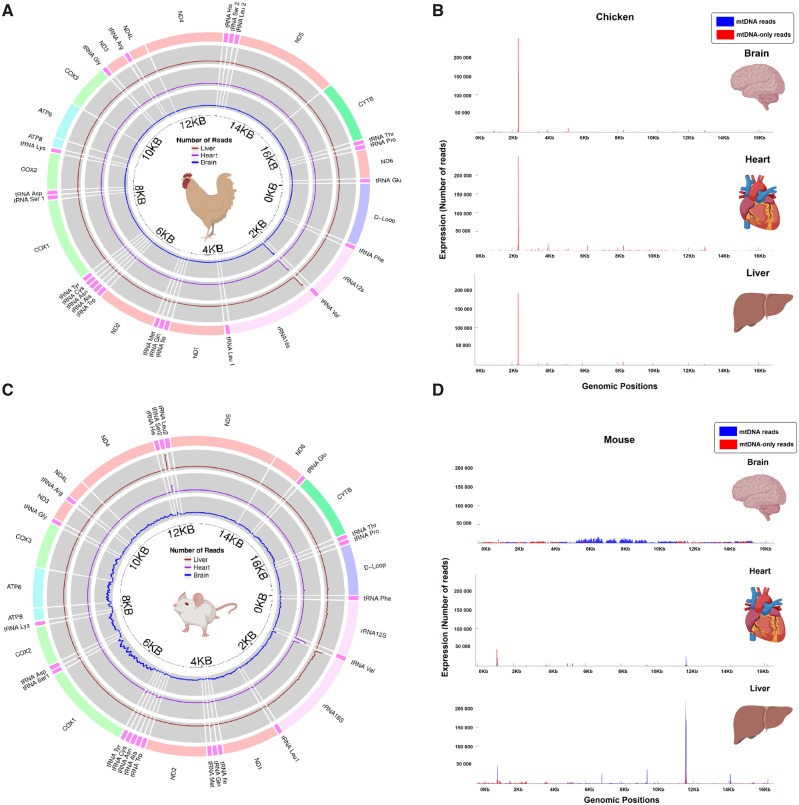
We next assessed whether differences in the abundances of NUMT and mtDNA-only reads across the two species are attributable to changes in the expression of small mitochondrial RNA molecules transcribed across many different regions of the mtDNA sequence, or by high levels of transcription at just a few specific sequence regions. In the chicken, almost all of the mtDNA reads align to the rRNA16S, aligning just after the 2 kb mark (fig. 5A and B). This peak aligns to mtDNA-only reads, consistent with expectation given the high percentage of mtDNA-only reads reported above for this species. Indeed, NUMT reads are almost entirely absent, and only minor peaks of mtDNA-only reads can be seen in the brain and heart samples.
Fig. 5.
—Expression profiles of small mitochondrial RNAs across three tissues of chicken (A & B) and mouse (C & D). Circular representations of (A) chicken mtDNA and (C) mouse mtDNA, in which three concentric circles show the number of reads aligning to each portion of the genome across the three tissue types. From the outer to the inner circle, we can see the expression of mtDNA reads in the liver (brown), heart (magenta), and brain (blue). The scale of each circle is not constant but changed accordingly to highlight the differences in expression among samples within a given tissue. However, within each tissue and species, the scale is constant across different regions of the mtDNA. The makers placed every 2 kb show the approximate location of the genes in the genome. These markers are placed to facilitate the comparison with the linear representations. Linear representations of the chicken (B) and mouse (D) mtDNA, in which small mitochondrial RNAs are mapped to the sequence position, with both mtDNA reads (blue) and mtDNA-only reads (red). The portion of the mtDNA reads not covered by the mtDNA-only reads represents the sequences that map both the mtDNA and the nucleus (NUMT reads). The median number of reads mapping to a specific portion of the mtDNA represents the level of expression in this plot.
In the mouse, hotspots of expression of mtDNA reads were observed in the heart and liver. This was not the case for expression patterns in the brain, in which many different transcripts across the coding, tRNA and rRNA regions of the mtDNA sequence were observed (fig. 5C and D). In particular, the abundance of mtDNA reads present in the mouse heart and liver is attributable to one hotspot of transcription (close to position 12 kb in the mtDNA) aligning to mt-tRNA Ser2 gene. Because of the similarity between mtDNA and NUMT sequence, this peak could technically be either mtDNA or NUMT encoded (they are NUMT reads). The second large peak, comprising a mix of NUMT and mtDNA-only reads, is present in all samples, aligning on mt-rRNA16s and mt-tRNA Val genes, suggesting very high conservation of these mtDNA reads across tissues in the mouse. The abundance of mtDNA reads in the brain samples show a diffuse pattern of expression of thousands of transcripts across the entire coding sequence of mtDNA, lacking specific hotspots of transcription, which is difficult to explain, based on our current knowledge.
Abundance of Small Mitochondrial RNAs Is Closely Associated with mtDNA Content
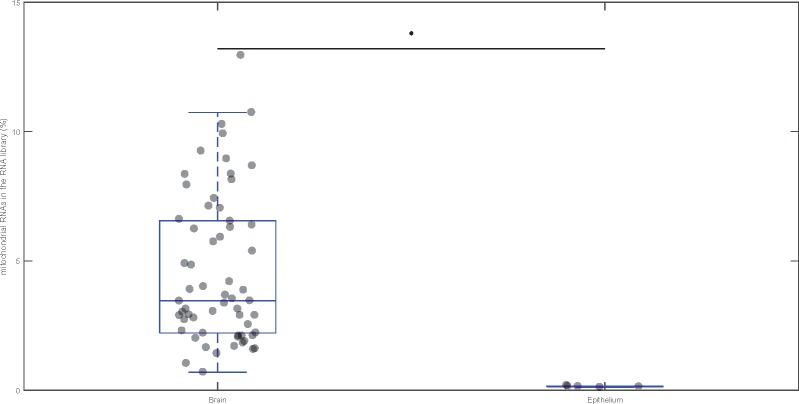
We extended our investigation, turning to RNA libraries from humans, to examine the correlation between the abundance of mtDNA reads and mtDNA content, across tissues that are likely to exhibit strong differences in their mitochondrial content. We collated small RNA sequencing data from three studies, two from the brain (a putatively mitochondria-rich tissue) and one from the bladder epithelium (a putatively mitochondria-poor tissue). We calculated the percentage of small RNA reads mapping to the mtDNA sequence across each of these tissues (fig. 6). The mitochondria-rich brain samples have higher levels of transcription of mtDNA reads (U = 1, n = 66, P = 0.0001). In fact, the mean abundance of mtDNA reads in the brain (3.46 ± 0.37%) is more than 30-fold more than the mean abundance in the bladder epithelium (0.15 ± 0.01%).
Fig. 6.
—The percentage of small mitochondrial RNAs mapping to the mtDNA in the brain (putatively mitochondria-rich) and bladder epithelium (mitochondria-poor) tissue of humans. The percentage of mtDNA reads in the bladder epithelium can barely be seen because the amount is so low when compared with the brain mtDNA reads (<0.5% of all aligned reads). The horizontal line in each box indicates the median of the distribution, and the light gray circles indicate individual data points. The distributions were tested using Mann–Whitney U test and significance (P < 0.05) is indicated with an asterisk.
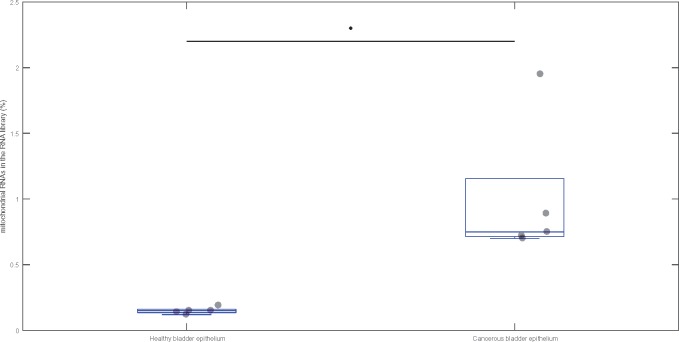
Finally, to further probe the relationship between expression profiles of mtDNA reads and mtDNA content, we leveraged data from a recent study examining the small RNA profiles of bladder samples taken from patients with bladder cancer relative to healthy patients (Itesako et al. 2014). It has previously been shown that cancerous bladder cells show higher levels of mtDNA than healthy bladder cells (Williams et al. 2015). The samples from cancerous tissue, with enriched mtDNA content, contained over 5-fold more mtDNA reads than the samples from healthy tissue (U = 0, n = 10, P = 0.008) (fig. 7). This result reinforces the observation that levels of small mitochondrial RNAs are tightly linked to the total content of mtDNA within a tissue type, thus supporting the contention that these RNAs are transcribed by the mtDNA, and not by NUMT sequence within the nuclear genome.
Fig. 7.
—The percentage of small mitochondrial RNAs mapping to the mtDNA in two different types of cells: Healthy (control) and cancerous (cancer). The horizontal line in each box indicates the median of the distribution, and the light gray circles indicate individual data points. The distributions were tested using Mann–Whitney U test and significance (P < 0.05) is indicated with an asterisk.
Discussion
Here, we present two lines of support for the hypothesis that the mtDNA is the primary source of small mitochondrial RNA transcription. Furthermore, our study demonstrates that the high levels of small mitochondrial RNA transcripts observed across the sampled small RNA data sets are largely attributable to a smaller set of tissue-specific transcripts; an observation that supports the contention that these transcripts are likely to confer a functional role.
The first line of support for the hypothesis comes from our observation that abundances of small mitochondrial RNA were not correlated to levels of NUMT sequence across species. One striking example of this point comes from the case of the opossum, a species with almost 10 times more NUMT sequence (∼2,093 kb) than the platypus, monkey, and human. Despite this order of magnitude difference in the level of NUMT reads, the proportion of mtDNA reads mapping to NUMT sequence was not tangibly higher across major tissues of the opossum relative to those of these other species. Furthermore, the presence of small mitochondrial RNAs in the chicken, a species with a distinct paucity of NUMT sequence, represents definitive evidence of the ability of the mtDNA to transcribe small mitochondrial RNAs. In this species, the transcripts mapping to the mtDNA do not map to any other part of the genome and their presence can thus only be explained by transcription from the mtDNA sequence. Indeed, across six species examined, we observed high proportions of reads mapping exclusively to the mtDNA sequence, confirming that transcription of small RNAs from the mitochondria occurs generally across vertebrates. Furthermore, we were able to confirm these general patterns by focusing on patterns in two species for which we had high levels of inferential power, given that RNA libraries were available from a number of independent studies for each of these species, and at high levels of replication.
The second line of support comes from analyses that show a tight link between expression levels of the small mitochondrial RNAs, and the mtDNA content of particular tissues. Our analyses are based on the assumption that a tissue with the high-energy demands, such as the brain (Veltri et al. 1990; Davey et al. 1998; Picard and McEwen 2014), will have more mitochondria than a tissue with putatively low demands, such as the epithelium (Gibson et al. 1996). On this, while there is evidence that brain tissue is rich in mitochondria (Davey et al. 1998; Picard and McEwen 2014), there is less direct evidence that epithelium tissue is low in mitochondria. However, despite direct measure of the amount of mitochondria, we argue that the difference in energy consumption between these two tissues should make our assumption reasonable. Under this assumption, if the mitochondria are the main source of mtDNA reads, we predict the mitochondria-poor tissue to express lower levels of these RNAs than their mitochondria-rich counterpart. Our predictions were supported. In fact, the abundance of mtDNA reads is strikingly higher in brain samples than in epithelium samples. Moreover, this result is supported by the analysis of cancerous cells naturally enriched in mtDNA (Williams et al. 2015; Vyas et al. 2016). The cancerous cells showed a 5-fold increase in their abundance of mtDNA reads, supporting the hypothesis that the mtDNA is a main player in the transcription of mtDNA reads.
We note that our lines of evidence support previous experiments performed in cell lines. In fact, Ro et al. (2013) investigated the origin of the small mitochondrial RNAs by analyzing the expression of cells with depleted mitochondria (Rho0). The authors found that Rho0 cells have strong downregulation of small mitochondrial RNAs transcription, thus adding a complementary layer of evidence that support our findings.
Further investigations into the origins of small mitochondrial RNAs should seek to leverage allelic differences between NUMT sequences and mtDNA. Over time, NUMT sequences are expected to diverge from their ancestral mtDNA sequence, accumulating SNPs that are unique to NUMT sequences and not found in the mtDNA. In theory, this divergence should provide an opportunity for future studies to investigate whether small mitochondrial RNAs that map exclusively to NUMT sequence exist. Currently, limitations in the available data preclude such an analysis. Given that different individuals are expected to harbor different SNPs in both mtDNA and NUMTs (i.e., natural allelic variation in these regions of sequence), such analysis would require both DNA and RNA data originating from the same individual (or from the same sets of individuals). Furthermore, small mitochondrial RNAs exist in very short sequences (∼30 nt), presenting small mutational targets in which is unlikely multiple mutations will occur. Therefore, such mutations would rarely lead to tangible molecular divergence between mtDNA and NUMT sequence. This will make future analyses based on molecular divergence challenging.
Considering the evidence presented here, we propose that the small mitochondrial RNA reads that map jointly to both mtDNA and NUMTs should be assumed to originate from the mtDNA. Moreover, we argue that we can safely assume that all small RNAs aligning to mtDNA, including in species not tested in our study, are likely to be transcribed by the mtDNA, and not by NUMTs. In fact, the lack of evidence that the NUMTs are transcribed, or play functional roles in any metazoan, aligns well with our hypothesis (Hazkani-Covo et al. 2010). Previous studies have identified and explored the putative function of small mitochondrial RNAs in other species (Mercer et al. 2011; Pozzi et al. 2017; Riggs et al. 2018). Our results provide strong support that their analyses and interpretations are unlikely to have been confounded by transcripts encoded by NUMTs present within the nuclear genome.
We have provided evidence to support the ability of the mtDNA to transcribe small RNAs in chickens, mice and humans. Thus from an evolutionary perspective, this allows us to date the evolution of this ability to at least the birth of the Amniota clade. However, while our study is the first to deeply investigate the origin of the small mitochondrial RNAs, it is not the first to have identified the presence of these molecules. In fact, prior to this study, previous studies had identified small mitochondrial RNAs in humans, chicken, mice, fish, and clams (Mercer et al. 2011; Ro et al. 2013; Pozzi et al. 2017; Bottje et al. 2017; Riggs et al. 2018; Larriba et al. 2018). This degree of conservation in the presence of these RNAs suggests that the ability to transcribe small RNAs from the mtDNA is conserved well beyond the Amniotes, extending to the origin of Protostomia around 600 Ma. Furthermore, we note a recent report of small mitochondrial RNAs in plants (Wu et al. 2015). Thus, we speculate that the ability of the mtDNA to transcribe small RNAs may date back to the early origins of eukaryogenesis, when the mitochondrial–eukaryote endosymbiosis was still in the incipient stages of evolution. While almost without evidence, such a possibility is intriguing, and worthy of further investigation.
Characteristics that are evolutionary conserved across many species tend to be linked to specific functions, since patterns of sequence conservation are shaped by strong selection (Margoliash 1963; Zuckerkandl and Pauling 1965; Kimura 1968). The fact that numerous metazoan species are able to transcribe small mitochondrial RNAs therefore provides hints of likely functionality. tRNAs are known for being one of the source for microRNA-like molecules named tRNA-derived RNA fragments (tRFs). These tRFs seem to be produced by both nuclear and organelle tRNAs. We hypothesize that such functionality would likely come through the involvement of these small RNAs in the RNAi mechanism. RNAi represents one of the most conserved molecular mechanisms among eukaryotes, the function of which is regulated by small noncoding nuclear RNAs of the same length of the small mitochondrial RNAs (Ha and Kim 2014). Importantly, our study shows that most small mitochondrial RNAs originate from the mt-tRNAs, aligning well with a previous study of human RNA that found evidence of a mt-tRNA binding to Ago2 (Ha and Kim 2014). Ago2 is a key protein involved in the regulation of RNAi, and which is present outside of the mitochondria and within the cytoplasm (Maniataki and Mourelatos 2005). Further connection to the tRNAs is represented by their general involvement with RNAi (Lee et al. 2009; Cloonan 2015). In fact, tRNAs encoded by the nucleus are a known source of small RNAs (∼30 nt) involved in RNAi, thus suggesting that the small mitochondrial RNAs might be generated in similar manner. Supporting this hypothesis, a recent study showed that organelles in plants also encode tRNA-derived small RNAs (Cognat et al. 2017). These results match the predictions made by Pozzi et al. (2017), who upon analysis of levels of expression, sequence lengths, and complementarity to target mRNAs, proposed a putative role for a subset of highly transcribed small mitochondrial RNAs in RNAi. Therefore, we argue that the presence of polymorphisms in sequences harboring small RNAs may alter their function through interfering with the complementarity between small RNA and mRNA target.
Conclusion
We have presented several lines of support for the hypothesis that the mitochondrial genome is able to consistently transcribe small RNAs, across species, and in a tissue-specific manner. The next frontier is now to home in on the question of whether these small RNAs play a functional role in the regulation of the organismal phenotype via mitochondrial–nuclear sequence interactions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We wish to acknowledge all researchers who made their data freely available, thus giving us the chance to perform this work. Furthermore, we would like to thank Paul Sunnucks for the insightful discussions that improved this work. The study was funded by the Australian Research Council, through grants DP170100165 and FT160100022 to D.K.D.
Author’s Contribution
A.P. and D.K.D. conceived the study, and A.P. performed all the computational analysis and prepared the figures. A.P. and D.K.D. interpreted the results and wrote the manuscript.
Data deposition: This project have been deposited at Sequence Reads Archive (https://www.ncbi.nlm.nih.gov/sra) under the accessions PRJNA174234, PRJNA232648, PRJNA283972, PRJNA326768, PRJNA203543, PRJNA401785, PRJNA407374, PRJNA219640, PRJNA314812, PRJNA421014, PRJNA396511, PRJNA434773, PRJNA204941, PRJNA272617, PRJNA394722.
Literature Cited
- Ambros V. 2004. The functions of animal microRNAs. Nature 431(7006):350–55. [DOI] [PubMed] [Google Scholar]
- Avise JC, et al. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Ann Rev Ecol Syst. 18:489–522. [Google Scholar]
- Avise JC. 2000. Phylogeography: the history and formation of species. Harvard University Press; https://play.google.com/store/books/details?id=lA7YWH4M8FUC [Google Scholar]
- Ballard JWO, Pichaud N.. 2014. Mitochondrial DNA: more than an evolutionary bystander. Edited by Charles Fox. Funct Ecol. 28(1):218–31. [Google Scholar]
- Ballard JWO, Whitlock MC.. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13(4):729–44. [DOI] [PubMed] [Google Scholar]
- Ballard JW, Kreitman M.. 1995. Is mitochondrial DNA a strictly neutral marker? Trends Ecol Evol. 10(12):485–88. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB.. 1995. Mitochondrial portraits of human populations using median networks. Genetics 141(2):743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baris, Tara Z, Dominique N. Wagner, David I. Dayan, Xiao Du, Pierre U. Blier, Nicolas Pichaud, Marjorie F. Oleksiak, and Douglas L. Crawford. 2017. Evolved Genetic and Phenotypic Differences due to Mitochondrial-Nuclear Interactions. PLoS Genetics 13 (3): e1006517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin E, Glémin S, Galtier N.. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312(5773):570–72. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Feldman MW, Petrov DA.. 2003. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 57(3):343–54. [DOI] [PubMed] [Google Scholar]
- Bottje WG, et al. 2017. Identification and differential abundance of mitochondrial genome encoding small RNAs (mitosRNA) in breast muscles of modern broilers and unselected chicken breed. Front Physiol. 8:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese FM, Simone D, Attimonelli M.. 2012. Primates and mouse NumtS in the UCSC genome browser. BMC Bioinformatics 13(Suppl 4):S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus MF, Wolff JN, Sgrò CM, Dowling DK.. 2017. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol Biol Evol. 34(10):2600–2612. [DOI] [PubMed] [Google Scholar]
- Chalkia D, et al. 2018. Mitochondrial DNA associations with East Asian metabolic syndrome. Biochim Biophys Acta. 1859(9):878–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N. 2015. Re-thinking miRNA–mRNA interactions: intertwining issues confound target discovery. Bioessays 37(4):379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognat V, et al. 2017. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 45(6):3460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa S, et al. 2016. Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways. Cardiovasc Res. 110(1):73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey GP, Peuchen S, Clark JB.. 1998. Energy thresholds in brain mitochondria: potential involvement in neurodegeneration. J Biol Chem. 273(21):12753–57. [DOI] [PubMed] [Google Scholar]
- Dowling DK. 2014. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim Biophys Acta. 1840(4):1393–1403. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Tompkins DM, Gemmell NJ.. 2015. The Trojan female technique for pest control: a candidate mitochondrial mutation confers low male fertility across diverse nuclear backgrounds in Drosophila melanogaster. Evol Appl. 8(9):871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estopinal CB, et al. 2014. Mitochondrial haplogroups are associated with severity of diabetic retinopathy. Investig Ophthalmol Vis Sci. 55(9):5589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florencia Camus M, Wolf JBW, Morrow EH, Dowling DK.. 2015. Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr Biol. 25(20):2717–22. [DOI] [PubMed] [Google Scholar]
- Gaziev AI, Shaikhaev GO.. 2010. Nuclear mitochondrial pseudogenes. Mol Biol. 44(3):358–68. [PubMed] [Google Scholar]
- Gibson PR, Anderson RP, Mariadason JM, Wilson AJ.. 1996. Protective role of the epithelium of the small intestine and colon. Inflamm Bowel Dis. 2(4):279–302. [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B.. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30(19):2811–12. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN.. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15(8):509–24. [DOI] [PubMed] [Google Scholar]
- Hao P, Waxman DJ.. 2018. Functional roles of sex-biased, growth hormone-regulated microRNAs miR-1948 and miR-802 in young adult mouse liver. Endocrinology 159(3):1377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Burton RS.. 2006. Tracing hybrid incompatibilities to single amino acid substitutions. Mol Biol Evol. 23(3):559–64. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W.. 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6(2):e1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO.. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6(10):e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JF, et al. 2017. Mitochondrial mutations drive prostate cancer aggression. Nat Commun. 8 (1):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoss AG, et al. 2015 Mar. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genomics. 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D.. 2009. The many pathways of RNA degradation. Cell 136(4):763–76. [DOI] [PubMed] [Google Scholar]
- Huang W, et al. 2018. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nature Commun. 9(1):700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF.. 2014. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 10(5):e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, et al. 2014. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun. 5:3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK.. 2011. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332 (6031):845–48. [DOI] [PubMed] [Google Scholar]
- Itesako T, et al. 2014. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One 9(2):e84311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JE, Piganeau G, Eyre-Walker A.. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217(5129):624–26. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larriba E, Rial E, Del Mazo J.. 2018. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genomics 19(1):634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A.. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23(22):2639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ.. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 39(2):174–90. [DOI] [PubMed] [Google Scholar]
- Malmevik J, et al. 2016. Distinct cognitive effects and underlying transcriptome changes upon inhibition of individual miRNAs in hippocampal neurons. Sci Rep. 6:19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z.. 2005. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the argonaute 2 protein. RNA 11(6):849–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR.. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 18(1):50–60. [Google Scholar]
- Margoliash E. 1963. Primary structure and evolution of cytochrome C. Proc Natl Acad Sci USA 50:672–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom S, Friger M, Mishmar D.. 2017. MtDNA meta-analysis reveals both phenotype specificity and allele heterogeneity: a model for differential association. Sci Rep. 7:43449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, et al. 2011. The human mitochondrial transcriptome. Cell 146 (4):645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, et al. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23 (1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales HE, et al. 2018. Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nat Ecol Evol. 2(8):1258–67. [DOI] [PubMed] [Google Scholar]
- Mossman JA, Biancani LM, Zhu C-T, Rand DM.. 2016. Mitonuclear epistasis for development time and its modification by diet in Drosophila. Genetics 203(1):463–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi JYY, et al. 2017. Identification of miR-34 regulatory networks in settings of disease and antimiR-therapy: implications for treating cardiac pathology and other diseases. RNA Biol. 14(5):500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M. 2010. Revision of the mtDNA tree and corresponding haplogroup nomenclature. Proc Natl Acad Sci USA. 107(11):E38–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazatos SP, et al. 2017. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry. 22 (5):760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, et al. 2016. A mitochondrial DNA hypomorph of cytochrome oxidase specifically impairs male fertility in Drosophila melanogaster. eLife 5:e16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McEwen BS.. 2014. Mitochondria impact brain function and cognition. Proc Natl Acad Sci USA. 111(1):7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi A, Plazzi F, Milani L, Ghiselli F, Passamonti M.. 2017. SmithRNAs: could mitochondria ‘bend’ nuclear regulation? Mol Biol Evol. 34(8):1960–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, et al. 2016. Mitochondrial DNA copy number variation across human cancers. eLife 5 https://elifesciences.org/articles/10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs CL, et al. 2018. Small non-coding RNA expression and vertebrate anoxia tolerance. Front Genet. 9:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, et al. 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328(5978):636–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HH, Griffiths-Jones S.. 2012. Mitochondrial pseudogenes in the nuclear genomes of Drosophila. PLoS One 7(3):e32593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney JP, et al. 2015. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol Biol. 1241:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, et al. 2013. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23(6):759–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MI, et al. 2011. RNA processing in human mitochondria. Cell Cycle. 10(17):2904–16. [DOI] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB.. 1965. An analysis of variance test for normality (complete samples). Biometrika 52(3/4):591–611. [Google Scholar]
- Singh KK, Kulawiec M.. 2009. Mitochondrial DNA polymorphism and risk of cancer. Methods Mol Biol. 471:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Fields PD, Havird JC.. 2015. Mitonuclear linkage disequilibrium in human populations. Proc Biol Sci/R Soc. 282(1815). https://royalsocietypublishing.org/doi/10.1098/rspb.2015.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink J, van den Heuvel L.. 2010. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol. 2010:737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G. 2002. An expanding universe of noncoding RNAs. Science 296(5571):1260–63. [DOI] [PubMed] [Google Scholar]
- Takasaki S. 2009. Mitochondrial haplogroups associated with Japanese Alzheimer’s patients. J Bioenerg Biomembr. 41(5):407–10. [DOI] [PubMed] [Google Scholar]
- Tourmente M, et al. 2017. mtDNA polymorphism and metabolic inhibition affect sperm performance in conplastic mice. Reproduction 154(4):341–54. [DOI] [PubMed] [Google Scholar]
- Vaught RC, Dowling DK.. 2018. Maternal inheritance of mitochondria: implications for male fertility? Reproduction 155(4):R159–68. [DOI] [PubMed] [Google Scholar]
- Veltri KL, Espiritu M, Singh G.. 1990. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 143(1):160–64. [DOI] [PubMed] [Google Scholar]
- Vyas S, Zaganjor E, Haigis MC.. 2016. Mitochondria and cancer. Cell 166(3):555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnefors M, et al. 2017. Sex-biased microRNA expression in mammals and birds reveals underlying regulatory mechanisms and a role in dosage compensation. Genome Res. 27(12):1961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Apitz J, Pohlheim F, Salinas-Hartwig A, Börner T.. 2009. Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in pelargonium. Mol Genet Genomics. 282(6):587–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, et al. 2015. Mitochondrial DNA content as risk factor for bladder cancer and its association with mitochondrial DNA polymorphisms. Cancer Prevent Res. 8(7):607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemichael BT, et al. 2016. The microRNA cluster miR-183/96/182 contributes to long-term memory in a protein phosphatase 1-dependent manner. Nat Commun. 7:12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Zhiqiang, James D. Stone, Helena Štorchová, and Daniel B. Sloan. 2015. High Transcript Abundance, RNA Editing, and Small RNAs in Intergenic Regions within the Massive Mitochondrial Genome of the Angiosperm Silene Noctiflora. BMC Genomics 16 (November): 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamtich J, Heo S-J, Dhahbi J, Martin DIK, Boffelli D.. 2015. piRNA-like small RNAs mark extended 3′UTRs present in germ and somatic cells. BMC Genomics 16:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, et al. 2017. High mitochondrial DNA copy number was associated with an increased gastric cancer risk in a Chinese population. Mol Carcinogenesis. 56(12):2593–2600. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. Academic Press; p. 97–166. https://www.sciencedirect.com/science/article/pii/B9781483227344500176?via%3Dihub [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.