Abstract
The mechanisms through which tumor cells genetically lose antigenicity and evade immune checkpoints remain largely elusive. Here, we report that tissue-specific expression of the human long-noncoding RNA LINK-A in mouse mammary glands initiated metastatic mammary gland tumors, which phenotypically resembled human triple-negative breast cancer (TNBC). LINK-A expression facilitated crosstalk between phosphatidylinositol-(3,4,5)-trisphosphate and inhibitory G-protein–coupled receptor (GPCR) pathways, attenuating protein kinase A (PKA)-mediated phosphorylation of the E3 ubiquitin ligase TRIM71. Consequently, LINK-A expression enhanced K48–polyubiquitination-mediated degradation of the antigen peptide-loading complex (PLC) and intrinsic tumor suppressors Rb and p53. Treatment with LINK-A-locked nucleic acids or GPCR antagonists stabilized the PLC components, Rb, and p53, and sensitized mammary gland tumors to immune checkpoint blockers (ICBs). Importantly, PD-1 blockade-resistant TNBC patients exhibited elevated LINK-A levels and downregulated PLC components. Hence, we demonstrated lncRNA-dependent downregulation of antigenicity and intrinsic tumor suppression, which may provide the basis for developing a therapeutic regimen of combinational immunotherapy and effective early prevention for TNBCs.
Introduction
The poor prognosis of triple-negative breast cancer (TNBC), hallmarked by the absence of estrogen receptor (ER), progesterone receptor (PR), and HER2 expression, and its resistance to standard chemotherapies have significantly hindered overall survival rates for this disease1, 2. Immunotherapy, including PD-1/PD-L1 blockade, has been demonstrated to inhibit cancer progression3. However, less than 20% of TNBC tissues are PD-L1 positive, and the overall response rate of PD-L1-positive TNBC patients to blockage strategies ranges from 10–18.5%4. These setbacks demand definition and genetic evidence of the molecular mechanisms of immunosuppression during tumor initiation.
One of the central roles of the immune system is the surveillance and elimination of malignant transformations5. To escape immunosurveillance, nascent malignant cells may develop diverse mechanisms, including reducing antigenicity so that anti-tumor lymphocytes fail to detect transformed cells, eliminating immunogenicity by upregulating immunoinhibitory molecules, and recruiting immunosuppressive cells to establish an immunosuppressive microenvironment6, 7. Mutation-derived tumor antigens, also known as neo-antigens, are produced through proteasome-mediated degradation, then transported into the endoplasmic reticulum (ER), where the antigenic peptides are loaded onto the newly synthesized major histocompatibility complex (MHC) I molecules and migrate to the cell surface to be recognized by cytotoxic T cells8. The presentation of neo-antigens derived from mutated proteins leads to tumor suppression9, indicating that mutation burden functions as a predictor of neo-antigens9 and sensitivity to immunotherapy10. However, how tumor cells lose antigenicity is unknown and therapeutic strategies that restore the antigen presentation pathway and sensitize cancers to immunotherapy are missing.
It has become increasingly apparent that many long-noncoding RNAs (lncRNAs) are aberrantly expressed in a broad spectrum of cancers and play key roles in promoting and maintaining cancer characteristics11, 12. An increased understanding of lncRNAs should stimulate new directions for future research and therapeutic options that focus on lncRNAs as novel prognostic markers and therapeutic targets for human cancer13. Although our previous data has indicated that a lncRNA, LINK-A (long intergenic non-coding RNA for kinase activation), is involved in breast cancer drug resistance and hypoxia14, 15, genetic mouse models of lncRNAs with spontaneous tumor development remain elusive and are crucial for developing a proof-of-concept that lncRNAs function as oncogenes that drive tumor initiation.
Here we investigated the role of LINK-A using a transgenic mouse model that represents human TNBC. LINK-A facilitated the association between PtdIns(3,4,5)P3 and inhibitory GCPRs, leading to reduced cyclic-AMP (cAMP) concentrations and PKA-mediated phosphorylation of a E3 ligase, TRIM71. As a consequence, TRIM71 catalyzed the K48-linked polyubiquitination and proteasome-mediated degradation of Rb, p53, and PLC components, thereby contributing to decreased immunosurveillance.
Results
LINK-A correlates with immunosuppression
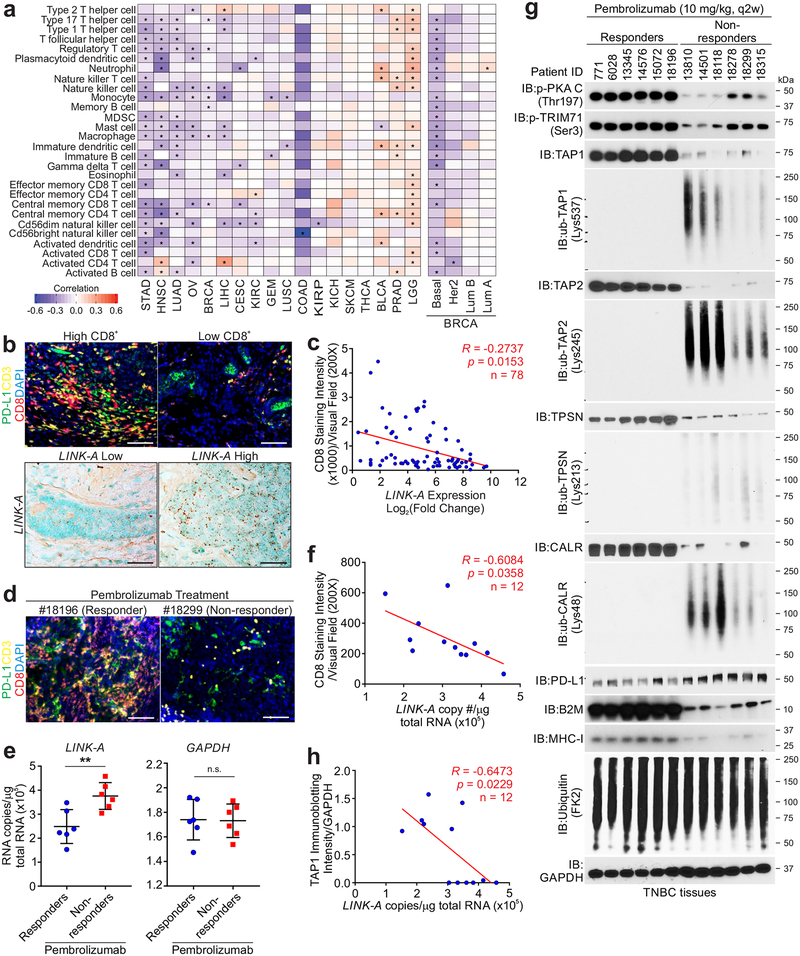
We previously demonstrated that LINK-A is upregulated in TNBC compared to non-TNBC breast cancer tissues and is correlated with poor outcomes for breast cancer patients. To investigate potential relationships between LINK-A and the immune microenvironment, we performed a TCGA pan-cancer analysis, finding that LINK-A is upregulated in multiple cancer types (Supplementary Fig. 1a). The expression of LINK-A was significantly correlated with relative immune cell abundance16 (see methods) and GZMB/CD8A mRNA expression ratio across multiple cancer types, and specifically anti-correlated with APC and CD8+ T cell abundance in basal-like breast cancer (Fig. 1a and Supplementary Fig. 1b). The top 25% of breast tumors with higher infiltration of activated CD8+ T cells and APC exhibited significantly reduced LINK-A expression compared to the bottom 25% of breast tumors (Supplementary Fig. 1c). Fluorescent multiplex (anti-PDL-1, CD3, CD8) immunohistochemistry staining and RNAscope® indicated that human breast cancer tissues with high LINK-A expression exhibited low CD8+CD3+ lymphocyte infiltration (Figs. 1b,c). Thus the expression of LINK-A is correlated with an immunosuppressive microenvironment.
Figure 1: LINK-A predicts immunosuppression and immunotherapy resistance.
a, Pearson correlation of LINK-A expression and relative immune cell abundance based on gene set variance analysis (GSVA) score across 18 cancer types (n=5662). The sample size for each cancer type is as follows: Bladder urothelial carcinoma [BLCA] (n=252), Breast invasive carcinoma [BRCA] (n=836), Cervical squamous cell carcinoma and endocervical adenocarcinoma [CESC] (n=196), Colon adenocarcinoma [COAD] (n=18), Glioblastoma multiforme [GBM] (n=154), Head and neck squamous cell carcinoma [HNSC] (n=426), Kidney chromophobe [KICH] (n=66), Kidney renal clear cell carcinoma [KIRC] (n=448), Kidney renal papillary cell carcinoma [KIRP] (n=198), Brain lower grade glioma [LGG] (n=486), Liver hepatocellular carcinoma [LIHC] (n=200), Lung adenocarcinoma [LUAD] (n=488), Lung squamous cell carcinoma [LUSC] (n=220), Ovarian serous cystadenocarcinoma [OV] (n=294), Prostate adenocarcinoma [PRAD] (n=374), Skin cutaneous melanoma [SKCM] (n=226), Stomach adenocarcinoma [STAD] (n=284), Thyroid carcinoma [THCA] (n=496). The sample size for each BRCA subtype: Basal (n=139), Her2 (n=67), LumA (n=417), LumB (n=191). b, Fluorescent multiplex immunohistochemistry labeling using indicated antibodies (upper panel) or RNAscope (bottom panel) of human breast cancer tissues. Three independent experiments were performed and yielded similar results. Scale bar: 100 μm. c, Pearson correlation between relative LINK-A expression and CD8 staining intensity per visual field. Six field per tissue were measured (n=78 tissues). d, Fluorescent multiplex immunohistochemistry labeling using indicated antibodies of human TNBC tissues upon Pembrolizumab treatment. Three independent experiments were performed and yielded similar results. Scale bar: 100 μm. e, Measurement of LINK-A RNA copy number (left), or GAPDH RNA copy number (right), of human TNBC tissues upon Pembrolizumab treatment (n=12 tissues) (n.s., p=0.925, **p=0.0062). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test. f, Pearson correlation of CD8 staining intensity with the LINK-A copy number of human TNBC tissues upon Pembrolizumab treatment (n=12 tissues). g, Immunoblotting detection using indicated antibodies of human TNBC tissues upon Pembrolizumab treatment. Three independent experiments were performed and yielded similar results. h, Pearson correlation of TAP1 immunoblotting intensities with the LINK-A copy number of human TNBC tissues upon Pembrolizumab treatment (n=12 tissues).
To demonstrate the potential prognostic value of LINK-A in TNBC patients needing immunotherapy, we determined the infiltration of CD8+ T cells in TNBC patients upon Pembrolizumab (anti-PD-1) treatment. The TNBC patients who responded to Pembrolizumab exhibited relatively lower expression of LINK-A and higher CD8+ T cell infiltration compared to non-responders (Figs. 1d,e and Supplementary Fig. 1d). CD8+ T cell infiltration in this cohort of TNBC patients negatively correlated with LINK-A expression (Fig. 1f).
The decreased APC infiltration in LINK-A-high TNBC suggested potentially impaired antigen-presentation machinery, prompting an investigation into the status of the PLC components within the tumors. The protein level of the PLC components, including TPSN, TAP1, TAP2, and CALR, were all downregulated in TNBC non-responders compared to responders upon Pembrolizumab treatment (Fig. 1g). Using modification-specific antibodies, we demonstrated that the K48-linked polyubiquitination (poly-Ub) of TPSN (Lys537), TAP1 (Lys245), TAP2 (Lys213), and CALR (Lys48) were upregulated in non-responders, although the level of total polyubiquitinated proteins remained unaltered (Fig. 1g), suggesting proteasome-regulated degradation of these components. Furthermore, the expression of LINK-A negatively correlated with the protein expression of TPSN, TAP1, TAP2, and CALR, as well as β2-microglobulin (β2M) and MHCI in this cohort (Fig. 1h and Supplementary Figs. 1e–i). These observations suggested the importance of LINK-A in modulating immune balance in favor of immunosuppression and that the expression of LINK-A potentially modulates the protein levels of the PLC.
LINK-A drives basal-like breast cancer
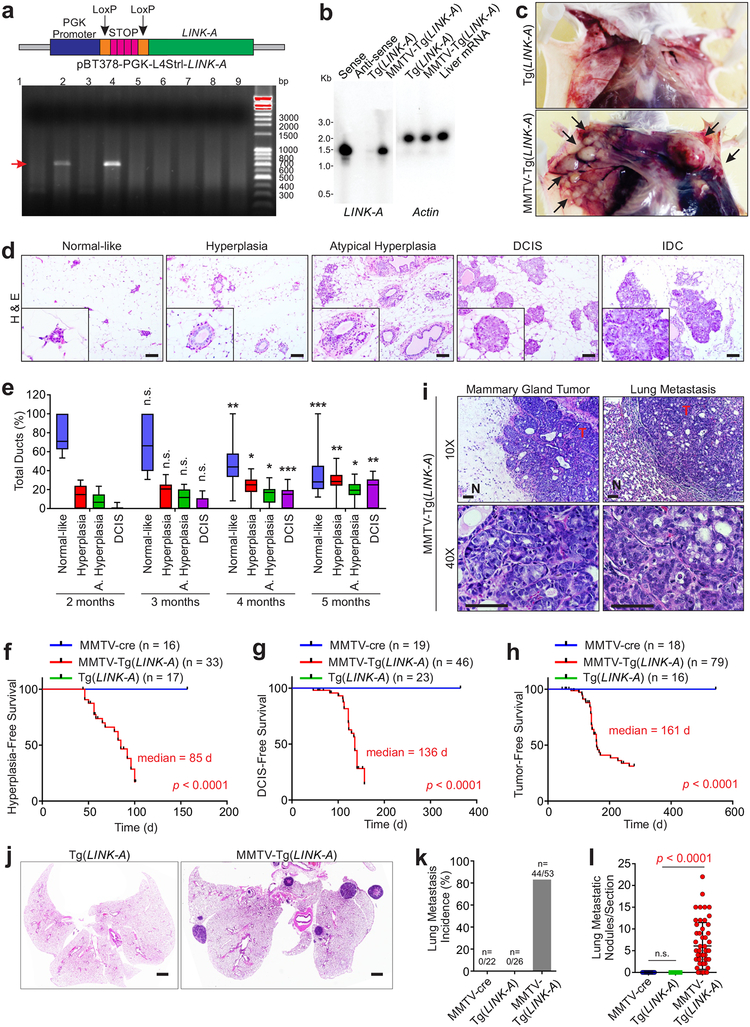
Previous studies using mouse mammary tumor virus (MMTV) long terminal repeat (LTR)-driven Neu (Erbb2), Ras, and Myc transgenic mice demonstrated the development of tumors in mouse mammary glands by 6–12 months17, 18, 19. Transgenic mice harboring a LINK-A-containing ‘flox-stop-flox’ cassette were bred with MMTV-Cre mice to induce expression of LINK-A in mammary tissues (referred to as MMTV-Tg(LINK-A) mice)20 (Fig. 2a, top). Two male founder transgenic animals were generated (Fig. 2a, bottom and Supplementary Fig. 1j) and both founder animals passed the transgene to their offspring in accordance with Mendelian inheritance. All the data reported are based on observations of the progeny of the two founder animals.
Figure 2: LINK-A initiates mammary gland tumors that models human TNBC.
a, Top panel: graphic illustration of tissue-specific expression of human LINK-A gene. Bottom panel: DNA agarose gel indication of Tg(LINK-A) genotyping. Three independent experiments were performed and yielded similar results. b, Northern blot detection of LINK-A or Actin using in vitro transcribed LINK-A sense, antisense or total RNA extracted from Tg(LINK-A) or MMTV-Tg(LINK-A) mammary gland. Three independent experiments were performed and yielded similar results. c, Representative images of Tg(LINK-A) or tumor-bearing MMTV-Tg(LINK-A) mice. Blue arrows: mammary gland tumors. Three independent experiments were performed and yielded similar results. d and e, H&E staining (d) or statistical analysis (e) of MMTV-Tg(LINK-A) mammary gland of animals at 2, 3, 4, or 5 month of age to indicate the development from normal-like breast epithelial cells through hyperplasia, atypical hyperplasia, DCIS and IDCs. Scale bar: 100 μm (d). Error bars (e): S.E.M. n=9, 15, 15, 11 animals respectively, unpaired Student’s t-test. f-h, Kaplan-Meier survival analysis of MMTV-cre, MMTV-Tg(LINK-A) or Tg(LINK-A) mice according to the presence of hyperplasia (f), DCIS (g) or IDC (h). n=16, 33, 17 (f) 19, 46, 23 (g) or 18, 79, 16 (h) animals respectively (log rank test). i, Representative images of MMTV-Tg(LINK-A) primary tumor and lung metastasis. T: tumor; N: adjacent normal tissue. Three independent experiments were performed and yielded similar results. Scale bar: 100 μm. j-l, Representative images of Tg(LINK-A) or MMTV-Tg(LINK-A) lung (j). Three independent experiments were performed and yielded similar results. Lung metastasis incidence of tumor-bearing mice (k) or metastatic nodules/section (l) of MMTV-cre, Tg(LINK-A) or MMTV-Tg(LINK-A) lung (n=18, 16, 53). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test. Scale bar: 1mm (j).
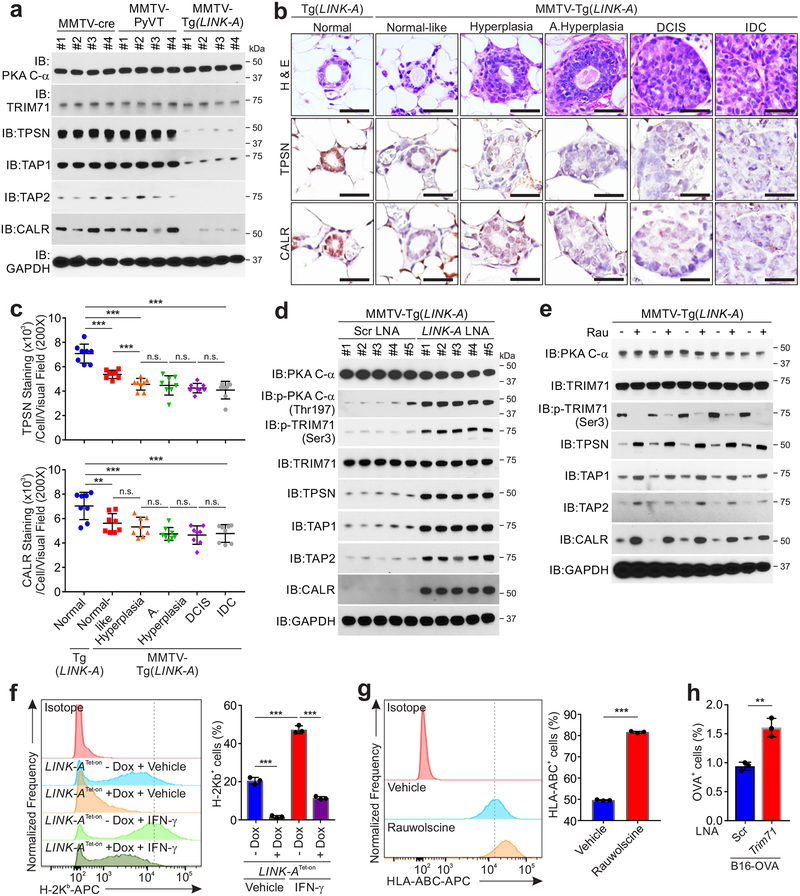
Moderate expression of LINK-A in mouse mammary glands following crossing with MMTV-cre was confirmed by northern blot (Fig. 2b). Virgin female mice with transgenic expression of LINK-A developed mammary gland adenocarcinomas, which involve the entire epithelium of the gland (Fig. 2c). Histologic evaluation indicated that at 8 weeks of age, the majority of mammary gland ducts appear normal (referred to as normal-like) (Fig. 2d). Mice that were 3, 4 and 5 months old exhibited a reduced percentage of normal-like mammary gland ducts with a concurrently increased percentage of hypoplasia, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC) over the time, respectively (Figs. 2d–e). The median time until development of hyperplasia, DCIS, and IDC in virgin female MMTV-Tg(LINK-A) animals was 85, 136, and 161 days, respectively (Figs. 2f–h). The primary mammary gland tumors metastasized to the lungs, which exhibited adenocarcinoma (Fig. 2i). 88.7% of tumor-bearing MMTV-Tg(LINK-A) animals developed lung metastasis, compared to no lung metastasis in the control MMTV-cre and Tg(LINK-A) animals (Figs. 2j–l). The copy number of LINK-A in MMTV-Tg(LINK-A) mouse tumors was comparable to human TNBC tissues (Supplementary Table 1 and Supplementary Figs. 1k–m). Immunohistochemistry indicated that MMTV-Tg(LINK-A) tumors exhibited similar expression of ER-alpha, PR-A/B, and HER2 as normal mammary glands (Supplementary Figs. 2a–b), suggesting that the MMTV-Tg(LINK-A) malignancies show no ER, PR, and HER2 amplification/upregulation.
We observed that MMTV-Tg(LINK-A) tumors harbored a similar number of missense somatic mutation burdens compared to human TNBCs (Supplementary Fig. 2c). Furthermore, MMTV-Tg(LINK-A) tumors harbored non-silencing somatic mutations on Trp53 and Pik3ca genes that are frequently mutated in human TNBCs21, 22 (Supplementary Fig. 2d). These observations suggested that the MMTV-Tg(LINK-A) tumors exhibit genetic similarity to human TNBCs, mimicking the bona fide tumor initiation and somatic mutation generation processes.
RNA-seq analyses of MMTV-Tg(LINK-A) tumors exhibited distinct transcriptional profiling compared to non-LINK-A–expressing Tg(LINK-A) mouse mammary glands (Supplementary Fig. 2e). The transcriptome of MMTV-Tg(LINK-A) tumors was co-clustered with human basal-like breast cancers compared to other subtypes (Supplementary Fig. 2e). Notably, up-regulated genes in tumors versus normal tissues were enriched in cell cycle and redox homeostasis, while down-regulated genes were enriched in lipid metabolism, T cell activation, and immunoresponse (Supplementary Fig. 2f).
Human TNBCs also exhibit a metabolic signature that is hallmarked by glycolysis and accumulation of redox23. Although the relative abundance of overall metabolites was comparable, MMTV-Tg(LINK-A) tumors were enriched with metabolites of glycolysis and redox homeostasis compared to normal tissues (Supplementary Figs. 2g–i). Hence, our findings suggested that the MMTV-Tg(LINK-A) tumors model human basal-like breast cancer metabolically.
LINK-A mediates PtdIns(3,4,5)P3-GPCR crosstalk
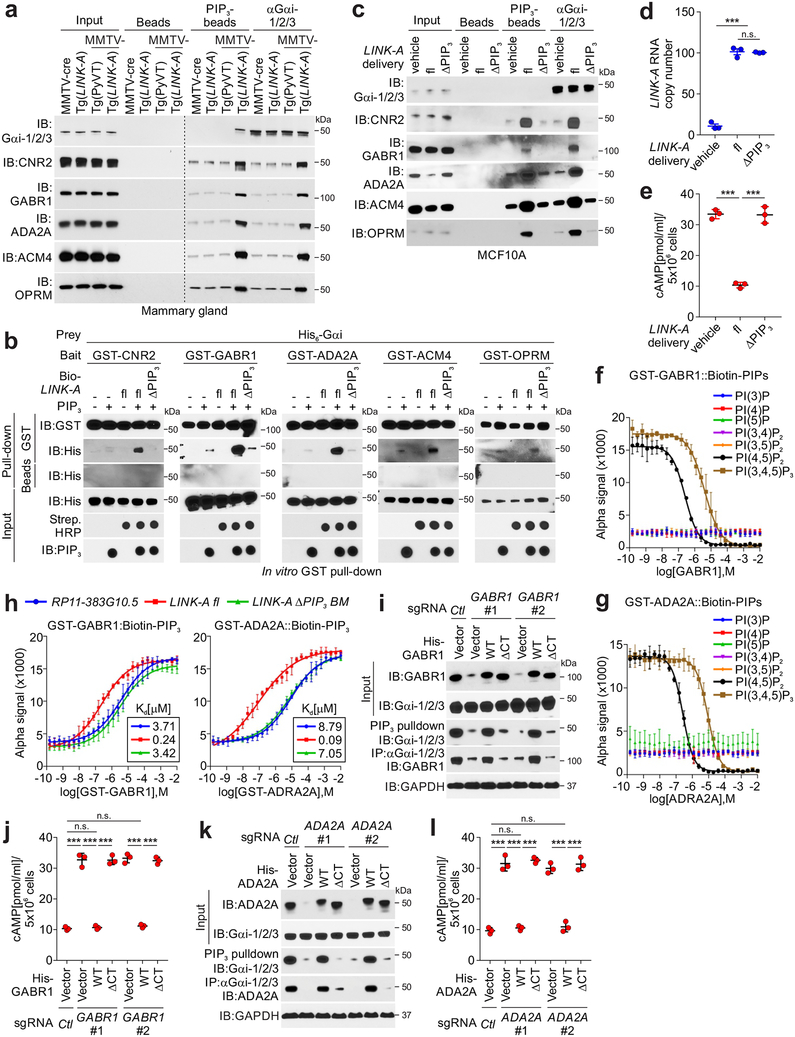
LINK-A associates with PtdIns(3,4,5)P3 and regulates the activation of AKT pathway15. To dissect the signaling pathway mediated by PtdIns(3,4,5)P3-bound LINK-A in MMTV-Tg(LINK-A) tumors, we surveyed the PtdIns(3,4,5)P3-binding proteins in Tg(LINK-A) mouse mammary glands and MMTV-Tg(LINK-A) tumors using Liquid chromatography–mass spectrometry (LC-MS) (Table 1 and Supplementary Table 2). Without Cre recombinase, PtdIns(3,4,5)P3 associated with a cohort of lipid-interacting proteins, which is consistent with previous findings24, 25, 26 (Table 1-green). Upon expression of LINK-A, PtdIns(3,4,5)P3 associated with GPCRs, which included cannabinoid receptor 2 (CNR2), γ-aminobutyric acid type B receptor subunit 1 (GABR1), α2A adrenergic receptor (ADA2A), muscarinic acetylcholine receptor M4 (ACM4), and Mu-type opioid receptor (OPRM) (Table 1-red). Upon ligand binding, GPCRs activated the associated G protein, in which the G protein alpha subunit (Gα) can be classified as Gαs, Gαi, Gαq, G12/1327. Interestingly, CNR2, GABR1, ADA2A, ACM4, and OPRM all associate with Gαi, leading to inhibition of adenylyl cyclase and reduced cAMP production upon ligand binding28. Consistent with this notion, PtdIns (3,4,5)P3 and Gαi exhibited robust interactions with GPCRs in MMTV-Tg(LINK-A) tumors compared to the mammary glands of FVB mice (background control mice) or MMTV-PyVT tumors (Fig. 3a), despite similar PtdIns(3,4,5)P3 enrichment (Supplementary Fig. 3a). The presence of exogenous ligands may further facilitate these interactions.
Table 1.
Summary of protein identification of PIP3 pulldown using Tg(LINK-A) normal mammary gland or MMTV-Tg(LINK-A) tumor.
| Experimental setting | MMTV-cre Mammary gland |
MMTV-Tg(LINK-A) Mammary gland tumor |
||
|---|---|---|---|---|
| Control Beads | PIP3 Beads | Control Beads | PIP3 Beads | |
| PtdIns(3,4,5)P3-Binding Proteins | AKT1 | AKT1 | ||
| ACAP1 | TEC | |||
| AKT2 | PHLB1 | |||
| TEC | ACAP1 | |||
| PHLB1 | AKT2 | |||
| ACAP2 | KPCD3 | |||
| Inhibitory GPCRs | CNR2 | |||
| GABR1 | ||||
| ADA2A | ||||
| ACM4 | ||||
| OPRM | ||||
Figure 3: LINK-A mediates crosstalk between PtdIns (3,4,5)P3 and GPCRs.
a, Immunoblotting detection using indicated antibodies of immunoprecipitates using indicated beads or antibodies using MMTV-cre or Tg(LINK-A) normal mammary gland, MMTV-PyVT tumor or MMTV-Tg(LINK-A) tumor lysates. Three independent experiments were performed and yielded similar results. b, GST-pulldown using His-tagged Gαi, GST-tagged GPCRs, and PtdIns (3,4,5)P3, in the presence of biotinylated LINK-A full length (fl) or ΔPtdIns (3,4,5)P3 mutant. Three independent experiments were performed and yielded similar results. c, Immunoblotting detection using indicated antibodies of immunoprecipitates using indicated beads or antibodies of MCF10A cells delivered with vehicle, LINK-A fl or ΔPtdIns (3,4,5)P3 mutant. Three independent experiments were performed and yielded similar results. d and e, Measurement of LINK-A copy number (d), or cellular cAMP level (e) of MCF10A cells delivered with vehicle, LINK-A fl or ΔPtdIns (3,4,5)P3 mutant (n.s., p=0.399, ***p<0.001). Results are mean ± s.e.m. of n=3 independent experiments yielding similar results, P values were determined by one-way ANOVA. f and g, Competition binding assay to determine Kd for interaction between GST-tagged Gαi (left top), or GST-tagged GABR1 (f) or –ADA2A (g) and biotinylated PIPs as indicated. The Kd value (μM) are shown. Results are mean ± s.d. of n=3 independent experiments yielding similar results. h, Competition binding assay to determine Kd for interaction between GST-tagged GPCRs and biotinylated PtdIns (3,4,5)P3 in the presence of RP11–383G10.5, LINK-A fl or LINK-A ΔPtdIns (3,4,5)P3 mutant. Results are mean ± s.d. of n=3 independent experiments. i-l, Immunoblotting detection using indicated antibodies in MDA-MB-231 cells harboring sgRNAs knocking out GABR1 (i, j), ADA2A (l, m) respectively, followed by expression of GPCRs wild type or ΔPtdIns(3,4,5)P3 mutants. The lysates were subjected to immunoblotting detection using indicated antibodies (i, k) or measurement of cAMP concentration (j, l) (n.s., p>0.05, ***p<0.001). Results are mean ± s.d. of n=3 independent experiments, P values were determined by one-way ANOVA.
To validate that the Gαi-GPCRs interaction is dependent on PtdIns (3,4,5)P3-bound LINK-A, we observed that full-length LINK-A facilitated the interactions between GST-tagged bacterially-expressed human GPCRs and Gαi (Fig. 3b). Conversely, the presence of a LINK-A PtdIns (3,4,5)P3-binding motif deletion mutant (nt. 1081–1140) (referred to as ΔPIP3)15 failed to do so (Fig. 3b). In LINK-A-low mouse/human mammary gland epithelial NMuMG/MCF10A cells, exogenous expression of the full-length lncRNA, but not the ΔPIP3 mutant, enhanced the recruitment of GPCRs to Gαi and PtdIns(3,4,5)P3 (Fig. 3c and Supplementary Fig. 3b). In LINK-A-high MDA-MB-231 cells with a LINK-A PtdIns (3,4,5)P3-binding motif depletion (referred to as PIP3-BM−/−)15, reintroduction of full-length LINK-A, but not the mutant, rescued these interactions (Supplementary Fig. 3c).
Activation of Gαi results in reduced cellular cAMP concentration28. MCF10A/NMuMG cells harboring ~100 copies of exogenous full-length LINK-A, but not the ΔPIP3 mutant, showed significantly reduced cAMP concentrations and PKA phosphorylation at Thr19729 with minimally altered cellular PtdIns(3,4,5)P3 (Figs. 3d–e and Supplementary Figs. 3d–i).
To determine the binding affinity of PtdIns(3,4,5)P3-GPCRs interactions, we applied an Alpha-Assay™. This assay utilizes “donor” and “acceptor” beads to capture interacting biomolecules in proximity so that leads to an energy transfer from one bead to the other, ultimately producing a luminescent/fluorescent signal. For these experiments, we used GST-tagged bacterially-expressed GPCRs and biotinylated-PtdIns(3,4,5)P3 as donor-acceptor pairs (Figs. 3f–h and Supplementary Figs. 3j–k). Gαi exhibited non-detectable interactions with PtdIns(3,4,5)P3; however, all five GPCRs showed moderate interactions with PtdIns(3,4,5)P3 (Kd 3.96–8.75 μM) and strong interactions with PtdIns(4,5)P2 (Kd 0.08–0.42 μM) (Figs. 3f–g and Supplementary Fig. 3j). Next, we determined the Kd value of PtdIns (3,4,5)P3-GPCRs interactions in the presence of LINK-A or a cardiolipin-binding lncRNA, RP11–383G10.515, as a control (Fig. 3h and Supplementary Fig. 3k). In the presence of full-length LINK-A, but not the ΔPIP3 mutant, the Kd values of PtdIns (3,4,5)P3-GPCRs interactions increased 15–18 fold compared to RP11–383G10.5 (Fig. 3h and Supplementary Fig. 3k).
The intracellular C-termini of GPCRs harbor hydrophobic acyl groups that facilitate the recruitment of GPCRs to membrane lipid rafts30. We reasoned that the intracellular C-termini of GPCRs are required for the crosstalk with LINK-A and PtdIns (3,4,5)P3. We used sgRNAs to deplete CNR2, GABR1, ADA2A, ACM4, or OPRM in the MDA-MB-231 cells (Figs. 3i–l and Supplementary Figs. 3l–q), followed by expression of wild-type or mutants with deletions in the C-terminal lipid raft binding domain (referred to as ΔCT) which had similar expression levels to endogenous GPCRs. Wild-type GPCRs, but not ΔCT mutants, rescued the PtdIns(3,4,5)P3-Gαi and GPCRs-Gαi interactions upon CNR2, GABR1, ADA2A, ACM4, or OPRM deletion (Figs. 3i,k and Supplementary Figs. 3l,n,p). As a consequence, cellular cAMP levels were increased upon depletion of the GPCRs and were restored upon expression of wild-type GPCRs but not the ΔCT mutants (Figs. 3j,l and Supplementary Figs. 3m,o,q). Expression of wild-type GPCRs or mutants showed minimal effect on cellular PtdIns (3,4,5)P3 levels or the expression of LINK-A (Supplementary Figs. 4a–b). It is unlikely that the cell membrane expression of these GPCRs was affected upon ΔCT. Taken together, our data indicated that LINK-A, upon association with PtdIns(3,4,5)P3, facilitates PtdIns(3,4,5)P3-GPCRs and Gαi-GPCRs interactions, resulting in reduced intracellular concentrations of cAMP.
GPCR antagonist represses tumor cell proliferation
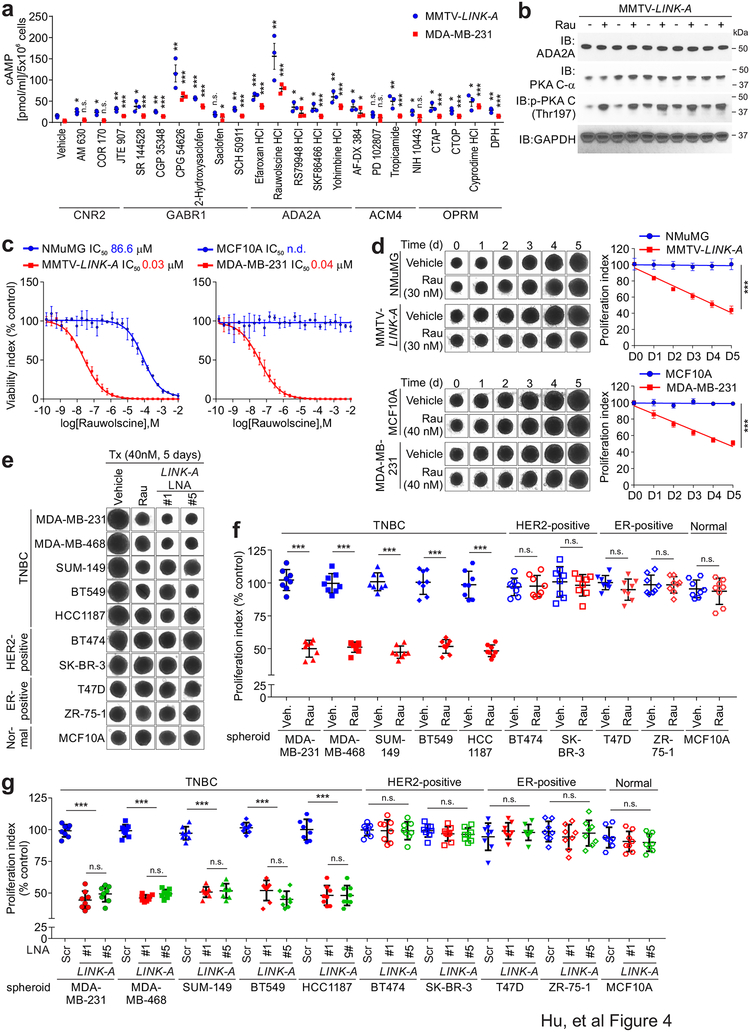
Our findings suggested that LINK-A-positive basal-like breast cancer may be addicted to GPCR signaling, which could be repressed with GPCRs antagonists or inhibitors. We screened and determined the effect of a cohort of GPCR antagonists/inhibitors using isolated tumor cells derived from MMTV-Tg(LINK-A) tumors (referred to as MMTV-LINK-A cells) and MDA-MB-231 (Fig. 4a). Compared to the vehicle, the ADA2A antagonist Rauwolscine exhibited potent effects on restoring cAMP concentrations in both MMTV-LINK-A and MDA-MB-231 cells (Fig. 4a). Rauwolscine is a natural alkaloid that acts as a selective and reversible ADA2A antagonist (Ki = 12 nM)31. The effect of Rauwolscine on restoring the phosphorylation of PKA (Thr197) was validated in MMTV-LINK-A cells derived from individual MMTV-Tg(LINK-A) tumors, in which the protein expression of ADA2A or the PKA catalytic subunit were unaltered (Fig. 4b).
Figure 4: Identification and characterization of GPCR antagonists.
a, Fold change of cellular cAMP concentration of MMTV-LINK-A and MDA-MB-231 cells treated with indicated GPCR antagonists (n.s., p>0.05, *p<0.05, **p<0.01, ***p<0.001). Results are mean ± s.e.m. of n=3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. b, Immunoblotting detection using indicated antibodies of tissue lysates of MMTV-Tg(LINK-A) tumor lysates subjected to vehicle or Rauwolscine treatment. Three independent experiments were performed and yielded similar results. c, Measurement of viability index of 3-dimentional spheroid formation assay of NMuMG and MMTV-LINK-A cells (left), or MCF10 A or MDA-MB-231 cells (right) in the presence of a serial 2-fold dilution of Rauwolscine. Results are mean ± s.d. of n=3 independent experiments. d, Representative images at day 0 to 5 (left) and measurement of proliferation index (right) of 3-dimentional spheroid formation assay of NMuMG and MMTV-LINK-A cells (top), or MCF10 A or MDA-MB-231 cells (bottom) in the presence of vehicle or Rauwolscine 30 nm (top) or 40 nM (bottom) (***p<0.001). Results are mean ± s.d. of n=3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. e and f, Representative images of day 8 (e) or measurement of proliferation index (f) of indicated normal or breast cancer cells in the presence of vehicle or Rauwolscine (Rau, 40 nM) (n.s., p>0.05, ***p<0.001). Results are mean ± s.d. of n=8 spheroids per experimental condition, P values were determined by one-way ANOVA. g, Measurement of proliferation index of indicated normal or breast cancer cells in the presence of scramble or LINK-A LNAs (5 nM) (n.s., p>0.05, ***p<0.001). Results are mean ± s.d. of n=8 spheroids per experimental condition, P values were determined by one-way ANOVA.
Rauwolscine exhibited minor effects on the cell viability of MCF10A/NMuMG cells but robust cell cytotoxicity against MMTV-LINK-A and MDA-MB-231 tumor cells with IC50 values of 30 and 40 nM, respectively (Figs. 4c–d). We then determined the efficacy of Rauwolscine in modulating cell proliferation using a panel of TNBC and non-TNBC breast cancer cell lines, finding that treatment with Rauwolscine specifically repressed the cell proliferation of TNBC cells, but not non-TNBC cells or normal cells (Figs. 4e–f). Treatment with LINK-A locked nucleic acids (LNAs) showed similar repression of cell proliferation in TNBC cells but not non-TNBC cells (Fig. 4g). We then tested combinatorial strategies against the growth of MDA-MB-468 cells, which do not respond to current anti-EGFR targeted therapies32. Individually, Rauwolscine and the CDK4/6 inhibitor Abemaciclib, but not Erlotinib, inhibited cell proliferation by 50%; however, a combined treatment consisting of Rauwolscine with Abemaciclib or Erlotinib significantly repressed cell proliferation (Supplementary Fig. 4c). These findings suggested that inhibiting GPCR signaling may improve the sensitivity of TNBCs to anti-CDK4/6 and anti-EGFR targeted treatments.
PKA stabilizes Rb and p53
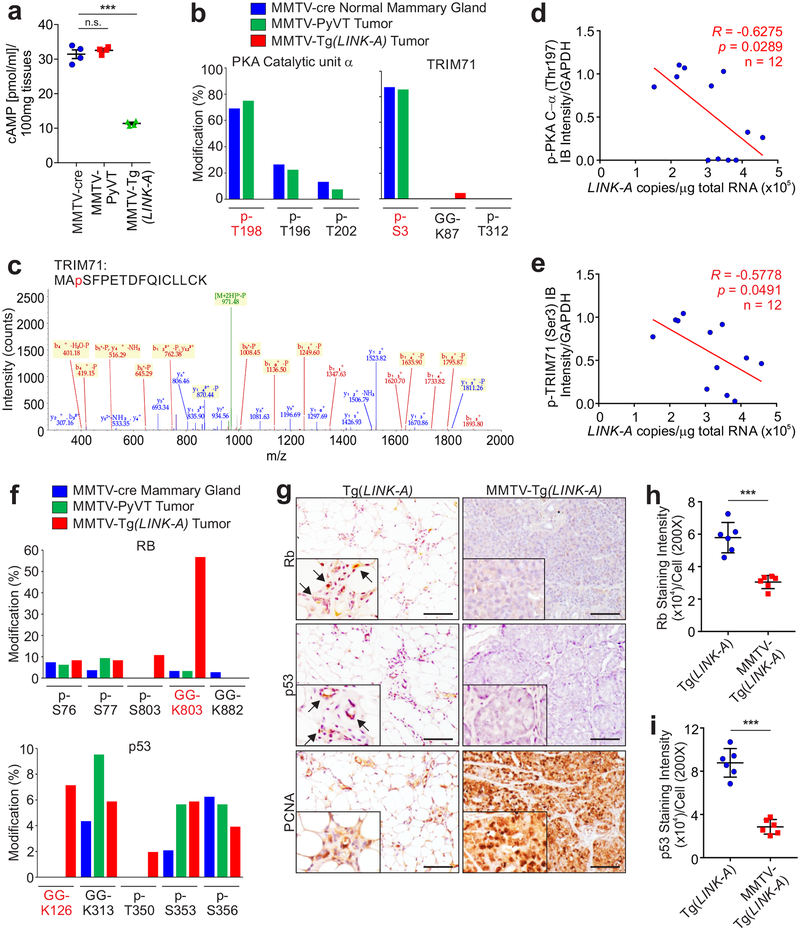
We demonstrated that LINK-A activates GPCRs, leading to downregulation of cAMP and inactivation of PKA. Although PKA depletion leads to carcinogenesis33, 34, the underlying molecular mechanism of how PKA prevents tumor initiation is unknown. Using LC-MS as an open-ended technology, we identified PKA-binding proteins and the post-translational modifications of these proteins in MMTV-Cre mammary glands, MMTV-PyVT tumors35, and MMTV-Tg(LINK-A) tumors (Table 2 and Supplementary Table 3). While the catalytic and regulatory subunits of PKA were detected in all three types of tissues (Table 2-green), PKA catalytic subunits (PKA-Cα, PKA-Cβ) exhibited phosphorylation at the Thr197 residues of both subunits in normal mammary glands and MMTV-PyVT tumors, but not in MMTV-Tg(LINK-A) tumors (Table 2-green). Consistently, cAMP levels were reduced in MMTV-Tg(LINK-A) tumors compared to MMTV-cre mammary glands and MMTV-PyVT tumors (Fig. 5a), validating the conclusion that expression of LINK-A inactivates the cAMP/PKA pathway.
Table 2.
Summary of protein identification and post-translational modification of PKA pulldown using MMTV-cre mammary glands, MMTV-PyVT tumors, or MMTV-Tg(LINK-A) tumors.
| Experimental setting | MMTV-cre Mammary gland |
MMTV-PyVT Mammary gland tumor |
MMTV-Tg(LINK-A) Mammary gland tumor |
|||
|---|---|---|---|---|---|---|
| IgG | α-PKA | IgG | α-PKA | IgG | α-PKA | |
| PKA Complex | KAP3 | KAP3 | KAP3 | |||
| p-PKA-C-α (Thr197) | p- PKA-C-α (Thr197) | PKA-C-α | ||||
| p- PKA-C-β (Thr197) | p- PKA-C-β (Thr197) | KAP2 | ||||
| KAP2 | KAP2 | KAP0 | ||||
| KAP0 | KAP0 | PKA-C-β | ||||
| KAP1 | KAP1 | KAP1 | ||||
| Ubiquitin Machinery | p-TRIM71 (Ser3) | p-TRIM71 (Ser3) | TRIM71 | |||
| Ubiquitin | ||||||
| UBA1 | ||||||
| UB2D3 | ||||||
| Ubiquitin Substrates | Rb | Rb | Ub-Rb (Lys803) | |||
| P53 | P53 | Ub-p53 (Lys126) | ||||
Figure 5: LINK-A represses PKA-dependent TRIM71 phosphorylation.
a, Measurement of cAMP concentration in MMTV-cre normal mammary gland, MMTV-PyVT or MMTV-Tg(LINK-A) tumor lysates (n.s., p>0.05, ***p<0.001). Results are mean ± s.e.m. of n=4 animals per experimental condition, P values were determined by one-way ANOVA. b, Percentage of modified vs. total number of peptides harboring indicated residues of PKA catalytic unit α (left) or TRIM71 (right) in MMTV-cre mammary gland, MMTV-PyVT or MMTV-Tg(LINK-A) tumors. c, Annotated MS/MS spectrum assigned to the TRIM71 peptide MApSFPETDFQICLLCK acquired from analysis of tryptic digest by high-sensitivity LC-MS/MS on an Orbitrap Elite high-resolution mass spectrometer. d and e, Pearson correlation between immunoblotting staining of p-PKA C-α (Thr197) (d) or p-TRIM71 (Ser3) (e) and LINK-A copy number of human TNBC tissues upon Pembrolizumab treatment (n=12 tissues). f, Percentage of modified vs. total number of peptides harboring indicated residues of Rb (top) or p53 (bottom) in MMTV-cre mammary gland, MMTV-PyVT or MMTV-Tg(LINK-A) tumors. g-i, Representative images (g) and statistical analysis (h and i) of immunohistochemistry staining using indicated antibodies of Tg(LINK-A) normal mammary gland and MMTV-Tg(LINK-A) tumors. Scale bars (g): 100 μm. (h and i) (***p<0.001). Results are mean ± s.d. of n=6 animals per experimental condition. P values were determined by unpaired two-tailed Student’s t test.
PKA associated with TRIM71, an E3 ligase36, in all three types of tissues; however, in MMTV-Tg(LINK-A) tumors, PKA associated with the ubiquitin pathway components: RL40 (ubiquitin precursor), UBA1 (E1), UB3D2 (E2) and TRIM71 (E3) (Table 2-black), suggesting that the ubiquitination pathway was activated in MMTV-Tg(LINK-A) tumors. Furthermore, TRIM71 was phosphorylated at Ser3 in normal tissues and MMTV-PyVT tumors but not in MMTV-Tg(LINK-A) tumors (Table 2-black, and Figs. 5b–c). This data suggested that TRIM71 is regulated by phosphorylation and that hypophosphorylated TRIM71 associates with ubiquitin machinery. We then tested the hypothesis that the phosphorylation of TRIM71 at Ser3 is catalyzed by PKA, finding that CRISPR-mediated ablation of the genes encoding PKA catalytic subunits using sgRNAs reduced the phosphorylation of TRIM71. The expression of the wild type PKA catalytic subunit, but not the kinase-dead mutant, K72H37, restored the phosphorylation of TRIM71 (Supplementary Figs. 4d–e). In TNBC Pembrolizumab treatment responders/non-responders, the status of the p-PKA C-α (Thr197) and p-TRIM71 (Ser3) negatively correlated with LINK-A expression (Figs. 5d–e). These observations suggested that LINK-A expression potentially inhibits PKA phosphorylation/activity and PKA-mediated phosphorylation of TRIM71 at Ser3.
Aiming to identify the ubiquitination substrates of LINK-A/PKA/TRIM71 signaling, we further analyzed our LC-MS data to identify the proteins with peptide adducts derived from ubiquitin. The C-terminus of the mature ubiquitin has the amino acid sequence KESTLHLVLRLRGG, in which the last Gly can be conjugated to lysine residues on target proteins. When the conjugated ubiquitin is cleaved with trypsin, it leaves Gly-Gly (GG) residues on the modified lysine residues of the target proteins. Rb and p53 exhibited GG modifications at Lys803 and Lys126, respectively, which suggested ubiquitination (Ub) modification of these proteins (Table 2-red and Fig. 5f). Ubiquitin contains 7 lysine residues: K6, K11, K27, K29, K33, K48, and K63, through which a specific polyubiquitin chain can be formed upon a target protein. To determine the types of poly-ubiquitination chains (poly-Ub) on the target proteins in MMTV-Tg(LINK-A) tumors, we analyzed which lysine residues of ubiquitin are predominantly conjugated with the GG di-peptide, finding that lysine 48 of ubiquitin is largely modified by GG (Supplementary Fig. 4f). This data indicated that Ub-Rb and -p53 are modified with K48-linked polyUb and are potentially subjected to proteasomal degradation38. Consistently, the protein status of Rb and p53, which have been shown to be downregulated in human breast tumors compared to normal adjacent tissues39, 40, were reduced in MMTV-Tg(LINK-A) tumors compared to mouse mammary glands (Figs. 5g–i). Taken together, our data suggested that expression of LINK-A downregulates intrinsic tumor suppressor barriers via the GPCR-PKA-TRIM71 signaling axis during tumorigenesis.
LINK-A-dependent degradation of PLC components
To address the hypothesis that LINK-A-mediated hypophosphorylation of TRIM71 in MMTV-Tg(LINK-A) tumors may catalyze poly-Ub chain formation in a panel of substrates, we identified TRIM71-bindng proteins with post-translational modifications using MMTV-Tg(LINK-A) tumors pre-treated with scramble (Scr) or LINK-A LNAs (Table 3 and Supplementary Table 4), which have been shown to efficiently knockdown LINK-A in vivo15. Although the regulatory and catalytic subunits of PKA were detected under both conditions, the catalytic subunits exhibited phosphorylated-Thr197 following LINK-A LNAs treatment (Table 3-green). Furthermore, the LINK-A LNAs treatment restored the phosphorylation of TRIM71 (Ser3), with concurrent disassociation of Ubiquitin, UBA1, and UB2D3 (Table 3-black). These observations suggested that the LINK-A LNAs treatment effectively reversed the inactivation of the cAMP/PKA pathway in MMTV-Tg(LINK-A) tumors.
Table 3.
Summary of protein identification and post-translational modification of TRIM71 pulldown of MMTV-Tg(LINK-A) tumor upon scramble (Scr) or LINK-A LNA treatment.
| Experimental setting | MMTV-Tg(LINK-A) tumor Scr LNA |
MMTV-Tg(LINK-A) tumor LINK-A LNA |
||
|---|---|---|---|---|
| IgG | α-TRIM71 | IgG | α-TRIM71 | |
| PKA Complex | KAP3 | KAP3 | ||
| KAP2 | KAP2 | |||
| KAP0 | KAP0 | |||
| PKA-C-α | p- PKA-C-α (Thr197) | |||
| PKA-C-β | p- PKA-C-β (Thr197) | |||
| KAP1 | KAP1 | |||
| Ubiquitin Machinery | TRIM71 | p-TRIM71 (Ser3) | ||
| Ubiquitin | ||||
| UBA1 | ||||
| UB2D3 | ||||
| Ubiquitin Substrates | Ub-Rb (Lys803) | Rb | ||
| Ub-p53 | P53 | |||
| Ub-TPSN (Lys213) | TPSN | |||
| Ub-TAP2 (Lys245) | TAP2 | |||
| Ub-CALR (Lys48) | TAP1 | |||
| Ub-TAP1 (Lys537) | CALR | |||
In addition to Rb and p53, TRIM71 associated with all six components of the PLC, namely TPSN, TAP1, TAP2, CALR, ERAP1, and PDIA3 in MMTV-Tg(LINK-A) tumors (Table 3-red). The PLC components facilitate the folding and loading of antigenic peptides and the transportation of the MHC I complex to the cellular surface41. TPSN, TAP1, TAP2, and CALR were all subjected to GG modification at the Lys213 (TPSN), Lys537 (TAP1), Lys245 (TAP2), and Lys48 (CALR) residues (Table 3-red), and all ubiquitin which associated with TRIM71 or TRIM71-binding proteins was K48-linked (Supplementary Fig. 4g). Upon LINK-A LNAs treatment, the GG modifications at Lys213 (TPSN), Lys537 (TAP1), Lys245 (TAP2), and Lys48 (CALR) were all abolished (Table 3-red and Supplementary Fig. 4h). These observations indicated that the TRIM71-associated PLC components were modified with K48-linked poly-Ub chains, which were diminished upon LINK-A knockdown. We developed modification-specific antibodies targeting ubiquitinated- (referred to as ub-) TPSN (Lys213), ub-TAP1 (Lys537), ub-TAP2 (Lys245), and ub-CALR (Lys48) (Supplementary Fig. 4i).
To validate these observations, we observed that the protein levels of TPSN, TAP1, TAP2, and CALR were downregulated in MMTV-Tg(LINK-A) tumors, but not in MMTV-PyVT tumors or normal mammary gland tissues (Fig. 6a). We further confirmed that in human breast cancer tissues (Duke Cohort, Supplementary Table 1), the expression of LINK-A negatively correlated with the protein levels of TPSN and CALR (Supplementary Fig. 5a). In TNBC resistant to anti-PD-1 blockage, ub-TPSN, -TAP1, -TAP2, and -CALR were all significantly increased compared to anti-PD-1 sensitive TNBC patients (Supplementary Fig. 5b).
Figure 6: LINK-A facilitates TRIM71-mediated ubiquitination and degradation of peptide-loading complex.
a, Immunoblotting detection using indicated antibodies in mammary glands of female MMTV-cre, breast tumors of MMTV-PyVT or MMTV-Tg(LINK-A) mice. Four animals per experimental conditions were examined. Three independent experiments were performed and yielded similar results. b and c, Representative images of H&E and immunohistochemistry staining using indicated antibodies in normal ducts of Tg(LINK-A) mammary gland (1st column) or normal-like, hyperplasic, atypical hyperplasic, DCIS or IDC of MMTV-Tg(LINK-A) mammary gland (2nd-6th column) (b) and statistical analysis of TPSN (c, top panel) or CALR (c, bottom panel) staining intensities per ductal cells per visual field. Six random fields per animal and eight animals per experimental condition were calculated (n.s., p>0.05, **p<0.01, ***p<0.001). Results are mean ± s.d. of n=8, 8, 8, 8, 8, 8 animals per experimental condition, P values were determined by one-way ANOVA. Scale bars: 40 μm. d, Immunoblotting detection using indicated antibodies in lysates extracted from MMTV-Tg(LINK-A) tumors with scramble or LINK-A LNA treatment (5 mg/kg, SubQ, every other day). 5 animals per experimental conditions were examined. Three independent experiments were performed and yielded similar results. e, Immunoblotting detection using indicated antibodies in lysates extracted from MMTV-Tg(LINK-A) tumors treated with vehicle or Rauwolscine (10 mg/kg, SubQ, daily). 5 animals per experimental conditions were examined. Three independent experiments were performed and yielded similar results. f, Flow cytometry detection of H-2Kb of B16F10 cells expressing Tet-on LINK-A vector with or without Doxycycline treatment followed by vehicle or IFN-γ stimulation (***p<0.001). Results are mean ± s.d. of n=3 independent experiments, P values were determined by one-way ANOVA. g, Flow cytometry detection of H-2Kb of B16F10 cells expressing blank vectors or Trim71 followed by vehicle or IFN-λ stimulation (***p<0.001). Results are mean ± s.d. of n=3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. h, Flow cytometry detection of OVA peptides of B16-OVA cells with knockdown of Trim71 (**p=0.0013). Results are mean ± s.d. of n=3 independent experiments. P values were determined by unpaired two-tailed Student’s t test.
To demonstrate that expression of LINK-A downregulates PLC components during tumor initiation, we took advantage of the mammary ductal transformation process of the normal-like, hyperplasia, DCIS, and IDC morphologies of MMTV-Tg(LINK-A) mice. Compared to the normal ducts of Tg(LINK-A) mice (Fig. 6b, first column), although the ducts of MMTV-Tg(LINK-A) mouse mammary glands at 8 weeks of age were morphologically similar to normal ducts, the protein status of TPSN and CALR were significantly downregulated in the epithelial cells of these normal-like ducts (Figs. 6b, second column and c), suggesting that the PLC complex is likely to be downregulated in mammary gland epithelial cells without substantial lymphocyte infiltration or accelerated cell division. In hyperplasic ducts, the protein status of TPSN and CALR was further downregulated compared to normal-like ducts, while the atypical hyperplasic ducts, DCIS, and IDCs all exhibited similar low-PLC statuses (Figs. 6b–c and Supplementary Figs. 5c–d). Hence, it is highly possible that one of the mechanisms by which malignant cells loss antigenicity during tumor initiation is through LINK-A-dependent downregulation of antigen presenting machinery.
Proteasome-mediated protein degradation occurs in the cytosol42; it is highly likely that the newly translated MHC1 complex and PLC components are subject to proteasome-medicated protein degradation before the assembly on the endoplasmic reticulum (ER) membrane. We addressed the hypothesis that the expression of LINK-A facilities the interactions between TRIM71 and PLC components, which could be reduced upon LINK-A knockdown using Duolink® proximity ligation assay (PLA) signals (Supplementary Fig. 5e). Robust PLA signals were detected in MDA-MB-231 cells harboring scramble LNAs, suggesting protein proximity between TRIM71:TAP1, TRIM71: CALR, TRIM71:TAP2, and TRIM71:TPSN, respectively, which were significantly reduced (Supplementary Fig. 5f). The majority of PLA signals did not overlap with ER markers, suggesting that a portion of PLC components could be subjected to TRIM71-mediated poly-ubiquitination and protein degradation on the exterior of the ER, which is consistent with previous literature suggesting that a portion of the MHC I complex overlaps with the ER marker43.
Tumor-bearing MMTV-Tg(LINK-A) mice treated with LINK-A LNAs showed restored phosphorylation of PKA (Thr197) and p-TRIM71 (Ser3) and elevated protein levels of TPSN, TAP1, TAP2, and CALR without affecting the total protein levels of PKA and TRIM71 (Fig. 6d and Supplementary Figs. 6a–b). To determine the poly-Ub types of the ubiquitinated-TPSN, TAP1, TAP2, and CALR, we performed denaturing immunoprecipitation (denaturing IP) to immunoprecipitate similar amounts of TPSN, TAP1, TAP2, and CALR proteins in MMTV-Tg(LINK-A) tumors. Upon scramble LNAs treatment, the immunoprecipitated proteins exhibited K48-linked poly-Ub chains, which were diminished upon LINK-A LNAs treatment (Supplementary Fig. 6b). Treatment with Rauwolscine efficiently restored the protein levels of the PLC components, as well as inhibited the K48-linked poly-Ub of these proteins without affecting LINK-A expression in these tumors (Fig. 6e and Supplementary Figs. 6c–d).
The PLC complex is vital for the stabilization and cellular presentation of the MHC I complex. To determine the functional role of LINK-A and the TRIM71-dependent molecular mechanism in antigen presentation, we determined the cellular surface MHC I complex of the B16F10 cells with Tet-on induced-expression of LINK-A, finding that expression of LINK-A suppressed the cellular surface expression of H-2Kb with or without IFN-γ stimulation (Fig. 6f and Supplementary Fig. 7a). Similarly, expression of exogenous TRIM71 led to reduced cell surface expression of H-2Kb (Supplementary Figs. 7b–c). Furthermore, Trim71 knockdown blocked the LINK-A-dependent cellular surface suppression of H-2Kb in B16F10 cells (Supplementary Fig. 7d). Exogenous expression of LINK-A in low LINK-A-expressing MCF7 cells led to reduced cell surface expression of B2M and HLA-ABC (Supplementary Figs. 7e–g); on the contrary, knockdown of LINK-A in high LINK-A-expressing MDA-MB-231 cells resulted in increased cell surface expression of B2M and HLA-ABC (Supplementary Figs. 7h–j). Treatment with Rauwolscine significantly improved the cellular surface expression of MHC1 and B2M (Fig. 6g and Supplementary Fig. 7k). Using B16-OVA cells44, we observed that knockdown of Trim71 increases cellular surface expression of the chicken OVA peptide (Fig. 6h). Hence, LINK-A downregulates PLC components and impairs antigen presentation, leading to tumor immunosurveillance evasion.
LINK-A LNAs suppresses tumor initiation and progression
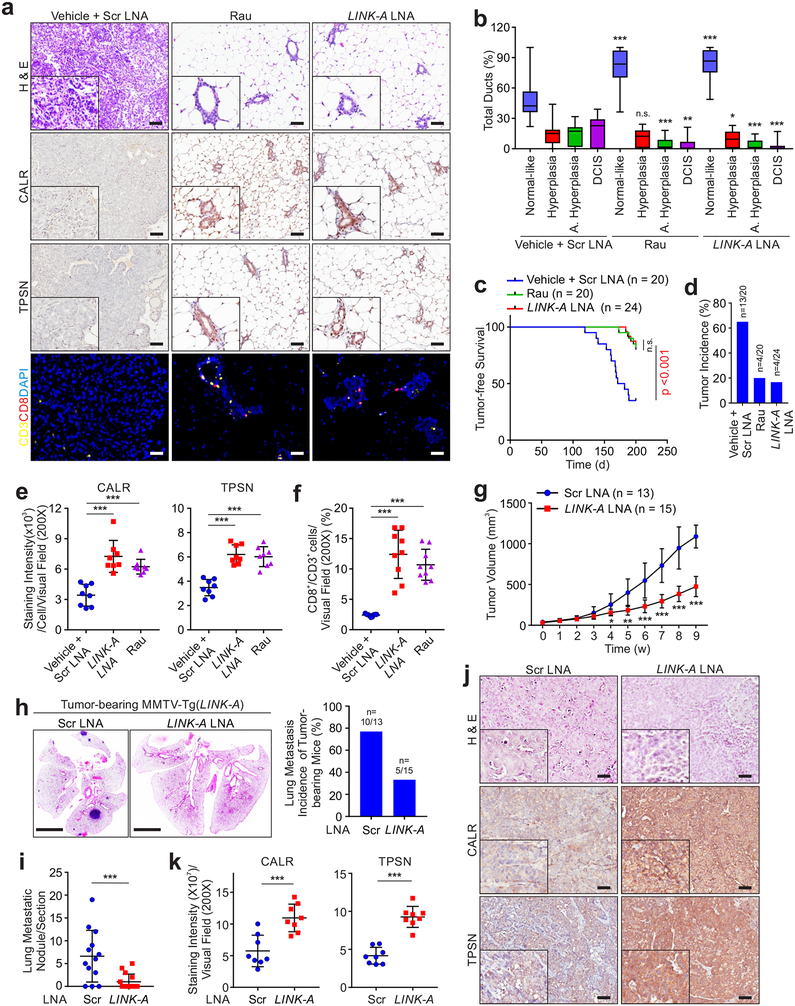
Administration of LINK-A or Rauwolscine exhibited minimal effects on the body weight, liver functions, and renal functions of the mice (Supplementary Figs. 8a–f). As a prevention trial, 12-week old female MMTV-Tg(LINK-A) mice without palpable mammary gland tumors were treated with LINK-A LNAs or Rauwolscine for a 16-week period of time. Mice given the vehicle treatment exhibited hyperplasia and atypical hyperplasia throughout their mammary glands with the presence of DCIS and IDCs (Figs. 7a–b). The ducts of mice subjected to LINK-A LNAs or Rauwolscine treatment exhibited a low degree of atypical hyperplasia and DCIS with more than 80% normal-like ducts (Figs. 7a–b). The LINK-A LNAs- or Rauwolscine-treated mice also exhibited reduced tumor incidence (Figs. 7c–d). LINK-A LNAs- or Rauwolscine-treated mammary glands exhibited restored protein statuses of CALR and TPSN in ductal epithelial cells and increased CD8+ T cell infiltration compared to the vehicle-treated group (Figs. 7a,e,f).
Figure 7: Targeting LINK-A prevents mammary gland tumor initiation and progression.
a, H&E staining, immunohistochemistry staining of CALR, TPSN and fluorescent multiplex immuno-histochemistry labeling of CD3 and CD8 in MMTV-Tg(LINK-A) mammary gland treated with indicated treatments. Six random fields per animal and twenty animals or twenty-four per experimental condition were analyzed. Scale bar: 50 μm. b, Percentage of normal-like, hyperplasic, atypical hyperplasic ducts and DCIS of MMTV-Tg(LINK-A) mammary gland with indicated treatments (n.s., p>0.05, *p<0.05, **p<0.01, ***p<0.001). Results are mean ± s.d. of n=20, 20, 24 animals. P values were determined by unpaired two-tailed Student’s t test. c and d, Kaplan-Meier survival analysis and tumor incidence of MMTV-Tg(LINK-A) mice with indicated treatments (n=20, 20, 24 animals) (n.s., p>0.05, ***p<0.001). e and f, Statistical analysis of staining intensities of CALR (e, left panel), TPSN (e, right panel), or percentage of CD8+ cells of CD3+ cells per visual field (f) of MMTV-Tg(LINK-A) mammary gland with indicated treatments. n=8, 8, 8 (e) or 8, 9, 9 (f) animals (***p<0.001). Results are mean ± s.d., P values were determined by one-way ANOVA. g, Tumor volume of MMTV-Tg(LINK-A) mice treated with scramble or LINK-A LNAs (n=13, 15 animals) (*p<0.05, **p<0.01, ***p<0.001). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test. h and i, Representative images of lung (h, left), lung metastasis incidence of tumor-bearing mice (h, right) or metastatic nodules/section (i) of MMTV-Tg(LINK-A) tumor-bearing mice treated with scramble or LINK-A LNAs (n=13, 15 animals). Scale bar (h): 4mm. Error bar (i), (***p<0.001). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test. j and k, Representative images (j) and statistical analysis (k) of CALR (k, left), and TPSN (k, right) staining intensities of MMTV-Tg(LINK-A) mice treated with scramble or LINK-A LNAs (n=8, 8 animals). (j) Scale bar: 100 μm. (k) (***p<0.001). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test.
In our regression model, the tumor-bearing MMTV-Tg(LINK-A) mice treated with LINK-A LNAs exhibited inhibited tumor growth and reduced lung metastasis compared to those treated with scramble LNAs (Figs. 7g–i and Supplementary Fig. 8g). The LINK-A LNAs-treated tumors showed increased protein levels of CALR and TPSN (Figs. 7j,k).
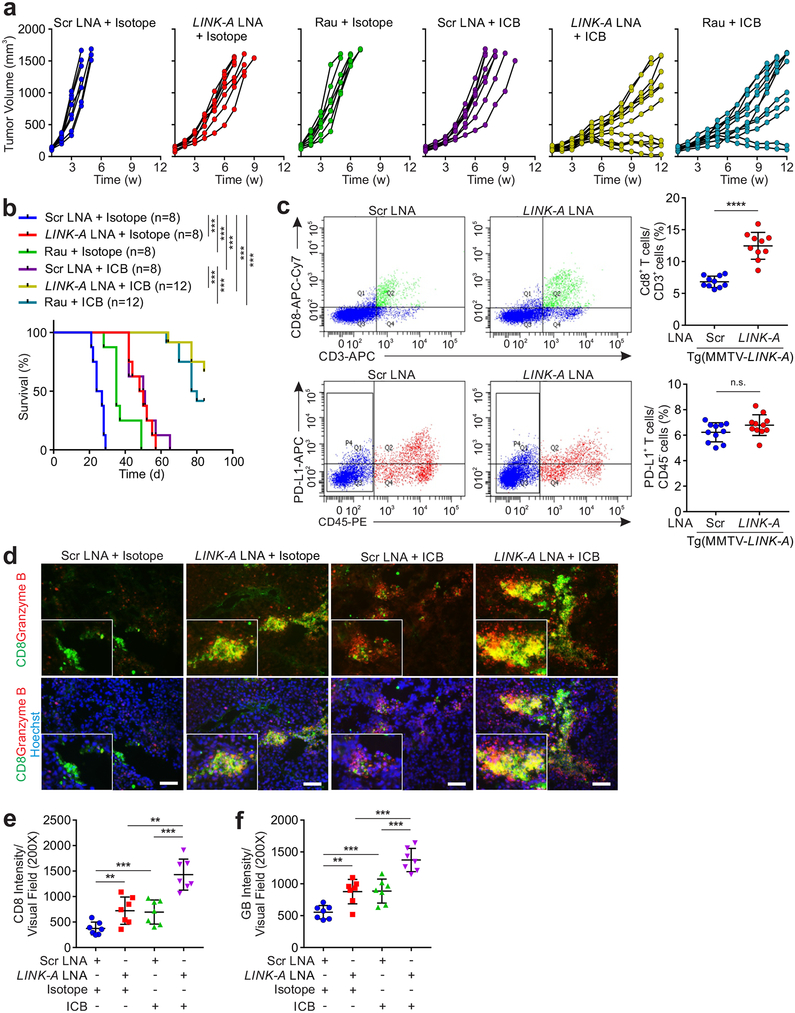
We then developed a syngeneic breast tumor model derived from MMTV-Tg(LINK-A) genetic mice using orthotopic transplantation of the mouse tumors to female FVB mammary glands. Treatment with LINK-A LNAs, Rauwolscine, or ICBs inhibited tumor growth in the syngeneic MMTV-Tg(LINK-A) tumors, which was synergistically suppressed by a combinatorial treatment of LINK-A LNAs/Rauwolscine and ICBs (Figs. 8a,b). The combined treatment significantly extended the survival time of the tumor-bearing mice (Fig. 8b). We then determined the anti-tumor effect of small molecular inhibitors against CNR2, GABR1, ADA2A, ACM4, and OPRM using JTE907 (CNR2 inhibitor), CPG54626 (GABR1 inhibitor), Rauwolscine (ADA2A inhibitor), Tropicamide (ACM4 inhibitor), and Cyprodime (OPRM inhibitor), respectively, finding that individual administration of these inhibitors significantly inhibited the growth of MMTV-Tg(LINK-A) syngeneic tumors (Supplementary Fig. 8h).
Figure 8: Targeting LINK-A sensitizes mammary gland tumor to immunotherapy.
a, Tumor volumes of syngeneic MMTV-Tg(LINK-A) mice treated with LINK-A LNA, Rauwolscine (Rau), ICB alone or in combination (n=8, 8, 8, 8, 12, 12 animals). b, Kaplan-Meier survival analysis of syngeneic MMTV-Tg(LINK-A) mice treated with LINK-A LNA, Rauwolscine (Rau), immune checkpoint blocks (ICB) alone or in combination (n=8, 8, 8, 8, 12, 12 animals), log rank test. c, Flow cytometry detection of CD3+/CD8+ cells (top panel) or CD45−/PD-L1+ cells (bottom panels) isolated from MMTV-Tg(LINK-A) tumors with scramble or LINK-A LNA treatment (5 mg/kg, SubQ, every other day, total of 7 doses) (n=10, 10 animals, top or 11,11 animals bottom) (****p<0.0001). Results are mean ± s.d.. P values were determined by unpaired two-tailed Student’s t test. d and f, Representative images (d) and statistical analysis of CD8 (e) and Granzyme B (GB) (f) of syngeneic MMTV-Tg(LINK-A) mice treated with LINK-A LNA, ICB alone or in combination (n=7, 7, 7, 7 animals). (d) Scale bars; 100μm. (e and f) (**p<0.01, ***p<0.001). Results are mean ± s.d., P values were determined by one-way ANOVA.
We then determined the status of tumor-resident CD8+ T cells in MMTV-Tg(LINK-A) tumors, finding that the LNAs treatment significantly improved CD8+/CD3+ T cell infiltration and cytotoxicity but exhibited minimal effects on the expression of PD-L1 in these tumors (Figs. 8c–f and Supplementary Fig. 8i). The LINK-A LNAs treatment showed minimal effects on the infiltration of CD8+ T cells, macrophages, and MDSCs in non-tumor-bearing mammary glands (Supplementary Figs. 8j–o). Taken together, we demonstrated a lncRNA-mediated molecular mechanism that modulates intrinsic tumor suppressors and antigen presentation in breast tumor initiation and progression (Supplementary Fig. 8p). Therefore, LINK-A may serve as a valuable biomarker for predicting the outcome of TNBC patients requiring immunotherapy, and targeting LINK-A further sensitizes breast tumors to immune checkpoint inhibitors.
Discussion
The battle between the immune system and malignant cells is constantly evolving, and cancer cells develop a variety of mechanisms to escape immunoediting. Although genetic evidence has indicated that malignant cells with restored antigenicity are subject to anti-tumor immunity, whether cancer cells passively or actively downregulate antigenicity remains elusive. We demonstrated that during breast cancer initiation, the transformed mammary gland epithelial cells downregulate antigen presentation machinery upon expression of LINK-A, illustrating one of the initial and important mechanisms through which cancer cells escape from immune checkpoints. We reasoned that mammary gland epithelial cells expressing LINK-A may exhibit antigenicity loss, which contributes to the survival and expansion of malignant cells. The peptide-loading complex plays vital roles in antigen presentation and transportation of MHC I. Post-translational modifications of PLC components in cancer cells may serve as an advantageous mechanism for downregulating antigenicity without losing achieved genomic mutations and mutation-derived growth advantages. Treatment with LINK-A LNAs or GPCR antagonists like Rauwolscine in vivo significantly improved the protein stability of the PLC components and MHC I complex, leading to sensitization of mammary gland tumors to immunotherapy. Most importantly, the enhanced CD8+ T cell infiltration was specific to tumor tissues; the LINK-A LNAs treatment did not affect the distribution of CD8+ T cells, macrophages, or MDSCs in normal mammary glands. Hence, our results suggested promising therapeutic strategies for improving antigen presentation and the efficacy of immunotherapy, which could be synergistic with immune checkpoint inhibitors.
Expression of LINK-A mediated the crosstalk between inhibitory GPCRs and PtdIns(3,4,5)P3, leading to inactivation of the cAMP/PKA pathway. The LINK-A-dependent suppression of PKA-dependent phosphorylation of TRIM71 resulted in the K48-linked polyubiquitination and protein degradation of the intrinsic tumor suppressors Rb and p53 and the components of peptide-loading complex. TRIM71 has been shown to modulate p53 degradation45. It is likely that LINK-A/TRIM71-dependent p53 ubiquitination is independent of MDM246, which is consistent with the notion that the major ubiquitination residues of p53 mediated by MDM2 are located within the tetramerization domain of p53 (Lys320, Lys321, Lys351, Lys357, Lys370, Lys372, Lys373)47, whereas LINK-A-dependent, TRIM71-mediated p53 poly-ubiquitination occurs at Lys126, which is within the DNA-binding domain of p53.
Despite the genome-wide identification of noncoding RNAs in human diseases, limited genetic evidence has demonstrated the biological importance of lncRNAs in cancer initiation and progression. Tissue-specific expression of LINK-A in mouse mammary glands led to mammary gland carcinogenesis, implicating it as an oncogene. Genome-wide analysis indicated that MMTV-Tg(LINK-A) tumors represent human TNBC genetically, transcriptionally, and metabolically. Hence, the MMTV-Tg(LINK-A) mouse model provides a valuable tool for studying the underlying molecular mechanisms of TNBC. The molecular mechanisms of LINK-A-driven breast tumors are mediated through multiple signaling pathways: we previously demonstrated that LINK-A expression mediates non-canonical HIF1α signaling and hyper-activation of the AKT pathway14, 15. With genetic evidence, our results suggested that LINK-A inactivates tumor suppressor pathways and downregulates antigen presentation through inactivation of PKA pathways, which is consistent with the previous notion that PKA knockout leads to carcinogenesis. While the total protein level of TRIM71 exhibited minimal changes under our experimental conditions, phosphorylated TRIM71 faithfully correlated with the outcome of TNBC patients treated with immunotherapy, suggesting that the LINK-A/PKA/TRIM71 signaling axis likewise correlates with patient outcomes. These results implicated the potential for these molecules to serve as biomarkers for predicting the outcome of cancer patients treated with immune checkpoint inhibitors.
Methods
In vivo murine models and treatment procedures
All animal-based research was conducted according to the guidelines and requirements set forth by the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals, the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act of 1966 as amended by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas M.D. Anderson Cancer Center (MDACC). Applied StemCell Inc.’s proprietary TARGATT™ transgenic mouse technology was utilized to generate the LINK-A transgenic mouse model. Briefly, the generated pBT378-PGK-L4StrL-LINK-A vector contains the transgene for ɸC31 integrase-mediated recombination. A mixture of the construct and ɸC31 integrase mRNA was microinjected into the pronucleus of each of 80 zygotes in the FVB genetic background using TARGATTTM Technology and the injected zygotes were implanted into four CD1 foster mice. Twenty-three mice were born from the microinjection. Successful integration from the founder mice was identified by PCR analyses of genomic DNA using primers targeting LINK-A. After screening, two male positive founders (Tg-LINK-A mice) were identified. All mice had a FVB genetic background. Tg-LINK-A mice were crossed with MMTV-cre mice [Tg(MMTV-cre)1Mam, The Jackson Laboratory] to produce mice with LINK-A transgene expression in the mammary glands. MMTV-Tg(LINK-A) and Tg(LINK-A) female mice were used as experimental and control mice, respectively. All of the mice genotyping primer sequences can be found in Supplementary Table 5.
For the prevention treatment, MMTV-Tg(LINK-A) mice started the LINK-A LNAs (5 mg/kg, SubQ, every other day) or Rauwolscine (5 mg/kg, IP, daily) treatment from 12 weeks of age, and the treatment was terminated at 28 weeks of age. After treatment, mice mammary gland tissues were collected for morphology and immunohistochemistry analysis. For the regression treatment, MMTV-Tg(LINK-A) mice bearing mammary tumors up to 150 mm3 were randomly assigned to treatment groups and injected with the following drugs: scramble LNAs or LINK-A LNAs (5 mg/kg, SubQ, every other day). Tumors were measured three times per week, and mice were euthanized once the ethical end point was reached [tumor volume of 1500 mm3, as determined by measuring the minimum and maximum tumor diameters using the following formula: [(minimum diameter)2(maximum diameter)/2)]. Mice mammary glands and lungs were collected for morphology and immunohistochemistry analysis.
For establishment of the syngeneic MMTV-Tg(LINK-A) model, MMTV-Tg(LINK-A) mice bearing mammary tumors up to 600 mm3 were euthanized, and the tumors were excised. Tumors were dissociated as a single cell using the gentle MACS Dissociator (Miltenui Biotec Inc) with a mouse Tumor Dissociation kit (Miltenui Biotec). A single-cell suspension was generated after filtration through a 70-μm cell strainer (BD Falcon). Single-cell suspensions were used for primary culture or transplant injection. Cells (40,000 per gland) were injected into the right inguinal fat pad of 4-week-old female FVB/N recipients. Three weeks after transplantation, when mammary tumors had reached a size of about 150 mm3, mice were randomly assigned to treatment groups and injected with combinations of the following drugs: scramble or LINK-A LNA (5 mg/kg, SubQ, every other day, QIAGEN), Rauwolscine hydrochloride (5 mg/kg, IP, daily, TOCRIS), JTE 907 (5 mg/kg, IP, daily, TOCRIS), CGP 54626 hydrochloride (5 mg/kg, IP, daily, TOCRIS), Tropicamide (5 mg/kg, IP, daily, TOCRIS), Cyprodime hydrochloride (5 mg/kg, IP, daily, TOCRIS), and anti-PD-1 (BioXcell, clone RMP1–14) and anti-CTLA-4 (BioXcell, clone UC10–4F10–11) as immune checkpoint blockers (ICB) 100 μg every 72 h, or an isotype control antibody (BioXcell, clone LTF-2) 200 μg every 72 h. Tumors were measured three times per week, and mice were euthanized once the ethical end point was reached. After treatment, mice tumors were collected for morphology, flow cytometry, and immunohistochemistry analysis.
Tissue samples
TNBC tissues from patients that responded or did not respond to Pembrolizumab (10 mg/kg, q2w, total of 3–4 months) were purchased from Boston Bioscource Inc. Fresh frozen breast carcinomas and their adjacent normal tissues were obtained from Duke University. The study protocol was approved by the Institutional Review Board of Duke University Health System. All tissue samples were collected in compliance with informed consent policy. Clinical information is summarized in Supplementary Table 1.
Immunohistochemistry, immunofluorescence and Duolink® proximity ligation assay
For multiplex immunohistochemistry staining, FFPE human tissues cut at a thickness of 5 μm were prepared, and the staining was conducted using a PD-L1, CD3ε, and CD8α Multiplex IHC Antibody Panel (Cell signaling technology; #65713) according to vendor’s instruction and imaged with a confocal microscope (Zeiss). Duolink® proximity ligation assays were performed following the manufacturer’s instructions (Sigma), using antibodies targeting TRIM71, TAP1, TAP2, CALR, and TPSN. Briefly, cells on round-cover glass slips were fixed in 4% PFA at 25 °C for 15 min after PBS washing. Cells were permeabilized in 0.5% Triton X-100 for 10 min and then treated in accordance with the duo-link assay kit instructions. Antibodies information is summarized in Supplementary Table 6. The confocal microscope (LSM700; Carl Zeiss) was used for image analysis. The number of PLA signals per cell was calculated. Immunofluorescence and immunohistochemistry were performed as previously described (Xing, 2014, cell). Antibody information is summarized in Supplementary Table 6.
Cell lines, transfection, treatments and cellular assays.
Human TNBC cell lines: MDA-MB-231, MDA-MB-468, BT549, HCC-1187; human HER2-positive breast cancer cell lines: BT474, SK-BR-3; human ER-positive breast cancer cell lines: MCF7, T47D, ZR-75–1; human normal mammary gland epithelial cell line: MCF10A and mouse mammary gland epithelial cell line: NMuMG were purchased from American Type Culture Collection (ATCC); SUM-149 (human TNBC cell line), B16-OVA (mouse melanoma cell line, a gift from H. Patrick), and B16F10 (mouse melanoma cell line) were maintained using standard media and conditions and the Characterized Cell Line Core Facility (MD Anderson Cancer Center). Lincode SMARTpool siRNA targeting Trim71 (636931) was used in this study, and the LNAs targeting LINK-A and the scrambled sequence were designed and synthesized by Exiqon. SiRNA, LNA, and plasmid transfections were performed using DharmaFECT4 (Dharmacon) and Lipofectamine 3000 (Life Technologies), respectively. B16F10 was constructed to stably express LINK-A by selection with G418 (1500 μg ml−1). To induce LINK-A overexpression, 50 ng/ml doxycycline (Sigma) was added to the culture medium at the indicated time. For the IFN-γ treatment, cells were treated with mIFN-γ (10 ng/ml). Human target PKAα cat, PKAβ cat, CNR2, ADA2A, GABR1, ACM4, and OPRM-specific sgRNA sequences are listed in Supplementary Table 5. MDA-MB-231 and MCF10A cells were constructed to stably express Cas9 and sgRNAs by selection with puromycin (1 μg ml−1). Single clones were obtained by serial dilution. LINK-A PtdIns(3,4,5)P3-binding motif-deficient cell lines were generated using the CRISPR/Cas9 genome editing system by the Gene Editing/Cellular Model Core Facility (MD Anderson Cancer Center). For pharmacologic inhibition, cells or tumor spheroids were treated with Abemaciclib (500 nM, Selleckchem), Erlotinib (10μM, Selleckchem), and Rauwolscine hydrochloride (10μM, TOCRIS) for the indicated times and at the indicated concentrations. Spheroid growth and invasion of indicated cells were conducted using a Cultrex® 3-D Spheroid Fluorometric Proliferation/Viability Assay kit (Trevigen) according to vendor’s instruction. For IC50 determination, a serial 2-fold dilution of the indicated compound starting at 0.4 mM to 0.05 nM was incubated with appropriated cells or spheroids, as indicated. IC50 values were derived by a Log (Rauwolscine) vs. response-Variable slope (four parameters) model for a competitive inhibition curve using GraphPad Prism 7 software. Total cellular cAMP was quantified using a cAMP Assay Kit (Competitive ELISA) (ab65355, Abcam) according to the manufacturer’s protocol.
Plasmid construction, protein recombination, and purification
Mammalian expression vectors for full-length LINK-A and the deletion mutant were constructed by subcloning the gene sequences into a pCDNA3.1 (+) backbone and pInducer20 inducible lentiviral expression vector (Life Technologies). Mammalian expression of full-length TRIM71, PKA C-α, CNR2, GABR1, ADA2A, ACM4, OPRM, and mutant vectors were constructed by subcloning the corresponding gene sequences into the His-tagged expression vector (pcDNA™-DEST40) using the Gateway system (Life Technologies). Bacteria expression of full-length CNR2, GABR1, ADA2A, ACM4, OPRM, and mutant vectors was constructed by subcloning the corresponding gene sequences into the GST-tagged expression vector pGEX-5X-1. All single-point and deletion mutations were generated using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Recombinant proteins were expressed in the Escherichia coli strain BL21-CodonPlus (DE3)-RIPL (Agilent Technologies) and purified using the Protein Purification Kit (Clontech).
Immunoprecipitation and immunoblotting
Cells, human tumor samples, mouse normal mammary gland tissues, and mouse breast tumor samples were homogenized in 1× RIPA buffer (EMD Millipore) supplemented with Protease/ Phosphatase Inhibitor Cocktail (Pierce, Thermo Scientific), Panobinostat (Selleck chemicals), and Methylstat (Sigma-Aldrich). Lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4 °C. Supernatants were analyzed for immunoblotting or immunoprecipitation with the indicated antibodies, and the immunoprecipitated proteins were subjected to either immunoblotting or protein identification by mass spectrometry. For the denaturing immunoprecipitation, tissues were lysed using TSD buffer (50 mM Tris-HCl, pH 7.5, 1% SDS, 5 mM DTT) boiled for 10 min. The cleared lysates were used for immunoprecipitation. The elutions were loaded on NuPAGE 4%–12% Bis-Tris Gel (GenScript) and then analyzed for immunoblotting with the indicated antibodies summarized in Supplementary Table 6.
RNA biology assays
Total RNA isolation and qRT-PCR were performed as previously described. (Xing, 2014, cell). Total RNAs in mouse normal mammary glands and mouse breast cancer samples were analyzed for LINK-A and Actb expression using biotin-labeled LNA probes (Exiqon) according to the instructions for the NorthernMax Kit (Ambion). Detection of LINK-A expression using RNAScope® probe (designed by Advanced Cell Diagnostics) and image quantification were performed as previously described using a RNAScope® 2.5 High Definition Assay kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). For DNA-FISH, single-cell suspensions from freshly harvested Tg(LINK-A) mammary gland epithelial cells or MMTV-Tg(LINK-A) mammary tumor cells were used to hybridize probes targeting full-length LINK-A using a FISH Tag DNA Kit according to the manufacturer’s instructions (Thermo Scientific, F32947). The RNA copy number was measured as previously described (Qingsong Hu, 2019, Cell Research). All of the primer and probe sequences are listed in the Supplementary Table 5.
Alpha assay
Alpha assays were performed in accordance with the manufacturer’s instructions (PerkinElmer)48. The Kd of the interaction between biotin-labeled phosphatidylinositol-phosphates and GST-tagged GPCRs recombinant proteins was determined in the Alpha indirect format using a competition experiment in which untagged GPCRs recombinant proteins were titrated from 0.4 mM to 0.05 nM. More specifically, triplicate samples containing the indicated GPCRs and phosphatidylinositol-phosphates at the indicated concentrations diluted in protein-lipid binding buffer (25 mM Tris-Cl, 150 mM NaCl, 0.1% Tween-20, 1% non-fat milk, 2 mM Ca2+ chloride, 1 mM Zn2+ sulfate) were transferred at a volume of 10 μl to each well of a 96-well assay plate then incubated at 25 °C for 1 h. 10 μl of Streptavidin AlphaLISA® Acceptor beads (100 μg/ml) were added to each well. The plate was placed on an orbital shaker for 10 min then incubated at 25 °C for 1 h. Following incubation, 10 μl of Alpha Glutathione Donor beads (100 μg/ml) were added to each well and incubated 30 min at 25 °C. The plate was read on the EnSpire Multimode Plate Reader (PerkinElmer) (wavelength: 615nm). The competitive inhibition curves were generated based on Alpha signal readings by fitting to a nonlinear regression “saturation binding” model and a “log (inhibitor) vs. response-Variable slope (four parameters)” model, respectively (GraphPad Prism 7 software).
Flow cytometry
Cell lines: 1 × 106 cells per condition were stained with the appropriate antibodies diluted in DPBS (Corning) plus 2% FBS (Gibco) for 30 min at 25 °C. Matched fluorescence minus one (FMO) staining for each condition was performed as a control. Mouse tissues and tumors: mouse tissues and tumors were dissociated as a single cell using the gentleMACS Dissociator (Miltenui Biotec Inc) with the mouse Tumor Dissociation kit (Miltenui Biotec). After lysis of red blood cells (RBC Lysis Buffer, BioLegend), single-cell suspensions were blocked with anti-CD16/32 (BioLegend) for 20 min on ice and then incubated with the appropriate antibodies for 30 min at 25 °C. Mouse antibodies: antibodies were purchased from BioLegend unless otherwise indicated: CD45, CD3, CD8, H-2Kb, H-2Kb bound to SINFEKL, β2-microglobulin, PD-L1, F4/80, CD11b, and Ly6G/Ly6C. Human antibodies: β2-microglobulin and HLA-A, -B, -C. To distinguish live/dead cells, Zombie Violet (BioLegend) fixable viability dyes were used. Flow cytometry was performed on an LSRII (BD Biosciences), and the data were analyzed using FlowJo (TreeStar). Antibodies information is summarized in Supplementary Table 6.
Mass spectrometry
To identify PtdIns (3,4,5)P3, PKA, or TRIM71-binding proteins, PtdIns(3,4,5)P3 pulldown or immunoprecipitation using the indicated antibodies was performed as previously described49. Briefly, Tg(LINK-A) mouse normal mammary gland tissues, MMTV-PyVT tumors, and MMTV-Tg(LINK-A) tumor tissue lysates were freshly prepared using a ProteaPrep Zwitterionic Cell Lysis Kit, Mass Spec Grade (Protea Biosciences) with an Anti-RNase, Protease/Phosphatase Inhibitor Cocktail supplemented in the lysis buffer. The eluted lncRNA-protein complexes were denatured, reduced, alkylated, and digested with immobilized trypsin (Promega) for MS analysis at the MD Anderson Cancer Center Proteomics Facility. To identify TRIM71-binding proteins, endogenous TRIM71 was immunoprecipitated followed by mass spectrometry analysis. Tumor tissue lysates of MMTV-Tg(LINK-A) tumors treated with scramble or LINK-A LNAs (5 mg/kg, SubQ, every other day, total of 7 doses) were freshly prepared using the ProteaPrep Zwitterionic Cell Lysis Kit, Mass Spec Grade (Protea) with an Anti-RNase, Protease/Phosphatase Inhibitor Cocktail supplemented in the lysis buffer. Immunoprecipitation was performed using a Dynabeads® Co-Immunoprecipitation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The eluted protein complexes were denatured, reduced, alkylated, and digested with immobilized trypsin (Promega) for MS analysis at the MD Anderson Cancer Center Proteomics Facility.
Metabolomics measurements
To measure the expression level of metabolites in Tg(LINK-A) mouse normal mammary gland tissues and MMTV-Tg(LINK-A) tumor tissues, Tg(LINK-A) mouse normal mammary gland tissues and MMTV-Tg(LINK-A) tumor tissues were freshly prepared for mass spectrometry. All metabolites were analyzed by the Baylor College of Medicine Proteomics Facility.
TCGA and RNA-seq analysis
Normalized gene expression data of 20 cancer types based on RSEM50 was downloaded from the TCGA data portal (http://gdac.broadinstitute.org/), and the normalized lncRNA expression based on Reads Per Kilobase Million (RPKM) data was downloaded from The Atlas of Noncoding RNAs in Cancer51 (TANRIC; http://bioinformatics.mdanderson.org/main/TANRIC). RNA sequencing was performed by Illumina Hiseq 2000 with a 75-bp paired-end read. All reads were aligned to the mouse reference genome (mm10) using hisat252 with the default setting. Stringtie53 was used to calculate the transcriptional expression level as fragments per kilobase per million (FPKM). Differential gene expression was defined if the fold change > 1.5 and p < 0.05 between tumor and normal samples.
TCGA data analysis and tumor-infiltration analysis
Normalized gene expression data of 20 cancer types based on RSEM was downloaded from TCGA data portal (http://gdac.broadinstitute.org/), and normalized lncRNA expression based on Reads Per Kilobase Million (RPKM) was download from The Atlas of Noncoding RNAs in Cancer (TANRIC; http://bioinformatics.mdanderson.org/main/TANRIC). We obtained gene sets of 28 subpopulations of tumor-infiltrating lymphocytes (TILs) from a previous study16, including cell types related to adaptive immunity (activated, central memory, effector memory CD4+ and CD8+ T cells, gamma delta T cells, T helper 1 (Th1) cells, Th2 cells, Th17 cells, regulatory T cells, follicular helper T cells, activated, immature, and memory B cells), as well as cell types related to innate immunity (macrophages, monocytes, mast cells, eosinophils, neutrophils, activated, plasmacytoid, and immature dendritic cells, natural killer cells, natural killer T cells, and Myeloid-derived suppressor cell). We used the gene set variance analysis (GSVA) program to calculate the absolute enrichment score of gene signatures for immune cells in each sample, which referred as relative immune cell abundance. Then, we calculated Pearson correlation between LINK-A expression level and GSVA score for each type of immune cell across cancer types or across different subtypes of breast cancer, considered FDR < 0.05 as statistical significance.
Mutation calling
Whole exome sequencing reads were aligned to the mouse reference (GRCm38_68) using Burrows-Wheeler Aligner (BWA 0.7.17)54. BAM files were processed using the Genome Analysis Toolkit55 to improve alignment accuracy. We identified somatic point mutations through four popular callers, including VarScan256, MuTect257, MuSE58, and SomaticSniper58, and only reported mutations called by at least two callers. To further reduce false positives and miscalled germline events, we removed any mutations called by MuTect2 in at least two normal samples59. We obtained TCGA triple-negative breast cancer (TNBC) mutations from http://gdac.broadinstitute.org/ as previously described60. Whole exome sequencing data was deposited to the NCBI Sequence Read Archive, with the ID PRJNA453620.
Statistics and reproducibility
The experiment was set up to use 3–8 samples/repeats per experiment/group/condition to detect a 2-fold difference with a power of 80% and a significance level of 0.05 using a two-sided test for significance studies. Each of these experiments was independently repeated 3 times. Analyses of relative gene expression were determined using the 2-ΔΔCt method with GAPDH as the internal reference gene. Results are reported as mean ± standard error of the mean (s.e.m.) or standard deviation (s.d) of at least three independent experiments, as indicated by the figure legends. Each exact n value is indicated in the corresponding figure legend. Statistical analysis was performed using GraphPad Prism 7 software. Comparisons were analyzed using unpaired Student’s t-test or one-way ANOVA test (n.s., p>0.05, *p<0.05, **p<0.01, and ***p<0.001), as indicated in the individual figures. Fisher’s exact test was implemented for statistical analyses of the correlation between markers and clinical parameters. Kaplan-Meier survival curves were compared using the log rank test.
Data availability
The breast cancer RNA-seq data used to analyze LINK-A expression were derived from the TCGA Research Network: http://cancergenome.nih.gov/, and the breast cancer RNA-seq BAM files were downloaded from the UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Source data for all human tissue experiments have been provided as Supplementary Table 1. Supplementary Tables 5,6 provide information about the oligonucleotides and antibodies used in this study, respectively. The raw RNA-seq data for this manuscript are available at GEO under the accession number (GSE113143). Whole exome sequencing data was deposited to the NCBI Sequence Read Archive, with the ID (PRJNA453620). All other data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
The Proteomics and Metabolomics Facility was supported in part by Cancer Prevention Research Institute of Texas (CPRIT) grant number RP130397 and NIH grant number 1S10OD012304–01 (D. H. Hawke). This project is partially supported by University of Houston Seq-N-Edit Core with funding from UH Division of Research; UH College of NSM and Department of Biology & Biochemistry; NRUF MINOR CORE 17 Grant to P. H. Gunaratne.; UH Small Core Equipment Program Grant to P. H. Gunaratne. We thank the core facilities at BCM: Metabolomics Core, (NIH P30CA125123), CPRIT Proteomics and Metabolomics Core Facility (N. Putluri), (RP170005), and Dan L. Duncan Cancer Center. This work was supported by R01CA216426, R01CA220297, U01CA214263, American cancer society 127430-RSG-15-105-01-CNE to N. Putluri. This project was partially supported by the NIH T32 Training Grant in Cancer Biology (5T32CA186892) (L.–C. Chan). This project was also supported by the grant of Cancer Prevention & Research Institute of Texas RR150085 (L. Han). This work was supported by NIH R01 award (1R01CA218025–01 and 1R01CA231011–01), CPRIT individual investigator research award (RP150094 and RP180259) and Department of Defense Breakthrough award (BC180196) to C. Lin. This work was supported by NIH R01 award (1R01CA218036–01), CPRIT First-time Faculty Recruitment Award (R1218), Department of Defense Breakthrough award (BC151465), The American Association for Cancer Research-Bayer Innovation and Discovery Grant (18-80-44) and Andrew Sabin Family Foundation Fellows Award to L. Yang.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Dent R et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research 13, 4429–4434 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE & Reis-Filho JS Triple-negative breast cancer. The New England journal of medicine 363, 1938–1948 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Chen L & Han X Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. The Journal of clinical investigation 125, 3384–3391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia H et al. Immunotherapy for triple-negative breast cancer: Existing challenges and exciting prospects. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy 32, 1–15 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Burnet FM The concept of immunological surveillance. Prog Exp Tumor Res 13, 1–27 (1970). [DOI] [PubMed] [Google Scholar]
- 6.Leach DR, Krummel MF & Allison JP Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P & Yamaguchi T Regulatory T cells: how do they suppress immune responses? Int Immunol 21, 1105–1111 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Johnsen AK, Templeton DJ, Sy M & Harding CV Deficiency of transporter for antigen presentation (TAP) in tumor cells allows evasion of immune surveillance and increases tumorigenesis. Journal of immunology 163, 4224–4231 (1999). [PubMed] [Google Scholar]
- 9.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR & Chinnaiyan AM The emergence of lncRNAs in cancer biology. Cancer Discov 1, 391–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C & Yang L Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends in cell biology 28, 287–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wapinski O & Chang HY Long noncoding RNAs and human disease. Trends in cell biology 21, 354–361 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Lin A et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nature cell biology 18, 213–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin A et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nature cell biology 19, 238–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charoentong P et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell reports 18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Muller WJ, Sinn E, Pattengale PK, Wallace R & Leder P Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105–115 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Nielsen LL, Discafani CM, Gurnani M & Tyler RD Histopathology of salivary and mammary gland tumors in transgenic mice expressing a human Ha-ras oncogene. Cancer research 51, 3762–3767 (1991). [PubMed] [Google Scholar]
- 19.Stewart TA, Pattengale PK & Leder P Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38, 627–637 (1984). [DOI] [PubMed] [Google Scholar]
- 20.Hippenmeyer S et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Network TCGA Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah SP et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaan YM et al. Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genomics Proteomics 11, 279–294 (2014). [PubMed] [Google Scholar]
- 24.Czech MP PIP2 and PIP3: complex roles at the cell surface. Cell 100, 603–606 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Felices M, Falk M, Kosaka Y & Berg LJ Tec kinases in T cell and mast cell signaling. Adv Immunol 93, 145–184 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Jackson TR et al. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. The Journal of cell biology 151, 627–638 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wettschureck N & Offermanns S Mammalian G proteins and their cell type specific functions. Physiological reviews 85, 1159–1204 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Birnbaumer L Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 alpha subunits plus betagamma dimers. Biochimica et biophysica acta 1768, 772–793 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg RA, Cauthron RD, Symcox MM & Shuntoh H Autoactivation of catalytic (C alpha) subunit of cyclic AMP-dependent protein kinase by phosphorylation of threonine 197. Molecular and cellular biology 13, 2332–2341 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chini B & Parenti M G-protein coupled receptors in lipid rafts and caveolae: how, when and why do they go there? Journal of molecular endocrinology 32, 325–338 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Perry BD & U’Prichard DC [3H]rauwolscine (alpha-yohimbine): a specific antagonist radioligand for brain alpha 2-adrenergic receptors. European journal of pharmacology 76, 461–464 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Normanno N et al. The MEK/MAPK pathway is involved in the resistance of breast cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol 207, 420–427 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Iglesias-Bartolome R et al. Inactivation of a Galpha(s)-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nature cell biology 17, 793–803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattabiraman DR et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herschkowitz JI et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome biology 8, R76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loedige I, Gaidatzis D, Sack R, Meister G & Filipowicz W The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic acids research 41, 518–532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer GH, Moore MJ & Taylor SS Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. The Journal of biological chemistry 280, 8800–8807 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Nandi D, Tahiliani P, Kumar A & Chandu D The ubiquitin-proteasome system. J Biosci 31, 137–155 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Pietilainen T et al. Expression of retinoblastoma gene protein (Rb) in breast cancer as related to established prognostic factors and survival. Eur J Cancer 31A, 329–333 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Lacroix M, Toillon RA & Leclercq G p53 and breast cancer, an update. Endocrine-related cancer 13, 293–325 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Hulpke S & Tampe R The MHC I loading complex: a multitasking machinery in adaptive immunity. Trends Biochem Sci 38, 412–420 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Arrigo AP, Tanaka K, Goldberg AL & Welch WJ Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature 331, 192–194 (1988). [DOI] [PubMed] [Google Scholar]
- 43.Harriff MJ et al. Endosomal MR1 Trafficking Plays a Key Role in Presentation of Mycobacterium tuberculosis Ligands to MAIT Cells. PLoS Pathog 12, e1005524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vianello F et al. Murine B16 melanomas expressing high levels of the chemokine stromal-derived factor-1/CXCL12 induce tumor-specific T cell chemorepulsion and escape from immune control. Journal of immunology 176, 2902–2914 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Nguyen DTT et al. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ 24, 1063–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda R, Tanaka H & Yasuda H Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS letters 420, 25–27 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Brooks CL & Gu W p53 regulation by ubiquitin. FEBS letters 585, 2803–2809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
- 48.Yasgar A, Jadhav A, Simeonov A & Coussens NP AlphaScreen-Based Assays: Ultra-High-Throughput Screening for Small-Molecule Inhibitors of Challenging Enzymes and Protein-Protein Interactions. Methods Mol Biol 1439, 77–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xing Z et al. lncRNA Directs Cooperative Epigenetic Regulation Downstream of Chemokine Signals. Cell 159, 1110–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu C et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nature methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pertea M et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan Y et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome biology 17, 178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng J et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun 8, 1533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Y et al. The Genomic Landscape and Pharmacogenomic Interactions of Clock Genes in Cancer Chronotherapy. Cell Syst 6, 314–328 e312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The breast cancer RNA-seq data used to analyze LINK-A expression were derived from the TCGA Research Network: http://cancergenome.nih.gov/, and the breast cancer RNA-seq BAM files were downloaded from the UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Source data for all human tissue experiments have been provided as Supplementary Table 1. Supplementary Tables 5,6 provide information about the oligonucleotides and antibodies used in this study, respectively. The raw RNA-seq data for this manuscript are available at GEO under the accession number (GSE113143). Whole exome sequencing data was deposited to the NCBI Sequence Read Archive, with the ID (PRJNA453620). All other data are available from the corresponding author upon reasonable request.