Abstract
The consolidation of long-term memories requires differential gene expression. Recent research has suggested that dynamic changes in chromatin structure play a role in regulating the gene expression program linked to memory formation. The contribution of histone methylation, an important regulatory mechanism of chromatin plasticity that is mediated by the counteracting activity of histone-methyltransferases and histone-demethylases, is, however, not well understood. Here we show that mice lacking the histone-methyltransferase myeloid/lymphoid or mixed-lineage leukemia 2 (mll2/kmt2b) gene in adult forebrain excitatory neurons display impaired hippocampus-dependent memory function. Consistent with the role of KMT2B in gene-activation DNA microarray analysis revealed that 152 genes were downregulated in the hippocampal dentate gyrus region of mice lacking kmt2b. Downregulated plasticity genes showed a specific deficit in histone 3 lysine 4 di- and trimethylation, while histone 3 lysine 4 monomethylation was not affected. Our data demonstrates that KMT2B mediates hippocampal histone 3 lysine 4 di- and trimethylation and is a critical player for memory formation.
Introduction
The consolidation of long-term memories critically involves differential gene expression and de novo protein synthesis (Kandel, 2001). Epigenetic mechanisms that mediate chromatin plasticity are key processes in the regulation of gene expression and especially post-translational histone modifications such as histone acetylation have been implicated with the transcriptional regulation of memory consolidation (Alarcón et al., 2004; Levenson et al., 2004; Chwang et al., 2006, 2007; Fischer et al., 2007; Vecsey et al., 2007; Lubin et al., 2008; Guan et al., 2009; Peleg et al., 2010). Histones undergo different types of chemical modifications at their N-terminal tails including acetylation and methylation (Strahl and Allis, 2000; Vaquero et al., 2003). Histone acetylation is mainly associated with active gene expression (Kurdistani et al., 2004; Li et al., 2007), whereas histone methylation can activate or repress gene expression depending on the residue it occurs (Shi and Whetstine, 2007; Shilatifard, 2008). For example, histone (H) 3 lysine 9 methylation inhibits transcription (Nakayama et al., 2001), while H3 lysine 4 methylation (H3K4me) is associated with transcriptional activation (Santos-Rosa et al., 2002). Although the role of histone acetylation in memory formation is well established, the contribution of histone methylation is less well understood.
Histone methylation is regulated in a specific manner by the activity of histone methyltransferases (HMTs) and histone demethylases and occurs in monomethylated, dimethylated, and trimethylated states (Santos-Rosa et al., 2002; Schneider et al., 2005), with each reaction being often catalyzed by a specific enzyme (Shi and Whetstine, 2007).
A potential role for histone methylation in memory function has been suggested by recent studies (Schaefer et al., 2009; Gupta et al., 2010; Gupta-Agarwal et al., 2012) and mutations in the H3K4me3-specific histone demethylase, JARID1C, have been identified in patients with mental retardation and autism (Adegbola et al., 2008). Indeed, patients suffering from autism show altered H4K3me3 in prefrontal cortex neurons (Shulha et al., 2012). The only enzyme mediating H3K4 methylation in yeast is SET1 (Roguev et al., 2001). Mammalian cells possess six Set1-related homologs that are capable of performing H3K4 methylation—KMT2A (MLL1), KMT2B (MLL2), KMT2C (MLL3), KMT2D (MLL4), Setd1a, Setd1b (Shilatifard, 2008; Austenaa et al., 2012).
In this study we investigate for the first time the role of Kmt2b in memory formation. Kmt2b, also known as Mll2, Mll4, or Wbp7, is highly expressed throughout development and also in adult tissue (Glaser et al., 2006). kmt2b knock-out in mice before E11.5 leads to embryonic lethality, whereas mice in which Kmt2b is deleted after that time point are viable and do not display any overt phenotype (Glaser et al., 2009). We used the Cre-lox system to delete kmt2b specifically from excitatory forebrain neurons of adult mice and found that animals lacking kmt2b exhibit impaired memory function while brain morphology remains intact. Microarray analysis revealed that loss of kmt2b leads to the downregulation of genes implicated with neuronal plasticity specifically in dorsal dentate gyrus. Downregulation of these genes was linked to reduced H3K4 di- and trimethylation in the corresponding promoter regions. In conclusion our data reveals a novel role for kmt2b in memory function and suggests a specific role of H3K4 di- and trimethylation in the regulation of plasticity genes in the dorsal dentate gyrus of the hippocampal formation.
Materials and Methods
Animals
Mice were housed in standard cages with food and water ad libitum. For experiments—which were performed according to the animal protection law and the District Government of Germany—3-month-old mice were used that were all of C57B/6J background. The generation of mice that lack kmt2b in the adult forebrain was achieved by crossing mice in which the exon 2 was flanked by loxP sites with camkii-cre mice as described previously (Minichiello et al., 1999; Glaser et al., 2006; Kuczera et al., 2011) (kmt2b cKO). In all the experiments, mice having the second exon “floxed” but not expressing the Cre transgene were used as controls and are referred to as “control.”
Behavioral analysis
Mice were habituated to the testing room for 1 week before the onset of behavioral experiments
Novel object recognition.
For the novel object recognition test mice were habituated to an empty plastic arena for 5 min for 2 consecutive days. For the next 2 d, mice were presented with two white boxes and were left to explore them for 5 min. On the training day the mice were introduced to two black cubes and left to explore them for 5 min. After that they were put back into the home cage. After 24 h the mice were reexposed to the plastic arena with one of the black cubes being exchanged to a red tape. Again they were allowed to explore the objects for 5 min. Relative preference to the novel object (i.e., small stone or red tape) over the old one (black cube) was used as an index for memory strength. For testing short-term memory the experiment was performed as described but after exposure to two similar objects mice were reexposed to the arena with a novel object already 5 min later. In contrast to the other behavioral tests, only male mice were analyzed in the novel object paradigm, since in our hands even female wild-type mice failed to perform this task.
Pavlovian fear conditioning.
Fear conditioning test was performed using TSE fear conditioning system. Mice were allowed to explore the context for 3 min after which they received a mild electric foot shock (constant current, 0.5 mA) for 2 s. The next day (i.e., 24 h later) they were introduced to the same context for 3 min without receiving a footshock and freezing behavior, indicative of associative memory, was analyzed.
Morris water maze.
The training was performed in a circular pool (diameter 1.2 m) filled with opaque water. A platform (11 × 11 cm) was submerged into the water in the center of one of the quadrants (target quadrant). The swimming behavior of the mice was recorded by a camera and was analyzed by VideoMot2 (TSE). At each training session the mice were placed into the maze subsequently from four random points and were allowed to swim (i.e., search for the platform) for 60 s. If during the 60 s the mouse failed to find the platform it was gently guided to it. Mice were allowed to stay on the platform for 15 s. Mice were subjected to the probe test 24 h after the last training session. In the probe test the platform was removed and the mice were placed into the maze and allowed to swim for 60 s.
Injection of viral particles
For the injection of virus particles (10E8 transducing units, Synapsin CRE AAV; University of Pennsylvania, Philadelphia, PA) mice were anesthetized and particles were bilaterally injected to the dorsal dentate gyrus as described before (Bahari-Javan et al., 2012) using a glass capillary (anteroposterior −1.70 mm relative to bregma; lateral ±1 mm; dorsoventral 2 mm from skull).
Immunostaining
Immunohistochemistry was performed as described previously (Fischer et al., 2007; Govindarajan et al., 2011) and was analyzed at a Leica SP2 confocal microscope. Anti-NeuN antibody was from Millipore Bioscience Research Reagents, anti-MAP2 antibody was from Synaptic Systems and anti-Synaptophysin antibody was from Sigma-Aldrich. Alexa Fluor 488- and Cy3-conjugated fluorescent secondary antibodies were purchased from Jackson ImmunoResearch.
Quantitative real time PCR
The dissection of dentate gyrus and CA regions was performed under a stereomicroscope (Motic) as described previously (Hagihara et al., 2009). The RNA from dorsal dentate gyrus and dorsal CA was isolated using TRIzol Reagent from Invitrogen. Quantitative reverse-transcriptase PCR (qPCR) was performed using a Roche 480 Light Cycler. cDNA was synthesized using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche). qRT-PCR using Roche Universal Probe Library (UPL) probes was performed to validate the results from DNA microarray and to detect Mll3 and Mill expression (Table 1). To confirm the loss of exon 2 in F/F CKII mice through recombination qRT-PCR using SYBR Green was performed. The results were normalized to the housekeeping gene hypoxanthine phosphoribosyltransferase (Hprt1).
Table 1.
Primers for qRT-PCR confirmation of DNA microarray results
| Primer | Sequence (5′-3′) | Tm° | UPL probe |
|---|---|---|---|
| Nkapl left | cacacctctcaagatgagaaacc | 63.9 | 38 |
| Nkapl right | agccattgctgcaccttc | 64.1 | 38 |
| Rab38 left | ccaaaacttctcctcgcact | 63.3 | 49 |
| Rab38 right | tcatgtttccaaatctttcttgac | 62.9 | 49 |
| Ap1s3 left | ggaccagcagcttcattgac | 64.8 | 17 |
| Ap1s3 right | tgcacagcaaaaatataaactagca | 63.3 | 17 |
| Acot4 left | atgcttcgacatccaaaggt | 63.4 | 17 |
| Acot4 right | ggaagccatgatcagacagac | 63.7 | 17 |
| Dusp2 left | gaagataaccagatggtggagataa | 63.0 | 79 |
| Dusp2 right | cccactattcttcaccgagtctat | 63.3 | 79 |
| Adcy5 left | cgggagaaccagcaacag | 64.6 | 18 |
| Adcy5 right | ctccatggcaacatgacg | 64.1 | 18 |
| Ptgr1 left | gactgagctcccacccttaaa | 64.3 | 18 |
| Ptgr1 right | gtaaggatccacagagaggaaca | 63.3 | 18 |
| Prkra left | gcgagcaaggctttaacataa | 63.0 | 17 |
| Prkra right | agacactgatactgtccgttgg | 63.0 | 17 |
| Gabrg3 left | ggctcactggatcaccaca | 65.1 | 17 |
| Gabrg3 right | ggcactctgcattgatagtgag | 63.7 | 17 |
| Car4 left | aaaccaaggatcctagaagcagt | 63.0 | 1 |
| Car4 right | gacaatgttgatgggggact | 63.6 | 1 |
| Ckap4 left | ggaggaggtccagcaggt | 64.5 | 7 |
| Ckap4 right | ttgcagggattggacctt | 63.1 | 7 |
| Stxbp2 left | tcttgtcatcctgttgtaaaatgtc | 62.9 | 9 |
| Stxbp2 right | ccgtttgttgatgtcttcca | 63.6 | 9 |
| Sypl2 left | tctatggggctggctaacct | 64.6 | 99 |
| Sypl2 right | cagcccacaggaagaagttg | 64.8 | 99 |
| Gkap1 left | cagaaggagtcacgggaaga | 64.4 | 94 |
| Gkap1 right | ttcaaacatttcagaggtcagc | 63.1 | 94 |
| Tpm4 left | cgaccgcaagtatgaggag | 63.4 | 108 |
| Tpm4 right | tcagatacctccgccctct | 63.8 | 108 |
| E2f1 left | tgccaagaagtccaagaatca | 64.6 | 5 |
| E2f1 right | cttcaagccgcttaccaatc | 63.5 | 5 |
DNA microarray
DNA microarray was performed as described previously (Peleg et al., 2010; Agis-Balboa et al., 2011).
Chromatin immunoprecipitation
Dorsal dentate gyri from two mice were pooled in one tube for chromatin immunoprecipitation (ChIP) analysis. Chromatin immunoprecipitation was performed using the Low Cell ChIP Kit from Diagenode. First, the beads were equilibrated in ChIP buffer and then left for 2 h incubating with the antibodies at 4°C. Antibody (4 μl; all antibodies used for ChIP analysis were from Millipore) were used in each experiment. Tissue was lysed in Dulbecco's PBS (Pan Biotech) containing protease inhibitor (Roche). Fixation of cells was performed with 37% Formaldehyde and was stopped by the addition of 1.25 m Glycine. After centrifugation and washing steps, the pellet was dissolved in Buffer B containing protease inhibitor (Diagenode) and the samples were subjected to sonication in Bioruptor (25 cycles; 30 s ON/OFF; 1 × 10 min, 3 × 5 min). After that 870 μl of ChIP buffer containing protease inhibitor (Diagenode) were added to the sheared chromatin. Diluted sheared chromatin (100 μl) was added to the antibody-conjugated beads and left for incubation at 4°C overnight. After that the beads were washed in ChIP buffer and once in Buffer C, and then were resuspended in 100 μl of DNA Isolation Buffer containing 1 μl of Proteinase K (DIB + Prot K). Moreover, 1 μl of sheared chromatin was dissolved in 99 μl of DIB + Prot K to be used as input DNA. The samples were first incubated at 55°C and then at 100°C, and finally centrifuged at 14,000 rpm for 5 min at 4°C. The supernatant was used for qRT-PCR analysis (Table 2).
Table 2.
Primers for ChIP–qPCR
| Primer | Sequence (5′- 3′) | Tm° |
|---|---|---|
| Acot4 reverse | ccacgtggtgttgtgaaagt | 63.6 |
| Adcy5 forward | gaggctctgttcgcctttc | 64.0 |
| Adcy5 reverse | cctgccagcattattttctt | 60.7 |
| Ap1s3 forward | gcgcaggtgtaagcactg | 63.4 |
| Ap1s3 reverse | tcctggcttcctcaaattgt | 63.5 |
| Car4 forward | catcttgccccaaatcaagt | 63.8 |
| Car4 reverse | cagggcttagaagcggagta | 63.3 |
| Ckap4 forward | ttcaaagcttttgcgaggat | 63.6 |
| Ckap4 reverse | ctctccacagctccagttc | 64.1 |
| Dusp2 forward | gacctcttggctaatcataccc | 63.8 |
| Dusp2 reverse | tgcacaatgaagacaacaaattc | 64.5 |
| E2f1 forward | ggctctggctacgaaagaaa | 63.3 |
| E2f1 reverse | cttcaggctcacctccaaag | 63.9 |
| Gabrg3 forward | cgtgttaattggggaaactcc | 64.1 |
| Gabrg3 reverse | gctctcgggagcgatcag | 66.5 |
| Gkap1 forward | agttttaaaaatgttaatgccaatg | 60.8 |
| Gkap1 reverse | gggttggaggacagaggag | 63.8 |
| Nkapl forward | gctcaaggtggggaatgtaa | 63.8 |
| Nkapl reverse | cgcaggcgcactagagac | 65.1 |
| Prkra forward | tgactactgcaggcgaaaga | 63.7 |
| Prkra reverse | ggcattgtcctacgtcacaa | 63.6 |
| Ptgr1 forward | ggcctcacgacaggaagtag | 63.8 |
| Ptgr1 reverse | gaggggtgtgtgtgtgtgtt | 63.6 |
| Rab38 forward | cagcttgacaggcagtagca | 64.4 |
| Rab38 reverse | ccttcctactctgcggactc | 62.9 |
| Stxbp2 forward | gtccgttcgagtccctgtc | 64.9 |
| Stxbp2 reverse | cgtggctctacgcgtcat | 65.2 |
| Sypl2 forward | gttctagtggggacccatcc | 64.4 |
| Sypl2 reverse | gtcaaacaatctggcggact | 64.0 |
| Tpm4 forward | aaaggctcccaggtaagtgc | 64.3 |
| Tpm4 reverse | ctgtcaggcccatgaggt | 64.0 |
Statistical analysis
Data were analyzed by unpaired Student's t test and two-way ANOVA when appropriate. Errors are displayed as SEM. Graphs were generated using GraphPad Prism.
Results
The expression of the full-length transcript of Kmt2b is decreased in the forebrain of Kmt2b cKO mice
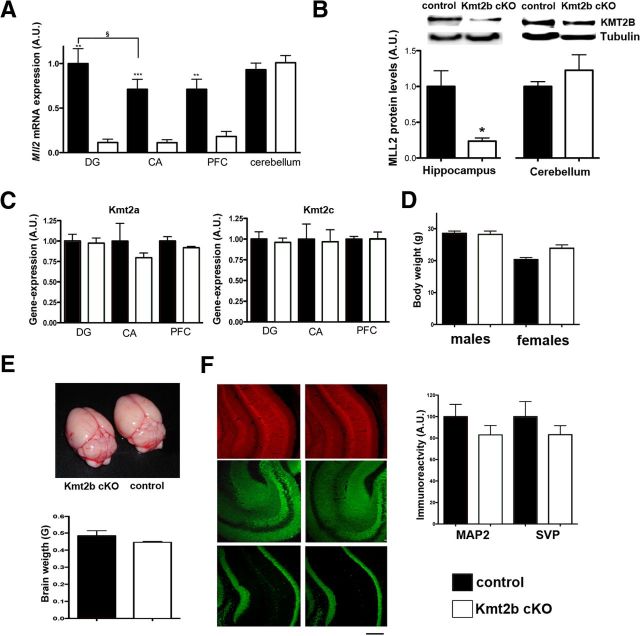
Mice lacking kmt2b from excitatory neurons of the adult forebrain were generated by crossing animal in which exon 2 of the Kmt2b gene were flanked by loxP sites (Glaser et al., 2006) with CamKII-Cre mice (kmt2b cKO mice). As described previously (Kuczera et al., 2011), for comparison we used mice harboring the floxed kmt2b gene without expression Cre recombinase (control). qPCR analysis revealed a significant downregulation of the Kmt2b gene in the hippocampal dentate gyrus and CA regions as well as in the prefrontal cortex (Fig. 1A). As expected kmt2b expression in the cerebellum was unaffected in kmt2b cKO mice (Fig. 1A). It is also interesting to note that kmt2b expression in control mice was significantly higher in the dentate gyrus compared with the CA regions, which is in line with expression data available at the Allen brain atlas (http://www.brain-map.org). Consistent with the gene-expression data we detected a significant downregulation of KMT2B protein levels in the hippocampus but not in the cerebellum of 3-month-old kmt2b cKO mice (Fig. 1B). We did not observe changes in expression levels for related MLL proteins such as MLL1 and MLL3 (Fig. 1C). kmt2b cKO mice were viable and displayed normal home cage behavior. Body size was also comparable among kmt2b cKO and control mice, although female kmt2b cKO showed a slightly increased body mass (Fig. 1D). Brain weight was not significantly altered when kmt2b cKO and control mice were compared indicating that kmt2b did not cause obvious changes in brain morphology (Fig. 1E). In line with these data, staining for MAP2, synaptophysin and NeuN, 3 well established markers for synaptic integrity, did not reveal any significant differences between kmt2b cKO and control mice (Fig. 1F).
Figure 1.
Mice lacking KMT2B from excitatory forebrain neurons show no overt phenotype. A, Loss of Kmt2b mRNA in the hippocampus and prefrontal cortex of Kmt2b cKO mice was confirmed via qPCR analysis. n = 9/group; **p < 0.01, ***p < 0.001, §p < 0.05. B, Immunoblot analysis shows that KMT2B protein levels are reduced in the hippocampus of kmt2b cKO mice (n = 4/group; *p < 0.05). C, qPCR analysis of kmt2a (mll1) and kmt2c (mll3) revealed no changes among kmt2b cKO and control mice (n = 5/group). D, Body weight of Kmt2b cKO and control mice was similar (n = 10/group). E, Brain weight of kmt2b cKO and control mice was similar (n = 8/group). F, Immunoreactivity for neuronal and synaptic marker proteins was similar in kmt2b cKO and control mice (n = 4/group). Left, Representative images. Scale bar, 300 μm. Right, Quantification (n = 4/group). DG, Dentate gyrus; CA, hippocampal CA region; PFC, prefrontal cortex; MAP2, microtubule-associated protein 2; SVP, synaptophysin; NeuN, Neuronal N. Error bars indicate SEM.
Loss of kmt2b in the forebrain leads to learning impairment
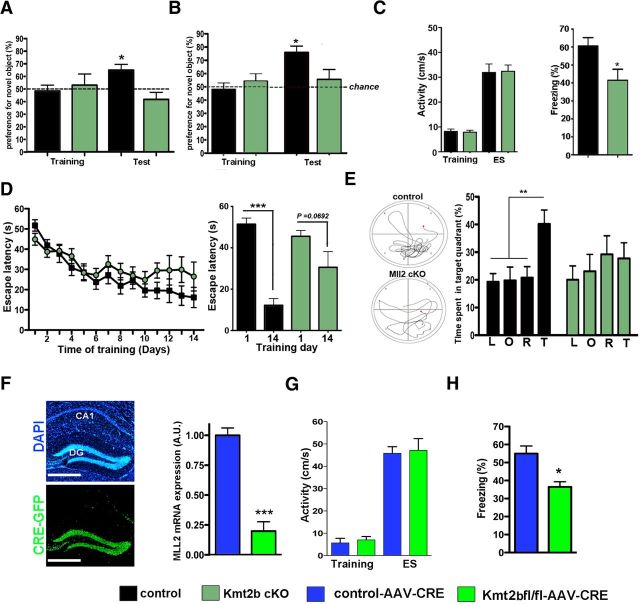
Next we studied cognitive function in kmt2b cKO and control mice. First, mice were subjected to novel object recognition learning to analyze short- and long-term memory. During the training session both groups of mice explored the two identical objects with a similar preference (Fig. 2A). During the memory test performed 5 min later to test for short-term memory the control group exhibited an increased preference for the novel object indicating intact short-term memory (Fig. 2A). kmt2b cKO mice did not show a significant preference for the novel object indicating impaired short-term memory in this paradigm (Fig. 2A, p < 0.05 for kmt2b vs control group). Additional groups of mice were subjected to the long-term memory version of this task. Twenty-four hours after exposure to the two identical objects animals where confronted with one novel and one familiar object. While the control group showed an increased preference for the novel object indicating successful long-term memory consolidation, kmt2b cKO mice showed no preference for the novel object indicating a severe impairment of object recognition learning (Fig. 2B). We also tested associative learning using the contextual fear-conditioning paradigm that critically depends on intact hippocampal function. Mice were subjected to the conditioning context followed by exposure to an electric foot-shock (0.5 mA, 2 s). The activity during the training and the response to the foot shock was similar among groups. Freezing behavior, which is indicative for associative memory consolidation, was tested 24 h later by reexposing the animals to the conditioning context. kmt2b cKO mice showed significantly less freezing behavior compared with the control group (Fig. 2C), indicating that hippocampus-dependent associative memory formation is impaired in kmt2b cKO mice. Male and female mice were grouped together in this test since two-way ANOVA did not yield any significant sex or interaction effects (sex: p value = 0.9746; interaction: p value = 0.7094), To test hippocampal memory function more specifically we subjected animals to the Morris water maze paradigm that allows the measurement of hippocampus-dependent spatial learning. While both groups improved in the escape latency throughout the training procedure, kmt2b cKO mice performed significantly worse compared with control animals (Fig. 2D,E). When subjected to the probe test 24 h after the last day of training control mice showed a significant preference for the target quadrant. In contrast no target preference was detected in kmt2b cKO mice (Fig. 2F). Two-way ANOVA revealed no sex or interaction effects (sex: p value = 0.3576; interaction: p value = 0.9128). Although the CamKII-Cre mice used in our study to delete Kmt2b from the adult forebrain have been used previously to delete target gene expression only postnatally (Minichiello et al., 1999; Kuczera et al., 2011; Görlich et al., 2012), it could be argued that subtle developmental effects might contribute to the observed behavioral phenotypes. To address this issue we used a viral mediated approach to delete kmt2b in 4 month-old mice. Adeno-associated virus (AAV) particles that mediate synapsin-driven expression of CRE fused to GFP were injected into the dorsal dentate gyrus of kmt2bfl/fl mice. Wild-type mice injected with the same AAV were used as control. qPCR analysis revealed the deletion of KMT2B 2 weeks after AAV injection in kmt2bfl/fl mice (Fig. 2F). In line with the data obtained from kmt2b cKO mice (Fig. 2C), viral mediated deletion of kmt2b in the dentate gyrus resulted in significantly impaired fear conditioning (Fig. 2G,H). There were no sex differences or sex vs genotype effects (sex: p value = 0.2505; interaction: p value = 0.7373) among mice. In summary, these data strongly suggest that loss of kmt2b leads to impaired hippocampus-dependent memory consolidation.
Figure 2.
Mice lacking KMT2B shown impaired memory function. A, Mice were subjected to the novel object recognition paradigm. Wild-type mice (n = 12) and kmt2b cKO mice (n = 9) explored both objects similarly during the training session (left). When subjected to short-term memory test 5 min later, wild-type but not kmt2b cKO mice showed a significant preference for the novel object (*p < 0.05). B, When subjected to long-term memory version of the novel object recognition test both groups of mice showed a similar preference to the two identical objects during the training (left; control: n = 14, kmt2b cKO: n = 10). B, During the long-term memory test for object recognition control (n = 14) but not kmt2b cKO (n = 10) showed a preference for the novel object (right; ***p < 0.001). C, Left, Activity during the fear conditioning training and during the foot shock was similar among groups (Control: n = 20, kmt2b cKO: n = 15). Right, Freezing behavior was significantly impaired in kmt2b cKO mice when analyzed 24 h after the training (*p < 0.05). D, Escape latency during the water maze training was impaired in kmt2b cKO (n = 23) mice compared with the control group (n = 19) during the first and the last day of training (right). E, Left, Representative swim path during the probe test. Right, During the probe test only control (n = 19) but not kmt2b cKO (n = 23) mice showed a significant preference for the target quadrant (**p < 0.01). F, Four-month-old kmt2bfl/fl and wild-type mice were injected with AAV particles mediating CRE expression. Left, Representative images showing the expression of CRE-GFP in the dentate gyrus. The corresponding DAPI staining is shown for orientation. Scale bar, 500 μm. Right, Another group of mice was injected with the AAV-CRE virus (Wild-type: n = 6, Kmt2bfl/fl: n = 3) and dorsal dentate gyrus tissue was isolated 2 weeks later. qPCR analysis revealed the deletion of Kmt2b 2 weeks after injection in AAV-CRE-injected kmt2bfl/fl mice. G, Four-month-old kmt2bfl/fl and wild-type mice were injected with AAV-CRE and subjected to contextual fear conditioning 2 weeks after (n = 9/group). Deletion of kmt2b in the dentate gyrus of 4 month-old kmt2bfl/fl mice did not affect activity during the training and during the electric footshock (ES). H, Contextual freezing was impaired in kmt2bfl/fl mice compared with wild-type mice (*p < 0.05), indicative of disrupted memory formation. Error bars indicate SEM.
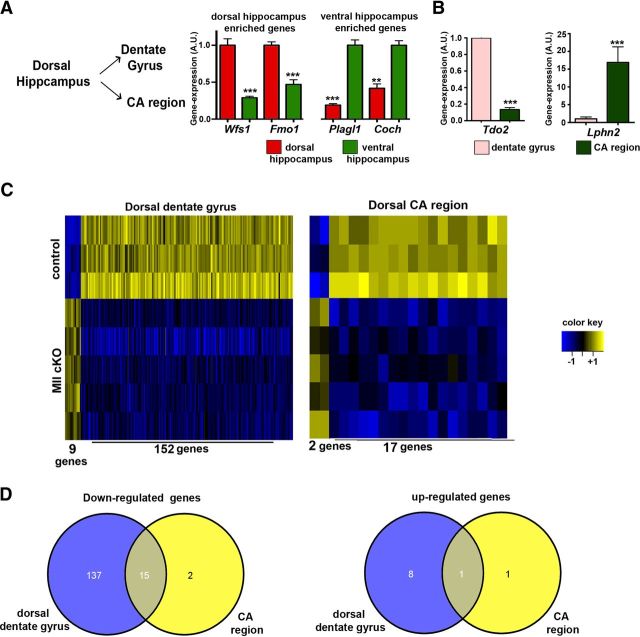
To elucidate the mechanisms by which kmt2b contributes to hippocampus-dependent memory formation we decided to analyze epigenetic gene expression in the hippocampal formation. Given that there exists a dorsoventral functional specification within hippocampus, with the dorsal hippocampus being especially responsible for spatial memory performance (Moser and Moser, 1998; Treves et al., 2008) we decided to focus our investigation on the dentate gyrus and CA regions of the dorsal hippocampus. First we confirmed the specificity of tissue isolation of the dorsal vs ventral hippocampus, as well as dentate gyrus vs CA regions via qPCR analysis of previously published marker genes specific to these regions (Thompson et al., 2008; Grayson et al., 2009; Ponomarev et al., 2010; Fig. 3A,B). Next we isolated RNA from the dorsal dentate gyrus and dorsal CA region from naive male Kmt2b cKO and control mice and performed a DNA microarray analysis.
Figure 3.
Loss of kmt2b leads to a deregulation of hippocampal transcriptome. A, RNA from the dorsal dentate gyrus and CA region was isolated. Specificity was confirmed by qPCR analysis for Wfs1 and Fmo1, two genes enriched in the dorsal hippocampus, and Plagl1 and Coch, two genes enriched in the ventral hippocampus (n = 4/group, **p < 0.01, ***p < 0.001). B, The dentate gyrus specificity was confirmed by qPCR analysis of the dentate gyrus-specific gene Tdo2 and the CA region-specific gene Lphn2 (n = 4/group; ***p < 0.001, **p < 001). C, Heat map showing differential gene expression among kmt2b cKO (n = 5) and control mice (n = 3) in the dorsal dentate gyrus and dorsal CA region. D, Venn diagram depicting downregulated and upregulated genes in the dorsal dentate gyrus and dorsal CA regions. Error bars indicate SEM.
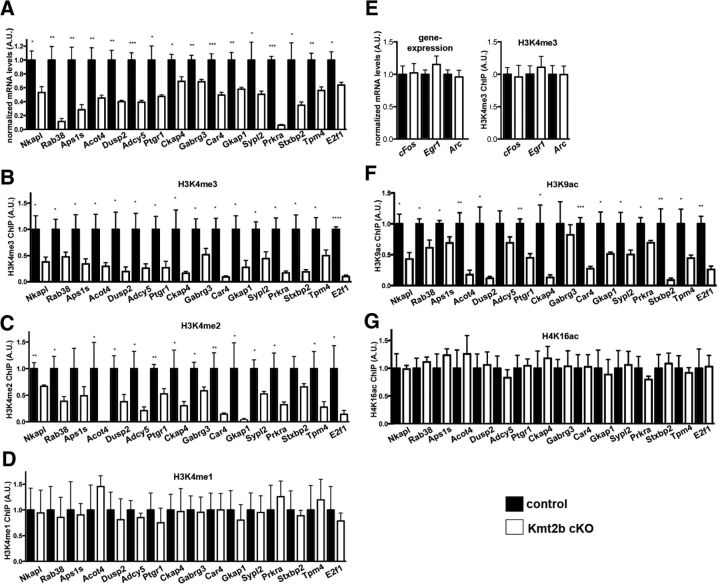
Data analysis revealed that 161 genes were differentially expressed in the dorsal dentate gyrus of kmt2b cKO mice compared with control mice (Fig. 3C, Table 3). In line with the fact that KMT2B mediates H3K4 methylation, which activates gene expression the majority of the differentially expressed genes (152) were significantly downregulated (fold change = 1.5; FDR = 0.05; Fig. 3C). In striking contrast to the data obtained for the dorsal dentate gyrus, only 22 genes (17 downregulated) were differentially expressed in the dorsal CA region when comparing kmt2b cKO and control mice (Fig. 3D, Table 4) suggesting a prominent role for kmt2b in the regulation of dentate gyrus-specific gene expression. Of152 genes downregulated in the dorsal dentate gyrus, many are linked to memory function and synaptic plasticity (Table 5) and we chose 16 of the most interesting genes for further analysis. We could confirm differential expression of all selected genes via qPCR analysis within the samples used for the gene array (data not shown) and within another independent experiment (Fig. 4A). Downregulation of selected genes was also confirmed in kmt2bfl/fl mice that were injected with AAV-CRE particles (data not shown), To further elucidate whether downregulation of these genes in kmt2b cKO mice is linked to altered H3K4 methylation we performed ChIP assays to measure H3K4 monomethylation (H3K4me1), dimethylation (H3K4me2), and trimethylation (H3K4me3) in the transcription start site regions of the selected genes. In all of the downregulated genes we observed significantly reduced H3K4me2 and H3K4me3 levels (Fig. 4B,C). In contrast, H3K4me1 levels did not change significantly among control and kmt2b cKO mice at any of the investigated genes (Fig. 4D). No changes in H3K4me3 were observed for selected genes that were unchanged in kmt2b cKO mice (Fig. 4E).
Table 3.
List of differentially regulated genes in dorsal dentate gyrus of kmt2b cKO mice (the genes in bold were confirmed by qRT-PCR in dorsal dentate gyrus and the genes in italic were confirmed in both dorsal dentate gyrus and dorsal CA regions)
| Gene symbol | Gene name | log2 (fold change) | Adjusted p value |
|---|---|---|---|
| Vsig8 | V-set and immunoglobulin domain containing 8 | −2.88 | 5.42 × 10−7 |
| Hfm1 | HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) | −2.62 | 1.32 × 10−7 |
| Efhc2 | EF-hand domain (C-terminal) containing 2 | −2.57 | 9.75 × 10−5 |
| Nkapl | NFKB activating protein-like | −2.52 | 1.04 × 10−5 |
| Dazl | deleted in azoospermia-like | −2.50 | 1.98 × 10−6 |
| Igsf1 | immunoglobulin superfamily, member 1 | −2.33 | 1.89 × 10−6 |
| Lor | loricrin | −2.33 | 5.94 × 10−11 |
| Hdx | highly divergent homeobox | −2.17 | 8.19 × 10−8 |
| Fbxl7 | F-box and leucine-rich repeat protein 7 | −2.13 | 1.02 × 10−5 |
| Wtip | WT1-interacting protein | −2.05 | 1.21 × 10−7 |
| Prss16 | protease, serine, 16 (thymus) | −2.02 | 2.40 × 10−4 |
| Zfp459 | zinc finger protein 459 | −1.96 | 6.67 × 10−6 |
| Wdr17 | WD repeat domain 17 | −1.92 | 1.63 × 10−8 |
| Rab38 | RAB38, member of RAS oncogene family | −1.90 | 1.31 × 10−4 |
| Tmem22 | transmembrane protein 22 | −1.89 | 3.44 × 10−7 |
| Lrrn4 | leucine rich repeat neuronal 4 | −1.89 | 4.97 × 10−6 |
| Prr15 | proline rich 15 | −1.89 | 3.49 × 10−6 |
| Ap1s3 | adaptor-related protein complex AP-1, sigma 3 | −1.84 | 5.45 × 10−7 |
| Gprc5c | G protein-coupled receptor, family C, group 5, member C | −1.81 | 8.53 × 10−5 |
| Nmnat3 | nicotinamide nucleotide adenylyltransferase 3 | −1.77 | 6.13 × 10−7 |
| Acot4 | acyl-CoA thioesterase 4 | −1.74 | 4.45 × 10−5 |
| Tceal1 | transcription elongation factor A (SII)-like 1 | −1.72 | 1.74 × 10−7 |
| Cd55 | CD55 antigen | −1.71 | 2.54 × 10−4 |
| BB557941 | expressed sequence BB557941 | −1.70 | 1.79 × 10−6 |
| Rimklb | ribosomal modification protein rimK-like family member B | −1.69 | 1.62 × 10−6 |
| Dusp2 | dual specificity phosphatase 2 | −1.67 | 8.38 × 10−7 |
| Tcf15 | transcription factor 15 | −1.66 | 1.16 × 10−6 |
| Tmem159 | transmembrane protein 159 | −1.66 | 1.72 × 10−5 |
| Artn | artemin | −1.61 | 5.81 × 10−5 |
| Naaladl2 | N-acetylated α-linked acidic dipeptidase-like 2 | −1.55 | 1.68 × 10−7 |
| Ltk | leukocyte tyrosine kinase | −1.55 | 3.62 × 10−5 |
| Zfp184 | zinc finger protein 184 (Kruppel-like) | −1.55 | 2.88 × 10−5 |
| Zfp647 | zinc finger protein 647 | −1.53 | 3.82 × 10−8 |
| Mfsd9 | major facilitator superfamily domain containing 9 | −1.52 | 2.48 × 10−7 |
| Zfp51 | zinc finger protein 51 | −1.48 | 3.01 × 10−7 |
| Six4 | sine oculis-related homeobox 4 homolog (Drosophila) | −1.46 | 7.90 × 10−5 |
| Rad51ap2 | RAD51 associated protein 2 | −1.46 | 2.69 × 10−5 |
| Mir17hg | MIR17 host gene 1 (non-protein coding) | −1.45 | 1.54 × 10−4 |
| Ccdc160 | coiled-coil domain containing 160 | −1.45 | 3.64 × 10−4 |
| Tusc1 | tumor suppressor candidate 1 | −1.44 | 1.47 × 10−6 |
| Nipsnap3b | nipsnap homolog 3B (C. elegans) | −1.43 | 1.14 × 10−5 |
| Haus4 | HAUS augmin-like complex, subunit 4 | −1.42 | 1.47 × 10−6 |
| Gm11818 | predicted gene 11818 | −1.36 | 9.85 × 10−5 |
| Zfp760 | zinc finger protein 760 | −1.35 | 3.55 × 10−5 |
| Wdsub1 | WD repeat, SAM and U-box domain containing 1 | −1.35 | 2.46 × 10−6 |
| Adcy5 | adenylate cyclase 5 | −1.34 | 9.63 × 10−6 |
| Fam117a | family with sequence similarity 117, memberA | −1.32 | 6.59 × 10−6 |
| Ccdc91 | coiled-coil domain containing 91 | −1.31 | 8.47 × 10−9 |
| Foxq1 | forkhead box Q1 | −1.31 | 2.02 × 10−5 |
| Metrnl | meteorin, glial cell differentiation regulator-like | −1.29 | 4.01 × 10−7 |
| Zfp641 | zinc finger protein 641 | −1.27 | 8.36 × 10−6 |
| Ccdc111 | coiled-coil domain containing 111 | −1.26 | 1.91 × 10−4 |
| Ptgr1 | prostaglandin reductase 1 | −1.25 | 1.89 × 10−4 |
| Neil2 | nei like 2 (E. coli) | −1.25 | 4.00 × 10−4 |
| Car10 | carbonic anhydrase 10 | −1.24 | 4.18 × 10−5 |
| Ikzf2 | IKAROS family zinc finger 2 | −1.24 | 2.23 × 10−4 |
| Prkra | protein kinase, interferon inducible double stranded RNA dependent activator | −1.22 | 7.15 × 10−7 |
| Immp2l | IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) | −1.22 | 2.31 × 10−7 |
| Gabrg3 | GABAA receptor, subunit γ3 | −1.22 | 3.64 × 10−4 |
| Zfp429 | zinc finger protein 429 | −1.20 | 1.71 × 10−5 |
| Xkrx | X Kell blood group precursor-related X-linked | −1.17 | 3.53 × 10−5 |
| Glyctk | glycerate kinase | −1.14 | 2.41 × 10−4 |
| Marveld1 | MARVEL (membrane-associating) domain containing 1 | −1.13 | 2.11 × 10−4 |
| Nt5dc1 | 5′-nucleotidase domain containing 1 | −1.13 | 5.42 × 10−5 |
| Syde2 | synapse defective 1, Rho GTPase, homolog 2 (C. elegans) | −1.13 | 2.31 × 10−5 |
| Ppic | peptidylprolyl isomerase C | −1.12 | 1.79 × 10−4 |
| Pole4 | polymerase (DNA-directed), epsilon 4 (p12 subunit) | −1.12 | 1.42 × 10−5 |
| Car4 | carbonic anhydrase 4 | −1.11 | 1.30 × 10−4 |
| Tox3 | TOX high mobility group box family member 3 | −1.11 | 3.61 × 10−4 |
| Larp1b | La ribonucleoprotein domain family, member 1B | −1.10 | 1.27 × 10−5 |
| Myom3 | myomesin family, member 3 | −1.10 | 1.56 × 10−5 |
| Sord | sorbitol dehydrogenase | −1.08 | 4.44 × 10−7 |
| Oxr1 | oxidation resistance 1 | −1.08 | 5.91 × 10−7 |
| Fah | fumarylacetoacetate hydrolase | −1.07 | 1.11 × 10−6 |
| Ckap4 | cytoskeleton-associated protein 4 | −1.07 | 6.12 × 10−7 |
| Paox | polyamine oxidase (exo-N4-amino) | −1.07 | 2.84 × 10−7 |
| Zfp612 | zinc finger protein 612 | −1.07 | 1.12 × 10−5 |
| Fbxl21 | F-box and leucine-rich repeat protein 21 | −1.07 | 4.96 × 10−5 |
| Stxbp2 | syntaxin binding protein 2 | −1.06 | 3.16 × 10−4 |
| Btg3 | B-cell translocation gene 3 | −1.06 | 4.53 × 10−5 |
| Kdelc2 | KDEL (Lys-Asp-Glu-Leu) containing 2 | −1.05 | 3.71 × 10−6 |
| Rimbp3 | RIMS binding protein 3 | −1.02 | 1.60 × 10−4 |
| Gm347 | predicted gene 347 | −1.00 | 6.61 × 10−5 |
| Irf1 | interferon regulatory factor 1 | −0.99 | 1.29 × 10−6 |
| B3gnt6 | UDP-GlcNAc:betaGal β-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) | −0.98 | 3.05 × 10−4 |
| Gpx7 | glutathione peroxidase 7 | −0.97 | 2.62 × 10−5 |
| Stbd1 | starch binding domain 1 | −0.97 | 5.00 × 10−5 |
| Rabepk | Rab9 effector protein with kelch motifs | −0.97 | 1.13 × 10−5 |
| Bag2 | BCL2-associated athanogene 2 | −0.97 | 5.25 × 10−7 |
| Ptgr2 | prostaglandin reductase 2 | −0.97 | 6.91 × 10−6 |
| Gm11110 | predicted gene 11110 | −0.96 | 1.70 × 10−4 |
| Parp11 | poly(ADP-ribose) polymerase family, member 11 | −0.94 | 8.40 × 10−7 |
| Gk5 | glycerol kinase 5 (putative) | −0.92 | 6.62 × 10−5 |
| Pole | polymerase (DNA directed), ϵ | −0.92 | 7.68 × 10−5 |
| Chst7 | carbohydrate (N-acetylglucosamino)sulfotransferase 7 | −0.92 | 1.05 × 10−4 |
| Echdc2 | enoyl Coenzyme A hydratase domain containing 2 | −0.92 | 6.50 × 10−6 |
| Bves | blood vessel epicardial substance | −0.91 | 2.06 × 10−5 |
| Sypl2 | synaptophysin-like 2 | −0.90 | 7.01 × 10−5 |
| Neu3 | neuraminidase 3 | −0.90 | 7.09 × 10−6 |
| Acot1 | acyl-CoA thioesterase 1 | −0.89 | 1.02 × 10−5 |
| Jag2 | jagged 2 | −0.89 | 1.84 × 10−4 |
| Cntln | centlein, centrosomal protein | −0.89 | 1.70 × 10−5 |
| Rab42-ps | RAB42, member RAS oncogene family, pseudogene | −0.89 | 2.84 × 10−4 |
| Nudt14 | nudix (nucleoside diphosphate linked moiety X)-type motif 14 | −0.87 | 3.32 × 10−5 |
| Lck | lymphocyte protein tyrosine kinase | −0.84 | 2.01 × 10−4 |
| Rell1 | RELT-like 1 | −0.84 | 7.22 × 10−6 |
| Slc1a6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | −0.84 | 1.88 × 10−4 |
| Ccdc138 | coiled-coil domain containing 138 | −0.83 | 7.65 × 10−6 |
| Galnt12 | UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 | −0.83 | 8.22 × 10−6 |
| Mina | myc induced nuclear antigen | −0.83 | 5.11 × 10−5 |
| Slc35f5 | solute carrier family 35, member F5 | −0.83 | 1.22 × 10−6 |
| Reep6 | receptor accessory protein 6 | −0.82 | 3.96 × 10−6 |
| Gkap1 | G kinase anchoring protein 1 | −0.82 | 4.27 × 10−5 |
| Pparg | peroxisome proliferator activated receptor gamma | −0.82 | 6.38 × 10−6 |
| Asl | argininosuccinate lyase | −0.78 | 6.34 × 10−5 |
| Churc1 | churchill domain containing 1 | −0.77 | 2.91 × 10−4 |
| Peci | peroxisomal delta3, delta2-enoyl-Coenzyme A isomerase | −0.76 | 7.54 × 10−6 |
| Prorsd1 | prolyl-tRNA synthetase domain containing 1 | −0.76 | 1.74 × 10−4 |
| Gm2954 | predicted gene 2954 | −0.75 | 1.40 × 10−4 |
| BC016495 | cDNA sequence BC016495 | −0.75 | 8.10 × 10−5 |
| Eda | ectodysplasin-A | −0.75 | 1.50 × 10−4 |
| Kctd15 | potassium channel tetramerization domain containing 15 | −0.75 | 3.52 × 10−4 |
| Crlf2 | cytokine receptor-like factor 2 | −0.74 | 1.70 × 10−4 |
| Ano10 | anoctamin 10 | −0.74 | 1.72 × 10−4 |
| Sfxn4 | sideroflexin 4 | −0.74 | 1.49 × 10−5 |
| Cdyl2 | chromodomain protein, Y chromosome-like 2 | −0.72 | 3.17 × 10−5 |
| Smo | smoothened homolog (Drosophila) | −0.71 | 4.38 × 10−6 |
| Tmem53 | transmembrane protein 53 | −0.71 | 1.66 × 10−4 |
| Slc38a6 | solute carrier family 38, member 6 | −0.71 | 2.77 × 10−4 |
| Zfp105 | zinc finger protein 105 | −0.71 | 1.40 × 10−4 |
| Ebf4 | early B-cell factor 4 | −0.70 | 1.53 × 10−4 |
| Cdca7 | cell division cycle associated 7 | −0.70 | 1.66 × 10−5 |
| Acadm | acyl-Coenzyme A dehydrogenase, medium chain | −0.69 | 1.31 × 10−5 |
| Tpm4 | tropomyosin 4 | −0.69 | 2.17 × 10−4 |
| Sh3bgrl2 | SH3 domain binding glutamic acid-rich protein like 2 | −0.67 | 2.91 × 10−4 |
| Cnga4 | cyclic nucleotide gated channel alpha 4 | −0.66 | 2.98 × 10−4 |
| Hddc2 | HD domain containing 2 | −0.66 | 5.03 × 10−6 |
| Bend3 | BEN domain containing 3 | −0.65 | 9.88 × 10−6 |
| Gpr125 | G protein-coupled receptor 125 | −0.65 | 2.30 × 10−5 |
| Galk1 | galactokinase 1 | −0.64 | 2.41 × 10−4 |
| Pttg1 | pituitary tumor-transforming gene 1 | −0.64 | 4.79 × 10−5 |
| Accs | 1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) | −0.64 | 1.70 × 10−4 |
| Ebpl | emopamil binding protein-like | −0.63 | 1.75 × 10−5 |
| Trmt2b | TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae) | −0.63 | 1.23 × 10−5 |
| Zfp948 | zinc finger protein 948 | −0.63 | 2.86 × 10−4 |
| Zfp945 | zinc finger protein 945 | −0.63 | 4.59 × 10−5 |
| Ebi3 | Epstein-Barr virus induced gene 3 | −0.62 | 2.14 × 10−4 |
| Tmsb15b1 | thymosin β 15b1 | −0.62 | 8.01 × 10−5 |
| Rangrf | RAN guanine nucleotide release factor | −0.61 | 8.53 × 10−5 |
| Gusb | glucuronidase, β | −0.61 | 2.18 × 10−4 |
| Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | −0.61 | 4.01 × 10−5 |
| E2f1 | E2F transcription factor 1 | −0.59 | 9.37 × 10−5 |
| Cabyr | calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrous heathin 2) | 0.64 | 2.26 × 10−4 |
| Angpt1 | angiopoietin 1 | 0.70 | 3.75 × 10−4 |
| Pcdhb22 | protocadherin β 22 | 0.80 | 3.74 × 10−7 |
| Ccdc88b | coiled-coil domain containing 88B | 0.95 | 1.46 × 10−5 |
| Pcdhb21 | protocadherin β 21 | 1.01 | 9.00 × 10−6 |
| Krt9 | keratin 9 | 1.01 | 6.28 × 10−6 |
| Chrdl2 | chordin-like 2 | 1.06 | 1.79 × 10−5 |
| Megf6 | multiple EGF-like-domains 6 | 1.22 | 8.98 × 10−5 |
| Dach2 | dachshund 2 (Drosophila) | 3.43 | 2.90 × 10−8 |
Table 4.
List of genes differentially regulated in dorsal CA region of kmt2b cKO mice (the genes in italic were confirmed by qRT-PCR)
| Gene symbol | Gene name | log2 (fold change) | Adjusted p value |
|---|---|---|---|
| Prss16 | protease, serine, 16 (thymus) | −3.41 | 9.51 × 10−7 |
| Dazl | deleted in azoospermia-like | −2.71 | 8.00 × 10−7 |
| Hfm1 | HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) | −1.78 | 1.14 × 10−5 |
| Lor | loricrin | −1.52 | 1.51 × 10−8 |
| Hdx | highly divergent homeobox | −1.50 | 6.37 × 10−6 |
| Zbtb8a | zinc finger and BTB domain containing 8a | −1.29 | 3.06 × 10−5 |
| Ap1s3 | adaptor-related protein complex AP-1, sigma 3 | −1.26 | 3.69 × 10−5 |
| Rimbp3 | RIMS binding protein 3 | −1.22 | 2.76 × 10−5 |
| Zfp647 | zinc finger protein 647 | −1.12 | 1.59 × 10−6 |
| Wdr17 | WD repeat domain 17 | −1.11 | 1.12 × 10−5 |
| Ccdc91 | coiled-coil domain containing 91 | −1.02 | 1.75 × 10−7 |
| Oxr1 | oxidation resistance 1 | −0.92 | 3.42 × 10−6 |
| Immp2l | IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) | −0.92 | 6.60 × 10−6 |
| Cmya5 | cardiomyopathy associated 5 | −0.89 | 4.09 × 10−5 |
| Rabepk | Rab9 effector protein with kelch motifs | −0.87 | 3.64 × 10−5 |
| Parp11 | poly (ADP-ribose) polymerase family, member 11 | −0.83 | 3.57 × 10−6 |
| Reep6 | receptor accessory protein 6 | −0.82 | 4.26 × 10−6 |
| 4930403L05Rik | protein phosphatase 1 regulatory subunit 2 pseudogene | 1.92 | 2.07 × 10−5 |
| Dach2 | dachshund 2 (Drosophila) | 3.04 | 1.26 × 10−7 |
Table 5.
List of genes that are differentially expressed in kmt2b cKO mice and have been implicated with processes linked to memory function
| Gene symbol | Gene name | Function | Publications/PubMed ID |
|---|---|---|---|
| Nkapl | NF-κB activating protein like | NF-κB pathway, synaptic plasticity | 8793106 |
| Rab38 | member of RAS oncogene family | G-protein signaling | 10611641 |
| Dusp2 | dual specificity phosphatase 2 | MAPK/ERK signaling pathway, synaptic plasticity | 17880897 |
| Adcy5 | adenylate cyclase 5 | cAMP/G-protein signaling | 8798683 |
| Prkra | protein kinase, interferon-inducible double stranded RNA dependent activator | protein phosphorylation | 9235897 |
| Gprc5c | G-protein-coupled receptor, family C, group 5, member C | G-protein-coupled receptor signaling | 11378403 |
| Gpr125 | G-protein-coupled receptor 125 | G-protein-coupled receptor signaling, neuropeptide signaling | 21658577 |
| 8429885 | |||
| 10467595 | |||
| 10844026 | |||
| 8845163 | |||
| Sypl2 | synaptophysin-like protein 2 | vesicle fusion, neurotransmitter release | 20434989 |
| Stxbp2 | syntaxin binding protein 2 | vesicle docking involved in exocytosis | 15355326 |
| Ckap4 | cytoskeleton associated protein 4 | intracellular trafficking | 16870134 |
| Ap1s3 | adaptor-related protein complex 1, sigma 3 subunit | endocytosis, vesicle transport | 9252190 |
| Rabepk | Rab9 effector protein with kelch motifs | vesicle docking involved in exocytosis, receptor-mediated endocytosis | 9425005 |
| 11797010 | |||
| 15066271 | |||
| 21844341 | |||
| 21849565 | |||
| Ptgr1 | prostaglandin reductase 1 | prostaglandin metabolism | 11917005 |
| Ptgr2 | prostaglandin reductase 2 | prostaglandin metabolism | 12604095 |
| Acot1 | acyl-CoA thioesterase 1 | lipid metabolism, fatty acid biosynthesis | 18540883 |
| Acot4 | acyl-CoA thioesterase 4 | lipid metabolism, fatty acid biosynthesis | 16099392 |
| 19012750 | |||
| 12037188 | |||
| 13130128 | |||
| 16077186 | |||
| 15653788 | |||
| 12037188 | |||
| 20810888 | |||
| E2f1 | E2f transcription factor 1 | transcriptional initiation, neurogenesis | 12401450 |
| Tceal1 | transcription elongation factor A-like 1 | transcriptional elongation | 7666170 |
| Oxr1 | oxidation resistance 1 | response to oxidative stress | 9063720 |
| Gabrg3 | GABA receptor gamma 3 | inhibitory neurotransmission | 18620045 |
| Car4 | carbonic anhydrase 4 | CO2 metabolism, synaptic plasticity | 20569240 |
| Car10 | carbonic anhydrase 10 | CO2 metabolism, synaptic plasticity | 22028674 |
| 11356917 | |||
| 11830265 | |||
| 11181911 |
Figure 4.
Downregulation of genes in kmt2b cKO mice is linked to altered H3K4 di- and trimethylation. A, Differential expression of selected genes identified in the gene array was confirmed via qPCR in an independent experiment (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.05 vs control). B–D, H3K4me3 (B), H3K4m2 (C), and H3K4me1 (D) levels for the selected genes was compared via ChIP analysis in kmt2b cKO and control mice (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.05 vs control). E, Three genes that were not altered in the gene array were used as control. Left, qPCR analysis confirmed similar expression of cFos, Egr-1, and Arc in Kmt2b cKO and control mice (n = 4/group). Right, ChIP analysis (n = 4/group) revealed that H3K4me3 at the TSS of c-Fos, Egr-1, and Arc genes was similar in kmt2b cKO and control mice. F, G, H3K9 acetylation (F) and H4K16 acetylation (G) levels for the selected genes were compared via ChIP analysis in Kmt2b cKO and control mice (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.001 vs control). Error bars indicate SEM.
Previous data suggested that H3K4 methylation at the transcription start site region (TSS) is a critical step to allow histone acetylation linked to active gene expression (Wang et al., 2009). Thus, we tested whether reduced H3K4 di- and trimethylation correlated with alterations in histone acetylation marks that had been associated with active gene transcription and memory function. For this purpose we used ChIP assays to measure H3K9 acetylation that has been associated with hippocampus-dependent memory consolidation (Levenson et al., 2004; Peleg et al., 2010). H4K16 acetylation, which does not change during memory formation (Peleg et al., 2010) was used as control. Interestingly, we observed decreased H3K9 acetylation (Fig. 4F) at the TSS of downregulated genes while H4K16ac levels were unaffected (Fig. 4G).
Discussion
In this study we analyzed the role of the HMT KMT2B in excitatory forebrain neurons of mice. While there is significant evidence that histone acetylation and the corresponding HATs and HDACs are implicated with memory function (Korzus et al., 2004; Levenson et al., 2004; Vecsey et al., 2007; Guan et al., 2009; Peleg et al., 2010), data on the role of histone methylation is comparatively rare. It was previously shown that complete loss of kmt2b from early developmental stages leads to embryonic lethality (Glaser et al., 2006). Therefore in our study we induced conditional knock-down of kmt2b in forebrain excitatory neurons by expressing Cre recombinase under CamKII promoter. Since the CamKII-Cre mouse line used in our study shows strong transgene expression only after postnatal day 19 and since CamKII promoter is only active in the forebrain excitatory neurons (Minichiello et al., 1999), loxP recombination of exon 2 of kmt2b gene is achieved only in forebrain excitatory neurons at the end of the third week after birth (Mayford et al., 1996; Rotenberg et al., 1996; Tsien et al., 1996; Minichiello et al., 1999). Although we cannot fully exclude developmental deficits, deleting kmt2b specifically from the postnatal forebrain did not cause an overt phenotype and brain morphology was similar in kmt2b cKO and control mice. In line with the loss of kmt2b expression in forebrain regions we observed reduced kmt2b protein level in the hippocampus of kmt2b cKO mice. Nevertheless, immunoblot analysis indicated that loss of kmt2b protein was not complete. This could be due to multiple reasons. For example, in our approach kmt2b is only deleted in excitatory neurons without affecting kmt2b levels in inhibitory neurons and glia cells. Moreover, it is established that CamKII-mediated gene-deletion in excitatory forebrain neurons allows knock down in the majority but not all cells (Valor et al., 2011). kmt2b cKO mice exhibited impaired memory consolidation in three different learning tests that require an intact hippocampus demonstrating that KMT2B function is required for hippocampus-dependent memory formation. Moreover, although we cannot fully exclude subtle developmental effects, learning impairment observed in adult kmt2bfl/fl mice after viral mediated cre expression also indicates that the behavioral effects of kmt2b knock-down are not due to developmental disturbance but rather are caused specifically by the knock-down of kmt2b at adult stage. These data are in line with a previous study that reported increased hippocampal H3K4 methylation in response to fear conditioning and furthermore showed that fear conditioning is impaired in mice heterozygous for MLL1 (Gupta et al., 2010). It has to be mentioned that loss of kmt2b also resulted in short-term memory impairment in novel object recognition task. Since short- and long-term memories in this task are linked to independent mechanisms (Izquierdo et al., 1999; Barker et al., 2006; Morice et al., 2008), it can be argued that Kmt2b is also required to mechanisms implicated with short-term memory.
In sum, our data underscore the role for histone methylation in learning behavior and is to our knowledge the first study analyzing brain-specific loss of a HMT in memory function. Since kmt2b mediates H3K4 methylation we wondered whether loss of kmt2b in the hippocampus would affect gene expression. DNA micro-array analysis revealed that loss of kmt2b leads to changes in gene expression within the dorsal hippocampus, a hippocampal region required for the formation of new memory traces (Treves et al., 2008). The fact that the large majority of the differentially expressed hippocampal genes were downregulated (96%) in kmt2b cKO mice is in line with the fact that H3K4 methylation is linked to the activation of genes (Shilatifard, 2008). These data also suggest that the identified genes are most likely direct targets of KMT2B function. However, since we could not identify a reliable KMT2B antibody for ChIP analysis the specific distribution of KMT2B along chromatin remains to be tested. Another interesting observation was the fact that 152 genes were differentially expressed in the dorsal dentate gyrus while only 19 genes were affected in the dorsal CA region suggesting a particularly important role for KMT2B in the dorsal dentate gyrus. In line with this we detected higher levels of kmt2b expression in the dentate gyrus compared with the CA region. Since the CA region is also required for memory formation and is highly responsive to learning stimuli, it is possible that KMT2B may only become important in this region upon exposure to a learning stimulus. Therefore, in future studies it would be interesting to analyze gene expression in dorsal dentate gyrus and CA regions of kmt2b cKO mice in response to a learning stimulus. Nevertheless most of the downregulated genes observed in the dentate gyrus of kmt2b cKO mice are linked to synaptic function and memory formation.
For example, Nkapl (NF-κB activating protein like) facilitates NF-κB signaling which has repeatedly been shown to be crucial for synaptic plasticity and memory function (Albensi and Mattson, 2000; Ahn et al., 2008). Similarly, adcy5 is an adenylate cyclase catalyzing synthesis of cAMP whose role in synaptic plasticity and learning and memory is well established (Barco et al., 2006). kmt2b may also impact on MAP Kinase signaling since a phosphatase (Dusp2) was downregulated in kmt2b cKO mice. dusp2 belongs to the dual specificity phosphatases that regulate MAP kinase activity (Charbonneau and Tonks, 1992) which is required for memory formation (Davis and Laroche, 2006). Interestingly, dusp1—a close homolog of dusp2—was shown to be differentially expressed upon LTP induction (Qian et al., 1993, 1994). Moreover a number of genes involved in vesicle fusion and vesicular transport such as syntaxin binding protein 2 (stxbp2), synaptophysin-like 2 (sypl2), or adaptor-related protein complex AP-1, σ 3 (ap1s3) were downregulated in kmt2b cKO mice.
Lipid signaling, especially the involvement of prostaglandins and prostaglandin synthesis, has many times been shown to be important for synaptic plasticity and learning and memory (Teather et al., 2002; Bazan, 2003, 2005; Murray and O'Connor, 2003; Chen and Bazan, 2005; Cowley et al., 2008; Yang et al., 2009; Koch et al., 2010). Thus it is interesting to note that several genes involved in lipid metabolism (prostaglandin reductases 1 and 2 and acyl-CoA thioesterase 1 and 2) were downregulated in kmt2b cKO mice.
Another set of downregulated genes previously shown to facilitate synaptic plasticity and learning and memory were the carbonic anhydrases (car4 and car10; Sun and Alkon, 2001). It was shown that carbonic anhydrase activators facilitate spatial learning by inducing a switch in GABAergic responses from inhibitory to excitatory transmission (Sun and Alkon, 2001). In line with this, the gene coding for GABA receptor subunit y3 (gabrg3) was also downregulated in dorsal dentate gyrus of kmt2b cKO mice.
Other genes that deserve attention are oxr1 and e2f1. OXR1 is involved in protecting cells against oxidative stress (Volkert et al., 2000; van Roessel et al., 2004; Durand et al., 2007). Increased oxidation is known to impair synaptic plasticity whereas reducing agents facilitate LTP (Gozlan and Ben-Ari, 1995; Schiller et al., 2008; Yang et al., 2010). E2F1, in turn, has been shown to be involved in adult neurogenesis (Cooper-Kuhn et al., 2002). Also it remains to be seen why Kmt2b specifically regulates the identified set of genes. There is substantial evidence that many of the hippocampal genes affected by reduced KMT2B function contribute to learning and memory and thus can explain the memory deficits observed in kmt2b cKO mice.
Sixteen of these genes were chosen for further analysis and it was found that downregulation in kmt2b cKO mice correlated with reduced dentate gyrus-specific H3K4 di- and trimethylation in the corresponding TSSs. This is in line with the finding that the TSSs of constitutively active genes are marked by H3K4 di- and trimethylation (Santos-Rosa et al., 2002). Interestingly, H3K4 monomethylation was not affected in kmt2b cKO mice. This is in agreement with earlier reports showing that Kmt2b knock-out in oocytes is required for bulk H3K4 di- and trimethylation but does not affect H3K4 monomethylation (Andreu-Vieyra et al., 2010). Although we cannot exclude the possibility that some potential monomethyltransferase activity of KMT2B is compensated in kmt2b cKO mice, these data strongly support a specific role for KMT2B for H3K4 di- and trimethylation.
An interesting observation was that reduced H3K4 di- and trimethylation at downregulated genes correlated also with decrease in H3K9 acetylation, which is dynamically regulated during memory consolidation (Peleg et al., 2010). This is in line with the finding that H3K4 trimethylation and H3K9 acetylation co-occur and are highly enriched at promoters of actively transcribed genes (Santos-Rosa et al., 2002; Schübeler et al., 2004). Furthermore, a previous study in lymphocytes suggests that H3K4 trimethylation plays a critical role in histone acetylation by serving as a mark that facilitates recruitment of histone acetyltransferases (HATs) to gene promoters (Wang et al., 2009). Interestingly, our data suggest that KMT2B-mediated H3K4 trimethylation affects histone acetylation in a specific manner since H4K16 acetylation, a modification not affected by memory consolidation (Peleg et al., 2010) was similar in kmt2b cKO and control mice. This finding is interesting since Kmt2a-mediated H3K4 trimethylation has been associated with H4K16 acetylation, while H3 acetylation was not affected by KMT2A (Dou et al., 2005). These data further confirm that KMT2B has a specific role in memory formation that is different from KMT2A.
In conclusion, our data show that Kmt2b is critical for memory consolidation and regulates the expression of hippocampal plasticity genes via H3K4 di- and trimethylation that is linked to H3K9 acetylation.
Footnotes
This work was supported by the EURYI Award, the Hans and Ilse Breuer Foundation, and funds from the DZNE to A.F. S.B.-J. was supported by the German Research Foundation Research Grant SA/1050/2-1. R.C.A.-B. was supported by an EMBO Fellowship. A.F.S. and A.K. were supported by the German Research Foundation (KR2154/3-1). We thank Dr. Stefan Bonn for reading this manuscript and for helpful comments.
References
- Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S, Haladyniak U, Agbemenyah HY, Zovoilis A, Salinas-Riester G, Opitz L, Sananbenesi F, Fischer A. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011;30:4071–4083. doi: 10.1038/emboj.2011.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou HC, Sweatt JD. c-Rel, an NF-kappaB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learn Mem. 2008;15:539–549. doi: 10.1101/lm.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Albensi BC, Mattson MP. Evidence for the involvement of TNF and NF-kappaB in hippocampal synaptic plasticity. Synapse. 2000;35:151–159. doi: 10.1002/(SICI)1098-2396(200002)35:2<151::AID-SYN8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8:e1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa L, Barozzi I, Chronowska A, Termanini A, Ostuni R, Prosperini E, Stewart AF, Testa G, Natoli G. The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity. 2012;36:572–585. doi: 10.1016/j.immuni.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bähr M, Burkhardt S, Delalle I, Kügler S, Fischer A, Sananbenesi F. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Bailey CH, Kandel ER. Common molecular mechanisms in explicit and implicit memory. J Neurochem. 2006;97:1520–1533. doi: 10.1111/j.1471-4159.2006.03870.x. [DOI] [PubMed] [Google Scholar]
- Barker GR, Bashir ZI, Brown MW, Warburton EC. A temporally distinct role for group I and group II metabotropic glutamate receptors in object recognition memory. Learn Mem. 2006;13:178–186. doi: 10.1101/lm.77806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan NG. Synaptic lipid signaling: significance of polyunsaturated fatty acids and platelet-activating factor. J Lip Res. 2003;44:2221–2233. doi: 10.1194/jlr.R300013-JLR200. [DOI] [PubMed] [Google Scholar]
- Bazan NG. Lipid signaling in neural plasticity, brain repair, and neuroprotection. Mol Neurobiol. 2005;32:89–103. doi: 10.1385/MN:32:1:089. [DOI] [PubMed] [Google Scholar]
- Charbonneau H, Tonks NK. 1002 protein phosphatases? Annu Rev Cell Biol. 1992;8:463–493. doi: 10.1146/annurev.cb.08.110192.002335. [DOI] [PubMed] [Google Scholar]
- Chen C, Bazan NG. Lipid signaling: sleep, synaptic plasticity, and neuroprotection. Prostaglandins Other Lipid Mediat. 2005;77:65–76. doi: 10.1016/j.prostaglandins.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Kuhn CM, Vroemen M, Brown J, Ye H, Thompson MA, Winkler J, Kuhn HG. Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol Cell Neurosci. 2002;21:312–323. doi: 10.1006/mcne.2002.1176. [DOI] [PubMed] [Google Scholar]
- Cowley TR, Fahey B, O'Mara SM. COX-2, but not COX-1, activity is necessary for the induction of perforant path long-term potentiation and spatial learning in vivo. Eur J Neurosci. 2008;27:2999–3008. doi: 10.1111/j.1460-9568.2008.06251.x. [DOI] [PubMed] [Google Scholar]
- Davis S, Laroche S. Mitogen-activated protein kinase/extracellular regulated kinase signalling and memory stabilization: a review. Genes Brain Behav. 2006;5:61–72. doi: 10.1111/j.1601-183X.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Durand M, Kolpak A, Farrell T, Elliott NA, Shao W, Brown M, Volkert MR. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol. 2007;28:13. doi: 10.1186/1471-2121-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory after neuronal loss is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of MLL2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K, Stewart AF. The histone 3 lysine 4 methyltransferase, MLL2, is only required briefly in development and spermatogenesis. Epigenetic Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich A, Zimmermann AM, Schober D, Böttcher RT, Sassoè-Pognetto M, Friauf E, Witke W, Rust MB. Preserved morphology and physiology of excitatory synapses in profilin1-deficient mice. PLoS One. 2012;7:e300068. doi: 10.1371/journal.pone.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan N, Agis-Balboa C, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimer's Disease. 2011;24:1–11. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- Gozlan H, Ben-Ari Y. NMDA receptor redox sites: are they targets for selective neuronal protection? Trends Pharmacol Sci. 1995;16:368–374. doi: 10.1016/s0165-6147(00)89077-x. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Chen Y, Dong E, Kundakovic M, Guidotti A. From trans-methylation to cytosine methylation: evolution of the methylation hypothesis of schizophrenia. Epigenetics. 2009;4:144–149. doi: 10.4161/epi.4.3.8534. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci. 2012;32:5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Toyama K, Yamasaki N, Miyakawa T. Dissection of hippocampal dentate gyrus from adult mouse. J Vis Exp. 2009;17:pii 1543. doi: 10.3791/1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo I, Medina JH, Vianna MR, Izquierdo LA, Barros DM. Separate mechanisms for short- and long-term memory. Behav Brain Res. 1999;103:1–11. doi: 10.1016/s0166-4328(99)00036-4. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Koch H, Huh SE, Elsen FP, Carroll MS, Hodge RD, Bedogni F, Turner MS, Hevner RF, Ramirez JM. Prostaglandin E2-induced synaptic plasticity in neocortical networks of organotypic slice cultures. J Neurosci. 2010;30:11678–11687. doi: 10.1523/JNEUROSCI.4665-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczera T, Stilling RM, Hsia HE, Bahari-Javan S, Irniger S, Nasmyth K, Sananbenesi F, Fischer A. The anaphase promoting complex is required for memory function in mice. Learn Mem. 2011;18:49–57. doi: 10.1101/lm.1998411. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Minichiello L, Korte M, Wolfer D, Kühn R, Unsicker K, Cestari V, Rossi-Arnaud C, Lipp HP, Bonhoeffer T, Klein R. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Morice E, Andreae LC, Cooke SF, Vanes L, Fisher EM, Tybulewicz VL, Bliss TV. Preservation of long-term memory and synaptic plasticity despite short-term impairments in the Tc1 mouse model of Down syndrome. Learn Mem. 2008;15:492–500. doi: 10.1101/lm.969608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8:608–619. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Murray HJ, O'Connor JJ. A role for COX-2 and p38 mitogen activated protein kinase in long-term depression in the rat dentate gyrus in vitro. Neuropsychopharmacology. 2003;44:374–380. doi: 10.1016/s0028-3908(02)00375-1. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Rau V, Eger EI, Harris RA, Fanselow MS. Amygdala transcriptome and cellular mechanisms underlying stress-enhanced fear learning in a rat model of posttraumatic stress disorder. Neuropsychopharmacology. 2010;35:1402–1411. doi: 10.1038/npp.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–547. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert M, Kandel ER. Temporal and spatial regulation of the expression of BAD2, a MAP kinase phosphatase, during seizure, kindling, and long-term potentiation. Learn Mem. 1994;1:180–188. [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg A, Mayford M, Hawkins RD, Kandel ER, Muller RU. Mice expressing activated CaMKII lack low frequency LTP and do not form stable place cells in the CA1 region of the hippocampus. Cell. 1996;87:1351–1361. doi: 10.1016/s0092-8674(00)81829-2. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller D, Cain CK, Curley NG, Schwartz JS, Stern SA, Ledoux JE, Phelps EA. Evidence for recovery of fear following immediate extinction in rats and humans. Learn Mem. 2008;15:394–402. doi: 10.1101/lm.909208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, Guo Y, Lessard A, Akbarian S, Weng Z. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun MK, Alkon DL. Pharmacological enhancement of synaptic efficacy, spatial learning, and memory through carbonic anhydrase activation in rats. J Pharmacol Exp Ther. 2001;297:961–967. [PubMed] [Google Scholar]
- Teather LA, Packard MG, Bazan NG. Post-training cyclooxygenase-2 (COX-2) inhibition impairs memory consolidation. Learn Mem. 2002;9:41–47. doi: 10.1101/lm.43602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, Cusick A, Riley ZL, Sunkin SM, Bernard A, Puchalski RB, Gage FH, Jones AR, Bajic VB, Hawrylycz MJ, Lein ES. Genomic anatomy of the hippocampus. Neuron. 2008;60:1010–2021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Treves A, Tashiro A, Witter MP, Moser EI. What is the mammalian dentate gyrus good for? Neuroscience. 2008;154:1155–1172. doi: 10.1016/j.neuroscience.2008.04.073. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci. 2011;31:1652–1663. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;14:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert MR, Elliott NA, Housman DE. Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc Natl Acad Sci U S A. 2000;97:14530–14535. doi: 10.1073/pnas.260495897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Wu PF, Long LH, Yu DF, Wu WN, Hu ZL, Fu H, Xie N, Jin Y, Ni L, Wang JZ, Wang F, Chen JG. Reversal of aging-associated hippocampal synaptic plasticity deficits by reductants via regulation of thiol redox and NMDA receptor function. Aging Cell. 2010;9:709–721. doi: 10.1111/j.1474-9726.2010.00595.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]