Abstract
A subpopulation of chick retinal projection neurons becomes tetraploid during development, an event prevented by blocking antibodies against p75 neurotrophin receptor (p75NTR). We have used an optimized flow cytometric assay, based on the analysis of unfixed brain cell nuclei, to study whether p75NTR-dependent neuronal tetraploidization takes place in the cerebral cortex, giving rise to projection neurons as well. We show that 3% of neurons in both murine neocortex and chick telencephalic derivatives are tetraploid, and that in the mouse ∼85% of these neurons express the immediate early genes Erg-1 and c-Fos, indicating that they are functionally active. Tetraploid cortical neurons (65–80%) express CTIP2, a transcription factor specific for subcortical projection neurons in the mouse neocortex. During the period in which these neurons are born, p75NTR is detected in differentiating neurons undergoing DNA replication. Accordingly, p75NTR-deficient mice contain a reduced proportion of both NeuN and CTIP2-positive neocortical tetraploid neurons, thus providing genetic evidence for the participation of p75NTR in the induction of neuronal tetraploidy in the mouse neocortex. In the striatum tetraploidy is mainly associated with long-range projection neurons as well since ∼80% of tetraploid neurons in this structure express calbindin, a marker of neostriatal-matrix spiny neurons, known to establish long-range projections to the substantia nigra and globus pallidus. In contrast, only 20% of tetraploid cortical neurons express calbindin, which is mainly expressed in layers II–III, where CTIP2 is absent. We conclude that tetraploidy mainly affects long-range projection neurons, being facilitated by p75NTR in the neocortex.
Introduction
Cumulative evidence indicates that a number of structures in the normal nervous system of higher vertebrates contains both glia (Westra et al., 2009) and neurons with double the normal amount of nuclear DNA (i.e., somatic tetraploidy). The neural structures in which somatic tetraploid neurons have so far been described include the human entorhinal cortex (Mosch et al., 2007), the mouse retina (Morillo et al., 2010) and the retina, optic lobes, dorsal root ganglia, cerebellum, and spinal cord of the posthatch chick (Morillo et al., 2010; López-Sánchez et al., 2011).
In the normal adult nervous system, somatic tetraploid neurons are likely generated during development. In the chick retina, these neurons derive from a subpopulation of differentiating retinal ganglion cells (RGCs) that reactivate the cell cycle and become tetraploid as they migrate out to the adult ganglion cell layer (Morillo et al., 2010). Cell cycle re-entry in these neurons is induced by the neurotrophin receptor p75 (p75NTR), a molecule with multiple functions including cell cycle regulation in neural cells (López-Sánchez and Frade, 2002). This was demonstrated by the capacity of blocking anti-p75NTR antibodies to prevent cell cycle re-entry and tetraploidy in differentiating RGCs (Frade, 2000; Morillo et al., 2010). The mechanism used by p75NTR to force cell cycle re-entry in these neurons has been shown to depend on its capacity to induce p38MAPK-dependent phosphorylation of the E2F4 transcription factor (Morillo et al., 2012).
In the chick retina neuronal tetraploidization occurs in a population of large RGCs innervating deep layers of the target tissue, the optic tectum (Morillo et al., 2010). Nevertheless, it is unclear whether tetraploidy affects long-range projection neurons in other structures of the normal nervous system as well, and whether p75NTR participates in neuronal tetraploidization in these structures.
We have focused on the mouse cerebral cortex to explore the presence of projection neurons with double the amount of DNA in their nuclei. We show that a small proportion of functionally active cortical neurons are tetraploid, and that most of these neurons express CTIP2, a transcription factor specific for long-range projection neurons (Arlotta et al., 2005). The presence of tetraploid CTIP2-positive neurons is evolutionarily conserved since we observed a similar proportion of these neurons in the chick telencephalon. We also provide genetic evidence for the involvement of p75NTR in neuronal tetraploidization since the proportion of these neurons is reduced in the cortex of the p75NTR−/− mice. This observation is consistent with the presence of double p75NTR-/βIII-tubulin-positive cells undergoing DNA synthesis in the cortical neuroepithelium during the period of neurogenesis of CTIP2-positive neurons, as previously described in the chick retina (Morillo et al., 2010). In contrast with this latter tissue, differentiating cortical neurons that reactivate the cell cycle were observed to express Rb. Finally, we show that in the striatum, neuronal tetraploidy is also associated with projection neurons, thus suggesting that neuronal tetraploidization in vertebrates mainly occur in this type of neurons.
Materials and Methods
Animals.
Original breeding pairs of mice heterozygous for a mutation targeted to the third exon of the Ngfr gene encoding a form of p75NTR that lacks its capacity to interact with its ligands (B6.129S4-Ngfrtm1Jae/J p75NTR+/− mice) (Lee et al., 1992) were obtained from The Jackson Laboratory. These functional knock-out mice were inbred to produce all three genotypes. Wild-type control (p75NTR+/+) mice and homozygous mice for the Ngfr mutation (p75NTR−/−) were used in the present study. Genotypes were determined by genomic PCR as previously described (Frade and Barde, 1999). Males and females were used interchangeably with no differences between genders. C57BL/6J mice (Harlan) were also used in this study. Pregnant females were identified by the presence of a vaginal plug. The day of plug observation was designed for embryonic day 0.5 (E0.5). Embryos were staged as described by Kaufman (1992). Fluorescent, ubiquitination-based cell cycle indicator (Fucci) mice [B6.B6D2-Tg(Fucci)504Bsi mice] (Sakaue-Sawano et al., 2008) were provided by Amalgaam. Fertilized eggs from White Leghorn hens were obtained from a local supplier (Granja Santa Isabel) and were incubated at 38.5°C in an atmosphere of 70% humidity. Posthatch chicks (7 d old) were used in this study. Experimental procedures were approved by the Consejo Superior de Investigaciones Científicas (CSIC) animal ethics committee, in accordance with the European Union guidelines.
Primary and secondary antibodies.
The rabbit polyclonal antiserum [9650] against the extracellular domain of human p75NTR (Massa et al., 2006) was a kind gift from Moses V. Chao (New York University), and it was diluted 1/800 for immunohistochemistry. The mouse monoclonal antibody (mAb) anti-βIII-tubulin (Millipore) was used at 1/1300 for immunohistochemistry. The rat mAb anti-CTIP2 antibody (Abcam) was used at 1/500 for immunohistochemistry and 1/400 for flow cytometry. The rabbit anti-calbindin-D28k (Swant) was used 1/800 for flow cytometry. Bromodeoxyuridine (BrdU) was visualized by immunohistochemistry with a 1/200 dilution of the rat mAb BU1/75 (ICR1; Serotec). The rabbit anti-Erg-1 mAb (clone T.126.1; Thermo Scientific) was used at 1/400 dilution for flow cytometry. The rabbit anti-c-Fos (9F6) antibody (Cell Signaling Technology) was used at 1/200 dilution for flow cytometry. The rabbit anti-Rb antibody raised against a peptide derived from the human Rb sequence around S780 (Abcam) was used at 1/75 dilution for immunohistochemistry. The rabbit anti-phosphoRb (Ser795) antibody (Cell Signaling Technology) was used at 1/100 dilution for immunohistochemistry. The rabbit polyclonal antibody 32A-III against Sox5 (Perez-Alcala et al., 2004) was kindly provided by J.A. Barbas (Cajal Institute, Madrid, Spain). This antibody was used at 1/1000 dilution for flow cytometry. All secondary antibodies were purchased from Invitrogen. The Alexa Fluor 594 goat anti-rabbit antibody was used at a 1/1000 dilution for immunohistochemistry. The Alexa Fluor 647 donkey anti-mouse and the Alexa Fluor 488 goat anti-rat antibodies were used at 1/1000 for immunohistochemistry and 1/500 for flow cytometry. The Alexa Fluor 488 goat anti-mouse, the Alexa Fluor 488 goat anti-rabbit, and the Alexa Fluor 647 goat anti-rabbit antibodies were diluted 1/500 for cytometric studies.
In vivo BrdU treatment.
Pregnant female C57BL/6J mice were intraperitoneally injected with BrdU in PBS (50 μg/g body weight) and killed 30 min later.
Immunohistochemistry.
For immunohistochemistry, mouse embryonic brains were fixed for 8–12 h at 4°C with 4% paraformaldehyde. Adult mice were perfused intracardially and the brain was then fixed for 18 h at the same conditions as above. Fixed tissue was cryopreserved overnight (O/N) at 4°C in PBS containing 30% sucrose (Merck), and embedded in the OCT compound Tissue-Tek (Sakura). Cryosections (12 μm, embryos; 20 μm, adults) were permeabilized and blocked for 1 h at room temperature (RT) in PBS containing 0.1% Triton X-100 (Sigma) and 10% fetal calf serum (FCS) (Invitrogen), and they were then incubated O/N at 4°C with the primary antibody in PBS/0.1% Triton X-100 plus 1% FCS. After five washes with PBS/0.1% Triton X-100, the sections were incubated for 1 h at RT with the secondary antibodies. The sections were finally washed five times in PBS/0.1% Triton X-100, and incubated with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma) in PBS before mounting in glycerol/PBS (1:1). BrdU labeling was revealed using a previously described protocol (López-Sánchez et al., 2011). Briefly, cryosections were permeabilized with 0.1% Triton X-100 in PBS for 10 min at RT, rinsed twice with PBS, and incubated for 30 min at 37°C in 100 mm Tris-HCl buffer, pH 8.5, containing 2.5 mm MgCl2, 0.5 mm CaCl2, and 10 μg/ml DNase I (Roche). Then, cryosections were rinsed twice with PBS and subjected to immunohistochemistry as described above.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling.
Apoptosis was analyzed in cryosections (20 μm) and isolated nuclei using the in situ cell death detection kit (ROCHE) following the manufacturer's instructions.
Cell nuclei isolation.
Fresh-frozen mouse and chicken tissues (one mouse hemicortex or one chick telencephalic hemisphere) were placed in 2.5 ml of ice-cold, DNase-free PBS containing 0.1% Triton X-100 (Sigma) and protease inhibitor mixture (Roche) (nuclear isolation buffer). Cell nuclei were then isolated by mechanical disaggregation using a dounce homogenizer. Undissociated tissue was removed by centrifugation at 200 × g for 1.5 min at 4°C. The supernatant was eightfold diluted with nuclear isolation buffer and centrifuged at 400 × g for 4 min at 4°C. Supernatant with cellular debris was discarded, and the pellet incubated at 4°C in 800–1000 μl of cold nuclear isolation buffer for at least 1 h, before mechanical disaggregation by gently swirl of the vial. In some cases, pellets were resuspended in ice-cold 70% ethanol/PBS and fixed O/N at 4°C as described by Westra et al. (2010). The quality and purity of the isolated nuclei was analyzed microscopically after staining with 100 ng/ml DAPI.
Cell isolation.
Fresh mouse cortex or striatum was placed in Ca2+-Mg2+-free PBS containing 3 mg/ml bovine serum albumin (BSA), and then treated with 0.5 mg/ml trypsin (Worthington) for 35 min at 37°C. Reactions were stopped by adding 0.5 mg/ml soybean trypsin inhibitor (Sigma). DNase I (10 μl) prepared at 1 μg/μl in PBS was then added; and the cells were subsequently dissociated by gentle trituration. Cells were centrifuged at 300 × g for 5 min at RT. Pellets were washed twice with PBS to remove cell debris, and then they were fixed O/N at 4°C in 70% ethanol/PBS.
Flow cytometry.
Nuclear immunostaining was performed by adding primary and secondary antibodies to 400 μl of isolated unfixed nuclei containing 5% FCS and 1.25 mg/ml BSA. In control samples, the primary antibodies were excluded. Finally, the reaction was incubated O/N at 4°C in the dark. Ethanol-fixed nuclei were centrifuged at 600 × g for 4 min at 4°C, washed twice with disaggregation buffer, and immunostained as described above. Immunostained nuclei (400 μl) were filtered through a 40 μm nylon filter, and the volume adjusted to 800–1000 μl with DNase-free PBS-0.1% Triton X-100 containing propidium iodide (PI; Sigma) and DNase-free RNase I (Sigma) at a final concentration of 40 and 25 μg/ml, respectively, and incubated for 30 min at RT. The quality of the nuclei and specificity of immunostaining signal was checked by fluorescence microscopy. Dissociated cells were immunolabeled and filtered as previously described by Morillo et al. (2010).
Flow cytometry was performed with a FACSAria cytometer (BD Biosciences) equipped with a double argon (488 nm) and helium-neon laser (633 nm). Data were collected by using a linear digital signal process. The emission filters used were BP 530/30 for Alexa 488, BP 616/23 for PI, and BP 660/20 for Alexa 647. Data were analyzed with FACSDiva (BD Biosciences) and Weasel 3.0.1 (Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia) software, and displayed using biexponential scaling. Electronic compensation for fluorochrome spectral overlap during multicolor immunofluorescence analysis was performed when needed as described by Herzenberg et al. (2006). Cellular debris, which was clearly differentiated from nuclei due to its inability to incorporate PI, was gated and excluded from the analysis. DNA content histograms were generated excluding doublets and clumps by gating on the DNA pulse area versus its corresponding pulse height (Nunez, 2001). The exclusion of doublets was confirmed by checking the DNA pulse area versus the pulse width of the selected population. The percentage of 4C nuclei was quantified by using the ModFit software package (Verity). A minimum of 15,000 and 20,000 nuclei were analyzed for the CTIP2-positive and NeuN-positive (including the double NeuN/Erg-1 analysis) populations, respectively. Approximately 15,000–20,000 cells were used for the analysis of tetraploidy in calbindin neurons.
Fluorescent in situ hybridization.
Isolated cell nuclei from cerebral cortex of 2-month-old mice were immunolabeled with anti-CTIP2 antibodies and counterstained with PI. Cell nuclei were then subjected to fluorescence-activated cell sorting (FACS) to isolate CTIP2-positive nuclei with 2C or 4C DNA content. Suspensions of nuclei were subsequently centrifuged at 300 ×g, and the pellets were then fixed with ethanol (Merck)/glacial acetic acid (Merck) (3:1) for 30 min. The fixed nuclei were dropped onto wet slides and dried overnight at room temperature. Fluorescent in situ hybridization (FISH) was performed using a Poseidon TK (11qE1) mouse probe (Kreatech Diagnostics) following the indications of the manufacturer. This probe recognizes mouse chromosome 11. Nuclei were finally stained with 100 ng/ml DAPI for 2 min. After dehydration with 70% ethanol, 90% ethanol, and 100% ethanol, coverslips were air dried and mounted with ProLong Gold Antifade Reagent (Invitrogen).
Slide-based cytometry.
Slide-based cytometry (SBC) is a well standardized method for DNA quantification in tissue sections (Mosch et al., 2007; Morillo et al., 2010). Mosch et al. (2007) have shown a close correlation between SBC and FISH or real-time PCR-dependent DNA quantification. The relative DNA content present in CTIP2-positive nuclei was analyzed by SBC in coronal cryosections (15 μm) obtained from cerebral cortex of E17.5 mouse embryos, a stage at which neurogenesis of layer V–VI has finished (Caviness, 1982). To this end, cryosections were immunostained with the anti-CTIP2 antibody as described above and labeled with 100 ng/ml DAPI prepared in PBS/0.1% Triton X-100 containing 1% FCS. The sections were finally washed five times in PBS containing 0.1% Triton X-100, once in PBS, and then they were mounted in glycerol/PBS (1:1). CTIP2-positive immunostaining and DAPI labeling was then recorded with a Nikon E80i microscope equipped with a DXM 1200 digital camera (Nikon), using a 20× magnification objective. The integral density of DAPI as well as the area of CTIP2-positive nuclei, obtained as arbitrary units, was quantified using the freely available ImageJ software (National Institute of Health, Bethesda, MD). Data were converted to fcs format using the CsvToFcs module available at the GenePattern Web site (http://genepattern.broadinstitute.org) (Reich et al., 2006), and analyzed with the flow cytometric software described above. In total, 8004 CTIP2-positive nuclei from the cerebral cortex of two different embryos were analyzed in this study (5860 cell nuclei from the prefrontal and motor cortex and 2144 from visual cortex).
Results
An optimized flow cytometric assay for the analysis of DNA content in fresh cell nuclei
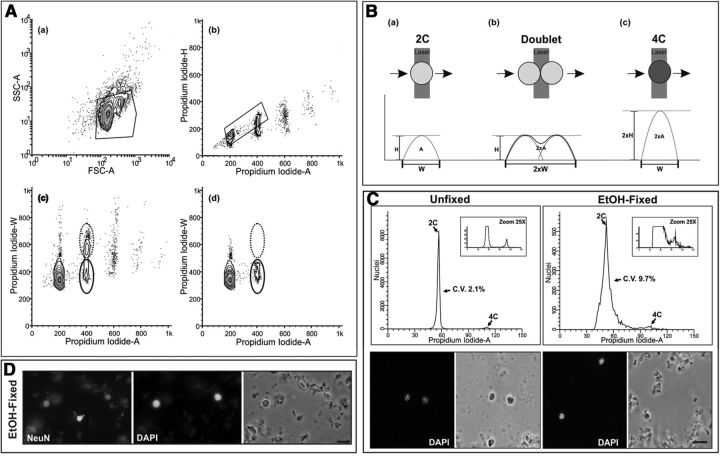
An optimized procedure for measuring DNA content in brain cells was developed based on flow cytometry performed with fresh cell nuclei (Fig. 1). For this analysis, it was first required to discriminate homogeneous nuclear populations from cell debris using forward scatter/side scatter biplots (Fig. 1Aa, polygonal line). Then, the proportion of tetraploid nuclei within the gated nuclear population was quantified. Doublets of diploid nuclei were discarded using the standard pulse processing method (for review, seeNunez, 2001), thus assuring that the gated events with a double amount of DNA accurately represent tetraploid nuclei. Doublet discrimination by the pulse processing method is based on the length of time the fluorescent signal requires to be recorded as well as its intensity (Fig. 1B). The time that elapses during the passage of a PI-labeled event through the laser beam is registered as the peak width (W), while the maximal intensity of the signal is recorded as the peak height (H). In addition, the integrated area of the peak is registered as the peak area (A). Therefore, tetraploid events can easily be discriminated from doublets of diploid events due to the higher H/A ratio of their PI signal (Fig. 1Ab,B). Our analysis included an additional criterion for tetraploidy based on the W/A ratio of the PI signal, which is higher in doublets of diploid events than in tetraploid events (Fig. 1B). This criterion confirmed that the gated nuclear population shown in Figure 1, Ab, polygonal line, does not contain doublets of diploid nuclei since the population with high W/A rate is absent (Fig. 1Ad, dashed line). Importantly, our analysis also allowed us to discriminate chick singlet/doublet nuclei, obtaining similar results with preparations of cell nuclei obtained from the posthatch chick telencephalon (data not shown).
Figure 1.
Electronic gating procedure for murine nuclear samples stained with PI and analyzed by flow cytometry. A, Fresh cell nuclei were isolated from the cerebral cortex of 2-month-old mice, stained with PI, and subjected to flow cytometric analysis. Nuclei were gated (polygonal box) on forward scattering area (FCS-A), a measure of particle size, and side scattering area (SSC-A), a measure of particle complexity (a). DNA content was then assessed from the gated nuclear population by plotting Propidium Iodide-H versus Propidium Iodide-A levels. Diploid and tetraploid nuclei were subsequently gated from these plots (polygonal box), while the doublets of diploid nuclei were discarded (b). DNA content was also assessed from the gated nuclear population in a by plotting Propidium Iodide-W versus Propidium Iodide-A levels. Tetraploid nuclei are surrounded by a solid line while doublets of diploid nuclei are surrounded by a dotted line (c). When DNA content is assessed from the gated nuclear population in b by plotting Propidium Iodide-W versus Propidium Iodide-A levels, doublets of diploid nuclei (dotted line) disappear (d), thus demonstrating that our gating procedure is an efficient method for analysis of diploid and tetraploid (solid line) nuclei. B, Scheme illustrating the method used for doublet discrimination based on the pulse-processing method. When a PI-labeled nucleus passes through the laser beam a fluorescent signal is converted into an electrical pulse defined by its width (W), its maximal intensity (H), and its integrated area (A). The signal recorded from a tetraploid nucleus (c) has a double H value compared with a diploid nucleus (a), whereas a doublet of diploid nuclei (b) results in a fluorescent signal with double W value compared with that of a diploid nucleus. Therefore, tetraploid nuclei can easily be discriminated from doublets of diploid nuclei due to the H/A and the W/A ratio of their PI signal. C, Cell nuclei isolated from the cerebral cortex of 2-month-old mice were either stained with PI and subjected to flow cytometric analysis (Unfixed) or fixed in ice-cold 70% ethanol overnight before staining with PI and flow cytometric analysis (EtOH-fixed). Bottom, Illustrates the aspect of the fresh and ethanol-fixed nuclei, counterstained with DAPI. Top, Illustrates representative DNA content histograms from either unfixed or ethanol-fixed nuclei. Flow cytometric analyses were performed in parallel. CV (C.V.) are shown. Note that the CV is lower when fresh nuclei are analyzed. D, Ethanol-fixed nuclei immunostained with an anti-NeuN antibody and counterstained with DAPI demonstrate a lot of staining background, which likely explains the increased CV. Scale bars: C, 10 μm; D, 20 μm.
In our hands, flow cytometric analyses performed with fresh nuclei consistently resulted in histograms with lower coefficients of variation (CV) compared with the analyses performed with ethanol-fixed nuclei (Fig. 1C, top). The low CV values observed with fresh nuclei are likely due to the absence in our preparations of cell debris both in solution and adhered to the nuclei (Fig. 1C, bottom). Moreover, nuclear marker immunostaining of ethanol-fixed cell nuclei systematically yielded higher backgrounds (Fig. 1D) than immunolabeling performed in fresh cell nuclei (Fig. 2A,F). This indicates that the use of fresh cell nuclei for flow cytometry represents a reliable method for the analysis of neuronal tetraploidy, provided that neuronal nuclear markers are available for immunolabeling.
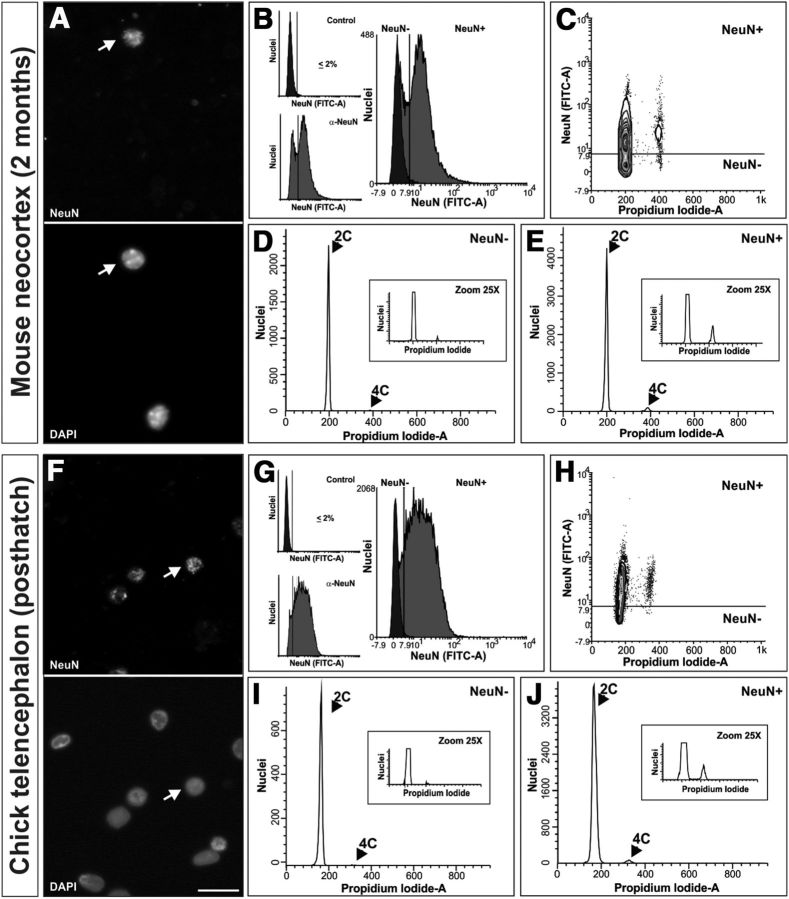
Figure 2.
Somatic tetraploid neurons in the mouse cerebral cortex and chick telencephalon. A, Cell nuclei isolated from the cerebral cortex of 2-month-old mice immunostained with an anti-NeuN-specific antibody and counterstained with DAPI. Arrow, An NeuN-positive nucleus. B, Cell nuclei immunostained with the anti-NeuN antibody (α-NeuN) or with the secondary antibody alone (Control) were PI stained and subjected to flow cytometric analysis. Right, Illustrates the threshold used for the discrimination of NeuN-positive nuclei. C, DNA content (Propidium Iodide-A) plotted against NeuN signal intensity (FITC-A) demonstrates that most nuclei with 4C content are positive for NeuN immunolabeling. D, DNA content histogram from NeuN-negative nuclei. E, DNA content histogram from NeuN-positive nuclei. See Table 1 for quantitative data. F, Cell nuclei isolated from the telencephalon of posthatch chicks immunostained with an anti-NeuN-specific antibody and counterstained with DAPI. Arrow, An NeuN-positive nucleus. G, Cell nuclei immunostained with the anti-NeuN antibody (α-NeuN) or with the secondary antibody alone (Control) were PI stained and subjected to flow cytometric analysis. Right, Illustrates the threshold used for the discrimination of NeuN-positive nuclei. H, DNA content (Propidium Iodide-A) plotted against NeuN signal intensity (FITC-A) demonstrates that most nuclei with 4C content are positive for NeuN immunolabeling. I, DNA content histogram from NeuN-negative nuclei. J, DNA content histogram from NeuN-positive nuclei. See Table 1 for quantitative data. Scale bar, 20 μm.
The mouse cerebral cortex contains tetraploid neurons
As indicated above, isolated unfixed nuclei can be immunolabeled with nuclear neuronal markers such as NeuN (Fig. 2A,B), and the DNA content present in singlet nuclei can be quantified by flow cytometry as a function of the intensity of PI labeling. In our experiments, nuclei were considered to be positive for NeuN only when the intensity of the signal was above that obtained with secondary antibody alone (Fig. 2B, dark gray). This procedure allowed us to reliably quantify the proportion of NeuN-positive nuclei in the cerebral cortex showing 4C DNA content (Fig. 2C). This analysis evidenced that most tetraploid nuclei were immunopositive for NeuN (Table 1; Fig. 2C–E), as previously observed in the retina (Morillo et al., 2010), thus demonstrating that tetraploidy in the murine cerebral cortex is mainly associated with neurons. Our results indicated that ∼3% of cortical neurons contained a double amount of DNA at both postnatal day 0 (P0) and 2 months (Table 1), suggesting that the final proportion of neuronal tetraploidy is already reached at P0 and that it does not change substantially during adulthood.
Table 1.
Percentage of tetraploid nuclei with NeuN-specific immunolabeling and NeuN-positive nuclei with tetraploid content in mouse cerebral cortex and chick telencephalon
| Tissue | % NeuN + nuclei in the 4C population (mean ± SEM) | % NeuN + nuclei with 4C DNA content (mean ± SEM) | n |
|---|---|---|---|
| Mouse neocortex (P0) | 98.66 ± 0.61 | 3.17 ± 0.15 | 8 |
| Mouse neocortex (2 month old) | 95.21 ± 0.71 | 3.12 ± 0.29 | 13 |
| Chick telencephalon (posthatch) | 98.30 ± 0.40 | 2.98 ± 0.22 | 5 |
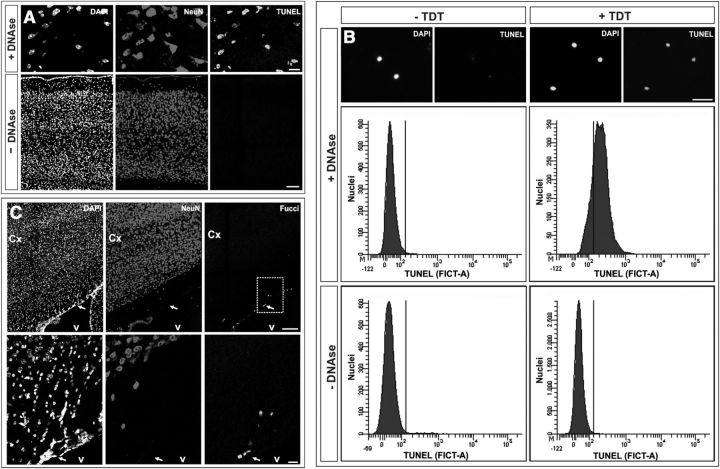
The stable proportion of neuronal tetraploidy as mice get older could also be explained in terms of continuous death of tetraploid neurons, which would be replaced by new diploid neurons undergoing DNA duplication. To test this alternative hypothesis, we first studied the presence of apoptotic cells in the cerebral cortex from mice of 1 month of age, an intermediate age between P0 and 2 months. This analysis demonstrated the absence of apoptosis in the cerebral cortex, as evidenced by terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) staining in either sections (n = 2 mice) (Fig. 3A, bottom) or fresh cell nuclei subjected to flow cytometry (n = 2 mice) (Fig. 3B, bottom). In contrast, positive controls treated with DNase showed high levels of TUNEL staining (Fig. 3A,B, top). We next tested the capacity of cortical neurons to reactivate the cell cycle and duplicate their DNA. Since BrdU can be toxic if administered for long periods (Kimbrough et al., 2011), we decided to perform this analysis in cortical sections from Fucci mice, immunolabeled with NeuN-specific antibodies. Fucci mice express human geminin fused to monomeric Azami Green (mAG) protein, and show green nuclei in cells that undergo S/G2 phase (Sakaue-Sawano et al., 2008). These mice contain a similar proportion of tetraploid neurons in the cerebral cortex as those observed in wild-type mice (data not shown). As expected, cells located in the subventricular zone along the lateral ventricles of 1-month-old Fucci mice often contained green nuclei (Fig. 3C), likely representing adult neuronal precursors. In contrast, no evidence for green labeling in NeuN-positive nuclei was found in the cerebral cortex of these mice (18 sections from two different mice) (Fig. 3C). Unfortunately, flow cytometry could not be used for the detection of green labeling in fresh cell nuclei from Fucci mice since the mAG-fused protein is lost during the nuclear isolation process (data not shown). Overall, we conclude that the cortical tetraploid neurons observed in the adult brain are likely those born during development, which can survive in the adult brain.
Figure 3.
Absence of tetraploid neuron turnover in 1-month-old mice. A, TUNEL staining in cryosections (12 μm) from the cerebral cortex of 1-month-old mouse treated with DNase (+DNAse) or left untreated (−DNAse), immunostained with an anti-NeuN antibody, and counterstained with DAPI. B, Flow cytometric analysis of fresh cell nuclei from 1-month-old mouse cortex treated with DNase (+DNAse) (positive control), or left untreated (−DNAse), and subjected to TUNEL staining either in the presence (+TDT) or absence (−TDT) (negative control) of terminal deoxynucleotidyl transferase. Quality of cell nuclei was evaluated using DAPI counterstaining (top). C, Cryosections (12 μm) from the cerebral cortex of 1-month-old Fucci mouse immunostained with an anti-NeuN antibody and counterstained with DAPI. Nuclei labeled with geminin/mAG (Fucci), representing cells in S/G2, can be observed in the subventricular zone (arrow). Bottom, High magnifications of the dashed rectangle from the top. Cx, cortex; v, ventricle. Scale bars: A top, B, 40 μm; A bottom, C, 100 μm.
The telencephalon of posthatch chick contains tetraploid neurons
To verify whether tetraploid neurons can also be detected in telencephalic derivatives of other species, a similar approach as that described above was performed with the telencephalon of the posthatch chick. Cell nuclei isolated from the telencephalon were immunostained with anti-NeuN antibodies (Fig. 2F,G), and subjected to flow cytometric analysis after PI labeling. As in the case of the mouse cerebral cortex, most nuclei showing 4C DNA content were immunopositive for NeuN (Table 1; Fig. 2H–J), and ∼3% of NeuN-positive nuclei isolated from the telencephalon of the posthatch chick contained a double amount of DNA (Table 1).
Tetraploid neurons in the murine cerebral cortex respond to environmental signals
The immediate early genes (IEG) Erg-1 and c-fos have been extensively used as markers for functional activity in neurons (Kaczmarek and Chaudhuri, 1997; Knapska and Kaczmarek, 2004), including aneuploid projecting neurons (Kingsbury et al., 2005). These IEGs are rapidly and transiently expressed in response to a variety of stimuli, including ongoing synaptic activity in the adult brain (Loebrich and Nedivi, 2009). To determine whether tetraploid neurons can express these markers, nuclei isolated from the cerebral cortex of 2-month-old mice were double labeled for NeuN and either Erg-1 (Fig. 4A) or c-fos (data not shown), labeled with PI, and then subjected to flow cytometric analysis discarding doublets and analyzing ploidy as described above. Figure 4, B–E, illustrates the gating procedure for identifying the different nuclear populations based on the expression of NeuN and Erg-1. For this analysis controls with only secondary antibodies, or secondary antibodies with each primary antibody, were used for determination of the threshold for each signal (Fig. 4B–D). DNA content analysis for each of the different populations demonstrated that 88.50 ± 1.76% (mean ± SEM; n = 12) of the tetraploid neurons expressed Egr-1 above background (Fig. 4G). This percentage was slightly higher than the proportion of diploid neurons showing Egr-1 expression (80.25 ± 3.56%; mean ± SEM; n = 7; p < 0.005, Student's t test) (Fig. 4G). Likewise, a similar analysis performed with antibodies against c-Fos demonstrated that 89.67 ± 6.40% (mean ± SEM; n = 3) of the tetraploid neurons expressed this IEG above background while c-Fos was detected only in 85.69 ± 5.81% (mean ± SEM; n = 3) of diploid neurons. Overall, these results indicate that tetraploid neurons are functionally active and can respond to environmental signals in the adult cerebral cortex.
Figure 4.
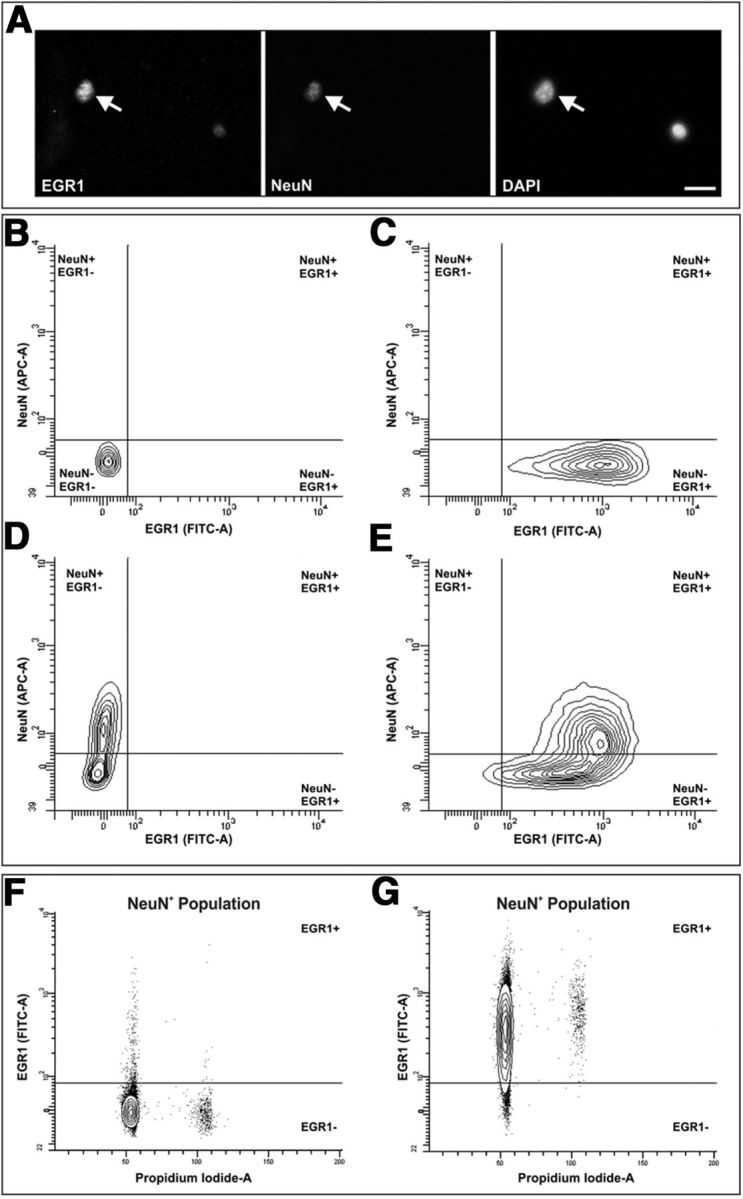
Analysis of Erg-1 expression in NeuN-positive cortical cell nuclei. A, Cell nuclei isolated from the cerebral cortex of 2-month-old mice coimmunostained with anti-Erg-1 (EGR1) and anti-NeuN (NeuN) antibodies, and counterstained with DAPI. Arrow, An NeuN/Erg-1-double positive nucleus. B, Egr-1 and NeuN intensity signals for control nuclei lacking primary antibodies. C–E, Egr-1 and NeuN intensity signals for nuclei labeled with anti-Egr-1 (C), anti-NeuN (D), or both (E) antibodies. Equal amounts of secondary antibodies were present in all situations illustrated in B–E. F, G, DNA content (Propidium Iodide-A) in cell nuclei immunostained either with anti-NeuN (F) or with anti-NeuN and anti-Egr-1 (G), plotted against Egr-1. Scale bar, 10 μm.
Most tetraploid neurons in the mouse cerebral cortex express CTIP2
To get insight into the identity of the tetraploid neurons present in the mouse cerebral cortex, and whether they constitute a population of long-range projection neurons, as it occurs in the retina (Morillo et al., 2010), flow cytometric analyses were performed after immunolabeling of isolated cortical cell nuclei with an antibody specific for CTIP2 (Fig. 5A,B). This antibody recognizes a transcription factor specific for subcortical projection neurons present in layers V and VI of the mouse neocortex (Arlotta et al., 2005). This analysis evidenced that ∼65% of tetraploid nuclei showed intense CTIP2 immunolabeling (Table 2; Fig. 5C), indicating that neuronal tetraploidy in the cerebral cortex is largely associated with projection neurons. Moreover, this analysis demonstrated that ∼5% of total CTIP2-positive nuclei were tetraploid at both P0 and 2 months (Table 2; Fig. 5C–E), thus demonstrating that only a subpopulation of projection neurons becomes tetraploid, as observed in the retina where most RGCs show a 2C DNA content in their nuclei (Morillo et al., 2010). These results also indicate that, as in the case of the NeuN-positive nuclei with 4C DNA content, the proportion of CTIP2-positive tetraploid nuclei did not change during adulthood.
Figure 5.
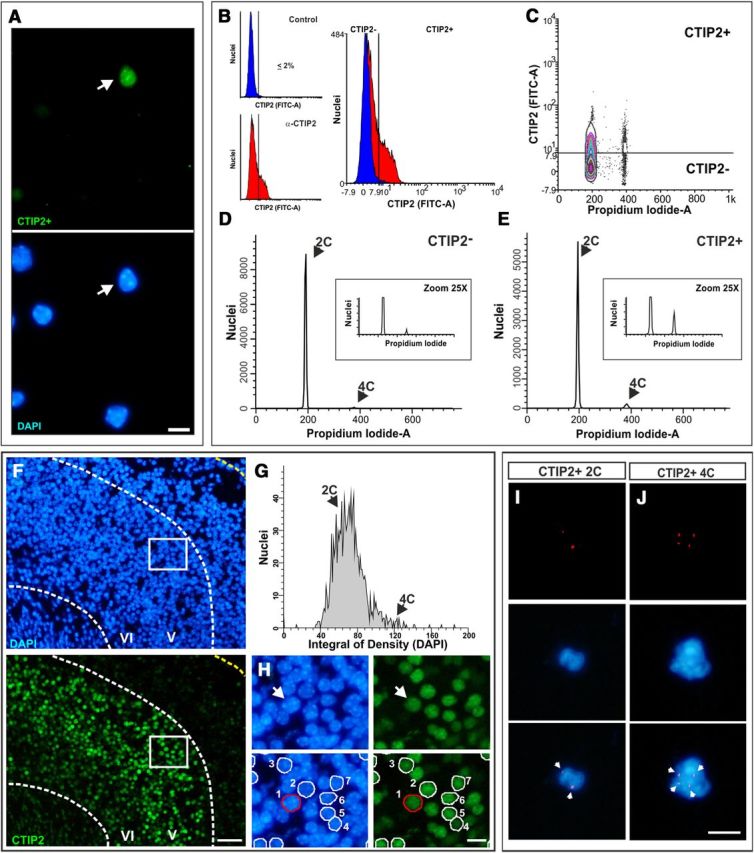
CTIP2-positive neurons with 4C DNA content in the mouse cerebral cortex. A, Cell nuclei isolated from the cerebral cortex of 2-month-old mice immunostained with an anti-CTIP2-specific antibody (green) and counterstained with DAPI (blue). Arrow, A CTIP2-positive nucleus. B, Cell nuclei immunostained with the anti-CTIP2 antibody (α-CTIP2) or with the secondary antibody alone (Control) were PI stained and subjected to flow cytometric analysis. Right, Illustrates the threshold used for the discrimination of CTIP2-positive nuclei. C, DNA content (Propidium Iodide-A) plotted against CTIP2 signal intensity (FITC-A) demonstrates that most nuclei with 4C content are positive for CTIP2 immunolabeling. D, DNA content histogram from CTIP2-negative nuclei. E, DNA content histogram from CTIP2-positive nuclei. See Table 2 for quantitative data. F, Coronal cryosections (15 μm) obtained from cerebral cortex of E17.5 mouse embryos were immunostained with and anti-CTIP2 antibody (green) and labeled with DAPI (blue) for DNA quantification. G, DNA content histogram in CTIP2-positive nuclei obtained by SBC. H, Top, A high magnification of box shown in F. Arrow, Nucleus with 4C DNA amount. Bottom, CTIP2-positive nuclei were used for DNA quantification only in those cases in which DAPI signal was clearly identified as a single nucleus (numbered nuclei). I, J, CTIP2-positive cell nuclei from the cerebral cortex of 2-month-old mice, isolated by FACS, subjected to FISH with a chromosome 11-specific probe (red), and counterstained with DAPI (blue). Arrows, Hybridization spots. Scale bars: A, H, I, J, 10 μm; F, 40 μm.
Table 2.
Percentage of tetraploid nuclei with CTIP2-specific immunolabeling and CTIP2-positive nuclei with tetraploid content in mouse cerebral cortex and chick telencephalon
| Tissue | % CTIP2 + nuclei in the 4C population (mean ± SEM) | % CTIP2 + nuclei with 4C DNA content (mean ± SEM) | n |
|---|---|---|---|
| Mouse neocortex (P0) | 64.78 ± 3.14 | 5.18 ± 0.44 | 11 |
| Mouse neocortex (2 month old) | 63.19 ± 2.52 | 4.79 ± 0.61 | 13 |
| Chick telencephalon (posthatch) | 80.40 ± 2.60 | 4.17 ± 0.22 | 5 |
The presence of CTIP2-positive cell nuclei with double the normal amount of DNA in the cerebral cortex was confirmed by SBC, a method able to reliably quantify DNA in tissue sections (Mosch et al., 2007; Morillo et al., 2010). This procedure provides spatial information about the areas in which CTIP2-positive nuclei showing 4C DNA content can be detected. To this aim, parasagittal cryosections including the cerebral cortex of mouse embryos were immunostained with an anti-CTIP2 antibody and counterstained with DAPI. We used embryos because of the lower background of DAPI staining when compared with that in adult tissues. E17.5 was chosen because at this stage all neurons from cortical layers V–VI and most neurons from other layers have already been generated (Caviness, 1982). Immunostaining with CTIP2 antibodies resulted in a dual pattern characterized by high levels of CTIP2 protein in neurons from layer V, while CTIP2 expression level in layer VI neurons was much lower (Fig. 5F), as previously described by McKenna et al. (2011) for postnatal cortical stages. The relative DNA content in the nuclei of CTIP2-positive neurons was estimated as a direct function of total DAPI intensity levels (see Materials and Methods). Using this approach we could observe CTIP2-positive nuclei with DNA content near 4C (Fig. 5G), located in both layer V (Fig. 5F,H) and VI (data not shown) of the cerebral cortex. Tetraploid cell nuclei were detected in the visual cortex (Fig. 5F–H) as well as in the other cortical areas that were studied such as the prefrontal and motor cortex (data not shown), indicating that tetraploid neurons are widely located throughout the whole cortex. Several areas within the different cortical regions analyzed were observed to contain an elevated proportion of overlapping nuclei, which forced us to exclude them from the analysis. Since we cannot be sure that these regions contain a similar proportion of tetraploid neurons as in the low nuclear density areas, we cannot provide a reliable quantification of neuronal tetraploidy in cortical CTIP2-positive nuclei using the SBC method.
Finally, FISH analysis performed on CTIP2-positive neuronal nuclei isolated by FACS further confirmed the presence of both diploid (Fig. 5I) and tetraploid nuclei (Fig. 5J) in these neurons.
The telencephalon of posthatch chick contains CTIP2-positive tetraploid neurons
Flow cytometry was also performed with cell nuclei isolated from the telencephalic derivatives of posthatch chick to evaluate the presence of tetraploid neurons expressing CTIP2 in the telencephalon of this species (data not shown). This analysis indicated that ∼80% of tetraploid nuclei showed intense immunolabeling for CTIP2 (Table 2). In addition, ∼4% of CTIP2-positive nuclei were observed to show 4C DNA content (Table 2). Therefore, the chick telencephalon has a similar proportion of CTIP2-positive tetraploid neurons as the mouse cerebral cortex.
p75NTR is expressed in the neuroepithelium of the mouse telencephalon during the generation of CTIP2 cortical neurons
We have previously shown that the use of blocking antibodies against p75NTR during the period of RGC genesis is able to reduce the proportion of tetraploid neurons in the developing chick retina (Morillo et al., 2010). In situ hybridization data available at GenePaint (http://www.genepaint.org) (Visel et al., 2004) indicate that the Ngfr gene encoding p75NTR is expressed in the telencephalic neuroepithelium of E10.5 mice (GENEPAINT Set ID MH318), a stage at which generation of layer VI neurons is about to start (Caviness, 1982). These observations suggest that p75NTR participates in the induction of neuronal tetraploidy in the murine cerebral cortex.
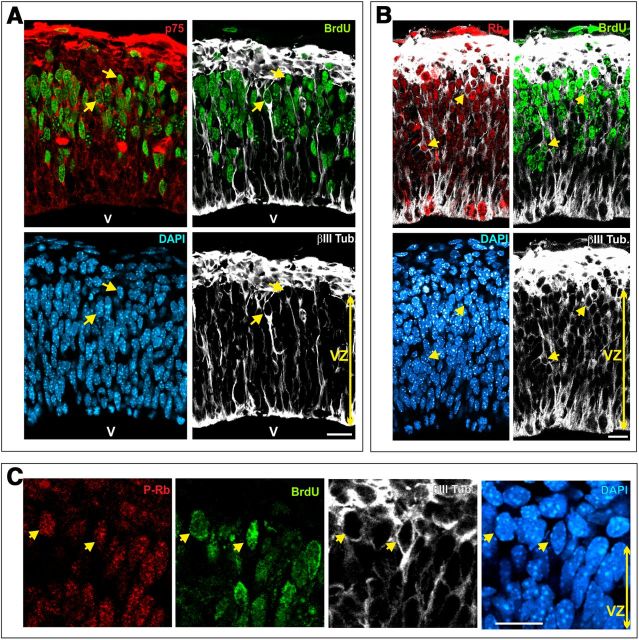
We therefore studied whether p75NTR is expressed in the telencephalic neuroepithelium during the period of generation of neurons from cortical layers V and VI, which in the mouse occurs from E11.5 to E13.5 (Caviness, 1982). To this aim, E12.5 mouse embryos were pulse labeled with BrdU for 30 min, fixed, and then subjected to cryosectioning. Coronal cryosections containing the cerebral cortex were then triple immunostained with specific antibodies against p75NTR, BrdU, and the early neuronal marker βIII-tubulin (Lee et al., 1990). Finally, cell nuclei were then counterstained with DAPI. This analysis demonstrated that p75NTR can be readily detected in scattered cells located within the ventricular zone (Fig. 6A), and that some of these cells show immunoreactivity for BrdU (Fig. 6A, arrows). Interestingly, p75NTR/BrdU-double positive cells are often expressing high levels of the early neuronal marker βIII-tubulin (Fig. 6A), a situation reminiscent of what can be observed during RGC tetraploidization in the chick retina (Morillo et al., 2010). Altogether, these results suggest that p75NTR may participate in the generation of tetraploid neurons in the cerebral cortex.
Figure 6.
p75NTR and Rb expression in mouse cortical newborn neurons that incorporate BrdU. Coronal cryosections (12 μm) from the cerebral cortex of E12.5 mouse labeled with BrdU for 30 min were triple immunostained with anti-βIII-tubulin (βIII tub.) (white), anti-BrdU (green), and either anti-p75NTR (p75) (A), anti-Rb (B), or anti-phospho-Rb (P-Rb) (C) (red) antibodies, and counterstained with DAPI (blue). Arrows, Neuroepithelial cells with high levels of βIII-tubulin immunolabeling coexpressing either p75NTR (A), Rb (B), or phospho-Rb (C) and labeled with the anti-BrdU antibody. V, ventricle; VZ, ventricular zone. Scale bars: A, B, 20 μm; C, 15 μm.
p75NTR-specific staining could also be detected in a population of CTIP2-positive postmitotic neurons, as evidenced by double immunostaining performed in sections from cerebral cortex of E12.5, E14.5, and E16.5 mouse embryos using antibodies specific for both p75NTR and CTIP2. At E12.5 only a small number of CTIP2-positive nuclei could be observed at the marginal zone, most of them being positive for p75NTR (Fig. 7A). At E14.5 and E16.5, p75NTR was observed in a subpopulation of CTIP2-positive cells (Fig. 7B,C). In contrast with the strong p75NTR labeling during development, the cerebral cortex of 1-month-old mice showed only background staining (Fig. 7D). In contrast, p75NTR was clearly observed in the basal forebrain of 1-month-old mice (Fig. 7E), as previously described by others (Yan and Johnson, 1989).
Figure 7.
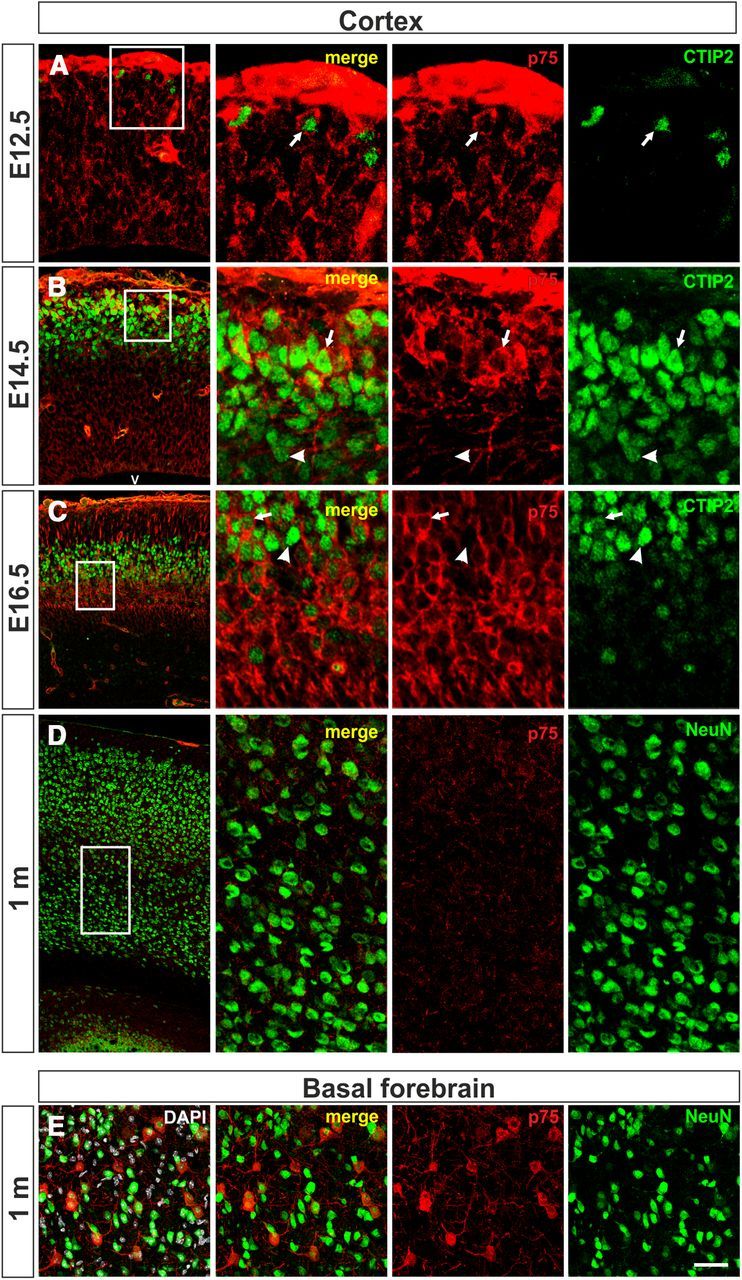
Expression of p75NTR in CTIP2-positive neurons. Coronal cryosections (12 μm) from the cerebral cortex (A–D) or basal forebrain (E) of mouse at the indicated ages were immunostained with anti-p75NTR (red) and anti-CTIP2 (green) antibodies. Left, Rectangles represent the area illustrated in middle and right parts. Arrows, CTIP2-positive neurons expressing p75NTR; arrowheads, CTIP2-positive neurons lacking p75NTR expression. V, ventricle. Scale bar: A–C left, 30 μm; A middle and right, 50 μm; B, C middle and right, 20 μm; D left, 200 μm; D middle and right, 60 μm; E, 60 μm.
Cell cycle-reactivating cortical neurons express Rb
In contrast with the chick retina, where RGCs becoming tetraploid lack retinoblastoma (Rb) protein (Morillo et al., 2010), differentiating cortical neurons that reactivate the cell cycle (i.e., βIII-tubulin-positive, BrdU-positive cells) were observed to express Rb (Fig. 6B). Therefore, the mechanism regulating neuronal tetraploidization in cortical neurons seems to differ from that observed in the chick retina. As expected from their capacity to incorporate BrdU, these neurons also showed immunoreactivity for Rb phosphorylated at Ser 795 (Fig. 6C), the preferred phosphorylation site of cdk4/cyclin D1 in the molecule (Pan et al., 1998). This observation is consistent with the known expression of cdk4 and cyclin D1 by cortical neurons (Sumrejkanchanakij et al., 2003).
p75NTR-positive subplate neurons (SPNs) are diploid
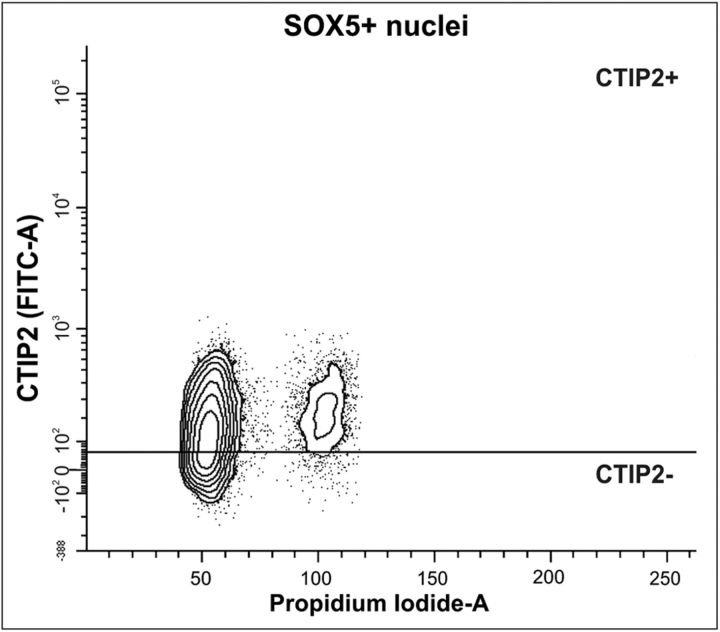
SPNs constitute a transient population of the first neurons that are born in the mouse cerebral cortex (McQuillen et al., 2002). These neurons express p75NTR (Allendoerfer et al., 1990), suggesting that they may be susceptible to become tetraploid during development, as it occurs with the RGCs (Morillo et al., 2010) and the CTIP2-positive cortical neurons (this study). SPNs are known to express Sox5 (Kwan et al., 2008; Kanold and Luhmann, 2010), a nuclear marker that can also be observed in layer V/VI neurons (Kwan et al., 2008). Therefore, we have taken advantage of this observation to identify the SPNs as those cortical cells expressing Sox5 but lacking CTIP2-specific immunostaining. By using this criterion we were able to analyze whether a subpopulation of SPNs shows 4C DNA content. To this aim, fresh cell nuclei were isolated from the cerebral cortex of P0 mice, a stage in which SPNs are still present (McQuillen et al., 2002). These cell nuclei were then immunolabeled with both anti-Sox5 and anti-CTIP2 antibodies, and subjected to flow cytometric analysis. This study demonstrated that the nuclei of the SPNs (i.e., Sox5-positive/CTIP2-negative population) were diploid, whereas layer V/VI neurons (i.e., Sox5-positive/CTIP2-positive population) contained both diploid and tetraploid nuclei (Fig. 8). We concluded that neuronal tetraploidy cannot be generalized to all neuronal populations that express p75NTR. Moreover, this result further stresses the specificity of our flow cytometric procedure, able to discriminate the presence of tetraploidy between different neuronal populations.
Figure 8.
Analysis of tetraploidy in SPNs. Cell nuclei isolated from the cerebral cortex of P0 mice were coimmunostained with anti-Sox5 and anti-CTIP2 antibodies, and subjected to flow cytometric analysis after labeling with PI. DNA content (Propidium Iodide-A) plotted against CTIP2 signal intensity (CTIP2 FITC-A) was evaluated in the Sox5-positive population. The horizontal line represents the threshold for CTIP2-specific staining. The Sox5-positive/CTIP2-negative population (i.e., SPNs) mainly contains diploid neurons.
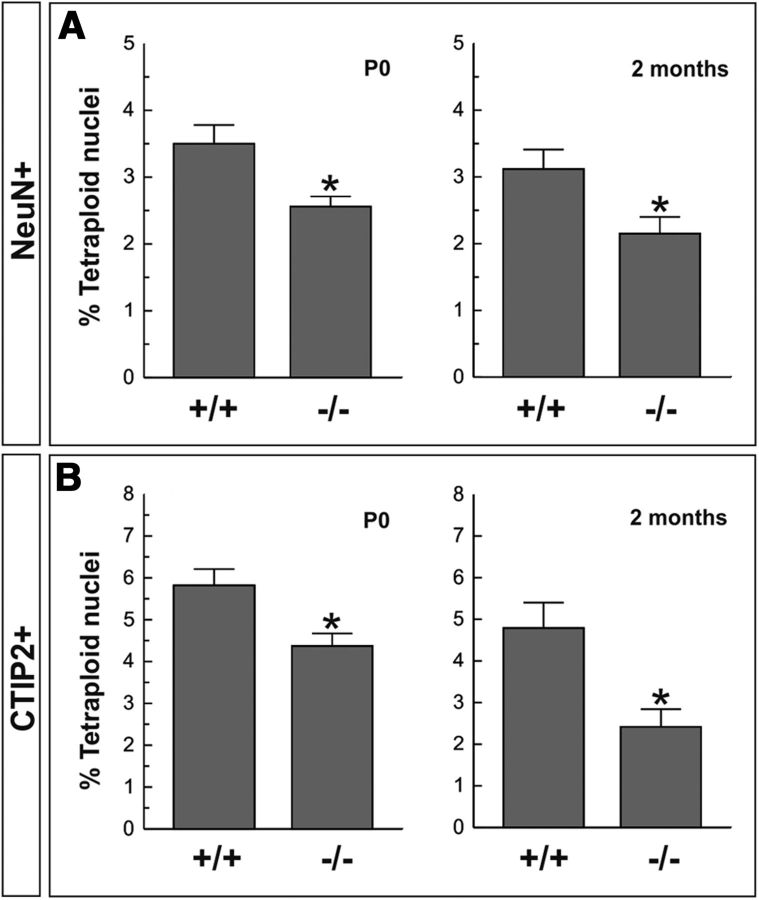
Neuronal tetraploidy is reduced in the cerebral cortex of p75NTR−/− mice
To directly study whether p75NTR participates in the creation of tetraploidy in murine cortical neuron, cell nuclei were isolated from the cerebral cortex of wild-type and p75NTR−/− littermates at both P0 and 2 months, and then they were immunostained with either anti-NeuN or anti-CTIP2 antibodies, followed by DNA labeling with PI. Flow cytometric analysis demonstrated that the proportion of NeuN-positive nuclei showing 4C DNA levels was significantly reduced in the p75NTR−/− mice compared with their wild-type littermates at P0, when the neurogenesis is just finished (Caviness, 1982), and in the adult mouse (Fig. 9A). Our study demonstrated that this is also the case for the subpopulation of CTIP2-positive neurons with 4C DNA content, since the proportion of these neurons is significantly reduced in the p75NTR−/− cerebral cortex compared with the cerebral cortex of the wild-type littermates at both P0 and 2 months (Fig. 9B).
Figure 9.
Percentage of tetraploid neurons in the cerebral cortex of wild-type and p75NTR−/− mice. A, Quantification of neuronal tetraploidy in the cerebral cortex of P0 and 2-month-old mice. The levels of tetraploidy in NeuN-positive nuclei were reduced in the cerebral cortex of p75NTR−/− mice. B, Quantification of neuronal tetraploidy in the cerebral cortex of P0 and 2-month-old mice. The levels of tetraploidy in CTIP2-positive nuclei were reduced in the cerebral cortex of p75NTR−/− mice. *p < 0.05 (Student's t test).
Tetraploidy is enriched in long-term projection neurons from the striatum
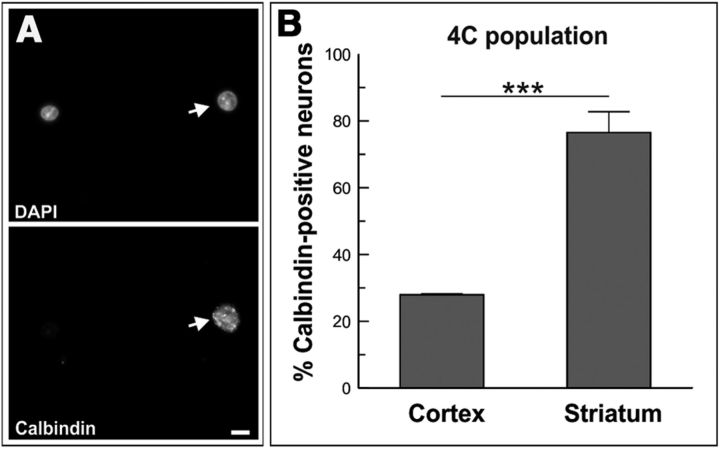
To confirm that tetraploidy is mainly associated with long-range projection neurons in other brain structures we focused on the neostriatal-matrix spiny neurons. These neurons establish long-range projections to substantia nigra and globus pallidus (Kawaguchi, 1997) and they express both CTIP2 (Arlotta et al., 2008) and calbindin (Kawaguchi, 1997), a calcium-binding protein specifically expressed in subsets of adult neurons (Liu and Graybiel, 1992). This contrasts with the neocortex, where calbindin is predominant in neurons from layers II–III (Hof et al., 1999), a structure containing cortical interneurons and short-range, corticocortical projection neurons. The cytoplasmic localization of calbindin forced us to perform flow cytometric analyses similar to those described by Morillo et al. (2010) for retinal neurons. To this aim, cells were dissociated from both cerebral cortex and striatum of adult mice (3- to 3.5-month-old mice). These cells were then ethanol fixed and immunostained with an anti-calbindin-specific antibody before labeling their DNA with PI. As expected, ∼80% of tetraploid striatal cells were observed to express calbindin (Fig. 10) whereas only ∼20% of tetraploid cortical cells showed immunoreactivity for this calcium-binding protein. Therefore, neuronal tetraploidy seems to be associated with long-range projection neurons also in the striatum. These results, along with our previous observations in the retina (Morillo et al., 2010), suggest that neuronal tetraploidy predominates in subpopulations of large projection neurons throughout the whole vertebrate nervous system.
Figure 10.
Enrichment of tetraploidy in calbindin-positive neurons in the mouse striatum. A, Cells isolated from the striatum of adult mice immunostained with an anti-calbindin-specific antibody and counterstained with DAPI. Arrow, A calbindin-positive neuron. B, Neurons immunostained with the anti-calbindin antibody, isolated from the cerebral cortex (Cortex) or striatum (Striatum) from P0 mice, were stained with PI and subjected to flow cytometry. The percentage of calbindin-positive neurons in the 4C population is shown for each tissue. ***p < 0.005 (Student's t test). Scale bar, 10 μm.
Discussion
We have optimized a flow cytometric method for DNA quantification in adult neurons, based on the analysis of unfixed isolated nuclei. Using this procedure we have demonstrated that the mouse cerebral cortex contains a small population of functionally active, tetraploid neurons. A substantial proportion of these neurons express CTIP2, a zinc finger transcription factor that specifies cortical neurons from layer V–VI projecting to subcortical targets (Chen et al., 2008). These observations, confirmed by SBC and FISH, were extended to the chick telencephalon, suggesting an evolutionarily conserved mechanism for the generation of CTIP2-positive tetraploid neurons. Tetraploid neurons are likely to be generated during early stages of cortical development through a p75NTR-dependent mechanism, thus explaining the reduction of tetraploid neurons in the cerebral cortex of p75NTR−/− mice. Finally, in the striatum neuronal tetraploidy is largely associated with neurons that project to long distance targets, further supporting the hypothesis that tetraploidy in the nervous system mainly takes place in subpopulations of long-range projection neurons.
So far, the analysis of adult brain cells by flow cytometry has been hindered by intrinsic difficulties of this tissue derived from the interconnection of neural cells and the existence of myelin debris, which impairs the recovery of single cell suspensions and increases the background. In contrast, the use of unfixed cell nuclei for flow cytometry has several advantages compared with previous procedures (Morillo et al., 2010; Westra et al., 2010) as it reduces tissue dissociation difficulties, gives better analytical resolution, and reduces the presence of artifacts. Due to the purity of the nuclear preparations, our procedure facilitates a reliable and quantitative analysis whenever a specific nuclear marker is available. Moreover, our method allows representative and reproducible neuronal sampling, thus resulting in strong statistical analyses. Our method, which discriminates actual tetraploid nuclei from diploid nuclear doublets, has demonstrated the existence of tetraploid neurons in both murine and avian telencephalic derivates. The specificity of our analysis is substantiated by the differential proportion of tetraploid neurons observed in cortical nuclei from wild-type mice versus p75NTR−/− mice.
The majority of cortical nuclei with 4C DNA content, both in mouse and in chicken, were observed to express NeuN, indicating that like in the retina (Morillo et al., 2010), most tetraploid cells are neurons. This observation agrees with previous studies showing tetraploid neurons in the human cerebral cortex (Mosch et al., 2007; Arendt et al., 2010). This also agrees with the study by Westra et al. (2009), who observed that, in humans, 17–48% of frontal cortical cells with 4C DNA content express neuronal markers. Surprisingly, these latter authors failed to observe neuronal markers in nuclei with duplicated pairs of chromosomes (Westra et al., 2009), an observation that contrasts with previous results from this same laboratory demonstrating that ∼1.1% of cortical and hippocampal neurons are tetrasomic for chromosome 21 (Rehen et al., 2005). Tetrasomic nuclei have also been detected in human cortical neurons (Mosch et al., 2007), and our results indicate that CTIP2-positive nuclei from the mouse cortex can contain four hybridization spots as revealed by FISH. Our results also indicate that a small proportion of tetraploid nuclei lacks NeuN-specific immunostaining. These cell nuclei could correspond to glial cells, in accordance with the presence of tetraploid and polyploid glia in humans (Westra et al., 2009) and Drosophila (Unhavaithaya and Orr-Weaver, 2012), respectively.
It is likely that the cortical neurons becoming tetraploid during development remain viable during adulthood since the same proportion of neurons with 4C DNA content were observed at P0 and 2 months. This view is consistent with the absence of dying cells in the cerebral cortex of 1-month-old mice, as previously observed by Angata et al. (2007). Furthermore, no evidence was found for the existence of cortical neurons undergoing S/G2 phase in Fucci mice, in agreement with previous studies in the cerebral cortex of adult mouse (Magavi et al., 2000) and rat (Dayer et al., 2005), thus ruling out a hypothetical turnover of dying tetraploid neurons being substituted by cortical neurons that duplicate their DNA. The existence of viable tetraploid neurons in the cerebral cortex was further substantiated by the observation that most tetraploid neurons express Erg-1 and c-Fos, two IEGs expressed in response to neuronal activity (Kaczmarek and Chaudhuri, 1997; Knapska and Kaczmarek, 2004). Therefore, tetraploid neurons appear to be functional and, probably, integrated into the brain circuits as occurs with aneuploid neurons (Kingsbury et al., 2005). The capacity of tetraploid neurons to integrate in brain circuits and innervate specific target areas is further substantiated by the experiments performed by Tompkins et al. (1984) using diploid/tetraploid frog chimeras. These authors demonstrated that the RGCs from a tetraploid eye grafted into a diploid frog can innervate broader regions of the target tissue as compared with the RGCs from the original diploid eye. Our data also indicate that a small proportion of both tetraploid and diploid neurons lack Erg-1 and c-Fos expression, an observation consistent with the variable expression of this transcription factor in the cerebral cortex (Loebrich and Nedivi, 2009).
Tetraploidy is usually associated with increased cell size (Edgar and Orr-Weaver, 2001; Ullah et al., 2009). Indeed, tetraploid neurons are known to contain larger cell somas, dendritic arbors, and innervation areas than the diploid counterparts (Tompkins et al., 1984; Szaro and Tompkins, 1987; Morillo et al., 2010). This increase in size likely facilitates the innervation of distant targets. Our results support the hypothesis that neuronal tetraploidy affects mainly to large projection neurons, usually born at initial stages of development. This is the case of the RGCs, the first neurons to be born in the retina (Sidman, 1961; Prada et al., 1991), which contains a subpopulation becoming tetraploid during development (Morillo et al., 2010). As in the case of the RGCs, neuronal tetraploidization in the cerebral cortex also affects to a subpopulation of early born neurons: the pyramidal cells from cortical layers V–VI (Caviness, 1982), but not to the SPNs.
We have demonstrated that p75NTR participates in neuronal tetraploidization in the cerebral cortex since the proportion of tetraploid nuclei expressing NeuN or CTIP2 becomes reduced in the cortex of 75NTR−/− mice. This observation is consistent with the expression of p75NTR by neuroepithelial cells at early stages of cortical development, when pyramidal cells from layer V–VI are born (Caviness, 1982). Some of these p75NTR-positive neuroepithelial cells are newborn neurons since they express the early neuronal marker βIII-tubulin at high levels. Importantly, a subpopulation of p75NTR-positive, newborn neurons were able to incorporate BrdU after a short 30 min pulse, suggesting that they were undergoing neuronal tetraploidization, as previously shown for tetraploid RGCs (Morillo et al., 2010). p75NTR-specific immunoreactivity was detected in a subpopulation of CTIP2-positive neurons at E14.5–E16.5, when these neurons project to their targets (De Carlos and O'Leary, 1992), extending previous observations by McQuillen et al. (2002), who reported that SPNs express this receptor. This raises the possibility that p75NTR could play a role in axonal projection in these neurons, as previously described to occur in differentiated RGCs (Yamashita et al., 1999). Only background levels of p75NTR were detected in the cerebral cortex of 1-month-old mice, as previously shown (Wang et al., 2011), suggesting that p75NTR is dispensable for long-lasting maintenance of subcortical projections.
Interestingly, a population of tetraploid neurons remain in the cerebral cortex of p75NTR−/− mice, like in the retina of chick embryos treated with blocking antibodies against p75NTR (Morillo et al., 2010). This suggests the existence of alternative mechanisms involved in the generation of tetraploid neurons, which may act synergistically with p75NTR to trigger neuronal tetraploidization in the normal and pathological nervous system (Frade and López-Sánchez, 2010).
Unlike in the chick retina (Morillo et al., 2010), newborn cortical neurons reactivating the cell cycle express Rb. In these neurons Rb was phosphorylated at Ser795, a known substrate for cdk4/cyclin D1 (Pan et al., 1998). This suggests that cell cycle reactivation in differentiating cortical neurons relies on the inhibition of Rb function. This hypothesis is consistent with the observations that differentiating cortical neurons incorporate BrdU in conditional Rb−/− mice (Ferguson et al., 2002) and that Rb−/− cortical cells become tetraploid in wild-type/Rb−/− chimeric mice (Lipinski et al., 2001); both effects are compatible with neuronal survival. Rb phosphorylation at Ser795 in differentiating cortical neurons is consistent with the expression of cyclin D1 and cdk4 in these cells (Sumrejkanchanakij et al., 2003). Nevertheless, phosphorylation of Rb in these neurons might also be triggered by p38MAPK (Wang et al., 1999), JNK (Chauhan et al., 1999), or ERK (Guo et al., 2005), which are known to be activated by p75NTR (Casaccia-Bonnefil et al., 1996; Susen et al., 1999; Morillo et al., 2012).
In sum, we provide evidence that somatic tetraploidization represent a common mechanism creating neuronal variability in the normal nervous system of vertebrates. This mechanism, which in part depends on p75NTR signaling, is mainly associated with long-range projection neurons, such as RGCs, pyramidal cells from cortical layers V-VI, as well as neostriatal-matrix spiny neurons. Further studies will be required to fully understand the actual mechanism leading to cell cycle reactivation and tetraploidization in cortical neurons.
Footnotes
This study was supported by grants from the “Ministerio de Ciencia e Innovación” (BFU2009–07671 and SAF2012-38316) and “Fundación Areces” (CIVP16A1815). N.L.-S. acknowledges a JAE-Doc contract (JAEDoc026, 2008 call) from the CSIC program “Junta para la Ampliación de Estudios” cofunded by the European Social Fund. We thank M.V. Chao for the antibody [9650], J.A. Barbas and A.V. Morales for the anti-Sox5 antibody, and M. Nieto for discussion about this work. The monoclonal antibody G3G4 (S. Kaufman) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa).
The authors declare no competing financial interests.
References
- Allendoerfer KL, Shelton DL, Shooter EM, Shatz CJ. Nerve growth factor receptor immunoreactivity is transiently associated with the subplate neurons of the mammalian cerebral cortex. Proc Natl Acad Sci U S A. 1990;87:187–190. doi: 10.1073/pnas.87.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata K, Huckaby V, Ranscht B, Terskikh A, Marth JD, Fukuda M. Polysialic acid-directed migration and differentiation of neural precursors are essential for mouse brain development. Mol Cell Biol. 2007;27:6659–6668. doi: 10.1128/MCB.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T, Brückner MK, Mosch B, Lösche A. Selective cell death of hyperploid neurons in Alzheimer's disease. Am J Pathol. 2010;177:15–20. doi: 10.2353/ajpath.2010.090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Carter BD, Dobrowsky RT, Chao MV. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature. 1996;383:716–719. doi: 10.1038/383716a0. [DOI] [PubMed] [Google Scholar]
- Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- Chauhan D, Hideshima T, Treon S, Teoh G, Raje N, Yoshihimito S, Tai YT, Li W, Fan J, DeCaprio J, Anderson KC. Functional interaction between retinoblastoma protein and stress-activated protein kinase in multiple myeloma cells. Cancer Res. 1999;59:1192–1195. [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer AG, Cleaver KM, Abouantoun T, Cameron HA. New GABAergic interneurons in the adult neocortex and striatum are generated from different precursors. J Cell Biol. 2005;168:415–427. doi: 10.1083/jcb.200407053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlos JA, O'Leary DD. Growth and targeting of subplate axons and establishment of major cortical pathways. J Neurosci. 1992;12:1194–1211. doi: 10.1523/JNEUROSCI.12-04-01194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/S0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Ferguson KL, Vanderluit JL, Hébert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM. Unscheduled re-entry into the cell cycle induced by NGF precedes cell death in nascent retinal neurones. J Cell Sci. 2000;113:1139–1148. doi: 10.1242/jcs.113.7.1139. [DOI] [PubMed] [Google Scholar]
- Frade JM, Barde YA. Genetic evidence for cell death mediated by nerve growth factor and the neurotrophin receptor p75 in the developing mouse retina and spinal cord. Development. 1999;126:683–690. doi: 10.1242/dev.126.4.683. [DOI] [PubMed] [Google Scholar]
- Frade JM, López-Sánchez N. A novel hypothesis for Alzheimer disease based on neuronal tetraploidy induced by p75NTR. Cell Cycle. 2010;9:1934–1941. doi: 10.4161/cc.9.10.11582. [DOI] [PubMed] [Google Scholar]
- Guo J, Sheng G, Warner BW. Epidermal growth factor-induced rapid retinoblastoma phosphorylation at Ser780 and Ser795 is mediated by ERK1/2 in small intestine epithelial cells. J Biol Chem. 2005;280:35992–35998. doi: 10.1074/jbc.M504583200. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- Hof PR, Glezer II, Condé F, Flagg RA, Rubin MB, Nimchinsky EA, Vogt Weisenhorn DM. Cellular distribution of the calcium-binding proteins parvalbumin, calbindin, and calretinin in the neocortex of mammals: phylogenetic and developmental patterns. J Chem Neuroanat. 1999;16:77–116. doi: 10.1016/S0891-0618(98)00065-9. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L, Chaudhuri A. Sensory regulation of immediate-early gene expression in mammalian visual cortex: implications for functional mapping and neural plasticity. Brain Res Brain Res Rev. 1997;23:237–256. doi: 10.1016/S0165-0173(97)00005-2. [DOI] [PubMed] [Google Scholar]
- Kanold PO, Luhmann HJ. The subplate and early cortical circuits. Annu Rev Neurosci. 2010;33:23–48. doi: 10.1146/annurev-neuro-060909-153244. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. San Diego: Academic; 1992. [Google Scholar]
- Kawaguchi Y. Neostriatal cell subtypes and their functional roles. Neurosci Res. 1997;27:1–8. doi: 10.1016/S0168-0102(96)01134-0. [DOI] [PubMed] [Google Scholar]
- Kimbrough A, Kwon B, Eckel LA, Houpt TA. Systemic 5-bromo-2-deoxyuridine induces conditioned flavor aversion and c-Fos in the visceral neuraxis. Learn Mem. 2011;18:292–295. doi: 10.1101/lm.2176611. [DOI] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci U S A. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Li E, Huber LJ, Landis SC, Sharpe AH, Chao MV, Jaenisch R. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell. 1992;69:737–749. doi: 10.1016/0092-8674(92)90286-L. [DOI] [PubMed] [Google Scholar]
- Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Macleod KF, Williams BO, Mullaney TL, Crowley D, Jacks T. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J. 2001;20:3402–3413. doi: 10.1093/emboj/20.13.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FC, Graybiel AM. Transient calbindin-D28k-positive systems in the telencephalon: ganglionic eminence, developing striatum and cerebral cortex. J Neurosci. 1992;12:674–690. doi: 10.1523/JNEUROSCI.12-02-00674.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89:1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez N, Frade JM. Control of the cell cycle by neurotrophins: lessons from the p75 neurotrophin receptor. Histol Histopathol. 2002;17:1227–1237. doi: 10.14670/HH-17.1227. [DOI] [PubMed] [Google Scholar]
- López-Sánchez N, Ovejero-Benito MC, Borreguero L, Frade JM. Control of neuronal ploidy during vertebrate development. Results Probl Cell Differ. 2011;53:547–563. doi: 10.1007/978-3-642-19065-0_22. [DOI] [PubMed] [Google Scholar]
- Magavi SS, Leavitt BR, Macklis JD. Induction of neurogenesis in the neocortex of adult mice. Nature. 2000;405:951–955. doi: 10.1038/35016083. [DOI] [PubMed] [Google Scholar]
- Massa SM, Xie Y, Yang T, Harrington AW, Kim ML, Yoon SO, Kraemer R, Moore LA, Hempstead BL, Longo FM. Small, nonpeptide p75NTR ligands induce survival signaling and inhibit proNGF-induced death. J Neurosci. 2006;26:5288–5300. doi: 10.1523/JNEUROSCI.3547-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillen PS, DeFreitas MF, Zada G, Shatz CJ. A novel role for p75NTR in subplate growth cone complexity and visual thalamocortical innervation. J Neurosci. 2002;22:3580–3593. doi: 10.1523/JNEUROSCI.22-09-03580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SM, Escoll P, de la Hera A, Frade JM. Somatic tetraploidy in specific chick retinal ganglion cells induced by nerve growth factor. Proc Natl Acad Sci U S A. 2010;107:109–114. doi: 10.1073/pnas.0906121107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SM, Abanto EP, Román MJ, Frade JM. NGF-induced cell cycle reentry in newborn neurons is triggered by p38MAPK-dependent E2F4 phosphorylation. Mol Cell Biol. 2012;32:2722–2737. doi: 10.1128/MCB.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A, Arendt T. Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. J Neurosci. 2007;27:6859–6867. doi: 10.1523/JNEUROSCI.0379-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez R. DNA measurement and cell cycle analysis by flow cytometry. Curr Issues Mol Biol. 2001;3:67–70. [PubMed] [Google Scholar]
- Pan W, Sun T, Hoess R, Grafstrom R. Defining the minimal portion of the retinoblastoma protein that serves as an efficient substrate for cdk4 kinase/cyclin D1 complex. Carcinogenesis. 1998;19:765–769. doi: 10.1093/carcin/19.5.765. [DOI] [PubMed] [Google Scholar]
- Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- Prada C, Puga J, Pérez-Méndez L, López R, Ramírez G. Spatial and temporal patterns of neurogenesis in the chick retina. Eur J Neurosci. 1991;3:559–569. doi: 10.1111/j.1460-9568.1991.tb00843.x. [DOI] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sidman RL. Histogenesis of the mouse retina studied with [3H]thymidine. In: Smelser GK, editor. The structure of the eye. London: Academic; 1961. pp. 487–506. [Google Scholar]
- Sumrejkanchanakij P, Tamamori-Adachi M, Matsunaga Y, Eto K, Ikeda MA. Role of cyclin D1 cytoplasmic sequestration in the survival of postmitotic neurons. Oncogene. 2003;22:8723–8730. doi: 10.1038/sj.onc.1206870. [DOI] [PubMed] [Google Scholar]
- Susen K, Heumann R, Blöchl A. Nerve growth factor stimulates MAPK via the low affinity receptor p75LNTR. FEBS Lett. 1999;463:231–234. doi: 10.1016/S0014-5793(99)01628-2. [DOI] [PubMed] [Google Scholar]
- Szaro BG, Tompkins R. Effect of tetraploidy on dendritic branching in neurons and glial cells of the frog, Xenopus laevis. J Comp Neurol. 1987;258:304–316. doi: 10.1002/cne.902580210. [DOI] [PubMed] [Google Scholar]
- Tompkins R, Szaro B, Reinschmidt D, Kaye C, Ide C. Effects of alterations of cell size and number on the structure and function of the Xenopus laevis nervous system. Adv Exp Med Biol. 1984;181:135–146. doi: 10.1007/978-1-4684-4868-9_11. [DOI] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell Cycle. 2009;8:1501–1509. doi: 10.4161/cc.8.10.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Nath N, Minden A, Chellappan S. Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. EMBO J. 1999;18:1559–1570. doi: 10.1093/emboj/18.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang X, Lu JJ, Li QX, Gao CY, Liu XH, Sun Y, Yang M, Lim Y, Evin G, Zhong JH, Masters C, Zhou XF. p75NTR regulates Aβ deposition by increasing Aβ production but inhibiting Aβ aggregation with its extracellular domain. J Neurosci. 2011;31:2292–2304. doi: 10.1523/JNEUROSCI.2733-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Barral S, Chun J. A reevaluation of tetraploidy in the Alzheimer's disease brain. Neurodegener Dis. 2009;6:221–229. doi: 10.1159/000236901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/S0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Yan Q, Johnson EM., Jr Immunohistochemical localization and biochemical characterization of nerve growth factor receptor in adult rat brain. J Comp Neurol. 1989;290:585–598. doi: 10.1002/cne.902900411. [DOI] [PubMed] [Google Scholar]