Abstract
Mutation in the GRN gene, encoding the progranulin (PGRN) protein, shows a dose-dependent disease correlation, wherein haploinsufficiency results in frontotemporal lobar degeneration (FTLD) and complete loss results in neuronal ceroid lipofuscinosis (NCL). Although the exact function of PGRN is unknown, it has been increasingly implicated in lysosomal physiology. Here we report that PGRN interacts with the lysosomal enzyme, glucocerebrosidase (GCase), and is essential for proper GCase activity. GCase activity is significantly reduced in tissue lysates from PGRN-deficient mice. This is further evidence that reduced lysosomal hydrolase activity may be a pathological mechanism in cases of GRN-related FTLD and NCL.
Introduction
Progranulin (PGRN), encoded by the GRN gene in humans, is a glycoprotein comprised of 7.5 conserved and highly disulfide-bonded homologous granulin domains connected by short linker regions [1–6]. While the exact function of PGRN remains elusive, it has been found to be involved in numerous normal physiologic and pathologic processes, including regulation of inflammation, wound healing, and tumorigenesis, and it has also been shown to function as a growth and neurotrophic factor [7–16]. Mutation in GRN has been linked to two neurodegenerative diseases with dose-dependent associations. Heterozygous mutation, resulting in PGRN haploinsufficiency, is known to cause frontotemporal lobar degeneration (FTLD), a clinically and pathologically heterogeneous disease often resulting in early-onset dementia [17–23]. Homozygous mutation, resulting in complete loss of PGRN, causes neuronal ceroid lipofuscinosis (NCL), a lysosomal storage disease [24, 25]. Although these two diseases are distinct, NCL-related phenotypes have been reported in FTLD patients with GRN mutation, suggesting that lysosomal dysfunction might serve as a common mechanism [26–28].
In addition to the connection to NCL, increasing lines of evidence indicate that PGRN is likely to be involved in lysosomal physiology. In addition to being lysosomally localized, having two independent trafficking pathways to the lysosome [29, 30], and being under transcriptional regulation with the majority of known lysosomal proteins [31], PGRN has recently been shown to be proteolytically processed to produce stable and functional granulin peptides in the lysosome [32–34], and is linked to the direct or indirect regulation of two lysosomal hydrolases, cathepsin D (CTSD) and glucocerebrosidase (GCase, encoded by the GBA gene). PGRN was shown to stabilize and directly modulate the activity of cathepsin D in vitro and PGRN deficiency results in a reduction of cathepsin D activity in vivo [28, 35, 36]. In another study, PGRN was purported to be a co-chaperone of glucocerebrosidase (GCase) [37, 38], a β-glucosidase mutated in Gaucher disease, in tandem with heat shock protein 70 (HSP70) [39]. Grn knockout mice with induced chronic inflammation display cellular phenotypes similar to those with Gaucher disease [37, 38].
To better understand the lysosomal role of PGRN, we performed a stable isotope labeling by amino acids in cell culture (SILAC)-based proteomic screen for PGRN protein interactors. Corresponding with the previous findings, one of the top hits was GCase. In this study, we demonstrate that PGRN loss results in a substantial decrease in GCase activity in mouse tissues without any changes in protein levels.
Material and methods
Primary antibodies and reagents
The following antibodies were used in this study: M2 mouse anti-FLAG (Sigma), M2 mouse anti-FLAG-conjugated beads (Sigma), mouse anti-myc (9E10), mouse anti-GAPDH (Proteintech Group), mouse anti-GBA (MilliporeSigma), rat anti-mouse LAMP1 (BD Biosciences), rabbit anti-calnexin (Abcam) and sheep anti-mouse PGRN (R&D Systems). Rabbit anti-mouse PSAP and PGRN antibodies were produced as previously described [30]. GFP-Trap and Myc-Trap beads were from ChromoTek. 4-Methylumbelliferyl β-D-glucopyranoside (4-MU), GCase substrate, were obtained from Sigma.
Plasmids
Human GBA cDNA in the pDONR223 vector from the human ORFeome 8.1 collection was a gift from Dr. Haiyuan Yu (Cornell University). GBA was cloned into pcDNA3.1/myc-His A vector (Thermo Fisher Scientific) after digestion with EcoRI and XhoI. Human PGRN in the pCMV-Sport6 vector was obtained as previously described [29]. GFP-PGRN was produced as previously described [30]. GFP-granulin peptides were produced as described [36]. Mouse GBA with C-terminal FLAG myc tag was obtained from Origene.
Protein production and purification
GST-granulin E, GST-granulin F and His-SUMO-saposin C proteins were expressed from Origami B(DE3) bacterial strains (MilliporeSigma) with 0.1 mM IPTG induction overnight at 18°C. Proteins were purified using GST or cobalt beads. His-PGRN was purified with cobalt beads from the culture media of HEK293T cells as previously described [30]. All purified proteins were concentrated and transitioned to PBS buffer with Centricon Centrifugal Filter Units (MilliporeSigma).
Mouse strains
C57/BL6 and Grn-/- mice [40] were obtained from The Jackson Laboratory. Gba-/- mice were generated as previously described [41]. Mixed male and female mice were used for this study.
Cell culture
HEK293T and BV2 cells were maintained in Dulbecco’s Modified Eagle’s Medium (Cellgro) supplemented with 10% fetal bovine serum (Gibco) in a humidified incubator at 37°C with 5% CO2. WT, Grn-/- and Gba-/- mouse fibroblasts were cultured as described [30].
Transfection, immunoprecipitation, and western blot analysis
Cells were transfected with polyethylenimine as previously described [42]. Cells were lysed in a cold, near-neutral pH solution containing 150 mM NaCl, 50 mM Tris pH 7.5 or 50mM sodium acetate pH5.3, 1% Triton X-100, 0.1% deoxycholic acid, 1X protease inhibitors (Roche). After centrifugation at 14,000 xg, for 15 minutes, at 4°C, supernatants were transferred to clean tubes on ice, to which rabbit anti-PGRN antibody-conjugated Affi-Gel 15 (Bio-Rad Laboratories), Myc-Trap or GFP-Trap beads were added, then rocked for 3–4 hours at 4°C. Samples were run on 12% polyacrylamide gels or 4–12% Bis-Tris gels (Invitrogen), then transferred to Immobilon-FL polyvinylidene fluoride membranes (Millipore Corporation) or nitrocellulose membranes (Millipore Corporation). Membranes were blocked with either 5% non-fat milk in PBS or Odyssey Blocking Buffer (LI-COR Biosciences) for 1 hour then washed with Tris-buffered saline with 0.1% Tween-20 (TBST) 3x for 5 minutes each. Membranes were incubated with primary antibodies, rocking overnight at 4°C, then washed as above, incubated with secondary antibodies for 2 hours at room temperature, then washed again. Membranes were scanned using an Odyssey Infrared Imaging System (LI-COR Biosciences). Densitometry was performed with Image Studio (LI-COR Biosciences).
Tissue preparation for enzyme assays and western blot
2-month- old WT and Grn-/- mice were perfused and tissues were dissected and snap-frozen with liquid nitrogen and kept at -80C. On the day of the experiment, frozen tissues were thawed and homogenized on ice with a glass Dounce homogenizer in a cold solution of either 1% (w/v) sodium taurocholate and 1% (v/v) Triton X-100, pH 5.2, for 4-MU activity assays or 0.2% (w/v) sodium taurocholate and 0.1% (v/v) Triton X-100 for MDW941 assays. Protein concentrations were determined via Bradford assay, then standardized.
Lysosome isolation
Lysosomes were isolated from liver tissues of 2-month- old WT and Grn-/- mice with lysosome isolation kit according to the instructions with the following optiprep gradients ((8%, 12%, 16%, 19%, 23%, 27%). Lysosomes are enriched in fraction #2 (12%-16%).
GCase activity assay with 4-MU substrate
Tissue lysates, prepared as described, were diluted in a cold buffer of 0.1 M citric acid/0.2 M disodium phosphate (pH 5.0), with 2 mg/mL bovine serum albumin added. Ten microliters of each sample were added to 75 μL of cold 10 mM 4-MU substrate in the same buffer and incubated at 37°C for 30 minutes. Reactions were stopped by the addition of 200 μL of a 0.3 M glycine/0.2 M sodium carbonate (pH 10.7) stop solution. Plates were read at 360 nm excitation/460 nm emission with an Infinite M1000 microplate reader (Tecan). To test for the direct activation of recombinant GCase, a 50 μL reaction mixture containing 1 mg/mL BSA, 0.1 M sodium acetate (pH 4.5), 0.02 U Cerezyme, 0.2 mM 4-MU substrate, 0.1% Triton X-100, and 1, 5, or 9 μM recombinant PGRN, Grn E, Grn F, saposin C, GST, or an equal volume of PBS was incubated at 37°C for 30 minutes. Reactions were stopped by the addition of 50μL of a 0.32 M glycine/0.32 M sodium carbonate (pH 10.4) stop solution. Plates were read at 340 nm excitation/420 nm emission with an Infinite M1000 microplate reader (Tecan).
Active GCase assessment with MDW941 probe
MDW941 was diluted to 100nM in tissue lysates, which were then incubated at 37°C for 30 minutes. Reactions were stopped by the addition of an equal volume of 2x Laemmli sample buffer with 10% β-Mercaptoethanol before heating at 95°C for 5 minutes. An equal amount of each sample (50μg total protein) was run on a 12% polyacrylamide gel, which was scanned at 532 nm excitation/580 nm emission with a Typhoon Imaging System (GE Healthcare), then western blot and assessment were performed as described above, with all values normalized to GAPDH.
MDW941 cell labeling, immunostaining, and confocal microscopy
WT and Grn-/- MEFs were cultured on glass coverslips overnight. The next day, MDW941 was diluted to 5 nM in culture media, then equal volumes were added to each well and the plate was incubated at 37°C for 2 hours. Cells were washed 2x with PBS, fixed with 3.7% paraformaldehyde for 15 minutes at room temperature, followed by 3 additional PBS washes. Cells were permeabilized with Odyssey Blocking Buffer LI-COR Biosciences) + 0.05% saponin for 30 minutes at room temperature. Primary antibodies were diluted in the same buffer and added to coverslips, which were incubated in a humidified chamber overnight at 4°C. Coverslips were washed 3x with PBS, for 5 minutes each, then secondary antibodies diluted in the same blocking/permeabilization solution were added to the coverslips, which were incubated at room temperature, in the dark, for 2 hours. After 3 additional PBS washes, coverslips were mounted on slides with Fluoromount-G (SouthernBiotech). Images were acquired with a CSU-X series spinning disc confocal microscope (Intelligent Imaging Innovations) with an HQ2 CCD camera (Photometrics) using a 100x objective.
SILAC and mass spectrometry
BV2 cells were infected with control lentivirus (pLenti-CRISPR2, Addgene) or lentivirus expressing Cas9 and guide RNA (sequence 5’-GCTCCCTGGGAGGCATCTGG-3’) and selected with puromycin. The cells were then grown a minimum of five generations in DMEM with 10% dialyzed fetal bovine serum (Sigma) supplemented with either heavy (C13, N15 arginine and lysine) amino acids or light (C12, N14 arginine and lysine). Cells were grown to confluency in two 15-cm plates, each, and lysed then immunoprecipitated as described above using homemade rabbit anti-PGRN antibodies bound to Affi-Gel 15 (Bio-Rad Laboratories). Samples were then mixed and boiled 5 minutes with 10mM DTT followed by alkylation by treating samples with a final concentration of 28 mM iodoacetamide. Proteins were precipitated on ice for 30 minutes with a mixture of 50% acetone/49.9% ethanol/0.1% acetic acid. Protein was pelleted and washed with this buffer, reprecipitated on ice, and dissolved in 8 M urea/50 mM Tris (pH 8.0) followed by dilution with three volumes of 50 mM Tris (pH 8.0)/150 mM NaCl. Proteins were digested overnight at 37°C with 1 μg of mass spectrometry grade Trypsin (Promega). The resulting peptide samples were cleaned up for mass spectrometry in a Sep-Pak C18 column (Waters) as follow: samples were acidified with 0.25% formic acid and 0.25% trifluoroacetic acid, loaded onto a preequilibrated C18 column, washed twice with 0.1% acetic acid and eluted with 80% acetonitrile/0.1% acetic acid into silanized vials (National Scientific). Samples were evaporated using a SpeedVac and then redissolved in 70% acetonitrile with ∼1% formic acid. Peptides were separated using hydrophilic interaction liquid chromatography on an Ultimate 300 LC (Dionex). Each fraction was evaporated in a SpeedVac and resuspended in 0.1% trifluoroacetic acid with spiked-in angiotensin II as an internal standard. Samples were run on a QE Orbitrap XL mass spectrometer (Thermo Fisher Scientific). XPRESS software, part of the Trans-Proteomic Pipeline (Seattle Proteome Center), was used to quantify all the identified peptides. Mann-Whitney U test was used to calculate the p value to generate the volcano plot.
Ethical approval and consent to participate
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The work under animal protocol 2017–0056 is approved by the Institutional Animal Care and Use Committee at Cornell University.
Results
PGRN interacts with GCase
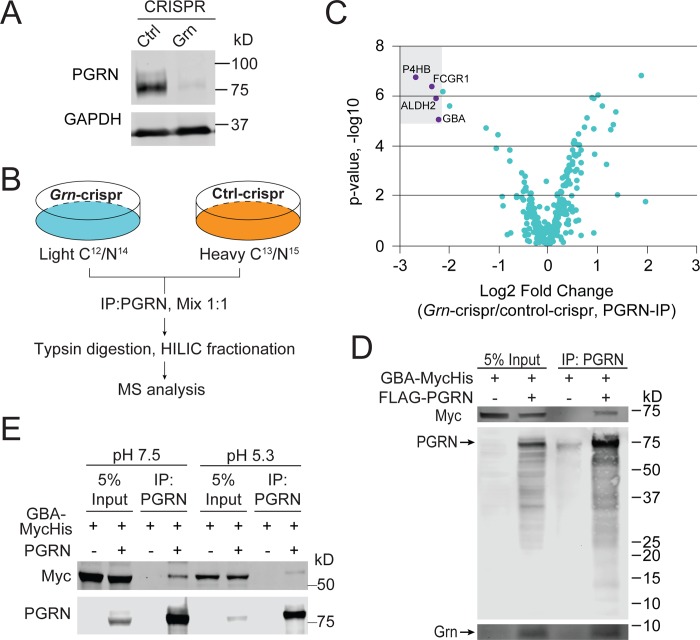
SILAC was performed in the murine microglia-derived BV2 cell line, with either normal PGRN expression or loss of PGRN expression after CRISPR/cas9 mediated genome editing [43, 44] (Fig 1A, 1B and 1C). GCase (encoded by the GBA gene) was one of the high-confidence hits for PGRN from this experiment (Fig 1C) (S1 Table).
Fig 1. PGRN interacts with GCase.
(A) Western blot analysis of PGRN levels in BV2 cells expressing control or guide RNA to the mouse GRN gene. (B) Schematic illustration of the SILAC experiment searching for PGRN interactors. (C) Volcano plot of SILAC hits. Top hits identified in the heavy fraction are highlighted. (D) Anti-PGRN co-immunoprecipitation of FLAG-PGRN and GBA-myc overexpressed in HEK293T cells. (E) Anti-PGRN co-immunoprecipitation of PGRN and GBA-myc overexpressed in HEK293T cells using buffers of pH7.5 and pH5.3.
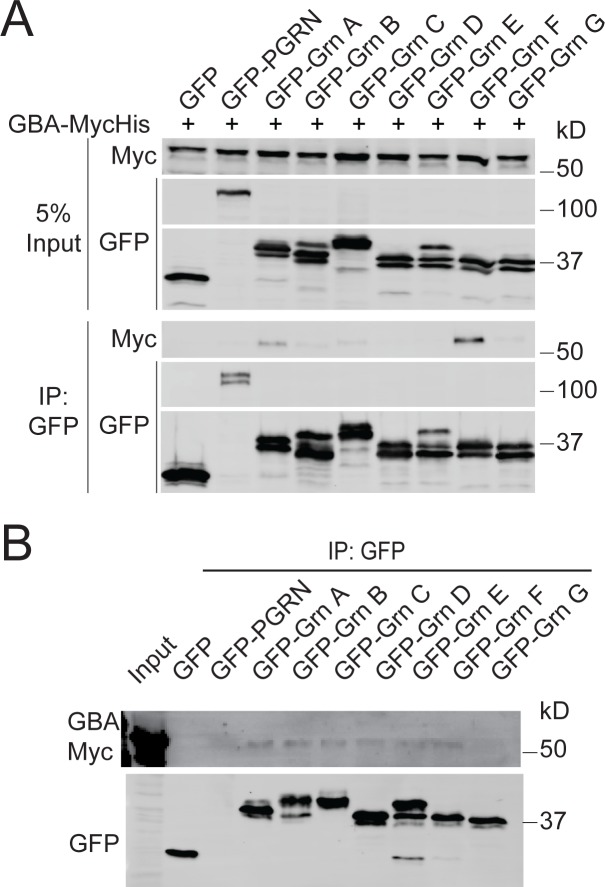
To verify the physical interaction between PGRN and GCase, FLAG-human PGRN and human GBA with a C-terminal myc tag were co-transfected in HEK293T cells, then anti-PGRN immunoprecipitation (IP) was performed. GBA-myc signal was detected in the IP products from the PGRN transfected, but not control samples, indicating a specific binding interaction (Fig 1D). This interaction persists at pH5.3 (Fig 1E). Similar interactions were detected with mouse PGRN and mouse GCase (S1 Fig). Additionally, PGRN has been shown to be processed into individual granulin peptides within the lysosome [32, 33] and two of these peptides, Grn E and Grn F, have been shown to interact with GCase [37, 38]. Since both PGRN and Grn peptides were pulled down in the anti-PGRN IP (Fig 1D), we tested whether GCase can bind specific Grn peptides. N-terminal GFP-tagged Grn peptides or GFP-PGRN were co-transfected with myc-tagged GBA, followed by anti-GFP IP. While there was no obvious binding of granulin E, granulin F and, to a lesser extent, granulin A did interact with GCase in the cell lysate (Fig 2A). However, it is not clear whether these granulin peptides are folded correctly when expressed individually. We repeated the experiments with secreted granulins, which, having proceeded through the entire secretory pathway, may be more likely to be properly folded. In this instance, all the granulins except Grn G shows weak binding to GCase (Fig 2B).
Fig 2. Interaction between GCase and granulin peptides.
(A) Anti-GFP co-immunoprecipitation of GFP-PGRN or individual GFP-granulins and GBA-myc overexpressed in HEK293T cells shows binding primarily to Grn F, with weaker binding to Grn A and other Grn peptides. (B) Anti-GFP co-immunoprecipitation of GFP-PGRN or individual GFP-granulins and GBA-myc from conditioned medium of transfected cells shows weaker binding to all the granulins except Grn G.
PGRN deficiency leads to reduced GCase activities
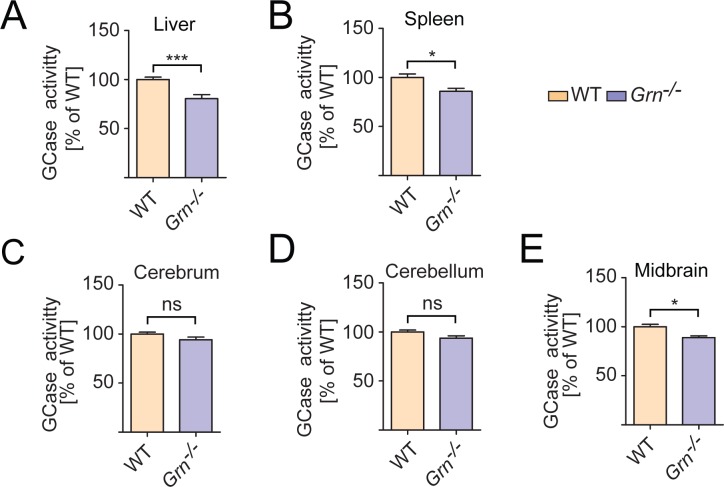
To determine whether PGRN regulates GCase activities, we performed an in vitro GCase activity assay using a well-established fluorogenic GCase substrate, 4-Methylumbelliferyl β-D-glucopyranoside (4MU-β-glc) [45–47] (Fig 3). GCase activity was measured in tissue lysates from 2-month-old WT and Grn-/- mice, before obvious lysosomal phenotypes were observed. Liver and spleen lysates from Grn-/- mice showed a significant decrease in GCase activity compared to WT controls (Fig 3A and 3B). While the cerebrum and cerebellum showed a trend toward decreased GBA activity, it was not significant (Fig 3C and 3D). However, the midbrain, where we tend to observe the most severe defects in our knockout mouse line, showed a significant decrease in GCase activity (Fig 3E). Thus PGRN is likely to have cell type and tissue specific effects on GCase activities (S2 Table).
Fig 3. PGRN deficiency results in decreased GCase activity in mouse tissues.
GCase activity was assessed in tissue lysates from 2-month-old WT and Grn-/- mice, as indicated, using the substrate 4-MU. (n = 5–6, ±SEM, *p value <0.05, **p-value <0.01, ns, not significant, Student’s t-test.
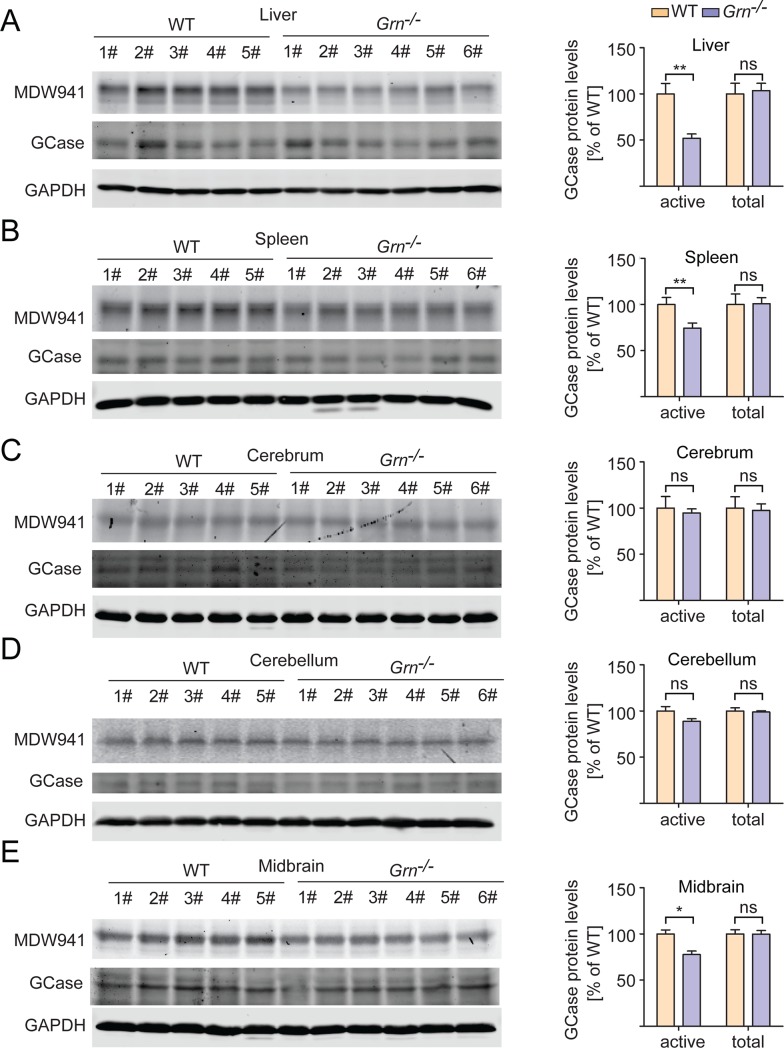
These results were confirmed using a recently developed ultrasensitive fluorescent probe, MDW941, which specifically reacts with active forms of the enzyme and has been shown to be sensitive enough to detect the activity of recombinant GCase in the attomolar range [48–50]. Using this probe, an even greater disparity between GBA activity in WT and Grn-/- tissues was observed (Fig 4A–4E, S2 Table). To verify that any changes in activity were not due to alterations in total GCase protein levels, SDS-PAGE and western blot of the tissue lysates were performed using antibodies specific to GCase (S2 Fig), with no significant differences seen between groups (Fig 4A–4E, S3 Fig, S2 Table).
Fig 4. PGRN deficiency results in decreased GCase activity without changes in GCase protein levels.
Tissue lysates from 2-month-old WT and Grn-/- mice were incubated with the MDW941 GCase activity probe. The samples were run on SDS-PAGE and MDW941 labeled GCase (active) was detected using a fluorescent scanner, then western blot was performed to assess total GCase protein levels. n = 5–6, ±SEM, *p value <0.05, **p-value <0.01, ns, not significant, Student’s t-test.
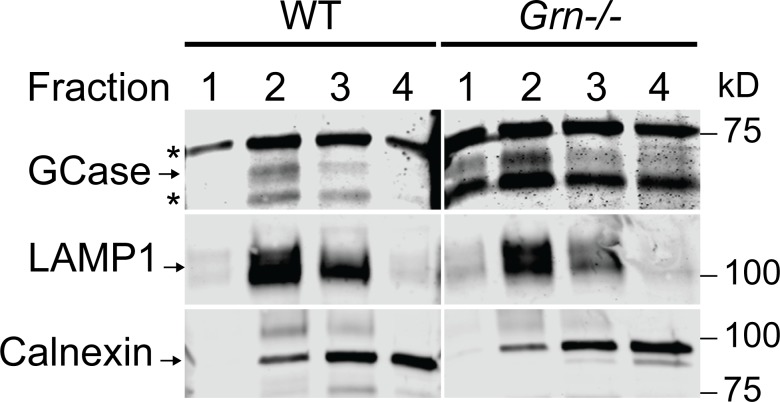
GCase was reported to aggregate in the cytosol in the absence of PGRN [37, 38]. To determine whether PGRN causes GCase trafficking defect, we performed lysosome isolation from WT and Grn-/- livers and blotted for GCase in the lysosome fraction (Fig 5), since none of commercially available antibodies for GCase gave specific signals in immunostaining. GCase is enriched in lysosome fractions in both WT and Grn-/- livers, suggstins that GCase lysosome trafficking is not affected by PGRN loss.
Fig 5. GCase is enriched in the lysosome fraction of WT and Grn-/- livers.
Lysosomes were isolated from liver tissues of 2-month-old WT and Grn-/- mice. The presence of GCase, LAMP1 and calnexin (marker for ER) was determined in the different fractions using Western blot analysis.
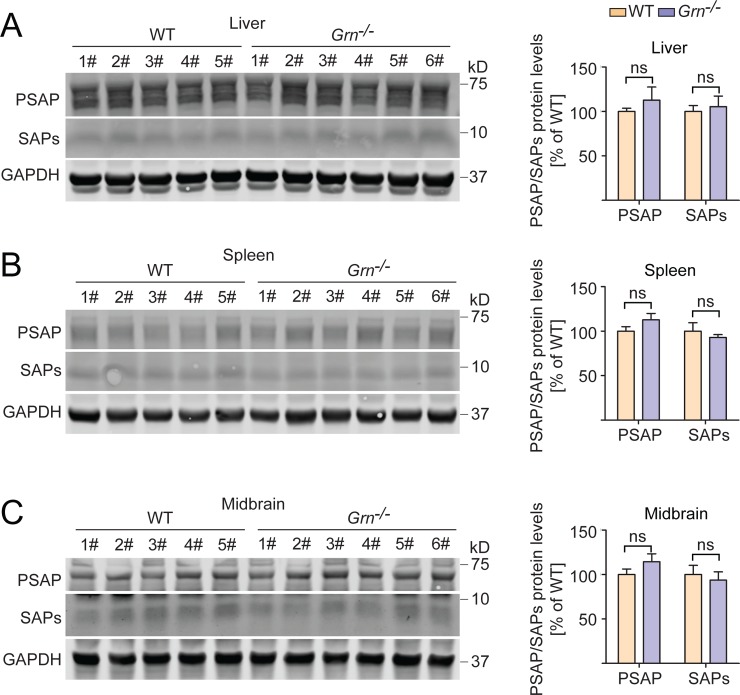
Previously we showed that PGRN binds to prosaposin (PSAP) to facilitate each other’s lysosomal trafficking [30, 51, 52]. PSAP is known to get cleaved in the lysosome to generate saposin peptides, of which saposins A and C are known activators of GCase [46, 53, 54]. This raises the possibility that the observed decrease in GCase activity with PGRN loss could be due to a decrease in PSAP or saposin peptides. However, we did not observe a significant change in PSAP or total saposins in the PGRN knockout tissues where GCase activity was reduced (Fig 6, S2 Table). Similarly, no obvious difference in saposin C levels in WT and PGRN deficient liver lysates were detected following immunoprecipitations using antibodies specific for saposin C (S4 Fig). Unfortunately we cannot test saposin A levels due to a lack of specific antibodies.
Fig 6. PSAP and total saposin levels are not changed in PGRN deficient mice.
Mouse tissue lysates from 2-month-old WT and Grn-/- mice were assessed for PSAP and total saposin peptide levels, which were normalized to GAPDH. n = 5–6, ±SEM, ns, not significant, Student’s t-test.
With the changes we observed in GCase activity, and because PGRN and Grn E have been shown to directly activate CTSD in vitro, we wanted to test whether PGRN, Grn E, or Grn F could directly augment the activity of GCase. We performed a 4-MU activity assay of a pharmaceutical grade recombinant GCase (Cerezyme) with the addition of recombinant His-PGRN, GST-Grn E, GST-Grn F, His-SUMO-saposin C, or GST control. While the addition of saposin C greatly enhanced GCase activity, such effects were not observed with PGRN or granulin peptides (S5 Fig).
Discussion
In this study, we demonstrate a physical interaction between the FTLD-related protein, PGRN, and the lysosomal enzyme, GCase, consistent with previous reports [37, 38]. Additionally, among the granulin peptides, granulin F and to a lesser extent, granulin A, but not the previously reported granulin E, were shown to bind GCase in the cell lysate (Fig 2A). All the secreted granulins except Grn G show weak binding to GCase (Fig 2B).
The previous studies examining the relationship between PGRN and GBA primarily utilized a chronic inflammation model based on the administration of ovalbumin (OVA) to WT and Grn-/- mice over the course of multiple weeks. The authors showed that PGRN interacts with GCase and acts as a co-chaperone of GCase and its trafficking receptor, lysosome membrane protein 2 (LIMP-2) [37, 38]. Furthermore, LIMP-2 and GCase were shown to aggregate in the cytoplasm in PGRN deficient macrophages with experimentally induced chronic lung inflammation models [37, 38]. Unfortunately, we were unable to find a single commercial antibody that specifically recognize the endogenous GCase protein in immunostaining using WT and Gba-/- mouse fibroblasts, thus we cannot test whether there is a GCase trafficking defect with PGRN loss. Nevertheless, MDW41 labeling shows normal localization of active GCase in PGRN knockout fibroblasts (S6 Fig). Furthermore, GCase is enriched in the lysosome fraction of Grn-/- liver similar to that of WT mice (Fig 5), suggesting that the decreased GCase activity in PGRN deficient liver lysates (Figs 3A and 4A) are not due to defects in GCase trafficking to lysosomes.
The relationship between PGRN and GCase is complicated by a series of interrelated factors. Two peptides derived from PSAP, saposin A and saposin C, are known activators of GCase [46, 53, 54]. We have previously shown that PGRN and PSAP share a lysosomal co-trafficking relationship, wherein PGRN can carry PSAP to the lysosome via the receptor, sortilin, and PSAP can carry PGRN to the lysosome via the cation-independent mannose-6-phosphate receptor (CI-M6PR) or the low-density lipoprotein receptor-related protein 1 (LRP1) [30, 51, 52]. Additionally, PGRN has recently been shown to bind and modulate the activity of the lysosomal protease, cathepsin D (CTSD), which is the major contributor to proteolytic PSAP processing. Because of these factors, it is possible that PGRN deficiency results in an alteration in PSAP processing and production of saposin peptides, thereby affecting GCase activation. However, PSAP, total saposin levels and saposin C levels do not appear to change in PGRN deficient tissue lysates (Fig 6, S4 Fig), although we cannot rule out the possibility that saposin A levels might be affected.
We failed to detect any changes in GCase activity with the addition of recombinant PGRN or granulin peptides, but this does not entirely rule out the possibility of a direct activation of GCase by PGRN. Recombinant PGRN and granulins might not fold correctly due to disulfide bond scrambling, or alternatively, further optimization of the conditions for the assay might be required.
Another possibility is that PGRN indirectly affects GCase activity by changing the lysosomal environment. PGRN deficiency has been reported to affect lysosomal pH [55], which could indirectly affect the activity of many lysosomal enzymes. Loss of PGRN also changes lipid contents of lysosomes [56], which could also indirectly affect GCase activity. While the exact mechanism of our findings is currently unknown, and more work needs to be performed to sift through the somewhat muddled relationships of the proteins involved, it is clear that PGRN deficiency leads to reduced GCase activity. This is significant, as lysosomal dysfunction is a commonality between NCL and FTLD with GRN mutation. It is possible that the decreased activity of multiple lysosomal hydrolases, including GCase and CTSD, accounts for lysosomal dysfunction with GRN mutations.
Mutations and polymorphism in the GRN gene have also been associated with Parkinson’s disease (PD)[57–60]. Also, parkinsonism is common in GRN mutation carriers with FTLD and occurs more frequently than in other forms of FTLD [61]. In addition, reduced serum levels of PGRN were found to be associated with PD risk [62]. Moreover, viral expression of PGRN was shown to protect midbrain dopaminergic neurons and improve the locomotor function in response to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) treatment to mimic PD in mouse models [63]. Notably, among different tested brain regions, midbrain, the major PD affected brain region, is the only brain region shows a significant reduction of GCase activity upon PGRN loss (Fig 3, Fig 4). Interestingly, heterozygous mutations in GBA is one of the genetic determinants of PD[64, 65]. Our work on the regulation of GCase activity by PGRN might provide a mechanistic explanation underlining the association of PGRN and PD.
Conclusions
Our data support that PGRN deficiency leads to a reduction of GCase activity in vivo.
Supporting information
HEK293T cells were transfected with mGBA-FLAG-myc and FLAG-mPGRN constructs as indicated and anti-myc (A) or anti-PGRN (B) immunoprecipitation experiments were carried out. The presence of PGRN and GCase in the immunoprecipitates were detected using Western blot analysis.
(TIF)
A) Primary fibroblasts from WT and Gba-/- mice. B) Lung tissue lysates from WT and Gba-/- mice. *indicates non-specific bands.
(TIF)
Dashed line indicated where the GCase bands are.
(TIF)
WT and PGRN-deficient liver lysates were immunoprecipitated using anti-saposin C antibodies and the IP products were analyzed by Western blot using polyclonal anti-PSAP antibodies.
(TIF)
(A) Commassie staining of recombinant PGRN, Grn E and Grn F proteins used in the activity assay. (B) Activity of recombinant GCase (Cerezyme) was measured with the addition of recombinant PGRN, Grn E, Grn F, or recombinant saposin C as a positive control.
(TIF)
WT and Grn-/- fibroblasts were labeled for 2 hours with MDW941 before fixation and immunostaining. Scale bar = 20 μm.
(TIF)
(XLSX)
Acknowledgments
We would like to thank Dr. Haiyuan Yu for his gifts of cDNA; and Xiaochun Wu for technical assistance. This work is supported by funding to F. Hu from National Institute of Aging and National Institute of Neurological Disorders and Stroke (R01NS095954; R01NS088448) and President's Council of Cornell Women Affinito-Stewart award. The authors declare no additional competing financial interests.
Abbreviations
- FTLD
frontotemporal lobar degeneration
- NCL
neuronal ceroid lipofuscinosis
- PGRN
progranulin
- GCase
glucocerebrosidase
- PSAP
prosaposin
- SAP
saposin
- SILAC
stable isotope labeling by amino acids in cell culture
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work is supported by funding to F. Hu from National Institute of Aging and National Institute of Neurological Disorders and Stroke (R01 NS095954, NS088448) and President's Council of Cornell Women Affinito-Stewart award.
References
- 1.Songsrirote K, Li Z, Ashford D, Bateman A, Thomas-Oates J. Development and application of mass spectrometric methods for the analysis of progranulin N-glycosylation. J Proteomics. 2010;73(8):1479–90. Epub 2010/03/02. 10.1016/j.jprot.2010.02.013 . [DOI] [PubMed] [Google Scholar]
- 2.Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, et al. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 2008;17(4):711–24. 10.1110/ps.073295308 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hrabal R, Chen Z, James S, Bennett HP, Ni F. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat Struct Biol. 1996;3(9):747–52. . [DOI] [PubMed] [Google Scholar]
- 4.Tolkatchev D, Ng A, Vranken W, Ni F. Design and solution structure of a well-folded stack of two beta-hairpins based on the amino-terminal fragment of human granulin A. Biochemistry. 2000;39(11):2878–86. Epub 2000/03/15. bi992130u [pii]. 10.1021/bi992130u . [DOI] [PubMed] [Google Scholar]
- 5.Vranken WF, Chen ZG, Xu P, James S, Bennett HP, Ni F. A 30-residue fragment of the carp granulin-1 protein folds into a stack of two beta-hairpins similar to that found in the native protein. J Pept Res. 1999;53(5):590–7. Epub 1999/07/29. . [DOI] [PubMed] [Google Scholar]
- 6.Palfree RG, Bennett HP, Bateman A. The Evolution of the Secreted Regulatory Protein Progranulin. PLoS ONE. 2015;10(8):e0133749 Epub 2015/08/08. 10.1371/journal.pone.0133749 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003;81(10):600–12. 10.1007/s00109-003-0474-3 . [DOI] [PubMed] [Google Scholar]
- 8.Shoyab M, McDonald VL, Byles C, Todaro GJ, Plowman GD. Epithelins 1 and 2: isolation and characterization of two cysteine-rich growth-modulating proteins. Proc Natl Acad Sci U S A. 1990;87(20):7912–6. 10.1073/pnas.87.20.7912 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plowman GD, Green JM, Neubauer MG, Buckley SD, McDonald VL, Todaro GJ, et al. The epithelin precursor encodes two proteins with opposing activities on epithelial cell growth. J Biol Chem. 1992;267(18):13073–8. . [PubMed] [Google Scholar]
- 10.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48(7):999–1009. 10.1177/002215540004800713 . [DOI] [PubMed] [Google Scholar]
- 11.Toh H, Cao M, Daniels E, Bateman A. Expression of the growth factor progranulin in endothelial cells influences growth and development of blood vessels: a novel mouse model. PLoS ONE. 2013;8(5):e64989 Epub 2013/06/07. 10.1371/journal.pone.0064989 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Z, Ong CH, Halper J, Bateman A. Progranulin is a mediator of the wound response. Nat Med. 2003;9(2):225–9. 10.1038/nm816 . [DOI] [PubMed] [Google Scholar]
- 13.Jian J, Konopka J, Liu C. Insights into the role of progranulin in immunity, infection, and inflammation. J Leukoc Biol. 2013;93(2):199–208. Epub 2012/10/24. 10.1189/jlb.0812429 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh H, Chitramuthu BP, Bennett HP, Bateman A. Structure, function, and mechanism of progranulin; the brain and beyond. J Mol Neurosci. 2011;45(3):538–48. Epub 2011/06/22. 10.1007/s12031-011-9569-4 . [DOI] [PubMed] [Google Scholar]
- 15.Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem. 2012;287(39):32298–306. Epub 2012/08/04. R112.399170 [pii] 10.1074/jbc.R112.399170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, Matsuwaki T, Yamanouchi K, Nishihara M. Exacerbated inflammatory responses related to activated microglia after traumatic brain injury in progranulin-deficient mice. Neuroscience. 2013;231:49–60. Epub 2012/12/04. 10.1016/j.neuroscience.2012.11.032 . [DOI] [PubMed] [Google Scholar]
- 17.Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51(6):1546–54. 10.1212/wnl.51.6.1546 . [DOI] [PubMed] [Google Scholar]
- 18.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–9. 10.1038/nature05016 . [DOI] [PubMed] [Google Scholar]
- 19.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–4. 10.1038/nature05017 . [DOI] [PubMed] [Google Scholar]
- 20.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15(20):2988–3001. 10.1093/hmg/ddl241 . [DOI] [PubMed] [Google Scholar]
- 21.Mackenzie IR, Baker M, Pickering-Brown S, Hsiung GY, Lindholm C, Dwosh E, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129(Pt 11):3081–90. 10.1093/brain/awl271 . [DOI] [PubMed] [Google Scholar]
- 22.Snowden JS, Pickering-Brown SM, Mackenzie IR, Richardson AM, Varma A, Neary D, et al. Progranulin gene mutations associated with frontotemporal dementia and progressive non-fluent aphasia. Brain. 2006;129(Pt 11):3091–102. 10.1093/brain/awl267 . [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JS, Shears S, et al. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60(3):314–22. 10.1002/ana.20963 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90(6):1102–7. Epub 2012/05/23. 10.1016/j.ajhg.2012.04.021 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida MR, Macario MC, Ramos L, Baldeiras I, Ribeiro MH, Santana I. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol Aging. 2016;41:200 e1–5. Epub 2016/03/30. 10.1016/j.neurobiolaging.2016.02.019S0197-4580(16)00178-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 26.Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, Taubes A, et al. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med. 2017;9(385). Epub 2017/04/14. eaah5642 [pii] 10.1126/scitranslmed.aah56429/385/eaah5642 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, et al. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014;127(6):845–60. Epub 2014/03/13. 10.1007/s00401-014-1262-6 . [DOI] [PubMed] [Google Scholar]
- 28.Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK, Krainc D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet. 2017;26(24):4861–72. Epub 2017/10/17. 10.1093/hmg/ddx364 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68(4):654–67. Epub 2010/11/26. S0896-6273(10)00776-2 [pii] 10.1016/j.neuron.2010.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210(6):991–1002. Epub 2015/09/16. 10.1083/jcb.201502029 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belcastro V, Siciliano V, Gregoretti F, Mithbaokar P, Dharmalingam G, Berlingieri S, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic Acids Res. 2011;39(20):8677–88. Epub 2011/07/26. gkr593 [pii] 10.1093/nar/gkr593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holler CJ, Taylor G, Deng Q, Kukar T. Intracellular Proteolysis of Progranulin Generates Stable, Lysosomal Granulins that Are Haploinsufficient in Patients with Frontotemporal Dementia Caused by GRN Mutations. eNeuro. 2017;4(4). Epub 2017/08/23. 10.1523/ENEURO.0100-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Paushter DH, Feng T, Sun L, Reinheckel T, Hu F. Lysosomal processing of progranulin. Mol Neurodegener. 2017;12(1):62 Epub 2017/08/25. 10.1186/s13024-017-0205-9 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CW, Stankowski JN, Chew J, Cook CN, Lam YW, Almeida S, et al. The lysosomal protein cathepsin L is a progranulin protease. Mol Neurodegener. 2017;12(1):55 Epub 2017/07/27. 10.1186/s13024-017-0196-6 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van Den Bosch L, et al. Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet. 2017. Epub 2017/04/30. 10.1093/hmg/ddx1623768439 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X, Paushter DH, Feng T, Pardon CM, Mendoza CS, Hu F. Regulation of cathepsin D activity by the FTLD protein progranulin. Acta Neuropathol. 2017;134(1):151–3. Epub 2017/05/12. 10.1007/s00401-017-1719-5 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jian J, Tian QY, Hettinghouse A, Zhao S, Liu H, Wei J, et al. Progranulin Recruits HSP70 to beta-Glucocerebrosidase and Is Therapeutic Against Gaucher Disease. EBioMedicine. 2016;13:212–24. Epub 2016/10/30. S2352-3964(16)30465-0 [pii] 10.1016/j.ebiom.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jian J, Zhao S, Tian QY, Liu H, Zhao Y, Chen WC, et al. Association Between Progranulin and Gaucher Disease. EBioMedicine. 2016;11:127–37. Epub 2016/08/16. S2352-3964(16)30353-X [pii] 10.1016/j.ebiom.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beutler E, Demina A, Gelbart T. Glucocerebrosidase mutations in Gaucher disease. Mol Med. 1994;1(1):82–92. Epub 1994/11/01. [PMC free article] [PubMed] [Google Scholar]
- 40.Yin F, Banerjee R, Thomas B, Zhou P, Qian L, Jia T, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207(1):117–28. Epub 2009/12/23. jem.20091568 [pii] 10.1084/jem.20091568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tybulewicz VL, Tremblay ML, LaMarca ME, Willemsen R, Stubblefield BK, Winfield S, et al. Animal model of Gaucher's disease from targeted disruption of the mouse glucocerebrosidase gene. Nature. 1992;357(6377):407–10. Epub 1992/06/04. 10.1038/357407a0 . [DOI] [PubMed] [Google Scholar]
- 42.Vancha AR, Govindaraju S, Parsa KV, Jasti M, Gonzalez-Garcia M, Ballestero RP. Use of polyethyleneimine polymer in cell culture as attachment factor and lipofection enhancer. BMC Biotechnol. 2004;4:23 Epub 2004/10/16. 1472-6750-4-23 [pii] 10.1186/1472-6750-4-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. Epub 2013/01/05. 10.1126/science.1231143 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. Epub 2013/01/05. 10.1126/science.1232033 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osiecki-Newman K, Fabbro D, Legler G, Desnick RJ, Grabowski GA. Human acid beta-glucosidase: use of inhibitors, alternative substrates and amphiphiles to investigate the properties of the normal and Gaucher disease active sites. Biochim Biophys Acta. 1987;915(1):87–100. Epub 1987/09/02. 0167-4838(87)90128-2 [pii]. 10.1016/0167-4838(87)90128-2 . [DOI] [PubMed] [Google Scholar]
- 46.Sun Y, Qi X, Grabowski GA. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem. 2003;278(34):31918–23. Epub 2003/06/19. 10.1074/jbc.M302752200 [pii]. . [DOI] [PubMed] [Google Scholar]
- 47.Qi X, Qin W, Sun Y, Kondoh K, Grabowski GA. Functional organization of saposin C. Definition of the neurotrophic and acid beta-glucosidase activation regions. J Biol Chem. 1996;271(12):6874–80. Epub 1996/03/22. 10.1074/jbc.271.12.6874 . [DOI] [PubMed] [Google Scholar]
- 48.Witte MD, Kallemeijn WW, Aten J, Li KY, Strijland A, Donker-Koopman WE, et al. Ultrasensitive in situ visualization of active glucocerebrosidase molecules. Nat Chem Biol. 2010;6(12):907–13. Epub 2010/11/17. 10.1038/nchembio.466 [pii]. . [DOI] [PubMed] [Google Scholar]
- 49.Ben Bdira F, Kallemeijn WW, Oussoren SV, Scheij S, Bleijlevens B, Florea BI, et al. Stabilization of Glucocerebrosidase by Active Site Occupancy. ACS Chem Biol. 2017;12(7):1830–41. Epub 2017/05/10. 10.1021/acschembio.7b00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herrera Moro Chao D, Kallemeijn WW, Marques AR, Orre M, Ottenhoff R, van Roomen C, et al. Visualization of Active Glucocerebrosidase in Rodent Brain with High Spatial Resolution following In Situ Labeling with Fluorescent Activity Based Probes. PLoS One. 2015;10(9):e0138107 Epub 2015/09/30. 10.1371/journal.pone.0138107 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Sun L, Bracko O, Choi JW, Jia Y, Nana AL, et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun. 2017;8:15277 Epub 2017/05/26. 10.1038/ncomms15277 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X, Sullivan PM, Sun L, Hu F. The interaction between progranulin and prosaposin is mediated by granulins and the linker region between saposin B and C. J Neurochem. 2017. Epub 2017/06/24. 10.1111/jnc.14110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morimoto S, Martin BM, Yamamoto Y, Kretz KA, O'Brien JS, Kishimoto Y. Saposin A: second cerebrosidase activator protein. Proc Natl Acad Sci U S A. 1989;86(9):3389–93. Epub 1989/05/01. 10.1073/pnas.86.9.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamargo RJ, Velayati A, Goldin E, Sidransky E. The role of saposin C in Gaucher disease. Mol Genet Metab. 2012;106(3):257–63. Epub 2012/06/02. 10.1016/j.ymgme.2012.04.024 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka Y, Suzuki G, Matsuwaki T, Hosokawa M, Serrano G, Beach TG, et al. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet. 2017;26(5):969–88. Epub 2017/01/12. 10.1093/hmg/ddx011 [pii]. . [DOI] [PubMed] [Google Scholar]
- 56.Evers BM, Rodriguez-Navas C, Tesla RJ, Prange-Kiel J, Wasser CR, Yoo KS, et al. Lipidomic and Transcriptomic Basis of Lysosomal Dysfunction in Progranulin Deficiency. Cell Rep. 2017;20(11):2565–74. Epub 2017/09/14. 10.1016/j.celrep.2017.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang KH, Chen CM, Chen YC, Hsiao YC, Huang CC, Kuo HC, et al. Association between GRN rs5848 polymorphism and Parkinson's disease in Taiwanese population. PLoS One. 2013;8(1):e54448 Epub 2013/01/24. 10.1371/journal.pone.0054448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Li S, Su L, Sheng J, Lv W, Chen G, et al. Association of progranulin polymorphism rs5848 with neurodegenerative diseases: a meta-analysis. J Neurol. 2015;262(4):814–22. Epub 2015/01/13. 10.1007/s00415-014-7630-2 . [DOI] [PubMed] [Google Scholar]
- 59.Rovelet-Lecrux A, Deramecourt V, Legallic S, Maurage CA, Le Ber I, Brice A, et al. Deletion of the progranulin gene in patients with frontotemporal lobar degeneration or Parkinson disease. Neurobiol Dis. 2008;31(1):41–5. Epub 2008/05/16. 10.1016/j.nbd.2008.03.004 . [DOI] [PubMed] [Google Scholar]
- 60.Carecchio M, Galimberti D, Fenoglio C, Serpente M, Scarpini E, Comi C, et al. Evidence of pre-synaptic dopaminergic deficit in a patient with a novel progranulin mutation presenting with atypical parkinsonism. J Alzheimers Dis. 2014;38(4):747–52. Epub 2013/09/26. 10.3233/JAD-131151 . [DOI] [PubMed] [Google Scholar]
- 61.Van Mossevelde S, Engelborghs S, van der Zee J, Van Broeckhoven C. Genotype-phenotype links in frontotemporal lobar degeneration. Nat Rev Neurol. 2018;14(6):363–78. Epub 2018/05/20. 10.1038/s41582-018-0009-8 . [DOI] [PubMed] [Google Scholar]
- 62.Mateo I, Gonzalez-Aramburu I, Pozueta A, Vazquez-Higuera JL, Rodriguez-Rodriguez E, Sanchez-Juan P, et al. Reduced serum progranulin level might be associated with Parkinson's disease risk. Eur J Neurol. 2013;20(12):1571–3. Epub 2013/02/13. 10.1111/ene.12090 . [DOI] [PubMed] [Google Scholar]
- 63.Van Kampen JM, Baranowski D, Kay DG. Progranulin gene delivery protects dopaminergic neurons in a mouse model of Parkinson's disease. PLoS One. 2014;9(5):e97032 Epub 2014/05/09. 10.1371/journal.pone.0097032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gegg ME, Schapira AHV. The role of glucocerebrosidase in Parkinson disease pathogenesis. FEBS J. 2018;285(19):3591–603. Epub 2018/02/01. 10.1111/febs.14393 . [DOI] [PubMed] [Google Scholar]
- 65.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med. 2009;361(17):1651–61. Epub 2009/10/23. 10.1056/NEJMoa0901281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HEK293T cells were transfected with mGBA-FLAG-myc and FLAG-mPGRN constructs as indicated and anti-myc (A) or anti-PGRN (B) immunoprecipitation experiments were carried out. The presence of PGRN and GCase in the immunoprecipitates were detected using Western blot analysis.
(TIF)
A) Primary fibroblasts from WT and Gba-/- mice. B) Lung tissue lysates from WT and Gba-/- mice. *indicates non-specific bands.
(TIF)
Dashed line indicated where the GCase bands are.
(TIF)
WT and PGRN-deficient liver lysates were immunoprecipitated using anti-saposin C antibodies and the IP products were analyzed by Western blot using polyclonal anti-PSAP antibodies.
(TIF)
(A) Commassie staining of recombinant PGRN, Grn E and Grn F proteins used in the activity assay. (B) Activity of recombinant GCase (Cerezyme) was measured with the addition of recombinant PGRN, Grn E, Grn F, or recombinant saposin C as a positive control.
(TIF)
WT and Grn-/- fibroblasts were labeled for 2 hours with MDW941 before fixation and immunostaining. Scale bar = 20 μm.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.