Abstract
We studied infection by Trypanosomatidae in bats captured in two areas with different degradation levels in the Atlantic Forest of Rio de Janeiro state: Reserva Ecológica de Guapiaçu (REGUA) and Estação Fiocruz Mata Atlântica (EFMA). Furthermore, we evaluated whether the diversity of trypanosomatids changes according to bat diversity and the different levels of preservation in the region. The results showed no influence of the level of preservation on bat species richness (15 and 14 species, respectively), with similar chiropterofauna and higher abundance of two common fruit-eating bat species in the tropics: Carollia perspicillata and Artibeus lituratus. Of the 181 bat specimens analyzed by LIT/Schneider hemoculture, we detected 24 infected individuals (13%), including one positive Sturnira lilium individual that was also positive by fresh blood examination. Molecular characterization using nested PCR targeting the 18 SSU rRNA-encoding gene fragment showed similar trypanosomatid infection rates in bats from the two areas: 15% in REGUA and 11% in EFMA (p = 0.46). Trypanosoma dionisii was the most frequently detected parasite (54%), followed by T. cruzi DTUs TcI and TcIV and Trypanosoma sp., in Neotropical phyllostomid bats (RNMO63 and RNMO56); mixed infections by T. dionisii/T. cruzi TcIII and T. dionisii/T. cruzi TcI were also observed. The T. cruzi DTUs TcI and TcIV are the genotypes currently involved in cases of acute Chagas disease in Brazil, and T. dionisii was recently found in the heart tissue of an infected child. Surprisingly, we also describe for the first time Crithidia mellificae, a putative monoxenous parasite from insects, infecting a vertebrate host in the Americas. Bats from the Atlantic Forest of Rio de Janeiro state harbor a great diversity of trypanosomatids, maintaining trypanosomatid diversity in this sylvatic environment.
Author summary
Among all mammals carrying trypanosomatids, bats are considered some of the most ancient hosts, and due to their good adaptation to different landscape features, they may play an important role in parasite dispersion. To assess whether the diversity of trypanosomatid species in bats is affected by the level of environmental disturbance, we collected blood from bats in two localities of Rio Janeiro state in Brazil: one with high and another with low human interference (more preserved). The collected blood was subjected to fresh blood tests and hemoculture. Using the 18S RNA gene as a molecular target, we identified 24 bats (13%) infected by trypanosomatids, mostly by T. dionisii, a trypanosome species commonly found in all Brazilian biomes and highly associated with bats. Three subpopulations (DTUs I, III and IV) of T. cruzi, the etiological agent of Chagas disease, were also found to infect bats. In addition, we detected for the first time a putative insect-associated parasite, Crithidia mellificae, in a mammal host in the Americas. Our work reveals the high diversity of trypanosomatids infecting bats of the Atlantic Forest of Rio de Janeiro state, reinforcing their importance as potential reservoirs of trypanosomatids.
Introduction
Trypanosomatids (Protozoa: Trypanosomatida: Trypanosomatidae) are parasitic uniflagellated protists that infect plants, invertebrates and vertebrates and have monoxenous or heteroxenous life cycles. The following monoxenous genera are currently recognized: Herpetomonas, Blastocrithidia, Crithidia, Leptomonas, Wallacemonas, Sergeia, Strigomonas, Angomonas, Rhynchoidomonas, Blechomonas, Paratrypanosoma, Kentomonas, Lotmaria, Lafontella, Jaenimonas, Zelonia, and Novymonas [1]. Heteroxenous genera include Endotrypanum, Phytomonas, Leishmania, and Trypanosoma [1]. Monoxenous trypanosomatids are described as being restricted to insects of the orders Diptera, Hymenoptera, Siphonaptera and Hemiptera. However, the monogenetic trait of these protists is not strict because heteroxenous life cycles have been reported for some, as follows: (i) Leptomonas seymouri observed in coinfections with Leishmania donovani in patients with visceral leishmaniasis [2,3]; (ii) Leptomonas sp. and Herpetomonas samuelpessoai mixed infection in an HIV-positive patient [4,5]; (iii) members of Herpetomonas infecting plants [6]; and (iv) a recent report of Blastocrithidia sp. infecting bats [7]. All these findings show that these parasites exhibit an unsuspected adaptability to new environments.
Trypanosomes (Trypanosoma spp.) belong to a monophyletic group of heteroxenous parasites that are well adapted to several vertebrate species and are transmitted by blood-sucking vectors, such as leeches, ticks and insects. Due to their public health importance, some of these parasite species have been studied extensively. Among them are the causative agents of Chagas disease and sleeping sickness, Trypanosoma cruzi and Trypanosoma brucei, respectively. T. cruzi is found in hundreds of New World mammal species; indeed, this highly heterogeneous parasite displays a complex life cycle involving high host diversity. T. cruzi genotypes have been grouped into six discrete typing units (DTUs), TcI–TcVI, in addition to a seventh one named Tcbat. All genotypes of T. cruzi are able to infect humans, and Chagas disease has possible severe clinical outcomes [8]. The geographical distribution of these DTUs varies according to the different scenarios of transmission in which their hosts are involved. T. cruzi is placed within the T. cruzi clade, which includes other trypanosome species that infect a diversity of mammals (including humans), bats in particular.
Unlike other mammals, bats (Chiroptera) have morphological and physiological adaptations for powered flight. Although these animals are key contributors to the value of nature to humans [9], bats may be distrusted due to their potential link to human diseases [10]. A trade-off between the evolution of flight and evolution of the immune system and their metabolism may have allowed bats to host many parasites, some of which are deadly to humans [11,12].
Bats from all over the world are parasitized by several species of Trypanosoma, with some exclusively infecting these mammals [7,13–17]. Furthermore, recent studies reveal high bat trypanosome diversity [13,17–20], clarifying the ancient origin of the bat-trypanosome interaction. Currently, the most accepted hypothesis explaining the origin of T. cruzi indicates that this parasite species evolved from an ancestral bat trypanosome [21].
The Atlantic Forest is the second-largest Brazilian biome, encompassing the east coast of the country. Despite severe fragmentation, the Atlantic Forest continues to show high levels of species diversity [22,23]. Although the bat diversity in the Atlantic Forest of Rio de Janeiro has been well studied, very little is known about the diversity of trypanosomatids circulating in bats and the role of bats in maintaining this diversity in the region. Moreover, approximately 70% of Rio de Janeiro’s population lives in the Atlantic Forest area [24], which reinforces the importance of including parasites that can potentially infect humans in studies of local biodiversity.
In this study, we surveyed the diversity of trypanosomatid species that circulate in bats from two areas with different conservation and connectivity levels in the Atlantic Forest of Rio de Janeiro. For the most preserved and larger area (Guapiaçu Ecological Reserve), there has been no indication of trypanosomatid infection. In contrast, the more degraded and smaller area (Fiocruz Atlantic Forest Biological Station) represents an important scenario in terms of the transmission of T. cruzi among wild mammals and Leishmania parasites, with human and canine cases of visceral leishmaniasis having been reported [25]. Our working hypothesis was that the diversity of trypanosomatids varies according to the local chiropterofauna, which is modulated by the level of preservation of the habitat.
Material and methods
Ethics statement
Bat samplings were conducted under permits of Instituto Chico Mendes de Conservação da Biodiversidade (19037–1). All collected bats received a number and were deposited in the mammal collection of the National Museum, administered by the Federal University of Rio de Janeiro, Rio de Janeiro, Brazil. Field procedures were approved by the Fiocruz’s Ethics Commission on Animal Use (licenses L-81/12, LW-68/12). All procedures were performed according to biosafety standards, and no environmental damage occurred.
Study areas and bat sampling
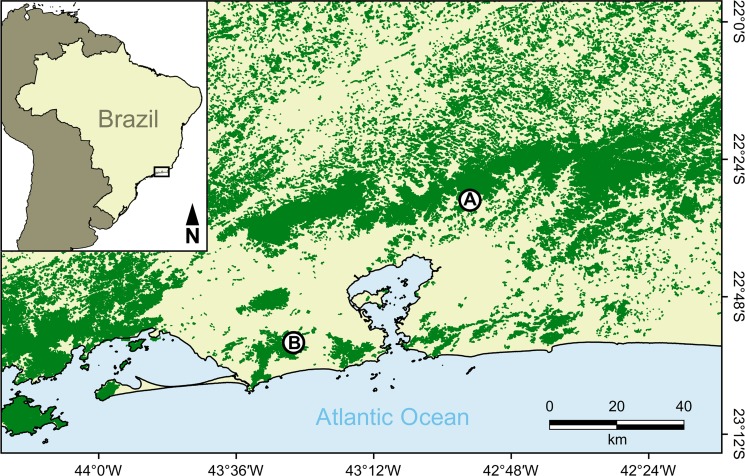
The study was performed in two areas, Reserva Ecológica de Guapiaçu (REGUA [Guapiaçu Ecological Reserve]) and Estação Biológica Fiocruz Mata Atlântica (EFMA [Fiocruz Atlantic Forest Biological Station]), both in the Atlantic Forest of Rio de Janeiro state (Fig 1). REGUA (22°25'53"S, 42°45'20"W) is a 4,000 ha private reserve in Guapiaçu village, Cachoeiras de Macacu municipality. This area is located in the mountainous region of Rio de Janeiro, 110 km away from the Rio de Janeiro municipality, and is widely connected to the largest remnant of Atlantic Forest in the state, with a total of more than 20,000 ha of continuous forests surrounded by matrixes of pastures and plantations. EFMA (22º56'23"S, 43º24'12"W) is located within the core of the Rio de Janeiro municipality, comprising 480 ha in the Pedra Branca Massif (~ 6,000 ha). The Pedra Branca Massif is fully isolated from other fragments and surrounded by low-income communities.
Fig 1.
Map of part of Rio de Janeiro state, southeastern Brazil, showing the Atlantic Forest remnants (green) where bats were sampled: Reserva Ecológica de Guapiaçu (A) and Estação Biológica Fiocruz Mata Atlântica (B).
Three sampling sites representing a forest structure gradient from the border to the interior of the forest were established in each area. These sites were defined as follows: (P1) forest edge with initial secondary forest and high anthropogenic intervention, (P2) initial secondary forest with low anthropogenic intervention, and (P3) late secondary forest with low anthropogenic intervention.
Bat and trypanosomatid samplings
From September to December 2015 and February to May 2016, bats were sampled every two months in each area, covering both dry and wet seasons. Ten mist nets (Zootech 9×3 m, 20 mm mesh) were used per night, with one night of sampling at each sampling site (totaling three nights per area every two months). Ground-level mist nets were placed along trails, in forest clearings and on watercourses, remaining open for five hours after sunset. Eight sampling campaigns were carried out, totaling 12 sampling nights for each area of capture. Bats were removed from the nets, sexed and measured [26]. Some bats were released after being marked with numbered bands on the forearm, and some were collected for the trypanosomatid survey according to the capture license. These bats were kept in individual cotton bags and processed the next day in the field laboratory.
In the following morning, the bats that were selected for the trypanosomatid survey were anesthetized using a combination of 10% ketamine hydrochloride and 2% acepromazine (9:1). After this procedure, the body was sanitized using bactericidal soap, iodized alcohol and 70% alcohol. Approximately 0.5–1 mL of blood was collected using heart puncture for the following procedures: (i) inoculation of one tube containing NNN/LIT (Novy-MacNeal-Nicolle/liver infusion tryptose) medium and another tube containing NNN/Schneider medium (0.2–0.4 mL in each tube) and (ii) a fresh blood examination. The anesthetized bats were euthanized after blood collection using direct application of potassium chloride to the heart. The collected specimens were deposited in the Museu Nacional, Universidade Federal do Rio de Janeiro. Bat identification followed Gardner [27], and nomenclature followed Nogueira et al. [28].
Parasitological procedures and DNA extraction
Fresh blood examination was performed in the field based on visual observation of a drop of blood on microscope slides using optical microscopy (400×). The samples that presented flagellates with a morphology similar to that of a Trypanosoma sp. were considered positive.
Hemocultures performed in the field were incubated at 28ºC for 10 days, after which the cultures were examined using an optical microscope every 14 days for up to 5 months. The positive blood cultures from which isolate flagellate parasites were identified were amplified and cryopreserved in the Collection of Trypanosoma from Wild and Domestic Mammals and Vectors (COLTRYP) (www.coltryp.fiocruz.br). Some positive blood cultures in which parasites were observed but for which cultures were not established were transferred to another tube (1.5 mL), centrifuged for 15 min, and stored at -20ºC for molecular characterization.
DNA extraction from positive hemocultures was performed using the phenol/chloroform method according to Sambrook and Russell [29]. At the end of this procedure, 150 μL or 40 μL of Tris-EDTA buffer solution (pH 8.0) was added to the DNA extracted from the hemocultures from which parasites were isolated or were not isolated, respectively.
Culture smears of the LBT7439 sample were stained with Giemsa and observed under a Zeiss Axioplan microscope (Oberkochen, Germany).
Molecular characterization and phylogenetic analyses
Parasite identification was performed using 18 SSU nested PCR according to Noyes et al [30]. In the first step, we used GoTaq Hot Start Green Master Mix (Promega), followed by the addition of 2 μL of DNA to 0.5 μL of each primer (Oligo Try R and Oligo Try F; TRY927F 5’GAAACAAGAAACACGGGAG3’ and TRY927R 5’CTACTGGGCAGCTTGGA3’) at 16 pmol, 12.5 μL of GoTaq Master Mix and ultra-pure water for a total volume of 25 μL. The reaction was performed under the following conditions: initial denaturation at 95ºC for 15 min and 30 cycles of 94ºC for 30 s, 55ºC for 60 s and 72ºC for 92 s. The expected fragment size for this step was 900 bp. In the second step, we used primers at the same concentrations (SSU561F 5’TGGGATAACAAAGGAGCA3’ and SSU561R 5’CTGAGACTGTAACCTCAAAGC3’). The product from the first step (2 μL) was used as the template for the second step. To obtain better results, we used 5 μL for all reactions for DNA samples derived from hemocultures from which no parasites were isolated. The expected fragment size for this step was 600 bp. Amplification using these primers did not occur for two DNA samples, and we thus tested the gGAPDH gene as the target according to Borghesan et al. [6]. To each 2 μL of sample DNA, 1 μL of each primer (0.4 μM; GAPTRY-F 5’GGBCGCATGGTSTTCCAG’3 and GAPTRYr-R 5’CCCCACTCGTTRTCRTACC’3) was added, in addition to 2.5 μL of buffer, 0.75 of MgCl2, 2 μL of dNTPs, 0.3 Taq Platinum and ultra-pure water for a total volume of 25 μL. The reaction was performed under the following conditions: initial denaturation at 94ºC for 3 min and 30 cycles of 94ºC for 1 min, 55ºC for 2 min and 72ºC for 2 min. The expected fragment size was approximately 800 bp.
The amplified PCR products were visualized using ethidium bromide-stained agarose gel electrophoresis (2%). Purification was performed for positive samples using illustra GFX PCR DNA/Gel Band Purification Kit (GE, Healthcare). DNA sequencing was performed with ABI 3730 BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, California, USA) using internal primers at 3.2 pmol and an Applied Biosystems DNA Analyzer at the Oswaldo Cruz Foundation Sequencing Platform (PDTIS/FIOCRUZ).
The obtained sequences were manually edited using the SeqMan (DNAstar) [31] program. To identify species, we used sequences with both coverage and identity equal to or greater than 98%. Sequence alignment was performed using BioEdit [32] software. For phylogenetic analysis, we applied the Kimura 2-parameters model (K2 + G) (Kimura, 1980) [33] for the 18S target and neighbor-joining (NJ) method (Saitou and Nei, 1987) [34] and the Tamura Nei model (TN 9 + G) (Tamura and Nei, 1993) [35] for the gGAPDH target, both using jModel Test program 2.1. The probabilistic method of maximum likelihood (ML) was employed for both 18S and gGAPDH. Only those sequences that matched equal or greater than 98% identity without double peaks in the electropherogram were included in the ensuing phylogenetic analyses.
Diversity indexes and statistical analysis
We utilized two diversity indexes to assess differences in species evenness between the sampling points (Shannon H and Simpson D), one to examine differences in richness (Margalef) and two to verify differences in dominance and equitability. We performed Kruskal-Wallis one-way analysis to evaluate distribution for the samples, whereby significance (p < 0.05) indicates that at least one sample stochastically dominated one other sample. To assess the association between the level of preservation and bat infection, we performed chi-square tests on contingency tables cross-classifying infection (infected/not infected) by the sampling points in each area; results with a p-value smaller than 0.05 were considered significant.
Results
A total of 181 bats (113 males and 68 females) of 18 species were surveyed for trypanosomatid infection in both areas. Among them, 116 bats (64%) were captured in REGUA and 65 (36%) in EFMA. Most species of bats included representatives of Phyllostomidae (17 species), in contrast to the Vespertilionidae (one species, Myotis riparius) family (Table 1). Carollia perspicillata and Artibeus lituratus were the most frequent species in both areas and represented the largest samples of specimens examined for trypanosomatids. Diversity indices indicated lower evenness and richness for most preserved sampling sites (P3). Despite differences in richness and diversity, Kruskal-Wallis tests did not show significant differences between the sampling areas (p = 0.51) or among the sampling sites within EFMA (p = 0.54) and REGUA (p = 0.24).
Table 1. Bat species examined at each area (Estação Biológica Fiocruz Mata Atlântica [EFMA] and Reserva Ecológica Guapiaçu [REGUA]) and sampling site (P1, P2 or P3).
| Area/sampling sites | Trophic guild (Kalko 1996)[36] |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bat species | EFMA | REGUA | |||||||
| P1 | P2 | P3 | P1 | P2 | P3 | ||||
| Anoura caudifer | – | – | 1 | 2 | 1 | – | Nectarivorous | ||
| Anoura geoffroyi | – | – | – | 2 | – | – | Nectarivorous | ||
| Artibeus fimbriatus | – | – | 1 | 1 | – | – | Frugivorous | ||
| Artibeus lituratus | 8 | 2 | 6 | 7 | 8 | 3 | Frugivorous | ||
| Artibeus obscurus | 1 | 1 | – | 1 | 1 | – | Frugivorous | ||
| Carollia perspicillata | 9 | 11 | 9 | 11 | 26 | 17 | Frugivorous | ||
| Chrotopterus auritus | – | – | – | 1 | – | – | Animalivorous | ||
| Desmodus rotundus | 1 | 2 | 1 | 1 | 4 | 1 | Hematophagous | ||
| Glossophaga soricina | – | 1 | – | – | – | – | Nectarivorous | ||
| Lonchophylla peracchii | – | 1 | – | – | – | – | Nectarivorous | ||
| Lonchorrina aurita | – | – | – | – | 1 | – | Insectivorous | ||
| Micronycteris microtis | – | 1 | – | – | – | – | Insectivorous | ||
| Myotis riparius | – | – | – | 1 | – | – | Insectivorous | ||
| Phyllostomus hastatus | 2 | – | – | 5 | – | – | Omnivorous | ||
| Platyrrhinus recifinus | - | – | 1 | – | – | 1 | Frugivorous | ||
| Sturnira lilium | 1 | 3 | – | 8 | 6 | 4 | Frugivorous | ||
| Sturnira tildae | 1 | – | – | – | – | 2 | Frugivorous | ||
| Vampyressa pusilla | – | 1 | – | – | 1 | – | Frugivorous | ||
| Total number of individuals | 23 | 23 | 19 | 40 | 48 | 28 | |||
| Total number of species | 7 | 9 | 6 | 11 | 8 | 6 | |||
Only one blood sample from an S. lilium individual (LBT7559) captured in REGUA presented flagellate parasites upon fresh blood examination. Infection of this bat was also confirmed by parasite isolation using both LIT and Schneider culture media, and the flagellates were characterized as Trypanosoma dionisii.
Overall, 24 bats (13%) were positive for trypanosomatids, with no significant difference between the studied areas: 15% (n = 17) and 11% (n = 7) for REGUA and EFMA, respectively (p = 0.46). Moreover, significant differences in infection rates among the sampling sites within each area were not observed (p = 0.5 for REGUA and p = 0.07 for EFMA) (Fig 2). Trypanosomatid infection was detected in 6 bat species: Carollia perspicillata, Artibeus lituratus, Sturnira lilium, Desmodus rotundus, Anoura caudifer and A. geoffroyi. Flagellate parasites were detected in LIT (42%, n = 10 bats), Schneider (17%, n = 4 bats) or both types (42%, n = 10 bats) of culture media. Parasites were isolated from 17 blood cultures and were cryopreserved and deposited in COLTRYP. Another 16 positive cultures did not yield in cryopreserved isolates but were molecularly analyzed by PCR using a single culture (LIT medium from LBT7663). T. cruzi-like flagellates were observed by light microscopy, but it was not possible to isolate flagellates to obtain sufficient DNA for molecular analysis (Table 2).
Fig 2. Trypanosomatid infection rate in bats captured at REGUA (Reserva Ecológica Guapiaçú) and EFMA (Estação Biológica Fiocruz Mata Atlântica), Rio de Janeiro state, Brazil.
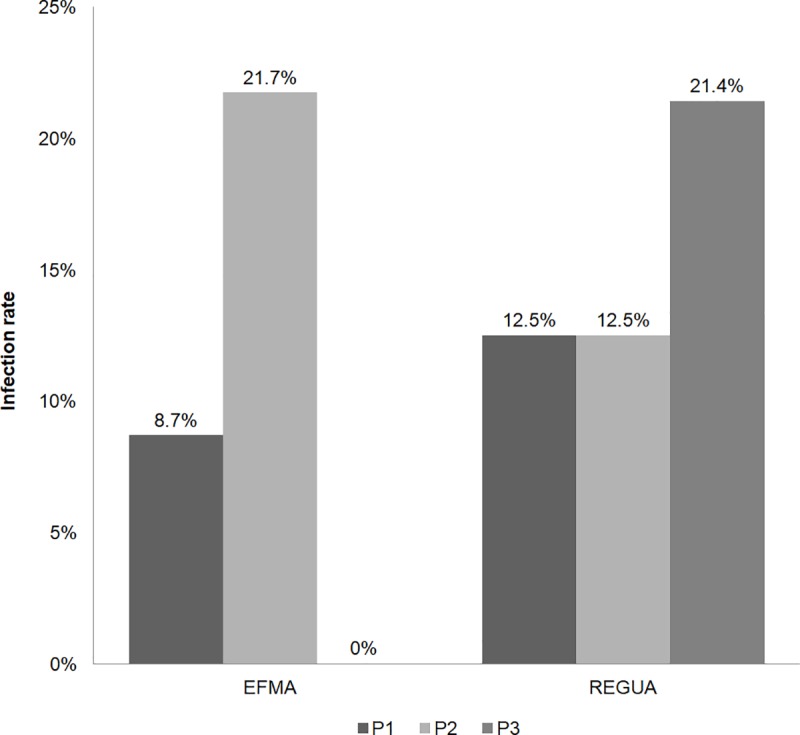
Infection rates by sampling site for each study area are shown in columns. Sites P1, P2 and P3 are represented by colors.
Table 2. Trypanosomatid species infecting bats.
This table shows the bat identification number, area (Reserva Biológica de Guapiaçu/REGUA or Estação Biológica Fiocruz da Mata Atlântica/EFMA), bat species, positive culture media, culture media in which the parasite was isolated and cryopreserved at COLTRYP (with their respective numbers) and the characterized parasite species and GenBank accession number.
| LBT | Area | Bat species | Media* | Isolate* | COLTRYP number | 18S SSU gene characterization |
GenBank accession number |
|---|---|---|---|---|---|---|---|
| 7439 | REGUA | Anoura caudifer | L | L | 685 | Crithidia mellificae** | MH469963 |
| 7149 | REGUA | Anoura geoffroyi | L/S | - | - | T. dionisii (L/S) | MH469960/MH469961 |
| 6873 | REGUA | Carollia perspicillata | L | - | - | Trypanosoma sp.1** | - |
| 6890 | REGUA | Carollia perspicillata | L | - | - | Trypanosoma sp.2 | MH469953 |
| 7578 | REGUA | Carollia perspicillata | L | - | - | Trypanosoma sp.1 | - |
| 6882 | REGUA | Carollia perspicillata | L | L | 642 | T. cruzi TcI | MH469942 |
| 7557 | REGUA | Carollia perspicillata | L | L | 706 | T. dionisii | MH469949 |
| 7137 | REGUA | Carollia perspicillata | S | - | - | T. dionisii | MH469958 |
| 7455 | REGUA | Carollia perspicillata | S | S | 698 | T. dionisii | MH469948 |
| 6889 | REGUA | Desmodus rotundus | L | L | 639 | T. dionisii | MH469940 |
| 7118 | REGUA | Desmodus rotundus | L/S | - | - | Trypanosoma sp.2 (L/S) | MH469955/MH469956 |
| 7123 | REGUA | Desmodus rotundus | S | - | - | Trypanosoma sp.2 | MH469957 |
| 6875 | REGUA | Sturnira lilium | L | L | 638 | T. dionisii | MH469939 |
| 7138 | REGUA | Sturnira lilium | L/S | - | - | Trypanosoma sp.1 (L/S) | - |
| 7148 | REGUA | Sturnira lilium | L/S | L | 693 | T. dionisii (L)/T. cruzi TcI(S) | MH469947/MH469959 |
| 7559 | REGUA | Sturnira lilium | L/S | L/S | 708 | T. dionisii (L/S) | MH469951/MH469952 |
| 7128 | REGUA | Sturnira lilium | S | - | - | T. cruzi | - |
| 6967 | EFMA | Artibeus lituratus | L/S | L | 645 | T. cruzi TcIII (L)/T. dionisii (S) | MH469946/MH469954 |
| 7668 | EFMA | Carollia perspicillata | L | - | - | T. cruzi TcIV | MH469962 |
| 7666 | EFMA | Carollia perspicillata | L | L | 709 | Trypanosoma sp.1 | - |
| 6968 | EFMA | Carollia perspicillata | L/S | L | 641 | T. dionisii (L/S) | MH469941 |
| 7663 | EFMA | Carollia perspicillata | L/S | S | 707 | T. dionisii (S) | MH469950 |
| 6973 | EFMA | Desmodus rotundus | L/S | L/S | 644 | T. dionisii (L/S) | MH469944/MH469945 |
| 6969 | EFMA | Sturnira lilium | L/S | L/S | 643 | T. dionisii (S)/Trypanosoma sp.1(L) | MH469943 |
* L = LIT; S = Schneider
** target gene—gGAPDH
1 Trypanosoma sp. not included in the phylogenetic tree due to the low identity observed by Blast search (<98%) or the presence of double peaks in sequence analysis.
2 Trypanosoma sp. included in the phylogenetic tree.
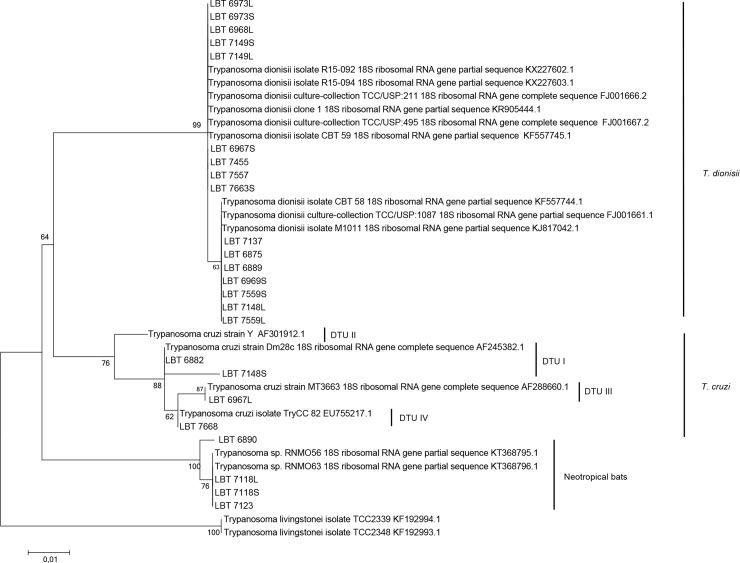
T. dionisii was found in the majority of the infected bats (54%; n = 13), which included C. perspicillata (n = 5), S. lilium (4), D. rotundus (2), A. lituratus (1) and A. geoffroyi (1), with eight from REGUA and five from EFMA. One individual of Sturnira lilium (LBT7148) and one of A. lituratus (LBT6967) presented mixed infections by T. cruzi DTUs I and III, respectively (Table 2).
Four samples of C. perspicillata and D. rotundus (LBT6890, LBT7118 and LBT7123) were infected with Neobat trypanosomes (Fig 3). Identity to sequences from species belonging to Trypanosoma was found for another five samples of C. perspicillata (n = 3) (LBT7578, LBT7666, and LBT6873) and S. lilium (n = 2) (LBT7138 and LBT6969); however, it was not possible to identify the species.
Fig 3. Molecular phylogenetic analysis of the parasite species detected in bats using maximum likelihood based on the 18S SSU rRNA gene with the K2 + G parameters and a bootstrap value of 1000 replicates.
T. livingstonei isolates TCC 2339 and TCC 2348 were used as outgroups.
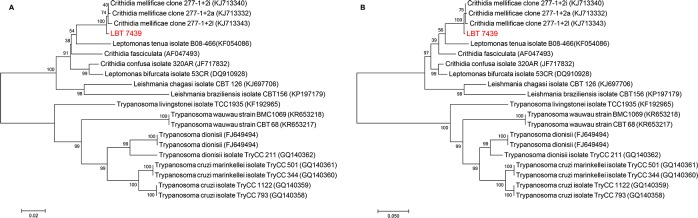
The parasites isolated from one individual of Anoura caudifer (LBT7439) from REGUA presented a peculiar choanomastigote morphology (Fig 4). Sequencing analysis of the products derived from 18S SSU gene PCR suggested a monoxenous parasite infection, but with low identity. The product revealed 88% identity with deposited Crithidia mellificae strains and with other putative insect parasites, such as C. expoeki, C. bombi and Leptomonas spp. Identification of this isolate as being the putative monoxenous trypanosomatid species C. mellificae was achieved with 99% identity using gGAPDH PCR (Table 2; Figs 3 and 5; S1 Text; S2 Text).
Fig 4. Choanomastigote forms observed in axenic culture of the LBT 7439 sample.
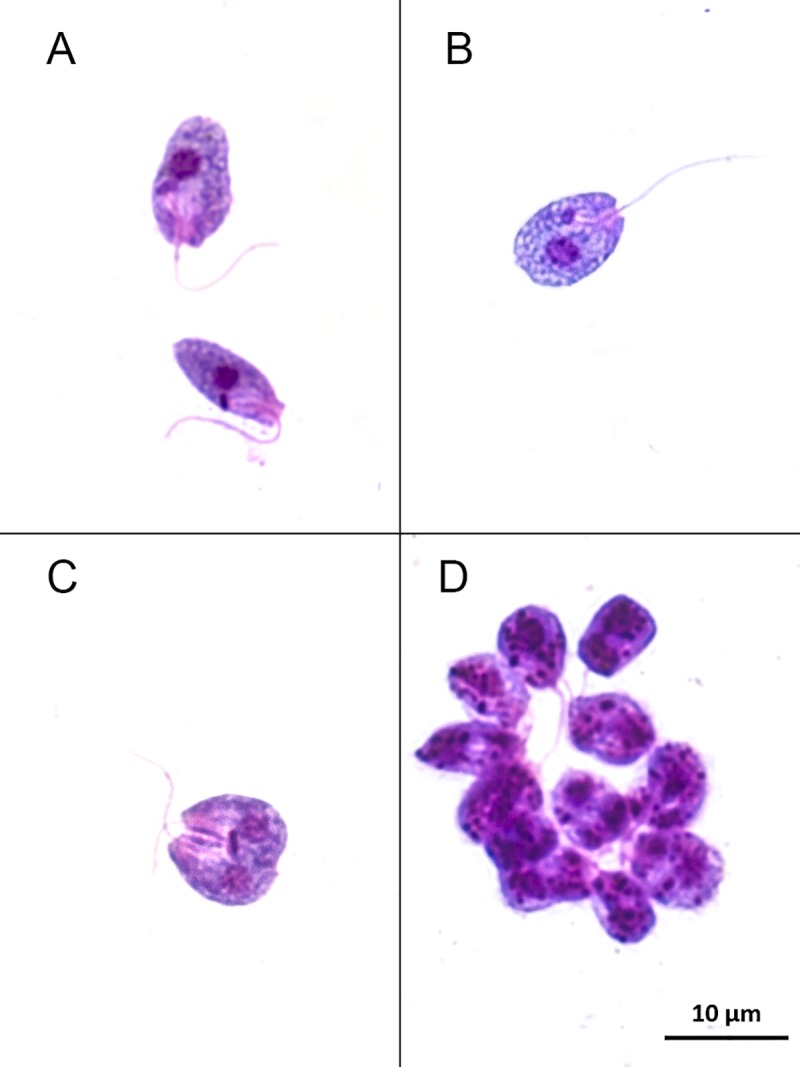
(A) Parasite cells show a typical collar-like extension through which a single flagellum emerges. (B) The kinetoplast is anterior to the nucleus and adjacent to the flagellar pocket where emergence of the flagellum can be observed. (C) Dividing forms. (D) Rosette of flagella.
Fig 5. Molecular phylogenetic analysis of the LBT 7439 sample.
(A) Analysis using neighbor-joining of the gGAPDH gene with the K2 parameter and a bootstrap value of 1000 replicates. (B) Analysis of the gGAPDH gene using maximum likelihood with Tamura-Nei (93 + G) and a bootstrap value of 1000 replicates.
Discussion
Bats have been described as hosts and/or potential reservoirs of trypanosomes and other microorganisms, such as viruses and bacteria [10,37,38]. Here, we compare trypanosomatid infection in bats from two areas with different levels of preservation and connectivity in the Atlantic Forest of Rio de Janeiro state.
Detection of parasites in one fresh blood examination (LBT 7559) and in blood cultures (LIT/Schneider) revealed that bats may maintain, at least for a time period, detectable parasitemia by the Trypanosoma species isolated in this study. Thus, bats may act as an infection source of these parasite species for vectors, as no more than 0.3 mL of blood was cultured and only one drop was used for the fresh blood examination. This also indicates the important role played by infected bats in the maintenance in nature of the trypanosomatid species detected [39]. We observed a sylvatic transmission cycle of distinct trypanosomatid species occurring among bats in REGUA and EFMA. Bats from the most preserved larger area (REGUA) showed slightly higher infection rates than did those from the most degraded and smaller areas (EFMA), 15% and 11%, respectively. The relationship between hemoparasite infection rates and bat species richness in response to fragmented or disturbed habitats was previously studied by Cottontail et al. [40] and Costa et al. [41], who reported a negative correlation between species richness and infection rate, supporting the Dilution Effect proposed by Keesing [42]. Thus, we would expect lower infection rates with higher bat species richness.
The shelters used by bats may reveal their parasite transmission dynamics. Twenty-three of 24 infected bats use a wide variety of shelters, including hollow trees, caves, palm leaves and artificial constructions. Only A. lituratus uses palm trees as shelter [43]. As Triatominae may also utilize these shelters, such behavior may increase the chances of occasional encounters between bats and T. cruzi vectors. A different scenario occurs in the Amazon biome, where the most important T. cruzi vectors are Rhodnius spp. [44–46]. The infection rate among bats roosting in palm trees appears to be higher in the Amazon compared to the Atlantic Forest [14,41,47].
The most frequent species in the study areas (C. perspicillata, S. lilium and D. rotundus [48,49]) presented the highest infection rates. These bats are well adapted to different landscapes. In South America, C. perspicillata has been reported as the species most infected by T. cruzi DTUs I, III, IV and V, as well as by T. c. marinkellei, T. dionisii, and T. rangeli [14,15,18,41,50–54]. In Brazil, S. lilium has been found to be infected by T. dionisii and T. cruzi [55,56]. In this sense, four distinct T. cruzi lineages able to infect humans were already associated with the most frequent bat species captured in the study areas. Nonetheless, these findings only reflect the high abundance of these bat species in the tropics, not necessarily the association between them and those parasites.
It is impossible to evaluate the impact of trypanosomatid infection on bat fitness and survival rates. Moreover, it is worth considering that all wild mammalian surveys include only those animals that preserved their ability to dislocate, which involves a unavoidable bias [54,57]. As expected, T. dionisii, described as infecting bats in both the New and Old Worlds, was the trypanosomatid species most detected, and the ability to infect a high diversity of bats, including at least 20 species representing five families and different trophic guilds, has been described for T. dionisii [20,53,54]. In Brazil, T. dionisii has been found to infect the phyllostomids C. perspicillata and S. lilium as well as noctilionids, molossids and vespertiolionids [54], though the means of transmission remains unclear. T. dionisii has also been reported to be a unique vector for the Old World parasite Cimex pipistrellus [58]. Conversely, bed bugs serve as vectors for another Trypanosoma species, T. vespertilionis, which is restricted to the Old World. Recently, our group detected T. dionisii infection in triatomines (Triatoma vitticeps) from the Guarapari municipality, Espírito Santo, Brazil [15]. This finding shows that T. cruzi and T. dionisii might share vectors in the New World.
We detected mixed infection by T. dionisii and T. cruzi TcI and TcIII in S. lilium and A. lituratus, respectively. Detection from blood samples reveals the capacity of the host to present high parasitemia, being a competent reservoir to transmit those parasites to a vector. We also detected single infections by T. cruzi DTUs TcI and TcIV in C. perspicillata. The DTUs TcI, TcIII and TcIV are often found in bats [54,59], even in mixed infections, such as TcI/TcIV, which is the second most common association in nature. The DTU TcI is considered to be the most widespread and frequent DTU in nature [60]. In recent acute cases of Chagas disease in Brazil (Chagas disease outbreaks), human isolates were genotyped as TcI and TcIV [61].
We also successfully isolated C. mellificae for the first time in South America. C. mellificae was first described as infecting bees (Apis mellifera) in Australia in 1967 and for many years was considered to be the main agent of honey bee colony losses around the world [62,63]. This monoxenic species appears to be able to infect a large diversity of representatives of the Hymenoptera order because it has been found in Vespula squamosa (wasp; Vespidae), Osmia cornuta (orchard bee; Megachilidae) and O. bicornis (mason bee; Megachilidae) in the United States [64].
We speculate two possible scenarios for explaining infection of Anoura caudifer, a primarily nectar-feeding bat [65]: (i) the bat may have become infected through ingestion of pollen contaminated with bee excreta, as this parasite is found in the posterior portion of the bee’s digestive tract; or (ii) an infected bee may have shed its stinger, which normally includes the posterior portion of its digestive tract, in the bat, and it then became infected by licking the sting site. Moreover, there are two unexpected and original aspects to consider: one is the first description of this monoxenic trypanosomatid in South America, and the other is the first isolation of a Crithidia species from a mammalian host. This highlights the necessity of (1) carefully reevaluating the current geographical distribution of C. mellificae, (2) attempting to understand whether this parasite is expanding its worldwide distribution or if it merely has never been previously noticed and (3) reevaluating the host specificity of trypanosomatids and their competence for spill over for other taxa. Regardless, the leap that constitutes adapting from the digestive tube of an insect to the circulatory system of a mammal is dramatic.
For both proposed scenarios, the infection would have occurred through the oral route, despite the rare opportunity for these animals to interact in nature because they have different foraging behaviors in terms of the timing of activity. A recent study conducted in Texas, USA, detected for the first time another monoxenous trypanosomatid, Blastocrithidia sp., infecting bats [7]. The authors described this parasite in the cardiac tissues of the insectivorous bats Tadarida brasiliensis (Molossidae) and Nycticeius humeralis (Vespertilionidae). Although not isolated, detection of this monoxenous parasite in cardiac tissue, along with our results, reveal the potential of these monoxenous parasites to successfully infect mammalian hosts.
Apart from the surprising isolation of C. mellificae, we still have scant information about the real diversity of trypanosomatid species that are able to infect bats. The pattern of infection by a parasite may change among its hosts, which is the reason why host biology should be important in parasitological studies. Considering the high diversity of trypanosomatid species and/or genotypes isolated from blood cultures, we conclude that bats are extremely resilient to infection by trypanosomatids and can be sources of infection to vectors, being able to maintain a high diversity of these parasites.
Our study provides an overview of trypanosomatid diversity in bats from two areas with different levels of human disturbance. In both areas, the most frequent species, Carollia perspicillata and Sturnira lilium, serve as reservoirs of T. cruzi and T. dionisii, both of which are able to infect humans. Moreover, the most frequently detected T. cruzi DTUs in bats (TcI and TcV) are most commonly associated with new Chagas disease cases in Brazil [61]. The finding of a putative monoxenous parasite (C. mellificae) infecting bats reinforces the urgency of discussing the concepts of parasitism and hosts among trypanosomatids. These bat species that were more frequent in the two areas exhibit plasticity in habitat usage, occurring in forests with different levels of preservation as well as in urban areas. Our data show that they are also flexible in their capacity to maintain a high diversity of trypanosomatid parasites.
Supporting information
Aligment of gGAPDH GenBank reference sequences used in Crithidia mellificae phylogenetic trees.
(TXT)
Aligment of 18S rRNA GenBank reference sequences used in Trypanosoma spp. phylogenetic trees.
(TXT)
Acknowledgments
We thank Nicholas Locke, Rachel Locke, Jorge Bizarro and all of the REGUA crew for their support in this research project and Isabela Bellizzi, Letícia Moraes, Lucas Slama, Rafael Koster, Raíssa Souza and Saulo Felix for fieldwork support in REGUA. We also thank Brunna Almeida, André Queiroz dos Santos and Fernanda Alves for fieldwork support in EFMA. We are also thankful to Carlos Ardé, Marcos Antônio dos Santos Lima and Marlon Vicente da Silva for technical support in the hemocultures and image processing; the DTIS/Fiocruz sequencing platform for sample sequencing; and Marina Rodrigues for support in the molecular analysis.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The study was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico/CNPq (PAPES VII - 401764/2015-0) to ALRR, Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro/FAPERJ (JCNE E-26/203.191/2016) to ALRR, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior do Brasil/CAPES (Finance Code 001) to DAR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.D’Avila-Levy CM, Boucinha C, Kostygov A, Santos HLC, Morelli KA, Grybchuk-Ieremenko A, et al. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Mem Inst Oswaldo Cruz. 2015;110: 956–965. 10.1590/0074-02760150253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraeva N, Butenko A, Hlaváčová J, Kostygov A, Myškova J, Grybchuk D, et al. Leptomonas seymouri: Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with Leishmania donovani. Tibayrenc M, editor. PLOS Pathog. Public Library of Science; 2015;11: e1005127 10.1371/journal.ppat.1005127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh S, Banerjee P, Sarkar A, Datta S, Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J Clin Microbiol. 2012;50: 2774–2778. 10.1128/JCM.00966-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morio F, Reynes J, Dollet M, Pratlong F, Dedet JP, Ravel C. Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. J Clin Microbiol. 2008;46: 3845–3847. 10.1128/JCM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacheco RS, Marzochi MCA, Pires MQ, Brito CMM, Madeira MDF, Barbosa-Santos EGO. Parasite Genotypically Related to a Monoxenous Trypanosomatid of Dog’s Flea Causing Opportunistic Infection in an HIV Positive Patient. Mem Inst Oswaldo Cruz. 1998;93: 531–537. 10.1590/s0074-02761998000400021 [DOI] [PubMed] [Google Scholar]
- 6.Borghesan TC, Ferreira RC, Takata CSA, Campaner M, Borda CC, Paiva F, et al. Molecular Phylogenetic Redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a Genus of Insect Parasites Associated with Flies. Protist. Urban & Fischer; 2013;164: 129–152. 10.1016/j.protis.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 7.Hodo CL, Goodwin CC, Mayes BC, Mariscal JA, Waldrup KA, Hamer SA. Trypanosome species, including Trypanosoma cruzi, in sylvatic and peridomestic bats of Texas, USA. Acta Trop. 2016;164: 259–266. 10.1016/j.actatropica.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12: 240–253. 10.1016/j.meegid.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 9.Díaz S, Pascual U, Stenseke M, Martín-López B, Watson RT, Molnár Z, et al. Assessing nature’s contributions to people. Science (80-). 2018;359: 270–272. 10.1126/science.aap8826 [DOI] [PubMed] [Google Scholar]
- 10.Moratelli R, Calisher CH. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem Inst Oswaldo Cruz. 2015;110: 1–22. 10.1590/0074-02760150048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brook CE, Dobson AP. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23: 172–180. 10.1016/j.tim.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DTS, Luis AD, et al. Bat Flight and Zoonotic Viruses. Emerg Infect Dis. 2014;20: 741–745. 10.3201/eid2005.130539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima L, Espinosa-Álvarez O, Hamilton PB, Neves L, Takata CS, Campaner M, et al. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasit Vectors. 2013;6: 221 10.1186/1756-3305-6-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.dos Santos FCB, Lisboa C V., Xavier SCC, Dario MA, Verde RDS, Calouro AM, et al. Trypanosoma sp. diversity in Amazonian bats (Chiroptera; Mammalia) from Acre State, Brazil. Parasitology. 2017;2016: 1–10. 10.1017/S0031182017001834 [DOI] [PubMed] [Google Scholar]
- 15.Dario MA, Lisboa CV, Costa LM, Moratelli R, Nascimento MP, Costa LP, et al. High Trypanosoma spp. diversity is maintained by bats and triatomines in Espírito Santo state, Brazil. Solari A, editor. PLoS One. 2017;12: e0188412 10.1371/journal.pone.0188412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto CM, Ocaña-Mayorga S, Tapia EE, Lobos SE, Zurita AP, Aguirre-Villacís F, et al. Bats, trypanosomes, and triatomines in Ecuador: New insights into the diversity, transmission, and origins of trypanosoma cruzi and chagas disease. PLoS One. 2015;10: 1–13. 10.1371/journal.pone.0139999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima L, Espinosa-Álvarez O, Pinto CM, Cavazzana M, Pavan AC, Carranza JC, et al. New insights into the evolution of the Trypanosoma cruzi clade provided by a new trypanosome species tightly linked to Neotropical Pteronotus bats and related to an Australian lineage of trypanosomes. Parasit Vectors. Parasites & Vectors; 2015;8: 657 10.1186/s13071-015-1255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dario MA, Moratelli R, Schwabl P, Jansen AM, Llewellyn S. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Negl Trop Dis. 2017;11: 1–15. 10.1371/journal.pntd.0005790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima L, Silva FM da, Neves L, Attias M, Takata CSA, Campaner M, et al. Evolutionary Insights from Bat Trypanosomes: Morphological, Developmental and Phylogenetic Evidence of a New Species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African Bats Closely Related to Trypanosoma (Schizotrypanum) cruzi and Allied Species. Protist. Elsevier GmbH.; 2012;163: 856–872. 10.1016/j.protis.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 20.Dario MA, Rodrigues MS, Helena J, Cristina S, Andrea PSD, Luiz A, et al. Ecological scenario and Trypanosoma cruzi DTU characterization of a fatal acute Chagas disease case transmitted orally (Espírito Santo state, Brazil). Parasit Vectors. Parasites & Vectors; 2016;9: 1–14. 10.1186/s13071-015-1291-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton PB, Teixeira MM, Stevens JR. The evolution of Trypanosoma cruzi: the 'bat seeding' hypothesis. Trends Parasitol. 2012;28(4):136–41. 10.1016/j.pt.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 22.Costa LP, Leite YLR, da Fonseca GAB, da Fonseca MT. Biogeography of South American Forest Mammals: Endemism and Diversity in the Atlantic Forest1. Biotropica. 2000;32: 872 10.1646/0006-3606(2000)032[0872:BOSAFM]2.0.CO;2 [DOI] [Google Scholar]
- 23.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. Nature Publishing Group; 2000;403: 853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- 24.Oliveira RR de. Os cenários da paisagem In: Oliveira RR de editor. As marcas do homem na floresta: História ambiental de um trecho urbano de mata atlântica. Rio de Janeiro: PUC- Rio; 2010. p. 230. [Google Scholar]
- 25.Marzochi MCDA, Fagundes A, Andrade MV De, Souza MB de, Madeira Ma de F, Mouta-Confort E, et al. Visceral leishmaniasis in Rio de Janeiro, Brazil: eco-epidemiological aspects and control Leishmaniose visceral no Rio de Janeiro, Brasil: aspectos eco-epidemiológicos e controle. Rev Soc Bras Med Trop. 2009;42: 570–580. 10.1590/S0037-86822009000500017 [DOI] [PubMed] [Google Scholar]
- 26.Kunz T. Methods of making bats In: Wilson DE, Russell-Cole F, Nichols JD, Rudran R FM, editor. Measuring and monitoring biological diversity: standart methods for mammals. Washington D.C: Smithsonian Press; 1996. pp. 304–310. [Google Scholar]
- 27.Gardner A. Mammals of South America, Volume I Marsupials, Xenarthrans, Shrews, and Bats. Chicago: The University of Chicago Press; 2008. [Google Scholar]
- 28.Nogueira M, Lima I, Moratelli R, Tavares V, Gregorin R, Peracchi A. Checklist of Brazilian bats, with comments on original records. Check List. 2014;10: 808–821. [Google Scholar]
- 29.Sambrook J, Russel DW. Molecular Cloning—A laboratory Manual. 3rd ed New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 30.Noyes HA, Stevens JR, Teixeira M, Phelan J, Holz P. A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int J Parasitol. 1999;29: 331–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/10221634 [DOI] [PubMed] [Google Scholar]
- 31.DNASTAR. Lasergene—SeqMan. -: Software, Version 7.0.0; 2006.
- 32.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT [Internet]. Nucleic Acids Symposium Series. 1999. pp. 95–98. citeulike-article-id:69177410780396 [Google Scholar]
- 33.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–20. Available: http://www.ncbi.nlm.nih.gov/pubmed/7463489 [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4: 406–25. Available: http://www.ncbi.nlm.nih.gov/pubmed/3447015 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10: 512–26. Available: http://www.ncbi.nlm.nih.gov/pubmed/8336541 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 36.Kalko EKV, Handley CO HD Jr. Organization, diversity, and long-term dynamics of a neotropical bat community,. In: Cody M.L. & Smallwood JA, editor. Long-term studies of vertebrate communities. Academic Press; 1996. pp. 503–553. [Google Scholar]
- 37.Volokhov D V., Becker DJ, Bergner LM, Camus MS, Orton RJ, Chizhikov VE, et al. Novel hemotropic mycoplasmas are widespread and genetically diverse in vampire bats. Epidemiol Infect. 2017;145: 3154–3167. 10.1017/S095026881700231X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballados-González GG, Sánchez-Montes S, Romero-Salas D, Colunga Salas P, Gutiérrez-Molina R, León-Paniagua L, et al. Detection of pathogenic Leptospira species associated with phyllostomid bats (Mammalia: Chiroptera) from Veracruz, Mexico. Transbound Emerg Dis. 2018; 1–9. 10.1111/tbed.13004 [DOI] [PubMed] [Google Scholar]
- 39.Sant’Anna MRV, Soares AC, Araujo RN, Gontijo NF, Pereira MH. Triatomines (Hemiptera, Reduviidae) blood intake: Physical constraints and biological adaptations. J Insect Physiol. 2016; 10.1016/j.jinsphys.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 40.Cottontail VM, Wellinghausen N, Kalko EK V. Habitat fragmentation and haemoparasites in the common fruit bat, Artibeus jamaicensis (Phyllostomidae) in a tropical lowland forest in Panamá. Parasitology. 2009;136: 1133–1145. 10.1017/S0031182009990485 [DOI] [PubMed] [Google Scholar]
- 41.da Costa AP, Nunes PH, Leite BHS, Ferreira JIG da S, Tonhosolo R, da Rosa AR, et al. Diversity of bats trypanosomes in hydroeletric area of Belo Monte in Brazilian Amazonia. Acta Trop. Elsevier B.V.; 2016;164: 185–193. 10.1016/j.actatropica.2016.08.033 [DOI] [PubMed] [Google Scholar]
- 42.Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol Lett. 2006;9: 485–498. 10.1111/j.1461-0248.2006.00885.x [DOI] [PubMed] [Google Scholar]
- 43.Reis NR dos Peracchi AL, Pedro WA, Lima IP de. Morcegos do Brasil. Reis NR dos, Peracchi AL, Pedro WA, Lima IP De, editors. Londrina: Nelio R. dos Reis; 2007. [Google Scholar]
- 44.Abad-Franch F, Monteiro FA. Biogeography and evolution of Amazonian triatomines (Heteroptera: Reduviidae): implications for Chagas disease surveillance in humid forest ecoregions. Mem Inst Oswaldo Cruz. 2007;102: 57–70. 10.1590/s0074-02762007005000108 [DOI] [PubMed] [Google Scholar]
- 45.Aguilar HM, Abad-Franch F, Dias JCP, Junqueira ACV, Coura JR. Chagas disease in the Amazon Region. Mem Inst Oswaldo Cruz. 2007;102: 47–56. 10.1590/s0074-02762007005000098 [DOI] [PubMed] [Google Scholar]
- 46.Coura JR. The main sceneries of chagas disease transmission. The vectors, blood and oral transmissions—A comprehensive review. Mem Inst Oswaldo Cruz. 2015;110: 277–282. 10.1590/0074-0276140362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szpeiter BB, Ferreira JIG da S, Assis FFV de, Stelmachtchuk FN, Peixoto Junior K da C, Ajzenberg D, et al. Bat trypanosomes from Tapajós-Arapiuns Extractive Reserve in Brazilian Amazon. Rev Bras Parasitol Veterinária. 2017;26: 152–158. 10.1590/s1984-29612017022 [DOI] [PubMed] [Google Scholar]
- 48.Esbérard CEL. Diversidade de morcegos em área de Mata Atlântica regenerada no sudeste do Brasil. Rev bras Zoociencias Juiz Fora. 2003;5: 189–204. [Google Scholar]
- 49.Peracchi AL, Reis NR. Mamíferos do Brasil. 2010. [Google Scholar]
- 50.Ramírez JD, Hernández C, Montilla M, Zambrano P, Flórez AC, Parra E, et al. Trypanosome species in neo-tropical bats: Biological, evolutionary and epidemiological implications. Infect Genet Evol. 2014;22: 250–256. 10.1016/j.meegid.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García L, Ortiz S, Osorio G, Torrico MC, Torrico F, Solari A. Phylogenetic Analysis of Bolivian Bat Trypanosomes of the Subgenus Schizotrypanum Based on Cytochrome b Sequence and Minicircle Analyses. Woodrow CJ, editor. PLoS One. Public Library of Science; 2012;7: e36578 10.1371/journal.pone.0036578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lourenço JLM, Minuzzi-Souza TTC, Silva LR, Oliveira AC, Mendonça VJ, Nitz N, et al. High frequency of trypanosomatids in gallery forest bats of a Neotropical savanna. Acta Trop. Elsevier; 2018;177: 200–206. 10.1016/j.actatropica.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 53.Acosta IDCL Da Costa AP, Gennari SM, Marcili A. Survey of Trypanosoma and Leishmania in Wild and Domestic Animals in an Atlantic Rainforest Fragment and Surroundings in the State of Espírito Santo, Brazil. J Med Entomol. 2014;51: 686–693. 10.1603/me13177 [DOI] [PubMed] [Google Scholar]
- 54.Cavazzana M, Marcili A, Lima L, da Silva FM, Junqueira AC V, Veludo HH, et al. Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int J Parasitol. Australian Society for Parasitology Inc.; 2010;40: 345–55. 10.1016/j.ijpara.2009.08.015 [DOI] [PubMed] [Google Scholar]
- 55.Nishimura SMM, Filho HO, Magalhães M. Trypanosomatids in Phyllostomids (Chiroptera, Phyllostomidae) from Perobas Biological Reserve, Paraná, Brazi l. Arq Ciênc Vet Zool UNIPAR, Umuarama. 2010;13: 87–91. [Google Scholar]
- 56.Lima L, Espinosa-Álvarez O, Ortiz PA, Trejo-Varón JA, Carranza JC, Pinto CM, et al. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Trop. Elsevier B.V.; 2015;151: 166–177. 10.1016/j.actatropica.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 57.Molyneux DH, Stiles JK. Trypanosomatid—vector interactions. Ann Soc Belg Med Trop (1920). 1991;71 Suppl 1: 151–66. Available: http://www.ncbi.nlm.nih.gov/pubmed/1793265 [PubMed] [Google Scholar]
- 58.Gardner RA, Molyneux DH. Schizotrypanum in British bats. Parasitology. 1988;97 (Pt 1): 43–50. Available: http://www.ncbi.nlm.nih.gov/pubmed/3174237 [DOI] [PubMed] [Google Scholar]
- 59.Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira ACV, et al. Comparative phylogeography of Trypanosoma cruzi TCIIc: New hosts, association with terrestrial ecotopes, and spatial clustering. Infect Genet Evol. 2009;9: 1265–1274. 10.1016/j.meegid.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 60.Jansen AM, Xavier SCC, Roque ALR. The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Trop. 2015;151: 1–15. 10.1016/j.actatropica.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 61.Zingales B. Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop. 2018;184: 38–52. 10.1016/j.actatropica.2017.09.017 [DOI] [PubMed] [Google Scholar]
- 62.Langride DF, Mcghee RB. Crithidia mellificae n. sp. an Acidophilic Trypanosomatid of the Honey Bee Apis mellifera. J Protozool. 1967;14: 485–487. [DOI] [PubMed] [Google Scholar]
- 63.Ravoet J, Maharramov J, Meeus I, De Smet L, Wenseleers T, Smagghe G, et al. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. Li Y, editor. PLoS One. 2013;8: e72443 10.1371/journal.pone.0072443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwarz RS, Bauchan GR, Murphy CA, Ravoet J, de Graaf DC, Evans JD. Characterization of Two Species of Trypanosomatidae from the Honey Bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J Eukaryot Microbiol. 2015;62: 567–583. 10.1111/jeu.12209 [DOI] [PubMed] [Google Scholar]
- 65.Barros M a. S, Rui AM, Fabian ME. Seasonal Variation in the Diet of the Bat Anoura caudifer (Phyllostomidae: Glossophaginae) at the Southern Limit of its Geographic Range. Acta Chiropterologica. 2013;15: 77–84. 10.3161/150811013X667876 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aligment of gGAPDH GenBank reference sequences used in Crithidia mellificae phylogenetic trees.
(TXT)
Aligment of 18S rRNA GenBank reference sequences used in Trypanosoma spp. phylogenetic trees.
(TXT)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.