Abstract
Objectives
We characterized clinical isolates of Campylobacter using whole-genome sequencing (WGS) for detection of virulence genes, antimicrobial resistance markers and phylogenetic analysis in order to increase the knowledge on the molecular epidemiology of Campylobacter in Ireland, where there are significant gaps due to the widespread in the use of culture independent methods for the diagnosis of campylobacteriosis.
Methods
WGS was applied to 122 Campylobacter human isolates collected over a 10-years period, from diarrhoeal stool samples submitted for routine enteric screening.
Results
Genes associated with cytotoxin production such as cdtA, cdtB and cdtC were found in 88%, 89% and 89% isolates, respectively; adherence, colonization and invasion genes such as cadF, dnaJ, racR, iam, virB11 and ciaB were found in 99%, 99%, 98%, 99%, 1% and 80% isolates, respectively. Genetic markers associated with resistance to quinolones (C257T in gyrA), beta-lactams (blaoxa-61) and tetracycline (tet(O)) were present in 43%, 71% and 25% isolates, respectively. The cmeABC operon was present in 94% of isolates. No macrolide or aminoglycoside resistance markers were detected. Phylogenetic analysis showed that 112 isolates were assigned to 29 sequence types grouped into 17 clonal complexes. Four clusters previously unidentified were detected. These results shown the similarity of Irish data compared to what has been described globally.
Conclusions
WGS has shown a high discriminatory power for cluster detection, demonstrating that its integration in routine laboratory surveillance could improve the detection and management of outbreaks. In addition we were able to demonstrate that virulence genes in clinical Campylobacter infections in Ireland were similar to those known previously. High prevalence of quinolone resistance markers has been found, which has implications for antimicrobial stewardship.
Introduction
Campylobacter is a leading cause of bacterial foodborne illnesses worldwide. In 2015, Campylobacter was the most commonly reported gastrointestinal bacterial pathogen in human in the European Union (EU) [1]. The species Campylobacter jejuni is responsible for 80–90% of cases, while Campylobacter coli accounts for 10–20% of human infections. The clinical manifestations caused by C. jejuni and C. coli infections are indistinguishable and comprise diarrhoea, abdominal pain and cramps as the most typical symptoms. Infection is mostly acute and self-limited. However, complications such as pancreatitis, cholecystitis, peritonitis and gastointestinal haemorrhage arise in less than 1% of cases and mostly in immunocompromised, the very young or very elderly individuals [2].
Campylobacter colonize the gastrointestinal tract of a wide variety of food-producing animals such as poultry, sheep, cattle and swine but also companion animals, wild animals and birds [3, 4]. Campylobacteriosis is a zoonosis, and humans acquire the infection by the consumption or handling of contaminated meat, poultry being the commonest source of infection. Water reservoirs have also been described as possible source of Campylobacter transmission [5, 6].
To date, the most commonly used typing methods of Campylobacter species are pulsed-field gel electrophoresis (PFGE) and the seven-locus multilocus sequence typing (MLST). However, these methods have limited discriminatory power and provide only typing information. WGS can provide information about subtypes, the presence of virulence genes and antimicrobial resistance markers using a single method, as well as providing higher discriminatory power to identify clusters of related strains which is important in public health laboratories [7].
In Ireland, as in the EU, campylobacteriosis is the commonest cause of gastroenteritis [8]. Currently, due to the widespread use of culture independent methods for the diagnosis of campylobacteriosis in most clinical laboratories since 2015, there are significant gaps in data on the molecular epidemiology of Campylobacter infection, with poor or no data on antimicrobial resistance markers circulating in humans isolates. Usually Campylobacter gastro enteric infections do not require treatment but in some severe cases treatment may be necessary, with macrolides, such as erythromycin, or quinolone, such as ciprofloxacin, the antibiotics of first and second choice.
The four priority antimicrobial drug resistance of C. jejuni and C. coli by ECDC-EFSA are ciprofloxacin, erythromycin, tetracycline and, since June 2016, gentamicin.[1].
Due to the high diversity of Campylobacter sp. the pathogenic mechanisms causing clinical symptoms are still not very well understood. Virulence properties found in Campylobacter include motility mediated by flagella, adherence and colonization of the intestinal mucosa, invasion capabilities and cytotoxin production (18).
The aim of this study was to perform a genomic characterization of clinical isolates of Campylobacter spp. using WGS in order to provide insights of the presence of 9 virulence genes previously identified as coding for virulence determinants, detection of antimicrobial resistance markers with regard to the priorities in the EU region and phylogenetic relationship of strains collected in the East region of Ireland.
Materials and methods
Isolates
A total of 122 human isolates of C. jejuni and C. coli isolated from diarrhoeal stool samples submitted to the regional Public Health Laboratory, Dublin-Health Service Executive, Ireland, for routine enteric screening from 2006 to 2016 were included in the study. The isolates had been stored frozen on beads. One bead of each isolate was streaked on blood agar and incubated at 42°C under microaerophilic conditions for 48 hours. Viable isolates were confirmed as Campylobacter spp. by real-time PCR using previously described primers targeting the 16SrRNA gene [9].
Whole-genome sequencing
DNA was extracted on MagNa Pure 96 (Roche Applied Science, Manheim, Germany) according to manufacturer’s instructions. The quantity of the DNA was measured using a Qubit fluorimeter 3.0 (Fisher Scientific, Massachusetts, USA) with the double-stranded DNA (dsDNA) assay HS kit. Libraries were prepared using the Nextera XT DNA kit, according to the manufacturer’s instructions (Illumina, Cambridge, UK). The quality of the library was assessed with a Bioanalyzer 2100 (Agilent Technologies, California, US). Paired-end sequencing was performed on the Illumina MiSeq platform using the v3 reagent kit.
Data analysis
FASTQ files from sequenced genomes were processed using BioNumerics v.7.6. (bioMérieux, Marcy-l'Étoile, France) calculation engine and the Multi Locus Sequence Type (MLST) client plug-in, which includes core genome MLST (cgMLST) analysis. Assembly-free and assembly-based allele detection analyses were performed for each isolate. The de novo assembly was performed using SPAdes integrated into the calculation engine. The quality of the assembly was assessed according to different parameters, the average quality score ranged from 31 to 37 (mean, 36), the average read coverage ranged from 31 to 467 (mean, 132), the N50 ranged from 6140 bp to 544758 bp (mean, 74417 bp), the number of contigs ranged from 11 to 734 (mean, 116) and the length of the sequences ranged from 1579481bp to 1844823 (mean, 1672005bp). The sequences included in the study were registered with the BioProject database under the BioProject ID PRJNA534408.
Identification of virulence genes
The detection of the different genes was performed using an in silico analysis using the Sequence Extraction plug-in in BioNumerics v.7.6 based on Basic Local Alignment Search Tool (BLAST, National Center for Biotechnology Information, US National Library of Medicine). The minimum sequence identity was set at 80% and the minimum length coverage was set at 90%. The sequences were downloaded in FASTA format from GenBank and then extracted from the de novo assemblies of each isolate. In this study the genes cdtA, cdtB and cdtC represented the pathogenic genes encoding for cytotoxin production; cadF, dnaJ and racR represented the pathogenic genes for the expression of adherence and colonization factors, and the genes virB11, iam and ciaB represented the pathogenic genes responsible for the expression of invasion [10, 11].
Identification of antimicrobial resistance markers
An in silico assessment of the presence of antimicrobial resistance markers was performed with the Sequence Extraction plug-in in BioNumerics v.7.6 as described above. For point mutations, ResFinder 3.0 (Center for Genomic Epidemiology) [12] and BLAST were used. The presence of cmeA, cmeB and cmeC as part of the cmeABC operon, a multidrug efflux pump conferring resistance to quinolones was studied. The presence of genes ermB (erythromycin resistance), blaoxa-61 (beta-lactam resistance), tet(O) (tetracycline resistance) and the cassette aadE-sat4-aphA3 (aminoglycoside resistance) was investigated. Point mutations such as mutation C257T in the DNA gyrase A gene (gyrA), associated with quinolone resistance, and mutation A2075G in the 23S rRNA gene, associated with macrolide resistance, were analysed.
Phylogenetic analysis
A core genome MLST (cgMLST) spanning tree was created in BioNumerics v.7.6. using categorical differences and the unweighted-pair group method with arithmetic mean (UPGMA). The cgMLST scheme had 1343 loci and 7 MLST for C. coli/jejuni (pubmlst.org) in BioNumerics v.7.6. and is based on the cgMLST by Cody et al. [13] and the 96 publicly available reference sequences of C. coli/ jejuni [14].
Results
Presence of virulence genes
The cdtABC operon was present in 105 (86%) isolates and there were no differences between C. jejuni and C. coli, with 86% and 89% of isolates presenting the operon, respectively. The genes cadF and dnaJ were present in 121 (99%) isolates and the gene racR was found in 119 (98%) isolates. Genes related to invasion were found as follows: iam in 121 (99%) isolates, virB11 in only 1 (1%) isolate and ciaB in 98 (80%) isolates. The prevalence of each gene and its role in pathogenesis are represented in Table 1.
Table 1. Prevalence of virulence-associated genes and antimicrobial resistance markers.
| Target | Gene | N* | % | |
|---|---|---|---|---|
| Virulence genes | Cytotoxin production | cdtA | 107 | 88 |
| cdtB | 108 | 89 | ||
| cdtC | 108 | 89 | ||
| Adherence and colonization | cadF | 121 | 99 | |
| dnaJ | 121 | 99 | ||
| racR | 119 | 98 | ||
| Invasion | virB11 | 1 | 1 | |
| iam | 121 | 98 | ||
| ciaB | 0 | 0 | ||
| Resistance markers | Multidrug efflux pump | cmeABC | 115 | 94 |
| Quinolones | gyrA mutation Thr86Ile | 52 | 43 | |
| Macrolides | 23S rRNA A2059G | 0 | 0 | |
| Erythromycin | ermB | 0 | 0 | |
| β- Lactams | blaoxa-61 | 87 | 71 | |
| Tetracycline | tet(O) | 30 | 25 | |
| Aminoglycoside | aadE-sat4-aphA3 | 0 | 0 | |
The presence of virulence-associated genes and antimicrobial resistance markers in C. jejuni and C. coli human isolates. PHL-Dublin, Ireland, 2006–2016.
* N = number of isolates.
Antimicrobial resistance genes and point mutations
The cmeABC operon was present in 115 (94%) isolates. The mutation Thr86Ile in gyrA was found in 52 (43%) isolates, while no evidence of the mutation A2075G in the 23S rRNA was found. The ermB was not detected in any isolates. The blaOXA-61 was present in 87 (71%) isolates and the tet(O) was detected in 30 (25%) isolates. The cassette aadE-sat4-aphA3 was not detected. The prevalence of resistance markers and its association with antimicrobial resistance are summarized in Table 1.
Phylogenetic analysis
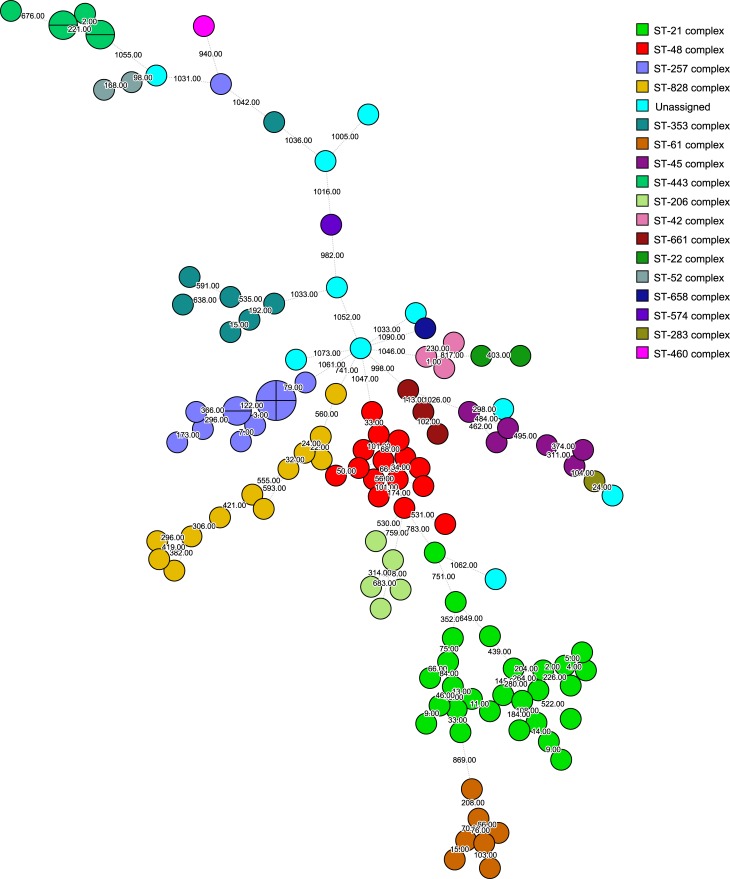
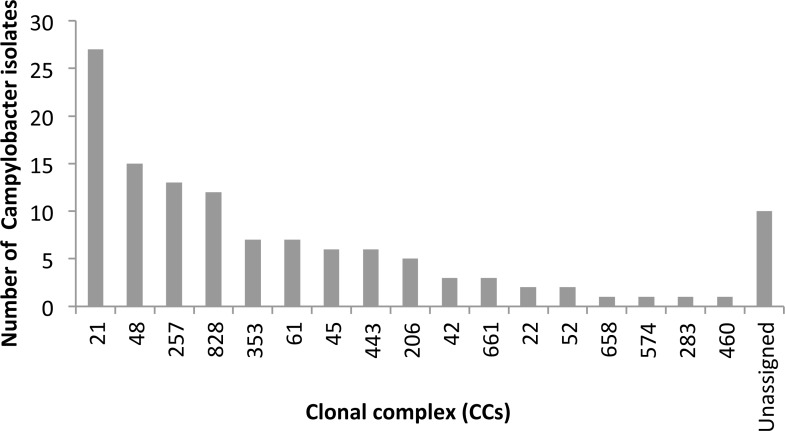
Of the 122 study isolates, 112 (92%) were assigned to 29 distinct sequence types (ST) grouped into 17 clonal complex (CC) (Fig 1). Ten isolates (8%) did not belong to a known CC but did include 4 ST (ST2153, ST4831, ST2274 and ST441). The most prevalent CCs (CC21, CC48, CC828, and CC257) accounted for 55% of isolates and the most prevalent STs were ST48, ST21 and ST257, with 14, 11 and 9 isolates, respectively (Fig 2). In addition, four clusters previously unidentified were detected, one with four isolates with no allele differences. There were other three clusters formed by two isolates in each with no allele differences. Further analysis of these clusters showed no obvious epidemiological link among the isolates since they were collected in different years and from different geographical regions.
Fig 1. Phylogenetic analysis.
Minimum spannin tree (MST) of cgMLST (1343 loci) data of Campylobacter human isolates collected from 2006 to 2016 at Public Health Laboratory, Dublin, Ireland. Each colour represents one clonal complex (CC). Isolates are represented by circles. Branches and numbers represent allelic differences between isolates.
Fig 2. Prevalence of clonal complex.
Distribution of CCs amongst 122 clinical Campylobacter spp. isolates obtained from 2006 to 2016, PHL-Dublin, Ireland.
Discussion
This is the first genomic study of Campylobacter in Ireland using WGS, providing insight into the prevalence of virulence genes, antimicrobial resistance markers as well as relatedness of human clinical isolates.
Pathogenesis of Campylobacter is still uncertain, with several virulence-associated genes identified but their role is not completely understood in the development of Campylobacter associated gastroenteritis [15]. In our present study, most isolates were positive for the presence of virulence-associated genes related with adherence, colonization and invasion. This was consistent with the findings in previous studies, where genes such as cadF, dnaJ, racR, iam and ciaB were also present in high proportion of the isolates studied [10, 16, 17]. The three genes cdtA, cdtB and cdtC are necessary for the functionality of the CDT operon [18] leading to toxin production and cell dystension, however its role in pathogenesis is still unclear. Previous studies in Bangladesh and Turkey have shown high prevalence of CDT in diarrhoeal samples indicating that the toxin production could increase the fluid secretion in the intestine and cause diarrhoea [10, 19]. All the isolates in our study were from diarrheic symptomatic patients, however we did not detect the three genes cdtA, cdtB and cdtC in all isolates (87%, 88% and 88%, respectively), suggesting that other virulence factors could contribute to the clinical presentation of gastroenteric campylobacteriosis, which was also observed in a previous study carried out in Denmark [20].
The gene virb11, encoded by the pVir plasmid [21], was found in only one isolate of C. jejuni. This result correlates with previous findings from Denmark, Bangladesh and Canada, where it was consistently low prevalence [10, 16, 22, 23], suggesting that virB11 is not an important virulence factor in human infection. Thus, the variability and uncertainty of what virulence factors are associated with clinical Campylobacter infection globally was also found in our Irish data. However, monitoring the presence of virulence genes could allow detection of emergent virulence markers (e.g. as a consequence of horizontal transmission), that could lead to more severe campylobacteriosis cases.
Although many limitations exist about reliability of antimicrobial susceptibility testing based on genomic methods compared to phenotypic methods [24], it is recognised that the advantage of knowing what molecular antimicrobial resistance markers are present in bacterial populations is crucial for future development of prevention and control strategies to fight antimicrobial resistance globally. However, such genomic antimicrobial resistance analysis needs validation and standardization prior its applicability in clinical settings as stated recently by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) Subcommittee [24]. In campylobacteriosis, the vast majority of cases have self-limiting symptoms and do not require antimicrobials. However, considering the volume of infections and the increasing acquisition of resistance mechanisms, genomic antimicrobial resistance detection is an essential surveillance tool for better understanding of the resistance markers in Campylobacter species circulating in a specific region. Such knowledge will improve the treatment options in the severe cases that do require systemic antimicrobials.
Ciprofloxacin resistance has been linked with the presence of two different markers. One is the presence of the cmeABC operon, which is the most common multidrug efflux pump found in Campylobacter and contributes to fluoroquinolone resistance by decreasing the amount of the drug in the cells [25, 26]. Other antimicrobial markers are point mutations in the quinolone-resistance determining region (QRDR) of DNA gyrase A (GyrA). In particular, the mutation C257T, which leads to the T86I substitution in GyrA, is the most frequently observed mutation in fluoroquinolone resistant isolates, conferring high-level resistance to these antimicrobials [26, 27]. The high level of fluoroquinolone resistance found in the Irish isolates (43%) is in line with what has been reported by other EU countries [1], and would preclude the use of these antimicrobials for empiric treatment of gastroenteric Campylobacter infections. Only if quinolone susceptibility is confirmed are such antimicrobials recommended for therapy if indicated.
Regarding macrolides, the most common class of antibiotics for Campylobacter infections treatment, two antimicrobial markers have been linked to resistance, the point mutation A2075G in the 23S rRNA gene and the ermB gene [6, 26, 27]. The absence of macrolide resistance in isolates of this study would make these antimicrobials the preferred Irish empiric treatment of choice for severe campylobacteriosis. However it is important to take into account that macrolide resistance has been mostly detected in C. coli isolates as confirmed by EFSA [1]. Since we only had 9 C. coli isolates in this study this macrolide data should be interpreted with caution.
Tetracycline resistance in Campylobacter is conferred by the gene tet(O) [26, 28] and it is of concern due to the widespread use of this antimicrobial in the poultry industry globally [29, 30]. The average percentage of Campylobacter tetracycline resistance in the EU is 68.8% in human isolates of C.coli and 44.6% of C. jejuni [1]. The Irish clinical isolates present lower levels (25%) of tetracycline resistance but continuous monitoring of this antimicrobial resistance genetic marker could aid in alerting the need for ‘One Health’ rapid implementation of reduction and control strategies in tetracycline usage, if an increase in resistance were observed in the future.
Since 2016, gentamicin resistance is included in the EU surveillance objectives to monitor invasive Campylobacter disease and trends in resistance in clinical human isolates [1]. In this study we analysed the presence of the gene cluster aadE-sat4-aphA3, which has been described in Campylobacter isolates from chicken and broilers in China and it is associated with resistance to multiple aminoglycosides. They have suggested that Campylobacter might have acquired the gene cluster from a Gram-positive organism [31]. Globally the resistance levels to gentamicin are still very low at EU level, with 0.8% and 1.6% C. jejuni and C. coli, respectively [1], correlating with the absence of such resistance in our Irish isolates. However, the emergence of aminoglycosides resistance in both US and China [31, 32] would require the continuous monitoring of such antimicrobial resistance, consistent with the EU strategy. The detection of 71% of Irish isolates with blaoxa-61 genes linked with beta-lactam resistance was not unexpected considering the usually high prevalence in Campylobacter isolates globally. However, this is not of significance considering that β-lactam antibiotics have limited application for treatment of Campylobacter infections.
In this study we have also highlighted the wide diversity of Campylobacter strains circulating in Ireland. The distribution of CCs among the sequenced isolates was very similar to what has been found in UK, with the CC-21 the most predominant CC, including more than 20% of the isolates, followed by CC-48, CC-257 and CC-828 [33]. These four CCs accounted for more than 50% of the isolates. The predominance of CC-21 has been also observed in other countries such as Israel [34]. In addition, comparing our human results with data from an earlier study with poultry isolates of Campylobacter collected in Ireland, there is an overlapping of the three most prevalent ST types [35] in both studies, suggesting a potential active transmission of Campylobacter through the food chain over time.
The isolates included in this study were not part of any notified outbreak, however the high discriminatory power of the WGS has brought to light the presence of 4 previously undetected clusters. Thus confirming the advantageous role of genomic characterisation of clinical Campylobacter isolates could play in aiding public health identification and investigation of linked cases.
This study has some limitations; it is not based on sentinel or continuous surveillance of clinical campylobacteriosis, but rather on a regional clinical laboratory database of stored isolates, which may not be representative nationally.
This study was confined to human isolates and does not include animal or food isolates, thus the postulated role of the food chain in transmission can only be hypothetical. However further ‘One Health’ Campylobacter studies are achievable and will help to inform future public health strategies for the reduction of campylobacteriosis. In addition, the data presented could not differentiate between C. jejuni and C. coli due to the unequal predominance of C. jejuni isolates compared to the low number of C. coli isolates (less than 10%) in the study dataset. Thus species-specific analysis particularly related to antimicrobial resistance could mislead and potentially leads to an underestimation of the antimicrobial resistance markers. Genotypic compared to historic phenotypic antimicrobial resistance data was not feasible because it was not comprehensive and not contemporaneous.
In conclusion, this WGS Campylobacter study has provided very useful additional information on the characteristics of Campylobacter isolates from human samples in Ireland. Thus confirming the presence of virulence genes in Campylobacter infections in humans. In addition, valuable information on genomic antimicrobial markers circulating in Ireland has been provided, which could contribute in the future to EU data. Finally, our study has shown the presence of clusters previously unidentified and the high diversity of Campylobacter circulating in the country, which along with future routine surveillance, could support the implementation of more effective One Health intervention strategies focused on the prevention and control of cases of campylobacteriosis.
Acknowledgments
We like to thank Dr Aura Andreasen and Dr Aftab Jasir for critically reviewing the manuscript.
Data Availability
All 122 sequences are available from the NCBI BioProject database (BioProject ID: PRJNA534408; Biosamples IDs 11489494 - 11489615).
Funding Statement
Dr Natalia Redondo was funded by European Centre for Disease Prevention and Control. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001;32(8):1201–6. 10.1086/319760 . [DOI] [PubMed] [Google Scholar]
- 3.Facciola A, Riso R, Avventuroso E, Visalli G, Delia SA, Lagana P. Campylobacter: from microbiology to prevention. J Prev Med Hyg. 2017;58(2):E79–E92. [PMC free article] [PubMed] [Google Scholar]
- 4.Wagenaar JA, French NP, Havelaar AH. Preventing Campylobacter at the source: why is it so difficult? Clin Infect Dis. 2013;57(11):1600–6. 10.1093/cid/cit555 . [DOI] [PubMed] [Google Scholar]
- 5.Ge B, McDermott PF, White DG, Meng J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005;49(8):3347–54. 10.1128/AAC.49.8.3347-3354.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, et al. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother. 2014;69(4):964–8. 10.1093/jac/dkt492 . [DOI] [PubMed] [Google Scholar]
- 7.Llarena AK, Taboada E, Rossi M. Whole-Genome Sequencing in Epidemiology of Campylobacter jejuni Infections. J Clin Microbiol. 2017;55(5):1269–75. Epub 2017/03/01. 10.1128/JCM.00017-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.HPSC. Campylobacter. Annual epidemiological report. 2016. [Google Scholar]
- 9.Linton D, Owen RJ, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996;147(9):707–18. . [DOI] [PubMed] [Google Scholar]
- 10.Talukder KA, Aslam M, Islam Z, Azmi IJ, Dutta DK, Hossain S, et al. Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J Clin Microbiol. 2008;46(4):1485–8. 10.1128/JCM.01912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Madi M, Behnke JM, Sharma A, Bearden R, Al-Banna N. Prevalence of Virulence/Stress Genes in Campylobacter jejuni from Chicken Meat Sold in Qatari Retail Outlets. PLoS One. 2016;11(6):e0156938 10.1371/journal.pone.0156938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cody AJ, McCarthy ND, Jansen van Rensburg M, Isinkaye T, Bentley SD, Parkhill J, et al. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J Clin Microbiol. 2013;51(8):2526–34. 10.1128/JCM.00066-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebure T, Bitar PD, Suzuki H, Stanhope MJ. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol. 2010;2:646–55. 10.1093/gbe/evq048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolton DJ. Campylobacter virulence and survival factors. Food Microbiol. 2015;48:99–108. Epub 2014/12/25. 10.1016/j.fm.2014.11.017 . [DOI] [PubMed] [Google Scholar]
- 16.Bang DD, Nielsen EM, Scheutz F, Pedersen K, Handberg K, Madsen M. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J Appl Microbiol. 2003;94(6):1003–14. . [DOI] [PubMed] [Google Scholar]
- 17.Zheng J, Meng J, Zhao S, Singh R, Song W. Adherence to and invasion of human intestinal epithelial cells by Campylobacter jejuni and Campylobacter coli isolates from retail meat products. J Food Prot. 2006;69(4):768–74. . [DOI] [PubMed] [Google Scholar]
- 18.Lara-Tejero M, Galan JE. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect Immun. 2001;69(7):4358–65. 10.1128/IAI.69.7.4358-4365.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findik A, Ica T, Onuk EE, Percin D, Kevenk TO, Ciftci A. Molecular typing and cdt genes prevalence of Campylobacter jejuni isolates from various sources. Trop Anim Health Prod. 2011;43(3):711–9. Epub 2010/11/23. 10.1007/s11250-010-9758-0 . [DOI] [PubMed] [Google Scholar]
- 20.Mortensen NP, Schiellerup P, Boisen N, Klein BM, Locht H, Abuoun M, et al. The role of Campylobacter jejuni cytolethal distending toxin in gastroenteritis: toxin detection, antibody production, and clinical outcome. APMIS. 2011;119(9):626–34. Epub 2011/06/21. 10.1111/j.1600-0463.2011.02781.x . [DOI] [PubMed] [Google Scholar]
- 21.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect Immun. 2000;68(8):4384–90. 10.1128/iai.68.8.4384-4390.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas D, Hannon SJ, Townsend HG, Potter A, Allan BJ. Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry. Int Microbiol. 2011;14(1):25–32. 10.2436/20.1501.01.132 . [DOI] [PubMed] [Google Scholar]
- 23.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003;52(Pt 4):345–8. 10.1099/jmm.0.05056-0 . [DOI] [PubMed] [Google Scholar]
- 24.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23(1):2–22. Epub 2016/11/23. 10.1016/j.cmi.2016.11.012 . [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46(7):2124–31. 10.1128/AAC.46.7.2124-2131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 2009;4(2):189–200. 10.2217/17460913.4.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payot S, Bolla JM, Corcoran D, Fanning S, Megraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006;8(7):1967–71. 10.1016/j.micinf.2005.12.032 . [DOI] [PubMed] [Google Scholar]
- 28.Moore JE, Barton MD, Blair IS, Corcoran D, Dooley JS, Fanning S, et al. The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 2006;8(7):1955–66. 10.1016/j.micinf.2005.12.030 . [DOI] [PubMed] [Google Scholar]
- 29.Luangtongkum T, Morishita TY, Martin L, Choi I, Sahin O, Zhang Q. Prevalence of tetracycline-resistant Campylobacter in organic broilers during a production cycle. Avian Dis. 2008;52(3):487–90. 10.1637/8181-112807-ResNote.1 . [DOI] [PubMed] [Google Scholar]
- 30.de Vries SPW, Vurayai M, Holmes M, Gupta S, Bateman M, Goldfarb D, et al. Phylogenetic analyses and antimicrobial resistance profiles of Campylobacter spp. from diarrhoeal patients and chickens in Botswana. PLoS One. 2018;13(3):e0194481 10.1371/journal.pone.0194481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, et al. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob Agents Chemother. 2012;56(10):5332–9. Epub 2012/08/06. 10.1128/AAC.00809-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, et al. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the USA. J Antimicrob Chemother. 2015;70(5):1314–21. Epub 2015/02/01. 10.1093/jac/dkv001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn SJ, Pascoe B, Turton J, Fleming V, Diggle M, Sheppard SK, et al. Genomic epidemiology of clinical Campylobacter spp. at a single health trust site. Microb Genom. 2018;4(10). Epub 2018/10/11. 10.1099/mgen.0.000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rokney A, Valinsky L, Moran-Gilad J, Vranckx K, Agmon V, Weinberger M. Genomic Epidemiology of. Front Microbiol. 2018;9:2432 Epub 2018/10/16. 10.3389/fmicb.2018.02432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Mahony E, Buckley JF, Bolton D, Whyte P, Fanning S. Molecular epidemiology of Campylobacter isolates from poultry production units in southern Ireland. PLoS One. 2011;6(12):e28490 10.1371/journal.pone.0028490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All 122 sequences are available from the NCBI BioProject database (BioProject ID: PRJNA534408; Biosamples IDs 11489494 - 11489615).