ABSTRACT
Salinity is a major abiotic stressor that limits the growth, development, and reproduction of plants. Our previous metabolic analysis of high salt-adapted callus suspension cell cultures from Arabidopsis roots indicated that physical reinforcement of the cell wall is an important step in adaptation to saline conditions. Compared to normal cells, salt-adapted cells exhibit an increased lignin content and thickened cell wall. In this study, we investigated not only the lignin biosynthesis gene expression patterns in salt-adapted cells, but also the effects of a loss-of-function of CCoAOMT1, which plays a critical role in the lignin biosynthesis pathway, on plant responses to high-salt stress. Quantitative real-time PCR analysis revealed higher mRNA levels of genes involved in lignin biosynthesis, including CCoAOMT1, 4CL1, 4CL2, COMT, PAL1, PAL2, and AtPrx52, in salt-adapted cells relative to normal cells, which suggests activation of the lignin biosynthesis pathway in salt-adapted cells. Moreover, plants harboring the CCoAOMT1 mutants, ccoaomt1-1 and ccoaomt1-2, were phenotypically hypersensitive to salt stress. Our study has provided molecular and genetic evidence indicating the importance of enhanced lignin accumulation in the plant cell wall during the responses to salt stress.
KEYWORDS: Arabidopsis, salt stress, salt adaptation, lignin biosynthesis, CCoAOMT1
Enhanced cell wall lignification has been observed in plants exposed to various environmental stresses.1 Salt stress strongly affects root lignification and cell wall solidification in vascular and xylem tissues.2 Salt stress also delays the differentiation of the primary xylem but accelerates the development of the secondary xylem in soybean roots.3 In wheat roots, salt stress led to considerable thickening of the cell walls in vascular tissues.4 In tomato roots, enhancement of the cell-to-cell pathway in response to salt stress led to a significant increase in lignin deposition in the vascular tissues.5 Lignin biosynthesis in response to salt stress is an irreversible process that greatly affects the structure and formation of the secondary cell wall.6
Many enzymes in the lignin biosynthetic pathway exhibit extensive substrate specificity, and many are multifunctional enzymes that catalyze multiple reactions.7 Various enzymes control the genetic manipulation of lignin. For example, phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate: CoA ligase (4CL), cinnamoyl CoA reductase (CCR), caffeoyl CoA O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeate 3-O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD) are involved in the synthesis of monolignols. Peroxidase (POD) and laccase (LAC) are involved in the polymerization of monolignols to yield the lignin polymer as a final product (Figure 1b).7 Although enhanced lignification has been observed in plants exposed to salt stress, the molecular functions and lignin biosynthetic gene expression patterns in plants exhibiting long-term adaptation to saline conditions are not fully understood.
Figure 1.
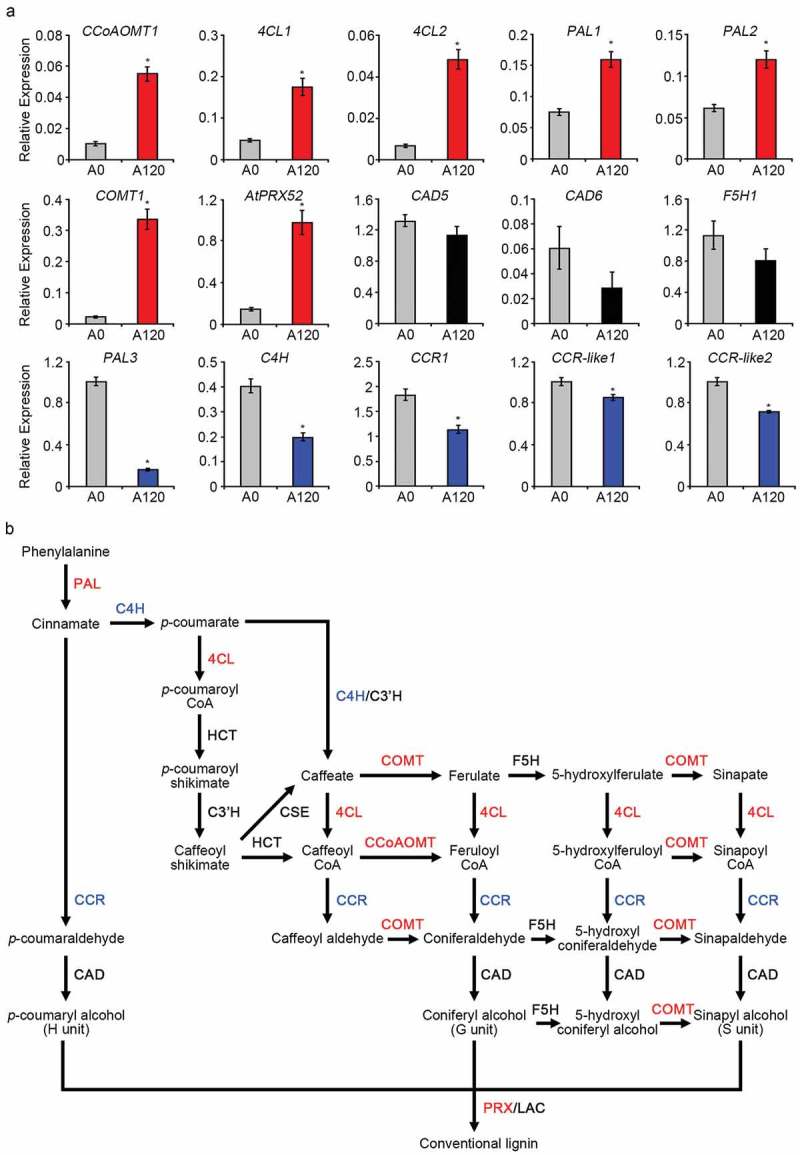
Lignin biosynthesis gene expression patterns in salt-adapted and normal cells.
(a) The mRNA levels of lignin biosynthesis genes, including CCoAOMT1, 4CL1, 4CL2, COMT1, PAL1, PAL2, PAL3, C4H, CAD5, CAD6, CCR1, CCR-like1, CCR-like2, F5H1, and AtPrx52 in normal (A0) and salt-adapted Arabidopsis root suspension cells (A120) were determined using quantitative real-time (qRT)-PCR with gene specific primers (Supplemental Table 1). The expression levels were normalized to the expression of TUBULIN2. Error bars represent the means ± standard deviations of three independent replicates. Asterisks indicate significant differences from the A0 (*; p-value ˂0.01, Student’s t-test). (b) Lignin biosynthesis pathway. This pathway has been modified based on a recently published paper.7 Red and blue coloring respectively indicate enzymes that were upregulated and downregulated in salt-adapted A120 cells relative to normal A0 cells. Enzymes that did not exhibit significant changes in expression are indicated in black.
Previously, we reported that Arabidopsis root suspension cells allowed to adapt to high-salt conditions over time exhibited thicker cell walls and an increased lignin content when compared with normal cells, indicating that physical reinforcement of the root cell wall is an important component of the long-term salt stress adaptation in this species.8 To investigate the molecular mechanisms underlying lignin biosynthesis in salt-adapted cells, we used quantitative real-time PCR to analyze the expression patterns of 15 genes involved in the lignin biosynthetic pathway (Figure 1a).7,9 Notably, we observed significantly upregulated expression of CCoAOMT1, 4CL1, 4CL2, COMT1, PAL1, PAL2, and AtPrx52 in salt-adapted cells (A120) relative to normal cells (A0) (Figure 1a). The AtPrx52 gene encodes a peroxidase involved in lignin polymerization.10 By contrast, we observed downregulated expression of PAL3, C4H, CCR1, CCR-like1, and CCR-like2 in salt-adapted cells and no significant differences in the expression of CAD5, CAD6, and F5H between salt-adapted and normal cells (Figure 1a). These results suggest that Arabidopsis root cells may adapt to salt stress via thickening of the cell walls, which induces the expression of many lignin biosynthetic genes under saline conditions. Moreover, the results suggest that each member of the gene family, such as PAL1, PAL2, and PAL3, may have a different function in lignin biosynthesis.
Previous studies have demonstrated that both the accumulation of lignin and strong expression of lignin biosynthetic genes are important factors in plants’ tolerance to salt stress.11–13 In white birch (Betula platyphylla), the overexpression of MYB46 and NAC012 led to enhanced salt tolerance by enabling the accumulation of lignin and upregulating the expression of genes encoding lignin biosynthetic enzymes that modulate the lignin composition.12,13 Transgenic Arabidopsis strains engineered to overexpress SOD from Potentilla atrosanguinea (35S::PaSOD) and APX from Rheum austral (35S::RaAPX) exhibited high lignin production in response to salt stress; however, the expression patterns of lignin biosynthetic genes differed between the transgenic strains.11 Specifically, the 35S::PaSOD plant exhibited reduced expression of CAD1, 4CL3, 4CL8, and CCoAOMT1, while the 35S::RaAPX plant exhibited increased expression of 4CL1, C4H, CCR2, CAD1, and CAD2 in response to salt stress.11 To evaluate the effects of lignin biosynthesis pathway suppression on the salt stress responses of plants, we analyzed the phenotypes of plants harboring mutations in the CCoAOMT1 gene, the product of which synthesizes feruloyl CoA from caffeoyl CoA towards guaiacyl (G) and sinapyl (S) lignin formation; this enzyme is also a key methyltransferase (OMT) in lignin biosynthesis under conditions of salt stress. It was previously reported that the Arabidopsis ccoaomt1 mutant exhibits lower lignin content than wild-type (WT) plants; moreover, the ccoaomt1 mutant shows a reduced amount of G monomer of lignin, but higher amounts of S and H monomers, relative to control plants.14 Furthermore, in our proteomics experiment, CCoAOMT1 protein was identified as one of the most highly induced proteins in salt-adapted cells compared to control cells (unpublished data).
We identified two homozygous T-DNA insertion mutants of CCoAOMT1 (At4g34050), ccoaomt1-1 (CS345726) and ccoaomt1-2 (SALK_151507), from the Arabidopsis Biological Resource Center (Figure 2A). To test the effect of a loss-of-CCoAOMT1 function on the responses of plants to salt stress, we compared root growth between the WT and two ccoaomt1 mutant lines under saline conditions. We first germinated seeds from all three plant lines on normal MS media, and subsequently transferred 4-day-old seedlings onto fresh media containing 0, 100, 125, or 150 mM NaCl. Under normal growth conditions, the WT, ccoaomt1-1, and ccoaomt1-2 plants exhibited similar root elongation (Figure 2(B,C)). Under saline conditions, however, both the ccoaomt1-1 and ccoaomt1-2 mutants exhibited significantly suppressed primary root elongation, compared to the WT plants. This suppression was most obvious in plants exposed to 125 mM NaCl (Figure 2(B,C)), indicating that a loss-of-function mutation in CCoAOMT1 increased the sensitivity of Arabidopsis to salt stress. Moreover, when compared to the WT plants, both the ccoaomt1-1 and ccoaomt1-2 mutants exhibited enhanced chlorosis in their cotyledons under saline conditions, indicating hypersensitivity to salt stress. These results suggest that CCoAOMT1 plays an important role in plant tolerance and salt stress adaptation by enhancing the physical strength of the plant cell walls.
Figure 2.
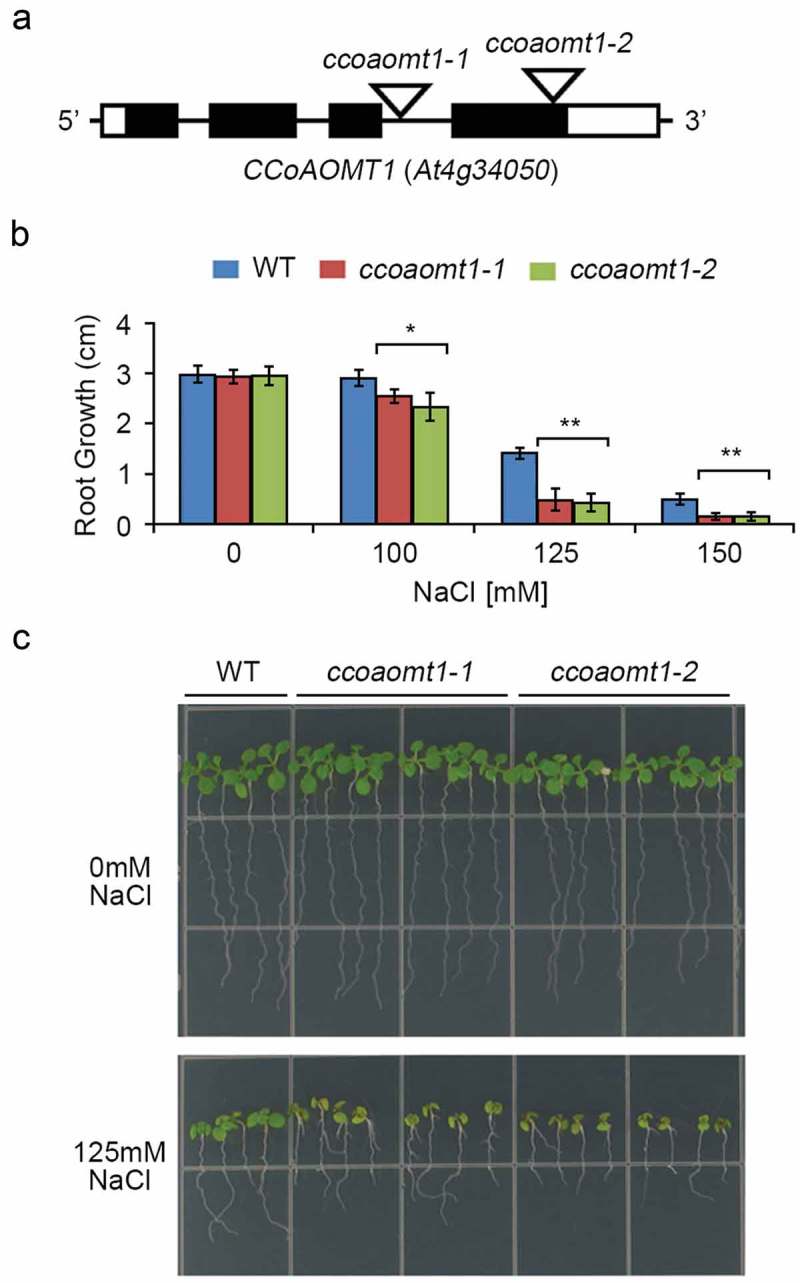
Arabidopsis ccoaomt1 mutants exhibiting hypersensitivity under saline conditions.
(a) Schematic representation of the T-DNA insertion sites in the ccoaomt1-1 and ccoaomt1-2 mutants. The exons, introns, UTR, and T-DNA insertion site are indicated by black boxes, black lines, white boxes, and white inverted triangles, respectively. (b) Four-day-old WT (Col-0), ccoaomt1-1 mutant, and ccoaomt1-2 mutant seedlings were transferred onto 1/2 MS medium (1.2% agar) containing 0, 100, 125, or 150 mM NaCl. At 7 days after transfer, the primary root lengths of WT and ccoaomt1 mutants were analyzed by using Image J software. Error bars represent the means ± standard deviations of three independent biological replicates of 6–8 seedlings per experiment. Asterisks represent significant differences from the WT (*; p-value ≤0.05, **; p-value ˂0.01, Student’s t-test). (c) Phenotypes of the WT and ccoaomt1 mutant seedlings under high-salt conditions. Four-day-old WT and ccoaomt1 mutants were transferred onto 1/2 MS medium (1.2% agar) containing 0 and 125 mM NaCl, respectively. The photograph was taken 7 days after transfer.
Recent proteomic analyses revealed increased levels of CCoAOMT protein in the roots of Arabidopsis, rice, wheat, barley, soybean, and tomato plants in response to salt stress.15–17 Additionally, SlPAL5 expression was strongly and rapidly induced in tomato plants under salt stress conditions.18 The observed involvement of CCoAOMT1 in the salt stress response has opened a new perspective to anatomizing the molecular mechanisms of lignin biosynthetic enzymes in the context of an abiotic stress response.
Our expressional and functional analyses of lignin biosynthesis genes in the responses of plants to salt stress have provided molecular, physiological, and genetic evidence supporting the importance of lignin biosynthesis in the cellular processes that endow plants with stress tolerance. Taken together, our previous metabolomics results of increased coniferin and lignin content in salt-adapted cells8 and our present results suggest that enhanced lignin biosynthesis is a critical factor in plant adaptation and tolerance to salt stress. Furthermore, it is suggested that the three lignin units, i.e., G, H, and S monomers, may have different functions in plant adaptation and tolerance to salt stress. To clarify this, we intend to analyze the amounts of G, H, and S monomers of lignin in salt-adapted suspension cells compared to normal cells in a future study.
Funding Statement
This work was supported by the Next Generation BioGreen21 Program (SSAC, grant number: PJ01318202), the Rural Development Administration Republic of Korea, and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant numbers: 2015R1A6A1A03031413, 2017R1D1A1B03029706).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental materials
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Moura JCMS, Bonine CAV, Vlana JOF, Dornelas MC, Mazzafera P.. Abiotic and biotic stresses and changes in lignin content and composition in plants. J Integr Plant Biol. 2010;52:360–376. [DOI] [PubMed] [Google Scholar]
- 2.Neves GYS, Marchiosi R, Ferrarese MLL, Siqueira-Soares RC, Ferrarese-Filho O. Root growth inhibition and lignification induced by salt stress in soybean. J Agron Crop Sci. 2010;196(6):467–473. doi: 10.1111/j.1439-037X.2010.00432.x. [DOI] [Google Scholar]
- 3.Hilal M, Zenoff AM, Ponessa G, Moreno H, Massa ED. Saline stress alters the temporal patterns of xylem differentiation and alternative oxidase expression in developing soybean roots. Plant Physiol. 1998;117(2):695–701. doi: 10.1104/pp.117.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jbir N, Chaïbi W, Ammar S, Jemmali A, Ayadi A. Root growth and lignification of two wheat species differing in their sensitivity to NaCl, in response to salt stress. C R Acad Sci III. 2001;324:863–868. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez-Aguayo I, Rodrigues-Galán JM, Garcia R, Torreblanca J, Pardo JM. Salt stress enhances xylem development and expression of S-adenosyl-l-methionine synthase in lignifying tissues of tomato plants. Planta. 2004;220(2):278–285. doi: 10.1007/s00425-004-1350-2. [DOI] [PubMed] [Google Scholar]
- 6.Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. Cell wall metabolism in response to abiotic stress. Plants (Basel). 2015. 16;4(1):112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front Plant Sci. 2018;9:1427. doi: 10.3389/fpls.2018.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun H, Baek D, Cho HM, Jung HS, Jeong MS, Jung WH, Choi CW, Lee SH, Jin BJ, Park MS, et al. Metabolic adjustment of Arabidopsis root suspension cells during adaptation to salt stress and mitotic stress memory. Plant Cell Physiol. 2019;60(3):612–625. doi: 10.1093/pcp/pcy231. [DOI] [PubMed] [Google Scholar]
- 9.Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM. Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 2005;168(1):123–140. doi: 10.1111/j.1469-8137.2005.01496.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Pérez F, Pomar F, Pedreño MA, Novo-Uzal E. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol Plant. 2015;154(3):395–406. doi: 10.1111/ppl.12310. [DOI] [PubMed] [Google Scholar]
- 11.Shafi A, Chauhan R, Gill T, Swarnkar MK, Sreenivasulu Y, Kumar S, Kumar N, Shankar R, Ahuja PS, Singh AK. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol Biol. 2015;87:615–631. doi: 10.1007/s11103-014-0262-1. [DOI] [PubMed] [Google Scholar]
- 12.Guo H, Wang Y, Wang L, Hu P, Wang Y, Jia Y, Zhang C, Zhang Y, Zhang Y, Wang C, et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol J. 2017;15(1):107–121. doi: 10.1111/pbi.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu P, Zhang K, Yang C. BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol. 2019;179(2):700–717. doi: 10.1104/pp.18.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226(5):1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- 15.Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002;2(9):1131–1145. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell. 2004;16(6):1378–1391. doi: 10.1105/tpc.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q, Zhang H, Wang T, Chen S, Dai S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J Proteomics. 2013;82:230–253. doi: 10.1016/j.jprot.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Guo J, Wang MH. Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L.). Mol Biol Rep. 2009;36(6):1579–1585. doi: 10.1007/s11033-008-9354-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


