ABSTRACT
Soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) are the key regulators control trafficking of cargo proteins to their final destinations and plays key role in plant development; however, their roles in plant defense remain largely unknown. R-SNARE VAMP727 and Qa-SNARE SYP22 were previously reported to associate with vacuolar protein deposition and brassinosteroids (BRs) receptor BRI1 plasma membrane targeting. Here, we identified that VAMP727 and SYP22 are induced by infection of root-knot nematode (RKN), a plant pathogen, which cause severe growth defect and yield loss. Furthermore, decreased root-knot nematode (RKN) invasion, growth and disease index were observed in bri1-5 and SYP22ND, a SYP22 negative dominant mutants when compared to control plants. Overall, our results suggest that VAMP727-SYP22 SNARE complexes regulate plant defense might be via control of abundances of BRI1 on the plasma membrane.
KEYWORDS: SNARE, VAMP727, SYP22, defense, plant
SNAREs are the key regulators of cargo protein intracellular trafficking in eukaryotic cells.1 SNARE proteins are classified into Qa-, Qb-, Qc-, SNAP25-like, and R-SNAREs based on conserved amino acids of SNARE motifs with coiled-coil helices. A functional SNARE complex consists of one set of Qa-, Qb-, Qc- and R-SNARE with a four-helical bundle assembled by four SNARE motifs.2,3 Fifteen R-SNARE members, including two Sec22-like, two Ykt6-like and seven VAMPs (vesicle-associated membrane proteins) have been identified in Arabidopsis.4 Our previous study identified that SYP22 and VAMP7272 interact with and control BRI1 plasma membrane targeting to regulate plant growth in Arabidopsis.5,6 Also, we identified that overexpression of cytosolic part of SYP22 exhibited negative dominant (ND) effect, and SYP22ND overexpressor exhibited dwarf phenotype similar with syp22/vamp727−/+ double mutant plants.6
Recent studies have demonstrated that BR signaling could positively regulate plant immune response to a broad range of pathogens.7,8 Root-knot nematode (RKN) generally induces formation of hook-like galls on roots and gradually impacts nutrient and water uptake.9 Interestingly, in the roots of RKN-infected rice, BRI1 mutants also showed a 30% reduction in gall numbers.10 In contrast, BR biosynthesis or silencing BR receptor increased susceptibility of tomato plants to RKN independent from salicylic acid and jasmonic acid signaling,11 thus suggesting that responses to RKN are related to BR signaling. In addition, BRI1 mutation enhances disease resistance in Brachypodium distachyon and barley (Hordeum vulgare) against necrotrophic and hemi-biotrophic pathogens showing short asymptomatic phase,12 suggesting that BRI1 is tightly associated with plant defense.
Since VAMP727 and SYP22 control BRI1 plasma membrane targeting to modulate BR signaling, the possible regulation of SYP22 and VAM272 on plant defense against RKN was investigated. The common RKN Meloidogyne incognita was inoculated to 2-week-old Arabidopsis Col-0 plants, and gene expression was monitored. The staining of RNK indicated that RNK was successfully invaded into plant roots after 24 h of inoculation (Figure 1(a,b)). qRT-PCR results indicated that VAMP727, SYP22, and PR1 were significantly induced by M. incognita infection (Figure 1(c,d)). To examine whether SYP22ND plants regulate plant resistance, SYP22ND #3, #5, and bri1-5 mutants were infected with M. incognita. At 18 days after infection with the second-stage juveniles (J2) of RKN, the gall and total RKN numbers in SYP22ND #3 and #5 or bri1-5 were lower than those in the corresponding control Col-0 and WS2 plants, respectively (Figure 2(a,b)). Moreover, J3 and J4 populations were lower in SYP22ND #3 and #5 or bri1-5, whereas higher J2 populations were observed compared with the corresponding controls (Figure 2(c)). These data indicate that RKN invasion, development, and disease index were all inhibited in the SYP22ND and bri1-5 mutants. SYP22 and VAMP727 were induced upon RNK inoculation, suggesting that accumulation of SYP22 and VAMP727 might inhibit plant defense via activation of BR signaling to promote RNK invasion and development in plants. We conclude that VAMP727 and SYP22 regulate plant defense to RKN might be through control BRI1 intracellular trafficking in Arabidopsis.
Figure 1.
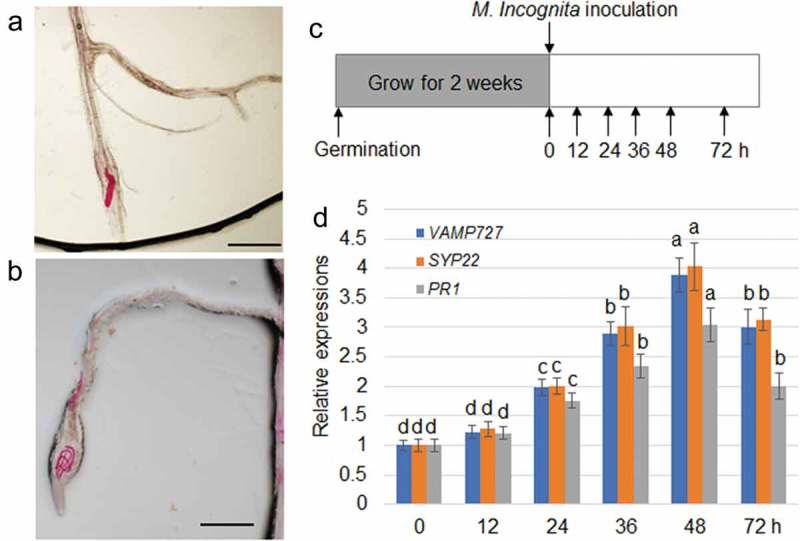
Expression of VAMP727, SYP22, and PR1 to M. incognita. The red lines (RNK) were observed in primary root tip (a) and lateral root apex (b). Bar = 100 µm. (c) Schematic showing M. incognita inoculation and time points for sampling expression of M. incognita-responsive genes. Col-0 plants were grown for 2 weeks, followed by inoculation of M. incognita. The whole roots were harvested after 0, 12, 24, 36, 48 and 72 h of the inoclution. (d) Expression levels of VAMP727, SYP22, and PR1 genes were examined using qRT–PCR. mRNA levels in the samples were normalized against those of Actin mRNA. Data are means ± s.e. (n = 3); different letters indicate significant differences between results (P < .05).
Figure 2.
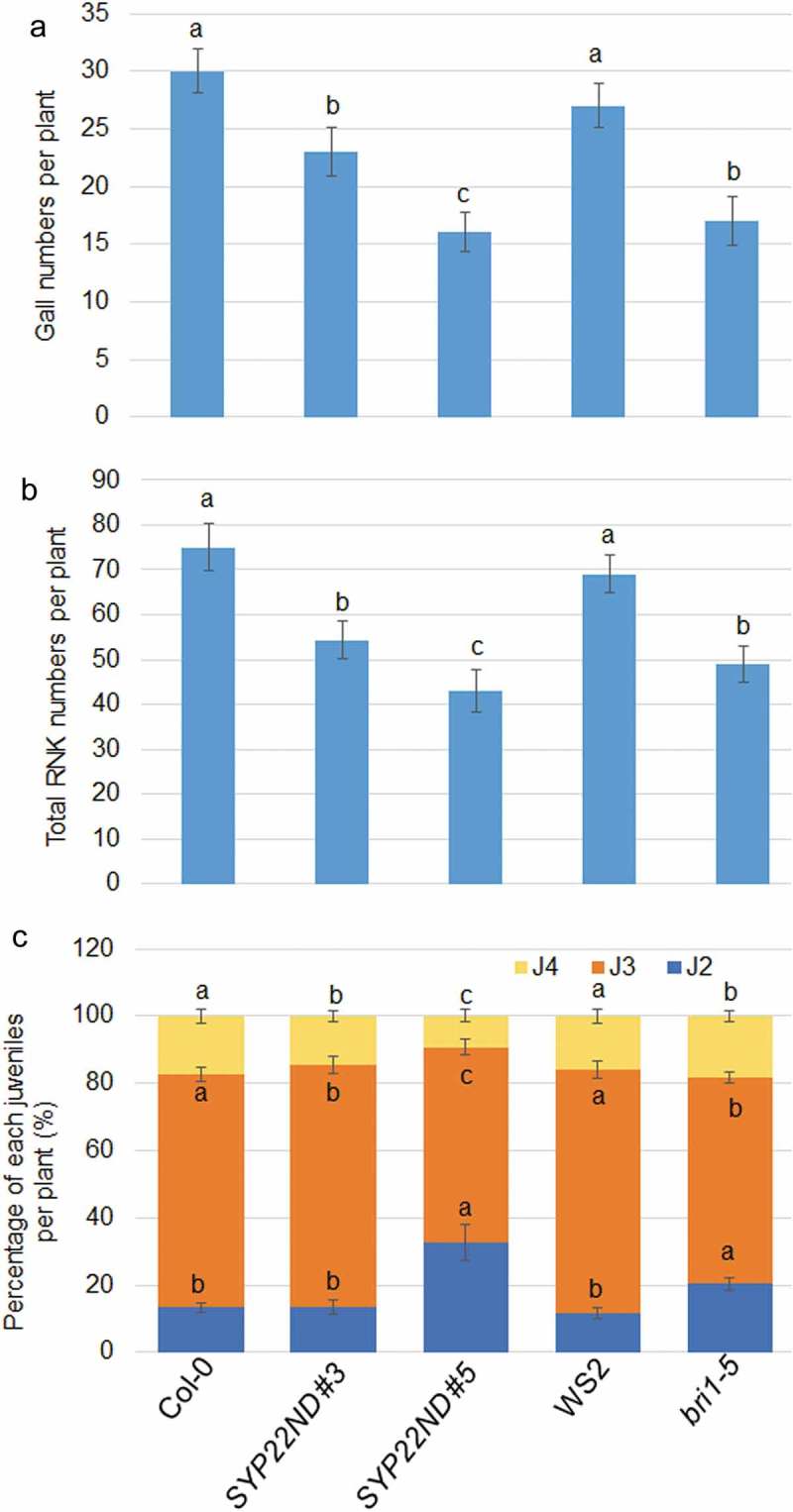
The numbers of gall, total RKN and each juvenile percentages of M. incognita in SYP22ND and BR insensitive mutants. (a) Number of galls in WT (Col-0 for SYP22ND mutants, WS2 for bir1-5), SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). (b) Total number of RKN in WT, SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). (c) The percentage of each juveniles (J2, J3, and J4) was calculated in WT, SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). Significant differences between groups, were analyzed via one-way analysis of variance (ANOVA), followed by Bonferroni’s multiple comparison tests. Differences among samples were considered significant at P < .05.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31571985]; Support Plan for Innovative Talents in Colleges and Universities of Liaoning Province [LR2017037].
Acknowledgments
We thank Prof. Zhi-yong Wang (Carnegie Institution for Science) for providing bri1-5 mutant seeds.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Cai H, Reinisch K, Ferro-Novick S.. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12(5):671–682. https://www.sciencedirect.com/science/article/pii/S1534580707001529?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 2.Fasshauer D, Sutton RB, Brunger AT, Jahn R.. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998; 95(26):15781–15786. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=9861047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(2):120–144. https://www.sciencedirect.com/science/article/pii/S0167488905000571?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 4.Lipka V, Kwon C, Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol. 2007;23:147–174. http://arjournals.annualreviews.org/doi/full/10.1146/annurev.cellbio.23.090506.123529?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed. [DOI] [PubMed] [Google Scholar]
- 5.Jones AM, Xuan Y, Xu M, Wang RS, Ho CH, Lalonde S, You CH, Sardi MI, Parsa SA, Smith-Valle E, et al. Border control–a membrane-linked interactome of Arabidopsis. Science. 2014;344(6185):711–716. http://science.sciencemag.org/content/344/6185/711.long. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Liu Y, Zhu XF, Jung JH, Sun Q, Li TY, Chen LJ, Duan YX, Xuan YH. SYP22 and VAMP727 regulate BRI1 plasma membrane targeting to control plant growth in Arabidopsis. New Phytol. 2019. doi: 10.1111/nph.15759. [DOI] [PubMed] [Google Scholar]
- 7.Bajguz A, Hayat S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol Biochem. 2009;47(1):1–8. https://www.sciencedirect.com/science/article/pii/S0981942808001873?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 8.Peng HC, Kaloshian I. The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS One. 2014; 9(3):e93302 http://dx.plos.org/10.1371/journal.pone.0093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson VM, Gleason CA. Plant-nematode interactions. Curr Opin Plant Biol. 2003; 6(4):327–333. https://linkinghub.elsevier.com/retrieve/pii/S1369526603000591. [DOI] [PubMed] [Google Scholar]
- 10.Nahar K, Kyndt T, Hause B, Hofte M, Gheysen G. Brassinosteroids suppress rice defense against root-knot nematodes through antagonism with the jasmonate pathway. Mol Plant Microbe Interact. 2013; 26(1):106–115. https://apsjournals.apsnet.org/doi/10.1094/MPMI-05-12-0108-FI. [DOI] [PubMed] [Google Scholar]
- 11.Song LX, Xu XC, Wang FN, Wang Y, Xia XJ, Shi K, Zhou Y-H, Zhou J, Yu J-Q. Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASE HOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ. 2018;41(5):1113–1125. https://onlinelibrary.wiley.com/doi/full/10.1111/pce.12952. [DOI] [PubMed] [Google Scholar]
- 12.Goddard R, Peraldi A, Ridout C, Nicholson P. Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol Plant Microbe Interact. 2014;27:1095–1106. doi: 10.1094/MPMI-03-14-0069-R. [DOI] [PubMed] [Google Scholar]


