Abstract
Introduction
Gemcitabine is an important component of pancreatic cancer clinical management. Unfortunately, acquired gemcitabine resistance is widespread and there are limitations to predicting and monitoring therapeutic outcomes.
Objective
To investigate the potential of metabolomics to differentiate pancreatic cancer cells that develops resistance or respond to gemcitabine treatment.
Results
We applied 1D 1H and 2D 1H-13C HSQC NMR methods to profile the metabolic signature of pancreatic cancer cells. 13C6-glucose labeling identified thirty key metabolites uniquely altered between wild-type and gemcitabine-resistant cells upon gemcitabine treatment. Gemcitabine resistance was observed to reprogram glucose metabolism and to enhance the pyrimidine synthesis pathway. Myo-inositol, taurine, glycerophosphocholine and creatinine phosphate exhibited a “binary switch” in response to gemcitabine treatment and acquired resistance.
Conclusion
Metabolic differences between naïve and resistant pancreatic cancer cells and, accordingly, their unique responses to gemcitabine treatment were revealed, which may be useful in the clinical setting for monitoring a patient’s therapeutic response.
Keywords: NMR metabolomics, pancreatic cancer, gemcitabine, drug resistance
Introduction
A drug has to pass through a variety of complex biological systems and must survive different cellular processes in order to reach its molecular target and exhibit a positive therapeutic response. In many instances, both the drug and the exposed cells undergo molecular change that either favors the desired outcome or leads to the development of resistance. Acquired resistance has been well-documented in the application of antibiotics and is commonly due to the overexposure or incorrect administration of the drugs. In this regard, bacteria rapidly acquire antibiotic resistance by altering their genetic expression, protein structure or metabolic processes. Since antimicrobial resistance is such a common problem, an array of therapeutic approaches and improved sensitivity tests have been developed to monitor a patient’s response to treatment and to improve therapeutic outcomes (Davies and Davies 2010; Housman et al. 2014). Similar efforts are on-going to combat acquired resistance for anticancer therapeutics. This has led to increased efforts to understand mechanisms of drug resistance and for the development of methods to test patient-specific drug sensitivity (Uhr et al. 2015). However, there are still serious unmet needs to achieving precision cancer therapies by identifying an optimal cancer-cell specific treatment (Bardin et al. 2014).
Cancer cells employ a variety of mechanisms to acquire resistance against chemotherapeutics (Rahman and Hasan 2015). In which, metabolic rewiring of tumors has been identified as a critical step in drug resistance. Cancer cells alter their metabolism by monitoring nutrient uptake (such as glucose and glutamine), which results in modulating certain metabolic pathways in response. Gemcitabine is an antimetabolite drug widely used in the treatment of breast, lung and pancreatic cancers. It is a prodrug activated by the effect of kinases in the cytoplasm. Gemcitabine is the first treatment of choice for pancreatic cancer patients, and is commonly used by its self or in combination therapies. Unfortunately, it is routine for patients to develop gemcitabine resistance shortly after beginning treatment (Fryer et al. 2011). We recently demonstrated that acquired resistance to gemcitabine results in the reprograming of glucose metabolism and an enhanced carbon-flow through the pyrimidine synthesis pathway (Shukla et al. 2017). This metabolic phenotype for pancreatic cancer cell lines is regulated by MUC1 and HIF1α cross-talk.
1D 1H NMR metabolomics is a versatile tool of systems biology that is routinely used to elucidate various metabolic alterations (Gebregiworgis and Powers 2012). Accordingly, we have utilized NMR metabolomics to study the impact of MUC1 overexpression (Chaika et al. 2012), to monitor tumor microenvironment alteration (Gebregiworgis et al. 2017), and as a means to reverse antibiotic resistance (Gaupp et al. 2015; Gardner et al. 2018). NMR metabolomics provides a detailed and specific analysis of metabolic perturbations by combining stable isotope labeling schemes (e.g., 13C glucose) with two dimensional (2D) NMR experiments (e.g., 1H-13C HSQC, HMBC, etc.). In this communication, we extend our investigation into the altered metabolism of gemcitabine resistance cell lines; and describe a global metabolic response to gemcitabine treatment.
Results and Discussion
Unique metabolic phenotype for gemcitabine resistant cells.
Briefly, ten replicates for each of the wild-type (WT, human pancreatic cancer cell line: T3M4 or Capan-1), gemcitabine resistant (GemR), WT treated with 10 nM of gemcitabine (WT+), and GemR treated with 10 nM of gemcitabine (GemR+) cell cultures were prepared as previously described for the 1D 1H NMR experiments (Shukla et al. 2017). As previously reported, the 10 nM gemcitabine dosage is significantly below a dosage (~1 μM) required to induce cell death in any of the cell lines (Shukla et al. 2017). Similarly, three additional replicates of WT, GemR, WT+, and GemR+ Capan-1 cells were prepared where the glucose in the culture media was replaced with 0.5 mM of 13C6-glucose. Please see the supplemental material for additional experimental details.
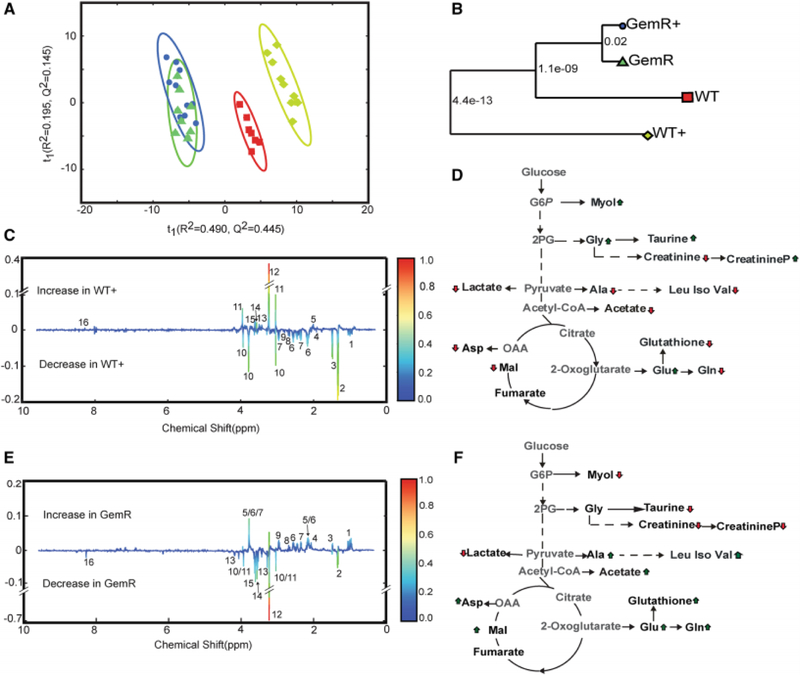
The 1D 1H NMR spectra collected from each cell lysate was analyzed using our MVAPACK metabolic toolkit (http://bionmr.unl.edu/mvapack.php) to generate a principal component analysis (PCA) model (Worley and Powers 2014). An unsupervised PCA model was used to illustrate the unique metabolic signature for each cell type, and to identify metabolic alterations that resulted from either gemcitabine resistance or from gemcitabine treatment (Gebregiworgis and Powers 2012). The resulting PCA scores plots and associated tree diagrams shown in Figures 1A, B and Figure S-1 clearly indicates that the metabolomes from the WT and WT-treated cells form distinct and separate groups. Conversely, the metabolomes from GemR and GemR-treated cells clustered together but separately from the gemcitabine-sensitive metabolomes.
Figure 1. Unique metabolic phenotype for gemcitabine resistant cells.
(A) PCA scores plot generated from 1D 1H NMR spectra from cell lysates of wild type T3M4 cells (WT,  n=8), WT cells treated with 10 nM of gemcitabine (WT+,
n=8), WT cells treated with 10 nM of gemcitabine (WT+,  n=9), gemcitabine-resistant (GemR,
n=9), gemcitabine-resistant (GemR,  n=10) cells, and GemR cells treated with 10 nM of gemcitabine (GemR+,
n=10) cells, and GemR cells treated with 10 nM of gemcitabine (GemR+,  n=8). Please see supplemental methods and Figure S-4 for explanation of excluded outliers. The ellipses correspond to 95% confidence intervals for a normal distribution. (B) Metabolic tree diagram generated from the PCA score plots. The number at each node is the p-value calculated from the Mahalanobis distance between each group. The coloring is identical to the PCA scores plot. (C) Back-scaled loadings plot generated from a validated OPLS-DA model (R2 0.99, Q2 0.90, CV-ANOVA p-value 7.94 × 10−7, permutation test p-value 0) comparing the WT and WT+ 1D 1H NMR datasets. Positive peaks indicate an increase in WT+ and negative peaks are a decrease in WT+. (D) Metabolic pathway summarizing the key metabolite differences between WT and WT+ as determined from the OPLS-DA back-scaled loadings plot in C. An up arrow indicates an increase in the metabolite in WT+ and a down arrow indicates a decrease in the metabolite in WT+. (E) Back-scaled loadings plot generated from a validated OPLS-DA model (R2 0.99, Q2 0.96, CV-ANOVA p-value 2.52 X 10−7, permutation test p-value 0) comparing the WT and GemR 1D 1H NMR datasets. Positive peaks indicate an increase in GemR and negative peaks are a decrease in GemR (F) Metabolic pathway summarizing the key metabolite differences between WT and GemR as determined from the OPLS-DA back-scaled loadings plot in E. An up arrow indicates an increase in the metabolite in Gem R and a down arrow indicates a decrease in the metabolite in GemR. The metabolite labeling in the OPLS-DA back-scaled loadings plots in C and E are numbered as follows: 1, branched chain amino acids (leucine, isoleucine, valine); 2, lactate; 3, alanine; 4, acetate; 5, glutamate; 6, glutamine; 7,glutathione; 8,malate; 9,aspartate; 10, creatinine; 11, creatinine phosphate; 12, glycerophosphocholine; 13, taurine, 14, glycine, 15, myo-inositol 16, AXP (AMP, ADP, and ATP).
n=8). Please see supplemental methods and Figure S-4 for explanation of excluded outliers. The ellipses correspond to 95% confidence intervals for a normal distribution. (B) Metabolic tree diagram generated from the PCA score plots. The number at each node is the p-value calculated from the Mahalanobis distance between each group. The coloring is identical to the PCA scores plot. (C) Back-scaled loadings plot generated from a validated OPLS-DA model (R2 0.99, Q2 0.90, CV-ANOVA p-value 7.94 × 10−7, permutation test p-value 0) comparing the WT and WT+ 1D 1H NMR datasets. Positive peaks indicate an increase in WT+ and negative peaks are a decrease in WT+. (D) Metabolic pathway summarizing the key metabolite differences between WT and WT+ as determined from the OPLS-DA back-scaled loadings plot in C. An up arrow indicates an increase in the metabolite in WT+ and a down arrow indicates a decrease in the metabolite in WT+. (E) Back-scaled loadings plot generated from a validated OPLS-DA model (R2 0.99, Q2 0.96, CV-ANOVA p-value 2.52 X 10−7, permutation test p-value 0) comparing the WT and GemR 1D 1H NMR datasets. Positive peaks indicate an increase in GemR and negative peaks are a decrease in GemR (F) Metabolic pathway summarizing the key metabolite differences between WT and GemR as determined from the OPLS-DA back-scaled loadings plot in E. An up arrow indicates an increase in the metabolite in Gem R and a down arrow indicates a decrease in the metabolite in GemR. The metabolite labeling in the OPLS-DA back-scaled loadings plots in C and E are numbered as follows: 1, branched chain amino acids (leucine, isoleucine, valine); 2, lactate; 3, alanine; 4, acetate; 5, glutamate; 6, glutamine; 7,glutathione; 8,malate; 9,aspartate; 10, creatinine; 11, creatinine phosphate; 12, glycerophosphocholine; 13, taurine, 14, glycine, 15, myo-inositol 16, AXP (AMP, ADP, and ATP).
The PCA scores plot demonstrates the overall metabolic impact of gemcitabine treatment on sensitive cells; and the corresponding lack of a response for resistant cells. Furthermore, the PCA scores plot identifies the presence of a metabolic adaptation for gemcitabine resistant cells. In effect, a distinct metabolic phenotype was observed for pancreatic cancer cells resistant to gemcitabine treatment. Furthermore, the observed alteration in metabolism may facilitate our understanding of the mechanism of gemcitabine resistance and provide a means to reverse the process (de Sousa Cavalcante and Monteiro 2014). Importantly, our results demonstrate a potential utility in precision medicine since the distinct metabolic phenotypes observed for WT and GemR cell lines may be leveraged for predicting a patient’s response to a gemcitabine treatment.
An orthogonal projection to latent structures - discriminant analysis (OPLS-DA) model was also generated from the 1D 1H NMR datasets to identify key metabolites that contribute to group specific separations (Figure S-2). Accordingly, the OPLS-DA model comparing the WT and WT-treated metabolome identified key-metabolic changes in response to gemcitabine treatment. The high-quality and statistical validity of the resulting OPLS-DA model from the WT/WT+ NMR dataset is evident by an R2 of 0.99, a Q2 of 0.90, a CV-ANOVA p-value of 7.9 × 10−7, and a permutation test p-value of 0. A back-scaled loadings plot generated from the OPLS-DA model is shown in Figure 1C. A total of sixteen metabolite changes were identified from the back-scaled loadings plot that significantly contributes to the metabolome differences between WT and WT-treated cells. The key metabolite changes between WT and WT+ are summarized in the pathway diagram shown in Figure 1D. An OPLS-DA model was similarly generated from the WT/GemR NMR dataset to identify key metabolite changes associated with gemcitabine resistance. The high-quality and statistical validity of the resulting OPLS-DA model from the WT/GemR NMR dataset is evident by an R2 of 0.99, a Q2 of 0.96, a CV-ANOVA p-value of 2.52 × 10−7, and a permutation test p-value of 0. Notably, the same set of sixteen metabolites w identified from the WT/GemR OPLS-DA back-scaled loadings plot as was observed in the WT/WT+ OLPS-DA model (Figures 1E, F). But, significantly, the relative metabolite trends are reversed when these two statistical models are compared. Specifically, acetate, alanine, and glutathione are increased in GemR cells compared to WT, while glycine, myo-inositol, taurine, glycerophospocholine, and creatinine phosphate are decreased in GemR cells relative to WT (Figure 1D and 1F). Again, these metabolic trends are completely reversed when WT-treated cells are compared to WT cells. Importantly, the metabolites involved in this metabolic “switch” have been previously linked to pancreatic cancer.
Myo-inositol and its metabolites regulate cancer cell proliferation, migration and the phosphatidylinositol-3-kinase (PI3K)/AKT signaling pathway (Vucenik and Shamsuddin 2003). Accordingly, many genes and pathways associated with myo-inositol synthesis and its biological activity have also been correlated with pancreatic cancer. Interestingly, natural and synthetic derivatives of myo-inositol have a demonstrated anticancer activity by reducing HIF1α expression and by decreasing the cellular concentration of nucleotides. Furthermore, the myo-inositol induced reduction in nucleotide concentrations also enhances gemcitabine efficacy (Raykov et al. 2014). Thus, drugs that target myo-inositol metabolism may also reverse gemcitabine resistance, and may be useful as part of a combination therapy.
Serine derived glycine is a precursor of single carbon metabolism and de novo purine nucleotide synthesis, which are critical processes in cancer pathogenesis (Yang and Vousden 2016). As noted above, a reduction in the cellular concentration of nucleotides enhances gemcitabine efficacy. Alternatively, an increase in glycine uptake and its metabolism has been observed to promote tumorigenesis (Amelio et al. 2014). Glycine is also an important metabolite in glutathione biosynthesis (Lu 2009). An increase in the expression of glutathione generating enzymes has been previously associated with gemcitabine resistance (Ju et al. 2015). Gemcitabine is known to increase reactive oxygen species and, presumably, an increase in the production of glutathione would negate the impact of this additional ROS. Consistent with these prior observations, we observed a decrease in glycine and a concurrent increase in glutathione in GemR. This could be attributed to an increase in the transformation of glycine into glutathione to combat gemcitabine-induced ROS. The reduction in glycine may also be a result of an increase of carbon-flow into nucleotide synthesis.
Alanine metabolism is a critical source of stromal cell derived fuel for cancer cell proliferation and survivability (Sousa et al. 2016). Thus, alterations in alanine metabolism may be an important process for acquiring gemcitabine resistance. Our observation that alanine metabolism was altered in both GemR and WT-treated cells are consistent with this viewpoint. Specifically, we observed a decrease in alanine in WT-treated cells and a corresponding increase in GemR cells. This result is also consistent with the observation that elevated alanine transaminase serum levels is linked to a poor therapeutic outcome and the development of gemcitabine resistance (Matsubara et al. 2010).
Acetate is a major carbon source for fatty acid and phospholipid biosynthesis, particularly in proliferating cancer cells and during metabolic stress. Acetyl-CoA synthetase 2 (ACSS2) catalyzes the conversion of acetate to acetyl-CoA. ACSS2 has been observed to inhibit tumor growth when depleted in hepatocellular carcinoma xenografts (Comerford et al. 2014). Our observation that acetate is depleted in WT-treated, but increased in GemR cells, highlights that acetate could serve as an important fuel for proliferating cancer cells.
Taurine has been previously observed to have an anti-proliferative effect on breast, colon, cervical, and hepatic cancers (Tu et al. 2015). In this regard, an increase in taurine concentration leads to apoptosis. Taurine was increased in the WT+ cells, but decreased in GemR. Our observations align with previous reports indicating that an acquired gemcitabine resistance inhibits gemcitabine-induced apoptosis.
A connection between creatine/creatine phosphate and pancreatic cancer has not been previously observed. Nevertheless, an alteration in the expression of creatine kinase, the enzyme that catalyzes the reversible conversion of creatine to creatine phosphate, has been strongly associated with other tumors such as small cell lung carcinoma (Gazdar et al. 1981).
A notable decrease in glycerophosphocholine was also observed in GemR cells with a corresponding increase in WT+ cells. This is consistent with abnormal choline metabolism that is a characteristic of multiple cancers and the associated changes in the expression of important choline metabolizing enzymes and choline transporters (Glunde et al. 2015). Since 11C Choline PET imaging is already used for diagnosing cancer (Hara et al. 1998), choline metabolism might yield a promising biomarker for gemcitabine-sensitive pancreatic cancers.
A receiver operating characteristics (ROC) analyses was applied to further asses the predictive power of the NMR metabolomics data to discriminate WT (T3M4 and Capan-1) cells from WT+ and GemR cells. The ROC curves (Figure S-3) show that the WT, WT+ and GemR cells were all confidently identified (AUC > 0.9) using only five spectral features from the 1D 1H NMR data sets. Notably, each of these spectral features was similarly identified from the OPLS-DA loadings plots. Thus, the essentially identical results obtain from both univariate and multivariate analysis for two different cell lines provides strong validation for the observed alteration in the cellular metabolomes.
Altered metabolism in gemcitabine resistant cells.
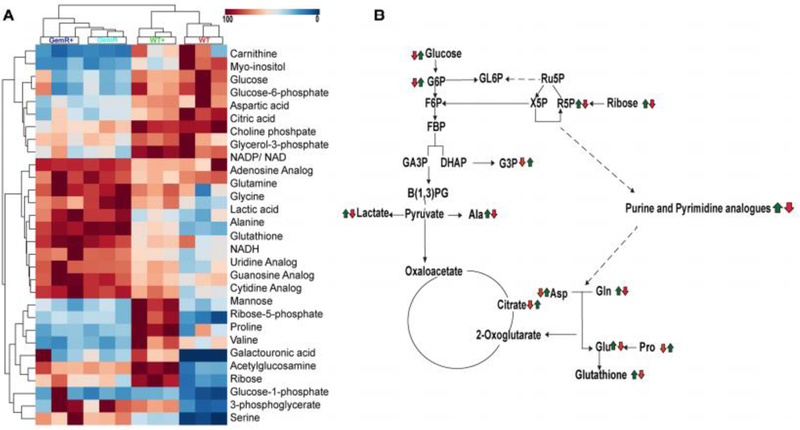
We have previously demonstrated that glucose metabolism is altered in gemcitabine resistant cells (Shukla et al. 2017). The application of stable isotope-resolved metabolomics (SIRM) techniques provides further confirmation for the role of metabolism in gemcitabine resistance. 2D 1H-13C heteronuclear single quantum coherence (HSQC) NMR spectra was used to analyze the metabolites derived from 13C6-glucose in WT Capan-1, WT-treated, GemR and GemR-treated cell lysates. A total of thirty metabolites were identified from the 2D 1H-13C HSQC NMR spectra. The relative metabolite concentrations are plotted in a heatmap with hierarchical clustering (Figure 2A). Consistent with the PCA analysis of the 1D 1H NMR dataset (Figures 1 and S-1), two major branches consisting of WT/WT+ and GemR/GemR+ was observed in the heatmap. Again, this indicates that a metabolic phenotype associated with gemcitabine-resistance was the major feature differentiating the four cell cultures. Furthermore, the gemcitabine sensitive cells formed two distinct branches based on gemcitabine treatment status. In contrast, the GemR cells formed a single branch irrespective of gemcitabine treatment.
Figure 2. Altered metabolism in gemcitabine resistant cells.
(A) Heatmap and hierarchical clustering analysis generated from 2D 1H-13C HSQC spectra of WT, WT+, GemR and GemR+ cell lysates. Peak intensities for each metabolite was normalized by the mean of all the peaks and then scaled by the maximum peak intensity for the metabolite across the four groups. The column clustering identifies group membership and the row clustering identifies metabolites with similar trends across the groups. (B) A metabolic pathway summarizing the metabolite differences between WT and GemR (first arrow); and between WT and WT+ (second arrow), respectively. A red arrow indicates a decrease in the metabolite in WT+ or GemR relative to WT. A green arrow indicates an increase in the metabolite in WT+ or GemR relative to WT.
In addition to clustering based on group membership, the metabolites in the heatmap were also hierarchically clustered (Figure 2A). As a result, the metabolites formed three distinct branches. The top branch contains metabolites that exhibited a decrease in concentration in response to acquiring gemcitabine resistance. This branch includes important metabolites such as glucose, glucose 6-phosphate, aspartate, and citric acids. The middle branch contains metabolites that all increased in concentration upon developing gemcitabine resistance. These metabolites include nucleotide analogs of adenine, cytidine, guanine and uracil along with amino acids such as glutamine, and alanine. The bottom cluster contains metabolites with distinct patterns dependent on the cell lines gemcitabine treatment. In effect, these metabolites primarily reflect the response of WT cells to treatment and are major contributors to distinguishing WT from WT+ cells.
The pathway shown in Figure 2B summarizes the metabolic changes that resulted from either gemcitabine resistance (first arrow) or from WT cell’s response to treatment (second arrow). The acquisition of gemcitabine resistance resulted in a major metabolic “switch”. This is further evident by the complete pairwise reversal in metabolite concentration changes as depicted in Figure 2B. Specifically, if a metabolite increased as a result of acquiring gemcitabine resistance, it was then observed to decrease in the WT cell’s response to gemcitabine treatment or vice-versa.
Our NMR metabolomics analysis indicated that GemR cells exhibited an altered nucleotide biosynthesis, which resulted from a redirection in carbon-flow from other major metabolic pathways. Specifically, metabolites from glycolysis and the pentose phosphate pathway were decreased in GemR. Presumably, this was a result of carbon flowing from glycolysis and PPP into nucleotide biosynthesis in order to increase the cellular concentration of nucleotides. This is consistent with prior observations that gemcitabine efficacy is affected by the nucleotide cellular pool (Raykov et al. 2014) and our observation that deoxycytidine triphosphate is a competitive-inhibitor of gemcitabine (Shukla et al. 2017). Again, the metabolic response of gemcitabine sensitive cells (e.g., wild-type cells) to a gemcitabine treatment was a complete reversal of the response in gemcitabine-resistant cells (e.g., GemR). In summary, GemR cells and WT-treated cells metabolize glucose differently. In GemR, glucose is primarily funneled into nucleotide synthesis to negate gemcitabine activity; whereas, glucose is primarily directed into aerobic glycolysis in a WT cell’s response to gemcitabine treatment.
Conclusion
A unique metabolic phenotype was identified for pancreatic cancer cells with an acquired resistance to gemcitabine. A metabolic “switch” was observed when comparing gemcitabine-resistant cells to a wild-type cell’s response to gemcitabine treatment. This metabolic switch enabled gemcitabine-resistant cells to funnel carbon from glucose into nucleotide biosynthesis, where the increased cellular pool of nucleotides function as a competitive inhibitor of gemcitabine. The distinct metabolic profiles for both a response to treatment and an acquired-drug resistance suggest a potential utility of metabolomics for monitoring a patient’s response to gemcitabine therapy.
Supplementary Material
Acknowledgments
This work was supported in part by funding from the National Institutes of Health grant (R01 CA163649, NCI) to P.K.S. and R.P.; the Redox Biology Center (P30 GM103335, NIGMS) to R.P.; the Nebraska Center for Integrated Biomolecular Communication (P20 GM113126, NIGMS) to R.P.; American Association for Cancer Research (AACR) -Pancreatic Cancer Action Network (PanCAN) Career Development Award (30-20-25-SING) to P.K.S.; the Specialized Programs for Research Excellence (SPORE, 2P50 CA127297, NCI) to P.K.S.; Pancreatic Tumor Microenvironment Research Network (U54, CA163120, NCI) to P.K.S.; and Fred & Pamela Buffett Cancer Center Support Grant (P30CA036727) to P.K.S. and R.P. The research was performed in facilities renovated with support from the National Institutes of Health (RR015468-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Compliance with Ethical Requirement
Conflict of Interest
Authors have no conflict of interest to declare.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Amelio I, Cutruzzola F, Antonov A, Agostini M, & Melino G (2014). Serine and glycine metabolism in cancer. Trends Biochem Sci, 39(4), 191–198, doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin C, Veal G, Paci A, Chatelut E, Astier A, Leveque D, et al. (2014). Therapeutic drug monitoring in cancer--are we missing a trick? Eur J Cancer, 50(12), 2005–2009, doi: 10.1016/j.ejca.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Chaika NV, Gebregiworgis T, Lewallen ME, Purohit V, Radhakrishnan P, Liu X, et al. (2012). MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc Natl Acad Sci U S A, 109(34), 13787–13792, doi: 10.1073/pnas.1203339109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford Sarah A., Huang Z, Du X, Wang Y, Cai L, Witkiewicz Agnes K., et al. (2014). Acetate Dependence of Tumors. Cell, 159(7), 1591–1602, doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, & Davies D (2010). Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev, 74(3), 417–433, doi: 10.1128/mmbr.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Cavalcante L, & Monteiro G (2014). Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol, 741, 8–16, doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- Fryer RA, Barlett B, Galustian C, & Dalgleish AG (2011). Mechanisms underlying gemcitabine resistance in pancreatic cancer and sensitisation by the iMiD lenalidomide. Anticancer Res, 31(11), 3747–3756. [PubMed] [Google Scholar]
- Gardner SG, Somerville GA, Marshall DD, Powers R, Daum RS, Daum RS, et al. (2018). Metabolic Mitigation of Staphylococcus aureus Vancomycin Intermediate-Level Susceptibility. Antimicrob Agents Chemother, 62(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupp R, Lei S, Reed JM, Peisker H, Boyle-Vavra S, Bayer AS, et al. (2015). Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. [ 10.1128/aac.00160-15]. Antimicrob. Agents Chemother, 59(7), 4226–4238, doi: 10.1128/aac.00160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Zweig MH, Carney DN, Van Steirteghen AC, Baylin SB, & Minna JD (1981). Levels of creatine kinase and its BB isoenzyme in lung cancer specimens and cultures. Cancer Res, 41(7), 2773–2777. [PubMed] [Google Scholar]
- Gebregiworgis T, & Powers R (2012). Application of NMR metabolomics to search for human disease biomarkers. Comb Chem High Throughput Screen, 15(8), 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebregiworgis T, Purohit V, Shukla SK, Tadros S, Chaika NV, Abrego J, et al. (2017). Glucose Limitation Alters Glutamine Metabolism in MUC1-Overexpressing Pancreatic Cancer Cells. J Proteome Res, 16(10), 3536–3546, doi: 10.1021/acs.jproteome.7b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunde K, Penet MF, Jiang L, Jacobs MA, & Bhujwalla ZM (2015). Choline metabolism-based molecular diagnosis of cancer: an update. Expert Rev Mol Diagn, 15(6), 735–747, doi: 10.1586/14737159.2015.1039515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Kosaka N, & Kishi H (1998). PET imaging of prostate cancer using carbon-11-choline. J. Nucl. Med, 39(6), 990–995. [PubMed] [Google Scholar]
- Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. (2014). Drug resistance in cancer: an overview. Cancers (Basel), 6(3), 1769–1792, doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju HQ, Gocho T, Aguilar M, Wu M, Zhuang ZN, Fu J, et al. (2015). Mechanisms of Overcoming Intrinsic Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma through the Redox Modulation. Mol Cancer Ther, 14(3), 788–798, doi: 10.1158/1535-7163.MCT-14-0420. [DOI] [PubMed] [Google Scholar]
- Lu SC (2009). Regulation of glutathione synthesis. Mol Aspects Med, 30(1–2), 42–59, doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara J, Ono M, Honda K, Negishi A, Ueno H, Okusaka T, et al. (2010). Survival prediction for pancreatic cancer patients receiving gemcitabine treatment. Mol Cell Proteomics, 9(4), 695–704, doi: 10.1074/mcp.M900234-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, & Hasan MR (2015). Cancer Metabolism and Drug Resistance. Metabolites, 5(4), 571–600, doi: 10.3390/metabo5040571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raykov Z, Grekova SP, Bour G, Lehn JM, Giese NA, Nicolau C, et al. (2014). Myo-inositol trispyrophosphate-mediated hypoxia reversion controls pancreatic cancer in rodents and enhances gemcitabine efficacy. Int J Cancer, 134(11), 2572–2582, doi: 10.1002/ijc.28597. [DOI] [PubMed] [Google Scholar]
- Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. (2017). MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell, 32(1), 71–87 e77, doi: 10.1016/j.ccell.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature, 536(7617), 479–483, doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Zhang X, Luo D, Liu Z, Yang X, Wan H, et al. (2015). Effect of taurine on the proliferation and apoptosis of human hepatocellular carcinoma HepG2 cells. Exp Ther Med, 10(1), 193–200, doi: 10.3892/etm.2015.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhr K, Prager-van der Smissen WJ, Heine AA, Ozturk B, Smid M, Gohlmann HW, et al. (2015). Understanding drugs in breast cancer through drug sensitivity screening. Springerplus, 4, 611, doi: 10.1186/s40064-015-1406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucenik I, & Shamsuddin AM (2003). Cancer inhibition by inositol hexaphosphate (IP6) and inositol: from laboratory to clinic. J Nutr, 133(11 Suppl 1), 3778S–3784S. [DOI] [PubMed] [Google Scholar]
- Worley B, & Powers R (2014). MVAPACK: A Complete Data Handling Package for NMR Metabolomics. [ 10.1021/cb4008937]. ACS Chem. Biol, 9(5), 1138–1144, doi: 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, & Vousden KH (2016). Serine and one-carbon metabolism in cancer. [ 10.1038/nrc.2016.81]. Nat. Rev. Cancer, 16(10), 650–662, doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.