Abstract
A multi-step NMR based screening assay is described for identifying and evaluating chemical leads for their ability to bind a target protein. The multi-step NMR assay provides structure-related information while being an integral part of a structure based drug discovery and design program. The fundamental principle of the multi-step NMR assay is to combine distinct 1D and 2D NMR techniques, in such a manner, that the inherent strengths and weakness associated with each technique is complementary to each other in the screen. By taking advantage of the combined strengths of 1D and 2D NMR experiments, it is possible to minimize protein requirements and experiment time and differentiate between non-specific and stoichiometric binders while being able to verify ligand binding, determine a semi-quantitative dissociation constant, identify the ligand binding site and rapidly determine a protein-ligand co-structure. Furthermore, the quality and physical behavior of the ligand is readily evaluated to determine its appropriateness as a chemical lead. The utility of the multi-step NMR assay is demonstrated with the use of PrgI from Salmonella typhimurium and human serum albumin (HSA) as target proteins.
Keywords: Multi-Step NMR Screen, Protein-Ligand Binding, line broadening, saturation transfer difference, 2D 1H-15N HSQC, Salmonella typhimurium, Protein PrgI, Human Serum Albumin
INTRODUCTION
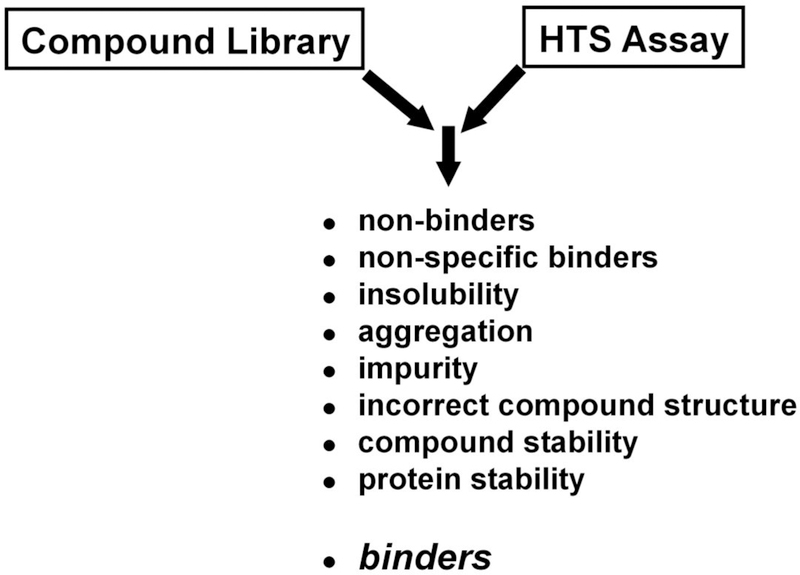
NMR has been demonstrated to be a useful addition to standard high-throughput screening (HTS) techniques to analyze small molecules for their ability to bind protein targets of interest.1,2 A typical HTS assay may yield a biological response upon addition of the inhibitor, but as a result of the complexity of the screening protocol and the mechanism of monitoring a response, it is generally not feasible to infer a binding interaction between the ligand and the protein of interest.3 There are numerous undesirable mechanisms resulting from poor physical behavior of the compound that will result in a positive response in an HTS screen (Fig. (1)).4–7 As a result, only a small percentage of the compounds identified by HTS may actually bind the protein target in a biologically relevant manner, where the remainder are false leads.8
Fig. (1).
The reality of screening compound libraries. A number of undesirable mechanisms may lead to a positive response in an HTS assay that is not a result of a specific interaction of the compound with the protein target.
NMR ligand affinity screens are routinely used to validate these HTS hits by providing direct evidence for a binding interaction between the ligand and protein target through a variety of NMR methodologies.9–14 Observation of a binding event may occur through: changes in line-width and/or peak intensity (T1 and T2 relaxation changes);15–18 changes in the measured diffusion coefficient for the ligand;19,20 chemical shift perturbations for either the ligand21,22 or protein;10,23,24 induced transferred NOEs (trNOE) for the ligand;25–27 a saturation transfer difference (STD) between either the protein or bulk solvent to the ligand;28–30 appearance of new NOEs and/or intermolecular NOEs between the ligand and protein.31–33 The information obtained from the NMR analysis can be used to identify the binding site, measure a dissociation constant34 and determine a co-structure of the protein with the ligand.25,35,36 NMR screens also provide critical information on the viability of a compound to be classified as a “good” lead candidate by verifying the ligand’s structure, purity and solubility.37,38
NMR ligand affinity assays are also being routinely used to identify novel chemical leads by screening small fragment-based chemical libraries.39 SHAPES40 and other related libraries41,42 typically contain a small collection of low molecular-weight compounds (150–250 Da) that correspond to fragments of known drugs or are diverse functional pharmacophores. The ultimate goal is to identify two or more ligands that can be chemically linked into a single inhibitor with a multiplicative improvement in relative binding affinities.43 Thus, this approach enables an extensive search of chemical space even though only <1000 compounds are experimentally screened by NMR.44
The NMR-based assays that are used to validate HTS hits and screen fragment-based libraries employ either ligand-detected NMR experiments45 or protein-detected NMR experiments.46 These fundamentally distinct NMR methods have different strengths and weaknesses related to the availability of protein material, the need for isotopically-labeled protein samples, the experiment time, the ability to differentiate between non-specific and stoichiometric binders and the ability to identify the ligand binding site. Ligand detected NMR screens utilize 1D NMR techniques, particularly relaxation measurements, diffusion-edited measurements, saturation transfer differences, NOE pumping, and transferred NOEs to identify complex formation from changes in the ligand’s NMR spectrum.33,47–49 These 1D NMR experiments eliminate the need for isotopically labeled protein samples while simultaneously minimizing protein sample requirements (nM-μM), decreasing data acquisition times (< 10 minutes) and increasing throughput. Unfortunately, these 1D NMR experiments do not provide information on the location of the ligand binding site and may not be able to differentiate between non-specific and stoichiometric binders. Thus, these methods are simply used to identify a binding interaction between the protein and ligand and are generally combined with modeling techniques, x-ray crystallography and other bioassays and biophysical techniques to optimize the chemical leads.50
Protein-detected NMR screens evaluate a small molecule’s ability to bind a protein from observed chemical shift perturbations (CSPs) in 2D 1H-15N HSQC43,51,52 or 2D 1H-13C HSQC protein NMR spectra.53 The observed CSPs also allow for the identification of the ligand binding site on the protein surface. Given, the use of 2D HSQC NMR spectra as a screen has some significant obstacles that limit its use in a high-throughput format. Mainly, the relatively low sensitivity of NMR requires significant quantities of isotope enriched protein54 (> 3mg/mL) and data acquisition time (>10 minutes) per sample. This drastically impacts the number of compounds that can be realistically screened.55 NMR cryoprobes and flow-through probes provide partial solutions to these issues through either a 3–4 fold increase in sensitivity or a method for increased throughput, respectively.48,56 Nevertheless, given the high resource requirement for protein-detected NMR screens and the routine reliance on x-ray crystallography to generate protein-ligand co-structures,57 ligand-detected NMR screens are generally the method of choice. This is especially true given the expanding impact of fragment-based screening on the drug discovery process.39
This common practice of using only ligand-detected or protein-detected experiments significantly limits the beneficial impact of NMR-based screens and ignores the natural synergy of the various NMR methods. Also, the routine combination of NMR-based screens with other bioassays and biophysical techniques unnecessarily complicates and delays the process of validating biologically relevant chemical leads. To circumvent these limitations, a multi-step NMR screen has been devised that combines a number of NMR techniques to accomplish the necessary goals of identifying chemical leads, measuring a dissociation constant (KD) and determining a rapid protein-ligand co-structure in a single integrated protocol. To illustrate the utility of this method, screening results for PrgI from Salmonella typhimurium and human serum albumin (HSA) will be discussed.
MATERIALS AND METHODS
Functional Compound Library:
Our screening library is composed of 414 compounds with known biological activity in a total of 113 mixtures comprising 3–4 compounds.58 The design of our screening library guarantees “drug-like” characteristics, while increasing the overall activity of the library. Overall hit rates of >4.5% have been observed compared to 0.1–0.5% hit rates for random libraries.59 Reference NMR spectra are collected for both the individual compounds and the mixtures. These spectra provide NMR assignments for the compounds, while verifying compound solubility, compatibility, and the presence of distinct NMR resonances attributed to each compound in the mixture to avoid deconvolution.60 Identical chemical shifts, coupling patterns and line-widths between the mixture and individual NMR spectra verify that no interaction is occurring between the compounds and that the compounds are equally soluble and stable in the mixture. All the compounds have been individually weighed, dissolved to a concentration of 20 mM in D2O or D6-DMSO (Sigma-Aldrich, St. Louis, MO), and stored in standard 2 ml 96-well plates at −80oC.
Proteins:
Two proteins were used for analysis of the multi-step NMR method. Human serum albumin (96% essentially fatty acid free, Sigma-Aldrich, St. Louis, MO) is an important secondary target for efficacy screening and a well-established system for monitoring protein-ligand interactions.61 HSA was used as a model system for saturation transfer difference (STD) and line broadening experiments. Salmonella typhimurium protein PrgI (generously provided by Dr. Roberto De Guzman from the University of Kansas) is an example of a protein with a well-defined function that was screened in our multi-step NMR assay as a starting point for drug discovery. This needle complex protrudes from the cell membrane of S. typhimurium to sense potential hosts. A solution structure for PrgI has recently been solved by NMR62 and the protein exhibits a helix-turn-helix motif.
Sample Preparation:
In general, 1D 1H NMR samples were screened in 20 mM deuterated bis-Tris (Cambridge Isotope, Andover, MA) at pH 7.0 (uncorrected), with 5% (v/v) dimethyl sulfoxide-d6 (DMSO-d6) (Sigma-Aldrich, St. Louis, MO) and 11 μM 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TMSP) (Cambridge Isotope, Andover, MA) screening buffer prepared in 99.98% deuterium oxide (Cambridge Isotope, Andover, MA). The 2D 1H-15N HSQC samples were prepared in a 20 mM deuterated bis-Tris screening buffer prepared in H2O at pH 7.0 and spiked with 5% deuterium oxide to obtain a lock signal. The protein (PrgI) and compound concentrations for each of the NMR experiments was 25 μM and 100 μM, respectively. For the HSA competition experiments the HSA concentration was increased from 2 μM to 20 μM.
For the non-competitive mixture screens, small molecule ligand samples were prepared in a 10 mL stock solution that contained 20 µM of each ligand (ibuprofen, cinoxacin, L-proline, and adenosine 5’-triphosphate disodium salt), 1% (v/v) DMSO-d6, 10 µM TMSP and a 50 mM potassium phosphate buffer at pH 7.0 (uncorrected) prepared in D2O.
Data collection:
All NMR spectra were collected at 298 K on a Bruker 500 MHz Avance spectrometer (Bruker Instruments, Billerica, MA) equipped with a triple-resonance, Z-axis gradient cryoprobe using a BACS-120 sample changer and IconNMR software for automated data collection.
The STD NMR spectra were collected using 512 transients, a sweep-width of 8992.8 Hz, and 16 K data points. A total time of 10 minutes was required to collect a single spectrum. The 1D line broadening NMR spectra for the non-competitive mixture titration experiments were collected using 512 transients, a sweep-width of 5482.6 Hz, 16 K data points, a relaxation delay of 2.0 s and the residual HDO resonance signal was suppressed using a composite pulse, solvent pre-saturation routine. A total time of 33 minutes was required to collect a single 1D spectrum including sample changing. The same NMR experimental parameters was used for screening the entire functional chemical library except 64 transients were collected per spectrum to reduce the total experiment time to 4.4 min per sample, including sample changing. The total 1D NMR acquisition time required to screen the entire functional chemical library was approximately 8 hrs. The 2D 1H-15N HSQC spectrum was collected with 16 transients, a sweep-width of 4734.85 Hz and 2 K data points in the direct dimension. The indirect dimension had a sweep-width of 1419.05 Hz with 256 data points. The total acquisition time for the secondary PrgI 2D 1H-15N HSQC screen of 6 samples was approximately 7.5 hrs.
Protein Ligand Co-structures
Rapid determination of a ligand bound protein co-structure with PrgI was completed using the molecular docking program AutoDock 4.063 and the previously elucidated protein structure for PrgI.62 AutoDock 4.0 was used to generate 100 docked, ligand bound co-structures, using the Lamarckian search algorithm, a population size of 300 with 500,000 energy evaluations. The chemical shift difference map from the 2D 1H-15N HSQC data was used to guide the ligand docking in AutoDock and our in house AutoDock Filtering program (ADF) was used to select the best co-structure consistent with the experimental chemical shift perturbations.64 On average, a protein-ligand co-structure is calculated in ~30–45 minutes on an Intel Xeon 3.06 GHz dual processor Linux workstation.
DISCUSSION
Description of the Multi-Step NMR Screen
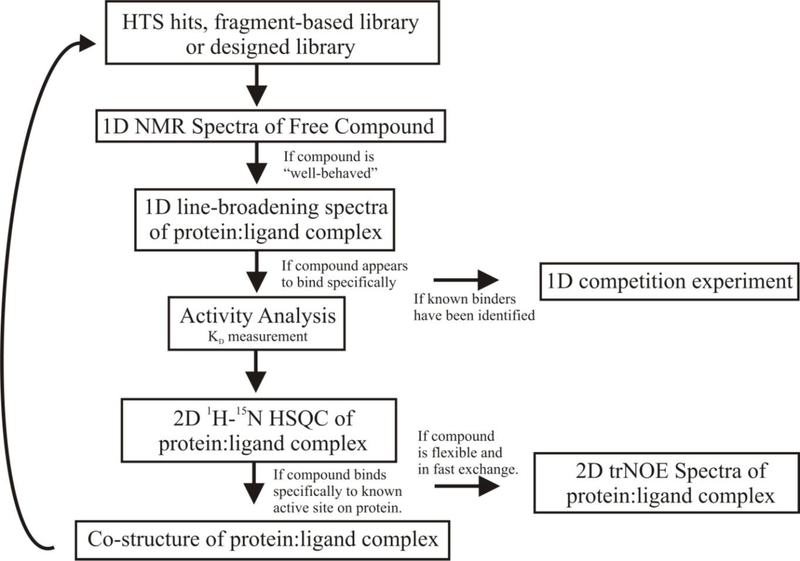
The overall protocol for the multi-step NMR screen is illustrated in Fig. (2). There are four major components to the process: (i) verify good physical properties for the compounds being screened, (ii) identify ligands that bind the protein target, (iii) determine a binding affinity to prioritize the chemical leads and (iv) determine protein-ligand co-structures. The tiered approached used in the multi-step NMR screen minimizes valuable resources, such as protein samples and NMR instrument time, while increasing throughput. The first ligand-detected NMR screening step is fast and minimizes sample requirements while simply identifying the compounds that bind the protein target. The second relatively resource intense protein-detected NMR screening step is only conducted on the positive hits from the first broad screen. In effect, the ligand-detected NMR screening step efficiently filters the screening library to identify binders for future evaluation. Similarly, the protein-detected NMR screening step filters out non-specific binders, confirms a stoichiometric interaction and identifies the ligand binding site. These experimental NMR spectra are then directly used to rapidly determine a protein-ligand co-structure, to measure a dissociation constant and to verify potential chemical leads. The direct outcome of these results from the multi-step NMR screen, which are essential for a drug discovery process, is a unique aspect of the protocol.
Fig. (2).
Flow diagram for the multi-step NMR screen.
Design of Chemical Library:
There are numerous and equally acceptable approaches to designing a chemical library utilized in the multi-step NMR screen. The variety of possible compound library designs has been described at length in the scientific community.65–69 In general, the source of the chemical library will originate from the results of a standard biological assay as part of a high-throughput screen70 or from a fragment-based chemical library.39,42 Future iterations of the screen may utilize a focused or combinatorial library designed based on initial hits.71 The multi-step NMR screen is equally amenable to utilizing either single compounds and/or mixtures.60 It is preferable to design the compound mixtures58 to avoid a necessary de-convolution step to optimize the efficiency of the NMR screen.60
To illustrate the multi-step NMR screen, PrgI from Salmonella typhimurium and human serum albumin were screened against our functional chemical library.58 This library was designed to contain compounds with “drug-like” characteristics based on the existence of known biological activity. Each compound in the library has a demonstrated binding affinity to a protein or protein class.
1D 1H NMR Spectra of Free Compound:
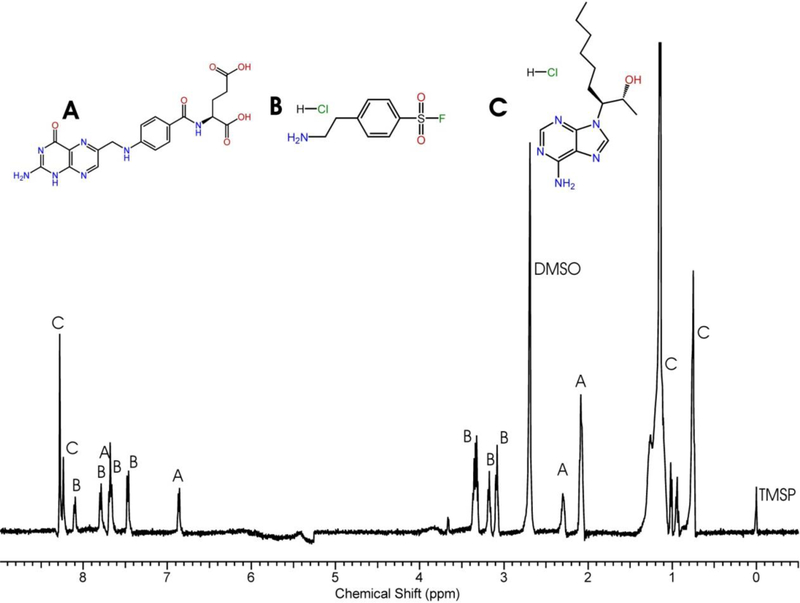
A reference 1D 1H NMR spectra for each individual compound in the chemical library is collected in a standard aqueous buffer and maintained as part of a database. Fig. (3) shows the mixture of folic acid, 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, and erythro-9-(2-hydroxy-3-nonyl) adenine hydrochloride. The 1D 1H NMR spectrum for the free compound indicates the relative aqueous solubility and stability of the compound, the compound’s tendency to form high-molecular weight aggregates or micelle-like structures and, in addition, it verifies the accuracy of the structure.6,7 Thus, the reference 1D 1H NMR spectra provides critical information to evaluate the utility of the compounds for screening in a drug discovery effort. The reference 1D 1H NMR spectra is also used to identify binding interactions based on spectral changes that are induced by the addition of the protein target.
Fig. (3).
An example of a reference 1H NMR spectrum for a compound mixture from the functional chemical library comprised of (A) folic acid, (B) AEBSF and (C) erythro-9-(2-hydroxy-3-nonyl)adenine hydrochloride. The DMSO solvent peak, TMSP reference peak and each peak corresponding to a compound in the mixture are labeled.
1D STD and 1D line-broadening NMR Spectra of Protein:Compound Complex:
The first goal of the multi-step NMR screen is to identify compounds that bind the protein target of interest while minimizing resources (protein and instrument time). The 1D STD experiment72 is a likely choice since it is commonly used in NMR ligand affinity screens.73 The 1D STD experiment utilizes unlabeled protein samples with concentrations as small as 1 nM and NMR acquisition times on the order of minutes. Additionally, the screening step may be done as mixtures where the deconvolution of hits is accomplished by the comparison of the resulting 1D STD spectrum with the reference spectra of the free compounds.
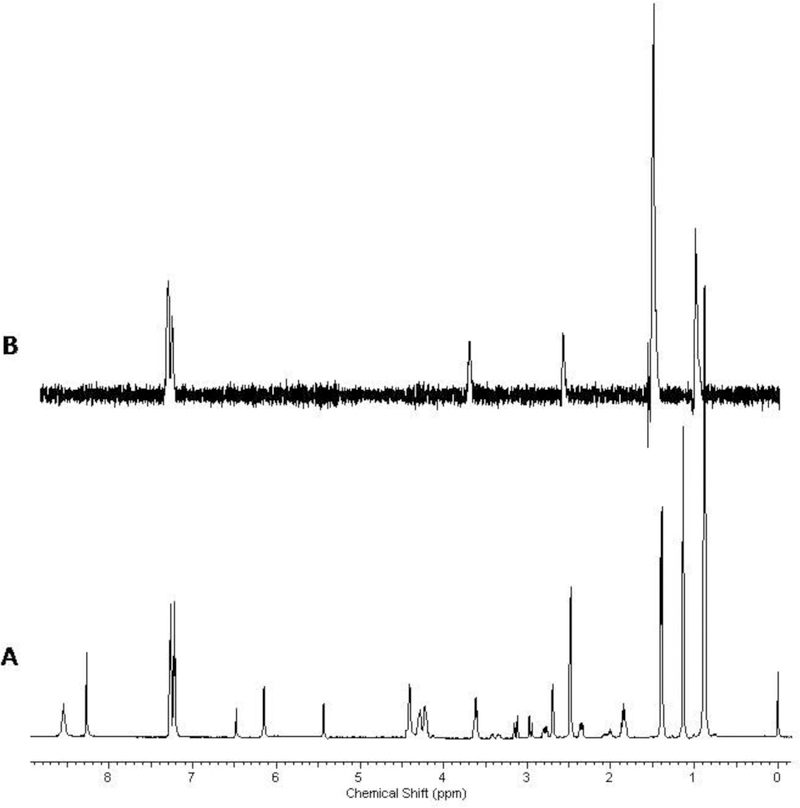
The basic principal of the STD approach is to observe binding between the protein and the ligand by the transfer of saturation from the protein to the ligand. The ligand is in large excess (~20–30:1) relative to the protein. Saturation occurs by selectively irradiating a region of the NMR spectrum that contains only protein resonances, usually in the vicinity of 0.0 ppm. In cases where no binding takes place, the resulting NMR spectrum for the compound is a null. If binding does occur between the protein and compound, then the resulting NMR spectrum would correspond to the spectrum of the free compound with potentially some protein background. Fig. (4) is an example of an STD experiment of a mixture of compounds, where only one compound exhibits binding to HSA. In the mixture of ibuprofen, cinoxacin, L-proline (Pro), and adenosine 5’-triphosphate disodium salt (ATP), only ibuprofen is a known HSA binder and is the only compound that exhibits a positive response in the STD experiment.
Fig. (4).
An example of a STD NMR experiment with a mixture of ibuprofen, cinoxacin, L-proline (Pro), and adenosine 5’-triphosphate disodium salt (ATP). The only peaks from ibuprofen in the STD experiment (B) are associated with the compound that bound to 2 μM of HSA and can be identified from the reference mixture spectrum (A) collected before the protein was added.
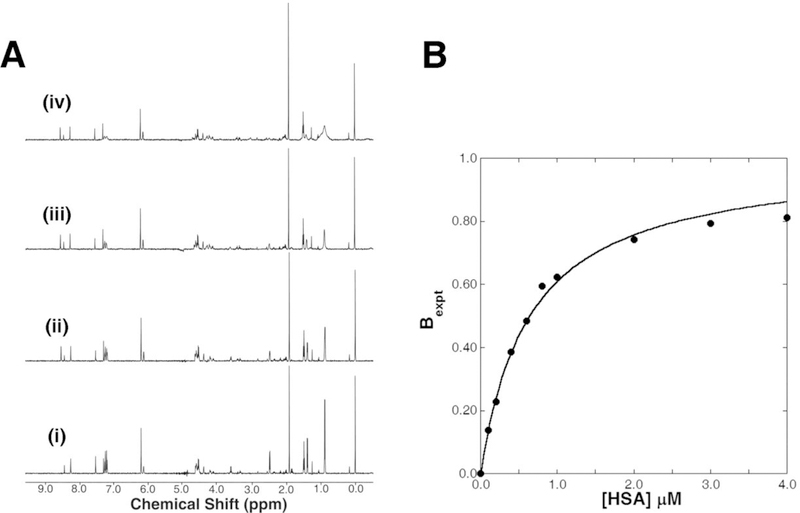
Despite the obvious appeal of the STD experiment, there are a number of serious drawbacks that significantly limit its value to the multi-step NMR screen. The method is overly sensitive to extremely weak (~mM) non-specific binders that result in a high percentage of false positives. Alternatively, tight binders (< 1 μM) are missed. The method is also not readily amenable to measuring a dissociation constant as part of the primary screen and requires significantly longer acquisition times relative to other 1D NMR experiments. An alternative approach to the 1D STD methodology is to measure a relaxation difference for the compound in the presence of the protein.74 NMR line-widths are directly related to the intrinsic T2 relaxation of the molecule, which in turn is directly correlated with the MW of the molecule. Thus, the NMR line-width of a compound bound to a protein will significantly increase due to the dramatic increase in the apparent MW of the compound. As an illustration of 1D 1H NMR line-broadening experiments, a mixture of four compounds was screened against HSA. Again, the mixture contained ibuprofen, cinoxacin, L-proline (Pro), and adenosine 5’-triphosphate disodium salt (ATP). Fig. (5) shows a series of typical 1H NMR spectra for a range of HSA concentrations. The NMR peaks associated with ibuprofen are incrementally broadened by the addition of HSA. These results clearly demonstrate the application of 1D 1H NMR line-broadening to identify binders. It also verifies that the presence of non-binding compounds within a mixture does not interfere with observing a positive binding event. Another advantage of the 1D 1H NMR line-broadening experiment to the multi-step NMR screen is the ability to directly measure a dissociation constant from the assay.
Fig. (5).
(A) A titration mixture of ibuprofen, cinoxacin, L-proline (Pro), and adenosine 5’-triphosphate disodium salt (ATP) with increasing concentrations (i) 0 μM, (ii) 0.4 μM, (iii) 1.0 μM, and (iv) 3.0 μM of HSA. (B) Only ibuprofen binds HSA and the decrease in peak intensity describes the binding curve from eq 1. The resulting equilibrium dissociation constant (KD) for ibuprofen was 0.46 ± 0.8 μM with a NMR line width ratio constant (c) of 31.8.
Semi-Quantitative Dissociation Constants Directly from 1D Line-Broadening Screens:
Typically, IC50 values obtained for each ligand from the HTS assay will provide an initial ranking of the chemical leads. A semi-quantitative dissociation constant (KD) can be obtained directly from the observed line-width changes in the multi-step NMR screen.34 The change in NMR line-widths of the bound ligand (Ib) relative to the free ligand (If) can be plotted against the added protein concentration to generate a typical binding curve (Fig. (5b)). The data can be fit to our newly derived binding isotherm to calculate a KD:34
| (1) |
[P]T and [L]T represent the total protein and ligand concentration, respectively, and the unit-less NMR line width ratio constant (c) accounts for the proportional change in the free ligand line width (νF) upon binding of a ligand to a protein (νB). In the context of the multi-step NMR screen, the binding isotherm can be simply rearranged to solve for the dissociation constant as a function of a single line-width change.
| (2) |
The free ligand line width is directly measured from a reference spectrum with minimum processing and generally ranges between 1 and 2 Hz. The bound ligand line width is approximated by the molecular weight of the protein (MWp) using our derived linear equation.34
| (3) |
For human serum albumin, the approximated bound ligand line width (νB) is 94.2 Hz. Ibuprofen was determined to bind HSA with a KD of 0.50 ± 0.1 μM (Fig. (5b)). The ability to directly measure a binding affinity as part of the 1D line-broadening step of the multi-step NMR screen enables a ranking and prioritization of these hits for further analysis in the assay.
1D Competition Experiment:
Depending on the specifics of the protein target that is being screened, there may be value in determining if the hits from the previous binding analysis exhibit competitive binding to known substrates, ligands or other hits (Fig. (6)). This is easily accomplished by the addition of a known binder to the protein:compound mixture to determine if the increase in line-width previously observed is now lost or reduced. This would suggest that the binding of the compound and known ligand is mutually exclusive and suggestive of a similar or overlapping binding site on the protein.
Fig. (6).
A competition experiment between two known binders to HSA. (A) 7-hydroxy-4-methyl coumarin alone, (B) 7-hydroxy-4-methyl coumarin in the presence of HSA, and (C) 7-hydroxy-4-methyl coumarin in the presence of HSA and warfarin. The 7-hydroxy-4-methyl coumarin (KA =2.1×104 M−1) is displaced by the addition of warfarin (KA =2.5×105 M−1).
Another important utilization of the 1D competition approach may be its application in the general multi-step NMR screening protocol to eliminate an unwanted class of compounds. Consider the situation where a known ligand exists for the protein target of interest, but it is undesirable to identify compounds that bind in a similar manner. By having the known ligand in molar excess relative to the compounds in the chemical library during the 1D line-broadening NMR screening step, competitors to this known class of binders will be severely diminished. This will minimize wasted effort in follow-up experiments for undesirable compounds. Additionally, this same approach may be used to explore alternative binding sites on the protein with the end goal of chemically linking compounds that interact in the distinct binding sites or simply identifying an alternative interaction mode.
As an illustration of an NMR competition experiment, Fig. (6) shows a coumarin-analogue being displaced from HSA by following the changes in the ligands NMR line-width from the addition of warfarin. Both 7-hydroxy-4-methyl coumarin (KA = 2.1×104 M−1) and warfarin (KA = 2.5×105 M−1) bind selectively to Sudlow Site I on HSA.75 The NMR resonances of the coumarin-analogue broaden (Fig (6a, b)) from the addition of HSA, consistent with the known affinity of the compound to HSA. The addition of warfarin to the coumarin-HSA complex (Fig. (6c)) results in a sharpening of the coumarin NMR line-widths as the compound is displaced from HSA by warfarin. Furthermore, the binding of warfarin to HSA is evident by the line-broadening of the warfarin NMR resonances.
2D 1H-15N HSQC Spectra of Protein:Compound Complex:
The next step in the multi-step NMR assay is the further evaluation of the hits from the 1D line-broadening NMR experiments. This is accomplished by collecting a 2D 1H-15N HSQC or 2D 1H-13C HSQC NMR experiment for the 13C or 15N –labeled protein target in the presence of each compound identified as a hit. For larger molecular-weight proteins (> 25 kDa) the TROSY version of the 2D 1H-15N HSQC experiment would be used.76 A complex is identified by the induced chemical shift perturbations in the protein spectrum caused by the addition of a ligand. Since the 2D HSQC NMR experiments are performed only on identified hits, greater care can be taken to maximize the quality of the NMR spectra and greater attention can be applied in the analysis of the data. This implies that a weak binding compound that may induce a minimal number of modest chemical shift perturbations has a less likelihood of being missed and that false positives resulting from pH or buffer changes may be eliminated. Furthermore, greater care can be used to monitor changes in peak intensity and the appearance or disappearance of peaks in 2D HSQC spectra, which is also indicative of a binding event.
In conjunction with previously determined NMR assignments and structure determination of the protein target, it is a straightforward procedure to map the amino acid residues exhibiting chemical shift perturbations and/or intensity changes onto the protein’s molecular surface to define the binding site of an identified hit. An observed clustering of amino acid residues in the same region of the protein surface provides a level of confidence that the inhibitor is binding specifically to the protein. Conversely, a random distribution or a complete lack of amino acids that incur chemical shift or intensity change is strongly suggestive of a non-specific binder.
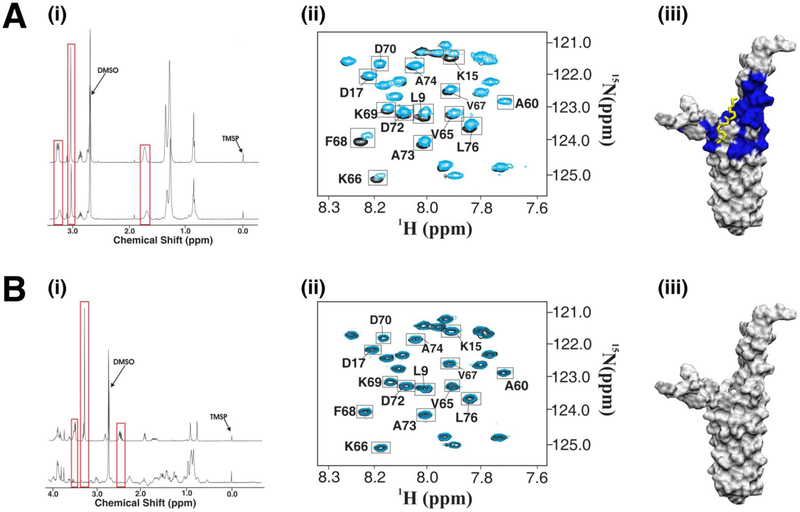
Four compounds from the functional chemical library were identified as binders to PrgI using the 1D 1H line-broadening experiment (Fig. (7i)). The 2D 1H-15N HSQC spectra clearly shows that only one compound exhibits specific binding to PrgI based on the clustering of chemical shift perturbations on the PrgI surface (Fig. (7ii, iii)). These results demonstrate fundamental differences in the application of 1D line-broadening and 2D 1H-15N HSQC spectra to differentiate between specific and non-specific binders. Essentially, the 2D HSQC NMR data is complimentary to and expands the information content obtainable from a 1D line-broadening screen. The 2D HSQC results confirm a specific binding interaction of the compound with the protein target while providing information on the binding site. Again, this illustrates the inherent strengths of the multi-step NMR screen. By integrating multiple NMR experiments, the strengths of one experiment compensates for the weakness of another.
Fig. (7).
The 1D line broadening and 2D 1H-15N HSQC multi-step NMR protocol was used to identify five compounds that bound to the Type III Secretion System (TTSS) protein PrgI of S. typhimurium. (A) One of the five compounds, didecyldimethylammonium bromide, showed minimal changes in line broadening experiments (i) but significant chemical shift changes in the 2D HSQC secondary screen (ii). These chemical shift changes were mapped to the surface of PrgI and didecyldimethylammonium bromide was docked to the region using AutoDock combined with our in house ADF program (iii). (B) The strongest binding ligand from the 1D NMR screens (i) showed no chemical shift changes in the 2D NMR screen (ii). This suggests the compound binds non-specifically to PrgI and therefore no binding site was mapped to the protein’s surface (iii).
Rapid Structure Determination and Iterative Design:
After verifying that the compounds bind selectively to the protein, the structure of the protein-ligand complex is rapidly elucidated by NMR.64 The chemical shift perturbations (CSPs) obtained from the 2D 1H-15N HSQC spectra are used to guide an AutoDock ligand docking calculation. The AutoDock 3D grid is reduced to a volume encompassing only the experimental binding site defined by the CSPs. Our AutoDock Filtering program (ADF) selects the best conformer(s) based on a consistency with the CSPs. Simply, the ligand is expected to be closer to residues that incurred larger chemical shift changes. On average, a protein-ligand co-structure is calculated within 35–45 minutes and exhibits an average RMSD of 1.17 ± 0.74 Å to high-resolution NMR or x-ray structures.
Chemical shift perturbations observed in the 2D 1H-15N HSQC spectrum for PrgI bound to didecyldimethylammonium bromide identified a ligand binding site corresponding to residues at the bifurcation point of the two helices (Fig (7Aii)). This binding site corresponds to residues S6, L9, S13, K15, and D17 of helix 1 and N59, V65, K66, V67, F68, K69, D70, D72, A73 and L76 of helix 2 and was used to guide the AutoDock simulation. The best-conformer selected based on consistency with the magnitude of CSPs is shown in Fig (7Aiii), where didecyldimethylammonium adopts an extended conformation that straddles both helices of PrgI.
Another option of the multi-step NMR screen is to obtain 2D trNOE spectra of the protein:compound complex with the goal of determining the bound conformation of the ligand. This information can then be used in combination with the binding site identified from the 2D HSQC data to aid in the rapid determination of the co-structure. The accuracy of an AutoDock calculation is often dependent on the number of torsional degrees of freedom in the ligand.77,78 Therefore, knowledge of the bound ligand’s conformation would limit the number of torsional angles permitted to undergo free rotation during the AutoDock simulation. The utility of this step depends on the size and conformational flexibility of the compound and is typically unnecessary.
Finally, the multi-step NMR protocol is amenable to an iterative approach where a library of structural analogs, based on the initial hits, can be used to further optimize the affinity and activity of the ligand.
CONCLUSION
The multi-step NMR assay is used in combination with HTS screens to expedite the validation and prioritization of lead compounds. The multi-step NMR methodology supplies information that is critical to a drug design program that is absent in traditional HTS assays. The multi-step NMR screen provides direct evidence that the compound of interest binds to the desired protein target in a biologically relevant manner. Additionally, information on the stoichiometry of binding, the dissociation constant, the identification of the ligand-binding site and a protein:ligand co-structure are all readily obtainable from the assay. The multi-step NMR easily provides information on the physical behavior of the ligand itself. The assay rapidly identifies such properties as relative solubility, purity and stability that are fundamental for a good lead compound. The strength of the multi-step NMR screen arises from the integration of ligand-detected and protein-detected NMR methodologies.
ACKNOWLEDGEMENTS
We would like to thank Dr. Roberto De Guzman from the University of Kansas for supplying the PrgI sample. This work was supported by a grant from the Nebraska Tobacco Settlement Biomedical Research Development Funds. Research was performed in facilities renovated with support from the NIH under grant RR015468-01.
REFERENCES
- (1).Shi Z; Tabassum S; Jiang W; Zhang J; Mathur S; Wu J; Shi Y Chembiochem 2007. [DOI] [PubMed]
- (2).Hasnat A; Bichenkova E; Yu X; Arnold JR; Fisher J; Fedorova O; Andrews J J Biomol Struct Dyn 2007, 25, 253–70. [DOI] [PubMed] [Google Scholar]
- (3).Barker J; Hesterkamp T; Schade M; Whittaker M 2007, 23, 19–22. [Google Scholar]
- (4).Rishton GM Drug Discovery Today 1997, 2, 382–384. [DOI] [PubMed] [Google Scholar]
- (5).Seidler J; McGovern SL; Doman TN; Shoichet BK J. Med. Chem 2003, 46, 4477–4486. [DOI] [PubMed] [Google Scholar]
- (6).McGovern SL; Helfand BT; Feng B; Shoichet BK J. Med. Chem 2003, 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- (7).McGovern SL; Caselli E; Grigorieff N; Shoichet BK J. Med. Chem 2002, 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- (8).Malo N; Hanley JA; Cerquozzi S; Pelletier J; Nadon R 2006, 24, 167–175. [DOI] [PubMed] [Google Scholar]
- (9).Peng JW; Moore J; Abdul-Manan N Prog. Nucl. Magn. Reson. Spectrosc 2004, 44, 225–256. [Google Scholar]
- (10).Zech SG; Olejniczak E; Hajduk PJ; Mack J; McDermott AE J. Amer. Chem. Soc 2004, 126, 13948–13953. [DOI] [PubMed] [Google Scholar]
- (11).Mittag T; Schaffhausen B; Gunther UL J. Amer. Chem. Soc 2004, 126, 9017–9023. [DOI] [PubMed] [Google Scholar]
- (12).Yao H; Costache AD; Sem DS J. Chem. Inf. Comput. Sci 2004, 44, 1456–1465. [DOI] [PubMed] [Google Scholar]
- (13).Golovanov AP; Blankley RT; Avis JM; Bermel W J Am Chem Soc 2007, 129, 6528–35. [DOI] [PubMed] [Google Scholar]
- (14).Krajewski M; Rothweiler U; D’Silva L; Majumdar S; Klein C; Holak TA J Med Chem 2007, 50, 4382–7. [DOI] [PubMed] [Google Scholar]
- (15).Shimotakahara S; Furihata K; Tashiro M Magn. Reson. Chem 2005, 43, 69–72. [DOI] [PubMed] [Google Scholar]
- (16).Eletsky AK, Alexander; Hilvert Donald; Pervushin Konstantin Biochemistry 2005, 44, 6788–6799. [DOI] [PubMed] [Google Scholar]
- (17).Zhu GX, Youlin; Lin Donghai; Gao Xialian. Methods Mol. Biol 2004, 278, 161–184. [DOI] [PubMed] [Google Scholar]
- (18).Yan JK, Allen D; Mo Huaping; Shapiro Michael J.; Zartler Edward R. J. Magn. Reson 2003, 163, 270–276. [DOI] [PubMed] [Google Scholar]
- (19).Larive W. H. O. a. C. K. J. Magn. Reson 2001, 153, 273–276. [DOI] [PubMed] [Google Scholar]
- (20).Jung D-ME, Susan E J. Agric. Food. Chem 2003, 51, 1988–1993. [DOI] [PubMed] [Google Scholar]
- (21).Tengel T; Fex T; Emtenäs H; Almqvist F; Sethson I; Kihlberg J Org. Biomol. Chem 2004, 2, 725–731. [DOI] [PubMed] [Google Scholar]
- (22).Wang BM, Kenneth M Jr. J. Amer. Chem. Soc 2005, 127, 5310–5311. [DOI] [PubMed] [Google Scholar]
- (23).Hemmi HK, Atsushi; Ito Shigeyasu; Suzuki Ryuichiro; Kaneko Satoshi; Hasegawa Tsunemi; Hirabayashi Jun; Kasai Ken-ichi. J. Biomol. NMR 2004, 30, 377–378. [DOI] [PubMed] [Google Scholar]
- (24).Peng C; Unger SW; Filipp FV; Sattler M; Szalma SJ Biomol. NMR 2004, 29, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lucas LH; Price KE; Larive CK J. Amer. Chem. Soc 2004, 126, 14258–14266. [DOI] [PubMed] [Google Scholar]
- (26).Johnson MAP, Mario B Carbohydr. Res 2004, 339, 907–928. [DOI] [PubMed] [Google Scholar]
- (27).Clement MJ; Fortune A; Phalipon A; Marcel-Peyre V; Simenel C; Imberty A; Delepierre M; Mulard LA J Biol Chem 2006, 281, 2317–32. [DOI] [PubMed] [Google Scholar]
- (28).Kover KE; Groves P; Jimenez-Barbero J; Batta G J Am Chem Soc 2007, 129, 11579–82. [DOI] [PubMed] [Google Scholar]
- (29).Cutting B; Shelke SV; Dragic Z; Wagner B; Gathje H; Kelm S; Ernst B Magn Reson Chem 2007, 45, 720–4. [DOI] [PubMed] [Google Scholar]
- (30).Wen XY, Yue; Kuntz Douglas A.; Rose David R.; Pinto B. Mario. Biochemistry 2005, 44, 6729–6737. [DOI] [PubMed] [Google Scholar]
- (31).Eichmuller CS, Wolfgang; Konrat Robert; Krautler Bernhard. J. Biomol. NMR 2001, 21, 107–116. [DOI] [PubMed] [Google Scholar]
- (32).Post CB Curr. Opin. Struct. Biol 2003, 13, 581–588. [DOI] [PubMed] [Google Scholar]
- (33).Reibarkh M; Malia TJ; Hopkins BT; Wagner G J Biomol NMR 2006, 36, 1–11. [DOI] [PubMed] [Google Scholar]
- (34).Shortridge MD; Hage DS; Harbison GS; Powers RJ Comb. Chem 2008, ACS ASAP. [DOI] [PMC free article] [PubMed]
- (35).Grant MA; Morelli XJ; Ridby AC Curr. Protein Pept. Sci 2004, 5, 235–248. [DOI] [PubMed] [Google Scholar]
- (36).Campbell A. R. P. a. I. D. Chemical Reviews 2004, 104, 3557–3565. [DOI] [PubMed] [Google Scholar]
- (37).Dalvit C; Caronni D; Mongelli N; Veronesi M; Vulpetti A Curr Drug Discov Technol 2006, 3, 115–24. [DOI] [PubMed] [Google Scholar]
- (38).Huth JR; Mendoza R; Olejniczak ET; Johnson RW; Cothron DA; Liu Y; Lerner CG; Chen J; Hajduk PJ J Am Chem Soc 2005, 127, 217–24. [DOI] [PubMed] [Google Scholar]
- (39).Hajduk PJ; Greer J Nat. Rev. Drug Discovery 2007, 6, 211–219. [DOI] [PubMed] [Google Scholar]
- (40).Lepre C Expert Opin. Drug Discovery 2007, 2, 1555–1566. [DOI] [PubMed] [Google Scholar]
- (41).Schuffenhauer A; Ruedisser S; Marzinzik AL; Jahnke W; Blommers M; Selzer P; Jacoby E Curr. Top. Med. Chem. (Sharjah, United Arab Emirates) 2005, 5, 751–762. [DOI] [PubMed] [Google Scholar]
- (42).Baurin N; Aboul-Ela F; Barril X; Davis B; Drysdale M; Dymock B; Finch H; Fromont C; Richardson C; Simmonite H; Hubbard RE J. Chem. Inf. Comput. Sci 2004, 44, 2157–2166. [DOI] [PubMed] [Google Scholar]
- (43).Shuker SBH, P.J.; Meadows RP; Fesik SW Science 1996, 274, 1531–1534. [DOI] [PubMed] [Google Scholar]
- (44).Lipinski C; Hopkins A Nature 2004, 432, 855–861. [DOI] [PubMed] [Google Scholar]
- (45).Zartler ER; Yan J; Mo H; Kline AD; Shapiro MJ Curr. Top. Med. Chem. (Hilversum, Netherlands) 2003, 3, 25–37. [DOI] [PubMed] [Google Scholar]
- (46).Zartler ER; Shapiro MJ Curr. Pharm. Des 2006, 12, 3963–3972. [DOI] [PubMed] [Google Scholar]
- (47).Hajduk PJ; Olejniczak ET; Fesik SW J. Am. Chem. Soc 1997, 119, 12257–12261. [Google Scholar]
- (48).Shapiro MJ; Wareing JR Curr. Opin. Drug Discovery Dev 1999, 2, 396–400. [PubMed] [Google Scholar]
- (49).Chen A; Shapiro MJ J. Am. Chem. Soc 1998, 120, 10258–10259. [Google Scholar]
- (50).Moore J; Abdul-Manan N; Fejzo J; Jacobs M; Lepre C; Peng J; Xie X J. Synchrotron. Radiat 2004, 11, 97–100. [DOI] [PubMed] [Google Scholar]
- (51).Matter HS, Manfred; Elshorst Bettina; Jacobs Doris M.; Saxena Krishna; Kogler Herbert. Bioorg. Med. Chem. Lett 2005, 15, 1779–1783. [DOI] [PubMed] [Google Scholar]
- (52).Horvath IH, Veronika; Perczel Andras; Palfi Villo; Nyitray Laszlo; Nagy Attila; Hlavanda Emma; Naray-Szabo Gabor; Ovadi Judit. J Biol Chem 2005, 280, 8266–8274. [DOI] [PubMed] [Google Scholar]
- (53).Vanwetswinkel S; Heetebrij RJ; van Duynhoven J; Hollander JG; Filippov DV; Hajduk PJ; Siegal G Chem. Biol 2005, 12, 207–216. [DOI] [PubMed] [Google Scholar]
- (54).Lian L-Y; Middleton DA Prog. Nucl. Magn. Reson. Spectrosc 2001, 39, 171–190. [Google Scholar]
- (55).Yee AA; Savchenko A; Ignachenko A; Lukin J; Xu X; Skarina T; Evdokimova E; Liu CS; Semesi A; Guido V; Edwards AM; Arrowsmith CH J Am Chem Soc 2005, 127, 16512–7. [DOI] [PubMed] [Google Scholar]
- (56).Hajduk PJ; Gerfin T; Boehlen J-M; Haeberli M; Marek D; Fesik SW J. Med. Chem 1999, 42, 2315–2317. [DOI] [PubMed] [Google Scholar]
- (57).Sharff A; Jhoti H Curr. Opin. Cell Biol 2003, 7, 340–345. [DOI] [PubMed] [Google Scholar]
- (58).Mercier KA; Germer K; Powers R Comb. Chem. High Throughput Screening 2006, 9, 515–534. [DOI] [PubMed] [Google Scholar]
- (59).Dove A; Marshall A Nat. Biotechnol 1999, 17, 859–863. [DOI] [PubMed] [Google Scholar]
- (60).Mercier KA; Powers RJ Biomol. NMR 2005, 31, 243–258. [DOI] [PubMed] [Google Scholar]
- (61).Peters T All About Albumin: Biochemistry, Genetics and Medicinal Applications; Academic Press: San Diego, 1996. [Google Scholar]
- (62).Wang Y; Ouellette AN; Egan CW; Rathinavelan T; Im W; De Guzman RN J Mol Biol 2007, 371, 1304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Morris GM; Goodsell DS; Halliday RS; Huey R; Hart WE; Belew RK; Olson AJ J. Comput. Chem 1998, 19, 1639–1662. [Google Scholar]
- (64).Stark J; Powers R J. Amer. Chem. Soc 2008, 130, 535–545. [DOI] [PubMed] [Google Scholar]
- (65).Xue L; Bajorath J Comb. Chem. High Throughput Screening 2000, 3, 363–372. [DOI] [PubMed] [Google Scholar]
- (66).Lewis RA; Pickett SD; Clark DE Rev. Comput. Chem 2000, 16, 1–51. [Google Scholar]
- (67).Willett P Curr. Opin. Biotechnol 2000, 11, 85–88. [DOI] [PubMed] [Google Scholar]
- (68).Spellmeyer DC; Grootenhuis PDJ Annu. Rep. Med. Chem 1999, 34, 287–296. [Google Scholar]
- (69).Gorse D; Lahana R Curr. Opin. Chem. Biol 2000, 4, 287–294. [DOI] [PubMed] [Google Scholar]
- (70).Jahnke W J. Biomol. NMR 2007, 39, 87–90. [DOI] [PubMed] [Google Scholar]
- (71).Beavers MP; Chen X J. Mol. Graphics Modell 2002, 20, 463–468. [DOI] [PubMed] [Google Scholar]
- (72).Mayer M; Meyer B Angew. Chem., Int. Ed 1999, 38, 1784–1788. [DOI] [PubMed] [Google Scholar]
- (73).Krishnan VV Curr. Anal. Chem 2005, 1, 307–320. [Google Scholar]
- (74).Hajduk PJ; Olejniczak ET; Fesik SW J. Amer. Chem. Soc 1997, 119, 12257–12261. [Google Scholar]
- (75).Zaton AML; Villamor JP Chem. Biol. Interact 2000, 124, 1–11. [DOI] [PubMed] [Google Scholar]
- (76).Pervushin K; Riek R; Wider G; Wuthrich K Proc. Natl. Acad. Sci. U. S. A 1997, 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Bursulaya BD; Totrov M; Abagyan R; Brooks CL 3rd J Comput Aided Mol Des 2003, 17, 755–63. [DOI] [PubMed] [Google Scholar]
- (78).Hetenyi C; Van Der Spoel D Protein Science 2002, 11, 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]