Abstract
Biased activation of G-protein-coupled receptors (GPCRs) is shifting drug discovery efforts and appears promising for the development of safer drugs. The most effective analgesics to treat acute pain are agonists of the μ opioid receptor (μ-OR), a member of the GPCR superfamily. However, the analgesic use of opioid drugs, such as morphine, is hindered by adverse effects. Only a few μ-OR agonists have been reported to selectively activate the Gi over β-arrestin signaling pathway, resulting in lower gastrointestinal dysfunction and respiratory suppression. Here, we discuss the strategies that led to the development of biased μ-OR agonists, and potential areas for improvement, with an emphasis on structural aspects of the ligand-receptor recognition process.
Introduction
From a pharmacological point of view, drug discovery campaigns aim for compounds with high potency, low toxicity, and few adverse effects. In opioid-based analgesia, high potency has been successfully achieved. However, opioid analgesics with reduced adverse effects have been elusive. Nevertheless, recent progress towards this goal has been forthcoming.
Nearly 40% of pharmacologically relevant targets are members of the GPCR superfamily [1]. These receptors interact with a variety of molecules, from peptides, hormones, nucleic acids, and lipids to external stimuli, such as odorants, flavors, or light (photons) [2,3]. Activation of GPCRs by agonists triggers key processes, such as desensitization, resensitization, downregulation, and internalization via downstream signaling through effector proteins, mainly heterotrimeric G proteins and β-arrestins [4].
Opioid receptors (OR), members of the GPCRs superfamily, are crucial in pain management, drug addiction, and mood disorders [5,6], with a central role in hedonic homeostasis and well-being [7]. There are four OR subtypes: μ-opioid receptor (μ-OR), δ-opioid receptor (δ-OR), κ-opioid receptor (κ-OR) and the opioid receptor-like 1 [OLR-1 or nociceptin receptor (NOP)]. Activation of μ-OR inhibits severe acute pain, modulating mechanical, chemical, and supraspinally controlled thermal nociception [8]. Given that μ-OR mediates rewarding properties of nonopioid drugs of abuse (i.e., cannabinoids, alcohol and nicotine), these receptors represent a molecular target for reward processing, contributing to the initiation of addictive behaviors [9]. Adverse effects of μ-OR agonists include nausea, vomiting, gastrointestinal constipation, respiratory depression, rewarding effects, tolerance, and dependence [10,11]. In the clinic, current strategies for minimizing these complications include the slow titration of opioids, dose reduction to achieve an equilibrium between analgesia and tolerable adverse effects, prevention of nausea, opioid rotation, and changing the route of administration [12]. In turn, κ-OR modulates spinally mediated thermal nociception and visceral pain. In addition to their analgesic properties, activation of κ-OR results in sedative, aversive, and hallucinogenic effects, and induces dysphoria and anhedonia [13]. By contrast, δ-OR weakly modulates acute nociception and has antidepressant and anxiolytic effects, but although its activation also appears to lead to convulsions [7].
Therefore, the search for new OR-based medications is the primary focus of a growing number of research groups, making use of the vast amount of structural and pharmacological information now available. Strategies to identify such molecules span a range of methodologies, from exhaustive searches, such as high-throughput screening (HTS), to structurally and pharmacologically oriented design. Notably, HTS campaigns have provided structurally novel ligands with potency comparable to that of endogenous peptides (e.g., endorphins) [14]. Another source of potent OR ligands are combinatorial chemistry libraries. Recent efforts in this field have rendered highly active and selective compounds [15]. From a pharmacological point of view, the use of positive allosteric modulators (PAM) has resulted in an increased potency and/or efficacy of the ligands binding to the orthosteric site [16]. Although the development of allosteric modulators is still in the early drug discovery stage, it shows promise in the development of analgesics with fewer adverse effects. Alternatively, agonists that activate both μ-OR/δ-OR and NOPR/μ-OR have demonstrated an increase in antinociceptive efficacy [17,18]. By contrast, molecules with dual but opposite effects acting as μ-OR agonists and δ-OR antagonists preferentially prevent or reduce tolerance and physical dependence [19]. Following this mechanism, opioid combination drugs [i.e., morphine (agonist)/naltrexone (antagonist) and oxycodone (agonist)/naloxone (antagonist)] have been developed and marketed for the treatment of moderate to severe pain. However, attempts to develop a new analgesic that acts as a mixed μ-OR agonist/δ-OR antagonist have not been fruitful. Nevertheless, a mixed μ-OR agonist/δ-OR antagonist, Viberzi®, has been marketed, although its primary indication is for the treatment of irritable bowel syndrome with diarrhea, rather than of acute or moderate pain [20]. Lastly, attention has recently focused on molecules capable of binding to μ-OR and transducing particular biological signals, a phenomenon called ‘functional selectivity’ or ‘biased agonism’ [21,22].
Opioid receptors and functional selectivity
Structural features of GPCRs
The growing number of high-resolution crystal structures of GPCRs [23] significantly expands our understanding of receptor activation and related signaling pathways. For example, GPCRs have high homology within the transmembrane (TM) domains (approximately 60% of sequence identity), but less in the extracellular regions, a feature that contributes to the selectivity between different ligands [24], particularly at extracellular loop 3 (ECL3) and the extracellular ends of TM6 and TM7 [25]. Differences in intracellular regions are involved in downstream signaling for the activation of different pathways, where conformational changes in TM7, helix 8 (H8), and ECL2 might have a role in β-arrestin recruitment, while changes in TM3, TM5, and TM6, as well as in the intracellular loop 2 (ICL2), might be related to G protein-mediated signaling [26]. This downstream preference is the basis of functional selectivity.
Realizing biased agonism is therapeutically relevant
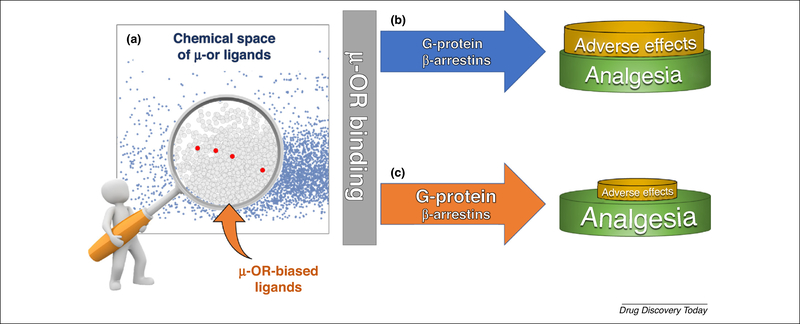
Functional selectivity, or biased agonism, explains the ability of a ligand to induce a specific conformation of the receptor. As a result, specific pathways are activated with differential efficacies. Such an effect can be induced either by the binding of the ligand to the orthosteric site of the receptor, or by the binding of allosteric modulators that induce biased signaling via the endogenous agonist [27,28]. This is achieved by fine-tuning the conformation of the receptor [27]. The key therapeutic feature of biased agonists is their ability to differentiate between specific signaling pathways, aiming to reduce the related adverse effects without compromising therapeutic value (Fig. 1a).
FIGURE 1.
Graphical representation of the functional selectivity concept at mu-opioid receptors (μ-ORs). (a) Chemical space of μ-OR ligands (blue dots) represented in a principal component analysis (PCA) plot according to relevant drug-likeness properties. Biased agonists are under-represented and fall in the same area (red dots). (b) Classical opiates, such as morphine, bind to the μ-OR and produce analgesia (mainly via the G-protein pathway) with the concomitant triggering of parallel signaling cascades (β-arrestin pathway) responsible for the adverse effects. (c) Biased agonists that are capable of preferentially activate the G-protein signaling pathway over the arrestin pathway produce analgesia with diminished adverse effects.
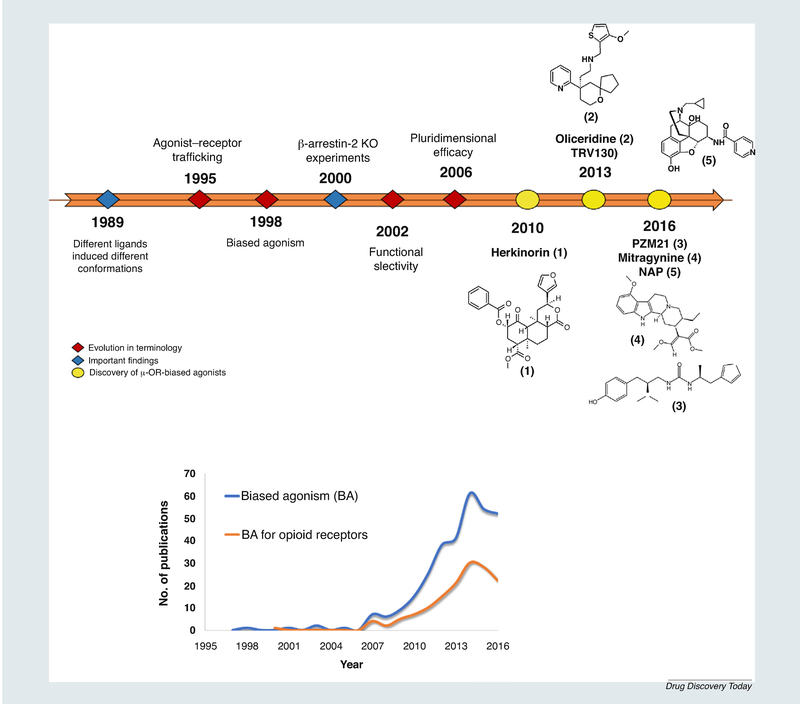
The discovery of molecules with the ability to selectively modulate GPCRs has been described extensively in the literature [27,29–34], supporting and validating the discovery of emerging functionally selective drugs. The increasing number of biased GPCR agonists with potential therapeutic impact is remarkable, as summarized in Table 1. This evidence builds on the generalizable idea of the selective modulation of GPCRs. Thus, as the number of new biased ligands increases, our ability to discover new therapeutic biased agents will also grow. Following that path, lessons learned from drug discovery efforts point towards the identification of features (structural, physicochemical, or based on the binding recognition process) responsible for the biased profiles to aid the design and discovery of new biased ligands. A general timeline for this field is depicted in Box 1, and shows research efforts documented in publications on this subject and in in vitro profiles [28]. Pharmacological data for μ-OR-biased ligands is rapidly increasing because of their potential applications in pain-related ailments. The number of compounds evaluated for binding affinity is in the millions, but only five molecules have been identified as μ-opioid biased ligands, as schematically depicted in Fig. 1b. The fact that only a few compounds are biased ligands (shown in red) does not imply that the rest are balanced ligands (i.e., nonbiased). In fact, it is possible that other μ-OR-biased compounds could be identified from the pool of compounds known to bind to μ-OR. Thus, investigations on the ligand-receptor recognition process will improve our understanding of the structural features behind the selective activation of either G proteins or β-arrestins [35]. As summarized in Table 1, positive therapeutic implications are achieved through the biased signaling of either the β-arrestins or the G protein pathway, depending on the receptor involved. In the particular case of μ-OR, the participation of β-arrestins in the mediation of adverse effects has been extensively supported by experiments in both cellular assays and β-arrestin-2-knockout (KO) mice. Upon coupling to GPCRs, β-arrestins hinder the G protein interaction with GPCRs at the ICL2 [36], resulting in receptor desensitization [37]. Consistently with a cellular study, β-arrestin-2 knockout mice are characterized by an elevated and prolonged morphine analgesia with impaired desensitization to elevated and prolonged morphine analgesia [38], as well as the attenuation of respiratory depression and acute constipation [39,40]. Therefore, selective modulation of G protein-signaling pathway by μ-OR ligands provides a means to produce analgesia with fewer adverse effects.
TABLE 1.
Currently known biased agonists for GPCRs and their therapeutic relevance
| GPCR | Biased ligand example | Biased signaling | Therapeutic implications | Refs |
|---|---|---|---|---|
| Angiotensin II type 1 receptor | SII, TRV120027, TRV120023 | β-arrestin-2 | Cardioprotective properties in vivo, reduction of blood pressure | [76–79] |
| Chemokine receptors (CCR1, CCR2, CCR4, CCR5, CCR7, CCR10, CXCR3) | CCL19 (CCR7) | β-arrestin-2 | Regulation of acute and chronic inflammatory responses in autoimmune diseases | [29,32,80] |
| Apelin receptor | CMF-019 | Gαi pathway | Increases cardiac contractility | [81] |
| Endothelin receptor | Macitentan | β-arrestin-2 | Blocks cellular responses associated with tumor progression | [82,83] |
| Adenosine A3 receptor | (N)-Methanocarba substituted derivatives | β-arrestin-2/Gi/o pathway | Protection in cardiac and lung ischemia and/or apoptosis | [84] |
| Adenosine A1 receptor | VCP746, capadenoson | Intracellular calcium mobilization | Retains cytoprotective signaling in absence of bradycardia | [85] |
| 5-HT1A receptor | F15599/F13714 | Cortical heteroceptors/raphe nuclei somatodendritic autoreceptors | Treatment of cognitive dysfunction and antidyskinetic pharmacotherapies | [86,87] |
| 5-HT2A receptor | Thioridazine, loxapine, methotrimeprazine Metergoline, pimetixene | Activate phospholipase A2 (PLA2) | Schizophrenia, hallucinations | [88,89] |
| Activation of phospholipase C (PLC) through Gαq | ||||
| 5-HT2C receptor | Benzofuran derivatives | Gq signalling | Weaker desensitization | [31] |
| Dopamine D1 receptor | Substituted benzapines | G-protein (agonist)/β-arrestin (antagonist) | Improved therapies for implicated diseases, such as Parkinson’s disease | [90,91] |
| Dopamine D2 receptor | MLS1547 | G-protein | Not described; could improve therapies for certain neuropsychiatric disorders | [92] |
| μ-opioid receptors | Oliceridine, herkinorin, PZM21, mitragynine | G-protein | Analgesia with diminished adverse effects | [30,46,55] |
| κ-Opioid receptors | Triazole 1.1, 6′-GNTI, collybolide, noribogaine | G-protein | Diminished dysphoric adverse effects | [93–96] |
| Beta adrenergic (B1AR) receptor | Carvedilol | β-arrestin-2 | Cardioprotection | [97] |
| Histamine H2R receptor | Ranitidine/tiotidine | β-arrestin-2 | Desensitization | [98,99] |
| GLP-1 receptor | P5 (exendin-4 analog) | G-protein | Improves glucose tolerance and chronic hyperglycemia control | [100] |
| Cannabinoid 1(CB1) receptor | Pyrrolidinyl analogs | β-arrestin-2 | Avoid undesirable psychotropic and psychiatric adverse effects | [101] |
BOX 1. Timeline of the discovery of biased ligands.
Probably the earliest hint of functional selectivity was proposed by Roth and Chuang In 1980 [71] to describe the phenomenon whereby one or more distinct signaling transductions could be activated by a single GPCR. Simultaneously, Kenakin and Morgan suggested that different ligands could induce different conformations of the same receptor [72]. For almost a decade from the mid-1990s onwards, there was evolution of the term. For example, based on the observations that a peptide in chemokine receptors induced a biased activity, Jarpe introduced the term ‘biased agonist’ [73]. It was not until the beginning of the 21 st century that the group of Mottola suggested the term ‘functional selectivity’ for dopamine receptor agonists [74]. After these findings, several research groups started to describe the apparent biased behavior of ligands in different GPCRs [75]. Even when assumptions about functional selectivity of opioid receptors were already known, it was not until work by Bohn on the pharmacology of morphine in β-arrestin2-KO versus wild-type mice that interest in their therapeutic potential as analgesics with safer profiles was triggered [38–40].
FIGURE I.
Timeline of the discovery of biased ligands.
Endogenous peptides represent relevant μ-OR ligands with a key role in functional selectivity of several GPCRs as well as opioid receptors [41]. Notably, μ-OR endogenous peptides (i.e., enkephalins, endorphins, dynorphins, and neoendorphins) and putative endogenous peptides (endomorphines) have shown promising analgesic effects compared with opiates [42], despite their high liability for proteolysis. Efforts in the development of endomorphin analogs have been conducted [43]. In addition to endogenous peptides, only a few compounds have been described as biased agonists of μ-OR. The first molecule reported with such activity was herkinorin, a selective μ-OR agonist derived from the natural product salvinorin A. This morphine-unrelated compound lacks the prototypical nitrogen atom found in most opioid ligands [44]. Whereas herkinorin activates G protein coupling and ERK1/2 in a naloxone reversible manner, the translocation of β-arrestin-2-GFP to the receptor is poor, even when GRK2 is overexpressed. By contrast, under GRK2 overexpression, morphine induces β-arrestin-2 recruitment. Thus, herkinorin is able to promote antinociceptive activity with reduced adverse effects, mainly as a result of its G-protein biased signaling preference [45–47]. Nevertheless, its poor efficacy following systemic administration precluded its use as a drug candidate; therefore, subsequent studies for improving its stability and solubility are required [46].
Discovery of μ-OR-biased agonists
Oliceridine (TRV130)
With information on the therapeutic applications of functional selectivity reported over the past decade, it is not surprising that the discovery of biased ligands had gained the attention of pharmaceutical companies. For example, screening an in-house chemical library, Trevena Inc., whose business model was the identification of biased agonists, reported the identification of oliceridine, a potent analgesic compound with efficacy comparable to that of morphine. Oliceridine (TRV130) is structurally unrelated to morphine or other previously described μ-OR agonists. TRV130 is a strong agonist for G protein signaling, with potency and efficacy higher than that of morphine, but weak β-arrestin-2 recruitment. It also exhibited reduced μ-OR internalization and significantly less receptor phosphorylation at serine 375 compared with morphine in a cell-based assay [34]. Compared with morphine, TRV130 produced equivalent analgesia but less respiratory depression and reduction of tolerance development in rodent models [48]. This is consistent with the evidence that morphine produces enhanced analgesia but reduced tolerance in β-arrestin-2 KO mice compared with wild-type mice, suggesting that β-arrestin-mediated signaling contributes to tolerance development [34,48]. However, the effects of TRV130 on gastrointestinal function were more complicated. One research group found that TRV130 caused less gastrointestinal dysfunction than morphine at equivalent analgesic doses [34]; another research group claimed that mice acutely or repeatedly treated with TRV130 at a dose of 10 mg/kg exhibited robust gastrointestinal inhibition in fecal accumulation assays [48]. This might correspond to the different TRV130 regimens and the different recorded time periods (4 h versus 1 h) reported in the two investigations. It could also indicate that, owing to its weak β-arrestin recruitment, TRV130 lacks sufficient bias to produce a reliable reduction in gastrointestinal inhibition [47]. Currently, TRV130 (OLINVO™) is undergoing Phase III clinical trials as a next-generation intravenous opioid analgesic for the management of moderate-to-severe acute pain. Under a 0.1 mg TRV130 regimen, the compound resulted in significantly lower rates of respiratory depression safety events but did not achieve the analgesic efficacy of morphine. Under 0.35 mg and 0.5 mg regimens, TRV130 produced rapid analgesia with efficacy superior to that of morphine in the modulation of moderate-to-severe pain; however, under a 0.5 mg regimen, the rates of constipation and respiratory events were not statistically different from those of morphine, which could be consistent with the conflicting rodent data and reflect its weak activation toward β-arrestins (www.trevena.com/OLINVO-development.php).
Thus, deciphering the uniqueness of oliceridine to avoid β-arrestin recruitment has become an urgent need. In an attempt to address this issue, Filizola and collaborators explored the allosteric communication between the binding pocket at the orthosteric site, and the intracellular region of the receptor through molecular dynamics simulations [49]. The analyses suggested that the recognition of the biased ligand starts at the vestibule region (an area between the extracellular part of the receptor and the orthosteric binding pocket). This metastable state is followed by interactions in the orthosteric binding site. Therefore, the kinetics of the ligand depends on the interactions of these two stages. Additionally, the authors proposed that, when oliceridine is bound to μ-OR, communication occurs between the residues in the oliceridine-binding pocket and the intracellular end of TM3, but not with residues at TM6. By contrast, a morphine-μ-OR model showed significant coupling between the binding pocket and both ends of TM3 and TM6. Key residues found to strongly contribute to the transmission of information from the orthosteric binding pocket to the intracellular side of the receptor include W3187.35, R1653.50, Y1493.34, F3478.54, and Y911.55.
PZM21
In an attempt to expand the incipiently explored chemical space of μ-OR-biased ligands, Manglik et al. performed a virtual screening of over 3 million molecules. This resulted in a μ-OR selective potent Gi activator with minimal β-arrestin-2 recruitment, PZM21, structurally unrelated to either oliceridine or morphine. In fact, at a maximal concentration of PZM21, its β-arrestin-2 recruitment was undetectable, making it difficult for a formal calculation of bias. Its activity in a β-arrestin-2 assay was minimal compared with DAMGO (H-Tyr-D-Ala-Gly-N-MePhe-Gly-OH) and morphine even in the presence of overexpressed GRK2. PZM21 also had a minimal level of μ-OR internalization compared with DAMGO and morphine, consistent with its inactive recruitment toward β-arrestin-2. Furthermore, the maximum effect of morphine is reached with 10 mg/kg, while PZM21 needed 40 mg/kg to reach the same effect, yielding an equi-analgesic response (87% versus 92%, respectively). According to Manglik et al., the analgesic profile of PZM21 appears to be unique, in the sense that, unlike morphine, it decreases affective pain perception with minimal effect on reflexive pain. Therefore, a minimal level of μ-OR internalization compared with that of DAMGO and morphine was observed. The molecular models presented bu the authors are in agreement with key interactions known to promote high affinity and selectivity, via D1473.32 and H2976.52, respectively [50].
Mytragynine
Natural products are an excellent source of bioactive molecules. The alkaloid mitragynine and its analog 7-hydroxymitragynine emerge as new μ-OR-biased ligands. Both compounds are biosynthesized by Mitragyna speciosa, a medicinal plant from the southeast region of Asia traditionally used both as stimulant to reduce fatigue, and as antinociceptive drug because of its opiumlike effect [51–53]. Despite their well-characterized μ-OR-mediated analgesic activity, it was not until recently that pharmacological investigations undertaken by Kruegel et al. suggested the G-protein-biased agonism of both alkaloids. Mitragynine alkaloids failed to recruit β-arrestin-2 at concentrations as high as 10 M, according to a bioluminescence resonance energy transference (BRET) assay [54]. Furthermore, the structurally related pseudoindoxyl mitragynine, a byproduct of the fungal metabolism of 7-hydroxymitragynine, also preserves the potent antinociceptive activity, although the diminished observed adverse effects might involve its μ-OR agonist/δ-OR antagonist behavior [55].
A μ-OR weak partial agonist/antagonist
Unlike the biased MOR agonists, NAP was characterized as a biased MOR antagonist rather than a weak partial agonist by Zhang et al. [26]. NAP has a morphine-like structure and exhibited weak partial agonist potency (23% of DMAGO) in a [35S]GTPγS binding assay with no apparent recruitment of β-arrestin-2 [56]. However, NAP showed no apparent activity in a calcium flux assay using hMOR-CHO cells transfected with chimeric Gαqi5, suggesting that it acted as an antagonist in this instance. Furthermore, NAP potently blocked μ-OR full agonist-induced β-arrestin-2 recruitment and translocation, denoting its ability to selectively antagonize the β-arrestin-2 pathway. This in vitro antagonist activity correlates with the results from in vivo studies, in which NAP failed to produce antinociception at doses up to 100 mg/kg but antagonized morphine analgesia in mice. Meanwhile, it also failed to induce muscle contractions of isolated distal and proximal colons from mice but significantly reversed the reduction of colon motility induced by morphine. Despite the apparent lack of analgesic effect, NAP seems to have the therapeutic potential to restore morphine-impaired intestinal motility, partially because of its latent biased competitive antagonism [35]. Nevertheless, further studies to confirm the supposed biased activity of NAP are required.
Binding models of biased ligands
The wealth of pharmacological information on OR has shown that binding of opioid ligands to the orthosteric site of μ-OR with high affinity drives the receptor equilibrium from an inactive conformation to an active form. Depending on the intrinsic activity of the ligand, it can result in a full response (full agonist) or a lesser response, even if there is a full occupancy of the binding site in the receptor (partial agonist). It has been suggested that partial agonists have a reduced ability to differentiate between active and inactive conformations, resulting in a lesser equilibrium towards the active form, or even more, to induce a different (and alternative) active conformation of the receptor, producing less activation of downstream signaling effectors. Similarly, functional selectivity is based on the paradigm of several active conformations of a receptor. However, this phenomenon is commonly simplified to active and inactive conformations of the receptor [16]. Thus, the analysis of the interactions at the molecular level of biased ligands with the crystal structures of both active and inactive conformations of μ-OR is informative, given that both structures represent snapshots of stabilized conformations.
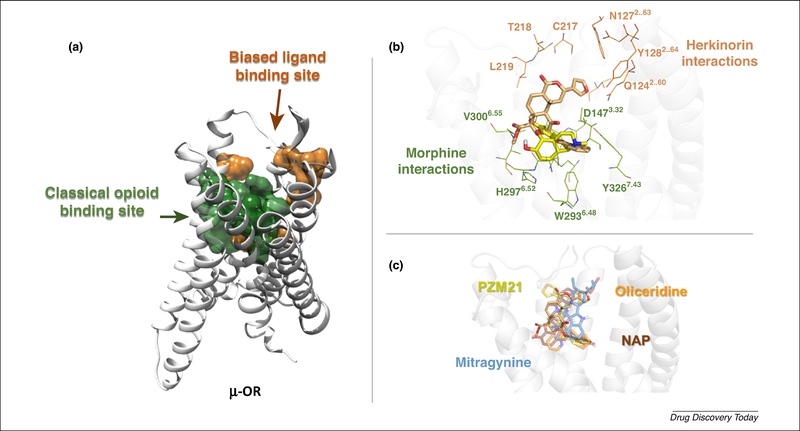
An estimate of binding models of oliceridine, PZM21, mitragynine, and NAP suggests common interactions with μ-OR among biased and nonbiased opioid ligands, as depicted in Fig. 2. Interactions observed in those models are located near the extracellular side of the receptor, involving TM2 (Q1242.60, N1272.63, and Y1282.64), ECL1 (W133), and ECL2 (D216, C217, T218, L219, and F221). Additionally, most of the interactions shared by μ-OR agonists, antagonists, and partial agonists are also shared by biased ligands, including the interaction with D1473.32, in agreement with previous modeling studies and site-directed mutagenesis experiments, which determined the crucial role of this residue in opioid ligand recognition [57]. These findings agree with the interacting regions close to TM2, TM3, and TM7 suggested by Schneider [49], involving two different ligand-binding stages. It has been proposed that the first step, or site of interaction, for the biased ligands is the so-called ‘vestibule region’, located between the orthosteric binding pocket and the extracellular side, before the ligand penetrates the orthosteric pocket. As mentioned above, this region could have a key role in the modulation of the receptor, with concomitant changes in the signaling profile. In opioid receptors, pharmacophoric features have been rationalized with the ‘message-address’ concept. In this context, the ‘address’ is the part of the ligand responsible for the receptor recognition and, therefore, its affinity;and the ‘message’ is part of the ligand responsible for the activity (efficacy) [58]. Similarly, an extension of the message-address concept for biased ligands could be proposed. For example, despite the structural diversity among biased ligands, the address core is represented mainly by those interactions near the orthosteric binding site, corresponding to the putative interactions of opiates. By contrast, interactions related to the side chains of the biased ligands with those residues mentioned above could represent the message transferred to the receptor: the furan ring for herkinorin, indole core for mitragynine, pyridine for NAP, or thiophene for PZM21 and oliceridine. Furthermore, the binding recognition process of herkinorin on μ-OR has recently been analyzed through molecular dynamics simulations and free energy calculations [59]. It was found that herkinorin reaches a previously noted allosteric site through interactions of the C2-benzyloxy group with N1503.35, which has a key role in preventing β-arrestin-2 signaling. Importantly, the mutation at N131A or N131V in δ-OR (homologous to N1503.35 in μ-OR) increases the activation levels for the β-arrestin-2 pathway compared with the wild-type protein [60]. Therefore, it is possible to hypothesize that molecules reaching this allosteric site by other structural means should also maintain this effect, as is the case with NAP, where the cyclopropyl group is oriented similar to the benzyloxy group of herkinorin at the allosteric site.
FIGURE 2.
Binding model of biased ligands and the classical opiates at mu-opioid receptors (μ-ORs). (a) Surface representation of the binding site for classical opiates (green) and biased ligands (orange). (b) Detail of binding model for biased ligands represented by herkinorin (light-orange sticks). Morphine is shown for reference (yellow sticks). (c) Predicted binding modes of biased ligands.
Several studies have explored the binding modes of different ligands at the μ, δ-, and κ-OR [5,61–63]. For instance, based on docking models, Noori et al. proposed two main binding regions in opioid receptors that might correlate with agonists and antagonists in terms of depth within the receptor, following a relationship between the affinity of the ligand and the proximity to the extracellular side [64]. Similarly, it could be hypothesized that the binding recognition profile of biased ligands might be characteristic and allow one to discriminate biased from nonbiased ligands. Further modeling and experimental studies to identify potential biased ligands are warranted.
Scaffolds and databases
From a structural point of view, no pharmacophore appears to be shared by currently known μ-OR-biased ligands. In addition, comparison of the chemical space (based on drug-like properties) of μ-OR ligands from relevant databases (i.e., GDD, GLASS, Pub-Chem, IUPHAR/BPS Guide to Pharmacology, and ChEMBL), illustrates that most of the active compounds share their densest area with the biased compounds (Fig. S1 in the Supplemental information online). Molecules were prepared and analyzed using the ChemAxon suit of programs. Clearly, the uniqueness of biased ligands was not captured by druglike properties used in this chemical space representation. This simple but informative comparison underscores the importance of considering features beyond the chemical description of the molecules, such as those involving ligand-receptor interactions.
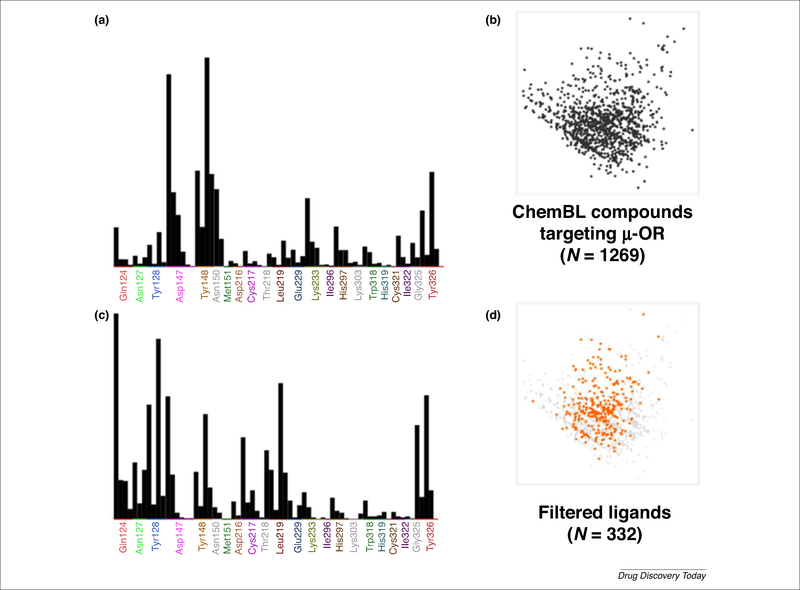
Figure 3 illustrates an analysis that is performed by the generation of protein-ligand interaction fingerprints (PLIF). These fingerprints are designed to capture the interactions between a ligand and a protein, providing a means for the rapid manipulation, storage, and analysis of the data in the binary string format [65]. The frequency count of the interactions, obtained from the PLIF analysis, was modeled for both biased and some opiate μ-OR ligands (Fig. S2 in the Supplemental information online). The profile allows the identification of interactions that were only observed for biased ligands, compared with agonists, as described above. The information collected from the PLIF analysis could be used as a first filter for the discovery of biased ligands, for example in virtual screening campaigns. Given that the basic requirement for a ligand to behave as a biased agonist is to have affinity for the receptor, a primary source of compounds are databases of known opioid ligands. In this context, affinity data of compounds towards μ-OR are available in several databases. A description of representative databases is shown in Table 2. While some of these databases are specific for GPCRs, others are more general. As an example of how the information described herein could be applied to the discovery of μ-OR-biased ligand candidates, we made a preliminary analysis of the μ-OR ligands in ChEMBL database. The search for compounds was set to those ligands targeting the human μ-OR with information on affinity data (Ki). Details are provided in supplementary information online. Briefly, the database was curated, energy minimized, and docked into the active and inactive state of μ-OR. The interactions were assigned as present (1) or absent (0). This binary digit (or bit) was characteristic for each protein-ligand complex in the data set and constituted its PLIF.
FIGURE 3.
Protein-ligand interaction fingerprints (PLIF) profile of mu-opioid receptor (μ-OR) ligands. (a) Population histogram and (b) chemical space of μ-OR ligands in the ChEMBL database. (c) Population histogram and (d) chemical space (orange dots) of filtered ligands from the ChEMBL database based on the proposed interactions corresponding to the biased profile.
TABLE 2.
GPCR databases with relevance in drug discovery
| Database (source) | GPCR specialized | Description | μ-OR ligands included |
|---|---|---|---|
| GLASS GPCR (http://zhanglab.ccmb.med.umich.edu/GLASS/) | Yes | GPCR-Ligand Association (GLASS) is a manually curated repository for experimentally validated GPCR-ligand interactions. Retrieves information from literature and public databases. Developed and maintained by the Zhang Lab at the University of Michigan, USA | ~5800 active molecules, different in vitro assays |
| GLIDA: GPCR-Ligand (http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/) | Yes | Database with information about both GPCRs and their known ligands. Enterable either by GPCR search or ligand search. Data are not available to batch-download. Maintained by the PharmacoInformatics Laboratory, Kyoto University | 235 agonists, 36 antagonists |
| GDD: GPCR Decoy (http://cavasotto-lab.net/Databases/GDD/) | Yes | A database created to help with GPCR docking simulations, along with the GPCR Ligand Library (GLL). For each ligand in GLL, there are 39 decoys from ZINC ensuring physical similarity of six properties, but structural dissimilarity. Provided by the Claudio N. Cavasotto Lab of the Instituto de Biomedicina de Buenos Aires- Max Planck Society Partner (IBioBA-MPSP) | Agonists: 140 molecules and 5460 decoys; antagonists: 27 compounds and 1053 decoys |
| PubChem (https://pubchem.ncbi.nlm.nih.gov/) | No | Contains biological information of small molecules (>2 000 000 compounds), retrieving data from more than 10 000 targets; it is organized as three linked databases (Substance, Compound and BioAssay) within the Entrez information retrieval system of the NCBI | ~400 active compounds, different in vitro assays |
| ChemBL (www.ebi.ac.uk/chembl/) | No | Bioactive drug-like small molecules (>2 000 000); includes 2D structures, calculated properties, and abstracted bioactivities (binding constants, pharmacology, and ADMET data). It is an easy-to-access database; searches can be defined by target (>11 000 targets) and browsed by activity type (EC50, Ki IC50, etc.). | ~6000 compounds assessed in human, rat, guinea pig, and mouse |
| IUPHAR/BPS Guide to pharmacology (www.guidetopharmacology.org/) | No | Provides quantitative information about drug targets, approved drugs, and experimental molecules of those targets. Depicts detailed data on targets. Created from a collaboration between The British Pharmacological Society (BPS) and the International Union of Basic and Clinical Pharmacology (IUPHAR) | 97 agonists, antagonists and allosteric modulators |
Frequency counts of the interactions obtained from the docking of 1269 compounds from ChEMBL, obtained from the PLIF analysis, are shown in Fig. 3a and the corresponding chemical space representation of this set is shown in Fig. 3b. From this set, 332 compounds satisfied the profile of residue interactions obtained for biased ligands (Fig. 3c, d). This analysis is preliminary but exemplifies the feasibility of identifying a characteristic profile of interactions for biased ligands. A consensus interacting profile obtained from a comprehensive study will enable the suggestion of putatively biased-driven μ-OR interactions that can then be used to search for new μ-OR-biased ligands.
Concluding remarks
Knowledge accumulated during the past few decades towards the discovery of the ideal pain-reliever has provided the first molecules with diminished adverse effects and enhanced analgesia, namely biased agonists. Selective modulation of specific signaling pathways is a complex and multicomponent process. Thus, it should be explored from different perspectives. In this review, we focused on structural aspects based on available yet limited information of biased ligands. Protein-ligand interacting patterns appear to provide valuable information that can be used to characterize these ligands. However, additional aspects should be taken into account. On the one hand, biased agonism is affected by experimental variables at different biological levels (tissue, cellular, or enzymatic), generally referred as system bias. These variables impact the biased effect measurement [28,66]. On the other hand, the sensitivity of the detection methods also affects the observed response, which is regarded as observational bias. Both types of deviation are called ‘apparent bias’. To suppress the contribution of the apparent bias, the data should be quantified using a method that excludes both system and observational bias [67]. A further step forward in the field of biased agonism was the quantification of the effect via a bias index [68,69]. Among the models to quantify biased agonism, two convenient methodologies are: the operational model of agonism (systematic independent quantification of agonist activity via the relative transduction ratio coefficient, or blig factor); and the intrinsic relative activity (RAi), which can be calculated from estimated parameters (EC50 and Emax). Yet, comparing the maximal effects (Emax) and potencies (EC50) of ligands for different signaling pathways is common [70]. Moreover, the strong dependence on apparent bias makes it hard to arrive at a validated and standardized methodology to evaluate the bias index.
However, detailed information about biased signaling at the molecular level is still limited and represents an open question. As described throughout this paper, an overall analysis of the information generated from the binding recognition models of biased ligands with the receptor could help in the design of new biased agonists. Hence, comprehensive molecular modeling and chemoinformatic studies are warranted. In this regard, refined docking studies and molecular dynamics simulations will increase our understanding of this field and help fill the gaps in the design of molecules with this promising pharmacological and therapeutic profile.
All in all, biased agonism profiling should be taken into account when working with GPCRs, especially for those receptors with known functional selectivity. Computational models, pharmacological information, and structural data are paving the way to reach this goal.
Supplementary Material
Acknowledgments
K.-M. thanks the Instituto de Química, UNAM for financial support. A.-M. acknowledges support from Dirección General de Asuntos del Personal Academico (DGAPA) for a postdoctoral fellowship at Universidad Nacional Autónoma de Mexico (UNAM) in 2016-2017. The authors thank the reviewers for insightful comments and ChemAxon for providing an academic license.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.drudis.2017.07.002.
References
- 1.Ho TT et al. (2017) Method for rapid optimization of recombinant GPCR protein expression and stability using virus-like particles. Protein Expr. Purif 133, 41–49 [DOI] [PubMed] [Google Scholar]
- 2.Perez DM and Karnik SS (2005) Multiple signaling states of G-protein-coupled receptors. Pharmacol. Rev 57, 147–161 [DOI] [PubMed] [Google Scholar]
- 3.Whalen EJ et al. (2011) Therapeutic potential of (β-arrestin- and G protein-biased agonists. Trends Mol. Med 17, 126–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pradhan AA et al. (2015) In vivo techniques to investigate the internalization profile of opioid receptors. Methods Mol. Biol 1230, 87–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang Y and Filizola M (2015) Opioid receptors: structural and mechanistic insights into pharmacology and signaling. Eur. J. Pharmacol 763, 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spampinato SM (2015) Overview of genetic analysis of human opioid receptors. Methods Mol. Biol 1230, 3–12 [DOI] [PubMed] [Google Scholar]
- 7.Pradhan AA et al. (2011) The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol. Sci 32, 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yongye AB and Martinez-Mayorga K (2012) Molecular aspects of opioid receptors and opioid receptor painkillers. In Pain Management—Current Issues and Opinions (Racz GB and Noe CE, eds), pp. 43–62, InTech [Google Scholar]
- 9.Kieffer BL and Evans CJ (2009) Opioid receptors: from binding sites to visible molecules in vivo. Neuropharmacology 56, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S-R et al. (2017) Discovery, structure-activity relationship studies, and antinociceptive effects of N-(1,2,3,4-tetrahydro-1-isoquinolinylmethyl)benzamides as novel opioid receptor agonists. Eur. J. Med. Chem 126, 202–217 [DOI] [PubMed] [Google Scholar]
- 11.Deekonda S et al. (2015) Design and synthesis of novel bivalent ligands (MOR and DOR) by conjugation of enkephalin analogues with 4-anilidopiperidine derivatives. Bioorg. Med. Chem. Lett 25, 4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maree TS and Wei HG (2012) Opioid analgesics. In Pain Management—Current Issues and Opinions (Racz GB and Noe CE, eds), pp. 288–306, InTech [Google Scholar]
- 13.Bruchas MR and Roth BL (2016) New technologies for elucidating opioid receptor function. Trends Pharmacol. Sci 37, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remesic M et al. (2016) Cyclic opioid peptides. Curr. Med. Chem 23, 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y et al. (2012) Fluorescent Mu selective opioid ligands from a mixture based cyclic peptide library. ACS Comb. Sci 14, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burford NT et al. (2015) Positive allosteric modulators of the μ-opioid receptor: a novel approach for future pain medications. Br. J. Pharmacol 172, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negus SS et al. (2009) Role of delta opioid efficacy as a determinant of mu/delta opioid interactions in rhesus monkeys. Eur. J. Pharmacol 602, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toll L et al. (2009) Comparison of the antinociceptive and antirewarding profiles of novel bifunctional nociceptin receptor/μ-opioid receptor ligands: implications for therapeutic Applications. J. Pharmacol. Exp. Ther 331, 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harland AA et al. (2015) Further optimization and evaluation of bioavailable, mixed-efficacy μ-opioid receptor (MOR) agonists/μ-opioid receptor (DOR) antagonists: balancing MOR and DOR affinities. J. Med. Chem 58, 8952–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating GM (2017) Eluxadoline: a review in diarrhoea-predominant irritable bowel syndrome. Drugs 77, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 21.Kelly E (2013) Efficacy and ligand bias at the-opioid receptor. Br. J. Pharmacol 169, 1430–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raehal KM et al. (2011) Functional selectivity at the mu-opioid receptor: implications for understanding opioid analgesia. Pharmacol. Rev 63, 1001–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katritch V et al. (2013) Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol 53, 531–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filizola M and Devi LA (2013) Grand opening of structure-guided design for novel opioids. Trends Pharmacol. Sci 34, 6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provasi D (2015) Computational structural biology of opioid receptors. Methods Mol. Biol 1230, 13–38 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y et al. (2015) 17-Cyclopropylmethyl-3, 14p-dihydroxy-4, 5a-epoxy-6β-(4′-pyridylcarboxamido)morphinan (NAP) modulating the mu opioid receptor in a biased fashion. ACS Chem. Neurosci 7, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hébert TE (2016) Biasing the odds: approaches to capturing, understanding and exploiting functional selectivity in GPCRs. Methods 92, 1–4 [DOI] [PubMed] [Google Scholar]
- 28.Kenakin T and Christopoulos A (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov 12, 205–216 [DOI] [PubMed] [Google Scholar]
- 29.Anderson CA et al. (2015) Biased agonism at chemokine receptors: obstacles or opportunities for drug discovery? J. Leukoc. Biol 99, 1–9 [DOI] [PubMed] [Google Scholar]
- 30.Chen X-T et al. (2013) Structure-activity relationships and discovery of a G protein biased m opioid receptor ligand, [(3-methoxythiophen-2-yl)methyl]({2-[(9R)-9-(pyridin-2-yl)-6-oxaspiro-[4.5]decan-9-yl]ethyl})amine (TRV130), for the treatment of acute severe pain. J. Med. Chem 56, 8019–8031 [DOI] [PubMed] [Google Scholar]
- 31.Cheng J et al. (2016) Design and discovery of functionally selective serotonin 2C (5-HT2C) receptor agonists. J. Med. Chem 59, 9866–9880 [DOI] [PubMed] [Google Scholar]
- 32.Corbisier J et al. (2015) Biased signaling at chemokine receptors. J. Biol. Chem 290, 9542–9554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa-Neto CM et al. (2016) A pluridimensional view of biased agonism. Mol. Pharmacol 90, 587–595 [DOI] [PubMed] [Google Scholar]
- 34.DeWire SM et al. (2013) A G protein-biased ligand at the μ-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J. Pharmacol. Exp. Ther 344, 708–717 [DOI] [PubMed] [Google Scholar]
- 35.Bologna Z et al. (2017) Biased G protein-coupled receptor signaling: new player in modulating physiology and pathology. Biomol. Ther 25, 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marion S et al. (2006) A β-arrestin binding determinant common to the second intracellular loops of rhodopsin family G protein-coupled receptors. J. Biol. Chem 281, 2932–2938 [DOI] [PubMed] [Google Scholar]
- 37.Ferguson SSG et al. (1996) Role of beta-arrestin in mediating agonist-promoted g protein-coupled receptor internalization. Science 5247, 363–366 [DOI] [PubMed] [Google Scholar]
- 38.Bohn LM (1999) Enhanced morphine analgesia in mice lacking β-arrestin 2. Science 5449, 2495–2498 [DOI] [PubMed] [Google Scholar]
- 39.Caron MG et al. (2000) -Opioid receptor desensitization by β-arrestin-2 determines morphine tolerance but not dependence. Nature 408, 720–723 [DOI] [PubMed] [Google Scholar]
- 40.Raehal KM et al. (2005) Morphine side effects in beta-arrestin-2 knockout mice. J. Pharmacol. Exp. Ther 314, 1195–1201 [DOI] [PubMed] [Google Scholar]
- 41.Thompson GL et al. (2015) Biased agonism of endogenous opioid peptides at the mu-opioid receptor. Mol. Pharmacol 88, 335–346 [DOI] [PubMed] [Google Scholar]
- 42.Rivero G et al. (2012) Endomorphin-2: a biased agonist at the μ-opioid receptor. Mol. Pharmacol 82, 178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zadina JE et al. (2016) Endomorphin analog analgesics with reduced abuse liability, respiratory depression, motor impairment, tolerance, and glial activation relative to morphine. Neuropharmacology 105, 215–227 [DOI] [PubMed] [Google Scholar]
- 44.Harding WW et al. (2005) Neoclerodane diterpenes as a novel scaffold for m opioid receptor ligands. J. Med. Chem 48, 4765–4771 [DOI] [PubMed] [Google Scholar]
- 45.Lamb K et al. (2012) Antinociceptive effects of herkinorin, a MOP receptor agonist derived from salvinorin A in the formalin test in rats: new concepts in mu opioid receptor pharmacology: from a symposium on new concepts in mu-opioid pharmacology. Drug Alcohol Depend. 121, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groer CE et al. (2007) An opioid agonist that does not induce -opioid receptor-arrestin interactions or receptor internalization. Mol. Pharmacol 71, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowan MP et al. (2014) Activation of mu opioid receptors sensitizes Transient Receptor Potential Vanilloid Type 1 (TRPV1) via β-arrestin-2-mediated cross-talk. PLoS One 9, e93688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altarifi AA et al. (2017) Effects of acute and repeated treatment with the biased mu opioid receptor agonist TRV130 (oliceridine) on measures of antinociception, gastrointestinal function, and abuse liability in rodents. J. Psychopharmacol 130, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider S et al. (2016) How oliceridine (trv-130) binds and stabilizes a β-opioid receptor conformational state that selectively triggers G protein signaling pathways. Biochemistry 55, 6456–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manglik A et al. (2016) Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto LT et al. (1999) Opioid receptor agonistic characteristics of mitragynine pseudoindoxyl in comparison with mitragynine derived from Thai medicinal plant Mitragyna speciosa. Gen. Pharmacol 33, 73–81 [DOI] [PubMed] [Google Scholar]
- 52.Shamima AR et al. (2012) Antinociceptive action of isolated mitragynine from mitragyna speciosa through activation of opioid receptor system. Int. J. Mol. Sci 13, 11427–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prozialeck WC et al. (2012) Pharmacology of Kratom: an emerging botanical agent with stimulant, analgesic and opioid-like effects. J. Am. Osteopath. Assoc 112, 792–799 [PubMed] [Google Scholar]
- 54.Kruegel AC et al. (2016) Synthetic and receptor signaling explorations of the mitragyna alkaloids: mitragynine as an atypical molecular framework for opioid receptor modulators. J. Am. Chem. Soc 138, 6754–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Váradi A et al. (2016) Mitragynine/Corynantheidine pseudoindoxyls as opioid analgesics with mu agonism and delta antagonism, which do not recruit β-arrestin-2. J. Med. Chem 59, 8381–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G et al. (2009) Design, synthesis, and biological evaluation of 6α- and 6β-N-heterocyclic substituted naltrexamine derivatives as mu opioid receptor selective antagonists. J. Med. Chem 52, 1416–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui X et al. (2013) Ligand interaction, binding site and G protein activation of the mu opioid receptor. Eur. J. Pharmacol 702, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dietis N et al. (2009) Simultaneous targeting of multiple opioid receptors: a strategy to improve side-effect profile. Br. J. Anaesth 103, 38–49 [DOI] [PubMed] [Google Scholar]
- 59.Marmolejo-Valencia AF and Martinez-Mayorga K (2017) Allosteric modulation model of the mu opioid receptor by herkinorin, a potent not alkaloidal agonist. J. Comput. Aided Mol. Des 31, 467–482 [DOI] [PubMed] [Google Scholar]
- 60.Katritch V et al. (2014) Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci 39, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui X et al. (2013) Ligand interaction, binding site and G protein activation of the mu opioid receptor. Eur. J. Pharmacol 702, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosberg HI et al. (2013) Opioid peptidomimetics: leads for the design of bioavailable mixed efficacyM opioid receptor (MOR) agonist/δ opioid receptor (DOR) antagonist ligands. J. Med. Chem 56, 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eguchi M (2004) Recent advances in selective opioid receptor agonists and antagonists. Med. Res. Rev 24, 182–212 [DOI] [PubMed] [Google Scholar]
- 64.Noori HR et al. (2014) A structural feature of the non-peptide ligand interactions with mice mu-opioid receptors. Curr. Comput. Aided Drug Des 10, 354–360 [DOI] [PubMed] [Google Scholar]
- 65.Marmolejo AF et al. (2015) Interaction fingerprints and their applications to identify hot spots. Methods Mol. Biol 1335, 313–324 [DOI] [PubMed] [Google Scholar]
- 66.Gundry J et al. (2017) A practical guide to approaching biased agonism at G protein coupled receptors. Front. Neurosci 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson GL et al. (2016) Systematic analysis of factors influencing observations of biased agonism at the mu-opioid receptor. Biochem. Pharmacol 113, 70–87 [DOI] [PubMed] [Google Scholar]
- 68.Winpenny D et al. (2016) Biased ligand quantification in drug discovery: from theory to high throughput screening to identify new biased m opioid receptor agonists. Br. J. Pharmacol 173, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stott LA et al. (2016) Unravelling intrinsic efficacy and ligand bias at G protein coupled receptors: a practical guide to assessing functional data. Biochem. Pharmacol 101, 1–12 [DOI] [PubMed] [Google Scholar]
- 70.Rajagopal S et al. (2011) Quantifying ligand bias at seven-transmembrane receptors. Mol. Pharmacol 80, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roth BL and Chuang DM (1987) Multiple mechanisms of serotonergic signal transduction. Life Sci. 41, 1051–1064 [DOI] [PubMed] [Google Scholar]
- 72.Kenakin TP and Morgan PH (1989) Theoretical effects of single and multiple transducer receptor coupling proteins on estimates of the relative potency of agonists. Mol. Pharmacol 35, 214–222 [PubMed] [Google Scholar]
- 73.Jarpe MB et al. (1998) D-Arg1, d-Phe5, d-Trp(7,9), Leu11 substance P acts as a biased agonist toward neuropeptide and chemokine receptors. J. Biol. Chem 273, 3097–3104 [DOI] [PubMed] [Google Scholar]
- 74.Mottola DM et al. (2002) Functional selectivity of dopamine receptor agonists: I. Selective activation of postsynaptic dopamine D-2 receptors linked to adenylate cyclase. J. Pharmacol. Exp. Ther 301, 1166–1178 [DOI] [PubMed] [Google Scholar]
- 75.Pupo AS et al. (2015) Recent updates on GPCR biased agonism. Pharmacol. Res 112, 49–57 [DOI] [PubMed] [Google Scholar]
- 76.Godin CM and Ferguson SSG (2012) Biased agonism of the angiotensin II type 1 receptor. Mini. Rev. Med. Chem 12, 812–816 [DOI] [PubMed] [Google Scholar]
- 77.Boerrigter G et al. (2012) TRV120027, a Novel β-arrestin biased ligand at the angiotensin II type I receptor, unloads the heart and maintains renal function when added to furosemide in experimental heart failure clinical perspective. Circ. Hear. Fail 5, 627–634 [DOI] [PubMed] [Google Scholar]
- 78.Tarigopula M et al. (2015) Cardiac myosin light chain phosphorylation and inotropic effects of a biased ligand, TRV120023, in a dilated cardiomyopathy model. Cardiovasc. Res 107, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toth AD et al. (2017) Angiotensin type 1A receptor regulates β-arrestin binding of the (2-adrenergic receptor via heterodimerization. Mol. Cell. Endocrinol 442, 113–124 [DOI] [PubMed] [Google Scholar]
- 80.Gaieb Z et al. (2016) Molecular mechanism of biased ligand conformational changes in CC chemokine receptor 7. J. Chem. Inf. Model 56, 1808–1822 [DOI] [PubMed] [Google Scholar]
- 81.Read C et al. (2016) Cardiac action of the first G protein biased small molecule apelin agonist. Biochem. Pharmacol 116, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maguire JJ (2016) Evidence for biased agonists and antagonists at the endothelin receptors. Life Sci. 159, 30–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosanò L and Bagnato A (2016) β-arrestin1 at the cross-road of endothelin-1 signaling in cancer. J. Exp. Clin. Cancer Res 35, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baltos J-A et al. (2016) Structure-activity analysis of biased agonism at the human adenosine A3 receptor. Mol. Pharmacol 90, 12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baltos JA et al. (2016) Quantification of adenosine A1 receptor biased agonism: Implications for drug discovery. Biochem. Pharmacol 99, 101–112 [DOI] [PubMed] [Google Scholar]
- 86.Becker G et al. (2016) Selective serotonin 5-HT1A receptor biased agonists elicit distinct brain activation patterns: a pharmacoMRI study. Sci. Rep 6, 26633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newman-Tancredi A (2011) Biased agonism at serotonin 5-HT1A receptors: Preferential postsynaptic activity for improved therapy of CNS disorders. Neuropsychiatry 1, 149–164 [Google Scholar]
- 88.Liu Y et al. (2015) Biased signalling: the instinctive skill of the cell in the selection of appropriate signalling pathways. Biochem. J 470, 155–167 [DOI] [PubMed] [Google Scholar]
- 89.Iglesias A et al. (2016) Development of a multiplex assay for studying functional selectivity of human serotonin 5-HT2A receptors and identification of active compounds by high-throughput screening. J. Biomol. Screen 21, 816–823 [DOI] [PubMed] [Google Scholar]
- 90.Conroy JL et al. (2015) Identification of G protein-biased agonists that fail to recruit β-arrestin or promote internalization of the d1 dopamine receptor. ACS Chem. Neurosci 6, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kenakin T (2011) Functional selectivity and biased receptor signaling. J. Pharmacol. Exp. Ther 336, 296–302 [DOI] [PubMed] [Google Scholar]
- 92.Free RB et al. (2014) Discovery and characterization of a G protein-biased agonist that inhibits β-arrestin recruitment to the D2 dopamine receptor. Mol. Pharmacol 86, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brust TF et al. (2016) Biased agonists of the kappa opioid receptor suppress pain and itch without causing sedation or dysphoria. Sci. Signal 9, ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zangrandi L et al. (2016) The G-protein biased partial (opioid receptor agonist 6′-GNTI blocks hippocampal paroxysmal discharges without inducing aversion. Br. J. Pharmacol 173, 1756–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gupta A et al. (2016) Collybolide is a novel biased agonist of e-opioid receptors with potent antipruritic activity. Proc. Natl. Acad. Sci. U. S. A 113, 6041–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maillet EL et al. (2015) Noribogaine is a G-protein biased kappa-opioid receptor agonist. Neuropharmacology 99, 675–688 [DOI] [PubMed] [Google Scholar]
- 97.Carr R et al. (2016) β-arrestin-biased signaling through the β2-adrenergic receptor promotes cardiomyocyte contraction. Proc. Natl. Acad. Sci. U. S. A 113, E4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alonso N et al. (2014) Signal transduction mechanism of biased ligands at histamine H2 receptors. Biochem. J 459, 117–126 [DOI] [PubMed] [Google Scholar]
- 99.Sadek B and Stark H (2016) Cherry-picked ligands at histamine receptor subtypes. Neuropharmacology 106, 56–73 [DOI] [PubMed] [Google Scholar]
- 100.Zhang H et al. (2015) Autocrine selection of a GLP-1R G-protein biased agonist with potent antidiabetic effects. Nat. Commun 6, 8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khurana L et al. (2017) Pyrimidinyl biphenylureas: identification of new lead compounds as allosteric modulators of the cannabinoid receptor CB 1. J. Med. Chem 60, 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.