Abstract
Background:
Clinically validated prognostic models for overall survival (OS) do not exist for patients with relapsed/refractory chronic lymphocytic leukaemia (CLL) on targeted therapies. A prognostic model is needed to identify high risk individuals not adequately served by available targeted therapies.
Methods:
We evaluated 28 candidate factors to derive and validate a risk model for OS in 2,475 previously treated patients with CLL from six randomized trials of ibrutinib, idelalisib, and venetoclax, and the Mayo Clinic CLL Database (MCCD). We applied univariate and multivariate analyses to derive the risk model in an ibrutinib/chemoimmunotherapy (CIT) training dataset (n=727). The primary endpoint was OS. We validated the model in an ibrutinib/CIT internal-validation (n=242) and three external-validations (idelalisib/CIT dataset, n=897; venetoclax/CIT dataset, n=389; MCCD, n=220), applying C-statistics (CS) as a measure of discrimination.
Findings:
The derived model consists of four factors (one point each; serum ß2-microglobulin ≥5mg/dL, lactate dehydrogenase >upper limit of normal, hemoglobin <110g/L for women or <120g/L for men, and time from initiation of last therapy <24 months), separating patients into low (score 0–1), intermediate (score 2–3), and high risk (score 4) groups. The model was prognostic for OS in the ibrutinib/CIT training dataset (CS=0·74, 95% CI 0·60–0·85, log-rank p<0·001), and confirmed in the internal-validation (CS=0·79, 95% CI 0·56–0·97, log-rank p<0·001) and in all three external-validations (idelalisib/CIT: CS=0·71, 95% CI 0·59–0·81, log-rank p<0·001; venetoclax/CIT: CS=0·76, 95% CI 0·66–0·85, log-rank p=0·01; MCCD: CS=0·61, 95% CI 0·56–0·66), log-rank p<0·001).
Interpretation:
We present the first validated risk model for OS in relapsed/refractory CLL. The model is applicable to patients treated with all currently approved targeted therapies: ibrutinib, idelalisib, and venetoclax. This identifies a well-defined cohort of previously treated CLL patients with an unmet clinical need suitable for prospective trials targeting these patients.
INTRODUCTION:
Rai classification and Binet stage incorporate clinical features and were the principal prognostic systems in chronic lymphocytic leukaemia (CLL) for 40 years.1,2 We have since learned that an unmutated immunoglobulin heavy chain variable region (IGHV) gene3 as well as deletion of chromosome 17p (del(17p)) and TP53 mutation (TP53M)4–7 are highly prognostic in CLL. Mutations in NOTCH1, SF3B1, ATM, and BIRC34,8,9 also appear important, and CD38 and ZAP70 expression might contribute prognostic value independent of their association with IGHV mutational status.7,10,11 Additionally, interactions exist between targeted therapies and biologic risk factors.
Investigators have proposed several prognostic models incorporating biologic risk factors, but none were derived in relapsed CLL or in the context of targeted therapies.12–14 The CLL-IPI is prognostic for survival in the relapsed/refractory setting, but differences in the distribution and relative weighting of individual adverse CLL-IPI factors limit application in its current form.15 We hypothesized that a risk model derived in a primary dataset of patients with relapsed/refractory CLL receiving targeted therapies would identify unique prognostic factors and reliably identify high risk patients not adequately served by available targeted therapies.
We therefore used a comprehensive model-building approach to derive a risk model for overall survival (OS) in a population of previously treated patients with CLL who received protocol-based therapy in ibrutinib phase 3 trials, and externally validated the risk model in three independent cohorts including a pooled dataset from idelalisib phase 3 trials, a venetoclax phase 3 trial, and the Mayo Clinic CLL Database (MCCD).
METHODS:
Patients
In collaboration with Janssen Pharmaceuticals, Gilead Sciences, Pharmacyclics, Genentech/Roche, and the Mayo Clinic, the analysis included six randomized phase 3 trials of ibrutinib, idelalisib, and venetoclax: ibrutinib plus bendamustine-rituximab (BR) vs placebo plus BR (HELIOS),16 ibrutinib vs ofatumumab (RESONATE),17 idelalisib plus BR vs placebo plus BR (Study 115);18 idelalisib plus rituximab vs placebo plus rituximab (Study 116);19 idelalisib plus ofatumumab vs ofatumumab (Study 119);20 venetoclax plus rituximab vs BR (MURANO);21 as well as the MCCD. All patients had CLL, were previously treated, and required treatment (International Workshop on CLL 2008 criteria).22 Clinical trials were approved by the Institutional Review Boards (IRB) of participating sites and registered at clinicaltrials.gov ( NCT01578707; NCT01611090; NCT01569295; NCT01539512; NCT01659021; NCT02005471). The MCCD was approved by the Mayo IRB. Written informed consent was obtained from all patients in accordance with Declaration of Helsinki and Guidelines for Good Clinical Practice.
The analysis included 2,475 patients. The model-building dataset included 969 patients from HELIOS (n=578)16 and RESONATE (n=391).17 A random sampling procedure stratified by event/censored status was performed to assign 75% of patients to the training dataset (727 patients with 193 events) and 25% to the internal-validation dataset (242 patients with 64 events), and median follow-up and distribution of candidate prognostic factors were similar between the training and internal-validation datasets. The idelalisib/chemoimmunotherapy external-validation dataset included 897 patients (299 events) from Study 115 (n=416),18 Study 116 (n=220),19 and Study 119 (n=261).20 The venetoclax/chemoimmunotherapy external-validation dataset included 389 patients (61 events) from MURANO.21 The real-world external-validation dataset included 220 patients (123 events) from the MCCD.
Candidate Prognostic Factors and Outcome Measures
The primary endpoint was OS and was calculated from date of randomization or initiation of relevant treatment (MCCD) to date of death. Patients were censored at date last known alive.
Twenty-eight candidate prognostic factors were identified based on literature review and collected at time of study enrollment in the dataset used for training and internal-validation. These included age, gender, ECOG performance status, number of prior therapies, time from initiation of last therapy, time from diagnosis, resistance to CD20 antibody plus purine analogue, Rai classification, Binet stage, largest diameter of longest lymph node by commuted tomography (CT), splenomegaly, hemoglobin, platelet count, absolute neutrophil count, ß2-microglobubulin (B2M), lactate dehydrogenase (LDH), IGHV mutational status, deletion 11q (del(11q)), trisomy 12 (+12), deletion 13q (del(13q)), del(17p), TP53 mutation (TP53M), NOTCH1 mutation, ATM mutation, CD38 expression, ZAP70 expression, and CpG-stimulated complex karyotype. Baseline characteristics are included in the Appendix (p. 4–7). Candidate factors with >30% missing data were excluded from the univariate analysis (complex karyotype, +12, and NOTCH1). TP53M data were missing for 367/969 (37·9%) patients in the training dataset. Hazard ratios of the univariate prognostic effect of the composite variable (del(17p) and/or TP53M; HR 1.52, 95% CI 1.04–2.20) and of del(17p) alone (HR 1.7, 95% CI 1.1.22–2.44) on OS were similar with overlapping 95% confidence intervals (Appendix p. 8), thus we used del(17p) for model building. Cytogenetic abnormalities were analyzed in a categorical manner.
Continuous and multilevel categorical covariates were evaluated without dichotomization throughout model building. Different cutoffs for dichotomized covariates in the final risk score were tested after the final factors were selected.
Risk Model Derivation
In the univariate analysis, a Cox proportional hazards regression model was performed in the training dataset with an interaction between treatment (ibrutinib vs no ibrutinib) and each candidate factor. If the interaction was significant (p<0·1), the factor was selected along with its interaction. Otherwise, a Cox proportional hazards regression model for the candidate factor as a main effect was performed. If the factor had a significant univariate effect on OS (<0·1), it was selected as a main effect. A complete case analysis was used for the univariate step. Presence of significant interactions between treatment and candidate factors would warrant consideration of separate risk models for patients treated with or without targeted therapies. We did not evaluate for interactions between the selected covariates, as we felt that this would reduce the usability and interpretability of the risk model.
Preliminary multivariate analyses were performed to determine whether to include Rai classification or its component covariates in the analysis. Next, a multivariate model was fit with selected factors from the univariate analysis, and factors that remained significant in the multivariate analysis (p<0·05) were included in the final model. The multiple imputation (MI) method was used to impute missing data in the multivariate analysis and included covariates with <30% missing data as well as the censoring indicator and log (survival time) to increase imputation accuracy.23 Five replicate imputations were conducted, and the chained equation method was used for MI.24
The strongest prognostic factors that remained significant in the final multivariate model (p<0·05) were selected by the project team based on their effect size, level of significance, and clinical relevance. We conducted a sensitivity analysis with complete case analysis at the final multivariate stage, and the results were similar. Cutoffs for dichotomized covariates in the final risk score were tested at this stage, and points were assigned to each factor based on effect size. Optimal risk groups were identified by visual assessment of Kaplan-Meier (KM) survival curves and by pairwise comparison of OS by consecutive scores.
A Cox regression was fit on the training dataset using the risk categories as the covariate to test the difference among the groups, and the C-statistic (CS) was calculated as a measure of discrimination. Given minimal missing data in the final multivariate model, a complete case analysis was used at this step.
Internal and External-validation
The risk model was applied to the validation datasets to calculate the C-statistic. The MI method was used to impute missing data in the internal-validation dataset. A complete case analysis was used for the external-validation datasets, wherein patients with sufficient available data for risk group ascertainment were included.
Role of the Funding Source
The funders (Lymphoma Research Foundation, Lymphoma Research Fund, and NCI Core Grant) had no role in the writing of the manuscript or the decision to submit it for publication. The project team (JDS, AN, ADZ) provided pseudocode for all statistical analyses and had full access to the data analyses. The project team including collaborating biostatisticians from Janssen Pharmaceuticals, Gilead Sciences, Pharmacyclics, Genentech/Roche, and the Mayo Clinic held regular video teleconferences to review raw data and statistical analyses. The project team was responsible for data analysis, data interpretation, and writing of the report. The corresponding author wrote the first draft of the manuscript and had final responsibility to submit for publication.
RESULTS:
Results of the Univariate Analysis
Results of the univariate analysis in the Training Dataset are shown in Table 1. In the univariate analysis, we identified an interaction between treatment (ibrutinib vs no ibrutinib) with del(13q) but with none of the other candidate factors. Factors associated with OS included age, ECOG performance status, number of prior therapies, time from initiation of last therapy, Rai classification, Binet stage, largest diameter of longest lymph node by CT, hemoglobin, platelet count, B2M, LDH, IGHV mutational status, and del(17p) (Table 1).
Table 1:
Results of the univariate analysis for overall survival.
| Interaction model1 | Main effect model2 | ||
|---|---|---|---|
| Prognostic Factor | P-value for prognostic factor by treatment interaction1 | P-value | Hazard Ratio (95% CI) |
| Sex (Female vs Male) | 0·63 | 0·36 | 0·866 (0·639–1·175) |
| ECOG performance status (0 vs 1) | 0·68 | 003 | 0·727 (0·542–0·974) |
| Rai stage | 1·00 | <0·001 | - |
| Stage 0 vs IV | -- | 0·10 | 0·190 (0·026–1·366) |
| Stage I vs IV | -- | <0·001 | 0·474 (0·317–0·709) |
| Stage II vs IV | -- | <0·001 | 0·420 (0·279–0·630) |
| Stage III vs IV | -- | 0·18 | 0·753 (0·496–1·141) |
| Binet stage | 0·18 | <0·001 | - |
| Stage A vs C | -- | 0·10 | 0·697 (0·452–1·076) |
| Stage B vs C | -- | <0·001 | 0·398 (0·281–0·563) |
| Enlarged spleen | 0·74 | 0·86 | 0·975 (0·730–1·302) |
| Del(17p) by FISH | 0·62 | 0·003 | 1·723 (1·202–2·470) |
| Del(11q) by FISH | 0·57 | 0·21 | 0·808 (0·582–1·124) |
| Del(13q) by FISH | 0·01 | 0·10 | 1·294 (0·956–1·751) |
| IGHV mutational status (unmutated vs mutated) | 0·12 | 0·07 | 1·458 (0·976–2·177) |
| CD38 expression | 0·58 | 0·48 | 0·894 (0·656–1·218) |
| ZAP70 expression | 0·82 | 0·73 | 0·946 (0·689–1·297) |
| Resistance CD20 mAb + purine analogue | 0·24 | 0·13 | 0·798 (0·595–1·071) |
| ATM mutation | 0·44 | 0·41 | 0·887 (0·667–1·179) |
| Age | 0·82 | 0·07 | 1·014 (0·999–1·030) |
| Largest diameter (mm) of longest lymph node | 0·42 | <0·001 | 1·009 (1·004–1·014) |
| Absolute lymphocyte count (per μl) | 0·97 | 0·43 | 0·999 (0·997–1·001) |
| Hemoglobin (g/l) | 0·60 | <0·001 | 0·978 (0·970–0·985) |
| Platelets (per μl) | 0·88 | 0·002 | 0·996 (0·993–0·998) |
| Neutrophils (per μl) | 0·20 | 0·53 | 1·010 (0·979–1·043) |
| LDH (units/l) | 0·42 | <0·001 | 1·001 (1·001–1·002) |
| B2M (mg/dl) | 0·63 | <0·001 | 1·157 (1·112–1·204) |
| Time from last therapy (months) | 0·38 | <0·001 | 0·983 (0·976–0·991) |
| Time from diagnosis (months) | 0·39 | 0·56 | 0·999 (0·997–1·002) |
| Number of prior therapies | 0·12 | <0·001 | 1·190 (1·115–1·271) |
Del17, deletion 17p; del(11q), deletion 11q; del(13q), deletion 13q; IGHV, immunoglobulin heavy chain variable region gene; LDH, lactate dehydrogenase; B2M, beta-2 microglobulin.
Interaction model includes the treatment (ibrutinib vs no ibrutinib), the candidate prognostic factor, and the interaction.
Main effect model includes only the prognostic factor.
Selection of Factors for the Multivariate Model
We excluded del(13q) to enable the development of a single model for relapsed/refractory CLL irrespective of treatment. Del(13q) was the only significant interaction effect, was only marginally significant as a main effect (p=0·10), and has not previously been associated with poor outcomes.5
Of Rai and Binet covariates, only hemoglobin remained independently prognostic (Appendix p. 9). Given that their prognostic effect was driven by hemoglobin alone, we excluded Rai classification and Binet stage from the multivariate analysis.
The eleven candidate prognostic factors selected for the multivariate model were age, performance status, number of prior therapies, time from initiation of last therapy, largest diameter of longest lymph node, hemoglobin, platelet count, B2M, LDH, IGHV mutational status, and del(17p).
Identification of the Risk Model
Six factors remained independently prognostic in the multivariate analyses of OS (p<0·05) and included B2M, LDH, hemoglobin, IGHV mutational status, number of prior therapies, and time from initiation of last therapy. Notably, del(17p) was not among the independent prognostic factors. Therefore, we undertook a multivariate logistic regression for del(17p), demonstrating that B2M, LDH, hemoglobin, and IGHV status were predictive of del(17p) (Appendix p. 10). We excluded number of prior therapies from the model, as its optimal cutoff might change with advances in CLL therapies. IGHV mutational status was excluded because it did not remain independently prognostic in the final model (p=0·06).
We used an established cutoff for LDH (>upper limit of normal; ULN) and selected the hemoglobin cutoff based on the gender-adjusted median hemoglobin, (<110 g/L for women or <120 g/L for men). We evaluated different cutoffs for B2M (≥4·5 and ≥5 mg/dL) and time from initiation of last therapy (<12 and <24 months), and identified the optimal B2M cutoff of ≥5 mg/dL and time from initiation of last therapy cutoff of <24 months.
Because the four adverse factors in the final model had similar hazard ratios, 1 point was allocated to each factor, and a derived total score of 0–4 was assigned to each patient (Table 2). Pairwise comparisons between consecutive scores in the training dataset revealed that OS was significantly different between patients with scores of 1 vs 2 (p=0·02) and 3 vs 4 (p=0·001), but not 0 vs 1 (p=0.20) and 2 vs 3 (p=0·18; Appendix p. 11). Risk grouping was also confirmed by visual assessment of KM survival curves (Appendix p. 12).
Table 2:
Results of the final multivariate model for the training dataset.
| Training Dataset1 (N=727) | Number of Events | Reference Group | Parameter Estimate | Hazard Ratio (95% CI) | P-value | Assigned Risk Score |
|---|---|---|---|---|---|---|
| B2M | 193 (27%) | ≥5mg/dL | −0·449 | 0·64 (0·47–0·88) | 0 005 | 1·0 |
| LDH | >ULN | −0·386 | 0·68 (0·50–0·92) | 001 | 1·0 | |
| Hemoglobin | <110 g/L for women <120g/Lformen |
−0·592 | 0·55 (0·40–0·76) | <0001 | 1·0 | |
| Time from last therapy | <24 months | −0·477 | 0·62 (0·46–0·78) | 0 002 | 1·0 |
B2M, beta-2 microglobulin; LDH, lactate dehydrogenase; ULN, upper limit of normal.
Parameter estimates are based on multiple imputation analysis.
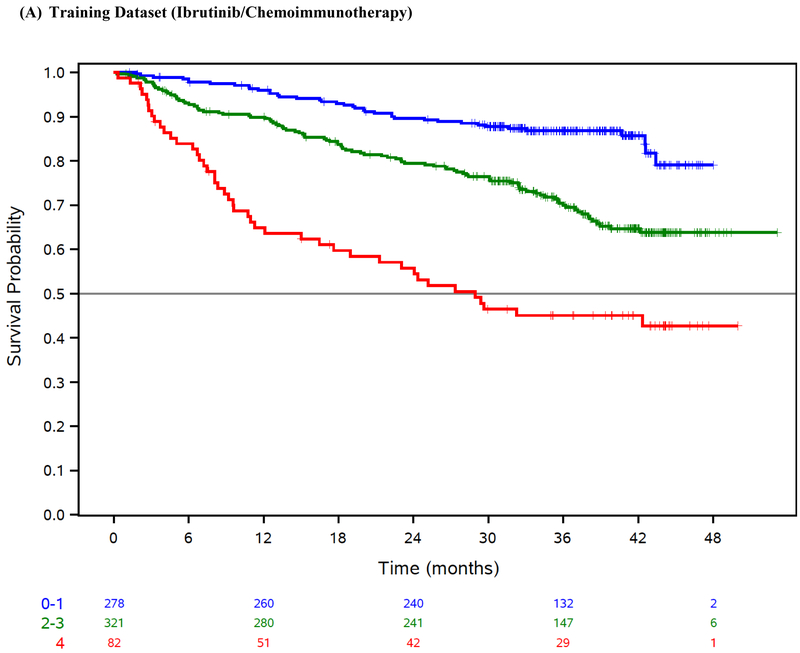
The final risk model consisted of 4 factors (Table 3), including B2M ≥5mg/L, LDH >ULN, hemoglobin <110 g/L for women or <120 g/L for men, and time from initiation of last therapy <24 months, and separated patients into low (score 0–1), intermediate (score 2–3), and high risk (score 4). Our risk score was prognostic for OS in the training dataset (log-rank test across risk groups p=<0·001; Table 3, Figure 1A) and exhibited robust discrimination (CS=0·74 [95% CI 0·60–0·85]).
Table 3:
Overall survival by risk group in the training and validation datasets.
| Score | n (%) | Comparison | Hazard Ratio (95% CI) | 24 Month OS (95% CI) | C-statistic (95% CI) | log-rank p-value | |
|---|---|---|---|---|---|---|---|
| Ibrutinib/CIT Training Dataset1 | n=727 | 0·74 (0·60–0·85) | <0001 | ||||
| Low | 0–1 | 278(38·2%) | — | — | 89·7% (85·4–92·7) | ||
| Intermediate | 2–3 | 321 (44·2%) | vs Low | 2·38 (1·65–3·45) | 79·5% (74·5–83·6) | ||
| High | 4 | 82 (11·3%) | vs Intermediate | 2·22 (1·56–3·16) | 55·8% (44·1–65·9) | ||
| vs Low | 5·29 (3·44–8·15) | ||||||
| Missing | — | 46 (6·3%) | — | — | |||
| Ibrutinib/CIT Internal-Validation2 | n=242 | 0·79 (0·56–0·97) | <0001 | ||||
| Low | 0–1 | 96 (39·7%) | — | — | 92·0% (83·9–96·1) | ||
| Intermediate | 2–3 | 119 (49·1%) | vs Low | 2·78 (1·45–5·33) | 76·4% (67·5–83·2) | ||
| High | 4 | 27 (11·2%) | vs Intermediate | 1·83 (1·00–3·33) | 66·7% (45·7–81·1) | ||
| vs Low | 508 (2·38–10·85) | ||||||
| Idelalisib/CIT External-Validation1 | n=897 | 0·71 (0·59–0·81) | <0001 | ||||
| Low | 0–1 | 278 (31·0%) | — | — | 82·6% (77·1–86·9) | ||
| Intermediate | 2–3 | 460 (51·3%) | vs Low | 2·71 (1·96–3·73) | 61·8% (56·9–66·4) | ||
| High | 4 | 147 (16·4%) | vs Intermediate | 1·44 (1·10–1·89) | 49·5% (40·7–57·8) | ||
| vs Low | 3·90 (2·71–5·62) | ||||||
| Missing | — | 12 (1·3%) | — | — | |||
| Venetoclax/CIT External-Validation1 | n=389 | 0·76 (0·66–0·85) | 001 | ||||
| Low | 0–1 | 170 (43·7%) | 951% (90·5–97·5) | ||||
| Intermediate | 2–3 | 180 (46·3%) | vs Low | 201 (1·14–3·56) | 84·6% (78·1–89·2) | ||
| High | 4 | 30 (7·7%) | vs Intermediate | 1·40 (0·65–3·02) | 82·2% (62·5–92·2) | ||
| vs Low | 2·82 (1·23–6·50) | ||||||
| Missing | — | 9 (2·3%) | |||||
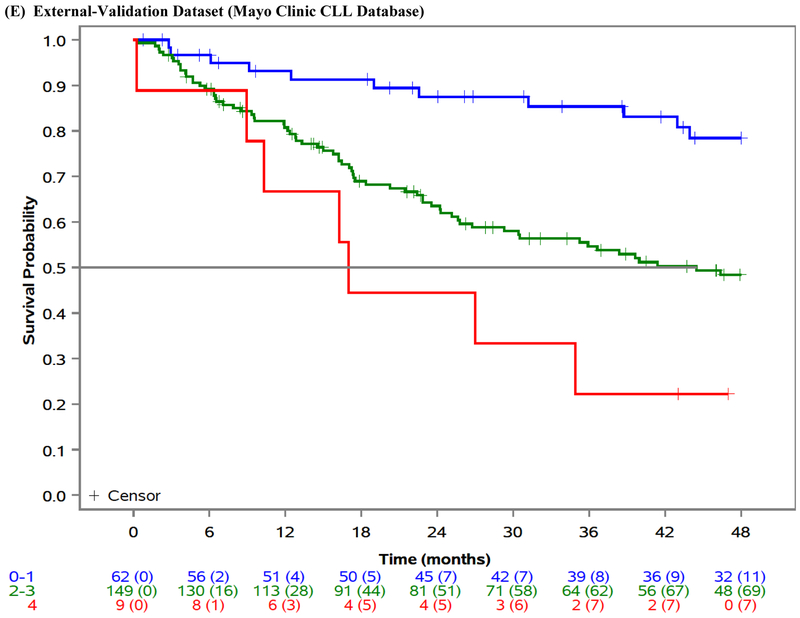
| MCCD External-Validation1 | n=220 | 0·61 (0·56–0·66) | <0001 | ||||
| Low | 0–1 | 62 (28·2%) | — | — | 87·5% (79·2–96·6) | ||
| Intermediate | 2–3 | 149 (67·6%) | vs Low | 2·95 (1·80–4·84) | 63·5% (56·0–72·1) | ||
| High | 4 | 9 (4·1%) | vs Intermediate | 2·02 (0·93–4·40) | 44·4% (21·4–92.3) | ||
| vs Low | 5·96 (2·46–14·41) |
OS, overall survival; CI, confidence interval; CIT, chemoimmunotherapy; MCCD, Mayo Clinic CLL Database;
Based on complete case analysis.
Based on multiple imputation analysis (imputed dataset whose C-statistic is the median of the 5 imputations is shown).
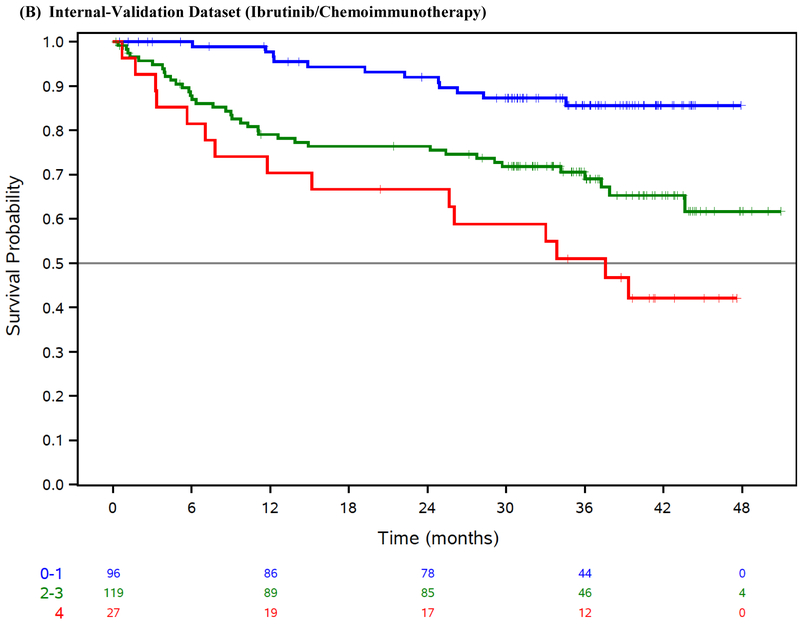
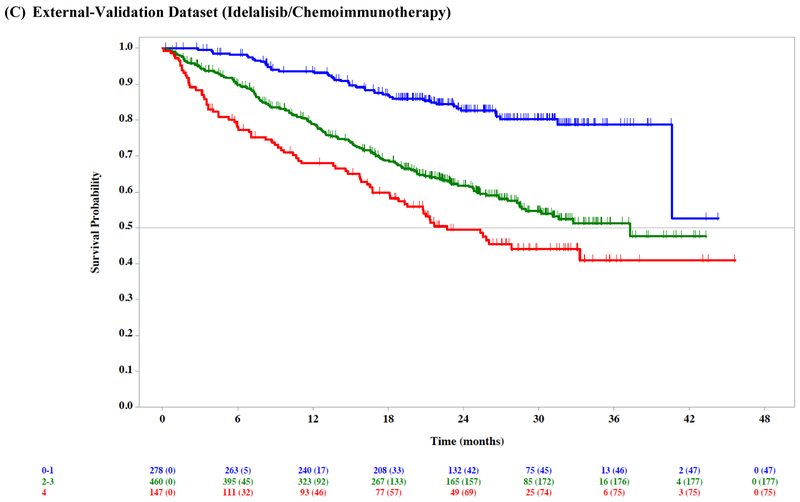
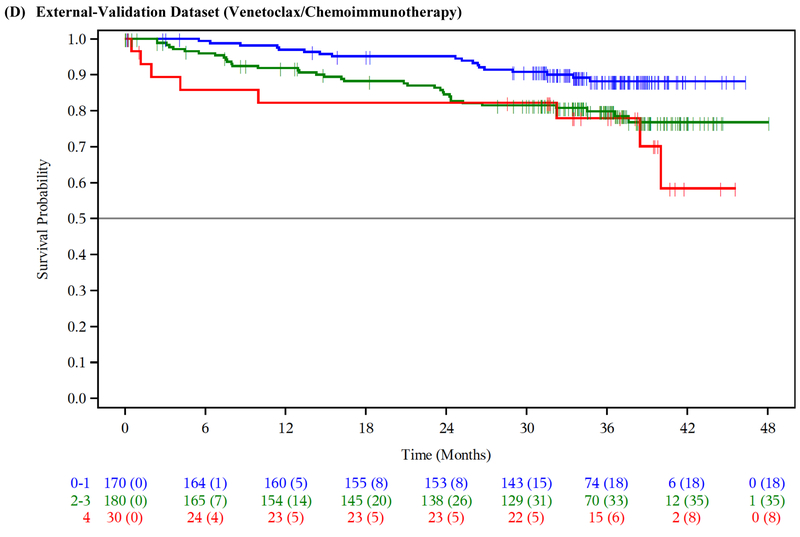
Figure 1: Overall survival by risk group.
Overall survival (OS) by risk group in the ibrutinib phase 3 trials training dataset (A), ibrutinib phase 3 trials internal-validation dataset (B), idelalisib phase 3 trials external-validation dataset (C), venetoclax phase 3 trial external-validation dataset (D), and Mayo Clinic CLL Database external-validation dataset (E).
We applied our risk model to the internal-validation dataset, and confirmed the model was prognostic for OS (log-rank test across risk groups p<0·001; CS=0·79 [95% CI 0·56–0·97]; Table 3, Figure 1B).
External-validation of the Risk Model
We externally validated our risk model in three independent cohorts of patients with relapsed/refractory CLL. The model separated patients into prognostic groups with different OS in the idelalisib/chemoimmunotherapy external-validation dataset (n=897; log-rank test across risk groups p<0·001; CS=0·71 [95% CI 0·59–0·81]; Table 3, Figure 1C), the venetoclax/chemoimmunotherapy external-validation dataset (n=389; log-rank test across risk groups p=0·01; CS=0·76 [95% CI 0·66–0·85]; Table 3, Figure 1D), as well as in the MCCD real-world external-validation dataset (n=220; log-rank test across risk groups p<0·001; CS=0·61 [95% CI 0·56–0·66]; Table 3, Figure 1E).
DISCUSSION:
We comprehensively evaluated prognostic factors in a pooled cohort of previously treated patients with CLL from ibrutinib, idelalisib, and venetoclax randomized phase 3 trials, and we present the first risk model for survival in relapsed/refractory CLL that is derived and fully validated in the context of all currently approved targeted therapies (ibrutinib, idelalisib, and venetoclax) and chemoimmunotherapy.
Of 28 widely reported clinical and biologic covariates explored in this analysis, we initially identified six factors that were independently prognostic for OS. Number of prior therapies was excluded because its optimal cutoff might change with advances in CLL therapy, and IGHV mutational status did not remain independently prognostic (p=0·06) in the multivariate analysis of the five remaining factors. The resulting four-factor model included B2M, LDH, hemoglobin, and time from initiation of last therapy.
This model can be remembered using the acronym BALL (B2M, Anemia, LDH, Last therapy), is easily ascertained from history and readily-available basic laboratory results in all geographic regions, and is available on Calculate by QxMD for iOS, Android and Windows at: https://qxcalc.app.link/CLL. Our model identified three distinct prognostic groups with significantly different OS and has been validated in an internal-validation dataset as well as in three independent external-validation datasets, which include patients treated with ibrutinib, idelalisib, venetoclax, and chemoimmunotherapy.
The primary goal of this effort was to identify patients with adverse disease risk not adequately served by currently available treatment options in the modern era. This model reliably identifies a group of patients at increased risk for death. By incorporating our risk model into trial eligibility criteria, investigators can address this unmet need by designing prospective trials targeting these patients with high risk disease. Our model may also be used to compare treated populations across prospective trials, and the relative frequency of the risk groups and their component covariates should be described in study reports.
Our risk model should not be used to guide whether to initiate therapy in the relapsed setting, which is described in the updated iwCLL Guidelines.25 However, our model has potential to inform clinical management by identifying patients for whom clinical trial participation should be encouraged. By routinely applying our model across all previously treated patients who require therapy, clinicians can now reliably identify patients expected to have suboptimal outcomes despite use of modern generally effective therapies. While we favor consideration of clinical trial participation for all patients, we propose that high and intermediate risk patients should be encouraged to participate in clinical trials since expected OS even with modern targeted therapies remains suboptimal. For example, patients may consider enrolling on trials incorporating novel treatment approaches such as rationale combinations of targeted therapies in CLL (e.g. BTK, PI3Kδ or BCL2), chimeric antigen receptor (CAR) T cell therapy, or response-adapted therapies targeting minimum residual disease negative responses. Additionally, many patients discontinue targeted therapies for toxicity and some are subsequently monitored off treatment. Whether a patient’s pre-treatment risk assessment may be used to guide management of patients in this setting is unknown but may be the subject of further evaluation.
That time from initiation of last therapy less than 24 months emerged as an independent predictor for OS among previously treated patients with CLL underscores the prognostic significance of the duration of response to previous therapy across hematologic malignancies in the relapsed setting (e.g., follicular lymphoma, diffuse large B-cell lymphoma, and Hodgkin lymphoma). Notably, time from initiation of first-line therapy less than 2 years was recently identified as a robust clinical prognostic factor for inferior OS in an independent analysis.26 Hemoglobin was among the earliest prognostic factors identified in CLL and has been incorporated into the Rai and Binet staging systems.1,2 B2M has been incorporated into previous prognostic systems (e.g., CLL-IPI).14 Recent analyses have identified LDH as independently prognostic of survival in a cohort of patients with del(17p) CLL treated with ibrutinib,27 and as an independent predictor for subsequent Richter’s transformation among ibrutinib-treated patients irrespective of del(17p) status.28
Of interest, IGHV mutational status and del(17p) were not independently prognostic for OS in our final multivariate model. While this is consistent with reports that IGHV mutational status is not independently prognostic for OS in the relapsed/refractory setting, as opposed to frontline therapy, these data contradict a recent report that del(17p) is independently prognostic among patients with relapsed/refractory CLL treated with ibrutinib.28,29 However, this analysis was performed in a small phase 2 study enriched for heavily pretreated patients (median 4 prior therapies). Their multivariate model included just two clinical factors (number of prior therapies and disease bulk) and none of the factors from our model. We performed a multivariate logistic regression for del(17p) in the training dataset which demonstrated that the proposed risk model subsumes the prognostic impact of del(17p). Because ZAP70 and CD38 did not have a univariate prognostic effect, neither was selected for inclusion in the multivariate analysis. Therefore, interactions between IGHV mutational status and ZAP70 or CD38 expression cannot explain the finding that IGHV mutational status is not independently prognostic for OS. With an expanding list of CLL therapies that are effective in patients whose CLL harbors an unmutated IGHV gene or del(17p), it is possible that the clinical factors included in the proposed risk model more directly represent tumor burden and treatment refractoriness.
One potential limitation of our study is that number of prior therapies was independently prognostic for survival in our multivariate analysis but was excluded in the final risk model because its optimal cutoff is changing with advances in CLL therapies. The number of prior therapies received should be considered in the management of patients with relapsed or refractory CLL, and the median number of prior therapies should be reported in future clinical trials. Another potential limitation is that several candidate prognostic factors including complex karyotype, SF3B1, BIRC3, and NOTCH1 were excluded from the univariate and multivariate analysis. These factors were not required in some of the included protocols, which led to a high rate of missing data for these factors. However, these factors are not widely available in clinical practice. Complex karyotype has emerged in previous analyses of ibrutinib- and venetoclax-treated patients as an independent predictor for progression-free survival.28,30,31 Whether these factors would contribute additional value in predicting overall survival remains unknown but may be the subject of further evaluation to attempt to increase the C statistic of the model. Our multivariate analyses included del(17p) alone rather than the composite covariate of del(17p) and/or TP53 mutation. This is a potential limitation because patients may harbor a TP53 mutation in the absence of del(17p). However, del(17p) and the composite covariate (del(17p) and/or TP53 mutation) had similar impact on OS with overlapping 95% confidence intervals, and we chose del(17p) alone as the data was more robust. We excluded del(13q) because it was the only significant interaction effect, was only marginally significant as a main effect (p=0.1) and has not previously been associated with poor outcomes. While this permitted the development of a single risk model for all patients with relapsed or refractory CLL, it remains unknown whether del(13q) might have contributed additional value. We included multiple external validations with heterogeneous patient populations and multiple alternative small molecular inhibitors, which confirms that our model can be applied broadly across heterogeneous patients with relapsed or refractory CLL.
Our study has other limitations. We used an intent-to-treat analysis which did not censor patients who crossed over from the comparator to ibrutinib arms, which might have diluted interaction effects between treatment and the candidate prognostic factors, but this was limited to the ibrutinib/chemoimmunotherapy training dataset. Because patients with ECOG performance status >2 at baseline were excluded from all of the phase 3 trials, the impact of impaired performance status on survival remains unknown. A limitation of the Mayo Clinic CLL Database external-validation dataset is the frequency of missing B2M data. This likely resulted in an underestimate of the frequency of high-risk patients in the Mayo Clinic CLL Database, as patients with one missing risk factor could only be evaluable in this complete case analysis in the low and intermediate risk groups (scores of 0 or 2).
In summary, our validated risk model consisting of four readily-available factors (B2M, LDH, hemoglobin, and time from initiation of last therapy) identifies three distinct prognostic groups with significantly different overall survival. This model reliably identifies patients with previously treated CLL at increased risk of death and has been validated for patients treated with all currently approved targeted therapies: ibrutinib, idelalisib, and venetoclax. Investigators can and should address this unmet need by designing prospective trials targeting these higher risk patients.
Supplementary Material
PUTTING RESEARCH INTO CONTEXT:
Evidence before this study:
Clinically validated prognostic models for survival exist for patients with previously untreated CLL treated with chemoimmunotherapy. Targeted therapies have largely replaced chemoimmunotherapy in the relapsed/refractory setting. We performed a literature search for prognostic factors and indices in CLL using PubMed, and we found that a clinically validated prognostic model for predicting outcomes in patients with relapsed or refractory CLL in the era of targeted therapies is lacking.
Added value of this study:
This is the first clinically validated prognostic model for survival derived in patients with relapsed or refractory CLL or on targeted therapies. We identified 28 widely reported clinical and biologic risk factors based on literature review, which we comprehensively evaluated to derive and validate the prognostic model. We used a pooled cohort of 2,475 previously treated patients with CLL from ibrutinib, idelalisib, and venetoclax randomized phase 3 trials, as well as the Mayo Clinic CLL Database. The results of this multivariate analysis led to a four-factor prognostic model which can be easily ascertained from history and readily-available basic laboratory results in all geographic regions.
Implications of all the available evidence:
This prognostic model identifies a well-defined cohort of patients with an increased risk of death despite modern targeted therapies, and investigators can address this unmet need by designing prospective trials targeting these higher risk patients.
Funding:
Lymphoma Research Foundation (JDS); Lymphoma Research Fund (ADZ); NIH/NCI Core Grant (P30CA008748).
DECLARATION OF INTEREST:
JDS receives research support from Genentech/Roche, BeiGene, and TG Therapeutics, and consultancy from Verastem. MD reports employment from Janssen. AL reports employment and stock ownership from Janssen. GX reports employment from Gilead. YM reports employement from Genentech. NEK reports research support from Acerta, Pharmacyclics, MEI Pharma, and Genentech. TDS reports research support from Pharmacyclics, Janssen, Genentech, Glaxo-SmithKline, Celgene, AbbVie, Cephalon, and Hospira; RRF reports consultancies from Abbvie, Acerta, AstraZeneca, Genentech, Janssen, Loxo Oncology, Pharmacyclics, Sunesis, TG Therapeutics, Verastem, and Gilead, and serves on data safety monitoring board for Incyte; PH reports research support and consultancies from Janssen, Pharmacyclics, Gilead, AbbVie, and Roche; JJ reports employment from Celgene, consultancies from Abbvie and Gilead; JFS reports research support from AbbVie, Celgene, Janssen, and Roche, consultancies from AbbVie, Acerta, Celgene, Janssen, Roche, and Takeda, and travel support and speaker fees from Abbvie, Celgene, and Genentech/Roche; JPS reports consultancies and research support from Gilead and TG Therapeutics; LF reports employment and stock ownership from Janssen; MM reports employment and stock ownership from Genentech/Roche; TS reports employment and stock ownership from Janssen and Genentech/Roche; VR reports employment from Pharmacyclics; LKD reports employment from Gilead; PB reports employment from Gilead; AH reports employment from Janssen; DFJ reports research funding from Acerta, employment and stock ownership from AbbVie/Pharmacyclics; ADZ reports consultancies from Genentech, Celgene, Janssen, Amgen, Novartis, Adaptive Biotechnology, MEI Pharma, Roche, AstraZeneca, MorphoSys, Abbvie, and Beigene Pharmaceuticals, research support from Gilead, MEI Pharma, and Roche, and data safety monitoring committee chairmanship from BeiGene. AN, KGR, JCB, and AAC have nothing to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jacob D. Soumerai, Massachusetts General Hospital Cancer Center, 55 Fruit Street, Boston, MA, 02114 Memorial-Sloan Kettering Cancer Center, 1275 York Avenue, New York, NY 10065.
Ai Ni, Memorial-Sloan Kettering Cancer Center, 1275 York Avenue, New York, NY 10065.
Guan Xing, Gilead Sciences, Inc., 199 East Blaine Street, Seattle, WA 98102.
Yong Mun, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080.
Neil E. Kay, Mayo Clinic, 200 1st St SW, Rochester, MN 55902
Tait D. Shanafelt, Mayo Clinic, 200 1st St SW, Rochester, MN 55902 Stanford University Medical Center, 300 Pasteur Drive, Suite 3215, Stanford, CA 94305.
Kari G. Rabe, Mayo Clinic, 200 1st St SW, Rochester, MN 55902
John C. Byrd, Ohio State University Comprehensive Cancer Center, 460 W 10th Avenue, Columbus, OH 43210
Asher A. Chanan-Khan, Mayo Clinic, 4500 San Pablo Rd S, Jacksonville, FL 32224
Richard R. Furman, Weill Cornell Medical College/New York Presbyterian Hospital, 520 East 70th St, Starr Pavilion, 3rd Floor, New York, NY 10021
Peter Hillmen, St James’s University Hospital, Level 3, Bexley Wing, Beckett Street, Leeds, LS9 7TF.
Jeffrey Jones, Ohio State University Comprehensive Cancer Center, 460 W 10th Avenue, Columbus, OH 43210.
John F. Seymour, Peter MacCallum Cancer Centre, Royal Melbourne Hospital, The University of Melbourne, Victoria 3010
Jeffrey P. Sharman, Willamette Valley Cancer Institute and Research Center, 1690 East 26th Ave, Eugene OR 97408
Lucille Ferrante, Janssen Pharmaceuticals, Inc., 800 Ridgeview Drive, Horsham, PA 19044.
Vijay Reddy, Pharmacyclics, LLC., 999 E Arques Ave, Sunnyvale, CA 94085.
Angela Howes, Janssen Pharmaceuticals, Inc., 800 Ridgeview Drive, Horsham, PA 19044.
REFERENCES
- 1.Rai KR, Sawitsky A, Cronkite E, Chanana A, Levy R, Pasternack B. Clinical staging of chronic lymphocytic leukemia. Blood. Blood 1975; 46: 219–34. [PubMed] [Google Scholar]
- 2.Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 1981; 48: 198–206. [DOI] [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig VH Genes Are Associated With a More Aggressive Form of Chronic Lymphocytic Leukemia. Blood 1999; 94 http://www.bloodjournal.org/content/94/6/1848.short (accessed March 29, 2017). [PubMed] [Google Scholar]
- 4.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 2014; 123: 3247–54. [DOI] [PubMed] [Google Scholar]
- 5.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–6. [DOI] [PubMed] [Google Scholar]
- 6.Döhner H, Fischer K, Bentz M, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood 1995; 85: 1580–9. [PubMed] [Google Scholar]
- 7.Grever MR, Lucas DM, Dewald GW, et al. Comprehensive Assessment of Genetic and Molecular Features Predicting Outcome in Patients With Chronic Lymphocytic Leukemia: Results From the US Intergroup Phase III Trial E2997. J Clin Oncol 2007; 25: 799–804. [DOI] [PubMed] [Google Scholar]
- 8.Rossi D, Rasi S, Fabbri G, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012; 119: 521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and Other Novel Cancer Genes in Chronic Lymphocytic Leukemia. N Engl J Med 2011; 365: 2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 Compared with Immunoglobulin Heavy-Chain Gene Mutation Status as a Predictor of Disease Progression in Chronic Lymphocytic Leukemia. N Engl J Med 2004; 351: 893–901. [DOI] [PubMed] [Google Scholar]
- 11.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–7. [PubMed] [Google Scholar]
- 12.Wierda WG, O’Brien S, Wang X, et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood 2007; 109: 4679–85. [DOI] [PubMed] [Google Scholar]
- 13.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013; 121: 1403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Group TC-IW. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol 2016; 17: 779–90. [DOI] [PubMed] [Google Scholar]
- 15.Soumerai JD, Ni A, Xing G, et al. Evaluation of the CLL-IPI in relapsed and refractory chronic lymphocytic leukemia in idelalisib phase-3 trials. Leuk Lymphoma 2018; : 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chanan-Khan A, Cramer P, Demirkan F, et al. Ibrutinib combined with bendamustine and rituximab compared with placebo, bendamustine, and rituximab for previously treated chronic lymphocytic leukaemia or small lymphocytic lymphoma (HELIOS): a randomised, double-blind, phase 3 study. Lancet Oncol 2015; 17: 200–11. [DOI] [PubMed] [Google Scholar]
- 17.Byrd JC, Brown JR, O’Brien S, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med 2014; 371: 213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zelenetz AD, Barrientos JC, Brown JR, et al. Idelalisib or placebo in combination with bendamustine and rituximab in patients with relapsed or refractory chronic lymphocytic leukaemia: interim results from a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol 2017; 18: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furman RR, Sharman JP, Coutre SE, et al. Idelalisib and Rituximab in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med 2014; 370: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones JA, Robak T, Brown JR, et al. Efficacy and safety of idelalisib in combination with ofatumumab for previously treated chronic lymphocytic leukaemia: an open-label, randomised phase 3 trial. Lancet Haematol 2017; 4: e114–26. [DOI] [PubMed] [Google Scholar]
- 21.Seymour JF, Mobasher M, Kater AP. Venetoclax–Rituximab in Chronic Lymphocytic Leukemia. N Engl J Med 2018; 378: 2141–4. [DOI] [PubMed] [Google Scholar]
- 22.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111: 5446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moons KGM, Donders RART, Stijnen T, Harrell FE. Using the outcome for imputation of missing predictor values was preferred. J Clin Epidemiol 2006; 59: 1092–101. [DOI] [PubMed] [Google Scholar]
- 24.van Buuren S Multiple imputation of discrete and continuous data by fully conditional specification. Stat Methods Med Res 2007; 16: 219–42. [DOI] [PubMed] [Google Scholar]
- 25.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018; 131: 2745–60. [DOI] [PubMed] [Google Scholar]
- 26.Ahn IE, Farber CM, Davids MS, et al. Early progression of disease as a predictor of survival in chronic lymphocytic leukemia. Blood Adv 2017; 1: 2433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones J, Mato A, Coutre S, et al. Evaluation of 230 patients with relapsed/refractory deletion 17p chronic lymphocytic leukaemia treated with ibrutinib from 3 clinical trials. Br J Haematol 2018; 182: 504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddocks KJ, Ruppert AS, Lozanski G, et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients With Chronic Lymphocytic Leukemia. JAMA Oncol 2015; 1: 80–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien S, Furman RR, Coutre S, et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood 2018; 131: 1910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson PA, O’Brien SM, Wierda WG, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer 2015; 121: 3612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson MA, Tam C, Lew TE, et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood 2017; 129: 3362–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.