Summary
The characterization of the architecture, structure and extracellular interactions of the CD6 glycoprotein, a transmembrane receptor expressed in medullary thymocytes and all mature T‐cell populations, has been enhanced by the existence of monoclonal antibodies (mAbs) that specifically recognize the various scavenger receptor cysteine‐rich (SRCR) domains of the ectodomain. Using engineered isoforms of CD6 including or excluding each of the three SRCR domains, either expressed at the membranes of cells or in soluble forms, we provide conclusive and definitive evidence that domain 2 of CD6, previously not identifiable, can be recognized by the CD6 mAbs OX125 and OX126, and that OX124 targets domain 3 and can block the interaction at the cell surface of CD6 with its major ligand CD166. Alternative splicing‐dependent CD6 isoforms can now be confidently identified. We confirm that following T‐cell activation there is a partial replacement of full‐length CD6 by the CD6Δd3 isoform, which lacks the CD166‐binding domain, and we find no evidence for the expression of other CD6 isoforms at the mRNA or protein levels.
Keywords: CD6, isoforms, scavenger receptor cysteine‐rich domain, T cell, T‐cell monoclonal antibodies
Introduction
The T‐cell surface glycoprotein CD6 has an impact on the regulation of T‐cell receptor‐mediated signalling and thymocyte maturation,1, 2, 3 and it has attracted renewed interest since it was found that CD6 is a susceptibility gene for multiple sclerosis.4 Furthermore, immunotherapy targeting CD6 with monoclonal antibodies (mAbs) has been attempted not only in mouse models but significantly also in human pathologies;5, 6 indeed, itolizumab has proven efficacy in the treatment of patients with rheumatoid arthritis and severe chronic plaque psoriasis.7, 8, 9
The interaction between CD6 and its widely expressed extracellular ligand CD166 is well characterized, with CD166 binding to the membrane proximal scavenger receptor cysteine‐rich (SRCR) domain (domain 3; d3) of CD6.10, 11 It has been speculated that itolizumab or other mAbs targeting d1 of CD6 could interfere with the binding of CD6 to CD166;12 however, this suggestion has not been substantiated experimentally. One alternative possibility to explain decreased T‐cell activation by CD6 mAbs is that CD6 is an inhibitory receptor and direct targeting of the molecule induces signalling repression.13 In an additional level of complexity, CD6 can display different alternative splicing‐dependent isoforms that arise during activation, namely the CD6∆d3 isoform that lacks the extracellular d3.14
It is therefore of the utmost importance that a thorough characterization of the binding specificities of CD6 mAbs is performed and the functional effects, such as ligand blocking, are described. Using engineered extracellular isoforms of CD6 containing or excluding each of the three SRCR domains of CD6, we analysed the specificities of several CD6 mAbs, their blocking efficacy, and their value as markers for CD6 isoforms. Importantly, we have also detected errors in the literature regarding the specificity of two CD6 mAbs.
Material and methods
Cells and cell lines
Jurkat E6.1 and Raji cell lines were grown in supplemented RPMI‐1640 and HEK293T cells in supplemented Dulbecco's modified Eagle medium.
Peripheral blood lymphocytes (PBLs) were obtained from buffy coats of healthy donors, provided by Serviço de Imunohemoterapia, Hospital São João (Porto, Portugal), by density‐gradient separation using Lympholyte‐H (Cedarlane Laboratories, Burlington, Ont., Canada) followed by exclusion of plastic‐adherent monocytes. For activation, 5 × 105 PBLs were stimulated with phytohaemagglutinin‐P (PHA‐P) at 7·5 μg/ml for different times. Flow cytometry was performed as previously described15 and analysed using a FACScanto 2 (BD Biosciences, San Jose, CA).
Monoclonal antibodies
Mouse anti‐human CD6 mAbs, OX124 (IgG1) and OX126 (IgG1) were supplied by Absolute Antibody (Redcar, UK) and together with OX125 (IgG2b) were also produced in house. Other CD6 mAbs used were MEM98 (EXBIO, Vestec, Czech Republic), BL‐CD6 (BioLegend, San Diego, CA), itolizumab (a kind gift from Kalet Leon, Centro de Imunologia Molecular, Havana, Cuba) and T12.1 (obtained from ATCC, Manassas, VA). OKT3 (CD3), and LN3 (HLA‐DR) were purchased from eBioscience (San Diego, CA), FN50 (CD69), BC96 (CD25) and OKT4 (CD4) from BioLegend, MEM233 (CD80) and BU63 (CD86) from EXBIO, 3A6 (CD166) from BD Pharmingen (San Diego, CA), N‐21 (CD166) and Y2/178 (CD5) from Santa Cruz Biotechnology (Dallas, TX).
cDNA constructs and lentiviral transduction
Wild‐type (WT)‐CD6 and isoform‐encoding sequences were amplified by polymerase chain reaction from pEGFP‐N1/CD6FL14 by removing exons 3, 4, 5 and 6, encoding d1, d2, d3 and stalk region (st), respectively, according to the annotated sequence NM_006725 (GenBank, NCBI), using exon specific primers (see Supplementary material, Table S1). Constructs were cloned in the lentiviral expression vector pHR using MluI and NotI restriction sites and transduced in E6.1 and HEK293T cell lines, as described previously.16
mRNA analysis of alternative splicing
Total RNA of 5 × 106 resting and activated PBLs was isolated using the TripleXtractor directRNA Kit (Grisp, Porto, Portugal). cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). The cDNA obtained was used to analyse the CD6 alternative splicing pattern by polymerase chain reaction with NZYTaq (NZYTech, Lisbon, Portugal). Primer sequences were the following: 5′‐acgcgtgccgcagcgacggga‐3′ (forward primer on exon 3), 5′‐gaggagcattagctcccgaga‐3′ (reverse primer on exon 7) and 5′‐ctgagcacaccgcgcccg‐3′ (reverse primer on exon 5).
Construction of CD166‐deficient Raji cells by CRISPR/Cas9
For the deletion of CD166 from Raji cells, the gRNA 5′‐TGAGGTACGTCAAGTCGGCA‐3′ was synthesized (Sigma‐Aldrich, St. Louis, MO) and cloned in pLentiCRISPRv2 (a gift from Feng Zhang; Addgene plasmid #52961; http://n2t.net/addgene:52961; RRID:Addgene_52961)17 using the BsmBI site. Raji cells were transduced with the lentiviral particles and selected with 2 μg/ml puromycin.
Recombinant proteins and tetramer assembly
Recombinant extracellular WT‐CD6 (GenBank AAA86419.1) and CD6∆d3 (GenBank ABH04237.1) were produced and biotinylated as described elsewhere;13 amplification of CD6∆d3 was from pEGFP‐N1/CD6∆D3.14
Tetramers were assembled by mixing 3 μg of biotinylated CD6 or CD6∆d3 with 0·75 μg of Streptavidin‐Alexa647 (Invitrogen) and incubating for 1 hr at 4° with agitation. The assembled mixture was added to 2·5 × 105 cells and allowed to interact for 45 min on ice. Cells were washed and analysed by flow cytometry.
In blocking experiments, CD6 mAbs were added to the assembled tetramers and incubated for 20 min on ice before cell staining.
Conjugate formation
Raji B cells were incubated with a mix of the superantigens (sAg) staphylococcal enterotoxins A, B, C3 and E (Toxin Technologies, Sarasota, FL), at 200 ng/ml each for 1 hr at 37°. Raji cells were centrifuged and resuspended in complete RPMI. PBLs or CD6‐expressing E6.1 cells (E6.1‐WT‐CD6) were added to Raji cells and allowed to interact at 37° for different times.
In blocking experiments, 1 μg of CD6 mAbs were added to E6.1‐WT‐CD6 cells and incubated for 20 min on ice before conjugate formation. Where indicated, Raji cells were pre‐incubated with 1 μg of N‐21 (CD166) mAb before sAg loading. Supernatants were collected at 48 hr and interleukin‐2 (IL‐2) was quantified by enzyme‐linked immunosorbent assay (R&D Duo Set human IL‐2; R&D Systems, Minneapolis, MN).
Surface plasmon resonance
Tissue culture supernatants containing CD6CD4d3d4 fusion proteins were produced in HEK293T cells.11, 18 Binding specificity was carried out using a T200 BIAcore at 25°. Monoclonal antibodies (100 μg/ml) were injected at 10 μl/min over CD6 fusion proteins immobilized via a CD4d3d4 mAb (OX68) coupled to a CM5 chip.11, 18
Results
The CD6 mAbs OX124 and OX126 are specific for d3 and d2, respectively
CD6 domain‐specific mAbs are valuable tools for monitoring the expression of specific extracellular domains and the architecture of particular isoforms. Most available CD6 mAbs bind to d1 of CD6 (see Supplementary material, Table S2). A separate set of mAbs, OX124, OX125 and OX126 were described to have been raised against the third extracellular SRCR domain of human CD6.18 These reagents were putatively useful to detect the presence of the domain that contacts CD166 and so directly inform on the availability of CD6 to bind the ligand.18
It was therefore surprising that E6.1 Jurkat cells where we artificially expressed the CD6Δd3 isoform were positively labelled by OX126 at similar levels to the E6.1 cells transfected and expressing WT‐CD6 (Fig. 1a). The described original immunogen included much of the stalk region sequence in addition to d3 of CD618; however, a CD6 isoform lacking d3 and the stalk (CD6Δd3Δst) was still recognized by OX126 (Fig. 1a). It was clear that the original characterization of OX12618 was incorrect.19
Figure 1.
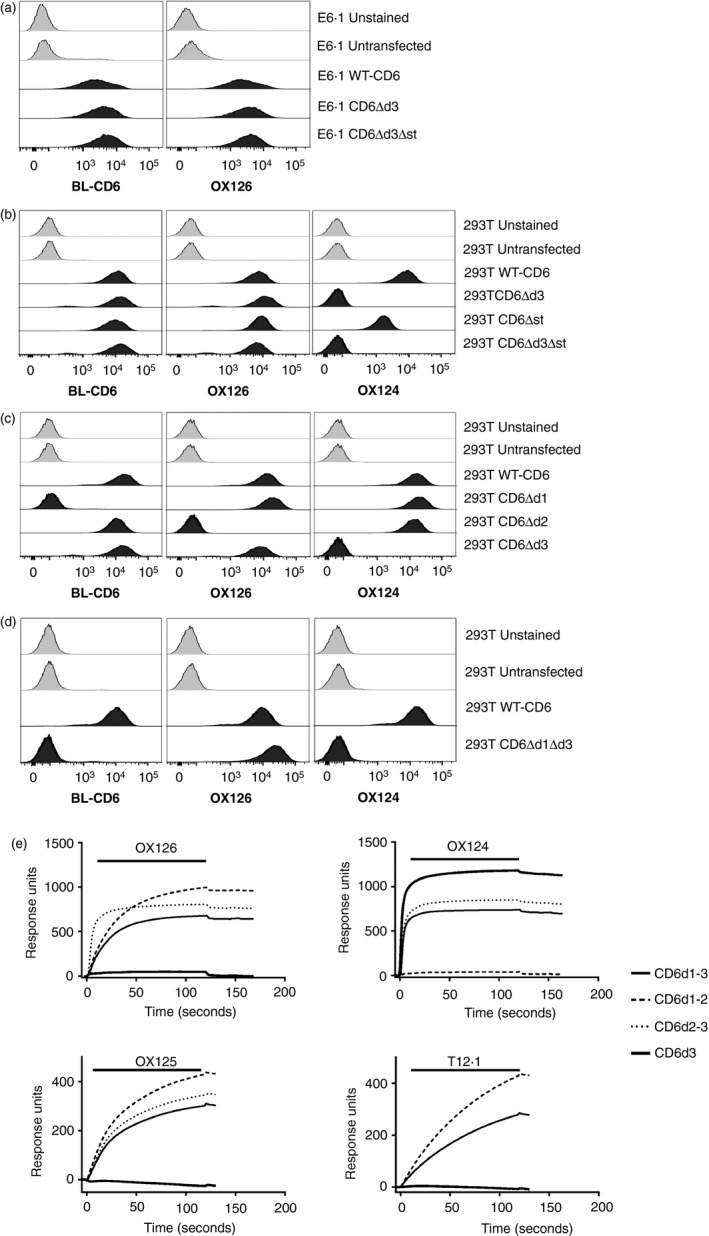
CD6 monoclonal antibodies (mAbs) OX126 and OX124 bind domain 2 (d2) and d3, respectively. Flow cytometry analysis of (a) Jurkat E6.1 and (b–d) HEK293T cells transfected with wild‐type (WT) ‐CD6 and domain deletion (Δ) mutants using CD6d1 mAb (BL‐CD6), OX126 and OX124. (a) OX126 binding does not depend on d3 and/or the membrane proximal stalk region of CD6. (b) OX124 binding depends on d3. (c) OX126 binding depends on d2. (d) OX126 binds directly to d2 without a contribution of other domains. (e) CD6 mAbs, OX126, OX124, OX125 and a d1 mAb (T12.1) were injected over the fusion proteins consisting of CD6 domains 1–3 (CD6d1–3), CD6d1–2, CD6d2–3 and CD6d3, immobilized on the surface of a CM5 chip. Sensorgram traces show that mAb binding depends on the presence of d2 for OX126 and OX125; d3 for OX124 and d1 for T12.1.
We undertook a thorough characterization of the domain specificity of OX124 and OX126. To avoid any interference by endogenous CD6, we expressed WT‐CD6 and different combinations of domains in HEK293T cells. HEK293T cells expressing WT‐CD6 were positive for a mAb likely to be specific for CD6 d1 (BL‐CD6),10 OX126 and OX124,18 but untransfected HEK293T cells were negative for these mAbs (Fig. 1b). However, whereas OX124 did not label HEK293T cells expressing the isoforms lacking d3, CD6Δd3 and CD6Δd3Δst, OX126 was still positive for all the cell lines analysed. The data showed that OX124 binds the SRCR d3 of CD6 and OX126 bound somewhere else in the CD6 ectodomain.
To define the domain specificity of OX126, we expressed CD6 isoforms each excluding one of the three SRCR domains in HEK293T cells and analysed the specificity of the different CD6 mAbs. As seen in Fig. 1(c), the absence of d1 and d3 correlated with the lack of BL‐CD6 and of OX124 binding, respectively, but OX126 labelling was lost only when d2 of CD6 was absent, only compatible with OX126 binding to d2. WT‐CD6 and CD6Δd3, but not CD6Δd1, were detected by BL‐CD6, confirming its specificity for d1.
To test whether OX126 bound directly to d2, we expressed a CD6 isoform that excluded both CD6d1 and CD6d3. Detection of this CD6Δd1Δd3 isoform by OX126 and not by OX124 definitively established that OX126 is specific for d2 (Fig. 1d).
OX125 and OX126 recognize different epitopes on CD6d2
We comprehensively characterized the domain specificity of all three CD6 mAbs, OX124, OX125 and OX126 by surface plasmon resonance using soluble recombinant CD6 isoforms (Fig. 1e). All mAbs were tested against isoforms containing either the full CD6 sequence from d1 to d3 (CD6d1–d3), omitting d3 (CD6d1–d2) or d1 (CD6d2–d3), or only containing d3 (CD6d3). Monoclonal antibodies were injected over the immobilized CD6 fusion proteins. OX126 bound to isoforms that contained CD6d2 (CD6d1–3, CD6d1–2 and CD6d2–3). OX124 binding was dependent on the presence of d3 (CD6d1–d3, CD6d2–d3 and CD6d3). OX125 displayed the same domain specificity as OX126. In sequential injections of mAbs over the same immobilized fusion proteins, OX126 did not prevent OX125 binding to CD6d2‐containing constructs, and vice versa (data not shown), indicating that OX125 and OX126 bind different epitopes on CD6d2. As expected, a CD6 d1 mAb (T12.1) bound only to CD6d1–d3 and CD6d1–d2.
In the current characterization of OX124, OX125 and OX126, the identity of each recombinant protein is confirmed in each experiment with domain‐specific antibodies. This was not done in the original description and it is now clear that the domain specificity of the mAbs was not proven.18 A labelling error was identified and the antibody preparations used in the original study were checked for specificity using the domain mutants as in Fig. 1(e). The current data indicate that the identities of OX124 and OX126 were switched in the original description.18, 19 Further scrutiny of the original data shows that these are consistent with recombinant CD6d1–3 being used both for binding analysis and immunization.18, 19
OX124 blocks CD6–CD166 interactions
Having clarified unequivocally the domain specificity of the CD6 mAbs, OX124 (d3), OX125 (d2) and OX126 (d2), we next investigated their potential to block CD6‐CD166 interactions. We assembled soluble extracellular WT‐CD6 and CD6Δd3 recombinant proteins as tetramers and tested their binding to Raji cells, expressing CD166 endogenously, or CRISPR/Cas9‐engineered CD166‐deficient Raji cells. Deletion of CD166 was confirmed by flow cytometry (Fig. 2a), together with the analysis of expression of the markers HLA‐DR, CD80 and CD86 (see Supplementary material, Fig. S1). WT‐CD6tet bound well to CD166‐expressing Raji cells but not to cells deficient in CD166 (Fig. 2b). As expected, CD6Δd3tet bound to neither cell line.
Figure 2.
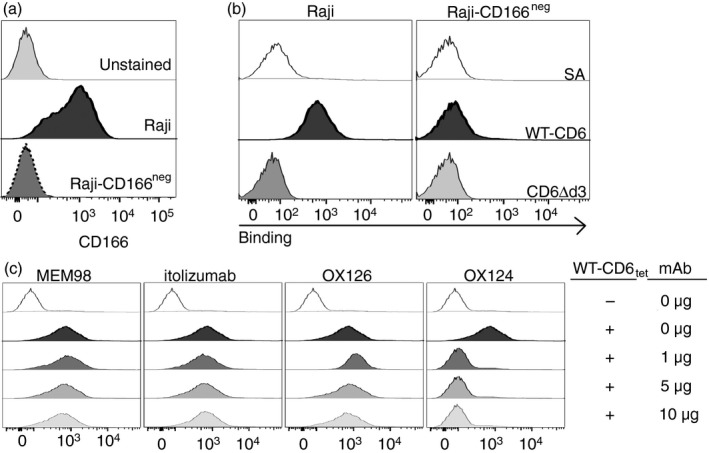
The CD6 monoclonal antibody (mAb) OX124, specific for domain 3 (d3), blocks the CD6–CD166 interaction. (a) Flow cytometry analysis of CD166 expression on Raji and CD166‐deficient Raji, edited by CRISPR/Cas9, confirms deletion of the CD6 ligand. (b) Fluorescent streptavidin‐tetramers of wild‐type (WT) ‐CD6, but not of CD6Δd3, bind to CD166‐expressing Raji cells and not to CD166neg Raji cells. Unbound streptavidin (SA) does not bind to cells. (c) Pre‐incubation of WT‐CD6 tetramers with increasing amounts of CD6d1 (MEM‐98 or itolizumab), d2 (OX126) or d3 (OX124) mAbs shows that only OX124 effectively blocks the CD6–CD166 interaction.
We next investigated which CD6 mAbs would interfere with the binding of WT‐CD6tet to Raji expressing CD166, and for that tested OX124, OX126 and the mAbs MEM‐98 and itolizumab, both binding to CD6d1 but reportedly not effective in interfering with CD6 ligand binding.11, 20 As seen in Fig. 2(c), only OX124 was effective in disrupting CD6 binding to CD166. However, this difference in the blocking capacity was not reflected in any disparities in the responses of E6.1‐WT‐CD6 cells, previously incubated with the mAbs, to antigen‐presenting cells. The presence of any of the CD6 mAbs (specific for d1, d2 or d3) resulted only in slightly decreased levels of IL‐2 production by E6.1‐WT‐CD6 cells interacting with sAg‐pulsed Raji cells (see Supplementary material, Fig. S2). On the other hand, the presence of a CD166 mAb induced an increase in IL‐2 production. These results show that the blocking potential of CD6 mAbs does not necessarily correlate with a particular functional effect on immune responses.
The alternative splicing‐generated CD6Δd3 isoform, but not CD6Δd2, is expressed upon activation
We have previously described the activation‐inducible CD6Δd3 isoform that does not bind to CD166,14, 21 and which is enriched in subjects containing the risk allele for multiple sclerosis.22 This isoform was then discovered not only by analysing the pre‐mRNA, but also by flow cytometry detecting T cells that were labelled with a CD6d1 mAb but were dim for the CD6 mAb, incorrectly named OX126 in the publication.14 The antibody preparation used in that study was traced back to the same material used in Hassan et al.,18 this was the d3 mAb, OX124.19 We re‐analysed the expression of different forms of CD6 on activation and for this stimulated PBLs and monitored the expression of WT‐CD6, CD6Δd3 and other possible isoforms. The proportion of CD6Δd3 was highest at 24 hr, as seen in flow cytometry by a decrease in the binding by OX124 (Fig. 3a,b). This coincided with the peak of cellular activation as reported by CD69 expression (Fig. 3a) and by the highest relative expression of an mRNA isoform lacking exon 5 (encoding d3; Fig. 3c). There was no evidence, however, for the appearance of a CD6Δd2 isoform at the protein or mRNA levels (Fig. 3).
Figure 3.
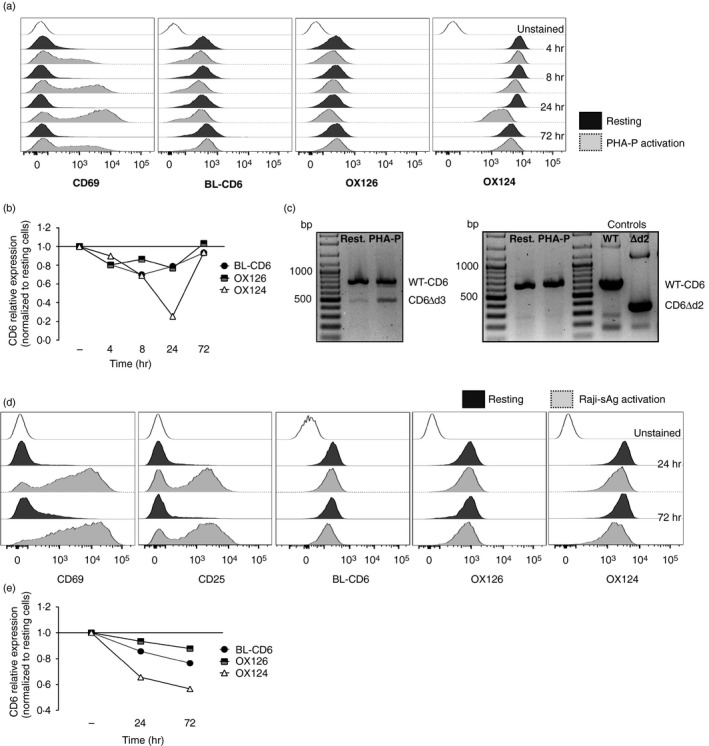
The CD6Δd3 alternative isoform, but not CD6Δd2, is induced upon activation. (a, b) Flow cytometry analysis of resting and phytohaemagglutinin‐P‐activated human peripheral lymphocytes, gated on CD3+ cells, stained with CD69, CD6d1 (BL‐CD6), OX126 and OX124 monoclonal antibodies (mAbs). (a) Expression of the activation marker CD69, and of CD6 domain 1 (d1), d2 and d3 upon stimulation with phytohaemagglutinin‐P for 4, 8, 24 and 72 hr. (b) Graphical representation of the kinetics of expression of CD6 d1, d2 and d3, assessed by normalization of the geometric mean fluorescence intensity of activated to resting cells. The expression of d3 is significantly reduced at 24 hr post activation, coinciding with the peak of CD69 expression shown in (a). (c) mRNA expression of CD6 isoforms in resting and 24 hr‐activated cells corroborates the results of the previous panels, with the increase of a transcript that encodes the CD6Δd3 isoform, which skips the d3‐encoding exon 5. Left panel: polymerase chain reaction amplification using a forward primer on exon 3 (encoding d1) and a reverse primer on exon 7 allowed the identification of a smaller isoform indicative of the skipping of a single domain‐encoding exon. Right panel: complementary polymerase chain reactions were performed to identify which of the exons 4 and 5 is skipped. Using a forward primer on exon 3 (d1) and a reverse primer on exon 5 (d3), transcripts corresponding to wild‐type (WT) ‐CD6, with a predicted size of 660 bp, were obtained from resting and 24‐hr phytohaemagglutinin‐P‐activated human peripheral lymphocytes. However, no mRNA corresponding to the CD6Δd2 isoform, having a predicted size of 348 bp, could be detected. WT‐CD6 and CD6Δd2 cDNAs were used as templates for positive control reactions. (d, e) Flow cytometry analysis and graphical representation of the expression of CD6 d1, d2 and d3 of human peripheral lymphocytes (gated on CD4+ cells) interacting with superantigen‐pulsed Raji cells (Raji‐sAg activation) or with unprimed Raji cells (resting). (d) T cells were stained with mAbs against CD69, CD25 and CD6 d1, d2 and d3 (BL‐CD6, OX126 and OX124, respectively) and the corresponding kinetics normalization (e) was as detailed in (b). Although the decrease of OX124 expression is less pronounced, d3‐mediated splicing is observed at 24 hr and more pronounced at 72 hr.
We further confirmed the development of the CD6Δd3 isoform in PBLs that were allowed to conjugate with sAg‐pulsed Raji cells (Fig. 3d,e). Although the kinetics of splicing were slower and the decrease in the levels of d3 was less pronounced, possibly because fewer T cells are engaged in activation and the stimulation is not as extensive as with phytohaemagglutinin‐P (PHA‐P), we observed a noticeable decrease in the labelling by OX124 that lasted until 72 hr after activation. Again, we found no evidence of diminished OX126 labelling and consequently of the generation of a CD6Δd2 isoform (Fig. 3d,e).
Discussion
Expression of different forms of CD6 has implications for the regulation of immune responses and how they are dysregulated in autoimmunity.14, 22 Characterization of the domain specificity of CD6 mAbs is crucial for distinguishing among different isoforms and there are now available mAbs that recognize each of the three extracellular SRCR domains of CD6. OX124 binds to CD6d3, OX125 and OX126 are non‐competitive mAbs that recognize CD6d2 and most of the other numerous CD6 mAbs bind to CD6d1.20 Our group had described the CD6Δd3 isoform based on the decrease of a CD6d3 mAb binding concomitant with the sustained levels of CD6d1.14 We extended that study using mAbs specific for d1, d2 and d3, and confirmed the induction on activation of CD6Δd3 and found no evidence for other isoforms.
Characterization of CD6 mAbs is important for the development of CD6‐based therapies. The CD6d3 mAb used in our previous study blocked interactions between T cells and CD166‐expressing Raji cells, consistent with OX124 being used in that study.14 A proposed mode of action of the therapeutic antibody, itolizumab, is steric hindrance of CD6–CD166 interactions; definitive characterization of the blocking d3 mAb OX124 is important for development of therapeutic CD6 mAbs. Itolizumab and other CD6d1 mAbs appear not to be effective at blocking CD6–CD166 interactions.20
CD6 is described as promoting cellular adhesion and facilitating T‐cell receptor scanning of antigens presented by antigen‐presenting cell‐expressed major histocompatibility complex, while an intrinsic inhibitory function of the cytoplasmic region may then restrain activation.2, 23 Disrupting the CD6–CD166 interaction without direct binding to CD6, such as that accomplished by CD166 mAbs, may result in diminished cellular adhesion and the absence of CD6‐mediated signalling inhibition, resulting in a higher level of activation (see refs 13, 14 and Supplementary material, Fig. S2). In contrast, the presence of any mAb that binds to CD6 may have the effect of potentiating the inhibitory function of CD6, such as the small decrease in IL‐2 secreted by E6.1‐WT‐CD6 cells interacting with sAg‐pulsed Raji cells that we describe here using mAbs against d1, d2 or d3, or a significant decrease in T‐cell proliferation, as was observed earlier.13 In an antigen‐specific recall response in peripheral blood, perturbation with the d3‐specific blocking antibody was immunosuppressive and the CD6 d2 mAb had no effect, leading to the conclusion that blocking CD6–CD166 interactions in this assay reduced an overall activating effect of CD6.18, 19 An alternative integrative interpretation is that in this case the d3 mAb caused the cumulative effect of acting as an inhibitory agonist while obtaining optimal blockage of the CD6–CD166 induced cell adhesion. These examples serve to illustrate the challenges in interpretation of the functional effects of CD6 mAbs and the requirement for further research.
In conclusion, our study confirms and extends our previous findings,13, 14 provides new data on the specificity and properties of CD6 mAbs, OX124, OX125 and OX126, and facilitates correction of the literature.18, 19
Disclosures
None.
Supporting information
Figure S1. CD166‐positive and CD166‐deficient RAJI cells express comparable levels of relevant surface markers.
Figure S2. Functional effects of CD6 and CD166 monoclonal antibodies on interleukin‐2 production by E6.1 cells conjugated with superantigen‐loaded Raji cells.
Table S1. Primers for CD6 constructs.
Table S2. Specific binding domains of anti‐CD6 antibodies.
Acknowledgements
This work was financed by FEDER – Fundo Europeu de Desenvolvimento Regional funds through COMPETE 2020 – Operacional Programme for Competitiveness and Internationalization (POCI), Portugal 2020, by Portuguese funds through FCT – Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior in the framework of the project POCI‐01‐0145‐FEDER‐032296 (PTDC/MED‐IMU/32296/2017), and by the CIU hybridoma fund, Oxford, UK. RFS is recipient of a PhD studentship from FCT, reference SFRH/BD/110691/2015. We thank Simon Davis for helpful discussions.
References
- 1. Santos RF, Oliveira L, Carmo AM. Tuning T cell activation: the function of CD6 At the immunological synapse and in T cell responses. Curr Drug Targets 2016; 17:630–9. [DOI] [PubMed] [Google Scholar]
- 2. Gonçalves CM, Henriques SN, Santos RF, Carmo AM. CD6, a rheostat‐type signalosome that tunes T cell activation. Front Immunol 2018; 9:2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orta‐Mascaró M, Consuegra‐Fernández M, Carreras E, Roncagalli R, Carreras‐Sureda A, Alvarez P et al CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med 2016; 213:1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeJager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT et al Meta‐analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009; 41:776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pinto M, Carmo AM. CD6 as a therapeutic target in autoimmune diseases: successes and challenges. BioDrugs 2013; 27:191–202. [DOI] [PubMed] [Google Scholar]
- 6. Li Y, Singer NG, Whitbred J, Bowen MA, Fox DA, Lin F. CD6 as a potential target for treating multiple sclerosis. Proc Natl Acad Sci USA 2017; 114:2687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez PC, Torres‐Moya R, Reyes G, Molinero C, Prada D, Lopez AM et al A clinical exploratory study with itolizumab, an anti‐CD6 monoclonal antibody, in patients with rheumatoid arthritis. Results Immunol 2012; 2:204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krupashankar DS, Dogra S, Kura M, Saraswat A, Budamakuntla L, Sumathy TK et al Efficacy and safety of itolizumab, a novel anti‐CD6 monoclonal antibody, in patients with moderate to severe chronic plaque psoriasis: results of a double‐blind, randomized, placebo‐controlled, phase‐III study. J Am Acad Dermatol 2014; 71:484–92. [DOI] [PubMed] [Google Scholar]
- 9. Aira LE, López‐Requena A, Fuentes D, Sánchez L, Pérez T, Urquiza A et al Immunological and histological evaluation of clinical samples from psoriasis patients treated with anti‐CD6 itolizumab. MAbs 2014; 6:783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowen MA, Bajorath J, Siadak AW, Modrell B, Malacko AR, Marquardt H et al The amino‐terminal immunoglobulin‐like domain of activated leukocyte cell adhesion molecule binds specifically to the membrane‐proximal scavenger receptor cysteine‐rich domain of CD6 with a 1:1 stoichiometry. J Biol Chem 1996; 271:17390–6. [DOI] [PubMed] [Google Scholar]
- 11. Chappell PE, Garner LI, Yan J, Metcalfe C, Hatherley D, Johnson S et al Structures of CD6 and its ligand CD166 give insight into their interaction. Structure 2015; 23:1426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bughani U, Saha A, Kuriakose A, Nair R, Sadashivarao RB, Venkataraman R et al T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS One 2017; 12:e0180088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oliveira MI, Gonçalves CM, Pinto M, Fabre S, Santos AM, Lee SF et al CD6 attenuates early and late signaling events, setting thresholds for T‐cell activation. Eur J Immunol 2012; 42:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castro M, Oliveira M, Nunes R, Fabre S, Barbosa R, Peixoto A et al Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J Immunol 2007; 178:4351–61. [DOI] [PubMed] [Google Scholar]
- 15. Bamberger M, Santos AM, Gonçalves CM, Oliveira MI, James JR, Moreira A et al A new pathway of CD5 glycoprotein‐mediated T cell inhibition dependent on inhibitory phosphorylation of Fyn kinase. J Biol Chem 2011; 286:30324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felce JH, Sezgin E, Wane M, Brouwer H, Dustin ML, Eggeling C et al CD45 exclusion‐ and cross‐linking‐based receptor signaling together broaden FcεRI reactivity. Sci Signal 2018; 11:eaat0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome‐wide libraries for CRISPR screening. Nat Methods 2014; 11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassan NJ, Simmonds SJ, Clarkson NG, Hanrahan S, Puklavec MJ, Bomb M et al CD6 regulates T‐cell responses through activation‐dependent recruitment of the positive regulator SLP‐76. Mol Cell Biol 2006; 26:6727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hassan NJ, Simmonds SJ, Clarkson NG, Hanrahan S, Puklavec MJ, Bomb M et al Second correction for Hassan et al., “CD6 regulates T‐cell responses through activation‐dependent recruitment of the positive regulator SLP‐76”. Mol Cell Biol 2019; 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garner LI, Hartland A, Breuning J, Brown MH. CD6 monoclonal antibodies differ in epitope, kinetics and mechanism of action. Immunology 2018; 155:273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. da Gloria VG, de Araujo MM, Santos AM, Leal R, de Almeida SF, Carmo AM et al T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J Immunol 2014; 193:391–9. [DOI] [PubMed] [Google Scholar]
- 22. Kofler DM, Severson CA, Mousissian N, De Jager PL, Hafler DA. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell proliferation. J Immunol 2011; 187:3286–91. [DOI] [PubMed] [Google Scholar]
- 23. Breuning J, Brown MH. A sequence conserved between CD5 and CD6 binds an FERM domain and exerts a restraint on T‐cell activation. Immunology 2019; 156:270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CD166‐positive and CD166‐deficient RAJI cells express comparable levels of relevant surface markers.
Figure S2. Functional effects of CD6 and CD166 monoclonal antibodies on interleukin‐2 production by E6.1 cells conjugated with superantigen‐loaded Raji cells.
Table S1. Primers for CD6 constructs.
Table S2. Specific binding domains of anti‐CD6 antibodies.


