Abstract
Despite being widely used in reporter technologies, bioluminescent systems are largely understudied. Of at least forty different bioluminescent systems thought to exist in nature, molecular components of only seven light-emitting reactions are known, and the full biochemical pathway leading to light emission is only understood for two of them. Here, we provide a succinct overview of currently known bioluminescent systems highlighting available tools for research and discussing future applications.
Keywords: Bioluminescence, Luciferin, Luciferase, Synthetic biology, Imaging
Introduction
Having evolved independently dozens of times, bioluminescence provides living organisms with a tangible advantage in certain ecological contexts. The ability to emit light in darkness has been observed in about 10,000 species from 800 genera, although this may well be an underestimation (Haddock et al. 2010). The exact benefit of light emission in various environments is far from being clear for a number of species, however, in most cases bioluminescence is thought to serve the purpose of visual communication to scare off predators, attract prey or in courtship behaviour (Ellis and Oakley 2016; Wainwright and Longo 2017; Verdes and Gruber 2017; Labella et al. 2017).
Evolution has stumbled upon and fixed numerous biochemical solutions for bioluminescence demonstrating that the ability to glow is accessible to living organisms in various points of genotype space, from bacteria to fungi and animals. Various luciferins, the small molecules prone to light emission upon oxidation, have been derived by evolution from unrelated biochemical pathways. Oxidation of these molecules is catalysed by non-homologous enzymes, luciferases, to create a palette of light-emitting reactions that are different in colour, catalysis rate, cellular localisation and dependence on ATP, NADH and other metabolites (Kaskova et al. 2016).
Although still largely understudied on a molecular level, hardly could such diversity of reactions with an easily measurable output escape becoming an essential part of modern reporter technologies. Luminescent reactions, where structures of both luciferin and luciferases have been discovered, are now utilised in vitro and in vivo in food testing (Shama and Malik 2013), environmental monitoring (Girotti et al. 2008), diagnostics (Frank and Krasitskaya 2014), drug screenings (Hasson et al. 2015; Kobayashi et al. 2010; Lampinen et al. 1995), and various kinds of biomedical research. Detailed reviews on chemistry and diversity of luciferins (Kaskova et al. 2016), luciferases (Kotlobay et al. 2019), and ecology of bioluminescence (Haddock et al. 2010; Widder 2010), as well as a comprehensive overview of all known bioluminescent systems (Shimomura and Yampolsky 2019), are available. In this article, we provide an intentionally succinct overview of light-emitting reactions where both luciferin and luciferase are known, highlighting their main features for practical applications.
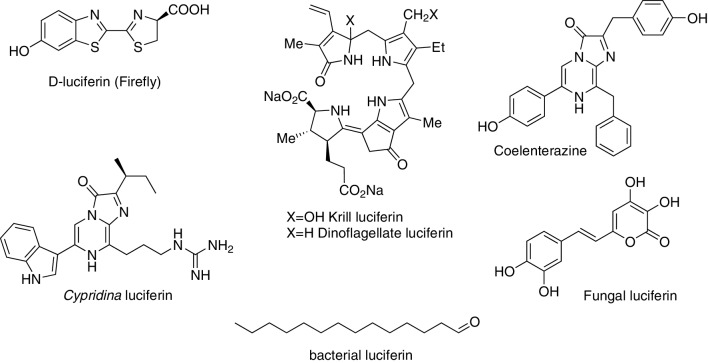
We will group bioluminescent systems by structures of their luciferins, as these compounds are the principal determinants of colour and properties of light-emitting reactions. Of at least forty bioluminescent systems thought to exist in nature (Haddock et al. 2010), structures of only nine luciferins are known, for seven of which at least a single luciferase gene has been discovered (Fig. 1).
Fig. 1.
Structure of luciferins from bioluminescent systems with known luciferases
Main text
Coelenterazine-dependent systems
The largest diversity of bioluminescent organisms is found in marine ecosystems. In the sea, it is often the same luciferin, coelenterazine, that serves as a substrate for numerous independently evolved luciferases in phylogenetically distant groups of organisms. Coelenterazine is a modified tripeptide produced from one phenylalanine and two tyrosine residues, however, the genes involved in its biosynthesis, as well as the exact biosynthetic route, are currently unknown. Most of marine organisms do not synthesise coelenterazine themselves, instead, they obtain it from food—the likely reason for a remarkable convergent evolution of bioluminescence in marine ecosystems. All coelenterazine-dependent systems from nature emit blue light, with emission maxima within the 450–500 nm range, and do not typically require any cofactors except for oxygen. In some cases, the colour of bioluminescence is altered by a fluorescent protein that interacts with the luciferase. Other characteristics such as molecular weight, pH-sensitivity, thermostability and catalysis rates of luciferases vary dramatically among coelenterazine-dependent systems. Below we highlight several practically important luciferases that utilize coelenterazine or its analogues.
Renilla luciferase: a medium-sized (36 kDa) cytosolic protein from a coral that produces a steady luminescent signal. Early discovery (Lorenz et al. 1991), as well as the availability of engineered versions with increased brightness and red-shifted spectra (Markova and Vysotski 2015), made this system popular for biomedical applications, in particular, in bioimaging and drug-screening (Hamdan et al. 2005; Prinz et al. 2006).
Gaussia luciferase: a small (20 kDa) secreted protein produced by a small crustacean from the group Copepoda, with high catalysis rate and exceptional thermostability. The activity of the protein depends on the formation of disulfide bonds making it unsuitable for certain heterologous systems. The signal of Gaussia luciferase scales linearly with the number of cells being assayed (Tannous et al. 2005; Chung et al. 2009) making this system useful for monitoring tumor progression and drug response.
Nanoluc luciferase: an engineered variant of a luciferase from the shrimp Oplophorus gracilirostris. This small (19 kDa) protein utilises a cell-permeable coelenterazine analogue, lacks disulfide bonds, and produces a bright signal suitable for a broad range of applications. Fusions with fluorescent proteins result in bright engineered bioluminescent constructs with red-shifted spectra facilitating single-cell and whole-body bioluminescent imaging in vivo (Saito et al. 2012). One of the drawbacks of this system is the high cost of reagents.
Cypridina luciferin-based system
Another modified tripeptide emitting blue light is cypridina luciferin, a metabolite found in the ostracod Cypridina and bioluminescent midshipman fish, Porichthys. Although the biosynthesis of this compound is also unknown, it was shown to be produced from tryptophan, isoleucine, and arginine (Oba et al. 2002). Among other applications, the Cypridina system has been widely used in bioimaging, in studies of circadian rhythms (Yamada et al. 2013; Watanabe et al. 2010; Noguchi et al. 2012), and in immunoassays (Miesenböck and Rothman 1997; Wu et al. 2007, 2009).
D-luciferin-dependent systems
Another practically important group of bioluminescent reactions, the most thoroughly studied one, has evolved in several lineages of beetles, including fireflies, click beetles and railroad worms. These reactions utilise a stable and nontoxic compound (Deluca 1976; Tiffen et al. 2010) known as d-luciferin to emit yellow, orange and in some cases red light, and represent another intriguing example of an independent origin of the same bioluminescent system (Fallon et al. 2018). Most luciferases oxidizing D-luciferin are proteins of ~ 60 kDa that depend on ATP and Mg2+ to catalyze a reaction similar to that of aminoacyl-tRNA synthetases and acetyl-CoA ligases. Dependence of light emission on the concentration of ATP has allowed to use the system as an ATP indicator in a variety of applications ranging from studies of cancer metabolism (Patergnani et al. 2014) to monitoring bacterial contaminants in water (Frundzhyan and Ugarova 2007) and diagnostics based on levels of ATP in blood (Abraham et al. 2003). Practically important enzymes include:
Firefly luciferase: very popular as a reporter molecule due to the early discovery, high quantum yield of bioluminescence, availability of thermostable mutant variants with enhanced spectral characteristics and ease of production in bacteria. Firefly luciferase has been extensively used in various in vitro and in vivo systems to detect pathogenic bacteria (Abe et al. 2012; Nakamura et al. 2011) and viruses (Zammatteo et al. 1995; Minekawa et al. 2013), to quantify protein–protein (Arai et al. 2002; Hattori et al. 2013; Radeck et al. 2017) and protein–ligand (Shekhawat and Ghosh 2011) interactions, and to assay metabolites involved in cell communication and cell signaling (Luker et al. 2008), among other applications.
Click beetle luciferases: the second most popular group of D-luciferin-dependent luciferases is derived from the click beetle Pyrophorus plagiophthalamus. This species emits light using four types of luciferases with emission maxima ranging from green (540 nm) to orange-red (593 nm). The color variability, tolerance to a wide range of pH conditions and the availability of engineered variants make click beetle luciferases attractive for numerous applications (Hall et al. 2018). Engineered variants are available commercially, for example, Chroma-Luc luciferases offered by Promega.
Tetrapyrrole-based luciferins
Dinoflagellates (protists) and Euphausiids (krill) utilise two very similar tetrapyrrole-based luciferins and form another large group of bioluminescent species. In dinoflagellates, bioluminescence is localised in special organelles, scintillons, and occurs as flashes of light triggered by electrical or mechanical stimulation. It is thought to serve as a defense mechanism making their attackers visible and attracting the attention of predators from higher trophic levels (Haddock et al. 2010). In Euphausiids, light emission occurs in complex organs with specialized lenses and ability to focus, yet the exact ecological role of this adaptation is unclear (Johnsen 2005). Luciferases from Dinoflagellates and Euphausiids are rarely used as tools in research due to the unavailability of synthetic luciferin.
Bacterial bioluminescent system
All bioluminescent bacteria utilise the same unique mechanism for light emission, where photons are produced in a set of reactions requiring flavin mononucleotide (FMN), myristic aldehyde, oxygen and nicotinamide adenine dinucleotide (NADH). In the course of reactions, myristic aldehyde is oxidised and is thus known as luciferin, although the true light source in bacterial bioluminescence is the FMN derivative. Bacterial luciferases consist of two polypeptide chains that form a complex (75 kDa) and are encoded in the lux operon together with enzymes catalyzing luciferin biosynthesis. In most cases, bioluminescence is blue (~ 490 nm), however, both natural (Daubner et al. 1987) and engineered (Ke and Tu 2011) red-shifted versions of the bacterial system exist.
The full pathway of luciferin biosynthesis has been known since late 80 s making the lux operon the only genetically encodable bioluminescent system available in the last three decades (Meighen 1991). This allowed to use the system to engineer autonomously glowing organisms, including other bacteria (Belas et al. 1982; Francis et al. 2000), yeasts (Gupta et al. 2003), mammalian cell lines (Patterson et al. 2005), plants (Krichevsky et al. 2010) and others. However, no brightly bioluminescent multicellular organisms have been created, perhaps due to toxicity or inefficiency of the system in eukaryotes (Hollis et al. 2001). Consequently, in most applications of the system, living bacteria are utilised as a light source. Among the main applications of the system are the studies of antimicrobial drugs, bacterial infections and environmental monitoring (Björkman and Karl 2001). The brightest version developed to date is iLux (Gregor et al. 2018).
Fungal bioluminescent system
In 2018, a biochemical pathway generating bioluminescence in fungi has been described in its entirety, providing the first genetically encodable pathway from eukaryotes (Kotlobay et al. 2018). Fungi utilise a simple α-pyrone 3-hydroxyhispidin that is oxidised by an insoluble luciferase in a reaction that only requires oxygen and results in the emission of green light (~ 520 nm). The wild-type Neonothopanus nambi luciferase, nnLuz, is functional in a variety of heterologous systems, with the performance similar to that of the firefly luciferase (Kotlobay et al. 2018), (Sarkisyan, unpublished data). It has been shown that the expression of as few as three genes from the fungal bioluminescent system is sufficient to engineer other glowing eukaryotes.
Final remarks
With no bioluminescent system suitable for any task and application, different light-emitting reactions occupy different niches in modern reporter technologies. In bioimaging, where applications of bioluminescence and fluorescence-based approaches overlap, the former is used in experiments that require high dynamic range, low background or deep-tissue imaging. Toxicity assays and studies of bacterial biology are typically based on the bacterial bioluminescent system while drug screenings often employ D-luciferin-dependent or coelenterazine-dependent systems. When selecting a luciferin–luciferase pair for a particular application, several criteria have to be taken into account including thermostability, pH optimum, protein size, cellular or extracellular location, aggregation properties, emission wavelength, intensity, rate of the reaction or dependence on ATP and other cofactors.
A recent discovery of a eukaryote-friendly genetically encodable pathway in fungi may stimulate the development of new bioluminescence-based technologies that would not require addition of the substrate. Expression of the fungal bioluminescent system may result in the generation of autonomously glowing animals and plants where light emission would be used to visualise development, report physiological changes, signal progression of pathological states, or simply serve esthetic purposes (Reeve et al. 2014; Kotlobay et al. 2018; Landau et al. 2009). We also envision that the discovery of the fungal pathway has the potential to bring autonomous bioluminescence beyond the use in reporter technologies towards engineering of light-based communications between cells, organisms or living and non-living systems.
The potential of bioluminescence-based tools in synthetic biology has only been marginally explored. Given the practical importance of light-emitting reactions, general appeal of glowing organisms and the scope of available methods in organic chemistry, metabolomics and genetics (Garrido-Cardenas and Manzano-Agugliaro 2017; Kaskova et al. 2016), the field of bioluminescence is surprisingly understudied. At the same time, with new insights into the photophysics, genetics and ecology of bioluminescence being made every year, engineering new light-emitting and light-communicating living systems is becoming more accessible than ever before.
Acknowledgements
This study was supported by the Russian Science Foundation project No. 17-74-10066. The authors thank Ilia Yampolsky for the feedback on the manuscript and Alexandra Tsarkova for the help with the preparation of the figure. The Synthetic biology Group is funded by the MRC London Institute of Medical Sciences (UKRI MC-A658-5QEA0).
Compliance with ethical standards
Conflict of interest
KSS is a co-founder of Planta, a company aimed to commercialize fungal bioluminescent system.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe K, Kumagai T, Takahashi C, Kezuka A, Murakami Y, Osawa Y, Motoki H, et al. Detection of pathogenic bacteria by using zinc finger protein fused with firefly luciferase. Anal Chem. 2012;84(18):8028–8032. doi: 10.1021/ac3018845. [DOI] [PubMed] [Google Scholar]
- Abraham EH, Salikhova AY, Hug EB. Critical ATP Parameters associated with blood and mammalian cells: relevant measurement techniques. Drug Dev Res. 2003;59(1):152–160. [Google Scholar]
- Arai R, Nakagawa H, Kitayama A, Ueda H, Nagamune T. Detection of protein–protein interaction by bioluminescence resonance energy transfer from firefly luciferase to red fluorescent protein. J Biosci Bioeng. 2002;94(4):362–364. doi: 10.1263/jbb.94.362. [DOI] [PubMed] [Google Scholar]
- Belas R, Mileham A, Cohn D, Hilman M, Simon M, Silverman M. Bacterial Bioluminescence: isolation and expression of the luciferase genes from Vibrio harveyi. Science. 1982;218(4574):791–793. doi: 10.1126/science.10636771. [DOI] [PubMed] [Google Scholar]
- Björkman KM, Karl DM. Novel method for the measurement of dissolved adenosine and guanosine triphosphate in aquatic habitats: applications to marine microbial ecology. J Microbiol Methods. 2001;47(2):159–167. doi: 10.1016/s0167-7012(01)00301-3. [DOI] [PubMed] [Google Scholar]
- Chung E, Yamashita H, Au P, Tannous BA, Fukumura D, Jain RK. Secreted Gaussia luciferase as a biomarker for monitoring tumor progression and treatment response of systemic metastases. PloS One. 2009;4(12):e8316. doi: 10.1371/journal.pone.0008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner SC, Astorga AM, Leisman GB, Baldwin TO. Yellow light emission of vibrio fischeri strain Y-1: purification and characterization of the energy-accepting yellow fluorescent protein. Proc Natl Acad Sci USA. 1987;84(24):8912–8916. doi: 10.1073/pnas.84.24.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca M. Firefly luciferase. Adv Enzymol Relat Areas Mol Biol. 1976;44:37–68. doi: 10.1002/9780470122891.ch2. [DOI] [PubMed] [Google Scholar]
- Ellis EA, Oakley TH. High rates of species accumulation in animals with bioluminescent courtship displays. Current Biology: CB. 2016;26(14):1916–1921. doi: 10.1016/j.cub.2016.05.043. [DOI] [PubMed] [Google Scholar]
- Fallon TR, Lower SE, Chang CH, Bessho-Uehara M, Martin GJ, Bewick AJ, Behringer M, et al. Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife. 2018 doi: 10.7554/eLife.36495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KP, John D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. Monitoring bioluminescent staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun. 2000;68(6):3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LA, Krasitskaya VV. Application of enzyme bioluminescence for medical diagnostics. Adv Biochem Eng/Biotechnol. 2014;144:175–197. doi: 10.1007/978-3-662-43385-0_6. [DOI] [PubMed] [Google Scholar]
- Frundzhyan V, Ugarova N. Bioluminescent assay of total bacterial contamination of drinking water. Lumin J Biol Chem Lumin. 2007;22(3):241–244. doi: 10.1002/bio.957. [DOI] [PubMed] [Google Scholar]
- Garrido-Cardenas JA, Manzano-Agugliaro F. The metagenomics worldwide research. Curr Genet. 2017;63(5):819–829. doi: 10.1007/s00294-017-0693-8. [DOI] [PubMed] [Google Scholar]
- Girotti S, Ferri EN, Fumo MG, Maiolini E. Monitoring of environmental pollutants by bioluminescent bacteria. Anal Chim Acta. 2008;608(1):2–29. doi: 10.1016/j.aca.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Gregor C, Gwosch KC, Sahl SJ, Hell SW. Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc Natl Acad Sci USA. 2018;115(5):962–967. doi: 10.1073/pnas.1715946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Patterson SS, Ripp S, Simpson ML, Sayler GS. Expression of the photorhabdus luminescens lux genes (luxA, B, C, D, and E) in Saccharomyces cerevisiae. FEMS Yeast Res. 2003;4(3):305–313. doi: 10.1016/S1567-1356(03)00174-0. [DOI] [PubMed] [Google Scholar]
- Haddock SHD, Moline MA, Case JF. Bioluminescence in the sea. Annu Rev Mar Sci. 2010;2:443–493. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- Hall MP, Woodroofe CC, Wood MG, Que I, Root MV, Ridwan Y, Shi C, et al. Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nature Commun. 2018;9(1):132. doi: 10.1038/s41467-017-02542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Audet M, Garneau P, Pelletier J, Bouvier M. High-throughput screening of G protein-coupled receptor antagonists using a bioluminescence resonance energy transfer 1-based β-arrestin2 recruitment assay. J Biomol Screen. 2005;10(5):463–475. doi: 10.1177/1087057105275344. [DOI] [PubMed] [Google Scholar]
- Hasson SA, Fogel AI, Wang C, MacArthur R, Guha R, Heman-Ackah S, Martin S, Youle RJ, Inglese J. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem Biol. 2015;10(5):1188–1197. doi: 10.1021/cb5010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Tanaka M, Takakura H, Aoki K, Miura K, Anzai T, Ozawa T. Analysis of temporal patterns of GPCR–β-arrestin interactions using split luciferase-fragment complementation. Mol bioSyst. 2013;9(5):957–964. doi: 10.1039/c2mb25443c. [DOI] [PubMed] [Google Scholar]
- Hollis RP, Lagido C, Pettitt J, Porter AJ, Killham K, Paton GI, Glover LA. Toxicity of the bacterial luciferase substrate, N-decyl aldehyde, to Saccharomyces cerevisiae and Caenorhabditis elegans. FEBS Lett. 2001;506(2):140–142. doi: 10.1016/s0014-5793(01)02905-2. [DOI] [PubMed] [Google Scholar]
- Johnsen S. The red and the black: bioluminescence and the color of animals in the deep sea. Integr Comp Biol. 2005;45(2):234–246. doi: 10.1093/icb/45.2.234. [DOI] [PubMed] [Google Scholar]
- Kaskova ZM, Tsarkova AS, Yampolsky IV. 1001 Lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem Soc Rev. 2016 doi: 10.1039/c6cs00296j. [DOI] [PubMed] [Google Scholar]
- Ke D, Tu SC. Activities, kinetics and emission spectra of bacterial luciferase-fluorescent protein fusion enzymes. Photochem Photobiol. 2011;87(6):1346–1353. doi: 10.1111/j.1751-1097.2011.01001.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Murakami T, Uchibori R, Chun NAL, Kobayashi E, Morita T, Ozawa K. Establishment and characterization of transplantable, luminescence labeled rat renal cell carcinoma cell lines. J Urol. 2010;183(5):2029–2035. doi: 10.1016/j.juro.2009.12.090. [DOI] [PubMed] [Google Scholar]
- Kotlobay AA, Sarkisyan KS, Mokrushina YA, Marcet-Houben M, Serebrovskaya EO, Markina NM, Somermeyer LG, et al. genetically encodable bioluminescent system from fungi. Proc Natl Acad Sci USA. 2018;115(50):12728–12732. doi: 10.1073/pnas.1803615115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlobay AA et al (2019) Natural luciferases: properties and prospects of applications in biomedical research. Acta Nat (under revision)
- Krichevsky A, Meyers B, Vainstein A, Maliga P, Citovsky V. Autoluminesc plants. PloS One. 2010;5(11):e15461. doi: 10.1371/journal.pone.0015461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella AM, Arahal DR, Castro D, Lemos ML, Borrego JJ. Revisiting the genus photobacterium: taxonomy, ecology and pathogenesis. Int Microb. 2017;20(1):1–10. doi: 10.2436/20.1501.01.280. [DOI] [PubMed] [Google Scholar]
- Lampinen J, Virta M, Karp M. Use of controlled luciferase expression to monitor chemicals affecting protein synthesis. Appl Environ Microbiol. 1995;61(8):2981–2989. doi: 10.1128/aem.61.8.2981-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau J, Cameron J, Worthington SS (2009) Avatar (film). 20th Century Fox
- Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and Expression of a cDNA Encoding Renilla Reniformis Luciferase. Proc Natl Acad Sci USA. 1991;88(10):4438–4442. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Gupta M, Luker GD. Imaging CXCR4 Signaling with firefly luciferase complementation. Anal Chem. 2008;80(14):5565–5573. doi: 10.1021/ac8005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova SV, Vysotski ES. Coelenterazine-dependent luciferases. Biochem Biokhimiia. 2015;80(6):714–732. doi: 10.1134/S0006297915060073. [DOI] [PubMed] [Google Scholar]
- Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55(1):123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G, Rothman JE. Patterns of synaptic activity in neural networks recorded by light emission from synaptolucins. Proc Natl Acad Sci USA. 1997;94(7):3402–3407. doi: 10.1073/pnas.94.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minekawa T, Takehara S, Takahashi M, Okamoto H. Development of a highly sensitive bioluminescent enzyme immunoassay for hepatitis B virus surface antigen capable of detecting divergent mutants. Clin Vaccine Immunol CVI. 2013;20(8):1255–1265. doi: 10.1128/CVI.00186-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Mie M, Kobatake E. Construction of a functional IgG-Binding luciferase fusion protein for the rapid detection of specific bacterial strains. Analyst. 2011;136(1):71–72. doi: 10.1039/c0an00460j. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y. A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PloS One. 2012;7(5):e37093. doi: 10.1371/journal.pone.0037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba Y, Kato SI, Ojika M, Inouye S. Biosynthesis of luciferin in the sea firefly, cypridina hilgendorfii: l-tryptophan is a component in cypridina luciferin. Tetrahedron Lett. 2002;43(13):2389–2392. [Google Scholar]
- Patergnani S, Baldassari F, Marchi ED, Karkucinska-Wieckowska A, Wieckowski MR, Pinton P. Methods to monitor and compare mitochondrial and glycolytic ATP production. Methods Enzymol. 2014;542:313–332. doi: 10.1016/B978-0-12-416618-9.00016-9. [DOI] [PubMed] [Google Scholar]
- Patterson SS, Dionisi HM, Gupta RK, Sayler GS. Codon optimization of bacterial luciferase (lux) for expression in mammalian cells. J Ind Microbiol Biotechnol. 2005;32(3):115–123. doi: 10.1007/s10295-005-0211-8. [DOI] [PubMed] [Google Scholar]
- Prinz A, Diskar M, Herberg FW. Application of bioluminescence resonance energy transfer (BRET) for biomolecular interaction studies. Chembiochem Eur J Chem Biol. 2006;7(7):1007–1012. doi: 10.1002/cbic.200600048. [DOI] [PubMed] [Google Scholar]
- Radeck J, Fritz G, Mascher T. The cell envelope stress response of bacillus subtilis: from static signaling devices to dynamic regulatory network. Curr Genet. 2017;63(1):79–90. doi: 10.1007/s00294-016-0624-0. [DOI] [PubMed] [Google Scholar]
- Reeve B, Sanderson T, Ellis T, Freemont P. How synthetic biology will reconsider natural bioluminescence and its applications. Adv Biochem Eng/Biotechnol. 2014;145:3–30. doi: 10.1007/978-3-662-43619-6_1. [DOI] [PubMed] [Google Scholar]
- Saito K, Chang YF, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, Yoshida Y, Matsuda T, Arai Y, Nagai T. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shama G, Malik DJ. The Uses and abuses of rapid bioluminescence-based ATP assays. Int J Hyg Environ Health. 2013;216(2):115–125. doi: 10.1016/j.ijheh.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein–protein interactions. Curr Opin Chem Biol. 2011;15(6):789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O, Yampolsky IV. Bioluminescence, chemical principles and methods. 3. Hackensack: World Scientific Publishing Co Pte Ltd; 2019. [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Therapy J Am Soc Gene Therapy. 2005;11(3):435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Tiffen JC, Bailey CG, Ng C, Rasko JEJ, Holst J. Luciferase expression and bioluminescence does not affect tumor cell growth in vitro or in vivo. Mol Cancer. 2010;9:299. doi: 10.1186/1476-4598-9-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdes A, Gruber DF. Glowing worms: biological, chemical, and functional diversity of bioluminescent annelids. Integr Comp Biol. 2017 doi: 10.1093/icb/icx017. [DOI] [PubMed] [Google Scholar]
- Wainwright PC, Longo SJ. Functional innovations and the conquest of the oceans by acanthomorph fishes. Curr Biol. 2017;27(11):R550–R557. doi: 10.1016/j.cub.2017.03.044. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Enomoto T, Takahashi M, Honma S, Honma KI, Ohmiya Y. Multichannel perfusion culture bioluminescence reporter system for long-term detection in living cells. Anal Biochem. 2010;402(1):107–109. doi: 10.1016/j.ab.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Widder EA. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science. 2010;328(5979):704–708. doi: 10.1126/science.1174269. [DOI] [PubMed] [Google Scholar]
- Wu C, Kawasaki K, Ogawa Y, Yoshida Y, Ohgiya S, Ohmiya Y. Preparation of biotinylated cypridina luciferase and its use in bioluminescent enzyme immunoassay. Anal Chem. 2007;79(4):1634–1638. doi: 10.1021/ac061754k. [DOI] [PubMed] [Google Scholar]
- Wu C, Irie S, Yamamoto S, Ohmiya Y. A Bioluminescent enzyme immunoassay for prostaglandin E(2) using cypridina luciferase. Lumin J Biol Chem Lumin. 2009;24(2):131–133. doi: 10.1002/bio.1086. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Nishide SY, Nakajima Y, Watanabe T, Ohmiya Y, Honma KI, Honma S. Monitoring Circadian time in rat plasma using a secreted cypridina luciferase reporter. Anal Biochem. 2013;439(2):80–87. doi: 10.1016/j.ab.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Zammatteo N, Moris P, Alexandre I, Vaira D, Piette J, Remacle J. DNA Probe hybridisation in microwells using a new bioluminescent system for the detection of PCR-amplified HIV-1 proviral DNA. J Virol Methods. 1995;55(2):185–197. doi: 10.1016/0166-0934(95)00050-5. [DOI] [PubMed] [Google Scholar]