Abstract
Phytophthora capsici (Leon.) is a globally prevalent, devastating oomycete pathogen that causes root rot in pepper (Capsicum annuum). Several studies have identified quantitative trait loci (QTL) underlying resistance to P. capsici root rot (PcRR). However, breeding for pepper cultivars resistant to PcRR remains challenging due to the complexity of PcRR resistance. Here, we combined traditional QTL mapping with GWAS to broaden our understanding of PcRR resistance in pepper. Three major-effect loci (5.1, 5.2, and 5.3) conferring broad-spectrum resistance to three isolates of P. capsici were mapped to pepper chromosome P5. In addition, QTLs with epistatic interactions and minor effects specific to isolate and environment were detected on other chromosomes. GWAS detected 117 significant SNPs across the genome associated with PcRR resistance, including SNPs on chromosomes P5, P7, and P11 that colocalized with the QTLs identified here and in previous studies. Clusters of candidate nucleotide-binding site-leucine-rich repeat (NBS-LRR) and receptor-like kinase (RLK) genes were predicted within the QTL and GWAS regions; such genes often function in disease resistance. These candidate genes lay the foundation for the molecular dissection of PcRR resistance. SNP markers associated with QTLs for PcRR resistance will be useful for marker-assisted breeding and genomic selection in pepper breeding.
Subject terms: Plant hybridization, Plant breeding
Introduction
Pepper (Capsicum annuum) is an economically important horticultural crop grown in tropical and temperate regions that is used as a fresh vegetable and as a processed food product. Phytophthora capsici (Leon.) is an invasive oomycete that poses a serious threat to pepper production across the globe1. P. capsici can infect pepper plants at any growth and developmental stage, causing damping off, root rot, stem rot, collar rot, fruit rot, and foliar blight. P. capsici root rot (PcRR) is the most devastating disease, causing up to 100% yield losses under warm (25–28 °C), humid environmental conditions2. The pathogen may enter the plant system through the roots or stem collar, causing water-soaked lesion formation and stem girdling, eventually leading to plant wilting and death. The broad range of host species of P. capsici, including Solanaceae, Fabaceae, and Cucurbitaceae, as well as its soil-born, random mating nature, makes it quite difficult to control PcRR. Cultural practices and chemical control measures for PcRR have proven to be ineffective and unsafe3. The use of cultivars resistant to PcRR represents the best control measure due to its eco-friendliness and cost-effectiveness.
Resistance to PcRR is influenced by several factors, including environmental cues, the virulence of the P. capsici isolates, their physiological races, and the source of resistance2,4. P. capsici isolates, with their short life cycles and rapid evolution of virulence, have a selective advantage over their slowly evolving plant hosts in the evolutionary arms race5,6. More than 45 physiological races have been described within the Phytophthora for each host plant species7. Three physiological races of P. capsici were identified in four pepper cultivars in Taiwan7, and recently, 24 new races were identified using recombinant inbred lines8. The virulence of P. capsici isolates is another factor that may affect the genetic analysis of resistance mechanisms. The presence of isolates with low virulence could lead to false resistance, whereas highly virulent isolates could overcome the resistance of plant lines9. Several resistance sources have been reported, including C. annuum lines CM334, PI201232, PI201234, AC2258, Perennial, and YCM3344,9,10. These resistant sources have been reported with varying levels of resistance to each P. capsici isolate11. Among these sources, CM334 has the highest resistance and has been extensively utilized in breeding programs12. Nevertheless, breeding for PcRR resistance is a complex process, and the resistance levels of commercial pepper cultivars are not comparable to those of the original resistance sources2,11. Therefore, the genetic improvement of pepper for increased resistance to PcRR is still an important objective of most pepper breeding programs.
In Solanaceae, resistance to Phytophthora is race specific and quantitative in nature13–15. The colocalization of single resistance genes and QTLs for resistance to Phytophthora has been reported in various Solanaceous species, indicating that homologous resistance genes control resistance to Phytophthora16. Two race-specific resistance genes have been reported in pepper: CaPhyto from PI201234 and PhR10 from CM334 on chromosomes P5 and P10, respectively9,17. In addition, major and minor resistance QTLs to PcRR have been mapped onto various chromosomes11,18,19. Major resistance QTLs to PcRR have been consistently identified in close proximity on chromosome P5 in various studies irrespective of the resistance sources or P. capsici isolates4,10,18,20,21. Based on meta-analysis of QTLs, the major effect QTL, MetaPc5.1 was localized to the 22.4–24.6 Mb region on chromosome P5, and MetaPc5.2 and MetaPc5.3 were localized to the 53.0–162.6 Mb and 9.7–13.3 Mb regions on chromosome P5, respectively9,21. Several minor and isolate-specific QTLs have been reported, but their positions are variable, depending on the genetic background and P. capsici isolate4,19,22. Several molecular markers linked to the major QTLs, such as Phyto5NBS1 and ZL6726, have been developed and utilized for marker-assisted selection (MAS) in pepper, but their use is limited to the selection of resistant plants to highly virulent and race nonspecific P. capsici isolates9,10.
Ultra-high-density linkage maps play a crucial role in genetic and genomics analyses. The emergence of next-generation sequencing (NGS) technologies and high-throughput genotyping tools has facilitated the rapid discovery of SNP markers. Among the numerous NGS technologies, genotyping by sequencing (GBS) is a simple, efficient, rapid approach with the potential to detect polymorphisms at relatively low cost. GBS has been widely used in several crops for biparental QTL mapping and genome-wide association study (GWAS) to uncover loci controlling various traits23–25.
Previous genetic studies of PcRR resistance in pepper have mainly focused on biparental QTL mapping, which primarily depends on the genetic diversity of the two parental lines. Therefore, the detected QTLs encompass large genomic regions with low map resolution, thus hampering the development of tightly linked markers and the identification of candidate genes10,24. These limitations could be overcome by employing GWAS, which allows QTL regions to be narrowed down to the candidate gene level using natural populations. However, GWAS has a high rate of false-positive QTL detection, requiring additional validation26,27. The shortcomings of biparental QTL mapping and GWAS can be overcome by combining these two approaches24. Such an approach had been successfully utilized for the genetic dissection of several important plant traits, including frost resistance, flowering time, panicle architecture, leaf architecture, pungency, and seed-related traits24,28–32.
In this study, we developed F7:8 recombinant inbred lines (RILs) derived from a cross between resistance source C. annuum cv. CM334 and susceptible cv. ECWR30 for QTL analysis of PcRR resistance to three P. capsici isolates in two environments. We then used previously reported core collections33 to evaluate PcRR resistance via GWAS. We used high-density SNP markers developed by GBS to identify and validate novel QTLs associated with PcRR resistance. By combining GWAS with QTL analysis, we successfully identified candidate genes for PcRR resistance.
Results
Evaluation of PcRR resistance in ECRILs and the GWAS core collection
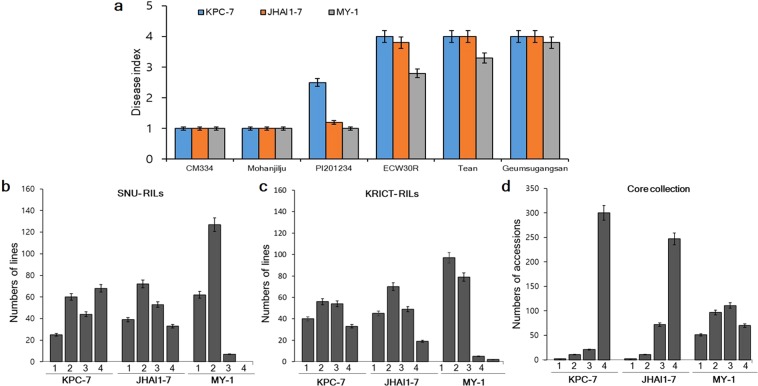
We evaluated 188 F7:8 ECRILs, parental lines, and additional resistant and susceptible controls in two environments (E1 and E2) for resistance to three P. capsici isolates (Fig. 1). The three isolates showed a significant difference in terms of symptom development and virulence. Susceptible controls (ECW30R, Tean, and Geumsugangsan) infected with the highly virulent isolate KPC-7, showed disease symptoms, including wilting and the appearance of water-soaked lesions at stem collars within 72 h of inoculation, and the plants completely wilted and died at 7 to 10 days post inoculation (DPI). By contrast, in plants infected with the moderately virulent isolate JHAI1–7, we noticed disease symptoms on the susceptible controls at 5 to 7 DPI, and the plants completely wilted and died at two weeks post inoculation. Susceptible controls infected with the isolate MY-1, with low virulence, symptoms were observed at 10 to 14 DPI, but complete plant death was not detected up to three weeks post inoculation. No visual symptoms of collar rot were observed on resistant controls cv. CM334 and Mohanjilju (commercial F1 hybrid, Syngenta Korea) after inoculation with all three isolates, even at three weeks post inoculation (Fig. 1a). However, PI201234 was partially resistant to isolates KPC-7 and JHAI1-7 and completely resistant to isolate MY-1 (Fig. 1a).
Figure 1.
Frequency distribution of PcRR in the parental lines, controls, ECRILs (SNU-RILs/KRICT-RILs), and GWAS (core collection). (a) Disease index of resistant controls (cv. CM334, Mohanjilju, and PI201234) and susceptible parents (cv. ECW30R, Tean, and Geumsugangsan) at three weeks post inoculation. (b) Frequency distribution of ECRILs against isolates KPC-7, JHAI1-7, and MY-1 in environment E1 (SNU-RILs). (c) Frequency distribution of ECRIL against isolates KPC-7, JHAI1-7, and MY-1 in environment E2 (KRICT-RILs). (d) Frequency distribution of GWAS (core collection) against isolates KPC-7, JHAI1-7, and MY-1. Bars denote the standard deviation.
We evaluated PcRR resistance in the ECRIL and GWAS core collection and scored disease severity on a scale of 1–4 at three weeks post inoculation in their respective environments, as described by36 (Supplementary Fig. S1). Ten plants from each ECRIL were grown and the resistance level of each plant scored individually. We calculated the frequency distribution based on the mean resistance values of each RIL (Fig. 1b,c). The frequency distribution of disease severity showed a normal distribution for KPC-7 and JHAI-7 in both environments, although more ECRILs showed susceptibility to KPC-7 in E1 than E2 (Fig. 1b,c). By contrast, the distribution of disease severity of the RILs skewed towards resistance to the low-virulence isolate MY-1, and no ECRILs reached the disease rating of 4 during the experiment in either environment, except for two ECRILs in environment E2 (Fig. 1b,c and Supplementary Fig. S2). Correlation analysis indicated a significant, positive correlation between the two environments for PcRR resistance; however, the correlations between different isolates varied (Supplementary Fig. S2).
We evaluated the GWAS accessions for PcRR resistance against the same three isolates as the ECRILs in two different environments (EA and E1). Among GWAS accessions, CM334 was completely resistant, whereas PI201234, YCM334, and nine other accessions were partially resistant, with slight disease symptoms at the stem collars upon infection with isolates KPC-7 and JHAI-7 (Fig. 1a,d and Supplementary Table S2). Most of these PcRR-resistant accessions include known resistance sources (Supplementary Table S2). The isolate JHAI1-7 showed slightly higher virulence compared to KPC-7 in a few lines of GWAS core collection, but a higher mortality rate was observed for KPC-7 (Fig. 1d and Supplementary Fig. S3). The disease response to the low-virulence isolate MY-1 was less severe, with 52 accessions showing no PcRR symptoms (Fig. 1d). Correlation analysis revealed significant, positive correlations between the KPC-7 and JHAI1-7 isolates and both environments for PcRR resistance (Supplementary Fig. S3). Highly virulent isolate KPC-7 for ECRILs in E2 and moderately virulent isolate JHAI1-7 for the GWAS accessions showed a minimum coefficient of variation (CV) of 38.55 and 28.09%, respectively (Table 1). Weak isolate MY-1 showed the highest (CV 57.0 to 62.02%) disease score in both screened populations in all environments (Table 1). The broad-sense heritability (H2%) was found to be high for all PcRR isolates. In E1, isolate KPC-7 showed the highest H2 (82.48%) among ECRILs, whereas isolate JAHI-7 showed an H2 of 87.40% and 84.16% for ECRILs in E2 and the GWAS accessions, respectively (Table 1).
Table 1.
Genetic variation among ECRILs and GWAS accessions inoculated with three isolates of P. capsici in different environments/years.
| Population (Environment-year) | Isolate | Mean ± SE | Range (Max-Min) | CV (%) | H² (Broad-sense) (%) | Expected genetic advancement (5%) |
|---|---|---|---|---|---|---|
|
SNU-ECRILs (E1-2017) |
KPC-7 | 2.12 ± 0.28 | 4–1 | 42.05 | 82.48 | 170.75 |
| JHAI-7 | 1.75 ± 0.25 | 4–1 | 46.10 | 80.22 | 171.30 | |
| MY-1 | 1.16 ± 0.21 | 2.09–1 | 57.00 | 52.45 | 89.32 | |
|
KRICT-ECRILs (E2-2018) |
KPC-7 | 2.38 ± 0.91 | 4–1 | 38.55 | 80.21 | 153.17 |
| JHAI-7 | 2,19 ± 0.84 | 4–1 | 38.60 | 87.40 | 124.16 | |
| MY-1 | 1.43 ± 0.36 | 4–1 | 56.29 | 75.82 | 178.83 | |
|
GWAS Population (EA-2017, E1-2018) |
KPC-7 | 2.86 ± 0.45 | 4–1 | 50.09 | 59.61 | 96.79 |
| JHAI-7 | 3.03 ± 0.26 | 4–1 | 28.09 | 84.16 | 122.39 | |
| MY-1 | 1.89 ± 0.37 | 4–1 | 62.06 | 72.85 | 178.73 |
SNP discovery through GBS and bin map construction for ECRILs
We performed genotyping of the ECRILs using GBS after EcoRI and MseI digestion. Sequencing of the prepared GBS libraries of 188 ECRILs and parental lines resulted in 525.62 million raw reads (data not shown). After trimming the raw reads, we obtained an average of approximately 4.4 million reads per sample (Table 2). After aligning the reads to the CM334 reference genome v1.6, 66,405 SNPs were detected (Fig. 2a). The SNPs were distributed more densely at the ends of the chromosomes than in the middle regions (Fig. 2a). After removing more than 90% of missing data and filtering unequally distributed SNPs, we obtained 13,021 high-quality SNPs that were equally distributed across the genome, which were used for bin linkage map construction (Tables 2 and 3 and Supplementary Fig. S4). To impute missing data and genotyping errors, we employed a sliding window approach23. Recombination breakpoints were determined by sliding 25 SNPs consecutively as one window. As a result, a high-density bin map consisting of 2,663 bins covering a total genetic distance of 1,428.8 cM was constructed (Table 3 and Supplementary Fig. S4). The average genetic distance between bins was estimated to be 0.6 cM (Table 3). Among the 12 linkage groups, maximum and minimum genetic distances of 195.6 and 44.7 cM were obtained for chromosomes P1 and P8, respectively (Table 3).
Table 2.
Genotyping data for GWAS and QTL mapping.
| ECRIL | GWAS core collection | |
|---|---|---|
| Number of accessions (lines) | 188 | 352 |
| Genotyping method | GBS (EcoRI/MseI) | GBS (PstI/MseI and EcoRI/MseI) |
| Average number of reads per sample | 4,450,271* | 3,326,422 |
| Total number of retained SNPs | 13,021 | 507,713 |
| Average distance between SNPs (bp) | 366,349 | 16,305 |
*Number of spots sequenced by paired-end reads.
Figure 2.
SNP density (number of SNPs per 1 Mbp). (a) SNP density for ECRILs digested with EcoRI/MseI. (b) SNP density for the GWAS (core collection) digested with PstI/MseI and EcoRI/MseI.
Table 3.
Comparison of the physical length and genetic distance of bins for biparental QTL mapping.
| Chr. | Number of SNPs | Number of bins | Physical length of bin (Mb) | Genetic distance of bin (cM) | ||
|---|---|---|---|---|---|---|
| Mean | Total | Mean | Total | |||
| P1 | 1551 | 329 | 1.2 | 272.7 | 0.6 | 195.6 |
| P2 | 925 | 190 | 1.0 | 171.1 | 0.5 | 94.1 |
| P3 | 887 | 275 | 1.1 | 257.9 | 0.6 | 162.9 |
| P4 | 613 | 191 | 1.5 | 222.6 | 0.6 | 125.6 |
| P5 | 1009 | 183 | 1.2 | 233.5 | 0.5 | 91.6 |
| P6 | 1034 | 219 | 1.1 | 236.9 | 0.5 | 114.4 |
| P7 | 1323 | 259 | 1.1 | 231.9 | 0.5 | 129.4 |
| P8 | 212 | 52 | 2.1 | 145.1 | 0.9 | 44.7 |
| P9 | 1864 | 260 | 1.2 | 252.8 | 0.5 | 119 |
| P10 | 808 | 207 | 1.3 | 233.6 | 0.5 | 115.3 |
| P11 | 2127 | 323 | 0.9 | 259.7 | 0.4 | 116.1 |
| P12 | 668 | 175 | 1.2 | 235.7 | 0.7 | 120.1 |
| Total | 13,021 | 2,663 | 1.2 | 2,753.5 | 0.6 | 1,428.8 |
PcRR resistance QTLs in the ECRILs
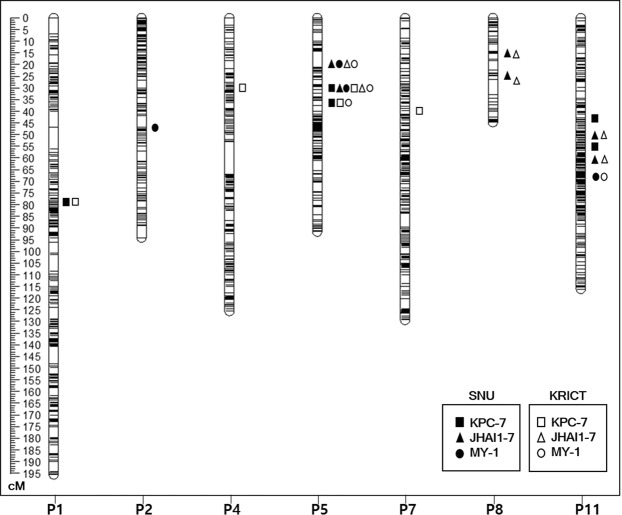
We identified QTLs for PcRR resistance to three isolates, KPC-7, JHAI1-7, and MY-1, using 188 ECRILs and the high-density SNP linkage map in two environments, E1 and E2 (Fig. 3 and Table 4). In total, 14 QTLs, including those with major and minor effects, those commonly detected, those specific to isolates, and those specific to environments were detected across the genome, explaining phenotypic variation (R2) ranging from 3.73 to 38.99% (Table 4). The PcRR resistance QTLs were detected on chromosomes P1, P2, P4, P5, P7, P8, and P11.
Figure 3.
Bin linkage chromosomal map showing the locations of PcRR resistance QTLs with the genetic distance shown in centimorgans (cM) for the ECRIL population evaluated with three P. capsici isolates in two respective environments.
Table 4.
PcRR resistance QTLs detected by composite interval mapping in the ECRIL population inoculated with three isolates evaluated in their respective environments.
| Isolate | Evaluating environment | QTLs | Chr. | Positiona (cM) | Locationb (Mb) | LODc | R2 (%)d | Additive effect |
|---|---|---|---|---|---|---|---|---|
| KPC-7 | E1-SNU*2017 | E1Kpc-1 | P1 | 77.61 | 9.5–9.63 | 3.97 | 4.57 | 2.27 |
| E1Kpc-5.2 | P5 | 28.91 | 27.3–29.2 | 21.32 | 33.77 | 6.26 | ||
| E1Kpc-5.3 | P5 | 34.61 | 34.6–37 | 14.87 | 25.40 | 5.41 | ||
| E1Kpc-11.1 | P11 | 41.40 | 49.5–50.5 | 4.78 | 5.59 | 2.53 | ||
| E1Kpc-11.2 | P11 | 52.01 | 74–76 | 4.20 | 4.94 | 2.40 | ||
| E2-KRICT*2018 | E2Kpc-1 | P1 | 77.61 | 9.5–9.63 | 6.69 | 7.81 | 4.42 | |
| E2Kpc-4 | P4 | 28.41 | 87–88.5 | 4.64 | 5.41 | 2.96 | ||
| E2Kpc-5.2 | P5 | 29.21 | 27.3–29.2 | 23.25 | 37.12 | 5.78 | ||
| E2Kpc-5.3 | P5 | 34.61 | 34.6–37 | 14.17 | 25.14 | 4.70 | ||
| E2Kpc-7 | P7 | 38.71 | 24.5–26.4 | 3.60 | 3.73 | 1.80 | ||
| JHAI1–7 | E1-SNU*2017 | E1Jha-5.1 | P5 | 22.61 | 18.7–19.5 | 18.30 | 27.31 | 4.80 |
| E1Jha-5.2 | P5 | 28.91 | 27.3–29.2 | 29.73 | 38.99 | 5.72 | ||
| E1Jha-8.1 | P8 | 13.31 | 126.4–126.6 | 7.25 | 6.85 | 2.40 | ||
| E1Jha-8.2 | P8 | 23.71 | 128.2–130.3 | 5.82 | 5.27 | 2.06 | ||
| E1Jha-11.1 | P11 | 48.41 | 61.2–63 | 9.01 | 8.47 | 2.62 | ||
| E1Jha-11.2 | P11 | 59.31 | 111.4–112.8 | 7.71 | 7.37 | 2.47 | ||
| E2-KRICT*2018 | E2Jha-5.1 | P5 | 22.61 | 18.7–19.5 | 18.13 | 26.62 | 4.54 | |
| E2Jha-5.2 | P5 | 28.91 | 27.3–29.2 | 29.66 | 38.21 | 5.44 | ||
| E2Jha-8.1 | P8 | 12.31 | 126.4–126.6 | 4.61 | 4.36 | 1.83 | ||
| E2Jha-8.2 | P8 | 19.51 | 128.2–130.3 | 4.57 | 4.26 | 1.77 | ||
| E2Jha-11.1 | P11 | 48.41 | 61.2–63 | 9.44 | 8.82 | 2.57 | ||
| E2Jha-11.2 | P11 | 60.51 | 111.4–112.8 | 7.34 | 7.03 | 2.33 | ||
| MY-1 | E1-SNU*2017 | E1My-2 | P2 | 45.91 | 129–131 | 4.30 | 6.99 | 1.00 |
| E1My-5.1 | P5 | 22.61 | 18.7–19.5 | 8.94 | 18.22 | 1.60 | ||
| E1My-5.2 | P5 | 28.61 | 27.3–29.2 | 10.80 | 19.68 | 1.66 | ||
| E1My-11 | P11 | 66.41 | 189–190 | 4.89 | 8.13 | 1.07 | ||
| E2-KRICT*2018 | E2My-5.1 | P5 | 22.31 | 18.7–19.5 | 4.97 | 10.02 | 1.19 | |
| E2My-5.2 | P5 | 28.61 | 27.3–29.2 | 6.36 | 12.61 | 1.35 | ||
| E2My-5.3 | P5 | 34.61 | 34.6–37 | 4.38 | 8.90 | 1.12 | ||
| E2My-11 | P11 | 64.41 | 189–190 | 3.33 | 6.17 | 1.50 |
aPositions of the markers on the linkage map in centimorgans (cM).
bPosition of right and left bins in the pepper genome. cMaximum log-likelihood (LOD) value of QTL. dPhenotypic variation (R2) explained by a QTL at the linked marker.
*E1-SNU, Environment 1, Seoul National University; *E2-KRICT, Environment 2, Korea Research Institute of Chemical Technology.
Two major QTLs to highly virulent isolate KPC-7 were detected on chromosome P5, E1Kpc-5.2 and E2Kpc-5.3, at 28.91 and 34.61 cM, corresponding to 27.3–29.2 Mb and 34.6–37 Mb, respectively, in the C. annuum ‘CM334’ reference genome. E1Kpc-5.2 explained 33.77% (LOD of 21.32) and 37.12% (LOD of 23.25) and E2Kpc-5.3 explained 25.40% (LOD of 14.87) and 25.14% (LOD of 14.17) of phenotypic variation (R2) in E1 and E2, respectively (Fig. 3 and Table 4). Three minor QTLs were detected in both environments, but only one QTL (on chromosome P1 at 77.61 cM [9.5–9.6 Mb]) was commonly detected in both environments (Fig. 3 and Table 4).
Two major QTLs to moderately virulent isolate JHAI1–7 on chromosome P5, E1Jha-5.1 and E2Jha-5.2, were detected at 22.61 and 28.91 cM, corresponding to 18.7–19.5 and 27.3–29.2 Mb, respectively, in the C. annuum ‘CM334’ reference genome. E1Jha-5.1 explained 27.31% (LOD of 18.30) and 26.62% (LOD of 18.13) and E2Jha-5.2 explained 38.99% (LOD of 29.73) and 38.21% (LOD of 29.66) of phenotypic variation (R2) in E1 and E2, respectively (Fig. 3 and Table 4). Four minor QTLs on P8 and P11 were detected in both environments (Fig. 3 and Table 4).
For low-virulence isolate MY-1, two major-effect QTLs on chromosome P5, E1My-5.1 and E2My-5.2, were detected at 22.61 and 28.61 cM, corresponding to 18.7–19.5 Mb and 27.3–29.2 Mb, respectively, in the C. annuum ‘CM334’ reference genome. E1My-5.1 explained 18.22% (LOD of 8.94) and 10.02% (LOD of 4.94) and E2My-5.2 explained 19.68% (LOD of 10.80) and 12.61% (LOD of 6.33) of phenotypic variation (R2) in E1 and E2, respectively (Fig. 3 and Table 4). One major QTL on chromosome P5, E1My-5.3, was detected in E2 only at 34.61 cM, corresponding to 34.6–37 Mb, which explained 8.90% (LOD 4.38) of phenotypic variation (R2). Two minor QTLs were detected in both environments, but only one minor QTL was commonly detected in both environments (on chromosome P11 at 66.41 cM [189–190 Mb]; Fig. 3 and Table 4). Notably, all QTLs associated with the MY-1 isolate showed lower R2 and LOD values when compared to the QTLs detected against the two other isolates used in this study (Fig. 3 and Table 4).
Taken together, three major QTLs on P5 were commonly detected for all three isolates in two environments, whereas minor QTLs on different chromosomes were isolate or environment specific (Fig. 3 and Table 4). Among the major QTLs, QTL5.2, which is located at 27.3–29.2 Mb, was commonly detected for all three isolates and was named according to its genomic position on chromosome P5. The major QTL at 18.7–19.5 Mb (common to JHAI1–7 and MY-1) was named QTL5.1. The major QTL at 34.6–37 Mb (common to KPC-7 and MY-1) was named QTL5.3. Even though minor QTLs were commonly detected on chromosome 11 for all three isolates, they were located at different positions. We detected isolate-specific minor QTLs on other chromosomes, such as the minor QTLs Kpc1 (for KPC-7) on P1, Jha8.1, Jha8.2, Jha11.1, and Jha11.2 (for JHAI1-7) on P8 and P11, and My11 (for MY-1) on P11.
Epistatic interactions of PcRR resistance QTLs
We detected additive-by-additive epistatic interactions against highly virulent isolate KPC-7 between major QTLs QTL5.2 and QTL5.3 and QTL5.2 and E1Kpc-11.1 in environment E1 and between QTLs QTL5.2 and QTL5.3 in environment E2 (Supplementary Table S3). We detected additive-by-additive epistatic interactions between QTL-5.2 and Jha8.1 and QTL5.2 and Jha11.2 in environment E1 and between QTLs QTL5.2 and Jha11.1 in environment E2 (Supplementary Table S3) against moderately virulent isolate JHAI1-7. By contrast, we detected additive-by-additive epistatic interactions only between QTL5.2 and My11 in both E1 and E2 (Supplementary Table S3) against low-virulence isolate. The total R2 effects of individual QTLs and their interactions ranged from 16.6 to 42.0% (Supplementary Table S3). The detection of epistatic interactions between loci on chromosomes P5 and P11 points to multilocus epistatic control of PcRR resistance in pepper.
Genome-wide association study of PcRR resistance
We aligned the sequences derived from GBS to C. annuum cv. CM334 reference genome v.1.6. GBS genotyping of 352 accessions using two sets of libraries derived from double digestion with restriction enzyme pairs PstI/MseI and EcoRI/MseI generated a total of 6,126,403 of SNPs. The SNPs were evenly distributed using this combination of restriction enzymes. SNPs in the GWAS core collection were relatively uniformly distributed across the chromosomes (Fig. 2b). We filtered out the SNPs with minor allele frequency >0.05, SNP coverage >0.6, and inbreeding coefficient >0.8, resulting in 507,713 high-quality SNPs distributed evenly on chromosomes P1 to P12 (Fig. 2b and Table 2).
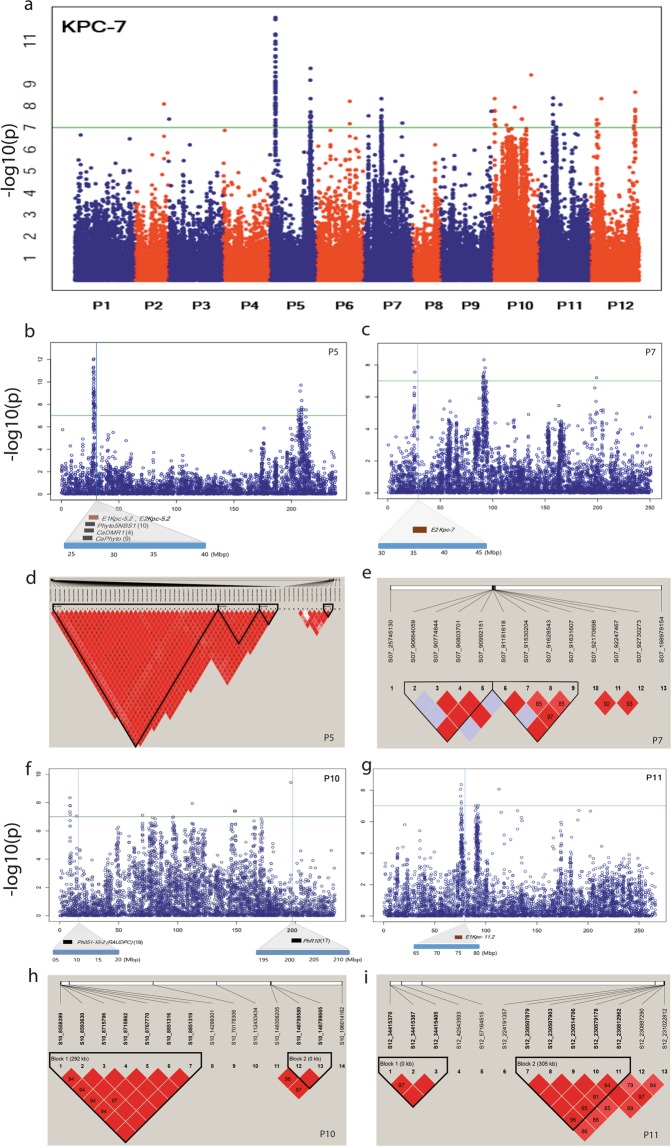
Of the SNPs association with resistance to PcRR against isolate KPC-7, 117 were significant SNPs, with -log10 (p) value > 7.0, explaining phenotypic variation (R2) > 0.2 (Fig. 4a and Supplementary Table S4). An association study revealed significant SNPs linked to PcRR resistance to KPC-7 on chromosomes P2 (1 SNP at 149 Mb), P3 (1 SNP at 5 Mb), P5 (64 SNPs at 27.7–29.7 and 206–212 Mb), P6 (2 SNP at 171 Mb), P7 (13 SNPs at 24 and 90 Mb), P9 (1 SNP at 263 Mb), P10 (14 SNPs at 8, 14, 70, 112, 148, and 196 Mb), P11 (8 SNPs at 75, 90, 92, and 112 Mb), and P12 (13 SNPs at 34, 42, 57, 224, and 230 Mb) (Fig. 4a and Supplementary Table S4). GWAS of isolate JHAI1-7 revealed an associated SNP peak on chromosome P5 with a similar position to that of the KPC-7 isolate (27.7 to 29.7 Mb), but only two SNPs showed a significance level above the adjusted threshold level at -log10(p) value > 7.0, explaining phenotypic variation (R2) > 0.2 (Supplementary Fig. S5a and Supplementary Table S4). The two significant SNPs were located at 28.2 to 28.6 Mb on chromosome P5. By contrast, GWAS against the low-virulence isolate MY-1 did not detect significant SNPs (Supplementary Fig. S5b). Notably, only a few or no significant SNPs were associated with isolates JHAI1-7 and MY-1, perhaps due to environment and genotype interactions or to the low virulence levels of the isolates.
Figure 4.
Manhattan plots based on GBS-GWAS showing the significant SNPs associated with PcRR resistance and haplotype analysis. (a) Significant SNPs associated with PcRR isolate KPC-7. (b,c,f,g) SNPs detected in common regions of GWAS and ECRIL QTL maps from this and previous studies. QTL positions are marked with blocks under the SNP positions. Brown and black blocks indicate QTLs detected in this and previous studies, respectively. (d,e,h,i) Haplotype blocks containing significant SNPs on chromosomes P5, P7, P10, and P11.
Importantly, significant SNPs associated with resistance to isolate KPC-7 on three regions of chromosomes P5, P7, and P11 colocalized with QTLs detected in this study (Fig. 4a and Table 4). On chromosome P5, 49 SNPs colocalized with commonly detected major QTL region QTL5.2 (27.3–29.2 Mb) (Fig. 4b and Table 4). One SNP detected on chromosome P7 at 25 Mb corresponded to the location of QTL E2Kpc-7 (Fig. 4c and Table 4). Another region on chromosome P11 at 75.2 Mb corresponded to QTL E1Kpc-11.2 (Fig. 4g and Table 4). The GWAS-SNPs at three regions on chromosomes P5, P10, and P11 colocalized with previously detected QTLs and linked markers (Fig. 4b,f). The GWAS-SNPs on chromosome P5 in a 2.5-Mb region at position 27.0–29.5 Mb colocalized with a QTL detected in earlier reports4,9,10,21 (Fig. 4b). SNP marker S10_14299301 located at 14,299 kb on chromosome P10 colocalized with the previously identified QTL Ph051-10-2 (RAUDPC) for PcRR resistance19 (Fig. 4f). Regions detected by GWAS with significant SNPs on chromosome P10 at 196 Mb (Fig. 4f) colocalized with the region containing PhR10, a race-specific PcRR resistance locus17. We identified 13 significant GWAS-SNPs linked to PcRR resistance at the end of the long arm (206–212 Mb) of chromosome P5 that were not detected in previous studies, emphasizing the involvement of this genomic region in PcRR resistance (Fig. 4a). We used the significant SNP S05_208290460, with a -log10(p) value of 9.72 identified from the GWAS-SNPs (Supplementary Table S4), to compare the resistance levels of the GWAS accessions. As shown in the box plots in Supplementary Fig. S6e, the homozygous genotype (S05_208290460) AA is associated with an increased resistance level against all three isolates compared to the alternative homozygous genotype GG.
We conducted extensive linkage disequilibrium (LD) analysis of the GWAS core collection (N = 352) based on all adjacent marker pairs within a chromosome or within a haplotype block (Supplementary Table S5). We identified 29,269 haplotypes, with an average of 2,439 per chromosome. The average haplotype was 81.2 kb in size, with an average of 16.3 SNPs per haplotype (Table 2 and Supplementary Table S5). Further haplotype block analysis carried out with significant SNPs on chromosome P5 revealed 4 blocks containing 64 significant SNPs (Fig. 4d and Supplementary Table S4). The three blocks on P5 colocalized with QTL 5.2 detected in this study and with previously identified QTLs4,9,10,21 (Fig. 4b,d). Two haplotype blocks were obtained on each of the chromosomes P7, P10 and P11, including regions with significant SNPs (Fig. 4e,h,i). These results indicate that the identified haplotype regions are strongly associated with the PcRR resistance.
Predicted candidate genes for PcRR
We looked for candidate PcRR resistance genes in the 1 Mb upstream and downstream flanking regions of all significant GWAS-SNPs and bin markers linked to PcRR QTLs. Detailed descriptions of the predicted genes in the target regions are provided in Supplementary Table S6. A major and colocalized genomic region detected for isolate KPC-7 via GWAS and biparental QTL mapping (QTL5.2) associated with PcRR resistance encompassing a 2.5 Mb region (27.0–29.5 Mb) (Fig. 3 and 4a) contains 20 predicted genes, including genes for three receptor-like kinase (RLK) domain-containing proteins, two nucleotide-binding site-leucine-rich repeat (NBS-LRR) domain-containing proteins, and one SAR8.2 precursor protein known to be associated with disease resistance (Table 5). Two NBS-LRR genes (PHT81215.1 and PHT81216.1) are located 618 and 319 kb upstream of the most significant SNP (S05_27703815), respectively. The most closely located RLK gene (PHT81221.1) was identified 144 kb upstream of a highly significant GWAS SNP. Two RLK genes (PHT81227.1 and PHT81229.1) were identified at 5.5 and 12.4 kb downstream of GWAS (SNP S05_28163627). One candidate gene, encoding the SAR8.2 precursor protein, was detected 148 kb upstream of GWAS SNP (S05_28825990) (Table 5 and Supplementary Table S6). These genes were detected by both QTL mapping and GWAS analysis, making them strong candidate genes for PcRR resistance.
Table 5.
Candidate resistance genes associated with the significant GWAS regions and QTLs for PcRR and their Gene Ontology (GO) descriptions.
| Chr. | Mapping strategy (GWAS/QTL) | Location (bp) | Peptide IDa | GO description |
|---|---|---|---|---|
| P5 | GWAS-QTL5.2 | 27085614–27086321 | PHT81215.1 | PREDICTED: disease resistance protein RPP13-like |
| P5 | GWAS-QTL5.2 | 27311924–27314491 | PHT81216.1 | PREDICTED: disease resistance protein RPP13-like |
| P5 | GWAS-QTL5.2 | 27558828–27565702 | PHT81221.1 | Inactive leucine-rich repeat receptor-like serine/threonine-protein kinase |
| P5 | GWAS-QTL5.2 | 28168701–28169191 | PHT81227.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | GWAS-QTL5.2 | 28172631–28176106 | PHT81228.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | GWAS-QTL5.2 | 28974813–28975852 | PHT81231.1 | SAR8.2 precursor |
| P5 | QTL5.1 | 18707644–18708354 | PHT81101.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | QTL5.1 | 18810592–18813153 | PHT81104.1 | PREDICTED: disease resistance protein RPP13-like |
| P5 | QTL5.1 | 18943734–18945083 | PHT81107.1 | PREDICTED: disease resistance protein RPH8A-like |
| P5 | QTL5.1 | 19074847–19077759 | PHT81108.1 | PREDICTED: disease resistance protein RPP13-like |
| P5 | QTL5.1 | 19094517–19097099 | PHT81109.1 | PREDICTED: disease resistance protein RPP13-like |
| P5 | QTL5.1 | 19153252–19169266 | PHT81110.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | QTL5.1 | 19288855–19292090 | PHT81111.1 | PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR |
| P5 | QTL5.1 | 19292463–19293858 | PHT81112.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | QTL5.1 | 19294140–19297023 | PHT81113.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | QTL5.1 | 19344986–19347916 | PHT81114.1 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase |
| P5 | QTL5.3 | 35411316–35411978 | PHT81306.1 | Kunitz trypsin inhibitor 2-like |
| P5 | QTL5.3 | 36088454–36089143 | PHT81317.1 | putative late blight resistance protein homolog R1B-17 |
| P5 | QTL5.3 | 36205447–36219154 | PHT81318.1 | PREDICTED: putative late blight resistance protein homolog R1C-3 |
aPeptide id given in the NCBI genome data base.
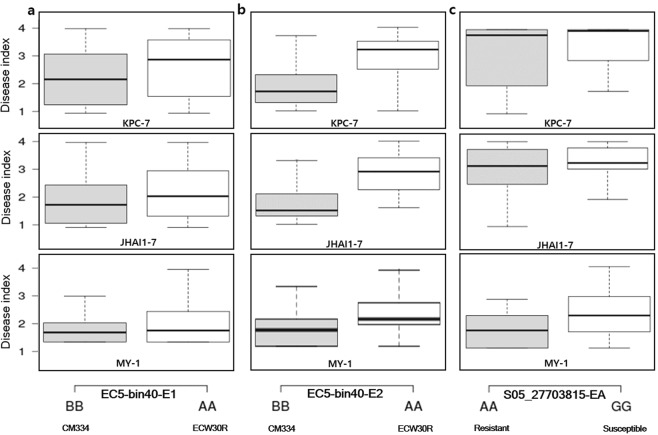
We used the highly significant GWAS SNP (S05_27703815), with -log10(p) value of 11.23 (Supplementary Table S4), located close to the candidate genes to compare the resistance levels of the GWAS accessions. As shown in the box plots in Fig. 5c, the homozygous genotype (S05_27703815) AA was associated with increased resistance compared to the alternative homozygous genotype GG among the three PcRR isolates examined. We used the tightly linked bin marker EC5-bin40 of QTL segment QTL5.2 overlapping with candidate genes to compare the resistance levels of the ECRILs. As shown in the box plots, the homozygous resistant genotype BB is associated with enhanced resistance compared to the homozygous susceptible genotype AA against all three isolates in both E1 and E2 (Fig. 5a,b).
Figure 5.
Box plots of tightly linked bins to QTLs in ECRILs and significantly associated GWAS-SNPs from chromosome P5. (a) ECRILs grouped based on the tightly linked bin to QTL5.2 in environment E1. (b) ECRILs grouped based on the tightly linked bin to QTL5.2 in environment E2. (c) Core collection GWAS (core collection) grouped based on the most significant SNP from QTL region 5.2.
The major QTL region QTL5.1, spanning 0.7 Mb, which was commonly detected against two isolates, JHAI1-7 and MY-1, contains 15 predicted genes (Supplementary Table S6), including genes associated with disease resistance, such as genes for six RLK domain-containing proteins and four NBS-LRR domain-containing proteins (Table 5). Finally, we used the tightly linked bin marker EC5-bin27 of QTL segment QTL5.1 overlapping with candidate genes to compare the resistance levels of the ECRILs. As shown in the box plots, the homozygous resistant genotype BB is associated with enhanced resistance compared to the homozygous susceptible genotype AA against all three isolates in both E1 and E2 (Supplementary Fig. S6a,b).
Finally, QTL5.3, a QTL region against isolates KPC-7 and MY-1, was detected at 34.6–37 Mb on chromosome P5, with 28 predicted genes. Among these, three predicted genes are linked to disease resistance proteins, including two NBS-LRR domain-containing proteins and one Kunitz trypsin inhibitor 2-like protein (Table 5). We used the tightly linked bin marker EC5-bin51 of QTL segment QTL5.3 overlapping with the candidate genes to compare the resistance levels of the ECRILs. As shown in the box plots, the homozygous resistant genotype BB is associated with enhanced resistance compared to homozygous susceptible genotype AA against all three isolates in both E1 and E2 (Supplementary Fig. S6c,d).
Discussion
In this study, we combined traditional QTL mapping with GWAS to increase our understanding of PcRR resistance in pepper and detected common and race-specific QTLs for this trait. Although many QTL studies have been performed to investigate PcRR using biparental QTL mapping4,9,10,20,21, this is the first comprehensive study of PcRR resistance loci for multiple P. capsici isolates in multiple environments using biparental QTL mapping and GWAS.
PcRR resistance QTLs
Due to the rapid genetic evolution and diversity of Phytophthora, there is an urgent need to breed pepper with resistance to multiple PcRR isolates4,35. Analysis of the differential responses of ECRILs and GWAS accessions against the three PcRR isolates provided us with valuable information about the range and frequency distribution of symptom development and virulence levels (Fig. 1). The mean disease scores of the resistant GWAS accessions were lower than those of the ECRILs (Supplementary Table S2), pointing to higher allelic diversity for PcRR resistance in the GWAS core collection. Furthermore, the high heritability of resistance (Table 1) points to the additive effects of genes, offering an excellent opportunity for improving this trait through selection.
Breeding for resistance to PcRR is quite challenging; even resistance incorporated into commercial pepper lines can be readily overcome by highly virulent P. capsici isolates2. CM334 is a well-known source of PcRR resistance. The inheritance of PcRR resistance in CM334 is complex9,10, and the genetic dissection of PcRR resistance would greatly facilitate the improvement of pepper cultivars. Therefore, in this study, we aimed to identify QTLs for different P. capsici isolates categorized based on virulence in two different environments. QTL mapping for PcRR resistance revealed significant QTLs, including three major-effect QTLs on chromosome P5 and several isolate-specific minor-effect QTLs on various chromosomes. Among the several QTLs identified for PcRR resistance, a major QTL on chromosome P5 has been consistently identified in several resistance sources irrespective of race, isolate, and level of virulence4,10. In agreement with this finding, two QTLs (QTL5.1 and QTL5.3) specific to single isolates and one QTL (QTL5.2) common to all three isolates of P. capsici were consistently detected on chromosome P5 in two environments. The location of QTL5.2 detected in this study coincides with that of the previously identified locus Pc5.14,10,21. The dominant resistance locus CaPhyto, conferring race-specific resistance to PcRR, was previously mapped to a genetic interval near Pc5.1 in a 3.3-cM region between two SSR markers, ZL6726 and ZL6970, in C. annuum accession PI201234.
Several minor-effect QTLs have previously been reported on various chromosomes4,11,18–20,36. The number of QTLs conferring PcRR resistance and their positions have varied among studies depending on the mapping populations used, disease screening methods, inoculum density, and isolate aggressiveness. In this study, we detected some minor QTLs with isolate specificity on chromosome P2 (Kpc1 for isolate KPC-7) and chromosome P8 (Jha8.1, Jha8.2, Jha11.1, and Jha11.2 for isolate JHAI1-7) and with no isolate specificity on chromosome P11 (Table 4). However, we could not compare the exact physical positions of these QTLs with previously detected QTLs due to the limited number of common markers and the lack of sequence information; instead, we used the closest markers for analysis.
In addition, epistatic interactions between PcRR resistance QTLs have been reported11,36. Consistent with these reports, we detected epistatic QTL interactions between resistance QTLs on chromosomes P5 and P11 for isolate KPC-7, on chromosomes P5 and P8 and chromosomes P5 and P11 for isolate JHAI1-7, and on chromosomes P5 and P11 for isolate MY-1 in both environments.
Validation of PcRR loci via GWAS
We took advantage of biparental QTL mapping together with GWAS to identify new PcRR resistance-related loci and to validate known QTL regions. This approach proved to be effective for the identification of PcRR resistance loci. We identified 117 significant SNPs related to PcRR isolate KPC-7 across the chromosomes. However, only SNPs identified on chromosomes P7, P5, and P11 colocalized with QTLs detected in this study (Fig. 4), perhaps because the ECRILs included only two parental alleles compared to the highly diverse GWAS accessions33. Thus, numerous biparental populations developed using different resistant genetic resources were likely captured among all resistant loci.
GWAS is a high-resolution, powerful technique, as it captures chronological recombination events that have accumulated inside an association panel. PcRR resistance loci have previously been mapped onto chromosomes P1, P3, P4, P6, P8, P9, P10, P11, and P124,11,18–20,36. To determine whether the GWAS-SNPs identified in this study are novel PcRR resistance SNPs, we compared their physical positions with the positions of previously reported QTLs. The GWAS-SNPs on chromosome P5 (at 27.0–29.5 Mb) coincided with major QTLs detected in previous studies4,10,21. Two GWAS-SNP peaks on chromosome P10 at 14.2 and 196 Mb colocalized with the previously detected QTL Ph051-10-2 (RAUDPC) and the race-specific locus PhR10, respectively17,19. However, due to the limited availability of marker sequences, it was not possible to compare and validate all GWAS-SNPs with previously reported resistance loci linked to PcRR. The GWAS-SNPs associated with PcRR identified in this study could serve as novel resistance loci that could be used to incorporate resistance into ongoing pepper-breeding programs.
Potential candidate genes for PcRR resistance
In this study, we identified several classes of genes associated with disease resistance in various QTL regions. A cluster of NBS-LRR domain-containing R genes was identified on chromosome P5: four NBS-LRR genes from QTL5.1 (18.7–19.5 Mb), two from QTL5.2 (27.3–29.2 Mb), and two from QTL5.3 (34.6–37 Mb). These candidate genes share high sequence identity with RPP13-like NBS-LRR protein genes and represent potential candidates for PcRR resistance in pepper. RPP13 is a CC (coiled-coil)-NBS-LRR domain-containing R gene that confers resistance to the oomycete pathogen Peronospora parasitica in Arabidopsis thaliana37.
Plant genomes generally contain numerous NBS-LRR class R genes, which are often clustered on specific chromosomes due to tandem and segmental duplications38–41. Several dominant R genes, such as R1 from potato42, Ph-3 from tomato43, and RpsUN1, RpsUN2, and Rpg1-b from soybean44,45, belong to NBS-LRR domain-containing complex R gene clusters. R gene clusters could function as hotspots for the generation of novel R genes and enhance the possibility of structural and copy number variation through various DNA recombination and rearrangement mechanisms, including duplications, unequal crossing over, gene conversion, and diversifying selection, which could account for the gain or loss of resistance45–47. The candidate R gene clusters identified in this study could account for enhanced durability and resistance to PcRR in pepper. In addition to the NBS-LRR R gene clusters, we identified six RLKs from QTL5.1 (18.7–19.5 Mb) and three from QTL5.2 (27.3–29.2 Mb) on chromosome P5. RLKs are extracellular surface (also called pattern recognition) receptors that play crucial roles in plant defense-related processes, including both host and non-host defense responses48. Therefore, the RLK genes identified in this study represent strong candidates for PcRR resistance genes.
We also identified SAR8.2 in QTL5.2 (27.3–29.2 Mb), a systemic acquired resistance (SAR)-related gene known as CASAR82A. CASAR82A localizes to the phloem and epidermal cells of pepper leaf and stem tissues infected by Colletotrichum coccodes and P. capsici or treated with salicylic acid48. The involvement of the pepper SAR8.2 gene in pathogen infection and environmental stress responses and its use as a marker of abiotic elicitors49 suggests it might also be an important candidate gene for PcRR resistance.
Kunitz trypsin inhibitor 2-like protein is involved in programmed cell death (PCD) in Arabidopsis. Arabidopsis serine protease (Kunitz trypsin) inhibitor (KTI1) appears to be a functional KTI when induced in bacteria and in planta. Atkti1 expression is induced at a late stage of infection in response to fungal and bacterial elicitors and to salicylic acid. RNAi silencing of AtKTI1 increased the rate of lesion development in leaf tissue after infiltration with the PCD-eliciting fungal toxin fumonisin B1 (FB1) or the avirulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 carrying avrB (Pst avrB)50. In this study, one gene encoding a Kunitz trypsin inhibitor 2-like protein was identified in a major QTL region on chromosome P5, QTL5.3 (34.6–37 Mb), suggesting it is a candidate gene for PcRR. However, to uncover the roles of intra- and extracellular immune receptors in disease resistance, further analyses of the candidate genes should be performed, including targeted knockdown of candidate genes and subsequent molecular analyses.
In this study, we combined the use of biparental QTL mapping and GWAS to identify QTLs and candidate genes for PcRR resistance in pepper. Based on the genetic map and phenotypic data, 14 significant QTLs were identified with R2 values ranging from 3.73 to 38.99% among the three PcRR isolates in their respective environments. Two of these QTLs on chromosome P5 showed major effects, with a combined R2 > 60%. GWAS identified 117 highly significant SNPs associated with PcRR across the genome for isolates KPC-7 and JHAI1-7. The additional loci detected through GWAS might enhance the accuracy of selection for PcRR resistance. The use of SNP markers associated with the candidate genes or QTLs could accelerate MAS and genomic selection in pepper breeding programs for PcRR resistance. The predicted candidate genes for PcRR resistance lay the foundation for the molecular dissection of PcRR resistance.
Materials and Methods
Plant materials and DNA extraction
The ECRIL mapping population comprised 188 F7:8 ECRILs developed from a cross between resistant C. annuum line CM334 and susceptible line ECW30R. The single seed descent method was used for ECRIL development, with shuttle breeding. The population was abbreviated as “EC”, based upon the first letter of parental accessions. The plants were grown in plastic pots in a greenhouse. GWAS was carried out on a core collection comprising 352 Capsicum accessions32. Both parents of the ECRIL population were included in the core collection. Genomic DNA was extracted from young leaf tissues of plants of the ECRILs and the association population at the seedling stage using the CTAB (cetyl trimethyl ammonium bromide) method as described previously51.
Disease assay and resistance scoring
Resistance against PcRR in the ECRILs was evaluated under two different environments: the Seoul National University (SNU) farm at Suwon, Republic of Korea (2017) (E1: Environment 1) and the Korea Research Institute of Chemical Technology (KRICT), Daejeon, Republic of Korea (2018) (E2: Environment 2). The GWAS core collection (consisting of 352 Capsicum accessions) was also evaluated in two different environments. Resistance screening was carried out in 2017 with isolate KPC-7 at Rural Development Authority (RDA), Jeonju, Republic of Korea (EA: Environment A), while isolates JHAI1-7 and MY-1 were utilized in 2018 at SNU (E1: Environment 1). P. capsici isolates MY-1, JHAI1-7, and KPC-7 (Supplementary Table S1) used in this study were kindly provided by Dr. Choi (KRICT). Isolates MY-1, JHAI1-7, and KPC-7 were reported to have low, moderate, and high levels of virulence, respectively10,52. To prepare the inoculum, the P. capsici isolates were cultured on V8-agar medium and incubated at 27 °C for mycelium growth for five days. The plates were flooded with distilled water to harvest the mycelia. The spore concentration was quantified using a hemocytometer, and the spore suspension density was adjusted to approximately 5 × 104 sporangia per mL prior to inoculation. The ECRILs (188 F7:8 plants) and GWAS panel of 352 core collection accessions were scored for disease resistance with 10 replications per line. The plants were grown in 50-cell plastic trays, and inoculation was carried out by drenching plants at the 4–6 true leaf stage with 5 mL of spore suspension at the base of the stem using a dispenser. The inoculated plants were maintained under protected conditions at 24–30 °C and frequently watered to facilitate disease establishment. C. annuum cultivars CM334, PI201234, and Mohanjilju (commercial F1 hybrid, Syngenta Korea) were used as the resistant controls, whereas C. annuum cultivars ECW30R, Tean, and Geumsugangsan (commercial F1 hybrid, Takii Korea) were used as the susceptible controls to compare the disease resistance levels. The disease indexes were scored at three weeks post inoculation based on a previously described disease scale of 1–4, where 1 = no visible symptoms, 2 = dark lesion visible at the base of the stem but surviving without wilting, 3 = wilting with a dark lesion at the base of the stem, and 4 = wilting and death of the whole plant34,53.
Genotyping-by-sequencing
Genotyping-by-sequencing was performed for the ECRILs and GWAS core collection as described previously23. Briefly, genomic DNA was diluted, adjusted to a concentration of 50 ng/µL per ECRIL, and digested with the restriction enzymes EcoRI and MseI using a SBG 100‐Kit v2.0 (Keygene N.V., Wageningen, the Netherlands). For the GWAS core collection population, GBS libraries were constructed manually using restriction enzymes PstI/MseI and EcoRI/MseI as described previously23,54. Following the ligation of selective adaptors, the libraries were amplified using selective primers containing “GA” for the ECRILs and “TA” for the GWAS core collection population. Amplified libraries consisting of 188 ECRILs and two replicates of susceptible parents were pooled into a single tube. Libraries consisting of the GWAS core collection populations were pooled into five tubes. The pooled libraries were sequenced on the Illumina HiSeq. 2000 platform at Macrogen (Macrogen, Inc. Seoul, Korea). Trimming and quality control of the GBS raw data were performed using a CLC Genomics Workbench v6.5 (Qiagen, Aarhus, Denmark) with a minimum read length of 80 bp and a minimum quality score of Q20. Filtered reads were aligned to the C. annuum cv. CM334 reference genome (chromosome v1.6, http://peppergenome.snu.ac.kr) using the Burrows-Wheeler Aligner (BWA)55. Filtering and SNP calling were performed using the Genome Analysis Toolkit (GATK) Unified Genotyper version 3.3-0. SNPs in the ECRILs were filtered with QUAL value > 20 and minimum read depth of 3. SNPs in the GWAS core collection population were filtered with minor allele frequency (MAF) > 0.03, calling rate >0.6, and inbreeding coefficient (F) > 0.8.
Construction of bin map and QTL analysis
SNPs with unequal segregation and > 10% missing data were excluded from genetic linkage map construction. For genetic linkage map construction, bins were treated as genetic markers. The sliding window approach was used to impute the missing data and to detect recombination breakpoints as described previously23. To assign genetic positions to the bins, arranged bins were mapped with a LOD (logarithm of the odds) threshold of 3.0 and a distance threshold of 30 cM using CarthaGene software. The Kosambi mapping function was used to infer genetic distances between markers in centimorgans (cM). QTL analysis was performed using Composite Interval Mapping (CIM) with Windows QTL Cartographer 2.556. The 1000-permutation test (P < 0.05) was performed to determine the LOD threshold for significance of each QTL. Explanations of the percentage of phenotypic variance [R2 (%)] and additive effects for each QTL were obtained using the same software. Epistatic effects and the interactions of major QTLs were examined using the Multiple Interval Mapping (MIM) option under the command “Scan through QTL mapping results file” and refined further through “Testing for existing QTLs” with a Bayesian Information Criterion (BIC-X) model using default options. The physical locations of the QTLs were compared with the genetic locations of bins linked to the significant QTLs. The C. annuum cv. CM334 reference genome (chromosome v1.6, http://peppergenome.snu.ac.kr) was used to compare the positions of significant SNPs in the QTLs and GWAS core collection identified in this study and with previously reported linked markers. The marker sequences of previous studies were retrieved using https://solgenomics.net/search/markers and BLAST analysis of the reference genomes. If the sequence information for a particular linked marker was not publically accessible, the closest marker was used.
Genome-wide association analyses for PcRR resistance
The 507,713 filtered SNPs discovered from the 352 core collection population were used for GWAS mapping. Population structure (PCA and kinship matrixes) and genome-wide association (GWAS) were estimated with the Compressed Mixed Linear Model (CMLM) using the R package Genomic Association and Prediction Integrated Tool (GAPIT)57 with default parameters. All probabilities generated in the association runs were transformed by -log10 P (FDR p-value < 0.05). Scores for an individual chromosome were then inspected in Manhattan plots to determine whether the SNPs reached the significance threshold. The -log10 p-values of SNPs from GWAS were adjusted by58 multiple test correction, and the adjusted cutoff value for accepting thresholds was set to -log10 (P) value ≥ 7.0. The haplotype blocks of the GWAS core collection were estimated using PLINK v1.9 with the following settings:–no-parents–allow-no-sex–blocks-max-kb 2000–blocks-inform-frac 0.9–blocks-strong-highci 0.85–blocks-recomb-highci 0.859. Haplotype block analysis for the chromosomes identified with significant SNPs was performed using Haploview 4.2 software60. The significant SNP data sets from GAPIT were used to calculate pair-wise LD between SNPs. A 1% cutoff was used. Haplotype blocks in the region were defined with the confidence interval method61.
Identification of candidate genes
To identify candidate genes associated with PcRR resistance, the positions of highly significant and linked bin markers within QTLs on the genetic map were compared with their physical positions on the pepper reference genome (v1.6), and 1-Mb upstream and downstream sequences were mined for genes associated with PcRR disease resistance. Similarly, 1-Mb sequence information for the highly significant SNPs associated with the PcRR resistance trait from the GWAS peak was used for BLAST (blastn) analysis against the C. annuum cv. CM334 reference genome database (chromosome v1.6, http://peppergenome.snu.ac.kr) to identify candidate PcRR resistance genes.
Statistical analysis to estimate genetic variability against PcRR
The mean disease scores of ten plants per ECRIL and GWAS core collection accessions were used to calculate the frequency distribution, coefficient of variation (CV%), range, and genetic variability parameters for three different isolates in their respective environments. Broad-sense heritability (H2 bs) was calculated with the formula (suggested previously62) , where, is the genetic variance and is the phenotypic variance. The Pearson correlation was calculated using Rstudio.
Supplementary information
Identifying candidate genes for Phytophthora capsici resistance in pepper (Capsicum annuum) via genotyping-by-sequencing-based QTL mapping and genome-wide association study
Acknowledgements
This research was supported by Golden Seed Project (213006-05-3-SB540), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS), Republic of Korea. This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01322901), Rural Development Administration, Republic of Korea.
Author Contributions
B.K. and M.I.S. conceived this research. B.K. and M.I.S. designed the experiments, M.I.S., H.L., K.H., N.R. and S.A.M. performed data analysis; M.I.S. developed the mapping population, performed genotyping and phenotyping; M.I.S. and J.V. drafted the manuscript; B.K., M.I.S, J.V. J.K., A.C. and A.S.P. revised the manuscript. All authors reviewed the manuscript and approved the final draft.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46342-1.
References
- 1.Bosland P, Lindsey D. A. seedling screen for Phytophthora root rot of pepper, Capsicum annuum. Plant Dis. 1991;75:1048–1050. doi: 10.1094/PD-75-1048. [DOI] [Google Scholar]
- 2.Foster J, Hausbeck M. Resistance of pepper to Phytophthora crown, root, and fruit rot is affected by isolate virulence. Plant Dis. 2010;94:24–30. doi: 10.1094/PDIS-94-1-0024. [DOI] [PubMed] [Google Scholar]
- 3.Lamour K, Hausbeck M. Mefenoxam insensitivity and the sexual stage of Phytophthora capsici in Michigan cucurbit fields. Phytopathology. 2000;90:396–400. doi: 10.1094/PHYTO.2000.90.4.396. [DOI] [PubMed] [Google Scholar]
- 4.Rehrig, W. Z., Ashrafi, H., Hill, T., Prince, J. & Van Deynze, A. CaDMR1 cosegregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). PlantGenome7 (2014).
- 5.Hausbeck MK, Lamour KH. Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 2004;88:1292–1303. doi: 10.1094/PDIS.2004.88.12.1292. [DOI] [PubMed] [Google Scholar]
- 6.Kamoun S, et al. The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant. Pathol. 2015;16:413–434. doi: 10.1111/mpp.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barchenger DW, Lamour KH, Bosland P. Challenges and strategies for breeding resistance in Capsicum annuum to the multifarious pathogen, Phytophthora capsici. Front. Plant. Sci. 2018;9:628. doi: 10.3389/fpls.2018.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barchenger DW, et al. Race characterization of Phytophthora root rot on Capsicum in Taiwan as a basis for anticipatory resistance breeding. Phytopathology. 2018;108:964–971. doi: 10.1094/PHYTO-08-17-0289-R. [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Wang L, Guo J, Yang W, Shen H. Molecular mapping of a gene conferring resistance to Phytophthora capsici Leonian race 2 in pepper line PI201234 (Capsicum annuum L.) Mol. Breeding. 2016;36:66. doi: 10.1007/s11032-016-0464-0. [DOI] [Google Scholar]
- 10.Liu WY, et al. Combined use of bulked segregant analysis and microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper. Theor. Appl. Genet. 2014;127:2503–2513. doi: 10.1007/s00122-014-2394-8. [DOI] [PubMed] [Google Scholar]
- 11.Thabuis A, et al. Comparative mapping of Phytophthora resistance loci in pepper germplasm: evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor. Appl. Genet. 2003;106:1473–1485. doi: 10.1007/s00122-003-1206-3. [DOI] [PubMed] [Google Scholar]
- 12.Candole BL, Conner PJ. Screening Capsicum annuum accessions for resistance to six isolates of Phytophthora capsici. HortScience. 2010;45:254–259. doi: 10.21273/HORTSCI.45.2.254. [DOI] [Google Scholar]
- 13.Vleeshouwers VGAA, et al. Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 2011;49:507–31. doi: 10.1146/annurev-phyto-072910-095326. [DOI] [PubMed] [Google Scholar]
- 14.Brouwer DJ, St. Clair DA. Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub NILs. Theor. Appl. Genet. 2004;108:628–638. doi: 10.1007/s00122-003-1469-8. [DOI] [PubMed] [Google Scholar]
- 15.Chen AL, et al. Reassessment of QTLs for late blight resistance in the tomato accession L3708 using a restriction site associated DNA (RAD) linkage map and highly aggressive isolates of Phytophthora infestans. PLoS ONE. 2014;9:e96417. doi: 10.1371/journal.pone.0096417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grube RC, Radwanski ER, Jahn M. Comparative genetics of disease resistance within the Solanaceae. Genetics. 2000;155:873–887. doi: 10.1093/genetics/155.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, et al. Mapping of a novel race specific resistance gene to phytophthora root rot of pepper (Capsicum annuum) using bulked segregant analysis combined with specific length amplified fragment sequencing strategy. PLoS ONE. 2016;11:e0151401. doi: 10.1371/journal.pone.0151401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnet J, et al. Are the polygenic architectures of resistance to Phytophthora capsici and P. parasitica independent in pepper? Theor. Appl. Genet. 2007;115:253–264. doi: 10.1007/s00122-007-0561-x. [DOI] [PubMed] [Google Scholar]
- 19.Truong H, et al. Identification of isolate‐specific resistance QTLs to phytophthora root rot using an intraspecific recombinant inbred line population of pepper (Capsicum annuum) Plant Pathol. 2012;61:48–56. doi: 10.1111/j.1365-3059.2011.02483.x. [DOI] [Google Scholar]
- 20.Kim HJ, et al. BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.) Theor. Appl. Genet. 2008;118:15–27. doi: 10.1007/s00122-008-0873-5. [DOI] [PubMed] [Google Scholar]
- 21.Mallard S, et al. A. key QTL cluster is conserved among accessions and exhibits broad-spectrum resistance to Phytophthora capsici: a valuable locus for pepper breeding. Mol. Breeding. 2013;32:349–364. doi: 10.1007/s11032-013-9875-3. [DOI] [Google Scholar]
- 22.Thabuis A, et al. Phenotypic and molecular evaluation of a recurrent selection program for a polygenic resistance to Phytophthora capsici in pepper. Theor. Appl. Genet. 2004;109:342–351. doi: 10.1007/s00122-004-1633-9. [DOI] [PubMed] [Google Scholar]
- 23.Han K, et al. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum) DNA Res. 2016;23:81–91. doi: 10.1093/dnares/dsv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han K, et al. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in. Capsicum. Plant Biotechnol. J. 2018;16:1546–1558. doi: 10.1111/pbi.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu F, et al. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017;7:4516. doi: 10.1038/s41598-017-04903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu C, Gore M, Buckler ES, Yu J. Status and Prospects of Association Mapping in Plants. Plant. Genome. 2008;1:5–20. [Google Scholar]
- 28.Brachi B, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowell S, et al. Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat. Commun. 2016;7:10527. doi: 10.1038/ncomms10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallam A, Arbaoui M, El-Esawi M, Abshire N, Martsch R. Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter faba bean. Front. Plant. Sci. 2016;7:1098. doi: 10.3389/fpls.2016.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015;13:211–221. doi: 10.1111/pbi.12249. [DOI] [PubMed] [Google Scholar]
- 32.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 33.Lee HY, et al. Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet. 2016;17:142. doi: 10.1186/s12863-016-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim BS, Ahn JH, Lee JM, Park DG, Kim HY. Restorer genotype for male sterile cytoplasm of genetic resources moderately resistant to Phytophthora capsici in Capsicum pepper. Korean J. Hortic. Sci. Technol. 2012;30:73–78. doi: 10.7235/hort.2012.11104. [DOI] [Google Scholar]
- 35.Naegele RP, et al. QTL mapping of fruit rot resistance to the plant pathogen Phytophthora capsici in a recombinant inbred line Capsicum annuum population. Phytopathology. 2014;104:479–483. doi: 10.1094/PHYTO-05-13-0143-R. [DOI] [PubMed] [Google Scholar]
- 36.Ogundiwin EA, et al. Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.) Genome. 2005;48:698–711. doi: 10.1139/g05-028. [DOI] [PubMed] [Google Scholar]
- 37.Rose LE, et al. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, et al. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- 39.Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 40.Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends. Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 41.McDowell JM, Simon SA. Recent insights into R gene evolution. Mol. Plant. Pathol. 2006;7:437–448. doi: 10.1111/j.1364-3703.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- 42.Ballvora A, et al. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C, et al. The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 2014;127:1353–1364. doi: 10.1007/s00122-014-2303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashfield T, et al. Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol. Plant Microbe Interact. 2003;16:817–826. doi: 10.1094/MPMI.2003.16.9.817. [DOI] [PubMed] [Google Scholar]
- 45.Li L, et al. Fine mapping and candidate gene analysis of two loci conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 2016;129:2379–2386. doi: 10.1007/s00122-016-2777-0. [DOI] [PubMed] [Google Scholar]
- 46.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 47.Nagy ED, Bennetzen JL. Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res. 2008;18:1918–1923. doi: 10.1101/gr.078766.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torii KU. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 2004;234:1–46. doi: 10.1016/S0074-7696(04)34001-5. [DOI] [PubMed] [Google Scholar]
- 49.Lee SC, Hwang BK. Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta. 2003;3:387–396. doi: 10.1007/s00425-002-0875-5. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Bradera GN, Palvaa ET. Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin B1 in Arabidopsis. Mol. Plant. 2008;3:482–495. doi: 10.1093/mp/ssn013. [DOI] [PubMed] [Google Scholar]
- 51.Yang HB, et al. Development and validation of L allele-specific markers in Capsicum. Mol. Breeding. 2012;30:819–829. doi: 10.1007/s11032-011-9666-7. [DOI] [Google Scholar]
- 52.Jo S-J, et al. Resistance of chili pepper cultivars to isolates of Phytophthora capsici. Korean J. Hortic. Sci. Technol. 2014;32:66–76. doi: 10.7235/hort.2014.13079. [DOI] [Google Scholar]
- 53.Siddique MI, et al. Resistance to Phytophthora capsici, restorer-of-fertility genotype for cytoplasmic male sterility and chemical quality components of breeding lines developed for improvement of subicho, a land race of pepper in Yeongyang. Hort. Sci. Technol. 2017;35:758–768. [Google Scholar]
- 54.Jo J, et al. Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays. Front. Plant. Sci. 2017;8:1606. doi: 10.3389/fpls.2017.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S., Basten, C. & Zeng, Z. Windows QTL cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University, USA. (2011).
- 57.Lipka AE, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 58.Bonferroni, C. E. Teoria statistica delle classi e calcolo delle probabilita. Florence (Libreria Internazionale Seeber; 1936).
- 59.Chang CC, et al. Second generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 61.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 62.Allard, R. W. Principles of plant breeding (John Wiley and Sons Inc.1960).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identifying candidate genes for Phytophthora capsici resistance in pepper (Capsicum annuum) via genotyping-by-sequencing-based QTL mapping and genome-wide association study