Abstract
By GWAS studies on celiac disease, gene expression was studied at the level of the whole intestinal mucosa, composed by two different compartments: epithelium and lamina propria. Our aim is to analyse the gene-expression and DNA methylation of candidate genes in each of these compartments. Epithelium was separated from lamina propria in biopsies of CeD patients and CTRs using magnetic beads. Gene-expression was analysed by RT-PC; methylation analysis required bisulfite conversion and NGS. Reverse modulation of gene-expression and methylation in the same cellular compartment was observed for the IL21 and SH2B3 genes in CeD patients relative to CTRs. Bioinformatics analysis highlighted the regulatory elements in the genomic region of SH2B3 that altered methylation levels. The cREL and TNFAIP3 genes showed methylation patterns that were significantly different between CeD patients and CTRs. In CeD, the genes linked to inflammatory processes are up-regulated, whereas the genes involved in the cell adhesion/integrity of the intestinal barrier are down-regulated. These findings suggest a correlation between gene-expression and methylation profile for the IL21 and SH2B3 genes. We identified a “gene-expression phenotype” of CeD and showed that the abnormal response to dietary antigens in CeD might be related not to abnormalities of gene structure but to the regulation of molecular pathways.
Subject terms: Gene expression, DNA methylation
Introduction
Celiac disease (CeD) is a systemic immune-mediated disease triggered by gluten ingestion in genetically susceptible individuals. It is the most common form of food intolerance, and its prevalence has increased over the last three decades1.
CeD has a strong genetic component, as suggested by our twin studies2. The primary genes associated with CeD are Major Histocompatibility Complex class II (MHC-II) genes encoding HLA-DQ2 (i.e. HLA-DQA1*05 and HLA-DQB1*02) or HLA-DQ8 (i.e. HLA-DQA1*03 and HLA-DQB1*03:02). These molecules consists of an alpha chain (HLA-DQα) and a beta chain (HLA-DQβ) that form a heterodimer, which is anchored to the cell membrane. There is an HLA gene-dose effect on disease risk, as individuals carrying two copies of HLA-DQ2 have a higher susceptibility for celiac disease than do those with only one copy3. Recently, it was shown that HLA-DQ7 represents an additive or independent CD-risk haplotype with respect to HLA-DQ2/DQ8 haplotypes4. However, these haplotypes are common in the general population, and not all carriers develop clinical disease; thus, they are not sufficient for disease development, accounting for approximately 40% of the heritability of CeD5.
Both MHC and non-MHC genetic factors influence CeD development, and since the first case/control Genome-Wide Association Study (GWAS) on celiac disease was published in 20076, a total of 57 non-HLA loci have been identified as associated with this disease. To date, 57 non-MHC variants have been estimated to account for 15% of CeD heritability, but the remaining 50% heritability of CeD remains unexplained7.
In a previous study, we aimed to improve the estimation of CeD risk in siblings by adding to HLA haplotype a small set of non-HLA genes. Applying a Bayesian approach, we improved the estimation of CeD risk among siblings over the HLA-based risk, providing a tool to predict the disease in at-risk individuals8.
The vast majority of CeD-associated SNPs do not map to exons but intersect with regulatory regions, implying that protein changes do not govern disease development. The analysis of expression Quantitative Trait Locus (eQTL) CeD-associated polymorphisms has shown that these anomalies often affect the expression of nearby genes in different cell types9,10; however, to date, they have been explored in small intestinal biopsiesonly, which contain multiple cell types, leading to results that are difficult to interpret. The intestinal mucosa is composed of two different compartments, the epithelium and the lamina propria.
The immune responses in the epithelium and lamina propria are separated by a basement membrane, which appears thinner and with more breaches in patients with active CeD than in patients on gluten-free diets or in non-CeD subjects11. The upstream events activating adaptive immune responses that occur in the lamina propria interact with the downstream events in the epithelium. However, how these immune responses in the lamina propria and the epithelium interact remains unclear12.
In our previous work, we showed that the combined expression of 4 non-HLA selected genes in peripheral blood monocytes enabled discrimination between CeD patients and controls (CTRs) and between CeD patients on a gluten-free diet and disease controls13. We then confirmed the importance of gene expression by showing that the expression of a small set of candidate genes, in peripheral blood mononuclear cells, can predict CeD at least 9 months before the appearance of any clinical and serological sign of disease in genetically at-risk infants14.
In the present study, we aim to analyse a set of candidate genes to explore both genetic and epigenetic alterations in isolated intestinal cell populations from both the epithelium and lamina propria.
The final aim of this work is to evaluate alterations in candidate gene expression; identify, at the level of distinct cell populations, potential alterations consistent with the gluten-induced damage in CeD; and describe the mechanisms of epigenetic regulation that underlie these alterations.The final aim of this work is to evaluate alterations in candidate gene expression; identify, at the level of single and distinct cell populations, potential alterations consistent with the gluten-induced damage in CeD; and describe the mechanisms of epigenetic regulation that underlie these alterations.
Results
Evaluation of sample purity
The efficacy of separation between the two compartments, epithelium and lamina propria, was evaluated by real-time PCR. We measured the Epithelial Cell Adhesion Molecule (EpCAM) expression level, as the specific marker of the epithelial cells, in both epithelial and lamina propria cells, normalized to the expression of an endogenous gene (GUSb) and used as reference sample in the epithelial compartment. As shown in Supplementary Fig. 1, a 98% purity of the epithelial compartment was achieved. In particular, the analysis of EpCAM expression generated a selection of 97.8% epithelial cells (CD326+) in celiac biopsies and 97.5% epithelial cells (CD326+) in the biopsies of controls (Supplementary Fig. 1).
Gene expression in the epithelium and lamina propria
The expression of 16 CeD-associated genes (Table 1) in each compartment was compared between CeD patients and controls. To simplify interpretation, we grouped the candidate genes into 4 putative functional groups: (1) genes directly involved in inflammation and damage, (2) “classical” candidate genes strongly associated with CeD of unpredictable function, (3) genes involved in the regulation of inflammation and damage, and (4) genes involved in the maintenance of cell adhesion and intestinal barrier integrity. These genes were selected based on their robust replication in several GWASs, our previous studies in several models and their likely putative roles in the gluten-induced abnormal immune response6–10,13,14.
Table 1.
List of genes analysed in the study, their functions, and the TaqMan Gene Expression assays used in the expression experiments (Life Technologies).
| Gene | Function | Functional Group | TaqMan Assay |
|---|---|---|---|
| IL12A | Pro-inflammatory cytokine | Inflammation/Damage Direct | Hs00222327_ml |
| IL21 | Pleiotropic cytokine | Inflammation/Damage Direct | Hs00168405_ml |
| NFKB1 | Regulation of autoimmunity and inflammation | Inflammation/Damage Direct | Hs00765730_m1 |
| C-REL | Subunit of the NF-kB transcription complex | Inflammation/Damage Direct | Hs00968436_m1 |
| TNFAIP3 | Negative feedback loop control of NF-kB | Inflammation/Damage Direct | Hs00234713_m1 |
| KIAA1109 | Located in the genomic region associated with CeD | Canidated/Associated | Hs00361070_ml |
| SH2B3 | Activates PI3-kinase | Inflammation/Damage Regulation | Hs00193878_m1 |
| RGS1 | Lymphocytes Homing | Inflammation/Damage Regulation | Hs00175260_m1 |
| TAGAP | Negative regulator of the immune response | Inflammation/Damage Regulation | Hs00611823_m1 |
| TNFRSF14 | Activation of NK intestinal and CD4 + T cells, “gut-homing cells” | Inflammation/Damage Regulation | Hs00998604_ml |
| TNFSF14 | Activation of NK intestinal and CD4 + T cells, “gut-homing cells” | Inflammation/Damage Regulation | Hs00542477_m1 |
| LPP | Extracellular matrix and cell-cell contact homeostasis | Cell Adhesion/Integrity of Intestinal Barrier | Hs00944352_m1 |
| TJP1 | Proteins of the tight junctions involved in maintaining the integrity of the intestinal barrier | Cell Adhesion/Integrity of Intestinal Barrier | Hs01551861_ml |
| PTPRK | Maintenance of cell junctions and participation in the modulation of EGFR activity, resulting in an inhibition of cell proliferation | Cell Adhesion/Integrity of Intestinal Barrier | Hs00267788_ml |
| ARHGAP31 | Regulation of cell migration, focal adhesion size and dynamics | Cell Adhesion/Integrity of Intestinal Barrier | Hs00393361_ml |
| C1orf106 | Involved in cell adhesion processes | Cell Adhesion/Integrity of Intestinal Barrier | Hs01009089_ml |
Figure 1A shows that the IL12A, IL21, c-REL, RGS1, SH2B3 genes, which are directly or indirectly involved in the inflammation process, were significantly up-regulated in the epithelial cells of CeD patients relative to controls.
Figure 1.
Gene expression analysis. In each of the epithelium and lamina propria, expression of 16 CeD-associated genes was compared between CeD patients and CTRs. The IL12A, IL21, c-REL, RGS1, and SH2B3 genes were significantly up-regulated in the epithelial cells of CeD patients relative to CTRs. In the lamina propria, the IL12A, IL21 and RGS1 genes were equally upregulated between CeD patients and CTRs. In addition,TNFSF14 and PTPRK were down-regulated in CeD patients relative to CTRs (B). *p < 0.05; **p < 0.01.
Similarly, IL12A, IL21, NFKB1 and RGS1 were equally up-regulated in the lamina propria of CeD patients relative to their corresponding expression in controls. In addition,TNFSF14 and PTPRK were down-regulated in the lamina propria of CeD patients relative to their expression in controls (Fig. 1B). Figures of the expression of each gene are provided in the supplementary data (Supplementary Fig. 2).
Differences between CeD patients and controls were evaluated by the rank-sum test (Mann-Whitney) because of the asymmetry in the data distributions.
Correlations in gene expression
The expression of genes in each compartment is the result of complex correlations among genes within specific and inter-related metabolic and signalling pathways.
Thus, it was of interest to examine the correlations among genes within a specific compartment in CeD patients and controls.
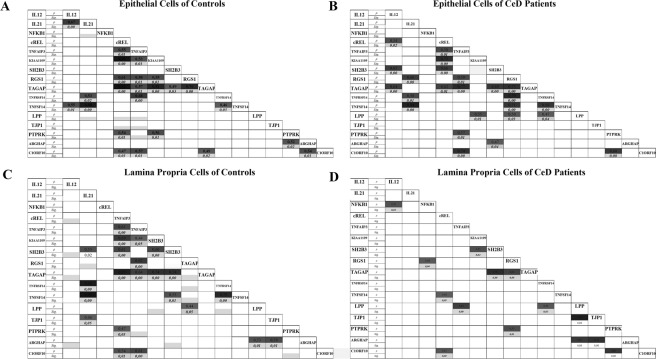
The results demonstrate the complexity of these correlations, which include not only bivariate correlations but also multi-dimensional correlations. Figure 2 shows the correlations of gene expression in the epithelium of CeD patients (B) and controls (A) and in the lamina propria of controls (C) and CeD patients(D).
Figure 2.
Patterns of correlations between genes in expression in the epithelium (A,B) and lamina propria (C,D) of celiacs and controls. For each pair of genes, the intensity of coloration of the box is proportional to the correlation between them, and the Pearson coefficient (ρ) is shown. Only those correlations significant at p < 0.05 are shown.
It may be observed that, based on the degree of darkness, some metabolic profiles were well correlated in both CeD patients and controls in both compartments, reflecting “mandatory” functional pathways. In contrast, other correlations that were strong among controls were absent in the epithelium and lamina propria of CeD patients. In the lamina propria compartment, 24 strong correlations (correlation coefficient (r) values > 0.5) were observed in the controls, whereas only 14 such correlations were observed in CeD patients. Specifically, in the lamina propria of controls, the expression of TNFAIP3 was strongly correlated with the expression other genes involved in the regulation of inflammation, including TAGAP, RGS1, cREL and C1orf106. In the lamina propria of CeD patients, there were no significant correlations among these genes, although a correlation between TNFAIP3 and LPP expression was observed.
In contrast, in the epithelium of CeD patients, no correlations were lost relative to those observed in the epithelium of controls, but several candidate genes showed stronger correlations in CeD patients than in controls. SH2B3 was strongly correlated with TAGAP, cREL and IL-12A only in CeD patients, suggesting a specific role of this gene in the gluten-induced immune response.
Signatures of CeD in the epithelium and lamina propria as evidenced from multivariate analysis
Multivariate analysis was performed to better understand the differential involvement of the candidate genes in the two compartments between CeD patients and CTR subjects.
Tables 2 and 3 show the results of the discriminant analysis for the epithelium: a small set of genes (IL21, TNFSF14 with TNFRSF14, NFKB1 and SH2B3) discriminated most CeD patients (13/18; 72.2%) from controls, whereas only 2/18 controls (11.1%) were incorrectly classified as CeD patients.
Table 2.
Stepwise discriminant analysis of gene expression in epithelial cell in epithelial cells.
| Step | Gene | Wilk’s Lambda | Variance Ratio F | |
|---|---|---|---|---|
| Statistic | p | |||
| 1 | IL21 EPI | 0,749 | 11,042 | 0.000 |
| 2 | TNFSF14 EPI | 0,643 | 8,878 | 0.000 |
| 3 | NFKB1 EPI | 0,558 | 8,183 | 0.000 |
| 4 | TNFRSF14 EPI | 0,508 | 7,268 | 0.000 |
| 5 | SH2B3 EPI | 0,456 | 6,926 | 0.000 |
Five genes (IL21, TNFSF14, NFKB1, TNFRSF14, and SH2B3) were selected for analysing discrimination capacity, with a p value less than 0.001.
Table 3.
Classification by discriminant equation of gene expression in epithelial cells.
| Status | Predicted Group | Total | ||
|---|---|---|---|---|
| CD | Not CD | |||
| Original Group | CeD | 13 (72.2%) | 5(27.8%) | 18 |
| Not CeD | 2 (11.1%) | 16 (88.9%) | 18 | |
Results of the prediction analysis: 88.9% of controls and 72.2% of celiac patients were correctly classified. Overall Correct Classification = 80.6%
In the lamina propria (Tables 4–5), the combinations of IL12 and IL21, TNFSF14 and PTPRK, and NFKB1 and KIAA1109correctlyclassified 90% (16/18 CeD patient and 17/19 CTRs)of individuals.
Table 4.
Results of discriminant analysis in lamina propria cells.
| Step | Gene | Wilk’s Lambda | Variance Ratio F | |
|---|---|---|---|---|
| Statistic | p | |||
| 1 | IL12 LP | 0,595 | 23,100 | 0.000 |
| 2 | NFKB1 LP | 0,442 | 20,823 | 0.000 |
| 3 | TNFSF14 LP | 0,400 | 16,018 | 0.000 |
| 4 | IL21 LP | 0,314 | 16,949 | 0.000 |
| 5 | PTPRK LP | 0,288 | 14,814 | 0.000 |
| 6 | KIAA1109 LP | 0,270 | 13,040 | 0.000 |
Six genes (Il12, NFKB1, TNFSF14, IL21, PTPRK, and KIAA1109) were selected for analysing discrimination capacity, with a p value less than 0.001.
Table 5.
Classification by discriminant equation of gene expression in lamina propria cells.
| Status | Predicted Group | Total | ||
|---|---|---|---|---|
| CD | Not CD | |||
| Original Group | CeD | 16 (88.9%) | 2 (11.1%) | 18 |
| Not CeD | 2(10.5%) | 17 (89.5%) | 19 | |
Results of the prediction analysis: 89.5% of controls and 88,9.2% of celiac patients were correctly classified. Overall Correct Classification = 89.2%.
TNFSF14 and IL21 with NFKB1 were efficient discriminators in both compartments. In the epithelium, the receptor of TNFSF14 (TNFRSF14) and SH2B3 provided further discrimination. In the lamina propria, PTPRK, KIAA1109 and IL12 contributed to better discrimination.
However, the actual “best profile”, obtained through a mathematical procedure, should be interpreted under a functional scenario. The specific pattern of variable selection through the stepwise procedure in the epithelium is not shown (Supplementary Table 1). At step 0, the gene producing the best F (variance) ratio between the CeD patients and CTRs was selected (IL21 F = 11.04): the second-best gene was IL12 (F = 8.22). However, at step 1, having included IL21 in the model, the F Ratio of IL12 decreased to 2.4. Thus, IL12 lost its ability to contribute to the discriminant model but certainly did not lose its function. In the lamina propria, RGS1 was a strong discriminator at step 0 (F = 4.6), but after the best gene,IL12, was included (with F = 23.1), RGS1 completely lost its discriminant ability (F declined to 0.35). However, it certainly did not lose its functional significance (Supplementary Table 2).
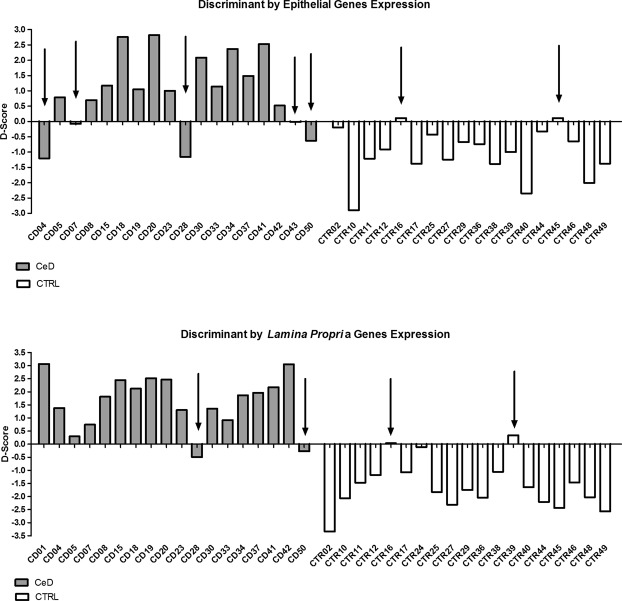
Through the model obtained by the analysis, a discriminating score (D-Score) was computed for each individual, related to the probability of membership among the cases or controls. Figure 3A,B show the pattern of probability of membership according to D-Score in CeD patients and controls. The gene expression signature obtained by the analysis produced an acceptable distinction of the celiac children from the controls. The classification might be slightly optimistic since we classified individuals by the coefficients obtained in the same cohort. However, when we applied a jack-knife method, classifying each individual through an auto-exclusion procedure, we still obtained 70% correct classification.
Figure 3.
D-Score graphs. For the epithelial cells, five CeD and two non-CeD samples (indicated by the arrows) were misclassified, yielding a total correct classification rate of 80.6% (A). For the lamina propria cells, two CeD and two non-CeD samples were misclassified, yielding a total correct classification rate of 89.2% (B).
Methylation analysis
The methylation analysis revealed several differences between CeD patients and controls in each compartment. Unfortunately, for RGS1 and PTPRK genes, the list of CpG islands was not available: they were not included in the methylation analysis. The differences in “mean level of methylation” for all candidate genes are shown in Fig. 4A,B. A mean value may show an aliasing bias since the methylation of specific CpG islands of the gene might be more important in regulating gene expression than is the average methylation through at least 20–40 CpG islands.
Figure 4.

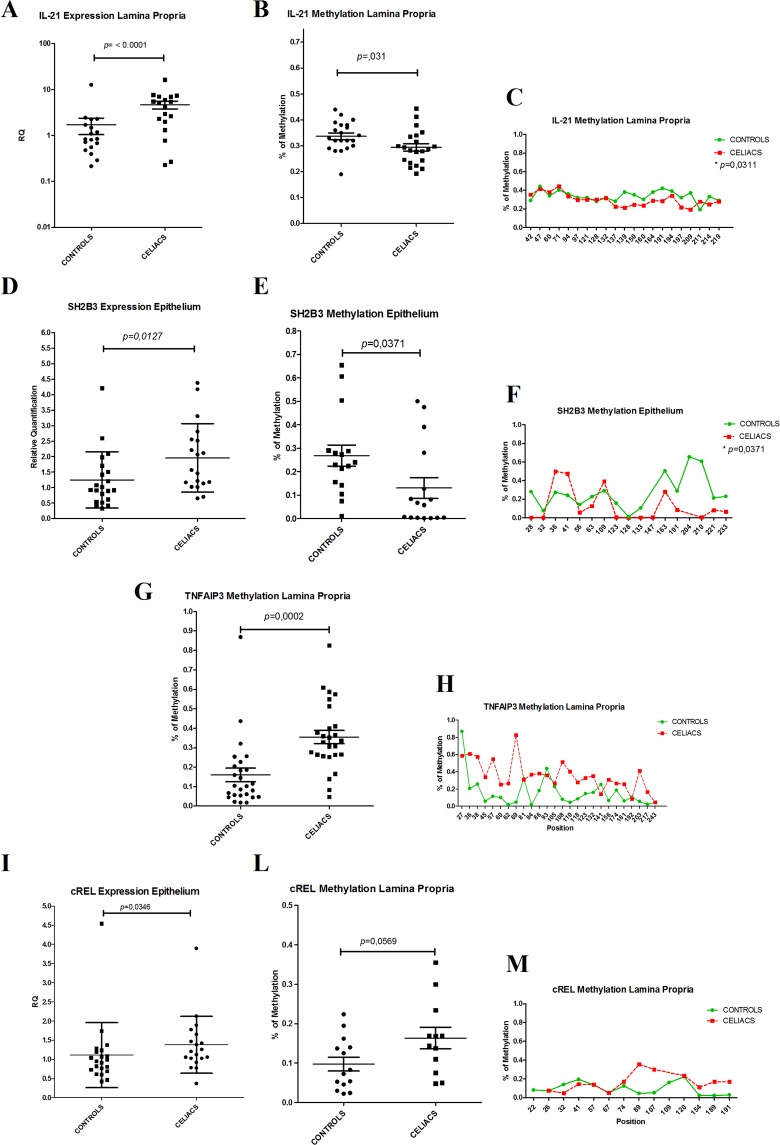
Average level of methylation for the candidate genes in the epithelium and lamina propria. In the epithelial cells (A), only SH2B3 was differentially methylated between CeD and CTR subjects (p = 0.003), whereas in the lamina propria (B) the genes IL21 (p = 0.03), TNFAIP3 (p < 0.001) and cREL (p = 0.005) showed differences in methylation level between CeD patients and CTRs. p < 0.05; **p < 0.01
Figure 4A,B shows the average level of methylation of the candidate genes in either the epithelium or the lamina propria. In epithelial cells, only SH2B3 was differentially methylated between CeD patients and controls (p = 0.003), whereas in the lamina propria, the genes IL21 (p = 0.03), TNFAIP3 (p < 0.001) and cREL(p = 0.005) showed differences in methylation level between CeD and CTRs.
Further investigation showed that IL21 expression in the lamina propria was greater in CeD patients than in controls (Fig. 5A):this gene was 20% less methylated in CeD patients than in controls. Figure 5B shows that several regions of the gene were differentially methylated. Figure 5C shows the methylation of each single nucleotide of the CpG island.
Figure 5.
Fine representation of methylation levels across the genes sequences. IL21(panel A) and SH2B3 (panel D) showed enhancer gene expression associated with lower methylation of the genes (panels B and E) in CeD patients than in controls. Conversely, TNFAIP3 (panel G) and cREL (panel L) showed higher methylation levels in CeD patients than in controls; higher gene expression in CeD patients was observed only for cREL. The methylation of each single nucleotide of the CpG island (position) is shown in panels C,F, H, and M.
Similarly, the lower methylation of the SH2B3 gene in CeD epithelial cells was associated with higher expression of the gene (Fig. 5D–F). When we explored this region, we observed that CpG island 132 was markedly differentially methylated between CeD patients and controls; the methylated nucleotides of that site coincided with the DNA regulation elements. By using the Epigenome Roadmap tool (http://www.roadmapepigenomics.org/), we identified the presence of several regulation elements as histone modifications H3K27ac and H3K4me3 in the small intestine and H3K4me1, H3K4me3, and H3K9ac in duodenum mucosa and a DNAse hypersensitive tract (Fig. 6).
Figure 6.
Bioinformatics analysis output related to the regulation elements of the SH2B3 gene: (A) Ref Seq analysis showed the presence of regulation elements in the SH2B3 gene region evidenced by the alteration of methylation levels in the duodenal mucosa. (B,C) Data analysed by the Epigenome Roadmap tool showed the presence of regulation elements DNase, H3K27ac, H3K4me3, and H3K9ac in the region.
c-REL and TNFAIP3 in the lamina propria showed higher methylation profiles in CeD patients than in controls (Fig. 5G,H–L). C-REL, which contributed to the differential gene expression profiles between CeD patients and controls, showed an inverse methylation profile (Fig. 5I–L,M).
Discussion
The increased incidence of CeD suggests a new epidemic of the current era. A strong genetic component has been confirmed. However, genes cannot explain a sudden increase in the incidence of a disease.
Several GWASs identified 57genes as associated with CeD, here referred to as “candidate genes”, each giving a small contribution to the CeD genetic risk. Together, these genes account for no more than 15% of the heritability of the disease7.Hence, half of the heritability remains to be explained.
None of the associated polymorphisms are in the coding region of the related gene, but at least half of these polymorphisms were related to the regulation of gene expression. We hypothesize that the study of epigenetic mechanisms could provide answers for some of the key questions in this field15.
In our previous studies, we explored the expression of candidate genes in the cells and tissues of several cohorts of CeD patients in the diagnostic phase of the disease or after treatment, in those of potential CeD patients and in those of a cohort of at-risk infants from CeD families13,14. These studies suggest an interesting scenario of interrelated expression of candidate genes and reinforce the actual selection of a small set of genes putatively implicated in the gluten-induced immune response in several cell compartments. Among the studied “candidate genes”, several genes were found to be differentially expressed in the peripheral monocytes between CeD patients and controls, and we developed multivariate models to discriminate between the two cohorts. We identified a small set of genes that enabled the correct classification (more than 90% correct classification) of the expression of “acute” CeD patients and controls without the need for clinical or serological data. Recently, we studied gene expression in peripheral blood cells in the first year of life and at least one year before the appearance of serum antibodies or clinical complaints. We found that the presence of 5 candidate gene polymorphisms together with the HLA “load” significantly increases the risk of developing the disease. Our findings enabled90% correct prediction of the outcome long before the appearance of any clinical or serological markers14. Tissue-related gene expression shows a cellular phenotype of an abnormal response to gluten, and its study may substantially contribute to our understanding of the specific cellular and immune responses to the offending agent in CeD15.
Thus, in this study, we explored gene expression in separate cellular compartments comprising a variety of cell types. A “single cell” approach might overcome some of the limitations of our work but will also add complexity to the interpretation of the results.
This is the first work to explore gene expression specifically in the purified epithelial tissue of the small intestine and the “non-epithelial” tissue of the mucosa16. Most previous reports present results obtained from isolated cells obtained from whole biopsies via tissue homogenization17 or from the isolation of epithelial cells by calcium chelants (EDTA)18. Unfortunately, these methods do not prevent cross contamination of different cell types from the epithelium and lamina propria.
Our microbead-based separation was efficient, with 98% purity of the epithelial compartment, as verified by RT-PCR. The separation of epithelial cells at high purity from the non-epithelial layer provides novel information about the function (expression) of candidate genes and insight into how the epithelial cells of CeD patients respond to gluten peptides via comparison with the same cells in other subjects that do not recognize gluten peptides.
Our results suggest that genes that are directly (NFKB, IL12A, IL21, and C-REL) or indirectly (SH2B3 and RGS1) involved in inflammation or damage processes are significantly up-regulated in CeD patients, in at least one cellular compartment. In contrast, the PTPRK gene, which is involved in the maintenance of cell junctions and the inhibition of cell proliferation, was down-regulated in CeD patients relative to controls.
A previous analysis of the correlations among candidate genes by Bilbao &colleagues16 showed that the pathway of normal correlation among this set of genes is grossly disrupted in CeD. The genes involved in inflammation do not function synergistically in CeD patients as they do in control subjects. It also appears that when we compared the correlations present in control cells with that observed in the cells of CeD subjects, new patterns appear in CeD: SH2B3 is co-regulated with other genes of inflammation (TAGAP ρ = 0.7, IL12 ρ = 0.65, cREL ρ = 0.76, ARHGAP31 = 0.47) only in CeD. We confirmed the presence of an altered correlation among tight junction genes, as observed by Jauregi-Miguel et al., in the mucosa of CeD patients on gluten-containing diets, which was restored after dietary treatment19. A good correlation among genes of the NFkB pathway was observed in the controls, whereas a significant disruption of these genes was observed in the CeD patients16. In contrast, co-methylation was stronger in CeD patients.
The present study is limited to the estimation of mRNA; hence, our understanding is incomplete since information on protein synthesis and regulation after the production of the messenger remains lacking.
The encouraging results of the differential gene expression analysis prompted us to explore the mechanisms of DNA methylation in the same set of candidate genes. The methylation data suggest modifications of the reading of the individual genome, which are unlikely to occur during the short-term development of the flat mucosa. Such modifications are generally considered to occur in the very early phase, including before birth.
DNA methylation is one of several mechanisms of epigenetic regulation of gene expression. We aimed to explore the regulation of interleukin IL21 and the SH2B3 gene, which play an unique roles in the pathogenesis of CeD. The multivariate discriminant analysis of the epithelium confirmed the pivotal roles of IL12-IL21 (in both cellular compartments) and the SH2B3 gene (in the epithelium). SH2B3 expression is higher in CeD patients than in CTRs in both the small intestine and peripheral blood cells before the appearance of the disease. This gene exerts multiple functions and establishes connections between immunity and inflammation20. The SH2B3 gene maps to chromosome12 at 12q24 and encodes a member of the Src homology 2-Binding (SH2-B) protein family, which is described as a negative regulator of T cell receptors, and it is implicated in T cell signalling21–23. SH2B3 is also a key regulator of haematopoietic cell lines, being a negative regulator of B cell lymphopoies is in the early phase of development and is expressed in hematopoietic stem cells (HSC) stem and hematopoietic progenitor cells (HPC) progenitor cell lines, the functions of which increase significantly in the absence of SH2B322,24.
SH2B3 is also involved in the three signalling pathways induced by erythropoietin (EPO)and thrombopoietin (TPO), which down-regulate JAK2 and stimulate HSC sand the production of megakaryocytes and erythrocytes. These data support the hypothesis that SH2B3 is a major negative regulator of HSC expansion and the production of blood cells through the modulation of growth factors and cytokines25–27. Outside the haematopoietic domain, SH2B3 is expressed in endothelial cells, phosphorylated by TNFa, and rapidly up-regulated either at the mRNA or protein level28. The ability of TNF to regulate SH2B3 has also been shown in human umbilical vein endothelial cells29.
Recently, it has been suggested that a lack of SH2B3 decreases the precursors of vascular cell adhesion molecule 1 (VCAM-1) on the membrane, suggesting a unique role of this gene in cell motility and adhesion30,31.
Despite knowledge of its multiple contributions, the complete pattern of SH2B3 regulation is not yet clear. This study, for the first time, shows that decreased methylation of a gene may modulate over-expression in the epithelium of CeD patients, suggesting an epigenetic regulation of the gene. Bioinformatics analysis showed that the differential methylation is centred in a genomic area of a DNA regulatory element and involves 4key histone modifications and a DNAse hypersensitive tract.
New types of experimental work, such as in vitro affinity tests and protein studies, are needed. Regardless, we suggest that the confirmed role of SH2B3 gene is well adapted to the impaired immune regulation observed after the gluten “offence” to CeD mucosa.
SH2B3 over-expression modifies the innate immune response, and the parallel induction of the gene by pro-inflammatory cytokines suggests the development of an inflammation “loop” induced by gluten peptides either at innate or induced levels.
In conclusion, we revealed the differential expression of candidate genes between CeD patients and controls in specific cell compartments of the intestinal mucosa. In addition, we identified a specific “gene expression phenotype” of CeD patients and showed that the abnormal response to dietary antigens might not be essentially related to abnormalities of gene structure but to the fine regulation of the pathways that respond to dietary antigens.
CeD patients appear to be “healthy and normal” people whose response to an abnormal dietary peptide is “physiologically” excessive and leads to inflammation and, eventually, severe cell destruction and clinical disease.
Methods
Patient enrolment
Expression studies of candidate genes in cells isolated from intestinal mucosa were performed on biopsy samples of duodenal mucosa from 19 patients (11 F and 8 M, median age 8 years, range 2–16 years) with CeD at the time of diagnosis and 21 (9 F and 12 M, median age 10 years, range 2–16 years) non-CeD CTRs (Supplementary Table 3). Written informed consent was obtained from the parents of the enrolled children, and the study was approved by The Independent Committee for Bioethics of I.R.C.C.S. Burlo Garofolo (Approval Number: CE/V-131). The patients were enrolled in the Department of Gastroenterology, Digestive Endoscopy and Clinical Nutrition of the Burlo Garofolo Hospital of Trieste, and the relevant biopsy samples of duodenal mucosa were collected by esophagogastroduodenoscopy (EGD).
Purification of intestinal epithelial cells
The biopsy samples were processed immediately after collection, and purification of the intestinal epithelium was performed using enzyme digestion followed by magnetic bead sorting as described previously32.
Extraction of nucleic acids and RNA reverse transcription
Total RNA was extracted from intestinal cells by using the All Prep DNA/RNA Kit Mini Kit (QIAGEN), which enables the simultaneous extraction of DNA and RNA from the same biological sample through a single procedure. The protocol provided by the manufacturer was followed. The kit uses column separation, and two nucleic acids are obtained by means of two different elutions. The quantities of RNA and DNA were measured using a Nanodrop® spectrophotometer, and RNA quality was analysed by agarose gel electrophoresis in Tris/Borate/EDTA buffer (TBE). RNA samples that failed quality control were excluded from further analysis.
The RNA (starting from 1 μg) was transcribed into cDNA by using the High-Capacity Reverse Transcription Kit (Applied Biosystems®).
Real-time PCR
Real-time PCR was performed by TaqMan methodology (7900T) using TaqMan Gene Expression (Life Technologies) probes. The relative expression of each gene was obtained by using the ΔΔct method and normalized to the expression of an endogenous gene (GUSb) as described elsewhere33.
The assays on demand, related to the candidate genes selected in our study, were provided by Applied Biosystems and are listed in Table 1. The candidates were divided into 4 categories based on their biological functions: (1)genes directly involved in inflammation/cell damage, (2) “classical” candidate genes strongly associated with CeD, (3)genes involved in the homing of lymphocytes and the regulation of inflammatory processes and cell damage, and 4)genes involved in cell adhesion and intestinal barrier integrity.
Methylation analysis
Methylation analysis of intestinal cells was performed for all enrolled subjects. DNA was extracted (as previously described) from the epithelial cells and the lamina propria isolated from the biopsy samples.
Conversion with bisulfite
The methods developed to detect and quantify DNA methylation use sodium bisulfite, through which unmethylated cytosines are deaminated and sulfonated for conversion into uracil, whereas the 5′-methyl-cytosines remain unchanged. The treatment therefore allows non-methylated cytosines to be discriminated from methylated ones, which are revealed by subsequent analyses. For the sodium bisulfite treatment, the Bisulfite Conversion kit from Active Motif® was used. The obtained samples were subsequently analysed by methylation-specific PCR.
A calibration curve was used to quantify the methylation status of each sample. The curve was constructed by using different concentrations of genomic DNA from HeLa cells, commercially available as both unmethylated and methylated DNA CpG. The following curve concentration levels were used: 0.12, 0.5, 25, 37.5, 50, 62.5, 75, 87.5 and 100%.
Methylation-specific PCR (MSP)
Methylation-Specific PCR (MSP) is a widely used technique for studying the methylation of CpG islands. The differences observed after Na-bisulfite treatment between methylated and non-methylated cytosines are at the basis of the functioning of MSP. The primers for MSP were designed by the MethPrimer program (http://www.urogene.org/methprimer/) based on CpG islands in the DNA sequence identified by Genome Browser (Supplementary Table 4).
We designed two sets of primers: one set to recognize sodium bisulfite-modified unmethylated DNA and a second set to identify methylated DNA. Using the Primer3 program, a third set of primers was developed to screen for unmodified DNA and assess the efficiency of bisulfite treatment. Na-bisulfite-treated DNA samples were amplified by three different probes provided with the KAPA2G Fast HotStartReadyMix PCR Kit (KAPABIOSYSTEMS®).The amplified samples were analysed by electrophoresis on a 2% agarose gel.
DNAclean up
The NucleoMag®96 PCR clean-up kit from MACHEREY-NAGEL® was used to purify the samples.
Sequencing
The processed samples were sent for next-generation high-throughput sequencing on an Illumina Nextera platform.
Statistics
Due to the small sample size, the gene expression levels were compared by Mann-Whitney rank test. Percentages were analysed by Chi-square test, with 1st degree error at 0.05.
To estimate the contribution of the expression of each gene in either the epithelium or the lamina propria to the differentiation of cases and controls, we adopted a stepwise discriminant analysis as in previous study14. The model was used to estimate the capacity of each gene to discriminate between cases and controls as indicated by Wilks’ Lambda, which ranged from 1 = complete overlap between groups to 0 = complete separation between groups. The variance ratio F was used to evaluate the significance of the contribution of each gene, taking into account the effects of all other genes.
By multiplying the standardized value of gene expression by the respective canonical discriminant coefficient, it was possible to obtain, for each individual, a probability of membership among the cases or the controls.
The percentage of correct classification provided an estimate of the reliability of the model in separating the two groups. Statistical analysis was performed with SPSS 21.0 (SPSS Inc., Chicago, IL).
Bioinformatics analysis
We analysed the genome sequences containing CpG islands, which were selected in the methylation analysis, using Epigenome Roadmap software34.This tool can analyse several key histone modifications, chromatin accessibility, DNA methylation and mRNA expression in a specific tissue or cell population; for this study, we selected small intestine and duodenum mucosa tissues. The Genomics and Proteomics facility of the University of the Basque Country sequenced the amplicons of the selected genes using pair-ended reads and an Illumina MiSeq platform. We employed different approaches to select the most optimal sequences for analysis: we used two mapping programs (BWA and Bowtie).In both cases, the reference sequence was the bisulfite-treated sequence. Bowtie achieved better mapping, and there were no differences between the two references, so we decided to use Bowtie and the “gene as chromosome” reference sequence to map all of the samples. However, when we analysed the percentage of Cs in CpG and no-CpG positions, we decided to take into account a CpG only if its surrounding no-CpGs had a C proportion <0.10. Then, using the CpGs that fulfilled this criterion, we performed Mann-Whitney U-tests between Celiac and No-Celiac values in CD326 positives and negatives.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from the parents of the enrolled children, and the study was approved by an independent ethical committee (CE/V-131).
Supplementary information
COMBINED ANALYSIS OF METHYLATION AND GENE EXPRESSION PROFILES IN SEPARATE COMPARTMENTS OF SMALL BOWEL MUCOSA IDENTIFIED CELIAC DISEASE PATIENTS’ SIGNATURES.
Acknowledgements
This work was funded by the FC-Grant2013. Programma di mobilità nell’ambito delle reti di eccellenza POR Campania FSE 2007–2013 Asse V. The authors thank the European Laboratory for Food-induced Disease (ELFID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
D.C. and M. G. conceptualized and designed the study, performed experiments, acquired data, analysed and interpreted the data, drafted the initial manuscript, performed critical revision of the manuscript,and approved the final manuscript as submitted. L.D.L., L.C. and A.T. recruited the patients and controls, performed clinical evaluation of the subjects, drafted the initial manuscript, performed critical revision of the manuscript, and approved the final manuscript as submitted. G.E.K. Performed bioinformatic analysis, analysed and interpreted the data. N.F.J. and T.N. performed critical revision of the manuscript for important intellectual content and approved the final manuscript as submitted. R.A. obtained funding, performed study supervision, analysed and interpreted the data, performed critical revision of the manuscript for important intellectual content, and approved the final manuscript as submitted. L.G. and J.R.B. performed study supervision, conceptualized and designed the study, drafted the initial manuscript, analysed and interpreted the data, performed statistical analysis, performed critical revision of the manuscript for important intellectual content, and approved the final manuscript as submitted.
Data Availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
D. Cielo and M. Galatola contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46468-2.
References
- 1.Catassi C, Gatti S, Fasano A. The new epidemiology of celiac disease. J Pediatr Gastroenterol Nutr. 2014;59(Suppl1):S7–9. doi: 10.1097/01.mpg.0000450393.23156.59. [DOI] [PubMed] [Google Scholar]
- 2.Auricchio R, et al. Respiratory Infections and the Risk of Celiac Disease. Pediatrics. 2017;4:140. doi: 10.1542/peds.2016-4102. [DOI] [PubMed] [Google Scholar]
- 3.Ploski R, Ek J, Thorsby E, Sollid LM. On the HLA‐DQ(α1*0501, β1*0201)‐associated susceptibility in celiac disease: A possible gene dosage effect of DQB1*0201. Tissue Antigens. 1993;41:173–7. doi: 10.1111/j.1399-0039.1993.tb01998.x. [DOI] [PubMed] [Google Scholar]
- 4.Tinto N, et al. High frequency of haplotype HLA-DQ7 in celiac disease patients from south Italy: Retrospective evaluation of 5,535 subjects at risk of celiac disease. PLoS One. 2015;10:e0138324. doi: 10.1371/journal.pone.0138324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaukinen K, Partanen J, Mäki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol. 2002;97:695–9. doi: 10.1111/j.1572-0241.2002.05471.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Heel DA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar V, et al. Systematic annotation of celiac disease loci refines pathological pathways and suggests a genetic explanation for increased interferon-gamma levels. Hum Mol Genet. 2015;24:397–409. doi: 10.1093/hmg/ddu453. [DOI] [PubMed] [Google Scholar]
- 8.Izzo V, et al. Improving the estimation of celiac disease sibling risk by non-HLA genes. PLoS One. 2011;6:e26920. doi: 10.1371/journal.pone.0026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MN, et al. Common Genetic Variants Modulate Pathogen-Sensing Responses in Human Dendritic Cells. Science. 2014;343(6175):1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verbeke S, et al. Basement membrane and connective tissue proteins in intestinal mucosa of patients with coeliac disease. J Clin Pathol. 2002;55:440–5. doi: 10.1136/jcp.55.6.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Bergen J, Mulder CJ, Mearin ML, Koning F. Local communication among mucosal immune cells in patients with celiac disease. Gastroenterology. 2015;148:1187–94. doi: 10.1053/j.gastro.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Galatola M, et al. Gene Expression Profile of Peripheral Blood Monocytes: A Step towards the Molecular Diagnosis of Celiac Disease? PLoS One. 2013;8:e74747. doi: 10.1371/journal.pone.0074747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galatola M, et al. Presymptomatic Diagnosis of Celiac Disease in Predisposed Children. J Pediatr Gastroenterol Nutr. 2017;65:314–20. doi: 10.1097/MPG.0000000000001519. [DOI] [PubMed] [Google Scholar]
- 15.Zilbauer M, et al. Epigenetics in Paediatric Gastroenterology, Hepatology, and Nutrition: Present Trends and Future Perspectives. J Pediatr Gastroenterol Nutr. 2016;62:521–9. doi: 10.1097/MPG.0000000000001053. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-jimenez N, et al. Coregulation and modulation of NFκB-related genes in celiac disease: Uncovered aspects of gut mucosal inflammation. Hum Mol Genet. 2014;23:1298–310. doi: 10.1093/hmg/ddt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontakou M, et al. Cytokine mRNA expression in the mucosa of treated coeliac patients after wheat peptide challenge. Gut. 1995;37:52–7. doi: 10.1136/gut.37.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiba M, Bartnik W, ReMine SG, Thayer WR, Shorter RG. Human colonic intraepithelial and lamina proprial lymphocytes: cytotoxicity in vitro and the potential effects of the isolation method on their functional properties. Gut. 1981;22:177–86. doi: 10.1136/gut.22.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jauregi-Miguel A, et al. Alteration of tight junction gene expression in celiac disease. J PediatrGastroenterolNutr. 2014;58:762–7. doi: 10.1097/MPG.0000000000000338. [DOI] [PubMed] [Google Scholar]
- 20.Devallire J, Charreau B. The adaptor Lnk (SH2B3): An emerging regulator in vascular cells and a link between immune and inflammatory signaling. BiochemPharmacol. 2011;82:1391–402. doi: 10.1016/j.bcp.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Li Y, Tanaka K, Moore KG, Hayashi JI. Cloning and characterization of Lnk, a signal transduction protein that links T-cell receptor activation signal to phospholipase C gamma 1, Grb2, and phosphatidylinositol 3-kinase. Proc Natl AcadSci USA. 1995;92:11618–22. doi: 10.1073/pnas.92.25.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaki S, et al. Characterization of Lnk: An adaptor protein expressed in lymphocytes. J Biol Chem. 1997;272:14562–70. doi: 10.1074/jbc.272.23.14562. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, He X, Schembri-King J, Jakes S, Hayashi J. Cloning and Characterization of Human Lnk, an Adaptor Protein with Pleckstrin Homology and Src Homology 2 Domains that Can Inhibit T Cell Activation. J Immunol. 2000;164:5199–206. doi: 10.4049/jimmunol.164.10.5199. [DOI] [PubMed] [Google Scholar]
- 24.Velazquez L, et al. Cytokine Signaling and Hematopoietic Homeostasis Are Disrupted in Lnk-deficient Mice. J Exp Med. 2002;195:1599–611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seita J, et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci. USA. 2007;104:2349–54. doi: 10.1073/pnas.0606238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong W, Lodish HF. Lnk Inhibits Tpo–mplSignaling and Tpo-mediated Megakaryocytopoiesis. J Exp Med. 2004;200:569–80. doi: 10.1084/jem.20040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bersenev A, Wu C, Balcerek J, Tong W. Lnk controls mouse hematopoietic stem cell self-renewal and quiescence through direct interactions with JAK2. J Clin Invest. 2008;118:2832–44. doi: 10.1172/JCI35808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitau J, Boulday G, Coulon F, Quillard T, Charreau B. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J Biol Chem. 2006;281:20148–59. doi: 10.1074/jbc.M510997200. [DOI] [PubMed] [Google Scholar]
- 29.Wan M, Li Y, Xue H, Li Q, Li J. Eicosapentaenoic acid inhibits TNF-alpha-induced Lnk expression in human umbilical vein endothelial cells: involvement of the PI3K/Akt pathway. JNutrBiochem. 2007;18:17–22. doi: 10.1016/j.jnutbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Takizawa H, et al. Enhanced engraftment of hematopoietic stem/progenitor cells by the transient inhibition of an adaptor protein, Lnk. Blood. 2006;107:2968–75. doi: 10.1182/blood-2005-05-2138. [DOI] [PubMed] [Google Scholar]
- 31.Moldenhauer G, Momburg F, Möller P, Schwartz R, Hämmerling GJ. Epithelium-specific surface glycoprotein of Mr 34,000 is a widely distributed human carcinoma marker. Br J Cancer. 1987;56:714–21. doi: 10.1038/bjc.1987.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenke AC, et al. DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile. PLoS One. 2013;8:e55636. doi: 10.1371/journal.pone.0055636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galatola M, Auricchio R, Greco L. Gene Expression Profiling of Celiac Biopsies and Peripheral Blood Monocytes Using Taqman Assays. Methods Mol Biol. 2015;1326:105–15. doi: 10.1007/978-1-4939-2839-2_11. [DOI] [PubMed] [Google Scholar]
- 34.Skipper M, et al. Presenting the epigenoma road map. Nature. 2015;518:313. doi: 10.1038/518313a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
COMBINED ANALYSIS OF METHYLATION AND GENE EXPRESSION PROFILES IN SEPARATE COMPARTMENTS OF SMALL BOWEL MUCOSA IDENTIFIED CELIAC DISEASE PATIENTS’ SIGNATURES.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.