Lanthanide metals, or rare earth elements, are abundant in nature and used heavily in technological devices. Biological interactions with lanthanides are just beginning to be unraveled. Until very recently, microbial mechanisms of lanthanide metal interaction and uptake were unknown. The TonB-dependent receptor LanA is the first lanthanum receptor identified in a methanotroph. Sequence homology searches with known metal transporters and regulators could not be used to identify LanA or other lanthanide metal switch components, and this method for mutagenesis and selection was required to identify the receptor. This work advances the knowledge of microbe-metal interactions in environmental niches that impact atmospheric methane levels and are thus relevant to climate change.
KEYWORDS: gene regulation, genetics, lanthanide, methanol dehydrogenase, methanotrophs, mutagenesis
ABSTRACT
Several of the metabolic enzymes in methanotrophic bacteria rely on metals for both their expression and their catalysis. The MxaFI methanol dehydrogenase enzyme complex uses calcium as a cofactor to oxidize methanol, while the alternative methanol dehydrogenase XoxF uses lanthanide metals such as lanthanum and cerium for the same function. Lanthanide metals, abundant in the earth’s crust, strongly repress the transcription of mxaF yet activate the transcription of xoxF. This regulatory program, called the “lanthanide switch,” is central to methylotrophic metabolism, but only some of its components are known. To uncover additional components of the lanthanide switch, we developed a chemical mutagenesis system in the type I gammaproteobacterial methanotroph “Methylotuvimicrobium buryatense” 5GB1C and designed a selection system for mutants unable to repress the mxaF promoter in the presence of lanthanum. Whole-genome resequencing for multiple lanthanide switch mutants identified several unique point mutations in a single gene encoding a TonB-dependent receptor, which we have named LanA. The LanA TonB-dependent receptor is absolutely required for the lanthanide switch and controls the expression of a small set of genes. While mutation of the lanA gene does not affect the amount of cell-associated lanthanum, it is essential for growth in the absence of the MxaF methanol dehydrogenase, suggesting that LanA is involved in lanthanum uptake to supply the XoxF methanol dehydrogenase with its critical metal ion cofactor. The discovery of this novel component of the lanthanide regulatory system highlights the complexity of this circuit and suggests that further components are likely involved.
IMPORTANCE Lanthanide metals, or rare earth elements, are abundant in nature and used heavily in technological devices. Biological interactions with lanthanides are just beginning to be unraveled. Until very recently, microbial mechanisms of lanthanide metal interaction and uptake were unknown. The TonB-dependent receptor LanA is the first lanthanum receptor identified in a methanotroph. Sequence homology searches with known metal transporters and regulators could not be used to identify LanA or other lanthanide metal switch components, and this method for mutagenesis and selection was required to identify the receptor. This work advances the knowledge of microbe-metal interactions in environmental niches that impact atmospheric methane levels and are thus relevant to climate change.
INTRODUCTION
Methane is present at a lower atmospheric concentration than carbon dioxide but has a 25- to 28-fold-higher global warming potential per molecule (1). Since methane levels have been rising in the past decade after a brief stabilization period, an understanding of natural and anthropogenic contributors to the methane cycle is necessary to assess and mitigate the effects of this potent greenhouse gas (2, 3). Methanotrophs, microorganisms that oxidize methane, are critical factors in determining which environments are methane sources and which are methane sinks (4). Methane utilizers and methanol utilizers, both termed methylotrophs, exist in communities in environments ranging from wetlands to lakes to grasslands (5). Research efforts over the past decade have revealed that most of these microorganisms contain alcohol dehydrogenase enzymes that are functionally dependent on rare earth elements, also called lanthanide metals (6–8).
To call them “rare” earth elements is a misnomer, as the lanthanides, a group of 15 metals ranging from atomic number 57 to 71, are relatively abundant in the earth’s crust (9). They are commonly used in many technological devices, including batteries, computers, catalysts, and magnets (10). Despite their important properties, mining difficulties and pollution concerns limit the number of mines worldwide, with most extraction sites currently in China (11). Because of these concerns, biometallurgy is an attractive possibility for lanthanide extraction from recycled materials or from contaminated sites. Methylotrophs that utilize lanthanides must have a mechanism for uptake or interaction and are therefore excellent candidates for this biometallurgy.
In the oxidation of methane to carbon dioxide by methanotrophic bacteria, it is a common theme for reactions to be catalyzed by two alternative enzymes, each with a preferred metal ion cofactor. For instance, many methanotrophs contain two types of methane monooxygenase (MMO), a membrane-bound or particulate MMO (pMMO) and a soluble MMO (sMMO) (12, 13). The pMMO is induced by the presence of copper (Cu), while the sMMO is strongly repressed by Cu, a phenomenon called the “copper switch.” This theme holds true for lanthanide metals and the methanol dehydrogenase switch in several methylotrophs, in addition to other alcohol dehydrogenase reactions in methylotrophs and at least one nonmethylotroph (7, 8, 14–16). In methylotrophs, lanthanides like lanthanum (La) and cerium (Ce) act at the transcriptional level to induce the XoxF methanol dehydrogenase, but they also repress the genes encoding the calcium-dependent MxaFI methanol dehydrogenase.
The type I gammaproteobacterial methanotroph “Methylotuvimicrobium buryatense” 5GB1C (formerly Methylomicrobium buryatense 5GB1C [17, 18]) is an excellent model for metal regulation studies, as it has only one copy of the gene cluster encoding the pMMO and one xoxF gene copy (19). However, the lanthanide metal regulatory mechanisms in this organism appear to be different from alphaproteobacterial methylotrophs. In the model methylotroph Methylorubrum extorquens AM1 (formerly Methylobacterium extorquens AM1 [20]), XoxF is required for mxaF transcription (21), and the MxcQE and MxbDM two-component systems activate mxaF in the absence of La (22). In the methanotroph M. buryatense 5GB1C, XoxF is not required for mxaF transcription (15), and homologs of the MxcQE and MxbDM two-component system genes are not present in the genome (19). However, a functional homolog to the orphan response regulator MxaB is located in the mxa gene cluster (15). MxaB is a LuxR-like transcription factor required for lanthanide-dependent repression of mxaF and activation of xoxF in M. buryatense 5GB1C, but only part of the regulation can be explained by MxaB (15). The integral membrane histidine kinase MxaY is also required for these phenotypes, and it is likely that these two proteins form a nontraditional two-component system for transducing the lanthanide signal (23). It is not known how or if lanthanides bind to MxaY, whether MxaB is a direct or indirect regulator of xoxF and mxaF, or how many additional components exist. There are also differences in the lanthanide switch between methanotrophs. For instance, in the alphaproteobacterial methanotroph Methylosinus trichosporium OB3b, it appears that the regulatory effect of Cu precludes that of La or Ce (16, 24), but this effect is not seen in M. buryatense 5GB1C (15).
Understanding the regulatory and uptake mechanisms of lanthanide metals in methanotrophs is critical to assess the microbiological impact on atmospheric methane levels and to apply these organisms to industrial bioproduction and biometallurgy. To discover components of lanthanide regulation other than MxaB and MxaY in M. buryatense 5GB1C, chemical mutagenesis techniques were developed for this organism. Most chemical mutagenesis techniques in methanotrophs have been unsuccessful, with only one report of success, using nitrosoguanidine (25). In the present study, nitrosoguanidine mutagenesis was applied to M. buryatense 5GB1C to generate mutants that were selected for expression from the mxaF promoter in the presence of La. All mutants resulting in a derepressed mxaF promoter contained mutations encoding premature truncations in a TonB-dependent receptor gene product, which we have named LanA. Deletion of this gene abolished mxaF repression and xoxF activation by La and also identified a small set of La-regulated genes. While cell-associated lanthanum was not affected by the deletion of the lanA gene, lanthanum-dependent growth on methane was abolished, suggesting that LanA is required for lanthanum uptake. These results reveal a new important player in the lanthanide metal switch and establish nitrosoguanidine as a useful mutagen for investigating gene regulation and other phenotypes in this methanotroph.
RESULTS
Nitrosoguanidine increases the mutation rate of Methylotuvimicrobium buryatense 5GB1C.
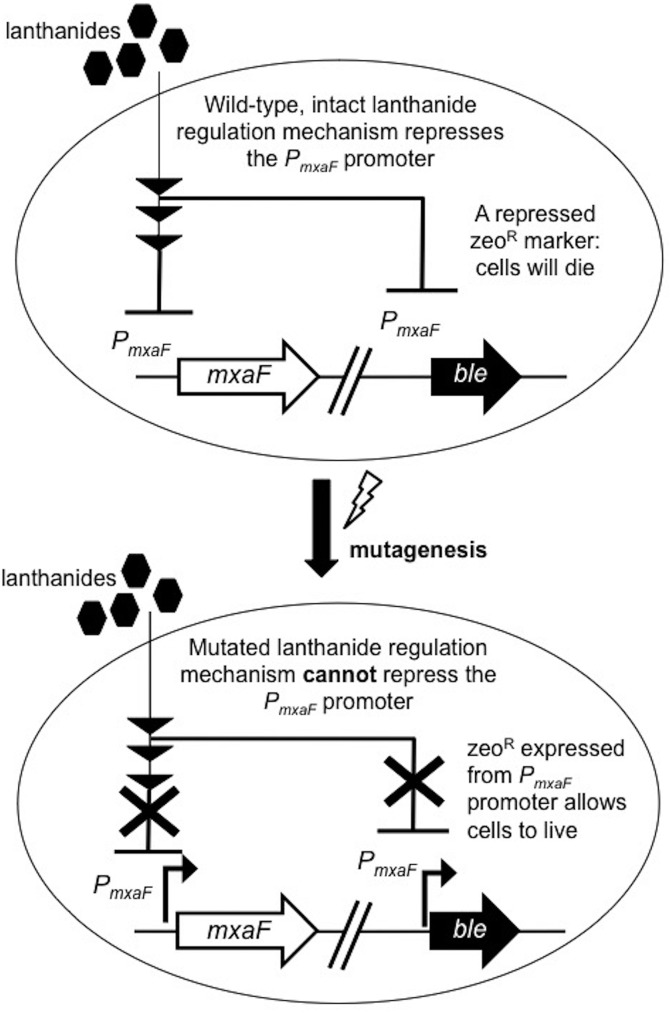
A reporter strain was created with the zeocin resistance gene ble driven by the PmxaF promoter (Fig. 1). The strain was chemically mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and plated onto solid medium with zeocin and lanthanum, using methane as the carbon source. The wild-type strain should not grow under these conditions, as PmxaF-ble is repressed by lanthanum, while mutants defective for lanthanide-mediated transcriptional repression of PmxaF are able to express ble in the presence of lanthanum.
FIG 1.
Scheme for the selection for lanthanide repression mutants. A strain with a zeocin resistance gene driven by the PmxaF promoter was chemically mutagenized with nitrosoguanidine. Mutants defective for lanthanide-mediated transcriptional repression of PmxaF are able to express the zeocin resistance gene in the presence of lanthanides and were selected on plates with zeocin and lanthanum chloride. mxaF, open reading frame encoding the MxaF methanol dehydrogenase large subunit; ble, zeocin antibiotic resistance gene; PmxaF, 300-bp upstream regulatory region of the mxaF gene.
Increasing the concentration of MNNG increased the number of zeocin-resistant colonies of the PmxaF-ble reporter strain of M. buryatense 5GB1C grown with lanthanum (see Fig. S1 in the supplemental material). The mutation frequencies were calculated for genes involved in the lanthanide switch. A baseline mutation frequency for the selection was observed in untreated samples, and the variation between replicates was high, but multiple biological replicates showed that the mutation frequency increased by 1 to 2 orders of magnitude when the cells were treated with MNNG.
Selection for abolishment of lanthanum repression at PmxaF yields mutations in a single gene that encodes a TonB-dependent receptor.
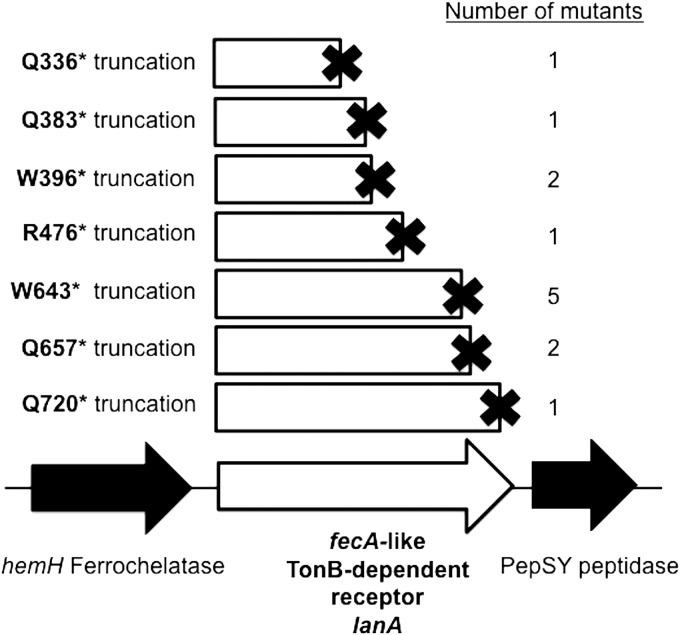
Mutants resistant to zeocin in the presence of lanthanum were isolated. Subsequent whole-genome resequencing revealed that each mutant contained 1 to 57 point mutations (see Tables S2 and S3 in the supplemental material), but all mutants contained a mutation in the same gene, encoding a TonB-dependent receptor with strong sequence similarity to the Escherichia coli ferric citrate transporter FecA (see Fig. S2 in the supplemental material). Because of the similarity, we have named this gene lanA (NCBI locus EQU24_02055; Genoscope locus MBURv2_130812). All of the mutants displayed point mutations that introduced premature stop codons into the lanA open reading frame (ORF), which should produce truncated gene products (Fig. 2).
FIG 2.
Lanthanide switch mutants all sustained point mutations in a gene encoding a TonB-dependent receptor. Independent mutations in strains resistant to zeocin in the presence of lanthanum were mapped to the M. buryatense 5G genome. The mutations, all causing premature stop codons, are shown in their respective locations on the open reading frame. The amino acid residue that became a stop codon in the particular mutant is indicated at the left of the truncation diagram. The number of mutations of each particular type among the 15 mutants sequenced is shown at the right.
LanA is present in many type I methanotrophs, yet compared to other TonB-dependent receptors it is most similar to the E. coli FecA protein, including other methylotrophic receptors (see Fig. S3 in the supplemental material). It does not exhibit high sequence similarity to the recently identified lanthanide TonB-dependent receptor from the methylotroph Methylobacterium extorquens PA1 (26), which clusters instead with a cerium-repressible receptor from the alphaproteobacterial methanotroph Methylosinus trichosporium OB3b (27). lanA and its type I methanotroph homologs are found upstream of genes encoding a PepSY/peptidase M4 (Fig. 2), but the function of such proteins with regard to TonB-dependent receptors is unknown. The hemH gene upstream of lanA, encoding a ferrochelatase, is not consistently found paired with lanA in other organisms (28).
Deletion of lanA leads to a broken lanthanide switch and the dysregulation of a small set of genes.
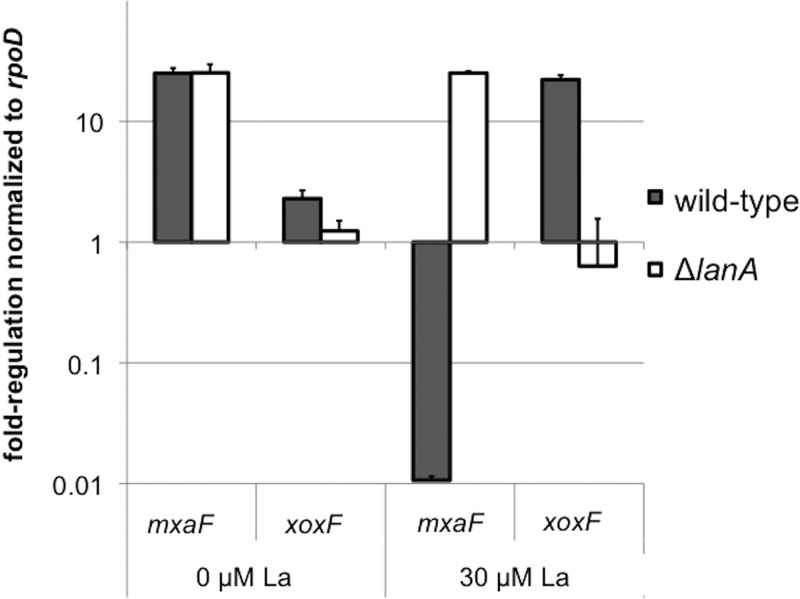
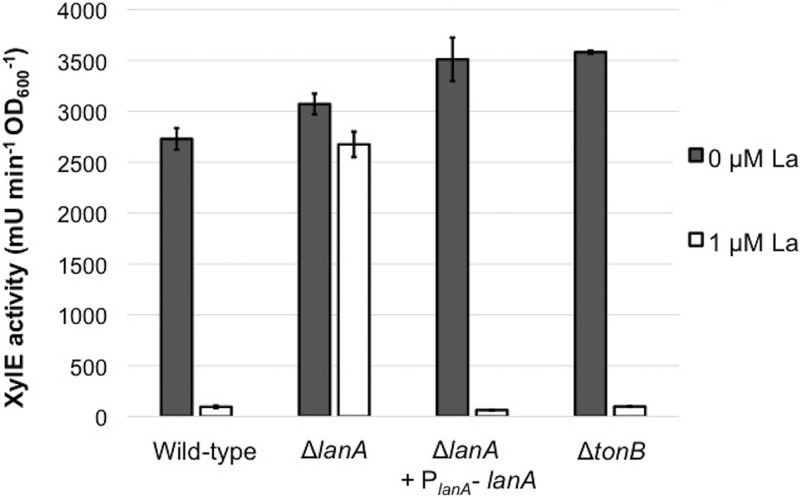
To verify that lanA did indeed abolish lanthanum repression, the open reading frame was deleted in a clean genetic background. As the lanthanum response is known to act at the transcriptional level to repress mxaF and activate xoxF (15), the methanol dehydrogenase genes were assessed with reverse transcriptase PCR (RT-PCR) in the wild-type and ΔlanA strains. In the mutant, the mxaF gene was no longer repressed in response to lanthanum, and the xoxF gene was no longer activated by lanthanum (Fig. 3). In addition to the wild-type strain, the ΔlanA mutation was created in a PmxaF-xylE reporter strain (15) for rapid assessment of the lanthanum repression phenotype. Complementation with a plasmid-borne PlanA-lanA cassette restored repression of PmxaF-xylE in the presence of lanthanum (Fig. 4).
FIG 3.
Reverse transcriptase PCR (RT-PCR) of lanthanide switch genes. Deletion of lanA results in a broken lanthanide switch for the methanol dehydrogenase genes. RT-PCR was performed on RNA harvested from M. buryatense wild-type and ΔlanA strains grown in the presence or absence of 30 μM lanthanum. Primers specific for the mxaF and xoxF methanol dehydrogenase genes were used to quantify the lanthanide switch. All cycle threshold values were normalized to the constitutive rpoD gene. Results show the averages ± standard deviations from two biological replicates (n = 2).
FIG 4.
PmxaF-xylE reporter activity with increasing LaCl3. Deletion of lanA abolishes lanthanum repression, but deletion of the lanthanum-repressed tonB gene does not. Whole-cell catechol-2,3-dioxygenase reporter gene assays were performed for wild-type and mutant strains. The wild-type was FC31 (PmxaF-xylE), and all mutants were created from this background strain. Cells were grown either without La or in the presence of 1 μM La. Results are the averages ± standard deviations from two biological replicates (n = 2).
To characterize the lanthanum response and the role of LanA in this response, we analyzed the transcriptomes of the wild type and the ΔlanA mutant in both the presence and absence of lanthanum. In order to improve the discovery power of these transcriptomes, the gapped M. buryatense 5GB1C reference genome sequence was closed using a combination of short- and long-read sequencing. A small set of genes were regulated by lanthanum (Table 1), and for some of them this regulation was broken by the deletion of lanA (Table 2). The full list of differentially regulated genes is reported in Table S4 in the supplemental material, along with raw transcripts per million (TPM) values. As expected, all of the genes involved in the MxaF methanol dehydrogenase are repressed by lanthanum, and this repression is lost in the mutant. Conversely, xoxF transcripts increase in response to lanthanum, and this activation is lost in the mutant. The mxaY histidine kinase gene previously identified as being required for the lanthanide switch is downregulated by lanthanum, but more subtly. In the wild type, a tonB gene (EQU24_21875) and clustered genes were downregulated in response to lanthanum, as was the lanA gene identified experimentally by mutagenesis in this study. The tonB cluster had the same expression pattern as the mxaF gene cluster in both the wild-type and ΔlanA strains. TonB complexes typically provide the energetics for the function of TonB-dependent receptors and physically interact with the receptors (29). TonB-dependent receptors are often downregulated by the substrate with which they interact (30).
TABLE 1.
RNA-Seq results for M. buryatense 5GB1C with and without La
| Gene annotation | Gene (Genoscope/NCBI locus no.) | Log2 fold change (with La/without La) | Adjusted P value |
|---|---|---|---|
| MxaF methanol dehydrogenase related | mxaF (MBURv2_210291/EQU24_18145) | −9.03 | <1.00E−300 |
| mxaJ (MBURv2_210292/EQU24_18140) | −8.92 | <1.00E−300 | |
| cytCL (MBURv2_210293/EQU24_18135) | −7.94 | <1.00E−300 | |
| mxaI (MBURv2_210294/EQU24_18130) | −7.07 | <1.00E−300 | |
| moxR (MBURv2_210295/EQU24_18125) | −6.28 | <1.00E−300 | |
| mxaA (MBURv2_210298/EQU24_18110) | −5.61 | 3.87E−296 | |
| mxaS (MBURv2_210297/EQU24_18115) | −5.56 | 1.31E−202 | |
| mxaP (MBURv2_210296/EQU24_18120) | −4.97 | 3.47E−158 | |
| mxaC (MBURv2_210299/EQU24_18105) | −4.87 | 3.97E−156 | |
| mxaK (MBURv2_210300/EQU24_18100) | −4.38 | 3.41E−132 | |
| mxaB (MBURv2_210290/EQU24_18150) | −4.30 | 1.47E−218 | |
| mxaL (MBURv2_210301/EQU24_18095) | −3.43 | 5.45E−83 | |
| mxaD (MBURv2_210287/EQU24_18160) | −3.27 | 1.36E−52 | |
| Unknown (MBURv2_210288/EQU24_18155) | −3.16 | 2.17E−65 | |
| TonB cluster | exbB (MBURv2_160247/EQU24_21890) | −2.14 | 1.07E−39 |
| exbD (MBURv2_160249/EQU24_21880) | −1.86 | 6.55E−18 | |
| Unknown (MBURv2_160246/EQU24_21895) | −1.76 | 3.10E−35 | |
| tonB (MBURv2_160250/EQU24_21875) | −1.70 | 6.57E−16 | |
| exbB (MBURv2_160248/EQU24_21885) | −1.01 | 3.77E−07 | |
| LanA TonB-dependent receptor | lanA (MBURv2_130812/EQU24_02055) | −1.71 | 1.33E−11 |
| MxaY histidine kinase | mxaY (MBURv2_120065/EQU24_06545) | −0.77 | 3.33E−07 |
| Conserved protein | Unknown (MBURv2_210432/EQU24_17470) | +0.90 | 3.66E−06 |
| Ferrous iron transcriptional regulator | feoC (MBURv2_20182/EQU24_14340) | +0.90 | 1.42E−04 |
| Conserved exported protein | Unknown (MBURv2_210064/EXU24_19210) | +1.54 | 9.94E−16 |
| XoxF methanol dehydrogenase | xoxF (MBURv2_210189/EXU24_18605) | +2.92 | 1.88E−80 |
TABLE 2.
RNA-Seq results for M. buryatense 5GB1C and its ΔlanA mutant with La
| Gene annotation | Gene (Genoscope/NCBI locus numbers) | Log2 fold change (ΔlanA mutant/5GB1C) | Adjusted P value |
|---|---|---|---|
| MxaF methanol dehydrogenase related | mxaJ (MBURv2_210292/EQU24_18140) | +8.74 | <1.00E−300 |
| mxaF (MBURv2_210291/EQU24_18145) | +8.70 | <1.00E−300 | |
| cytCL (MBURv2_210293/EQU24_18135) | +7.86 | <1.00E−300 | |
| mxaI (MBURv2_210294/EQU24_18130) | +7.19 | <1.00E−300 | |
| moxR (MBURv2_210295/EQU24_18125) | +6.18 | <1.00E−300 | |
| mxaA (MBURv2_210298/EQU24_18110) | +5.42 | 5.81E−277 | |
| mxaS (MBURv2_210297/EQU24_18115) | +5.30 | 2.42E−183 | |
| mxaP (MBURv2_210296/EQU24_18120) | +4.82 | 6.42E−149 | |
| mxaC (MBURv2_210299/EQU24_18105) | +4.64 | 1.40E−141 | |
| mxaK (MBURv2_210300/EQU24_18100) | +4.16 | 9.86E−119 | |
| mxaB (MBURv2_210290/EQU24_18150) | +4.10 | 1.01E−197 | |
| mxaL (MBURv2_210301/EQU24_18095) | +3.13 | 4.01E−69 | |
| Unknown (MBURv2_210288/EQU24_18155) | +3.04 | 4.76E−60 | |
| mxaD (MBURv2_210287/EQU24_18160) | +2.81 | 2.20E−38 | |
| TonB cluster | exbB (MBURv2_160247/EQU24_21890) | +1.74 | 6.31E−26 |
| Unknown (MBURv2_160246/EQU24_21895) | +1.65 | 2.56E−30 | |
| tonB (MBURv2_160250/EQU24_21875) | +1.35 | 1.48E−09 | |
| exbD (MBURv2_160249/EQU24_21880) | +1.32 | 1.83E−08 | |
| exbB (MBURv2_160248/EQU24_21885) | +0.74 | 1.36E−03 | |
| Fructosamine kinase | FN3K (MBURv2_120061/EQU24_06560) | +0.83 | 1.41E−04 |
| EnvC domain-containing protein | Unknown (MBURv2_80054/EQU24_07250) | −0.94 | 1.28E−05 |
| NusG transcriptional antiterminator | nusG (MBURv2_190042/EQU24_19965) | −1.04 | 1.67E−11 |
| Conserved exported protein | Unknown (MBURv2_210064/EXU24_19210) | −1.14 | 3.18E−09 |
| XoxF methanol dehydrogenase | xoxF (MBURv2_210189/EXU24_18605) | −3.04 | 8.17E−87 |
Besides the expected mxaF and xoxF genes, several conserved genes of unknown function showed altered expression in the presence of lanthanum and/or upon deletion of lanA. The gene encoding an unknown exported protein, EQU24_19210, is regulated in a manner similar to xoxF, i.e., induced by lanthanum in the wild type and uninducible by lanthanum in the ΔlanA mutant. It is also relatively highly expressed compared to the rest of the genome. The open reading frame is conserved and syntenic with an ABC transporter in many type I methanotrophs, including Methylomicrobium and Methylobacter species (28) (see Fig. S4 in the supplemental material). Interestingly, it is flanked by inverted repeats that could indicate transposition at some point in its evolutionary history. Another gene induced by lanthanum yet not regulated by lanA, EQU24_17470, is conserved and syntenic with genes encoding proteins of unknown function and a NUDIX hydrolase in various type I methanotrophs (see Fig. S5 in the supplemental material). The function of this gene encoding a hypothetical protein is unknown. EQU24_14340 is upregulated by lanthanum and is conserved and syntenic (and likely cotranscribed) with the FeoAB ferrous iron transporter in several type I methanotrophs, including Methylomicrobium, Methylobacter, Methylomarinum, and Methylomonas strains, implying a potential cross talk between the systems for the uptake and regulation of these metals (see Fig. S6 in the supplemental material). Finally, the EQU24_07250 gene is upregulated in the ΔlanA strain. While the product has a conserved EnvC peptidase domain at its N terminus, its function is unknown. Homologs are apparent only in closely related type I methanotroph genomes, in which it resides next to a gene encoding a fatty acid hydroxylase. A fatty acid hydroxylase gene was found to be lanthanide inducible in Methylomicrobium alcaliphilum 20ZR, presumably affecting the remodeling of membranes that house the pMMO (31).
LanA is required for lanthanum-dependent growth on methane.
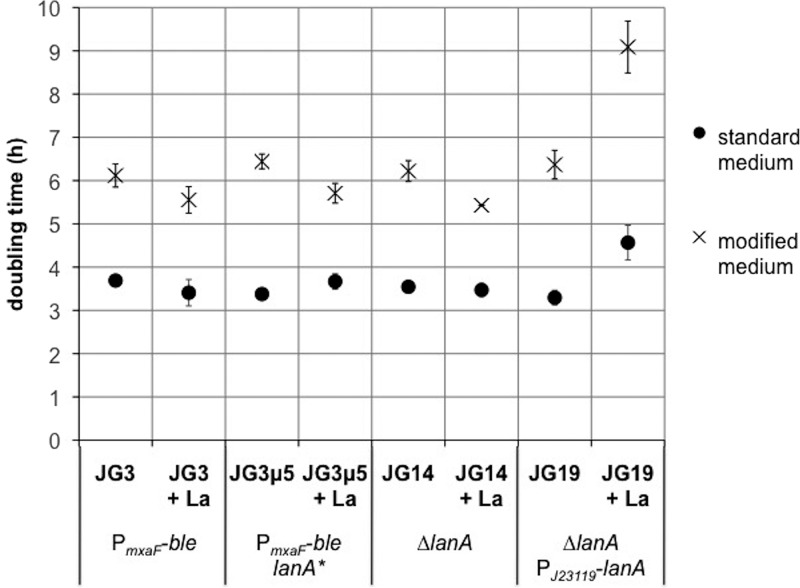
Lanthanide uptake has been measured in the methanotroph Methylosinus trichosporium OB3b (24, 32), but detailed mechanisms of lanthanide uptake in methanotrophs and specifically in type I methanotrophs have not been reported. We measured cell-associated lanthanum to test whether the LanA TonB-dependent receptor or the lanthanum-repressible TonB protein is involved in metal uptake. These measurements required a modified medium to minimize metal precipitation. The growth rate in this medium was found to be ∼50% lower than that of cells grown in the standard NMS2 medium (Fig. 5; see Fig. S7 in the supplemental material). Inductively coupled plasma mass spectrometry (ICP-MS) of supernatants compared to cell pellets showed that there was no measurable difference in cell-associated lanthanum between the wild-type and the mutant strains, either in early exponential phase or at the end of growth (see Fig. S8 in the supplemental material).
FIG 5.
Growth phenotypes of mutants with and without lanthanum. Mutation of lanA does not affect growth, but overexpression of lanA results in slower growth. M. buryatense 5GB1C derivatives were grown in either standard medium or lanthanum uptake medium without or with lanthanum; 30 μM La was used for standard medium and 2 μM La was used for modified medium, to be consistent with other experiments. Doubling times were calculated from 3 time points in exponential growth phase. All strains were grown in biological triplicate (n = 3).
Since cell-associated lanthanum might reflect external binding rather than uptake, a different approach was used to test whether the LanA transporter was required for lanthanum-dependent growth of M. buryatense 5GB1C on methane. A zeocin resistance-conferring linear DNA deletion construct for the mxaF gene was electrotransformed into the wild-type strain and several mutants in the presence of lanthanum to generate ∆mxaF mutants. Mutants deficient in MxaF require active XoxF in order to grow on methane. Thus, strains with MxaF deleted can grow only if they can take up lanthanum to generate active XoxF, and double mutations causing deficiency in lanthanum uptake and in MxaF would be lethal. We found that ∆lanA ∆mxaF double mutants could not be generated by electrotransforming the ΔlanA strain with ΔmxaF::ble, while the wild-type strains, two ΔlanA strains complemented with lanA, and the ΔtonB strain could all be transformed to zeocin resistance and deleted for mxaF (Table 3). The ΔlanA strain could readily be transformed with other gene deletion constructs besides ΔmxaF::ble (data not shown). These results strongly suggest that LanA is required for lanthanum transport, while the lanthanum-repressible TonB protein is not.
TABLE 3.
LanA is required for the deletion of the mxaF methanol dehydrogenase gene
| M. buryatense strain | Genotype | Medium | CFU/μg ΔmxaF::ble DNA (mean ± SD; n = 2) growing on zeocin after electrotransformation |
|---|---|---|---|
| 5GB1C | Wild type | 30 μg/ml zeocin, 30 μM La | 245 ± 135 |
| JG15 | ΔlanA PmxaF-xylE | 30 μg/ml zeocin, 30 μM La | 0 ± 0 |
| JG19 | ΔlanA PJ23119-lanA | 30 μg/ml zeocin, 30 μM La | 216 + 141 |
| JG20 | ΔlanA PmxaF-xylE + PlanA-lanA (Kanr) | 30 μg/ml zeocin, 30 μM La, 50 μg/ml kanamycin | 63 ± 58 |
| JG28 | ΔtonB PmxaF-xylE | 30 μg/ml zeocin, 30 μM La | 1,100 ± 850 |
Interestingly, the overexpression of LanA led to a lanthanum-dependent growth defect (Fig. 5 and S7). A strain wild type at the lanA locus, a lanA* truncation mutant, and a ΔlanA mutant grow similarly on NMS2 medium with or without lanthanum. The ΔlanA mutant complemented with a wild-type lanA gene driven by the J23119 Anderson promoter exhibited a much longer lag phase and a slower doubling time when lanthanum was added (Fig. 5 and S7). Whereas the endogenous lanA gene is expressed at low levels and transcriptionally repressed by lanthanum (Table S4), the J23119 promoter is constitutively and moderately expressed and is not affected by lanthanum (see Fig. S9 in the supplemental material). These results suggest that an excess of LanA results in a burden on the cell, possibly by allowing too much lanthanum to cross the outer membrane.
Deletion of additional suspected lanthanide switch components does not abolish lanthanum regulation.
We used the lanthanide switch reporter strain carrying PmxaF-xylE to assess the lanthanide switch in multiple mutants. The tonB gene, which was downregulated in response to lanthanum (Table 1), may be involved in some function of LanA, but the wild-type lanthanum repression in this strain shows that it is not required for the signal transduction of the lanthanide switch (Fig. 5). As stated above, this tonB gene is also not required for lanthanum-dependent growth on methane (Table 3). TonB-dependent receptors often make physical connections with regulatory complexes to transduce a transcriptional program, which is the case for E. coli FecA with the anti-sigma factor/sigma factor pair FecR and FecI (33). There are three clear fecR homologs in M. buryatense 5GB1C, two of which are near lanA on the chromosome (28). If one of these were required for lanthanide signal transduction, fecR mutations would result in abolishment of the downstream repression on PmxaF. Deletions of each fecR-like gene individually and a triple deletion had no effect on lanthanide repression (see Fig. S10 in the supplemental material). The genome also contains three FecI-like sigma factor homologs, none of which affected lanthanum repression (Fig. S10). Additionally, reporter assay results and RT-PCR tests show that the three FecI homologs do not have critical roles in lanthanum activation of the xoxF gene under these growth conditions (see Fig. S11 in the supplemental material).
DISCUSSION
Chemical mutagenesis of a type I methanotroph coupled to a selection scheme revealed that the LanA TonB-dependent receptor is required for the lanthanide signaling cascade, adding to the known signaling components MxaY (23) and MxaB (15). While targeted selection for mutations in methane monooxygenase genes has produced useful mutants (34–36), most chemical mutagenesis attempts in type I methanotrophs have been unsuccessful. One notable exception is that nitrosoguanidine led to a small increase in the mutation frequency to streptomycin resistance in Methylomonas albus (25). Nitrosoguanidine mutagenesis applied to M. buryatense 5GB1C was successful in increasing the mutation rate by 1 to 2 orders of magnitude, allowing the selection of mutants defective for the lanthanide switch. For this selection, the spontaneous mutagenesis baseline was near the top of the range commonly reported for E. coli (1 in 10−6), but this could reflect the variety of mutations, such as point mutations and frameshifts, that would allow the zeocin resistance phenotype to occur (37).
Eight zeocin-resistant mutants were subjected to whole-genome resequencing, and 15 mutants in total were sequenced at lanA, mxaY, mxaB, and the PmxaF-ble cassette. The fact that the mutagenic selection consistently resulted in mutations in lanA could reflect a variety of causes. For instance, it is possible that the expression of the zeocin resistance protein from the strong mxaF promoter caused a fitness defect that biased the screen. Alternatively, the selection may be too tight, such that the only possibility to derepress the zeocin resistance cassette sufficiently to allow growth is by completely precluding lanthanum from exerting its repressive effect at the top of the signaling cascade. Gain-of-function mutations in the known components of the lanthanide switch mxaY or mxaB in the presence of lanthanides might not allow enough expression from PmxaF for survival at the inhibitory zeocin concentration. Of the 15 lanA mutant alleles sequenced, there were 8 unique truncations, with 3 of the truncation variants being hit multiple times in the set. Most of the strains were not siblings: 6 of the 8 mutant strains subjected to whole-genome sequencing had their own unique sets of additional mutations relative to the reference genome, ranging from 0 to 56 point mutations besides the common lanA mutation (see Tables S2 and S3 in the supplemental material). Further components of the lanthanide switch might be identified by decreasing the zeocin concentration in the lanthanum repression selection or by focusing on mutations that alter the activity of the xoxF promoter rather than the mxaF promoter.
Transcriptomics revealed some new untested components of the lanthanum response (Tables 1 and 2), in addition to confirming the importance of the lanA gene for methanol dehydrogenase gene transcription, potentially by controlling the access of lanthanides to the periplasm. The mxaY gene, encoding an integral membrane histidine kinase, is clearly lanthanum repressed, but <2-fold. This is consistent with its role in the lanthanide switch (23). However, its regulation does not change in the ΔlanA strain, suggesting either that these two components are not in the same regulatory circuit or that LanA functions downstream of MxaY in the regulatory circuit. The latter would not be consistent with the proposal that LanA should be at the top of the regulatory circuit. Curiously, a nearby ORF, EQU24_06560, is upregulated in the ΔlanA strain. This ORF is homologous to the fructosamine kinase gene, and its colocalization with mxaY is conserved in many type I methanotrophs (28).
Further analysis of the lanthanide transcriptional response in M. buryatense 5GB1C, a gammaproteobacterial type I methanotroph, demonstrated a more restricted set of coregulated genes than the published alphaproteobacterial methylotroph lanthanide responses (27, 38, 39). When grown with lanthanum, M. extorquens AM1 similarly represses the MxaF complex genes and activates xoxF, but it also activates downstream metabolic genes encoding functions of formaldehyde and formate oxidation (38), which was not observed in M. buryatense 5GB1C. Besides the typical methanol dehydrogenase regulation, growth with cerium in M. trichosporium OB3b clearly represses a TonB-dependent transporter and also upregulates several ABC transporters and a cluster of genes for the synthesis and transport of the iron siderophore pyoverdine (27). The presence of copper and cerium together in M. trichosporium OB3b further changes the effect of lanthanide metal, including inducing formaldehyde oxidation enzymes. Again, these effects have some commonalities with M. buryatense 5GB1C, but lanthanide-dependent regulation is broader in the alphaproteobacterial methylotrophs.
The transcriptional fold changes in M. buryatense 5GB1C are more consistent with the lanthanum versus calcium transcriptome of the related type I methanotroph “Methylotuvimicrobium alcaliphilum” 20ZR (31). Still, that study reports the upregulation of formaldehyde-activating enzyme and formate dehydrogenase genes in M. alcaliphilum 20ZR, as seen in M. extorquens AM1 but not in M. buryatense 5GB1C. This difference could be a result of cultivation in a bioreactor rather than in closed vials, or it could be an actual result of metabolic differences between the organisms. One example of bioreactor versus vial physiological difference in M. buryatense 5GB1C is increased acetate excretion in O2-limited vials but not in an O2-limited bioreactor (40).
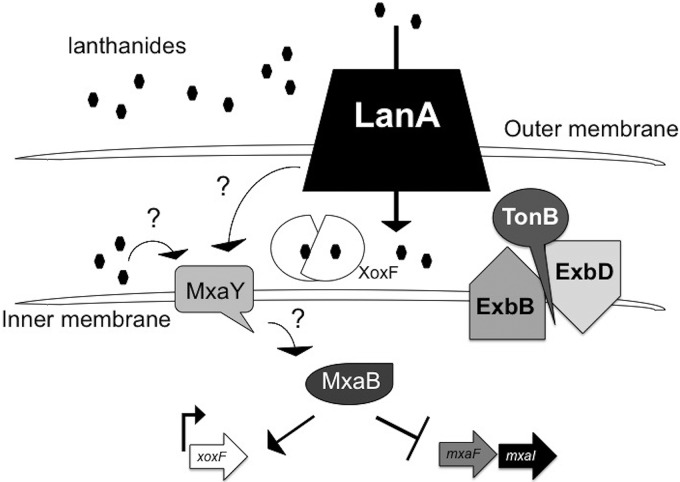
The combination of mutant analysis and transcriptomics in this study provide an updated model for the lanthanide metal switch circuitry (Fig. 6). In analogy to other TonB-dependent receptor-mediated regulatory circuits, LanA likely is involved in lanthanide recognition and signal transduction. The cognate signal transduction protein is not clear at this time, but since MxaY is an integral membrane histidine kinase known to affect lanthanide signaling, it is possibly involved in this cascade. In the E. coli ferric citrate uptake system, the FecI sigma factor is required for full and selective activation of the fecA gene, encoding the TonB-dependent transporter for ferric citrate (41). FecR is an anti-sigma factor complex in the inner membrane that contacts FecA through the periplasm. When the iron citrate substrate is bound by FecA, the FecI sigma factor is released from FecR to enact the iron citrate-responsive transcriptional program (42). While our results show that the FecRI homologs do not affect lanthanum regulation (see Fig. S10 and S11 in the supplemental material), it is possible that a particular FecR and FecI have a role in the regulation of lanA itself, which is expressed at very low levels (see Table S4 in the supplemental material), or that their effects are not as drastic as the transcriptional repression of mxaF. As shown in Fig. 6, TonB-dependent receptors typically pair with a TonB complex, which provides the energetics for the function of the receptor (29), and so a TonB complex (unknown at this time) is included in the model. The lanthanum-repressible tonB gene was not required for any of the tested functions of LanA, but there are at least seven TonB homologs encoded by M. buryatense 5GB1C (28). A recent report on iron TonB-dependent transporters has shown redundancy of TonB function when multiple paralogs exist in the genome (43).
FIG 6.
Working model for the lanthanide metal switch in M. buryatense 5GB1C. LanA is required for lanthanide recognition and signal transduction, but this signal transduction is still not clear. Lanthanide metal in the presence of an intact LanA protein results in the repression of mxaF and the activation of xoxF. FecR-like and FecI-like anti-sigma/sigma factor homologs are not players in signal transduction of the lanthanide metal switch. It is possible that LanA interacts with the integral membrane histidine kinase MxaY directly to relay the signal to the methanol dehydrogenase genes mxaF and xoxF or that lanthanides imported into the periplasm by LanA cause signal transduction via MxaY and the response regulator MxaB.
While lanthanide gene regulatory mechanisms have been described in many organisms, possible mechanisms of lanthanide uptake have only recently been found in two closely related alphaproteobacterial methylotrophs. In Methylorubrum extorquens AM1, a highly selective periplasmic lanthanide-binding protein called lanmodulin has been identified and studied in vitro (44, 45). No homolog for lanmodulin exists in M. buryatense 5GB1C, so the binding mechanism must be different. It is likely that lanthanides function in the periplasm, where XoxF resides (14, 46), but a recent study has shown that lanthanum exists in the cytoplasm (47). Copper is known to enter the bacterial cytoplasm in other organisms, upon which it is pumped out by multiple efflux ATPases, which are specific for loading particular periplasmic cuproenzymes (48). A similar situation could be true for lanthanide metals.
A TonB-dependent transporter with function similar to that of LanA has been recently identified in Methylobacterium extorquens PA1. Deletion of the transporter gene in a ΔmxaF genetic background resulted in the inability to grow on methanol with supplemental lanthanum, implicating the encoded transporter in lanthanum uptake (26). Although the M. extorquens PA1 transporter is not a close homolog to M. buryatense 5GB1C LanA (see Fig. S3 in the supplemental material), our experiments show clearly that LanA is involved in lanthanide signal transduction. Furthermore, because an intact lanA gene is required for lanthanum-dependent growth with the XoxF methanol dehydrogenase in a ΔmxaF mutant (Table 3), it appears that LanA likely has both transporter and transducer functions despite ΔlanA mutants not having a measurable difference in cell-associated lanthanum. It is likely that lanthanide metals simply adhere to ligands on the cell wall, as observed in other organisms (49–51), and the amount that is taken up and utilized by the bacterium cannot be measured accurately with the cell association assay. Additionally, the slower growth in a lanA overexpression mutant suggests that tight control of LanA is important for proper management of lanthanum by the cell (Fig. 5; see Fig. S7 in the supplemental material). If an excess of LanA is present, it could allow too much lanthanum into the periplasm, causing mismetallation of metal-binding sites, as is seen with Cu toxicity (52), or other problems for the cell.
It is clear that LanA is a crucial component of the lanthanide switch in this methanotroph, most likely involved in interacting with lanthanides at the cell surface and coupled at some point in the regulatory circuit to MxaY in the inner membrane and MxaB in the cytoplasm. It might also be coupled to other regulatory components, ultimately responsible for lanthanide-dependent control of which methanol dehydrogenase is expressed in the cell. The details of lanthanide uptake and regulatory switch systems are important for understanding methane uptake and utilization in the environment, as well as for pursuing applications for industrial bioproduction and biometallurgy.
MATERIALS AND METHODS
Bacterial growth and medium composition.
Methylotuvimicrobium buryatense 5GB1C and its derivatives were grown aerobically on NMS2 medium (53) at 30°C under an atmosphere of 25% methane and 75% air. Cells were grown in either 28-ml stoppered tubes or 250-ml stoppered vials with shaking at 200 rpm. For lanthanide uptake experiments, the medium was altered to prevent the precipitation of lanthanum, which has been previously observed for NMS medium (27). To avoid lanthanum precipitation, several alterations were required: (i) the phosphate concentration was decreased 20-fold from that in the published medium, (ii) the medium was buffered at pH 8.2 with 50 mM Trizma (Sigma-Aldrich, St. Louis, MO) rather than at pH 9.5 with carbonate buffer, and (iii) the 500× concentrated trace metal stock was chelated with 300 mM sodium citrate (Sigma-Aldrich, St. Louis, MO) rather than disodium EDTA. This decreased the lanthanum precipitation to <200 nM added lanthanum. Each of these media was used for growth experiments. Reporter gene assays were performed in modified medium for comparison with cell-associated lanthanum assays. E. coli was grown in Luria-Bertani broth supplemented with 50 μg/ml kanamycin when selecting for the presence of a plasmid.
Construction of linear fragments and plasmid vectors.
iProof high-fidelity polymerase (Bio-Rad, Hercules, CA) was used for the PCR for both linear constructs and Gibson assembly of plasmid vectors, according to the manufacturer’s instructions. Linear electroporation fragment cJDG1 was created by overlap PCR by mixing 5′ and 3′ flanking regions previously used for genomic insertion (15), the native mxaF regulatory region, and the zeocin antibiotic resistance cassette ble (54). The reaction included 15 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 1 min, after which primers for amplification of the 1.798-kb electroporation fragment were added and 30 cycles were performed at 98°C for 10 s, 60°C for 30 s, and 72°C for 1 min (primers are listed in Table S1 in the supplemental material). Linear knockout fragments cJDG3 and cJDG13 were created in a similar fashion, but linking ∼800 to 1,000 bp of the 5′ and 3′ flanking regions of the deletion target to the kanamycin resistance cassette. The linear fragments at the correct sizes were excised from a 0.7% (wt/vol) agarose gel using the Zymo Research gel extraction kit (Zymo Research, Irvine, CA).
Plasmids were created with the Gibson Assembly master mix (NEB, Ipswich, MA) according to manufacturer’s instructions (primers for assembly are listed in Table S1). Reaction products were transformed into house-made E. coli Top10 electrocompetent cells, and verified plasmids were transformed into house-made E. coli S17-1 λpir by heat shock to prepare E. coli strains for mating with M. buryatense 5GB1C. Plasmids are listed in Table 4.
TABLE 4.
Strains, plasmids, and linear DNA constructs used in this study
| Strain, plasmid, or construct | Description | Reference |
|---|---|---|
| M. buryatense strains | ||
| 5GB1C | Variant of M. buryatense 5GB1 cured of native plasmid | 53 |
| FC31 | 5GB1C EQU24_01645::PmxaF-xylE | 23 |
| JG3 | 5GB1C EQU24_01645::PmxaF-ble; Zeor | This work |
| JG3μ5 | JG3 lanA* (nonsense point mutation in EQU24_02055) | This work |
| JG8x | FC31 ΔfecI1::Kanr (ΔEQU24_01900::Kanr) | This work |
| JG14 | 5GB1C ΔlanA (ΔEQU24_02055) | This work |
| JG15 | FC31 ΔlanA (ΔEQU24_02055) | This work |
| JG16 | FC31 ΔfecR1 (ΔEQU24_01905) | This work |
| JG17 | FC31 ΔfecR2 (ΔEQU24_01890) | This work |
| JG18 | FC31 ΔfecR3 (ΔEQU24_11875) | This work |
| JG19 | JG14 EQU24_01645::PJ23119-lanA | This work |
| JG20 | JG15 + pJDG49 PlanA-lanA; Kanr | This work |
| JG26 | FC31 ΔfecR1 ΔfecR2 ΔfecR3 | This work |
| JG27 | FC31 ΔfecI3::Kanr (ΔEQU24_11895::Kanr) | This work |
| JG28 | FC31 ΔtonB (ΔEQU24_21875) | This work |
| JG35 | FC31 ΔfecI2 (ΔEQU24_01885) | This work |
| JG41 | 5GB1C EQU24_01645::PxoxF-xylE | This work |
| JG45 | JG41 ΔfecI2 (ΔEQU24_01885) | This work |
| SF8 | 5GB1C + pSMF8 PJ23119-xylE; Kanr | This work |
| Plasmids | ||
| pAWP45 | pCM433-based knockout vector for glgA1; Kanr | 53 |
| pAWP78 | IncP broad-host-range plasmid, minimized from pCM66 | 53 |
| pFC30 | Chromosomal insertion vector at EQU24_01645 containing PmxaF-xylE; Kanr | 23 |
| pJDG2z | pAWP78 with PmxaF-ble; Zeor Kanr | This work |
| pJDG14 | pAWP45 containing flanks to delete lanA (EQU24_02055); Kanr | This work |
| pJDG16 | pAWP45 containing flanks to delete fecR1 (EQU24_01905); Kanr | This work |
| pJDG17 | pAWP45 containing flanks to delete fecR2 (EQU24_01890); Kanr | This work |
| pJDG18 | pAWP45 containing flanks to delete fecR3 (EQU24_11875); Kanr | This work |
| pJDG27 | pFC30 chromosomal insertion vector containing PJ23119-lanA; Kanr | This work |
| pJDG34 | pAWP45 containing flanks to delete tonB (EQU24_218750); Kanr | This work |
| pJDG49 | pAWP78 with PlanA-lanA; Kanr | This work |
| pJDG51 | pAWP45 containing flanks to delete fecI2 (EQU24_01885); Kanr | This work |
| pJDG52 | pFC30 chromosomal insertion vector containing PxoxF-xylE; Kanr | This work |
| pSMF8 | pAWP78 with PJ23119-xylE; Kanr | This work |
| Constructs | ||
| cJDG1 | EQU24_01645|PmxaF-ble|EQU24_01640; Zeor | This work |
| cJDG3 | EQU24_01905| kan| EQU24_01895; Kanr | This work |
| cJDG13 | EQU24_11890| kan| EQU24_11900; Kanr | This work |
| ΔmxaF::ble | EQU24_18140| Ptac-ble| EQU24_18150 | 23 |
Genetic manipulation of M. buryatense 5GB1C.
Electroporation was performed as previously described (54). Fresh cell biomass was spread onto NMS2 plates. The next day, biomass was scooped up with a loop, washed twice with 50 ml room temperature water, and resuspended in the smallest volume of water possible. Linear DNA construct (500 ng to 1 μg) was mixed with a 50-μl aliquot of cell suspension, and the suspension was electroporated in a 2-mm-gap cuvette with an exponential pulse using a Bio-Rad Gene Pulser (2.5 kV, 25 μF, 200 Ω). Cells were immediately recovered in 10 ml NMS2 at 30°C for 3 to 4 h with shaking at 200 rpm. The cultures were harvested at 4,500 × g, and the pellets were resuspended in 500 μl NMS2. One hundred microliters of cell suspension was plated onto each plate of NMS2 with 30 μg/ml zeocin (Alfa Aesar, Haverhill, MA) and 30 μM LaCl3 (Sigma-Aldrich, St. Louis, MO) for cJDG1 and ΔmxaF::ble (15) or with 50 μg/ml kanamycin for cJDG2 and cJDG13. Colonies formed after 4 to 5 days and were picked into the same liquid medium for frozen stock preparation or DNA isolation. Deletion mutants were screened with both external and internal PCR primers to ensure the absence of wild-type sequences.
Conjugation for plasmid-based transformation and gene deletion was performed as previously described (53). Briefly, biomass for the strain of interest was spread onto an NMS2 mating plate and grown overnight under a methane atmosphere at 30°C. The next day, biomass of E. coli S17-1 λpir harboring the plasmid to be transformed was mixed with the fresh M. buryatense 5GB1C biomass and allowed to grow for 2 days at 30°C, and then a thin layer of mixed biomass was spread onto a minimal NMS2 plate supplemented with antibiotic (kanamycin at 50 μg/ml). After 4 days of growth, colonies were streaked onto NMS2 supplemented with kanamycin and 30 μg/ml rifamycin SV (Sigma-Aldrich, St. Louis, MO) to select against the E. coli mating strain. Once grown, biomass was spread first onto NMS2 and subsequently onto NMS2 supplemented with 2.5% (wt/vol) sucrose to counterselect for the sacB marker. Sucrose-resistant colonies were screened by PCR for the desired gene deletions.
Mutagenesis with nitrosoguanidine.
Cells were mutagenized with a protocol modified from previous published mutagenesis protocols for bacteria (55). M. buryatense JG3 (PmxaF-ble) was grown in 50 ml NMS2 supplemented with 30 μM LaCl3 to an optical density at 600 nm (OD600) of ∼0.4 to 0.5. Eleven milliliters was harvested at 4,500 × g for 7 min at 25°C and then washed with 100 mM citrate buffer, pH 5.5. Cells were again pelleted at 4,500 × g for 7 min at 25°C and then resuspended in 300 μl citrate buffer and split into 100-μl aliquots for nitrosoguanidine treatment in Eppendorf tubes. N-Methyl-N′-nitro-N-nitrosoguanidine (MNNG) was added at final concentrations of 10 μg/ml and 30 μg/ml with shaking at 30°C for 10 min. After incubation with MNNG, cells were resuspended and washed with 500 μl 10 mM phosphate buffer, pH 7.0. Cells were finally resuspended in 500 μl NMS2 medium, 50 μl was taken for immediate dilution plating to calculate cell killing, and the remaining 450 μl was inoculated into 5 ml NMS2 supplemented with 30 μM LaCl3 for recovery and mutation fixation. Cells were plated once the cultures reached an OD600 of ∼0.2, after approximately 20 h. Cultures were harvested at 4,500 × g for 7 min at 25°C and resuspended in 300 μl for plating, and then 270 μl of this was plated onto streptomycin when applicable (see Table S2 in the supplemental material) and 30 μl was diluted to 10−1 for plating onto NMS2 plus 30 μM LaCl3 plus 30 μg/ml zeocin for lanthanide repression mutants and to either 10−4 or 10−5. The calculation for mutation rate (Fig. 1) was as follows: (10 × Zeor colonies on 30 μM La)/colonies on NMS2 × 104) = mutation rate.
Whole-genome resequencing.
Genomic DNA from zeocin-resistant colonies was isolated with a GeneJET genomic DNA isolation kit (Thermo Scientific, Waltham, MA). This DNA was sent to Genewiz (South Plainfield, NJ) for 2 × 150-bp paired-end sequencing on the Illumina (San Diego, CA) MiSeq sequencing platform. The paired-end reads for each condition were pooled and processed with breseq version 0.27.1 (56) against the MaGE (28) MBURv2 annotation of M. buryatense 5G from 26 November 2013.
Protein sequence alignment and phylogenetic tree construction.
The National Center for Biotechnology Information (NCBI) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to search for previously identified TonB-dependent transporter protein sequences from E. coli, Pseudomonas aeruginosa, and Yersinia enterocolitica (30). Pairwise alignments were performed with EMBOSS Needle with default settings (57). The Sequence Manipulation Suite was used to visualize the pairwise alignment (58). Clustal Omega was used for multiple-sequence alignment with default settings (59, 60). RAxML was used to build maximum-likelihood trees and determine posterior probabilities from the multiple-sequence alignment, with default settings and 1,000 bootstrap iterations (61). FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the phylogenetic trees generated by RAxML.
Hybrid sequencing of the Methylotuvimicrobium buryatense 5GB1C genome.
DNA for short-read sequencing was isolated from stationary-phase M. buryatense 5GB1C using phenol-chloroform extraction and ethanol precipitation. This DNA was sent to Genewiz for 2 × 150-bp paired end sequencing on the Illumina HiSeq sequencing platform. The resulting sequencing data were trimmed using trimmomatic v0.36 with the command “java -jar trimmomatic -0.36.jar PE -threads 18 -trimlog trim.log {forward_reads}_R1_001.fastq.gz {reverse_reads}_R2_001.fastq.gz R1_P.fastq R1_U.fastq R2_P.fastq R2_U.fastq ILLUMINACLIP:/work/software/trimmomatic/adapters/NexteraPE-PE.fa:3:27:9 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.” After trimming, fully redundant reads were removed with fastuniq v1.1 with the command “fastuniq -i fastq.list -o R1_P_uniq.fastq -p R2_P_uniq.fastq,” where “fastq.list” contained the sample names R1_P.fastq and R2_P.fastq. After these processing steps, 17,329,619 unique read pairs remained (>1,000× coverage).
Long reads were generated from a separate DNA isolation using the MasterPure complete DNA and RNA purification kit (Epicentre, Madison, WI) from 500 ml of stationary-phase culture. The sequencing library was prepared using the Oxford Nanopore Rapid Sequencing (SQK-RAD004) kit (Oxford Nanopore Technologies, Oxford, UK) and immediately loaded onto the Oxford Nanopore MinION long-read sequencer. Bases were called from the raw data using MinKNOW v1.15.1. Adapter sequences were removed using porechop v.0.2.3 with the command “porechop -i long_reads.fastq -o trimmed_long_reads.fastq −t 10.” Both short and long reads were included in a hybrid assembly using Unicycler v0.4.7 with the command “python unicycler-runner.py -1 R1_P_uniq.fastq -2 R2_P_uniq.fastq -l trimmed_long_reads.fastq -o Assembly,” resulting in a single 4,998,879-bp circular contig.
RNA isolation.
Starter cultures were grown to late logarithmic phase (OD600 of ∼0.8), and a 2% (vol/vol) inoculum was transferred into 50 ml NMS2 medium under an atmosphere of 25% methane and 75% air. Wild-type and ΔlanA cells were growth in biological duplicate in either 0 μM or 30 μM lanthanum, for a total of 8 samples. Once samples reached an OD600 of ∼0.6 to 0.7, 2.5 ml of a stop solution of 5% (vol/vol) phenol in ethanol was added to 22.5 ml of culture. Cells were pelleted for 15 min at 4°C and resuspended in RNA extraction buffer (a 1:3 ratio of 5% cetrimonium bromide in 2.5 M NaCl to 0.1 M phosphate buffer at pH 5.8). An equal volume of phenol-chloroform-isoamyl alcohol (25:24:1 ratio) was added, and sodium dodecyl sulfate and N-lauroylsarcosine sodium salt were each added at 0.5% (vol/vol). Cells were lysed in a bead beater with 0.1-mm zirconia-silica beads (Biospec Products, Bartlesville, OK) for 4 min and then centrifuged at 14,000 × g for 5 min. The aqueous fraction was then washed with chloroform-isoamyl alcohol (24:1 ratio), and the aqueous fraction from this wash was precipitated overnight with 150 mM sodium acetate (pH 5.5), 1.5 mM MgCl2, and 50% isopropanol at −80°C. Precipitated RNA was harvested at 14,000 × g for 30 min at 4°C and treated with DNase I (Ambion, Foster City, CA) before purification with the RNeasy minikit and on-column DNase (Qiagen, Hilden, Germany).
qRT-PCR assays.
cDNA was produced using 500 ng of RNA with the SensiFAST cDNA synthesis kit (Bioline, Boston, MA). Quantitative PCR (qPCR) was performed with 400 μM primers, the SensiFAST SYBR No-Rox kit (Bioline), cDNA at a 10−1 dilution factor, and water filled to 10 μl. Real-time qPCRs were performed on a LightCycler 2.0 (Roche Diagnostics, Indianapolis, IN) in 48-well plates (Bio-Rad, Hercules, CA). Cycle threshold (CT) values were measured using Opticon Monitor 3 software (Bio-Rad), and all gene expression values were normalized to rpoD CT values. Primers for qRT-PCR are listed in Table S2.
Transcriptome sequencing (RNA-Seq) analysis.
RNA samples were converted to cDNA libraries and sequenced by Genewiz (South Plainfield, NJ) using Illumina HiSeq 2 × 150 (pair-ended) reads. The raw reads from the sequencing facility were aligned to the newly assembled Methylotuvimicrobium buryatense 5GB1C genome. Alignment was performed using BWA version 0.7.12-r1044 using the BWA-MEM algorithm and default parameters (62). The alignments were postprocessed into sorted BAM files with SAMtools version 1.2-232-g87cdc4a (63). Reads were attributed to ORFs using the htseq-count tool from the “HTSeq” framework version 0.6.1p1 in the “intersection-nonempty” mode (64). Differential abundance analysis was performed with DESeq2 1.2.10 (65) using R 3.3.1. Benjamini-Hochberg-adjusted P values of less than 1.0 × 10−3 were regarded as significant. Log2 fold changes of >1 were regarded as significant, with a few notable exceptions (Tables 1 and 2).
xylE reporter gene assay.
M. buryatense 5GB1C PmxaF-xylE and its derivatives were grown to mid-exponential phase and subcultured to an OD600 of 0.01 in 5 ml NMS2 medium. The cultures were grown to an OD600 of ∼0.6 to 1.0 and normalized to an OD600 of 0.5, at which point catechol-2,3-dioxygenase activity was assayed in whole cells by measuring the rate of change in absorbance at 375 nm in the presence of 1 mM catechol (Sigma-Aldrich, St. Louis, MO) with a plate reader (Molecular Devices SpectraMax 190) as previously described (15, 66).
Cell-associated lanthanum assay.
Assay for lanthanum uptake with whole cells was performed similarly to in previous studies with M. trichosporium OB3b, M. extorquens AM1, and Methylacidiphilum fumariolicum SolV (24, 67), except that a modified medium was required to minimize metal precipitation, as described above. Fifty-milliliter cultures of FC31, JG15, and JG28 were inoculated at 2% (vol/vol) in modified medium as described above; 0.4% (vol/vol) methanol was used as a carbon source so that cells could be grown in plastic Erlenmeyer flasks, since lanthanide metals stick to glass and interfere with uptake calculations. Five-milliliter samples were taken at OD600s of ∼0.3 and ∼1.3 to assess uptake at early exponential phase and early stationary phase. Cells were harvested, and supernatants were saved for analysis. Cell pellets were resuspended in 500 μl NMS2 with 20% nitric acid, lysed at 95°C for 2 h, and diluted to a final concentration of 2% nitric acid. Lanthanum concentrations in cell pellets and supernatants were analyzed by inductively coupled plasma mass spectrometry (ICP-MS) on an Elan DRC-e instrument (PerkinElmer, Shelton, CT) with yttrium as an internal standard.
Data availability.
Final gene annotations were created through submission to the National Center for Biotechnology Information (NCBI) WGS submission portal, accession number CP035467. The whole-genome resequencing reads have been deposited through the NCBI Sequence Read Archive (SRA) with accession number PRJNA535342. All RNA-Seq reads and read counts per gene were uploaded to the NCBI Gene Expression Omnibus (GEO) under accession number GSE125909.
Supplementary Material
ACKNOWLEDGMENTS
Research support was provided by discretionary funding to Mary Lidstrom from the University of Washington.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Ludmila Chistoserdova, Aaron Puri, and David Beck for useful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00120-19.
REFERENCES
- 1.Forster P, Ramaswamy V, Artaxo P, Berntsen T, Betts R, Fahey DW, Haywood J, Lean J, Lowe DC, Myhre G, Nganga J, Prinn R, Raga G, Schulz M, Van Dorlan R, 2007. Changes in atmospheric constituents and in radiative forcing, p 129–234. In Solomon S. et al. (ed), Climate change 2007: the physical science basis. Contribution of Working Group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 2.Saunois M, Jackson RB, Bousquet P, Poulter B, Canadell JG. 2016. The growing role of methane in anthropogenic climate change. Environ Res Lett 11:120207. doi: 10.1088/1748-9326/11/12/120207. [DOI] [Google Scholar]
- 3.Kirschke S, Bousquet P, Ciais P, Saunois M, Canadell JG, Dlugokencky EJ, Bergamaschi P, Bergmann D, Blake DR, Bruhwiler L, Cameron-Smith P, Castaldi S, Chevallier F, Feng L, Fraser A, Heimann M, Hodson EL, Houweling S, Josse B, Fraser PJ, Krummel PB, Lamarque J-F, Langenfelds RL, Quéré C, Naik V, O’Doherty S, Palmer PI, Pison I, Plummer D, Poulter B, Prinn RG, Rigby M, Ringeval B, Santini M, Schmidt M, Shindell DT, Simpson IJ, Spahni R, Steele PL, Strode SA, Sudo K, Szopa S, van der Werf GR, Voulgarakis A, van Weele M, Weiss RF, Williams JE, Zeng G. 2013. Three decades of global methane sources and sinks. Nature Geosci 6:813–823. doi: 10.1038/ngeo1955. [DOI] [Google Scholar]
- 4.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck D, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina del Rio T, Ivanova N, Lidstrom ME, Chistoserdova L. 2013. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 1:e23. doi: 10.7717/peerj.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova L. 2016. Lanthanides: new life metals?. World J Microbiol Biotechnol 32:1–7. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 7.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez CN. 2016. Pyrroloquinoline quinone-containing ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multi-carbon substrates. J Bacteriol 198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. 2017. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8:e00570-17. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen HJM. 1979. Environmental chemistry of the elements. Academic Press, New York, NY. [Google Scholar]
- 10.Martinez-Gomez NC, Vu HN, Skovran E. 2016. Lanthanide chemistry: from coordination in chemical complexes shaping our technology to coordination in enzymes shaping bacterial metabolism. Inorg Chem 55:10083–10089. doi: 10.1021/acs.inorgchem.6b00919. [DOI] [PubMed] [Google Scholar]
- 11.Nancharaiah YV, Mohan VS, Lens PNL. 2016. Biological and bioelectrochemical recovery of critical and scarce metals. Trends Biotechnol 34:137–155. doi: 10.1016/j.tibtech.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 13.Larsen Ø, Karlsen OA. 2016. Transcriptomic profiling of Methylococcus capsulatus (Bath) during growth with two different methane monooxygenases. Microbiol Open 5:254–267. doi: 10.1002/mbo3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez CN, Skovran E. 2016. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol 198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu F, Lidstrom ME. 2016. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farhan Ul Haque M, Kalidass B, Bandow N, Turpin EA, DiSpirito AA, Semrau JD. 2015. Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microbiol 81:7546–7552. doi: 10.1128/AEM.02542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orata FD, Meier-Kolthoff JP, Sauvageau D, Stein LY. 2018. Phylogenomic analysis of the gammaproteobacterial methanotrophs (order Methylococcales) calls for the reclassification of members at the genus and species levels. Front Microbiol 9:3162. doi: 10.3389/fmicb.2018.03162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyuzhnaya MG, Khmelenina V, Eshinimaev B, Sorokin D, Fuse H, Lidstrom M, Trotsenko Y. 2008. Classification of halo(alkali)philic and halo(alkali)tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int J Syst Evol Microbiol 58:591–596. doi: 10.1099/ijs.0.65317-0. [DOI] [PubMed] [Google Scholar]
- 19.Khmelenina VN, Beck DAC, Munk C, Davenport K, Daligault H, Erkkila T, Goodwin L, Gu W, Lo CC, Scholz M, Teshima H, Xu Y, Chain P, Bringel F, Vuilleumier S, DiSpirito A, Dunfield P, Jetten MSM, Klotz MG, Knief C, Murrell JC, den Camp OHJM, Sakai Y, Semrau J, Svenning M, Stein LY, Trotsenko YA, Kalyuzhnaya MG. 2013. Draft genome sequence of Methylomicrobium buryatense strain 5G, a haloalkaline-tolerant methanotrophic bacterium. Genome Announce 1:e00053-13. doi: 10.1128/GenomeA.00053-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green PN, Ardley JK. 2018. Review of the genus Methylobacterium and closely related organisms: a proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int J Syst Evol Microbiol 68:2727–2748. doi: 10.1099/ijsem.0.002856. [DOI] [PubMed] [Google Scholar]
- 21.Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME. 2011. XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol 193:6032–6038. doi: 10.1128/JB.05367-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer AL, Morris CJ, Lidstrom ME. 1997. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology 143:1737–1744. doi: 10.1099/00221287-143-5-1737. [DOI] [PubMed] [Google Scholar]
- 23.Chu F, Beck D, Lidstrom ME. 2016. MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ 4:e2435. doi: 10.7717/peerj.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W, Haque M, DiSpirito AA, Semrau JD. 2016. Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol Lett 363:1–6. doi: 10.1093/femsle/fnw129. [DOI] [PubMed] [Google Scholar]
- 25.Williams E, Shimmin MA, Bainbridge BW. 1977. Mutation in the obligate methylotrophs Methylococcus capsulatus and Methylomonas albus. FEMS Microbiol Lett 2:293–296. doi: 10.1016/0378-1097(77)90054-4. [DOI] [Google Scholar]
- 26.Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld‐Miller M, Hattendorf B, Vorholt JA. 2019. Use of rare‐earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol 111:1152–1166. doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Semrau JD. 2017. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 101:8499–8516. doi: 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 28.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Smith A, Weiman M, Médigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci 33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Akberdin IR, Collins DA, Hamilton R, Oshchepkov DY, Shukla AK, Nicora CD, Nakayasu ES, Adkins JN, Kalyuzhnaya MG. 2018. Rare earth elements alter redox balance in Methylomicrobium alcaliphilum 20ZR. Front Microbiol 9:2735. doi: 10.3389/fmicb.2018.02735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semrau JD, DiSpirito AA, Gu W, Yoon S. 2018. Metals and methanotrophy. Appl Environ Microbiol 84:e02289-17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enz S, Mahren S, Stroeher UH, Braun V. 2000. Surface signaling in ferric citrate transport gene induction: Interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol 182:637–646. doi: 10.1128/JB.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPheat WL, Mann NH, Dalton H. 1987. Isolation of mutants of the obligate methanotroph Methylomonas albus defective in growth on methane. Arch Microbiol 148:40–43. doi: 10.1007/BF00429645. [DOI] [Google Scholar]
- 35.Nicholaidis AA, Sargent AW. 1987. Isolation of methane monooxygenase-deficient mutants from Methylosinus trichosporium OB3b using dichloromethane. FEMS Microbiol Lett 41:47–52. doi: 10.1111/j.1574-6968.1987.tb02139.x. [DOI] [Google Scholar]
- 36.Phelps PA, Agarwal SK, Speitel GE Jr, Georgiou G. 1992. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl Environ Microbiol 58:3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenstadt E, Carlton BC, Brown BJ. 1994. Gene mutation, p 297–315. In Gerhardt P. (ed), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 38.Good NM, Walser ON, Moore RS, Suriano C, Huff AF, Martinez-Gomez N. 2018. Investigation of lanthanide-dependent methylotrophy uncovers complementary roles for alcohol dehydrogenase enzymes. bioRxiv 10.1101/329011. [DOI]
- 39.Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilman A, Fu Y, Hendershott M, Chu F, Puri AW, Smith A, Pesesky M, Lieberman R, Beck D, Lidstrom ME. 2017. Oxygen-limited metabolism in the methanotroph Methylomicrobium buryatense 5GB1C. PeerJ 5:e3945. doi: 10.7717/peerj.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeda H, Jishage M, Nomura T, Fujita N, Ishihama A. 2000. Two extracytoplasmic function sigma subunits, sigma E and sigma FecI, of Escherichia coli: promoter selectivity and intracellular levels. J Bacteriol 182:1181–1184. doi: 10.1128/JB.182.4.1181-1184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastiaansen KC, Bitter W, Llamas MA. 2012. ECF sigma factors: from stress management to iron uptake, p 59–86. In Filloux AAM. (ed), Bacterial regulatory networks. Caister Academic Press, Poole, UK. [Google Scholar]
- 43.Dong Y, Geng J, Liu J, Pang M, Awan F, Lu C, Liu Y. 2019. Roles of three TonB systems in the iron utilization and virulence of the Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl Microbiol Biotechnol 103:4203–4215. doi: 10.1007/s00253-019-09757-4. [DOI] [PubMed] [Google Scholar]
- 44.Cook EC, Featherston ER, Showalter SA, Cotruvo JA. 2018. Structural basis for rare earth element recognition by Methylobacterium extorquens lanmodulin. Biochemistry 58:120–125. doi: 10.1021/acs.biochem.1028b01019. [DOI] [PubMed] [Google Scholar]
- 45.Cotruvo JA, Featherston ER, Mattocks JA, Ho JV, Laremore TN. 2018. Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J Am Chem Soc 140:15056–15061. doi: 10.1021/jacs.8b09842. [DOI] [PubMed] [Google Scholar]
- 46.Bayer ME, Bayer MH. 1991. Lanthanide accumulation in the periplasmic space of Escherichia coli B. J Bacteriol 173:141–149. doi: 10.1128/jb.173.1.141-149.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattocks JA, Ho JV, Cotruvo JA. 2019. A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J Am Chem Soc 141:2857–2861. doi: 10.1021/jacs.8b12155. [DOI] [PubMed] [Google Scholar]
- 48.Stewart LJ, Thaqi D, Kobe B, McEwan AG, Waldron KJ, Djoko KY. 2019. Handling of nutrient copper in the bacterial envelope. Metallomics 11:50–63. doi: 10.1039/c8mt00218e. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi Y, Yamamoto M, Yamamoto Y, Tanaka K. 2010. EXAFS study on the cause of enrichment of heavy REEs on bacterial cell surfaces. Geochim Cosmochim Acta 74:5443–5462. doi: 10.1016/j.gca.2010.07.001. [DOI] [Google Scholar]
- 50.Ozaki T, Suzuki Y, Nankawa T, Yoshida T, Ohnuki T, Kimura T, Francis AJ. 2006. Interactions of rare earth elements with bacteria and organic ligands. J Alloys Compounds 408–412:1334–1338. doi: 10.1016/j.jallcom.2005.04.142. [DOI] [Google Scholar]
- 51.Takahashi Y, Châtellier X, Hattori KH, Kato K, Fortin D. 2005. Adsorption of rare earth elements onto bacterial cell walls and its implication for REE sorption onto natural microbial mats. Chem Geol 219:53–67. doi: 10.1016/j.chemgeo.2005.02.009. [DOI] [Google Scholar]
- 52.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puri AW, Owen S, Chu F, Chavkin T, Beck DAC, Kalyuzhnaya MG, Lidstrom ME. 2015. Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol 81:1775–1781. doi: 10.1128/AEM.03795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan X, Chu F, Puri AW, Fu Y, Lidstrom ME. 2016. Electroporation-based genetic manipulation in type I methanotrophs. Appl Environ Microbiol 82:2062–2069. doi: 10.1128/AEM.03724-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foster PL. 1991. In vivo mutagenesis. Methods Enzymol 204:114–125. doi: 10.1016/0076-6879(91)04007-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq, 165 In Sun L, Shou W (ed), Engineering and analyzing multicellular systems: methods and protocols. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 58.Stothard P. 2000. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 59.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2014. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 66.Puri AW, Schaefer AL, Fu Y, Beck DAC, Greenberg PE, Lidstrom ME. 2016. Quorum sensing in a methane-oxidizing bacterium. J Bacteriol 199:e00773-16. doi: 10.1128/JB.00773-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hogendoorn C, Roszczenko-Jasińska P, Martinez-Gomez CN, de Graaff J, Grassl P, Pol A, den Camp HJM, Daumann LJ. 2018. Facile arsenazo III-based assay for monitoring rare earth element depletion from cultivation media for methanotrophic and methylotrophic bacteria. Appl Environ Microbiol 84:e02887-17. doi: 10.1128/AEM.02887-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Final gene annotations were created through submission to the National Center for Biotechnology Information (NCBI) WGS submission portal, accession number CP035467. The whole-genome resequencing reads have been deposited through the NCBI Sequence Read Archive (SRA) with accession number PRJNA535342. All RNA-Seq reads and read counts per gene were uploaded to the NCBI Gene Expression Omnibus (GEO) under accession number GSE125909.