O antigen is an important virulence factor presented on the cell surface of Gram-negative bacteria that is critical for bacterial physiology and pathogenesis. However, some aspects of O antigen biosynthesis, such as the mechanisms for determining polysaccharide chain length, are poorly understood. In this study, we identified unique regions in the O antigen chain length regulators (termed Wzz) of the problematic opportunistic pathogen Pseudomonas aeruginosa. We show that these regions are critical for determining O antigen chain length, which provides new insight into the model of the Wzz mechanism. Ultimately, our work adds knowledge toward understanding an important step in the biosynthesis of this virulence factor, which is applicable to a wide range of Gram-negative pathogens.
KEYWORDS: chain length, O antigen, Pseudomonas aeruginosa, lipopolysaccharide, polysaccharide co-polymerase
ABSTRACT
The outer leaflet of the outer membrane of nearly all Gram-negative bacteria contains lipopolysaccharide (LPS). The distal end of LPS may be capped with O antigen, a long polysaccharide that can range from a few to hundreds of sugars in length. The chain length of the polysaccharide has many implications for bacterial survival and consequently is tightly controlled. In the Wzx/Wzy-dependent route of O antigen synthesis, one or more Wzz proteins determine the chain length via an unknown mechanism. To gain insight into this mechanism, we identified and characterized important regions of two Wzz proteins in Pseudomonas aeruginosa serotype O13, which confer the production of “long” (Wzz1) and “very long” (Wzz2) chain lengths, respectively. We found that compared to Wzz1, Wzz2 has distinct amino acid insertions in the central α-helices (insα6 and insα7) and in membrane-distal (insL4) and -proximal (insIL) loops. When these regions were deleted in Wzz2, the mutant proteins conferred drastically shortened chain lengths. Within these regions we identified several conserved amino acid residues that were then targeted for site-directed mutagenesis. Our results implicate an RTE motif in loop 4 and a “hot spot” of charged and polar residues in insα7 in the function of Wzz2. We present evidence that the functionally important residues of insα7 are likely involved in stabilizing Wzz through coiled-coil interactions.
IMPORTANCE O antigen is an important virulence factor presented on the cell surface of Gram-negative bacteria that is critical for bacterial physiology and pathogenesis. However, some aspects of O antigen biosynthesis, such as the mechanisms for determining polysaccharide chain length, are poorly understood. In this study, we identified unique regions in the O antigen chain length regulators (termed Wzz) of the problematic opportunistic pathogen Pseudomonas aeruginosa. We show that these regions are critical for determining O antigen chain length, which provides new insight into the model of the Wzz mechanism. Ultimately, our work adds knowledge toward understanding an important step in the biosynthesis of this virulence factor, which is applicable to a wide range of Gram-negative pathogens.
INTRODUCTION
The dominant molecule in the outer leaflet of the outer membrane of nearly all Gram-negative bacteria is lipopolysaccharide (LPS), which plays integral roles in interactions between bacteria and their environment. LPS is a tripartite molecule, but minimally it can be found on the bacterial cell envelope comprised of two domains: lipid A, which anchors the glycolipid in the outer membrane, and a short core oligosaccharide. A third moiety, termed O antigen, may then cap the molecule by covalent linkage to the core. O antigen is characterized by its heterogeneity in the composition of its sugar constituents and in the length of the polymer, even within the same species. Various lengths of O antigen polymer have been shown to benefit bacteria in numerous ways, including resistance to serum (1–4), bacteriophage infection (5), and bile acid (6, 7). Further, regulation of O antigen chains to specific lengths is typically required for full virulence of many organisms (1, 2, 7–9), and the length of the O antigen chain is also an important consideration in the development of effective glycoconjugate vaccines against diseases caused by bacterial pathogens (10). Therefore, understanding the mechanisms by which bacteria regulate O antigen chain length is central to understanding bacterial survival and treatment of infections.
The synthesis of O antigen follows one of four known routes: an ABC transporter-dependent pathway, a synthase-dependent pathway, a Wzk-dependent pathway, or a Wzx/Wzy-dependent pathway (11). In the Wzx/Wzy-dependent pathway, O antigen is synthesized from short oligosaccharide building blocks, often termed O repeats or O units. O units are built on an undecaprenyl phosphate (Und-P) moiety at the inner face of the inner membrane, flipped to the periplasmic face by Wzx, and then polymerized into long chains by Wzy (12). Polymerization of O units by Wzy is relatively inefficient unless accompanied by another class of proteins called polysaccharide copolymerases (PCPs), which are designated Wzz according to standard nomenclature (13, 14). A given Wzz protein determines a range of polysaccharide lengths that is referred to as the modality, or modal lengths, and seems to be inherent to each Wzz protein, such that heterologous expression of a Wzz in a recipient strain will result in the production of LPSs of lengths similar to those of the strain that is the source of the gene (2, 10, 15–18). Remarkably, the lengths of the O antigen conferred by different Wzz proteins can range from a few to >100 O units, and many bacteria express more than one Wzz, resulting in the production of various populations of O antigen chains in a wide range of sizes (8, 19, 20).
The topology of Wzz proteins consists of an N- and C-terminal transmembrane helix with a large periplasmic domain between them. The structures of the periplasmic domains of several Wzz proteins have been solved, including WzzB and FepE from Salmonella enterica serovar Typhimurium, WzzB from Shigella flexneri, and WzzE from Escherichia coli (21–23). These structural studies have revealed that Wzz proteins share conserved protomeric and oligomeric structures; the periplasmic domain contains a membrane-proximal α/β base domain followed by a long α-helix that extends approximately 100 Å into the periplasm and leads into a flexible loop (loop 4) at the apex of the protein. This loop is followed by several helical and coiled structures that back to the membrane, leading into the second transmembrane helix (21). The protomers of Wzz form large oligomers that resemble a bell (see Fig. S1 in the supplemental material), although the number of subunits within this oligomer varies in the different published reports, apparently due to the methods used for structural determination (21, 23, 24). Recently, the first structural information for the transmembrane domains (TMDs) of WzzB in S. Typhimurium was reported, and the authors determined that the membrane helices of the same protomer form helical bundles but do not interact with the bundles of other protomers. This results in the TMDs resembling an open claw in the oligomer (25). The authors proposed that Wzy might interact with Wzz between these TMDs, which is supported by biochemical and genetic evidence of Wzy-Wzz interaction (26–29).
Even though the aforementioned structural data have provided insight into Wzz architecture, the details of how O antigen chain length is regulated by this protein are unknown. The most recent model suggests that chain length is determined by a combination of so-called “stopwatch” and “ruler” mechanisms (25). The proposed interaction of Wzy and Wzz via their TMD is the stopwatch: when this association breaks, polymerization rapidly terminates. The likelihood of dissociation would decrease as the polymer increases in length since this interaction would be stabilized by Wzz interactions with the polysaccharide. The ruler is defined by a given capacity of Wzz to bind polysaccharide; i.e., when this capacity is reached, polymerization ceases. While the ruler appears to be dependent on the combined contributions of positions throughout the Wzz protein, specific regions or amino acids have been implicated in playing a greater role in the regulation of O antigen chain length. First, studies on two Wzz proteins of S. flexneri and E. coli (WzzB and FepE, respectively) revealed that insertion of small amino acid linkers or specific amino acid changes in a small loop at the base of the proteins could increase or decrease the chain length conferred by Wzz (30, 31). Second, studies on FepE indicated that deletions and specific amino acid replacements in loop 4 (situated at the top of the protein) drastically reduced O antigen chain length (22, 31, 32). However, since there is low sequence similarity of Wzz between species, whether these regions are important in other Wzz proteins is not readily apparent. Further, biochemical characterization of Wzz in other systems is necessary for a comprehensive understanding of Wzz-mediated chain length regulation of O antigen.
In this study, we investigated O antigen chain length regulation in the important opportunistic pathogen Pseudomonas aeruginosa. P. aeruginosa can colonize individuals with compromised host defenses, causing life-threatening infections. It is also the etiological agent of chronic lung dysfunction and death in patients with cystic fibrosis (33). One O antigen glycoform produced by P. aeruginosa is the O-specific antigen (OSA), which is the basis of the classification of this bacterium into 20 unique serotypes depending on the sugar identities and linkages in the polysaccharide chain (34). In those serotypes studied to date, OSA may be regulated to long and very long chain lengths by two Wzz proteins, Wzz1 and Wzz2, respectively (4, 17, 20, 35). Importantly, the long chains endow P. aeruginosa with resistance to serum, and the loss of both Wzz1- and Wzz2-mediated O antigen chain length regulation results in attenuation of the bacterium in a mouse model of pneumonia (4, 9). A previous report by Kintz and Goldberg showed that expressing variants of Wzz2 with mutations at amino acid residue 321 in serotype O11 altered the OSA chain length, indicating that this position is important for Wzz2 function (36). However, to date, further biochemical characterization of Wzz1 and Wzz2 is lacking. Here we report on the discovery of key sequence/structural elements involved in Wzz1 and Wzz2 function in a previously uncharacterized serotype, O13. Specifically, we discovered that the sequence of Wzz2 contains unique insertions and deletions compared to the sequence of Wzz1 and that these anomalies are critical to the function of Wzz2. We also identified conserved amino acids in these areas of the protein that, when mutated, have varied effects on the production of Wzz2-mediated chain lengths. We provide evidence that these functionally important amino acids are responsible for maintaining the stability of Wzz2 but are not necessary for oligomerization. Finally, our results lead us to suggest a role for each of the described regions in the mechanism of Wzz.
RESULTS
Confirmation of the role of Wzz1 and Wzz2 in serotype O13.
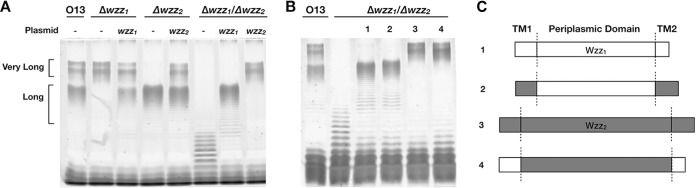
The IATS serotype O13 strain was chosen for this study because we found that this strain produces an LPS banding profile that consistently stains well by silver staining. This is critical when investigating changes to the Wzz-mediated OSA chain lengths, which may be subtle or drastic. Another advantage of using serotype O13 is that all O antigen bands can be attributed to OSA since this serotype does not produce common polysaccharide antigen, which is another O antigen produced by a separate pathway in some strains (37). Finally, probing the role of Wzz function in LPS biosynthesis in O13 offered an opportunity to investigate P. aeruginosa OSA chain length regulation in a serotype where this had yet to be examined. We first characterized the function of Wzz1 and Wzz2 in O13 by creating clean/in-frame deletions of each of the corresponding genes. When wzz1 was deleted from the O13 chromosome, the LPS banding pattern in silver-stained SDS-PAGE gels showed that the bands corresponding to long chain lengths of OSA were completely abrogated, leaving only two distinct very long OSA bands (Fig. 1A). Deleting wzz2 caused the mutant bacteria to produce LPS totally lacking these very long OSA bands, whereas the long chain length ladder-like bands were not affected. In the case of the double mutant in which wzz1 and wzz2 were both deleted, the bacteria produced unregulated OSA chain lengths. These results are consistent with previous studies of Wzz proteins in P. aeruginosa, i.e., Wzz1 regulates the long chains and Wzz2 regulates the very long chains.
FIG 1.
Characterization of the Wzz proteins of P. aeruginosa serotype O13. (A) Gene deletions of wzz1 and wzz2 affect OSA chain length. The results indicate that Wzz1 regulates the long-chain OSA, while Wzz2 regulates the very-long-chain OSA in serotype O13. (B) Reciprocal swaps of the periplasmic and TM/cytosolic domains of Wzz1 and Wzz2. Lane 1, Wzz1; lane 2, Z2-Z1(prp)-Z2 (Wzz2 TM/cytosolic domains with Wzz1 periplasmic domain); lane 3, Wzz2; lane 4, Z1-Z2(prp)-Z1 (Wzz1 TM/cytosolic domains with Wzz2 periplasmic domain). (C) The constructs were expressed in the O13 Δwzz1 Δwzz2 double-mutant background. The results indicate that the periplasmic domain is the main determinant of chain length, while the TM/cytosolic regions play a minor role.
A previous study by the Cygler group on two Wzz proteins from E. coli O157, namely, WzzB and FepE, showed that the periplasmic domain, but not the transmembrane (TM)/cytosolic domain, of these two proteins was responsible for the observed differences in the LPS chain lengths they confer (32). Hence, we aimed to test the hypothesis that the periplasmic domain of the Wzz proteins in P. aeruginosa O13 plays a similar role in LPS chain length regulation. To test this, we constructed reciprocal swaps of the periplasmic and TM/cytosolic domains of Wzz1 and Wzz2 and expressed them in the O13 Δwzz1 Δwzz2 double mutant (Fig. 1B). When a chimera of the Wzz1 periplasmic domain and the Wzz2 TM/cytosolic regions was expressed, the resulting OSA chains produced long LPS chain lengths that were nearly identical to the LPS banding pattern regulated by wild-type Wzz1. However, a very slight increase in the average LPS chain length was observed. Similarly, a construct of the chimera expressing the Wzz2 periplasmic domain fused with the Wzz1 TM/cytosolic regions imparted an OSA modality similar to that conferred by Wzz2, except for a slight decrease in chain length. These experiments verified that the periplasmic domains of the P. aeruginosa Wzz proteins play a predominant role in determining O antigen chain length. However, the TMS/cytosolic regions also play a role in determining O antigen chain length, albeit to a lesser extent.
Wzz2 contains unique insertions and deletions compared to Wzz1.
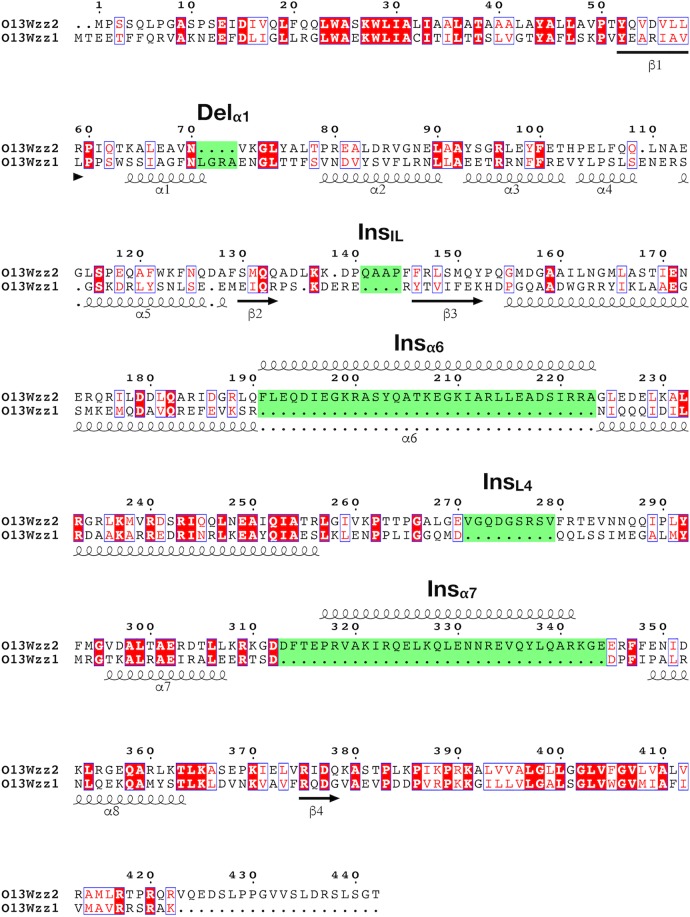
Due to the obvious role of the periplasmic domains in determining chain length, we sought to identify any sequence features in this particular region that might be unique to either Wzz1 or Wzz2. Wzz2 of O13 is 93 amino acids (aa) longer than Wzz1 (442 aa versus 349 aa), and alignment of these two protein sequences identified several gaps (see Fig. S2 in the supplemental material). Further, the level of sequence identity between the two proteins is low, at only 29%. To accurately determine the locations of the gaps, we needed to analyze many homologs of these proteins in a multiple-sequence alignment (MSA). We compiled a diverse set of homologs of Wzz1 and Wzz2 from the genus Pseudomonas (Table S3) by subjecting the amino acid sequences to a BLAST search. Intriguingly, results from the MSA clearly showed that the sequences of the Wzz proteins included in the analysis could be separated into two groups with clearly defined insertions and deletions between them. These indels were at very different positions compared to the alignment involving only the sequences of Wzz1 and Wzz2. The first group, containing Wzz2, had several insertions and one deletion compared to the second group, containing Wzz1 (Fig. S3). The largest insertions in the Wzz2 sequence were at aa 191 to 223 and aa 313 to 344. Two smaller insertions were also present at aa 141 to 144 and aa 271 to 279, and a small deletion (4 amino acids) was localized to Wzz2 N70/V71, which aligned with Wzz1 N72/E77 (Fig. 2).
FIG 2.
Alignment of Wzz1 and Wzz2 protein sequences reveals distinct insertions and deletions. Sequence alignment of Wzz1 and Wzz2. The alignment was extracted from a larger alignment of Wzz1, Wzz2, and their homologs in various Pseudomonas spp. The secondary structure based on the Wzz1 homology model is displayed below the alignment, and the predicted structures of regions exclusively found in Wzz2 are displayed above the alignment. The relevant insertions and deletions are highlighted in green.
To determine how these indels contribute to the overall protein structure, we created models of both Wzz1 and Wzz2 using the protein structure prediction program I-TASSER (38). As anticipated, the server generated an overall structure of Wzz1 that was nearly identical to the top template, FepE (PDB code 3B8M; root mean square deviation [RMSD] = 0.81) (Fig. 3). Using this structural model, we were able to map the start of each large insertion of Wzz2 (aa 191 to 223 and aa 313 to 344) to the long central α-helix (α6) and the coiled region between α7 and α8, which runs antiparallel to α6. In the model of the Wzz2 protomer generated by I-TASSER, these insertions were shown to protrude from the central axis (Fig. 3). Since the published structures of Wzz revealed that it is oligomeric, the protrusions would most likely disrupt interactions between neighbouring protein subunits. Further, this model does not agree with the conserved protomeric structure described previously (21) (Fig. S1). Therefore, we deemed this model to be not representative of the true structure of this region. Instead, we propose that the insertions in α6 and between α7/8 would extend the Wzz2 oligomers along the vertical axis. This interpretation is rational because the insertion sequences in question are essentially the same size (33 aa versus 32 aa). Presumably, any extension of the long central helix would require an equal extension in the opposite direction to maintain the overall symmetry of the Wzz structure. The insertions at aa 141 to 144 and aa 271 to 279 were localized to the loop between β2 and β3 (within the internal cavity of the protein) and L4 (at the apex of the protomer), respectively. Finally, the 4-aa deletion was within α1 (Fig. 3). Here we refer to these indels as insα6, insα7, insIL, insL4, insα1, and delα1 accordingly.
FIG 3.
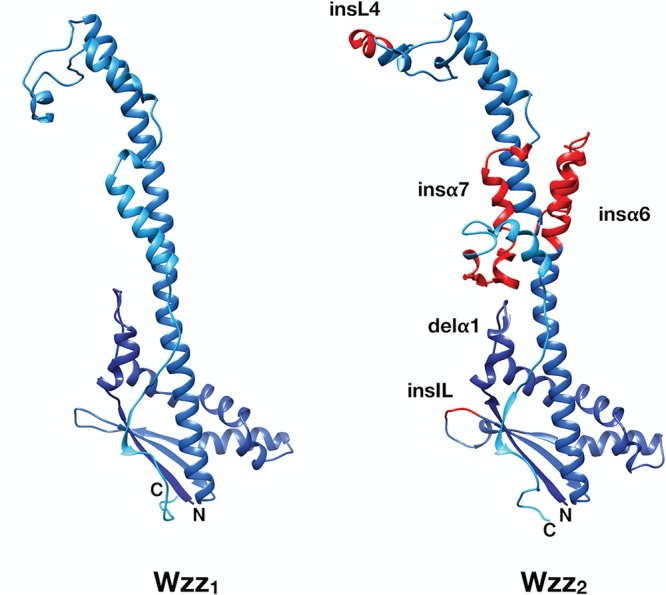
I-TASSER structural prediction of Wzz1 and Wzz2. The models are shaded dark to light blue from the N to the C terminus. The predicted structure and locations of the Wzz2 insertions (“ins”) and deletions (“del”) are indicated in red.
Deletion of insIL, insL4, or insα6/7 alters the chain length conferred by Wzz2.
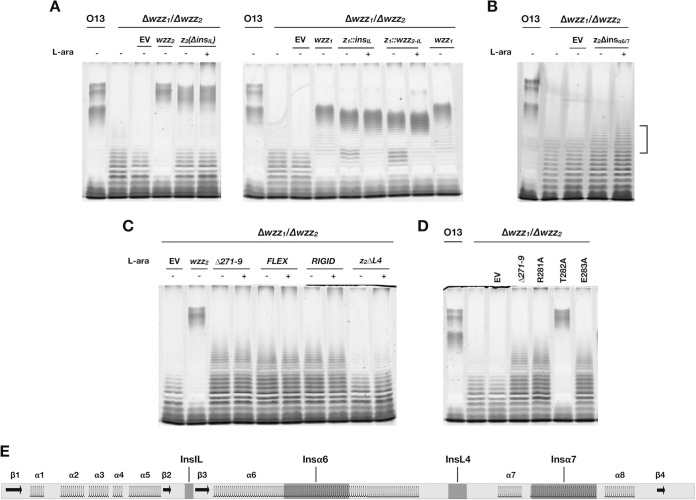
To investigate the role played by insIL, insL4, and insα6/7 in OSA chain length regulation, we prepared constructs with various deletions of these regions in Wzz2 and observed changes to the LPS banding profiles when each of these proteins was expressed in the double-knockout mutant O13 Δwzz1 Δwzz2. Deletion of the four amino acids of insIL caused a decrease in the LPS chain length that is normally imparted by Wzz2 (Fig. 4A). Moreover, this deletion caused a change to the overall banding pattern of OSA in SDS-PAGE analysis. Rather than the usual bimodal banding pattern conferred by Wzz2, Wzz2ΔinsIL conferred an LPS banding pattern showing equal distribution between the minimum and maximum chain length bands. Interestingly, this broad pattern is more similar to the pattern conferred by Wzz1, which lacks this insertion. A previous study by Papadopolous and Morona showed that two different mutants with 5-aa insertions in the analogous region in WzzB increased the chain length (30). This led us to test the effect on OSA chain length when insIL was introduced into Wzz1. When expressed in the O13 Δwzz1 Δwzz2 background, this bacterial strain produced an OSA banding pattern that was similar to that imparted by Wzz1 but was shifted toward shorter chain lengths. We hypothesized that a more drastic change in this region could have a greater effect on chain length; this prompted us to construct a recombinant protein whereby the entire loop of Wzz1 was swapped with the loop from Wzz2. This construct resulted in an effect on Wzz1-mediated LPS chain length regulation similar to that of the 4-aa insertion (Fig. 4A). This indicates that this particular region is more amenable to changes in amino acid sequence without significantly perturbing Wzz1 function and that an increase in the length of this loop alone does not translate to an increase in OSA chain length. In Wzz2, a small change to this loop appears to have a more drastic effect on LPS chain length regulation.
FIG 4.
Manipulation of the indels of Wzz2 modulates chain length determination. The mutants were expressed in the O13Δwzz1 Δwzz2 double mutant in the absence (−) or presence (+) of L-arabinose. (A) Deletion of insIL in Wzz2 [z2(ΔinsIL)] results in a shorter, broader chain length pattern. Insertion of insIL into Wzz1 (z1::insIL) reduces chain length. Replacement of the entire Wzz1 internal loop with that of Wzz2 (z1::wzz2-IL) similarly reduces chain length. (B) Deletion of insα6/7 in Wzz2 (z2Δinsα6/7) results in severely reduced chain length determining activity (bracket). (C) Deletion of residues 271 to 279 of insL4 in Wzz2 (Δ271-9) significantly reduces the OSA chain length. Replacing this region with flexible (FLEX) or rigid (RIGID) peptide sequences has a similar effect. Deletion of L4 (ΔL4) results in the greatest reduction in chain length-determining activity. (D) L4 R281A and E283A variants result in a very short chain length phenotype similar to that observed when insL4 is deleted. (E) Schematic representation of the periplasmic domain of Wzz2. The location of each region investigated is in dark gray. EV, empty vector.
As described above, the location of insα6 and insα7 suggests that both of these two insertions needed to be present to maintain symmetry of the protomeric structure of Wzz2. This led us to delete both of these insertions simultaneously, resulting in the construct Wzz2Δinsα6/7. When this construct was expressed in the O13 Δwzz1 Δwzz2 double mutant, it produced a significantly reduced, or “short,” chain length (Fig. 4B). This indicates that these two insertions are critical for mediating very long OSA chain lengths. We attempted a similar experiment in which we inserted insα6 and insα7 into Wzz1 to test if this would cause Wzz1 to confer longer chain lengths. We were unable to create a functional protein, which could be due to the low sequence identity of Wzz1 and Wzz2 (29%) (data not shown). Previous researchers have also been unsuccessful in constructing chimeric proteins from dissimilar Wzz proteins of the same strain (32), which highlights the difficulty of this particular experiment. We also tested the importance of delα1 in Wzz2 function. Since this region contains a 4-aa deletion between N70 and V71 compared to Wzz1, we prepared a construct with the corresponding amino acids from Wzz1 inserted into Wzz2 (Wzz2N70::LGRA). This construct did not exhibit any chain length-determining activity when expressed in the double-knockout mutant (data not shown). Comparison of the location of delα1 to the FepE structure suggests that this region may be involved in contact with either α6 or α8 and that insertion of amino acids at this site disrupts the protein’s fold (21). With this thought in mind, we made the construct Wzz2(N70::LGRAΔinsα6/7), but this protein also did not confer any chain length-regulating activity (data not shown). This suggests that other contacts may be disrupted, but these were difficult to predict from our sequence alignment and protein model alone, which hindered further investigation of this region.
Next we investigated the role of insL4, which extends L4 of Wzz2 by nine amino acids compared to Wzz1. We prepared a Wzz2 construct with a deletion of aa 271 to 279 (Wzz2Δ271-9) and expressed it in O13 Δwzz1 Δwzz2. The resulting OSA banding profile from LPS prepared from this recombinant bacterium was drastically shorter than that conferred by the wild-type Wzz2. Interestingly, Wzz2Δ271-9 regulated the OSA chains to a maximum length similar to those imparted by Wzz1, which does not contain this insertion (Fig. 4C). However, the most intense bands produced by the Wzz2Δ271-9 construct were only a few O units long, or “very short.” In contrast, a construct of Wzz2 with a full deletion of L4 appeared to exhibit almost negligible chain length-determining activity, as shown by some faint banding that was not present in the double mutant expressing the empty plasmid alone (Fig. 4C). These results are consistent with those reported by Kalynych et al. wherein a deletion of residues 256 to 273 in L4 of FepE caused a significant decrease in the LPS chain length regulated by that protein (32). In another study, expression of an engineered FepE that had residues 258 to 265 replaced with a short GSG flexible linker conferred an LPS banding pattern that was similar to the one displayed by the deletion construct (22). In those experiments, the length of the GSG linker did not match the number of amino acids that it was replacing. Based on the aforementioned observations, we hypothesized that the length of L4 could be important for determining chain length. Hence, we replaced aa 271 to 279 of Wzz2 with a flexible linker with an equal number of amino acids (sequence: GGGGSGGGG) (39). We also tested the importance of flexibility in this region by inserting a physically more rigid peptide at this site with the sequence AAEAAAKAA (39). Remarkably, when either construct was expressed in O13 Δwzz1 Δwzz2, the chain length was indistinguishable from that imparted by Wzz2Δ271-9 (Fig. 4C). This suggests that the specific amino acids in this region are important for determining chain length, rather than the flexibility and length of the loop alone.
Conserved residues in insL4 and insα6/α7 are critical for determining OSA chain length.
Since we determined that insIL, insL4, and insα6/α7 were important for Wzz2 function, we next targeted individual amino acids in these regions for site-directed mutagenesis to investigate the effects on Wzz2-mediated chain length regulation. Guided by the MSA of Wzz homologs, we searched for conserved charged or polar amino acids since these classes of amino acids were previously suggested to be involved in determining chain length (22). We found that insIL had very little conservation among the Wzz2-like group and were unable to identify any obvious candidate amino acids to target. Similarly, there was little conservation in insL4 except for an Arg-Thr-Glu motif at positions 281 to 283 (Fig. S4). We switched each of these amino acids to alanine to investigate their role in determining chain length. The T282A variant had no appreciable effect on the chain length-determining activity of Wzz2 when expressed in the O13 Δwzz2 mutant background. However, replacement of R281 or E283 with alanine caused a loss of very long OSA chains and the appearance of a new modality that ranged from very short to short and was centered on OSA that was only a few O units long (data not shown). This phenotype appeared very similar to that of the Wzz2Δ271-9 construct. To clearly compare the three mutations, we analyzed the OSA profiles conferred by each construct in the O13ΔΔwzz1 Δwzz2 background. This analysis revealed that Wzz2 with either mutation R281A or E283A conferred chain lengths that were identical to those conferred by Wzz2Δ271-9 (Fig. 4D).
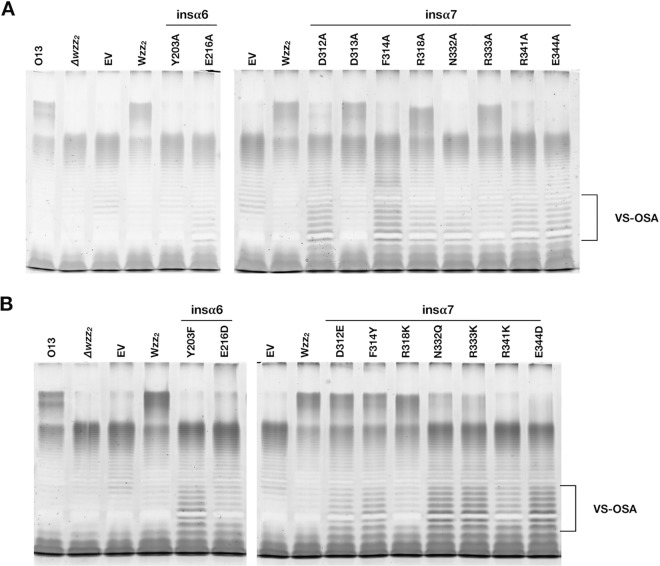
In insα6 and insα7 we identified several charged, polar, and aromatic amino acids that were highly conserved among the Wzz2-like group (Fig. S4). Interestingly, insα7 contained many more such conserved residues than did insα6 (9 amino acids versus 3 amino acids). We targeted these residues for alanine replacement and analyzed the resulting LPS profiles when expressed in the O13 Δwzz2 mutant. Ten of the 12 amino acids targeted in this manner affected the ability of Wzz2 to regulate the length of the very long OSA chains. Specifically, the following amino acid changes abolished chain length control by Wzz2 when expressed at basal levels from the pBAD promoter of pHERD20T: Y203A, E216A, D312A, F314A, N332A, R341A, and E344A (Fig. S5). Interestingly, two particular amino acid changes altered the Wzz2-mediated banding profile: the R318A and R333A variants both caused a decrease in the average chain length conferred by Wzz2. The R333A variant also caused a loss of the precise double banding pattern characteristic of the wild-type Wzz2 and instead resulted in a broad population of chain lengths. When the expression of these single-amino-acid variants was increased by the addition of 0.1% l-arabinose to the growth medium, the resulting banding profiles were similar to those for the cognate uninduced samples except that we detected small amounts of very long OSA in cultures expressing the D312A and F314A variants (Fig. 5A). Further, in these two site-specific mutants, we also observed an increase in the amount of very short OSA. The results are summarized in Table 1. Due to the conservation of these functionally important residues, we tested the capacity of like amino acids at these positions to restore Wzz2 function. The D312E and F314Y mutations were the only conservative amino acid switches that resulted in wild-type Wzz2-mediated chain lengths when expressed at basal levels, whereas the E216D, R318K, N332Q, R333K, R341K, and E344D mutations did not restore Wzz2 function (Fig. S5). The R318K variant had a chain length profile similar to that of the R318A mutant, wherein a broader and shorter population of very long chain lengths was produced. Surprisingly, the R333K variant was more detrimental to Wzz2 than the more drastic R333A mutation. When induced with 0.1% arabinose, some very long OSA was detected in cultures expressing Wzz2 N332Q and R333K variants (Fig. 5B). Once again, in the LPS preparations from the induced culture, we noticed that many Wzz2 variants had an increased production of very short OSA, namely, the Y203F, D312E, F314Y, N332Q, R333K, R341K, and E344D variants (Fig. 5B and Table 1). Intriguingly, this resulted in a novel phenotype for the D312E, F314Y, N332Q, and R333K variants since they clearly mediated the simultaneous production of both very short and very long OSA. Together, these results point to the importance of a specific amino acid chemistry at certain positions and identifies insα7 as a hot spot for amino acids critical to Wzz2 function. We have also described a novel Wzz2 phenotype: specific amino acid changes result in the production of two populations of OSA of vastly different lengths.
FIG 5.
Replacement of conserved residues in insα6/7 affects chain length regulation in Wzz2. (A) Effect of replacing conserved amino acids with alanine. All variants were expressed in the O13 ΔWzz2 background. The R318A and R333A variants alter the Wzz2-mediated banding pattern, while the Y203A, E216A, N332A, R341A, and E344A mutations abolish chain length control. The D312A and F314A variants produce increased amounts of very short OSA and some very long OSA. (B) Effect of substituting functionally important residues with like amino acids in Wzz2. The Y203F, R341K, and E344D variants produce very short OSA, while the D312E, F314Y, N332Q, and R333K variants simultaneously mediated the production of both very short (VS) and very long OSA.
TABLE 1.
Summary of LPS profiles conferred by Wzz2 insα6/7 point mutantsa
| Construct | Location of mutation | Production of: |
||
|---|---|---|---|---|
| Very short OSA | Very long OSA | Protein | ||
| Wzz2 | NA | − | + | + |
| Y203A variant | insα6 | − | − | − |
| E216A variant | insα6 | − | − | − |
| D312A variant | insα7 | + | +/− | +/− |
| F314A variant | insα7 | + | +/− | +/− |
| R318A variant | insα7 | − | + | + |
| N332A variant | insα7 | − | − | − |
| R333A variant | insα7 | − | + | + |
| R341A variant | insα7 | − | − | +/− |
| E344A variant | insα7 | − | − | − |
| D313A variant | insα7 | − | + | NA |
| Y203F variant | insα6 | + | − | NA |
| E216D variant | insα6 | − | − | NA |
| D312E variant | insα7 | + | + | NA |
| F314Y variant | insα7 | + | + | NA |
| R318K variant | insα7 | − | + | NA |
| N332Q variant | insα7 | + | +/− | NA |
| R333K variant | insα7 | + | +/− | NA |
| R341K variant | insα7 | + | − | NA |
| E344D variant | insα7 | + | − | NA |
+, appreciable production; +/− intermediate production; −, no appreciable production; NA, not applicable.
Wzz2 mutants are expressed at reduced levels but are able to form higher-order oligomers.
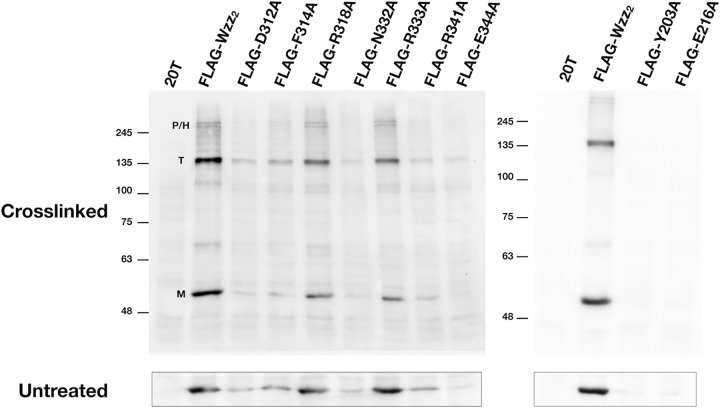
We hypothesized that some of the described mutations may affect the expression of Wzz2. To test this, we prepared N-terminally FLAG-tagged versions of these proteins to allow for their detection by Western immunoblotting. To ensure that the FLAG tag had no effects on the phenotypes conferred by the Wzz2 site-specific mutants, we reanalyzed the LPS profiles of O13 Δwzz2 expressing these FLAG-tagged variants. The chain lengths conferred by these constructs were indistinguishable from those conferred by the corresponding untagged mutants (Fig. S6). Immunoblots of whole-cell lysates revealed that Wzz2 harboring single mutations D312A, F314A, N332A, R341A, and E344A were expressed at levels much lower than that of the wild-type control, while proteins with single mutations R318A and R333A were expressed at levels similar to those of wild-type Wzz2 (Fig. 6). Negligible amounts of Wzz2 with mutations Y203A and E216A were detected. To test if any of these constructs were defective in the ability to oligomerize, we performed in vivo formaldehyde cross-linking. In samples expressing FLAG-wzz2, several bands appeared after cross-linking that were not present in the untreated or plasmid-only controls. The two most intense bands had apparent molecular masses of 53 kDa and 143 kDa, which corresponded closely with the expected size of the Wzz2 monomer and trimer (expected masses of 50 kDa and 150 kDa, respectively). Two less intense bands that migrated in the SDS-PAGE gel above the 245-kDa molecular mass standard had apparent molecular masses of 263 kDa and 281 kDa and likely correspond to pentamers and hexamers (expected masses of 250 kDa and 300 kDa, respectively). As expected, the cross-linked bands of constructs that expressed poorly were less intense than the wild-type protein. However, we were still able to detect banding profiles similar to those of FLAG-Wzz2 in the cross-linked samples of the mutant proteins (except for the Y203A and E216A mutants). In all samples we detected a band consistent with a Wzz2 trimer, but the bands corresponding to the pentamer and hexamer were detected only in the F314A, R318A, and R333A mutants. Overall, this suggests that these mutants were capable of forming higher-order oligomers.
FIG 6.
Stability of Wzz2 insα6/7 mutants determined by whole-cell protein expression and in vivo cross-linking. (Bottom) Whole-cell lysates of O13Δwzz2 expressing FLAG-tagged variants of Wzz2 were subjected to SDS-PAGE and immunoblotting with anti-FLAG antibody and detected using chemiluminescence. Several variants express at levels lower than for the wild-type control. (Top) Corresponding whole-cell cross-linked samples of FLAG-tagged Wzz2 variants. Bands were detected that approximately correspond to the monomeric (M), trimeric (T), and penta-/hexameric (P/H) forms of FLAG-Wzz2. Numbers on the left are molecular masses, in kilodaltons.
DISCUSSION
In this study, we have thoroughly characterized important regions of the OSA chain length regulator, Wzz2, from P. aeruginosa serotype O13 by constructing deletions, chimeras, and single amino acid changes guided by the alignment of a diverse set of Wzz sequences. We established that the differences in the chain lengths imparted by Wzz1 and Wzz2 are mostly due to the variations in their periplasmic domains. However, our results showed that chain length was partially dependent on the TM/cytosolic domain (Fig. 1B), which is in contrast to a previous study performed by Kalynych et al. (32). These authors recombinantly fused the periplasmic region of FepE of E. coli O157 to the TM/cytosolic domain of WzzB of the same strain, which resulted in the production of O antigen chains identical to those mediated by the wild-type FepE. This led them to conclude that the periplasmic region solely determines the chain length in this protein. It was recently suggested that the space between the TMDs of Wzz protomers could be a binding pocket for Wzy (25). We postulate that the variations in chain length we observed could be due to differential affinity of Wzy to the TMDs of Wzz1 and Wzz2, which affects the time until release of Wzy, i.e., the molecular stopwatch.
Our analysis of Wzz1 and Wzz2 homologs from the genus Pseudomonas has led to the observation that there are distinct sequence insertions in Wzz2 compared to Wzz1, specifically in the loop between β2 and β3, in L4, and in the two antiparallel α-helical regions. The results of our study indicate that the loop between β2 and β3 has only a minor role in chain length regulation and that Wzz can accommodate drastic changes to the amino acid sequence at this site. In fact, attempts to replace specific regions of Wzz1 with the cognate segments from Wzz2 to create a functional chimera were unsuccessful except for the constructs presented here, Wzz1::insIL and Wzz1::Wzz2-IL. Indeed, there is a lack of conservation in this loop, which supports the notion that the specific amino acid identities of this region are not entirely essential to Wzz1 or Wzz2 function. This is in accordance with the study of Papadopolous and Morona (30), in which the insertion of random 5-aa linkers into the β2-β3 loop of WzzBSF resulted in an increase in the chain length conferred by this protein, even though mutations in the α/β-base domain of Wzz proteins typically result in a knockout or abrogated chain length phenotype. In that study, the length of the β2-β3 loop was suggested to control the Wzz barrel size and thus partly determine chain length; i.e., a larger loop increases barrel size and consequently the chain length. We found that deletion of amino acids in this loop in Wzz2 did in fact cause a decrease in chain length, which would be consistent with their hypothesis. However, the reciprocal experiment of inserting amino acids into Wzz1 decreased rather than increased the chain length conferred by this protein. Importantly, the experimental designs of our study and the study of Papadopoulous and Morona are comparable; the insertion we made in Wzz1 was at the predicted end of the loop just before the start of β3, which is the same position as one of the insertions (i131) that was reported to increase the chain length imparted by WzzBSF (30). Notably, the sequence of our insertion (QAAP) and the sequence of i131 (LRPQP) both terminate in proline. We analyzed the solved crystal structures of WzzB and FepE to investigate if the β2-β3 loop made any significant inter- or intraprotomer contacts that might explain how mutations in this region could affect chain length, but this exercise did not reveal any obvious interactions. Based on the full-length cryo-electron microscopy (cryo-EM) structure of WzzBSF, Collins et al. hypothesized that Wzy and/or the polysaccharide could be contained within the barrel of Wzz (25). Therefore, we suggest that this loop is more likely to be involved with interaction with the polysaccharide chain or Wzy and that variations in this sequence alter the affinity for either of these molecules, rather than affecting the size of the Wzz barrel.
We were surprised to observe that the Wzz2 constructs with a deletion of aa 271 to 279 of L4 or replacement with either a flexible or rigid peptide sequence produced similar chain lengths that ranged from very short to long. Since experimental evidence indicates L4 is highly flexible (21–23, 32), we expected replacement with a sterically hindered peptide sequence (i.e., AAEAAAKAA) to have a greater effect on Wzz function than a more flexible one (i.e., GGGGSGGGG). One possibility is that we removed important amino acids from region 271 to 279 when constructing these replacements. However, we did not find any obvious residues to target for site-directed mutagenesis to explore this hypothesis. Another explanation is that manipulation of this area caused a displacement of a required orientation of the adjacent residues R281 and E283, which we showed had similar chain length phenotypes when individually mutated to alanine compared to the flexible or rigid amino acid insertions. To explore possible roles for these residues, we draw upon previous mutagenesis and structural studies. One report identified residue D268 in L4 of FepE to be important for chain length control since several amino acid changes at this position resulted in altered O antigen lengths (31). This residue also aligns with another residue, D226, in WzzBSF (31). Analysis of the solved crystal structures of FepE (PDB code 3B8M) and WzzBSF (PDB codes 4ZM1 and 4E2H) indicates that these aspartate residues are involved in putative salt bridge interactions with lysine residues in α7 (FepE) and α3 and α4 (WzzB). Interestingly, these interactions appear to be disrupted in the structure of a FepE L4 variant conferring shorter chain lengths (PDB code 4E2L) (see Fig. S7 in the supplemental material). Our analysis of these structures indicates that the charged residues of L4 have a role in mediating inter- and intraprotomer contacts with other charged residues in the helical regions of Wzz proteins. Therefore, we predict that this is the role of R281 and E283 in Wzz2. However, without an experimentally derived structure of Wzz2, these potential interactions are difficult to discern from sequence alone.
Wzz1 is comparable in size to FepE, which is the largest Wzz for which structural information is available. Since Wzz2 is larger than these two proteins, our results suggest how the structure of Wzz2 might differ from the previously characterized chain length regulators. The most notable difference is in the extra amino acids in α6 and α7. Hence, we hypothesized that the two large insertions identified in this study, insα6 and insα7, extend the Wzz2 protomer along the central axis. This may be one of the reasons that Wzz2 mediates the production of longer chain lengths than for Wzz1; the extension of the protomer creates an oligomer that can accommodate a longer polysaccharide chain. This is supported by the observation that simultaneous deletions of insα6 and insα7 resulted in a Wzz2 protein that conferred drastically shortened chain lengths. The fact that these two large regions could be deleted and a functional protein could still be obtained also suggests that the majority of the inter- and intraprotomer contacts made by insα6 and insα7 either are to each other or are nonspecific interactions. However, we cannot dismiss the idea that deletion of these sequences may have disrupted specific interactions, which could have also contributed to the shortened chain length phenotype.
Replacement of conserved amino acids in insα6 and insα7 revealed several residues that are absolutely essential to producing Wzz2-regulated chain lengths. We found that most of these variants of Wzz2 were expressed at lower levels than the wild-type protein, implying a role in conferring stability or the proper folding of Wzz2. Interestingly, several of the residues we replaced (R318, N332, R333, R341, E344, and E345) are found within a predicted coiled coil that spans aa 318 to 347. Notably, OSA chain length and Wzz2 protein stability were previously reported by Kintz and Goldberg to be dependent on the residue at position 321, which they identified to be within this predicted coiled coil (36). Similar to our findings, the authors described that various amino acid changes at this site decreased protein expression and/or altered the OSA chain length. However, the only change that abolished chain length control was a complete deletion of this position. This is in contrast to our study: we found that replacement of many residues in insα7 with alanine or even a similar amino acid abrogated Wzz2 function, suggesting that the amino acid identity at these positions is more critical. Consistent with this hypothesis, residues N332, R333, and E344 are found in positions e and g of the predicted coiled-coil heptad, which often form stabilizing interactions with the corresponding positions of another helix. Importantly, the e and g positions are typically occupied by charged or polar residues, which is consistent with the physiochemical properties of Asn, Arg, and Glu. In contrast, residue 321 occupies the c position, which is typically less essential in the formation of the coiled coil (40). Although the number of predicted coiled coils in Wzz proteins was reported to have a positive correlation with modal length (14), it should be noted that the Wzz structures solved to date do not show that these proteins form extensive coiled-coil interactions. However, when applying the algorithm of the Network Protein Server Analysis (NPS@) online platform to predict coiled-coil domains (41), Wzz2 has the highest score of those Wzz proteins with experimentally determined structures (data not shown). It would be useful to determine the structure of Wzz2 to examine the extent of coiled-coil interactions compared to other PCPs and how the residues from this study are positioned in the Wzz2 oligomer. We also found that residues of insα6 and insα7 that were outside of the predicted coiled-coil region were essential for Wzz2 function, namely, Y203, E216, D312, and F314. The decreased expression of alanine mutants at these positions suggests that they also have a role in maintaining the stability of Wzz2 but is likely not due to coiled-coil interactions.
Despite the reduced expression of Wzz2 variants with mutations in insα6 and insα7, we did not detect any differences in the cross-linked protein profiles of this protein from cultures expressing these mutants, which is consistent with experiments performed with other organisms (31, 32, 36, 42). We suggest that in vivo, the mutants described here are able to oligomerize but cannot maintain this state and are degraded faster than the wild-type Wzz2. However, during the cross-linking experiments, this transient oligomer is covalently captured. Our results also suggest that some mutants are able to maintain a biologically relevant oligomer in a complex with Wzy long enough to regulate chain length. Specifically, the Y203F, D312A, D312E, F314A, F314Y, N332Q, R333K, R341K, and E344D variants produced some very short OSA when overexpressed. We propose that these oligomers cannot withstand the force of the growing polysaccharide chain and disassemble before longer chains can be made. In the D312E, F314Y, N332Q, and R333K variants, we observed the distinct production of both very short and very long chain lengths, suggesting that in these cases, the oligomer might be stabilized once the polysaccharide reaches a certain length, thereby allowing polymerization to proceed.
Based on the data presented here and by others, we propose that the capacity of the Wzz “ruler” is a combination of the following: (i) Wzz oligomer size, (ii) oligomer stability, and (iii) nonspecific interactions between the polysaccharide and inside of the Wzz barrel. As polymerization begins, the polysaccharide would be sequestered within the barrel of Wzz and form nonspecific stabilizing interactions with amino acids of the β2-β3 loop and residues facing the inside of the Wzz cavity (30, 31). During polymer elongation, the pressure of containing the polysaccharide builds and increases the likelihood of Wzz oligomer disassembly (Fig. S8). In this model, Wzz proteins with larger cavities would accommodate longer polysaccharides, and those that form stronger inter- and intraprotomer contacts would be able to maintain the oligomeric state for more rounds of polymerization. These stabilizing contacts would be made throughout the Wzz protomer and include the interactions made by L4 and the coiled-coil regions. Future studies to determine the structure of Wzz2 should be pursued to test the extent of these interactions in this protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacteria were cultured at 37°C in LB. When necessary, cultures were supplemented with the appropriate antibiotics at the following concentrations: 100 μg/ml of ampicillin and 10 μg/ml of gentamicin for E. coli or 300 μg/ml of carbenicillin and 300 μg/ml of gentamicin for P. aeruginosa. P. aeruginosa IATS serotype O13 (ATCC 33360) was acquired from our lab collection.
General DNA methods and construction of wzz1 and wzz2 deletions.
PCR was performed using either Phusion Hot Start DNA polymerase or DreamTaq Green polymerase (Thermo). PCR and plasmid purification were performed using GeneJet purification kits (Thermo). Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs. All enzymes and kits were used according to the manufacturer’s instructions. The wzz1 and wzz2 deletion constructs were cloned into pEX18Gm by gene splicing by overlap extension (SOEing) (43). Briefly, two amplicons corresponding to the upstream and downstream regions of wzz1 or wzz2 were amplified in separate reactions. The PCR products were designed to contain a short region of homology to each other to facilitate their annealing in a second PCR, wherein the final construct was amplified. The PCR products were cloned into the EcoRI and XbaI sites of pEX18Gm. P. aeruginosa O13 was transformed with pHERD20T plasmids using the electroporation protocol described by King et al. (44). For the construction of point mutations, insertions, and deletions in wzz2-pHERD20T, either the QuikChange mutagenesis protocol (Agilent) or the in vitro assembly method was followed (45). All primers used in this study are listed in Table S1 and Table S2 in the supplemental material.
Bioinformatics analyses.
Homologs of Wzz1 and Wzz2 were identified using blastp (46). The search was limited to the Pseudomonas genus (excluding P. aeruginosa), and the maximum number of target sequences was set to 1,000. Only hits with a sequence identity to Wzz1 or Wzz2 greater than 29% were considered. The list of homologs was manually curated to remove partial sequences and those representing non-Wzz polysaccharide copolymerases. Sequences were aligned with Clustal Omega (47) and visualized with Jalview V2.10.0b1 (48). The final alignment was generated by removing redundant protein sequences (70% identity) based on the Jalview “remove redundancy” algorithm. All alignment images were generated using the ESPript 3.0 server (49).
Visualization of LPS.
LPS was prepared from overnight cultures according to the method of Hitchcock and Brown (50). Samples (5 μl) were electrophoresed on 10% SDS-PAGE gels and visualized by the rapid silver staining method of Fomsgaard et al. (51).
In vivo cross-linking.
In vivo formaldehyde cross-linking was performed as described previously (20), with minor modifications. Overnight cultures induced with 0.1% L-arabinose were washed twice in phosphate-buffered saline (PBS) and equilibrated to the equivalent of a 0.5-ml culture with an optical density at 600 nm (OD600) of 0.7. The bacteria were suspended in either 0.5 ml of PBS or 0.5 ml of PBS containing 0.5% formaldehyde and incubated at room temperature for 1 h. Following treatment, bacteria were pelleted and suspended in 50 μl of SDS-PAGE loading dye. For Western immunoblotting, samples (15 μl) were electrophoresed on 8% SDS-PAGE gels and transferred to nitrocellulose. The primary antibody used was monoclonal mouse anti-FLAG, and the secondary antibody was goat anti-mouse antibody conjugated to horseradish peroxidase (HRP). Blots were developed using the Illuminata HRP substrate (EMD Millipore). The molecular mass of the bands was predicted using the Quantity One one-dimensional (1D) analysis software from Bio-Rad.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by operating grants from CIHR awarded to C.M.K. and J.S.L. S.M.H. is the recipient of an Ontario Graduate Scholarship.
We thank Chris Whitfield for critical review of the work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00165-19.
REFERENCES
- 1.Hong M, Payne SM. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol 24:779–791. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 2.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, Blondel CJ, Zaldívar M, Valvano MA, Contreras I. 2008. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J Med Microbiol 57:938–946. doi: 10.1099/jmm.0.47848-0. [DOI] [PubMed] [Google Scholar]
- 4.Kintz E, Scarff JM, DiGiandomenico A, Goldberg JB. 2008. Lipopolysaccharide O-antigen chain length regulation in Pseudomonas aeruginosa serogroup O11 strain PA103. J Bacteriol 190:2709–2716. doi: 10.1128/JB.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cota I, Sánchez-Romero MA, Hernández SB, Pucciarelli MG, García-Del Portillo F, Casadesús J. 2015. Epigenetic control of Salmonella enterica O-antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genet 11:e1005667. doi: 10.1371/journal.pgen.1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May JF, Groisman EA. 2013. Conflicting roles for a cell surface modification in Salmonella. Mol Microbiol 88:970–983. doi: 10.1111/mmi.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Bäumler AJ. 2012. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog 8:e1002918. doi: 10.1371/journal.ppat.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov IE, Kintz EN, Porter LA, Goldberg JB, Burnham NA, Camesano TA. 2011. Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J Bacteriol 193:1259–1266. doi: 10.1128/JB.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegerle N, Bose J, Ramachandran G, Galen JE, Levine MM, Simon R, Tennant SM. 2018. Over-expression of O-polysaccharide chain length regulators in Gram-negative bacteria using the Wzx/Wzy-dependent pathway enhances production of defined modal length O polysaccharide polymers for use as haptens in glycoconjugate vaccines. J Appl Microbiol 125:575–585. doi: 10.1111/jam.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 12.Islam ST, Lam JS. 2014. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen IT, Beness AM, Saier MH. 1997. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 14.Morona R, Van Den Bosch L, Daniels C. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146:1–4. doi: 10.1099/00221287-146-1-1. [DOI] [PubMed] [Google Scholar]
- 15.Daniels C, Morona R. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol Microbiol 34:181–194. doi: 10.1046/j.1365-2958.1999.01591.x. [DOI] [PubMed] [Google Scholar]
- 16.Klee SR, Tzschaschel BD, Timmis KN, Guzmán CA. 1997. Influence of different rol gene products on the chain length of Shigella dysenteriae type 1 lipopolysaccharide O antigen expressed by Shigella flexneri carrier strains. J Bacteriol 179:2421–2425. doi: 10.1128/jb.179.7.2421-2425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrows LL, Chow D, Lam JS. 1997. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Rol). J Bacteriol 179:1482–1489. doi: 10.1128/jb.179.5.1482-1489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol 174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson G, Kessler A, Reeves P. 1995. A plasmid-borne O-antigen chain length determinant and its relationship to other chain length determinants. FEMS Microbiol Lett 125:23–30. doi: 10.1111/j.1574-6968.1995.tb07330.x. [DOI] [PubMed] [Google Scholar]
- 20.Daniels C, Griffiths C, Cowles B, Lam JS. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ Microbiol 4:883–897. doi: 10.1046/j.1462-2920.2002.00288.x. [DOI] [PubMed] [Google Scholar]
- 21.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Féthière J, Matte A, Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol 15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 22.Kalynych S, Yao D, Magee J, Cygler M. 2012. Structural characterization of closely related O-antigen lipopolysaccharide (LPS) chain length regulators. J Biol Chem 287:15696–15705. doi: 10.1074/jbc.M112.354837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalynych S, Cherney M, Bostina M, Rouiller I, Cygler M. 2015. Quaternary structure of WzzB and WzzE polysaccharide copolymerases. Protein Sci 24:58–69. doi: 10.1002/pro.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larue K, Kimber MS, Ford R, Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J Biol Chem 284:7395–7403. doi: 10.1074/jbc.M809068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins RF, Kargas V, Clarke BR, Siebert CA, Clare DK, Bond PJ, Whitfield C, Ford RC. 2017. Full-length, oligomeric structure of Wzz determined by cryoelectron microscopy reveals insights into membrane-bound states. Structure 25:806–815. doi: 10.1016/j.str.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 26.Nath P, Morona R. 2015. Detection of Wzy/Wzz interaction in Shigella flexneri. Microbiology 161:1797–1805. doi: 10.1099/mic.0.000132. [DOI] [PubMed] [Google Scholar]
- 27.Islam ST, Huszczynski SM, Nugent T, Gold AC, Lam JS. 2013. Conserved-residue mutations in Wzy affect O-antigen polymerization and Wzz-mediated chain-length regulation in Pseudomonas aeruginosa PAO1. Sci Rep 3:3441. doi: 10.1038/srep03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor VL, Udaskin ML, Islam ST, Lam JS. 2013. The D3 bacteriophage α-polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J Bacteriol 195:4735–4741. doi: 10.1128/JB.00903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J Bacteriol 188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadopoulos M, Morona R. 2010. Mutagenesis and chemical cross-linking suggest that Wzz dimer stability and oligomerization affect lipopolysaccharide O-antigen modal chain length control. J Bacteriol 192:3385–3393. doi: 10.1128/JB.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran ENH, Morona R. 2013. Residues located inside the Escherichia coli FepE protein oligomer are essential for lipopolysaccharide O-antigen modal chain length regulation. Microbiology 159:701–714. doi: 10.1099/mic.0.065631-0. [DOI] [PubMed] [Google Scholar]
- 32.Kalynych S, Ruan X, Valvano MA, Cygler M. 2011. Structure-guided investigation of lipopolysaccharide O-antigen chain length regulators reveals regions critical for modal length control. J Bacteriol 193:3710–3721. doi: 10.1128/JB.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 34.Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 1:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bélanger M, Burrows LL, Lam JS. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 36.Kintz EN, Goldberg JB. 2011. Site-directed mutagenesis reveals key residue for O antigen chain length regulation and protein stability in Pseudomonas aeruginosa Wzz2. J Biol Chem 286:44277–44284. doi: 10.1074/jbc.M111.273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam MYC, McGroarty EJ, Kropinski AM, MacDonald LA, Pedersen SS, Hoiby N, Lam JS. 1989. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol 27:962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Zaro JL, Shen WC. 2013. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason JM, Arndt KM. 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 41.Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem Sci 25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 42.Purins L, Van Den Bosch L, Richardson V, Morona R. 2008. Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 154:1104–1116. doi: 10.1099/mic.0.2007/014225-0. [DOI] [PubMed] [Google Scholar]
- 43.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 44.King JD, Mulrooney EF, Vinogradov E, Kneidinger B, Mead K, Lam JS. 2008. lfnA from Pseudomonas aeruginosa O12 and wbuX from Escherichia coli O145 encode membrane-associated proteins and are required for expression of 2,6-dideoxy-2-acetamidino-l-galactose in lipopolysaccharide O antigen. J Bacteriol 190:1671–1679. doi: 10.1128/JB.01708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.García-Nafría J, Watson JF, Greger IH. 2016. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci Rep 6:27459. doi: 10.1038/srep27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fomsgaard A, Freudenberg MA, Galanos C. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol 28:2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.