Abstract
The activation mechanism of chimeric antigen receptor (CAR)-engineered T cells may differ substantially from T cells carrying native T cell receptor, but this difference remains poorly understood. We present the first comprehensive portrait of single-cell level transcriptional and cytokine signatures of anti-CD19/4-1BB/CD28/CD3ζ CAR-T cells upon antigen-specific stimulation. Both CD4+ helper T (TH) cells and CD8+ cytotoxic CAR-T cells are equally effective in directly killing target tumor cells and their cytotoxic activity is associated with the elevation of a range of TH1 and TH2 signature cytokines, e.g., interferon γ, tumor necrotic factor α, interleukin 5 (IL5), and IL13, as confirmed by the expression of master transcription factor genes TBX21 and GATA3. However, rather than conforming to stringent TH1 or TH2 subtypes, single-cell analysis reveals that the predominant response is a highly mixed TH1/TH2 function in the same cell. The regulatory T cell activity, although observed in a small fraction of activated cells, emerges from this hybrid TH1/TH2 population. Granulocyte-macrophage colony stimulating factor (GM-CSF) is produced from the majority of cells regardless of the polarization states, further contrasting CAR-T to classic T cells. Surprisingly, the cytokine response is minimally associated with differentiation status, although all major differentiation subsets such as naïve, central memory, effector memory, and effector are detected. All these suggest that the activation of CAR-engineered T cells is a canonical process that leads to a highly mixed response combining both type 1 and type 2 cytokines together with GM-CSF, supporting the notion that polyfunctional CAR-T cells correlate with objective response of patients in clinical trials. This work provides new insights into the mechanism of CAR activation and implies the necessity for cellular function assays to characterize the quality of CAR-T infusion products and monitor therapeutic responses in patients.
Keywords: Single-cell transcriptomics, Single-cell proteomics, CAR-T, T cell activation
Introduction
Adoptive transfer of anti-CD19 chimeric antigen receptor (CAR) T cells has demonstrated remarkable efficacy in treating patients with B cell acute lymphoblastic leukemia (B-ALL), chronic lymphocytic leukemia (CLL), and other indolent lymphomas [1], [2], [3], [4]. Despite the demonstrated success, there exist large variation of responses and unpredictable toxicity in patients [5], [6], which could be attributed in part to inter-patient and intra-population heterogeneity of CAR-T infusion product. This makes it imperative to develop a high-resolution approach to characterize not only phenotypic composition but also the function of CAR-T cells at the systems level. The single-chain variable fragment (scFv) ectodomain of CAR binds to CD19 expressed on the surface of a target tumor cell and transmits signal via the transmembrane linker to the intracellular signaling domain such as CD3ζ to elicit T cell activation. This process is independent of the signaling from T cell receptor (TCR)-mediated binding to the peptide major histocompatibility complex (p-MHC) [7]. Incorporating a co-stimulator domain such as CD28 or 4-1BB (CD137) in the second generation CAR further enhanced proliferation, persistence, and potency [8]. Therefore, the mechanism of CAR-T activation could differ substantially from that of classic T cells. Nonetheless, this speculation remains inadequately tested as of today. The questions like how different CAR-T cell subsets, such as CD4+ helper T cells and CD8+ cytotoxic cells, respond to CAR stimulation, how the polarization subtypes, such as TH1, TH2, and regulatory T (Treg) cells, differentially control CAR-T cell responses, and how the differentiation status could affect the activation state, are all yet to be fully elucidated.
Current methods for evaluating CAR-T cell activation include the measurement of interferon (IFN)-γ secretion by ELISA or the detection of IFNγ-secreting cells by ELISpot [9], [10]. Multiparameter flow cytometry was used for immunophenotyping of CAR-T cells, which is one of the mainstay techniques used for monitoring CAR-T product manufacturing, but the number of markers and functions (cytokines) it can measure is limited [11], [12]. Intra-cellular cytokine staining for flow cytometric analysis is not a true secretion assay and often leads to over-estimation of cytokine-secreting cells. Recently, Xue et al used single-cell multiplex cytokine profiling to measure the cytokine output of anti-CD19/4-1BB/CD3ζ CAR-T cells using CD19-coated beads and revealed a diverse polyfunctional response upon activation [13]. However, the cytokine profile of a CAR-T cell is yet to be directly correlated to cytotoxicity, subtype, and signaling in order to elucidate the underlying mechanisms.
Herein, we use high-throughput single-cell 3′ mRNA transcriptome sequencing [14], [15], single-cell multiplex cytokine secretion assay [13], [16], [17], together with live cell imaging of cytotoxic activity to interrogate third-generation anti-CD19/4-1BB/CD28/CD3ζ (CD19-BB-28-3z) CAR-T cells, yielding the first comprehensive portrait of single-cell level transcriptional [18] and cytokine signatures of CAR-T cells upon antigen-specific stimulation. The predominant response is found to be a highly mixed TH1/TH2 function with Treg activity emerges from a small fraction of this hybrid TH1/TH2 population. The cytokine response is minimally associated with differentiation states such as naïve, central memory, effector memory, and effector cells. All the results suggest that the activation of CAR-engineered T cells is a canonical process associated with a highly mixed multi-functional response, supporting the notion that polyfunctional CAR-T cells correlate with objective response of non-Hodgkin’s lymphoma patients reported in a CD19 CAR-T clinical trial [19].
Results and discussions
Single-cell level measurement of transcriptional, cytokine, and cytotoxic function
We combined a set of single-cell techniques to interrogate CAR-T cells upon antigen specific stimulation (Figure 1A). This allowed us to quantitatively dissect the activation states related to subtypes, differentiation, and other factors such as intracellular signaling cascades, co-stimulators and immune checkpoints. CAR-T cells used in this study were manufactured through ex vivo transduction of autologous T cells with a CD19-BB-28-3z CAR construct, expansion for ∼10 days using CD3/CD28 Dynabeads, and then purification by bead removal and enrichment for CAR expression. T cells were isolated from three healthy donors and for the purpose of this study the human B cell lymphoma Raji cell line was used as a target. Single-cell 3′ mRNA transcriptome profiling was performed using a massively parallel cell barcoding method called scFTD-seq implemented in a bead-in-a-well microchip [14]. Single CAR-T cell cytolytic activity was measured by co-seeding CAR-T cells and target tumor cells in the microwell array to image the uptake of SYTOX Green nucleic acid dye [20] indicative of target cell lysis by CAR-T cells. Single-cell multiplex cytokine secretion assay was performed using a previously developed antibody barcode microchip assay [16], [17], [21], which was further modified in this study to co-measure cytotoxicity by seeding both CAR-T and tumor cells in the same single-cell protein secretion assay microchamber. Integrating all these single-cell analyses tools provides an unprecedented resolution to correlate the activation states such as anti-tumor cytotoxicity and cytokine response in single CAR-T cells to their phenotype, differentiation, signaling, and other characteristics.
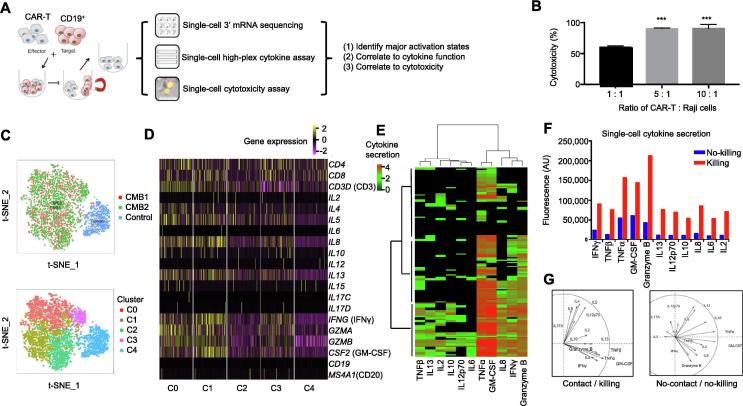
Figure 1.
Single-cell integrative analysis reveals varying degrees of CAR-T cell activation with direct correlation to cytotoxicity
A. Depiction of the workflow. Single-cell transcriptome, cytokine secretion, and live cell tracking of single-cell cytolytic activity analyses were used to characterize CAR-T cells. B. LDH assay confirms CAR-T cells used in this study are cytotoxic to the target CD19-expressing Raji cells (tumor cells). Percentage of cell death were measured by uptake of SYTOX Green, calculated as 60.02 ± 2.41% at 1:1, 90.1 ± 1.36% at 5:1, and 90.52 ± 6.51% at a 10:1 ratio of CAR-T:Raji cells. Data are shown as mean ± SD. One-way ANOVA was performed for statistical analysis and *** indicates significant difference in cell death between cell ratio of 1:1 with that of 5:1 and 10:1 (P < 0.01). C. t-SNE analysis of single-cell transcriptomes from a duplicate activation experiment (CMB1 and CMB2) vs. unstimulated control (control), showing highly consistent and efficient activation of >95% CAR-T cells and four major clusters/states (C0–C3) with varying degrees of activation compared to control (C4). D. Normalized gene expression of the main cytokines in single cells from major clusters (C0–C4). E. Single-cell multiplex cytokine secretion assay revealing three ties of activity, each of which is a different combination of cytokines shown by hierarchical clustering using Manhattan distance. Each row represents a microwell with a successful target cell killing event and each column corresponds to a cytokine of interest. Color intensity correlates with log scale of normalized fluorescence intensity of cytokine detected. F. Cytokine secretion intensity is elevated in microchambers where a CAR-T cell killed target tumor cells. The fluorescence intensity of detected cytokines was normalized against background as described previously [17], [21] and shown as arbitrary unit. G. PCA showing distinct grouping patterns of cytokines secreted between killing and no-killing cases. CAR, chimeric antigen receptor; t-SNE, t-distributed stochastic neighbor embedding; PCA, principal component analysis; IL, interleukin; TNF, tumor necrotic factor; IFN, interferon; GM-CSF, granulocyte–macrophage colony stimulating factor.
CAR activation leads to heterogeneous responses involving a range of cytokines, correlating directly to cytotoxicity
We first performed a population lactate dehydrogenase (LDH) release assay to detect the impact of CAR-T cells on the target Raji cells by co-culturing these cells for 6 h. As shown in Figure 1B, the CAR-T cells are highly cytotoxic (60% and 90% at the ratios of 1:1 and 5:1 for CAR-T:Raji cells, respectively). This observation is also confirmed by time-lapse imaging of SYTOX Green uptake (Figure S1A). The cytotoxicity was concurrently correlated with the secretion of a panel of cytokines, e.g., IFNγ, tumor necrotic factor α (TNFα), GM-CSF, interleukin 4 (IL4), IL5, IL8, and IL13, as detected with a protein microarray assay (Figure S1B).
We then conducted a single-cell massively parallel 3′ mRNA sequencing using unstimulated (control) and stimulated CAR-T cells (replicates: CMB1 and CMB2). The stimulated cells were prepared by co-culturing with CD19-expressing target cells for 6 h and then purified by magnetic cell sorting to remove target cells. The sequencing data were of high quality (Figures S2 and S3) and 3817 single CAR-T cell transcriptomes were obtained, which were visualized with t-distributed stochastic neighbor embedding (t-SNE) plots (upper panel, Figure 1C). We observed high consistency between replicates and highly efficient activation, with <0.5% of activated CAR-T cells (CMB1 in red and CMB2 in green) found in the control cluster (blue). Differential gene expression analysis (lower panel, Figure 1C and Figure S4) further identified four clusters C0–3 within the activated population that show varying degrees of cytokine responses (Figure 1D). Most cytokines measured are highly expressed in C0 and C1, moderately expressed in C2 and C3, as compared to the unstimulated control (C4). Single-cell 14-plex cytokine secretion assay confirms the heterogeneous activation with 3 clusters identified that secret most, nearly half, or very few cytokines, respectively (Figure 1E).
To correlate cytokine function directly to cytotoxicity, live cell imaging of SYTOX Green uptake was performed in the CAR-T:Raji cell pairs co-loaded in single-cell cytokine secretion microchambers over 12 h. The cytokine profiles are separated into no-killing and killing groups (Figure 1F and Figure S5). Rather than specific cytotoxic effector cytokines such as granzymes, the levels of a range of cytokines are elevated upon CAR activation and directly correlate to cytotoxic activity. Principal component analysis (PCA) revealed that upon CAR activation, the cytokines are grouped into two clusters, one dictated by TNFα, IFNγ, granzyme B, GM-CSF, and IL8, and the other dictated by IL4, IL5, IL6, and IL17 (left panel, Figure 1G). In contrast, the cytokine profile from single CAR-T cells that did not kill tumor cells showed no distinguishable grouping (right panel, Figure 1G). A diverse landscape of antigen-specific response was observed in CD19-BB-3z CAR-T cells [13]. Their dominant polyfunctional cells also produce granzyme B, GM-CSF, IL8, TNFα, IL13, and IFNγ, in concordance with our observation in this study using the third-generation CAR-T cells.
Both CD4+ and CD8+ CAR-T cells are highly cytotoxic and produce similar combination of cytokines at the proteomic and transcriptional level
In addition to the differentiated CD4+ subsets discussed above, the cytotoxic CD4+ T cells (CD4+ CTL), which can secrete granzyme B and perforin, have been observed as a transient state during viral infections or in antitumor and chronic inflammatory responses [22]. Although CD4+ CTL cells within the whole CD4+ T cell population are supposedly very rare, 66% CAR-T cells in our study can secrete granzyme B, regardless CD4+ or CD8+ subtypes. To examine whether the varying degrees of CAR-T cell activation is a consequence of varying CD4+ helper cell phenotype present in the CAR-T product, we conducted a SYTOX Green assay with target cells loaded in microfabricated wells (100 μm × 100 μm) together with CD4+ or CD8+ CAR-T cells, respectively (Figure 2A and Movies S1 and S2). CD4+ and CD8+ CAR-T cells are equally efficient in killing target cells [23], both requiring similar time and distance to find and lyse target tumor cells. Our single-cell cytokine secretion (proteome) and mRNA expression (transcriptome) data showed that the frequency and levels of cytokine expression were nearly indistinguishable between CD4+ and CD8+ subsets, except a slight increase in the level of GZMB in CD8+ cells and a slight upregulation in the expression of IL4, IL5, IL13, and IL10 in CD4+ cells (Figure 2B). Both subsets are highly polyfunctional, since more than 50% CAR-T cells co-secreted >5 cytokines, with a slightly higher polyfunctionality found in the CD4+ subset (Figure 2C). PCA further confirms a similar pattern of cytokine grouping in both subsets (Figure 2D). This is consistent with previous findings [13] and supports the hypothesis that CAR-T cell activation is largely independent of class I or class II p-MHC signaling. CAR-T cells with defined CD4:CD8 composition have been investigated in treating B-ALL patients and demonstrated high potency, allowing for delineating factors correlated with expansion, persistence, and toxicity of the CAR-T cells [24]. Our results confirm the importance of CD4+ cells in CAR-T product and the necessity to fully evaluate the role of these “helper” T cells in immunotherapy.
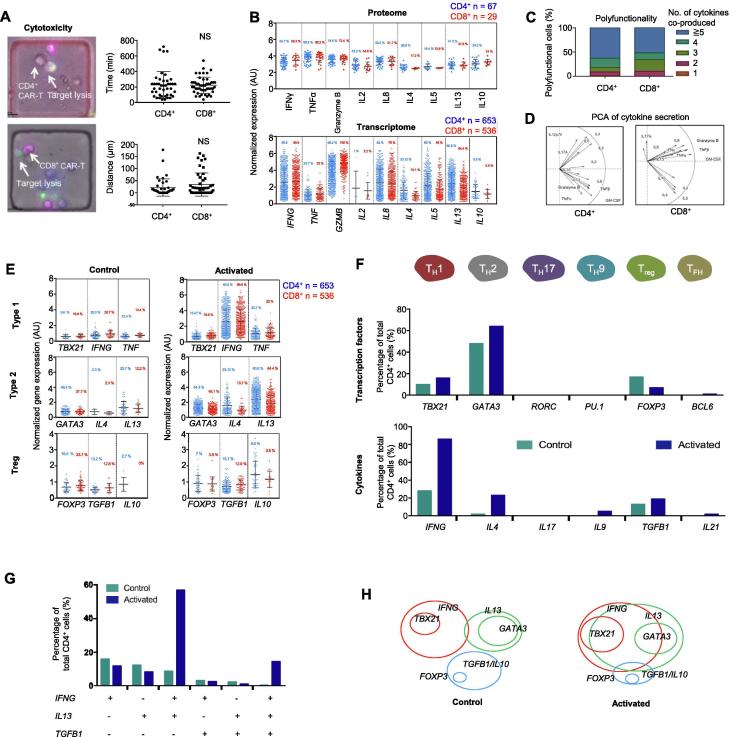
Figure 2.
Mapping single-cell data to phenotypes reveals a highly mixed TH1/TH2 cell response
A. Single-cell cytolytic activity assay revealing that both CD4+ and CD8+ CAR-T cells are cytotoxic (on the left) and show insignificant difference in anti-tumor reaction (on the right). B. Comparing cytokine protein secretion (proteome) and gene expression (transcriptome) in single CAR-T cells between CD4+ and CD8+ subsets. C. Polyfunctionality, a term that defines the ability of a single cell to co-secrete multiple cytokines, is slightly higher in CD4+ than in CD8+ CAR-T cells. D. PCA revealing similar clustering of major cytokines secreted between CD4+ and CD8+ cells. E. Expression of type 1, type 2, and Treg cell signature genes in single CAR-T cells upon activation. F. Quantification of CD4+ T cell subsets according to the expression of signature transcription factor and cytokine genes. G. Analysis of co-expression of TH1, TH2, and Treg cytokine genes in single CAR-T cells. H. Diagram showing phenotypic composition of CD4+ CAR-T cells and the alteration upon antigen-specific activation. NS, not significant; AU, arbitrary unit; TH, helper T cell; Treg, regulatory T cell; TFH, follicular helper T cell. t-test was performed for statistical analysis.
Mapping into subtypes reveals a predominantly mixed TH1/TH2 response in conjunction with Treg activity in the same CAR-T cells
To assess whether the observed heterogeneous activation depends on CAR-T cell polarization and subtypes, we examined the expression of master transcription factors (TFs) and signature cytokines. We found increased gene expression of type 1 and type 2 TFs, as well as the respective signature cytokine genes (Figure 2E). Expression of FOXP3, a master TF gene for Treg cells, was detected in unstimulated cells and the frequency of FOXP3+ cells decreased upon CAR-T cell activation, which was associated with the elevated expression of TGFB1 and IL10, the genes encoding regulatory cytokines TGFβ and IL10. All these alterations are minimally dependent on CD4 or CD8 subtypes, despite a slightly higher type 2 activity in CD4+ cells. Further mapping of all single CD4+ cell transcriptomes into TH1, TH2, TH17, TH9, Treg, and follicular helper T cell (TFH) subtypes showed that TH1 and TH2 subtypes were dominant and the Treg response was observed (Figure 2F). Although expression of TBX21 (Tbet) was detected in 15.5% of cells, IFNG (IFNγ) was expressed in 85.6% of cells, suggesting that the IFN response is also elicited or amplified through T-bet-independent pathways, for example, STAT1 signaling. GATA3 is expressed in 64.3% of cells, but the gene encoding predominant TH2 cytokine IL4 was expressed in 23.1% of these cells, with other TH2 cytokines (66% cells are IL5+ and 80.6% cells are IL13+) being more predominant. These data are indicative of a prevalent but slightly skewed TH2 response, consistent with the previous report that IL13 rather than IL4 is the dominant TH2 cytokine observed in CD19-BB-3z CAR-T cell activation [13]. Thus, for further analysis, IL13 rather than IL4 was chosen as the TH2 dominant cytokine gene.
To further answer if these are stringent polarization subtypes as in classic T cell biology, we quantified the frequency of dominant TH1 (IFNG), TH2 (IL13), and Treg (TGFB1) responses in the same single cells. It turned out that the ∼76% of CD4+ CAR-T cells upon activation showed a mixed TH1 (IFNG) and TH2 (IL13) response, and 75% of Treg (TGFB1) – like cells are triple positive for TH1 (IFNG) and TH2 (IL13) (Figure 2G). Therefore, CAR-T cell activation appears to differ substantially from classic T cells in that the predominant response is a highly mixed TH1/TH2 phenotype, within which a fraction of cells further exhibits regulatory characteristics, presumably to counteract over-activation (Figure 2H). Hegazy et al has reported that interferons can direct TH2 cell reprogramming to generate a hybrid subset with combined TH2 and TH1 cell functions [25]. This could be a mechanism attributable in our CAR-T cells, given that GATA3 is prevalently expressed and interferon response is the most profound, which together may confer TH1 function to GATA3+ CAR-T cells. Other mechanisms reported previously include stochastic cytokine expression leading to the formation of mixed helper T cell states [26]. Peine et al reported that stable TH1/TH2 hybrid cells arise in vivo from naïve precursors to limit immunopathologic inflammation [27]. This may explain in part the observation that CAR-T cells in general show a balanced response in anti-tumor effect and inflammatory toxicity, despite the unusually wide range of cytokines co-produced per cell.
GM-CSF is highly prevalent in activated CAR-T cells and co-produced with types 1 and 2 cytokines
GM-CSF is a potent pro-inflammatory cytokine involved in the recruitment, maturation, and activation of myeloid cells [28]. The helper T cells producing GM-CSF have been identified and recently reported to serve a nonredundant function in autoimmune pathogenesis, arguably representing a unique helper T cell subset [29]. The secretion of GM-CSF, a stimulator factor for monocytes and macrophages, may activate myeloid cells in vivo and amplifies systemic immunotoxicity [28]. Our data showed that >80% of activated CD4+ cells express CSF2, the gene encoding GM-CSF, which was confirmed independently by single-cell cytokine secretion (proteome) and single-cell mRNA sequencing (transcriptome) (Figure S6). We observed that 89% of TBX21+ cells and 83.5% of GATA3+ cells express CSF2. Previously, it was found that STAT5 programs a distinct subset of GM-CSF-producing Th cells in neuro-inflammation and autoimmunity [30]. In this study, 65.2% of CSF2-expressing CD4+ CAR-T cells were negative or low for STAT5 expression. Instead, we observed the expression of a wide range of STAT genes, but STAT1 is predominant, which may contribute to the profound TH1 interferon response despite a modest level of TBX21 (T-bet) expression. Our gene set enrichment analysis (GSEA) confirmed the relevance of STAT5 signaling and also revealed the importance of IL6/STAT3 (Figure S7), which has been observed as a determinant of CAR-T memory phenotype and therapeutic persistence in patients [31]. The JAK/STAT signaling relies on phosphorylation of STATs and their nuclear translocation. The mechanism leading to a prevalent GM-CSF-positive phenotype in activated CAR-T cells is yet to be further investigated.
Cytokine production in CAR-T cells upon antigen-specific activation is minimally dependent on differentiation status
To examine if the activation state of CAR-T cells is dependent on the differentiation status, we stratified the transcriptomes of single cells (both control and activated) into four subsets: naïve (TN), central memory (TCM), effector memory (TEM), and effector (TEFF) T cells, based on the expression of marker genes CD45RA and CCR7 [32], [33]. T cell subsets have been shown to persist following transduction with the CAR construct and antigen stimulation [34], [35]. The TN-like subset herein may also contain T stem-cell memory (TSCM) phenotype. We found that both CD4+ and CD8+ populations mainly comprised naïve-like and effector-like cells based on the expression of maker genes, but central memory and effector memory phenotypes were also observed (Figure 3A). CAR-T cell activation led to the increased frequencies of TCM-like and TEM-like cells and as a result a reduced frequency of TN-like cells. However, when we analyzed the correlations between the cell differentiation status and the expression of a range of stimulatory and cytotoxic cytokine genes that are involved in TH1, TH2, and Treg phenotypes in CD4+ cells, we found no or minimal dependence on differentiation status (Figure 3B). A closer look revealed a slight increase in TNF expression and a modest decrease in the expression of TH2 cytokine genes (e.g., IL4 and IL13) in the TCM-like subset (red bars in Figure 3B). CAR-T product containing central memory cells has been reported to be correlated with in vivo expansion, persistence, and potency [34]. Thus, the role of TCM-like subset in CAR-T cell activation requires further investigation.
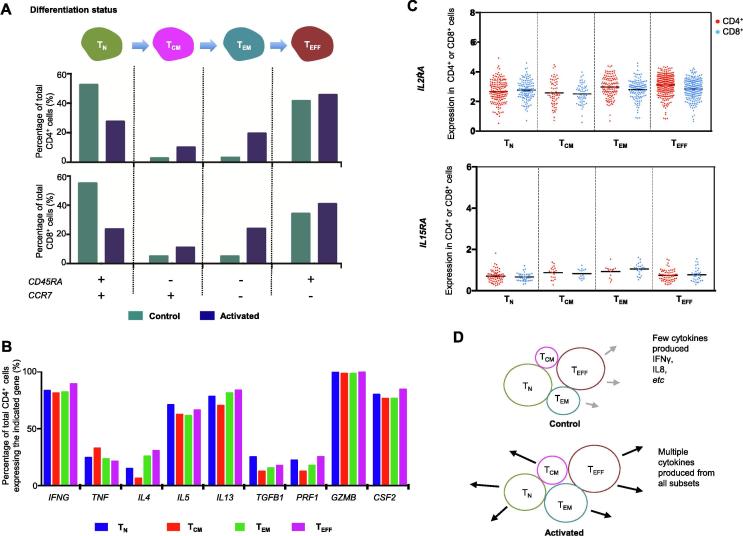
Figure 3.
Correlating differentiation status of CAR-T cells to cytokine functions reveals minimal dependence
A. Stratification of unstimulated and stimulated CAR-T cells into different differentiation status using a pair of gene markers CD45RA and CCR7. B. Correlating cytokine gene expression in CD4+ cells to differentiation status. C. Expression of T cell growth factor receptor genes IL2RA and IL15RA in different subsets of CAR-T cells upon activation. D. A model describing the activation of CAR-T cells to produce effector cytokines, largely independent of differentiation status. TN, naïve T cell; TCM, central memory T cell; TEM, effector memory T cell; TEFF, effector T cell.
In addition, we examined the expression pattern of IL2RA and IL15RA, the genes encoding T cell growth factor receptors IL2RA (CD25) and IL15RA, in the four aforementioned subsets of CD4+ and CD8+ CAR-T cells. There is no statistically significant difference found either (Figure 3C). Interestingly, IL2RA is also a marker gene for Treg cells. CD4+CD25+ T cells are considered as immunosuppressive and often used as a surrogate for Foxp3+ Treg cells [36]. In this study, nearly 100% of CAR-T cells upon activation were IL2RA-positive (Figure 3C) but apparently did not exert the suppressor function, highlighting the importance to characterize the function of CAR-T cells following activation in addition to surface marker phenotyping. A model to describe CAR-T activation as a function of differentiation states is shown in Figure 3D. It is worth noting that the differentiation status of CAR-T cells manufactured from healthy donors and patients with B cell malignancies could differ substantially and the patient samples can contain significantly higher percentage of central and effector memory cell subsets [24]. However, the fundamental mechanism revealed in this study regarding CAR-T cell activation as a function of differentiation status (e.g., TN, TCM, TEM, and TEFF) should still hold true.
CAR-T cell activation leads to elevated expression of CTLA4 that correlates with upregulation of IL10 in CD4+ and TGFB1 in CD8+ subsets
To investigate the possible relationship between CAR-T cell activation and the changes in the expression of co-stimulators or immune checkpoints, we examined their expression in single-cell transcriptomes. We observed that CTLA4 is the most upregulated genes among all immune checkpoint genes upon CAR-T cell activation (Figure 4A). PDCD1, the gene encoding checkpoint protein programmed cell death protein 1 (PD-1), demonstrates a similar trend. However, PDCD1 was lowly expressed and its expression was only detected in 9.8% CD4+ and 3.7% CD8+ subsets. Given PD-1 is also the primary marker of T cell exhaustion [37], our data suggest an infrequent incidence of acute stimulation-induced exhaustion following CAR-T cell action.
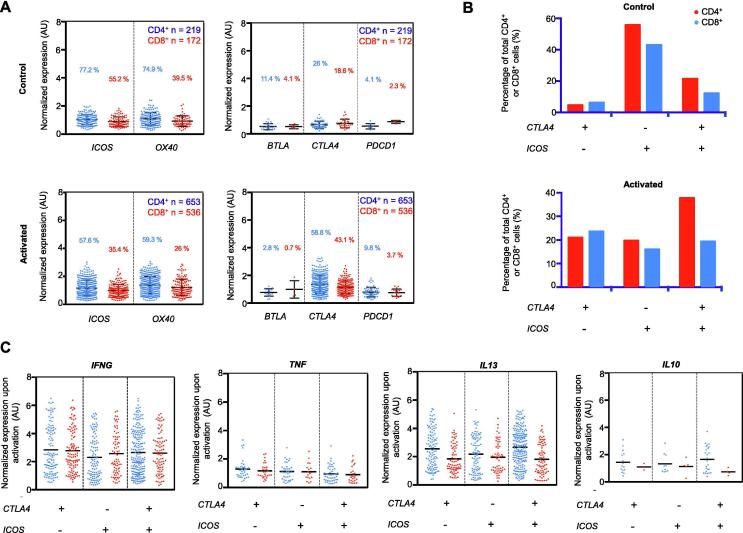
Figure 4.
Effect of activation state on the gene expression of co-inhibitors and co-stimulators
A. Gene expression profiles of common co-stimulators (ICOS and OX40) and co-inhibitors (BTLA, CTLA4, and PDCD1) in CD4+ and CD8+ subpopulations of activated and control CAR-T cells. In the CD4+ subset population, CTLA4 expression doubles from the control (26%) to the activated state (58.8%) while there is a slight decrease in the co-stimulatory ICOS expression from 77.2% in the control state to 57.6% in the activated state. B. Percentage of cells expressing three different combinations of CTLA4 and ICOS expression are plotted and compared. C. Gene expression of TH1 (IFNG and TNF), TH2 (IL13), and Treg (IL10) markers in the three cell populations shown in B.
Concurrently, there is a decreased frequency of ICOS and OX40 expression but slightly increased expression levels of these genes in a fraction of cells (Figure 4A). Correlation analysis revealed that unstimulated cells are predominantly ICOS+CTLA4−, whereas all three expression status combinations of these two genes were readily observed in the activated cells, and the ICOS+CTLA4+ subset was dominant in CD4+ CAR-T cells (Figure 4B). All three subsets of cells were capable of producing effector cytokines after activation (Figure 4C). Finally, the question is whether the significant upregulation of CTLA4 correlates or may contribute to the observed cytokine response heterogeneity. Comparing CTLA4-high (top 10%) cells to CTLA4-low (bottom 10%) cells for a range of cytokines (Figure S8), we observed significant increases in IL10 and TGFB1 expression in CD4+ subset, as well as TGFB1 expression in CD8+ subset, whereas expression of other cytokine genes was minimally or insignificantly correlated with the CTLA4 expression level. These findings suggest a possible mechanism for CAR-T cells to control immune homeostasis without developing distinct regulatory subtypes, which has important implications in combining checkpoint blockage and CAR-T targeted immunotherapies [38], [39], [40].
Conclusions
In this study, we employ high-throughput single-cell 3′ mRNA transcriptome sequencing, multiplexed single-cell cytokine secretion assay, together with live cell imaging of cytolytic activity, to interrogate third-generation anti-CD19 4-1BB/CD28/CD3ζ (CD19-BB-28-3z) CAR-T cells at the systems level [41] upon antigen-specific activation. CD4+ and CD8+ CAR-T cells are found to be equally effective in direct killing of target tumor cells and the cytotoxic activity is associated with the elevated co-production of a wide range of cytokines. Both TH1 and TH2 responses are prevalent, as confirmed by the expression of master TF genes TBX21(T-bet) and GATA3, as well as signature cytokine genes (proteins) including IFNG (IFNγ), TNF (TNFα), IL5 (IL5), and IL13 (IL13). Treg cell activity, although detected in a small fraction of CAR-T cells, is associated with elevated TGFB1 and IL10 expression. Unexpectedly, all these responses are often observed in the same CAR-T cells rather than distinct subsets, supporting the notion that polyfunctional CAR-T cells correlate with objective response of patients in clinical trials [14]. GM-CSF is produced from the majority of CAR-T cells regardless of the polarization state, further contrasting CAR-T cells to conventional T cells. We find that the cytokine response is minimally dependent on differentiation status although all major subsets such as naïve-like, central memory-like, effector memory-like, and effector-like cells are all detected.
Antigen-specific activation increases the level and frequency of cells expressing immune checkpoints such as CTLA4 and PDCD1, and slightly reduces the frequency of co-stimulator expression, which correlates with elevated expression of immunosuppressive cytokine genes like IL10 and TGFB1. In summary, the activation states of these CAR-T cells are highly mixed with TH1, TH2, Treg, and GM-CSF-expressing T cell responses in the same single cells and largely independent of differentiation status.
This study provides the first comprehensive portrait of CAR-T cell activation states at the single-cell level. Although CAR-T cell activation states may differ between patients, our work does reveal some common mechanisms regarding how different subtypes of CAR-T cells respond to antigen-specific challenge. It provides valuable information about the third-generation CD19-BB-28-3z CAR-T cells, which are being used in clinical trials to further improve therapeutic efficacy for non-responding patients. Our work sheds new insights on the biology of CAR-T cell activation and a rational path to develop single-cell approaches for CAR-T infusion product quality assurance and to monitor the changes of CAR-T cells in patients post infusion in clinical trials.
Methods
Cell culture and labeling
Human anti-CD19scFv-CD28-4-1BB-CD3ζ (Catalog No. PM-CAR-1002, Promab, Richmon, CA) T cells were cultured in complete X-Vivo 10 (Catalog No. 04-743Q, Lonza, Morristown, NJ) medium supplemented with IL2 (10 ng/ml, Catalog No. 589102, BioLegend, Dedham, MA) at a concentration of 5 × 105 cells/ml. To identify the CD4+ and CD8+ subsets, 1 × 105 CAR-T cells were pelleted and resuspended in 100 μl of PBS containing anti-human CD4 FITC (Catalog No. 130-113-213, Miltenyi Biotec, Somerville, MA) and anti-human CD8 Alexa Fluor 647 (Catalog No. 557708, BD Biosciences, Billerica, MA). The labeling reaction was incubated for 15 min at room temperature in the dark. The cells were then rinsed twice in PBS and once in complete RMPI medium (Catalog No. 11875085, Thermo Fisher, Waltham, MA) with 10% FBS (v/v; Catalog No. SER-500, Zenbio, Research Triangle, NC). Raji cells (Catalog No. CCL-86, ATCC, Manassas, VA) were labeled with Vybrant DiD (Catalog No. V22887, Thermo Fisher) following the manufacturer’s instructions. Briefly, the Raji cells were resuspended at a density of 1 × 106 cells/ml in serum-free RPMI containing 5 μl/ml of Vybrant DiD solution and incubated for 20 min at 37 °C. The labeled cells were then washed three times in complete RMPI medium and kept at 4 °C before use.
CAR-T cell stimulation with target tumor cells
CAR-T cells and Raji cells (5 × 104 cells/ml concentration) were incubated for 6 h in a round bottom well plate. Afterward, Raji cells were separated from co-culture by positive selection using a protocol of B cell selection with MagCellect (Catalog No. MAG997, R&D Systems, Minneapolis, MN). Briefly, CAR-T:Raji cell co-culture was centrifuged and the cell pellet was resuspended in serum-free RPMI medium and labeled with anti-CD19 biotinylated antibodies (Catalog No. 302204, BioLegend). Following manufacturer’s protocol, MagCellect Streptavidin Ferrofluid was added to the solution and the mixture was incubated for the recommended amount of time according to the product manual. The Streptavidin Ferrofluid beads bound to the anti-CD19 biotinylated Raji cells were collected on the side of the test tube by applying a magnet and then the CAR-T cells were pipetted out of the reaction. This procedure is repeated twice to make sure that most Raji cells were separated. The isolated CAR-T cells were then added to the microwell array device we fabricated for mRNA capture and transcriptome sequencing. Unstimulated CAR-T cells (5 × 104 cells/ml) were incubated for 6 h alone before subjected to single-cell RNA sequencing (scRNA-seq).
Bulk LDH cytotoxicity assessment
Cytotoxicity was assessed using the Pierce LDH Cytotoxicity Assay kit (Catalog No. 88953, Thermo Fisher). The effector cells and target cells were mixed at three ratios, i.e., 1:1, 5:1, and 10:1. Target and effector cells were incubated for 6 h in complete RPMI medium at a final concentration of 7 × 105 cells/ml. LDH release was measured in the supernatant according to manufacturer’s instructions. Maximum LDH release was obtained by incubating target cells in the provided 10× lysis buffer. Target cell cytotoxicity was calculated using the following formula: % of cytotoxicity = 100 × [(CAR-T:target cells − CAR-T cells alone − target cells alone)/(maximum target cell lysis − target cells alone without lysis buffer)].
Population SYTOX Green cytotoxicity assay
Effector CAR-T cells and the labeled target Raji cells were co-cultured in a 96 well U-bottom plate at three ratios, i.e., 1:1, 5:1, and 10:1. Raji cells at the concentration of 1 × 104 cells/ml were mixed with CAR-T cells at the concentrations of 1 × 104, 5 × 104, and 1 × 105 cells/ml to obtain the ratios mentioned above. To distinguish cell death, SYTOX Green (Catalog No. S34860, Thermo Fisher) was added to each well according to manufacturer’s instructions. Images were taken at 0, 4, 6, and 10 h of co-culture using the Nikon Eclipse Ti microscope.
Single CAR-T cell cytotoxicity assay in a microwell array
100 μm × 100 μm nanowell array was prepared by curing poly(dimethylsiloxane) (PDMS; Dow Corning Sylgard 184, Catalog No. 184 SIL ELAST KIT 3.9 KG, Ellsworth Adhesives, Germantown, WI) on a silicon wafer master with the etched array design. The nanowell array was cut to fit within the well of a 24-well plate and it was surface-treated with oxygen plasma prior to cell loading to decrease PDMS hydrophobicity [42]. Labeled Raji (target) and CAR-T cells (effector), as described above, were seeded at a density of 1 × 105 cells/ml. Stochastic distribution of the cells within the array allowed for a wide range of effector:target cell ratios. Images were acquired on a Nikon Eclipse Ti microscope fitted with an incubation chamber (37 °C and 5% CO2) using a 10×/0.3 objective. Automated multi-loci images were taken at 15 min intervals for a total duration of 10 h. Images were processed using the built-in Nikon software.
Fabrication of antibody barcode array slides and single-cell microchamber array chip
A panel of 14 anti-human antibodies is listed in Table S1 (IL2, IL4, IL5, IL6, IL8, IL10, IL12p70, IL13, IL17a, granzyme B, TNFα, TNFβ, IFNγ, and GM-CSF). FITC-BSA serving as the alignment control (Catalog No. A23015, Thermo Fisher), was patterned onto a poly-l-lysine glass slide using an air-pressure driven flow through a PDMS mold with 20 microfluidic channels. The PDMS mold was fabricated in-house as previously described [16], [17]. The subnanoliter microchamber array for cell capture contains 14 columns of 220 wells and was fabricated into a PDMS slab. Prior to cell loading, the microchamber PDMS slab was treated with oxygen plasma and then topped with RPMI medium.
Single-cell cytokine secretion profiling
Single-cell antibody barcode chips (SCBCs) for single-cell cytokine secretion profiling were developed in our laboratory as described previously [16], [17], [21], [43]. Following labeling, CAR-T and Raji cells were incubated at the ratio of 1:1 for 15 min in RPMI medium that was supplemented with SYTOX Green at a final concentration of 0.2 μM. The suspension was then gently pipetted onto the microchamber array. The SCBC device was assembled by overlaying the 14-plex antibody barcode glass slide and secured using our clamping system. The cell-loaded device was imaged (Nikon Eclipse Ti microscope) twice at 0 h and 14 h, respectively after incubation (37 °C and 5% CO2). Number of CD4+ CAR-T, CD8+ CAR-T, and Raji cells, as well as cell-to-cell contact and Raji cell death were recorded for each microchamber from the phase-contrast and fluorescent images. After the 14-h incubation, the antibody barcode chip was developed using an ELISA sandwich immunoassay as previously described [16]. The developed slides were scanned using the GenePix 4200A microarray scanner (Molecular Devices, Downingtown, PA).
Single-cell cytokine profiling data analysis
The fluorescent signals were processed using the GenePix software and a custom Excel macro was used to determine the average fluorescent signal for each secreted cytokine. The threshold gate (background) used for cytokine intensity normalization was calculated according to the formula: average of raw mean fluorescence intensity of a given cytokine for the 0-cell wells + 2× (standard deviation of raw mean fluorescence intensity of a given cytokine for the 0-cell wells) [44]. Secretors were defined as cells in the microchambers where the corresponding fluorescence cytokine signal intensity was higher than the threshold. We performed log transformation on the normalized cytokine intensity values (raw fluorescence intensity of a secretor for each cytokine – threshold gate for each cytokine) and applied single-cell analysis visualization tools to represent such large-scale high-dimensional data with PCA and viSNE [45]. All single-cell secretomic analysis was analyzed using Excel, R, and MATLAB.
Population-level cytokine secretion measurement of CAR-T:Raji cell pairs
CAR-T cells were co-incubated with target cells, both at the density of 1 × 105 cells/ml, in triplicates in a u-bottom 96-well plate with the effector:target cell ratio of 1:1. CAR-T cells alone and Raji cells alone were included in the same plate at the same concentration as the control. After a 10-h incubation, the cell suspension was pelleted at 200 g and the supernatant was removed and the population cytokine assay was performed as previously described [16], [17].
High-throughput scRNA-seq using a microwell array chip and data analysis
The massively parallel 3′ mRNA sequencing of single cells (called scFTD-seq) [14], [46] was based upon a closed microwell array system for co-isolation of single cells and DNA barcode beads. The chemistry was modified from the previous publications [47]. Briefly, the cell suspension prepared as described above was pipetted onto the inlet of the flow cell of our microchip, in which the entire bottom surface of the flow cell was covered by around 15,000 microwells (40 μm in diameter) to allow for trapping of single cells. The cells were withdrawn into the device using a negative pressure applied to the outlet. Once the microwell array was filled with the cell solution, the fluid flow was stopped and cells were allowed to settle by gravity into the wells. Excessive cells were washed out by PBS and mRNA capture beads were then loaded similarly to the cells. Excessive beads were also washed out by PBS and then lysis buffer [14], [46] was introduced into the device. Fluorinated oil (Fluorinert FC-40, Catalog No. 86508-42-1, Sigma–Aldrich, St. Louis, MO) was withdrawn into the device to seal the wells. Following lysis, the microfluidic device was incubated for 1 h inside a humidity chamber to allow mRNA capture onto the beads. Reverse transcription, library construction, and sequencing were performed as previously discussed [14], [46]. Sequencing libraries were constructed using Nextera XT according to the manufacturer’s protocol (Illumina). The finished libraries were sequenced on the HiSeq 2500 sequences (Illumina). The transcripts were then aligned to the reference human transcriptome hg19 using STAR v2.5.2b. The resulting gene expression matrices were analyzed using the Seurat package in R studio, custom built algorithms in R, MATLAB, as well as Excel and GraphPad Prism were used for plotting.
Statistical analysis
One-way ANOVA and t-tests were used to determine P values and difference with P < 0.05 is assumed to be statistically significant. Data were analyzed using GraphPad Prism V.7.0 and presented as mean ± STD.
Data availability
The single-cell RNA-seq data are available in the Gene Expression Omnibus (GEO) as GEO: GSE129007.
Authors’ contributions
RF conceived and designed the project; IX, BD, and GHL conducted experiments; DK and YX performed validation and support studies; IX, BD, GHL, and RF performed data analysis. IX and RF wrote manuscript with inputs from all other authors. All authors read and approved the final manuscript.
Acknowledgments
This research was supported by the Packard Fellowship for Science and Engineering, the CAREER award from the National Science Foundation (NSF), United States (Grant No. CBET-1351443), the grants from National Institutes of Health (NIH), United States (Grant No. U54 CA193461 and Sub-Project 7297 of Grant No. U54 CA209992), the Co-Pilot Grant from Yale Cancer Center, United States, to RF. Sequencing was performed at the Yale Center for Genome Analysis (YCGA) facility, United States. We used the Core facilities at the Yale Cooperative Center of Excellence in Hematology, supported by National Institutes of Health, United States (NIH U54DK106857). The molds for microfluidic devices were fabricated in the Yale School of Engineering and Applied Science cleanroom. We graciously thank Michael Power, Chris Tillinghast, and James Agresta for their help with fabrication process.
Handled by Yun-Gui Yang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.03.002.
Supplementary material
The following are the Supplementary data to this article:
Cytotoxic potential and cytokine secretion from CAR-T cells, Raji cells, and co-cultures A. Fluorescent imaging of cytolytic activity. The target Raji cells were prestained in red lipophilic dye. SYTOX Green dye was added in the co-culture and accumulated in Raji cells (yellow) as they were killed and lysed by the effector cells (CAR-T cells). B. Population-level assay of cytokine secretion using a pin-spotted antibody microarray, showing the cytokine secretion from CAR-T cells only (green), Raji cells only (red), and their co-culture (orange).
Quality assessment of scRNA-seq data from unstimulated and antigen-stimulated CAR-T cells A. Scatter violin plots showing total number of genes, transcripts (UMI), and mitochondrial genes detected in single cells of unstimulated CAR-T cells (control) and antigen-stimulated CAR-T cells (activated). Cells are filtered based on the percentage of mitochondrial genes (less than 10%) and gene counts to exclude cell multiplets and low quality cells. B. Rank ordered list of genes contributing to PC1 and PC2 after performing PCA of all single cell transcriptomes combined both activated (CMB) and control groups. C. Visualization of the expression of specific genes of interest in t-SNE plots. These genes include pan-T cell marker CD3D (encoding CD3), T cell checkpoint PDCD1 (encoding PD-1), housekeeping gene ACTB (encoding beta actin), as well as genes encoding effector proteins/cytokines GZMB (encoding granzyme B), PRF1 (encoding perforin), and IFNG (encoding IFNγ). Color (blue) intensity correlates with the level of gene expression. UMI, unique molecular identifier.
Computational identification of pure CAR-T cell data and removal of residual tumor cell data from scRNA-seq A. t-SNE plot of stimulated CAR-T cells following magnetic bead-based removal of target Raji cells. A subpopulation emerged (cluster 5, circled), distinct from the larger cell cluster. B. Expression of B cell markers (IGHM encoding Ig Mu and CD19) and T cell markers (CD3D and GZMB) to verify the identity of each cluster. Cluster 5 was identified as residual B cells with markedly increased expression of B cell markers and removed computationally before further analysis.
Heatmap of top-ranked DEGs between the unstimulated and antigen-stimulated CAR-T cells A range of activation-specific genes encoding cytokines and proteins appeared with various effector, stimulatory, and inflammatory functions such as PRF1 (encoding perforin), GZMB (encoding granzyme B), IL5, IL13, and CSF2 (encoding GM-CSF), as well as IL8 and IFNG (encoding IFNγ), as indicated by arrows. This analysis was performed in Seurat (https://satijalab.org/seurat/) using single-cell gene expression data.
Multiplexed measurement of cytokine secretion from single CAR-T cells and CAR-T:Raji cell interactions CAR-T cells and the target Raji cells were co-cultured and multiplexed measurement of cytokine secretion was performed with a microchamber array chip integrated with high-density antibody barcodes. A. Scanned microchamber images of CAR-T cells (no staining) and Raji cells (labeled with red fluorescent membrane dye) co-loaded to sub-nanoliter microchambers. B. Fluorescent signal detected corresponding to cytokine secretion in individual microchambers. C. Overlaid optical and fluorescent images. The optical image provides information on cell number, cell type, and cell integrity (tracked with SYTOX) that are correlated to fluorescent signals corresponding to cytokine secretion.
GM-CSF expression in activated CAR-T cells A. At the single-cell protein secretion level, 87% of CD4+ subset cells (blue) and 78.5% of CD8+ subset (red) secreted GM-CSF. Similarly, at the transcriptional expression level, 81% of CD4+ cells and 76.9% of CD8+ CAR-T cells expressed CSF2 (encoding GM-CSF). B. Gene expression level of CSF2 compared to that of the genes encoding signature cytokines including IFNG (IFNγ) in TH1, IL5 in TH2, as well as IL10 and TGFB1 (TGFβ) in Treg cells. C. Gene expression level of CSF2 compared to that of the genes encoding TFs, including TBX21 (T-bet) in TH1, GATA3 in Th3, and FOXP3 in Treg cells, as well as STAT5. Expression of CSF2 has a high correlation with that of cytokine genes in TH1 (71.7 % for IFNG) and TH2 (54.8% for IL5) cells, as well as TF genes in TH2 cells (53.8% for GATA3). D. Gene expression of STAT1, STAT3, STAT4, STAT5, and STAT6 genes was examined and the normalized expression values are plotted for CD4+ cells (blue) and CD8+ (red) CAR-T cells after activation. E. The percentage of CD4+ cells expressing different STAT genes alone and together with STAT5.
Gene set enrichment analysis of antigen-stimulated CAR-T cells vs. unstimulated CAR-T cells A ranked gene list is created based on genes differentially expressed between antigen-stimulated CAR-T cells and unstimulated CAR-T cells. A positive ES indicates that the expressed gene is at the top of the ranked gene list, whereas a negative ES indicates that the gene is at the bottom of the ranked gene list. A. Top ranked gene sets or pathways. B. Graphical view of enrichment scores for different gene sets. GS, gene set; ES, enrichment score; NES, normalized enrichment score; FWER, family-wise error rate.
Correlating CTLA4 gene expression to cytokine gene expression in single cells A. Top 10% and bottom 10% of CTLA4-expressing cells with the highest and lowest expression levels were classified as high expression and low expression groups and separated to evaluate the relationships between cytokine functions and CTLA4 expression. Normalized expression of TH1 (IFNG and TNF), TH2 (IL13 and IL5), and Treg (IL10 and TGFB1) signature cytokine genes in CD 4+ T cells with high (gray) and low (purple) CTLA4 expression. B. Normalized expression of TH1 (IFNG and TNF), TH2 (IL13 and IL5), and Treg (IL10 and TGFB1) signature cytokine genes in CD 8+ T cells with high (gray) and low (purple) CTLA4 expression. Elevated IL13 (TH2) and IL10 (Treg) expression was correlated with the increase in CTLA4 expression in CD4+ cells. Elevated TGFB1 (Treg) expression was correlated with the increase in CTLA4 expression in CD8+ cells. t-test was performed for statistical analyses. #P ≥ 0.1; *P < 0.1; **P < 0.05; ****P < 0.01.
List of antibodies used for cytokine assays
Live cell tracking of single CAR-T cell killing a target tumor cell using the SYTOX Green uptake assay
Live cell tracking of multiple CAR-T cells attacking a target tumor cell using the SYTOX Green uptake assay
References
- 1.Sadelain M. CAT therapy: the CD19 paradigm. J Clin Invest. 2015;125:3392–3400. doi: 10.1172/JCI80010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sadelain M. CAT therapy: the CD19 paradigm. J Clin Invest 2015;125:3392–400. [DOI] [PMC free article] [PubMed]
- 2.Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. New Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]; Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. New Engl J Med 2011;365:725–33. [DOI] [PMC free article] [PubMed]
- 3.Park J.H., Riviere I., Gonen M., Wang X.Y., Senechal B., Curran K.J. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. New Engl J Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park JH, Riviere I, Gonen M, Wang XY, Senechal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. New Engl J Med 2018;378:449–59. [DOI] [PMC free article] [PubMed]
- 4.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. New Engl J Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]; Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. New Engl J Med 2017;377:2531–44. [DOI] [PMC free article] [PubMed]
- 5.June C.H., Warshauer J.T., Bluestone J.A. Is autoimmunity the Achilles' heel of cancer immunotherapy? Nat Med. 2017;23:540–547. doi: 10.1038/nm.4321. [DOI] [PubMed] [Google Scholar]; June CH, Warshauer JT, Bluestone JA. Is autoimmunity the Achilles' heel of cancer immunotherapy? Nat Med 2017;23:540–7. [DOI] [PubMed]
- 6.Davila M.L., Riviere I., Wang X.Y., Bartido S., Park J., Curran K. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]; Davila ML, Riviere I, Wang XY, Bartido S, Park J, Curran K, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 2014;6:224ra25. [DOI] [PMC free article] [PubMed]
- 7.Sadelain M., RiviSre I., Riddell S. Therapeutic T cell engineering. Nature. 2017;545:423–431. doi: 10.1038/nature22395. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sadelain M, RiviSre I, Riddell S. Therapeutic T cell engineering. Nature 2017;545:423–31. [DOI] [PMC free article] [PubMed]
- 8.Sadelain M. CD19 CAR T cells. Cell. 2017;171:1471. doi: 10.1016/j.cell.2017.12.002. [DOI] [PubMed] [Google Scholar]; Sadelain M. CD19 CAR T cells. Cell 2017;171:1471. [DOI] [PubMed]
- 9.Kalyuzhny A.E. Chemistry and biology of the ELISPOT assay. Methods Mol Biol. 2005;302:15–31. doi: 10.1385/1-59259-903-6:015. [DOI] [PubMed] [Google Scholar]; Kalyuzhny AE. Chemistry and biology of the ELISPOT assay. Methods Mol Biol 2005;302:15–31. [DOI] [PubMed]
- 10.Wilkie S., Picco G., Foster J., Davies D.M., Julien S., Cooper L. Retargeting of human T cells to tumor-associated MUC1: the evolution of a chimeric antigen receptor. J Immunol. 2008;180:4901–4909. doi: 10.4049/jimmunol.180.7.4901. [DOI] [PubMed] [Google Scholar]; Wilkie S, Picco G, Foster J, Davies DM, Julien S, Cooper L, et al. Retargeting of human T cells to tumor-associated MUC1: the evolution of a chimeric antigen receptor. J Immunol 2008;180:4901–9. [DOI] [PubMed]
- 11.Jena B., Moyes J.S., Huls H., Cooper L.J. Driving CAR-based T-cell therapy to success. Curr Hematol Malig Rep. 2014;9:50–56. doi: 10.1007/s11899-013-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jena B, Moyes JS, Huls H, Cooper LJ. Driving CAR-based T-cell therapy to success. Curr Hematol Malig Rep 2014;9:50–6. [DOI] [PMC free article] [PubMed]
- 12.Kaiser A.D., Assenmacher M., Schroder B., Meyer M., Orentas R., Bethke U. Towards a commercial process for the manufacture of genetically modified T cells for therapy. Cancer Gene Ther. 2015;22:72–78. doi: 10.1038/cgt.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kaiser AD, Assenmacher M, Schroder B, Meyer M, Orentas R, Bethke U, et al. Towards a commercial process for the manufacture of genetically modified T cells for therapy. Cancer Gene Ther 2015;22:72–8. [DOI] [PMC free article] [PubMed]
- 13.Xue Q., Bettini E., Paczkowski P., Ng C., Kaiser A., McConnell T. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J Immunother Cancer. 2017;5:85. doi: 10.1186/s40425-017-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xue Q, Bettini E, Paczkowski P, Ng C, Kaiser A, McConnell T, et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J Immunother Cancer 2017;5:85. [DOI] [PMC free article] [PubMed]
- 14.Dura B., Choi J.Y., Zhang K., Damsky W., Thakral D., Bosenberg M. scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3′ mRNA profiling. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1173. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dura B, Choi JY, Zhang K, Damsky W, Thakral D, Bosenberg M, et al. scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3' mRNA profiling. Nucleic Acids Res 2019;47:e16. [DOI] [PMC free article] [PubMed]
- 15.Deng Y., Finck A., Fan R. Single-cell omics analyses enabled by microchip technologies. Annu Rev Biomed Eng. 2019;21:365–393. doi: 10.1146/annurev-bioeng-060418-052538. [DOI] [PubMed] [Google Scholar]; Deng Y, Finck A, Fan R. Single-cell omics analyses enabled by microchip technologies. Annu Rev Biomed Eng 2019;21:365–93. [DOI] [PubMed]
- 16.Lu Y., Chen J.J., Mu L.Y., Xue Q., Wu Y., Wu P.H. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;85:2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu Y, Chen JJ, Mu LY, Xue Q, Wu Y, Wu PH, et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem 2013;85:2548–56. [DOI] [PMC free article] [PubMed]
- 17.Lu Y., Xue Q., Eisele M.R., Sulistijo E.S., Brower K., Han L. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci U S A. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci U S A 2015;112:E607–15. [DOI] [PMC free article] [PubMed]
- 18.Yu P.J., Lin W. Single-cell transcriptome study as big data. Genomics Proteomics Bioinformatics. 2016;14:21–30. doi: 10.1016/j.gpb.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu PJ, Lin W. Single-cell transcriptome study as big data. Genomics Proteomics Bioinformatics 2016;14:21–30. [DOI] [PMC free article] [PubMed]
- 19.Rossi J., Paczkowski P., Shen Y.W., Morse K., Flynn B., Kaiser A. Polyfunctional anti-CD19 CAR T cells determined by single-cell multiplex proteomics associated with clinical activity in patients with advanced non-Hodgkin’s lymphoma. Cancer Res. 2017;77:nr2990. [Google Scholar]; Rossi J, Paczkowski P, Shen YW, Morse K, Flynn B, Kaiser A, et al. Polyfunctional anti-CD19 CAR T cells determined by single-cell multiplex proteomics associated with clinical activity in patients with advanced non-Hodgkin’s lymphoma. Cancer Res 2017;77:nr2990.
- 20.Jones L.J., Singer V.L. Fluorescence microplate-based assay for tumor necrosis factor activity using SYTOX Green stain. Anal Biochem. 2001;293:8–15. doi: 10.1006/abio.2001.5116. [DOI] [PubMed] [Google Scholar]; Jones LJ, Singer VL. Fluorescence microplate-based assay for tumor necrosis factor activity using SYTOX Green stain. Anal Biochem 2001;293:8–15. [DOI] [PubMed]
- 21.Xue Q., Lu Y., Eisele M.R., Sulistijo E.S., Khan N., Fan R. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci Signal. 2015;8:ra59. doi: 10.1126/scisignal.aaa2155. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xue Q, Lu Y, Eisele MR, Sulistijo ES, Khan N, Fan R, et al. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci Signal 2015;8:ra59. [DOI] [PMC free article] [PubMed]
- 22.Takeuchi A., Saito T. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front Immunol. 2017;8:194. doi: 10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front Immunol 2017;8:194. [DOI] [PMC free article] [PubMed]
- 23.Varadarajan N., Liadi I., Romain G., Singh H., Cooper L.J.N. Quantitative single-cell functional characterization of CD19-specific CAR+ T cells for immunotherapy. Cancer Res. 2013;73:nr4745. [Google Scholar]; Varadarajan N, Liadi I, Romain G, Singh H, Cooper LJN. Quantitative single-cell functional characterization of CD19-specific CAR+ T cells for immunotherapy. Cancer Res 2013;73:nr4745.
- 24.Turtle C.J., Hanafi L.A., Berger C., Gooley T.A., Cherian S., Hudecek M. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]; Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 2016;126:2123–38. [DOI] [PMC free article] [PubMed]
- 25.Hegazy A.N., Peine M., Helmstetter C., Panse I., Frohlich A., Bergthaler A. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]; Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 2010;32:116–28. [DOI] [PubMed]
- 26.Fang M.Q., Xie H.M., Dougan S.K., Ploegh H., van Oudenaarden A. Stochastic cytokine expression induces mixed T helper cell states. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001618. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fang MQ, Xie HM, Dougan SK, Ploegh H, van Oudenaarden A. Stochastic cytokine expression induces mixed T helper cell states. PLoS Biol 2013;11:e1001618. [DOI] [PMC free article] [PubMed]
- 27.Peine M., Rausch S., Helmstetter C., Frohlich A., Hegazy A.N., Kuhl A.A. Stable T-bet+GATA-3+ Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peine M, Rausch S, Helmstetter C, Frohlich A, Hegazy AN, Kuhl AA, et al. Stable T-bet+GATA-3+ Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol 2013;11:e1001633. [DOI] [PMC free article] [PubMed]
- 28.Becher B., Tugues S., Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity. 2016;45:963–973. doi: 10.1016/j.immuni.2016.10.026. [DOI] [PubMed] [Google Scholar]; Becher B, Tugues S, Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity 2016;45:963–73. [DOI] [PubMed]
- 29.Herndler-Brandstetter D., Flavell R.A. Producing GM-CSF: a unique T helper subset? Cell Res. 2014;24:1379–1380. doi: 10.1038/cr.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]; Herndler-Brandstetter D, Flavell RA. Producing GM-CSF: a unique T helper subset? Cell Res 2014;24:1379–80. [DOI] [PMC free article] [PubMed]
- 30.Sheng W., Yang F., Zhou Y., Yang H., Low P.Y., Kemeny D.M. STAT5 programs a distinct subset of GM-CSF-producing T helper cells that is essential for autoimmune neuroinflammation. Cell Res. 2014;24:1387–1402. doi: 10.1038/cr.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sheng W, Yang F, Zhou Y, Yang H, Low PY, Kemeny DM. STAT5 programs a distinct subset of GM-CSF-producing T helper cells that is essential for autoimmune neuroinflammation. Cell Res 2014;24:1387–402. [DOI] [PMC free article] [PubMed]
- 31.Fraietta J.A., Lacey S.F., Orlando E.J., Pruteanu-Malinici I., Gohil M., Lundh S. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24:563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med 2018;24:563–71. [DOI] [PMC free article] [PubMed]
- 32.Riddell S.R., Sommermeyer D., Berger C., Liu L., Balakrishnan A., Salter A. Adoptive therapy with chimeric antigen receptor–modified T cells of defined subset composition. Cancer J. 2014;20:141–144. doi: 10.1097/PPO.0000000000000036. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riddell SR, Sommermeyer D, Berger C, Liu L, Balakrishnan A, Salter A, et al. Adoptive therapy with chimeric antigen receptor–modified T cells of defined subset composition. Cancer J 2014;20:141–44. [DOI] [PMC free article] [PubMed]
- 33.Golubovskaya V., Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel) 2016;8:E36. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]; Golubovskaya V, Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel) 2016;8:E36. [DOI] [PMC free article] [PubMed]
- 34.Wang X.L., Popplewell L.L., Wagner J.R., Naranjo A., Blanchard M.S., Mott M.R. Phase 1 studies of central memory-derived CD19 CAR T-cell therapy following autologous HSCT in patients with B-cell NHL. Blood. 2016;127:2980–2990. doi: 10.1182/blood-2015-12-686725. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang XL, Popplewell LL, Wagner JR, Naranjo A, Blanchard MS, Mott MR, et al. Phase 1 studies of central memory-derived CD19 CAR T-cell therapy following autologous HSCT in patients with B-cell NHL. Blood 2016;127:2980–90. [DOI] [PMC free article] [PubMed]
- 35.Schmueck-Henneresse M., Omer B., Shum T., Tashiro H., Mamonkin M., Lapteva N. Comprehensive approach for identifying the T cell subset origin of CD3 and CD28 antibody–activated chimeric antigen receptor-modified T cells. J Immunol. 2017;199:348–362. doi: 10.4049/jimmunol.1601494. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schmueck-Henneresse M, Omer B, Shum T, Tashiro H, Mamonkin M, Lapteva N, et al. Comprehensive approach for identifying the T cell subset origin of CD3 and CD28 antibody–activated chimeric antigen receptor–modified T cells. J Immunol 2017;199:348–62. [DOI] [PMC free article] [PubMed]
- 36.Cohen J.L., Trenado A., Vasey D., Klatzmann D., Salomon B.L. CD4+CD25+ immunoregulatory T cells: new therapeutics for graft-versus-host disease. J Exp Med. 2002;196:401–406. doi: 10.1084/jem.20020090. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cohen JL, Trenado A, Vasey D, Klatzmann D, Salomon BL. CD4+CD25+ immunoregulatory T cells: new therapeutics for graft-versus-host disease. J Exp Med 2002;196:401–6. [DOI] [PMC free article] [PubMed]
- 37.Kato T., Nishida T., Murase M., Murata M., Naoe T. Exhaustion of CMV specific T cells with enhanced PD-1 expression in persistent cytomegalovirus infection after allogeneic stem cell transplantation. Blood. 2010;116:3912. [Google Scholar]; Kato T, Nishida T, Murase M, Murata M, Naoe T. Exhaustion of CMV specific T cells with enhanced PD-1 expression in persistent cytomegalovirus infection after allogeneic stem cell transplantation. Blood 2010;116:3912.
- 38.Cherkassky L., Morello A., Villena-Vargas J., Feng Y., Dimitrov D.S., Jones D.R. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J Clin Invest. 2016;126:3130–3144. doi: 10.1172/JCI83092. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cherkassky L, Morello A, Villena-Vargas J, Feng Y, Dimitrov DS, Jones DR, et al. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J Clin Invest 2016;126:3130–44. [DOI] [PMC free article] [PubMed]
- 39.Fedorov V.D., Themeli M., Sadelain M. PD-1-and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci Transl Med. 2013;5:215ra172. doi: 10.1126/scitranslmed.3006597. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fedorov VD, Themeli M, Sadelain M. PD-1-and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci Transl Med 2013;5:215ra172. [DOI] [PMC free article] [PubMed]
- 40.Chong E.A., Melenhorst J.J., Lacey S.F., Ambrose D.E., Gonzalez V., Levine B.L. PD-1 blockade modulates chimeric antigen receptor (CAR)-modified T cells: refueling the CAR. Blood. 2017;129:1039–1041. doi: 10.1182/blood-2016-09-738245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chong EA, Melenhorst JJ, Lacey SF, Ambrose DE, Gonzalez V, Levine BL. PD-1 blockade modulates chimeric antigen receptor (CAR)-modified T cells: refueling the CAR. Blood 2017;129:1039–41. [DOI] [PMC free article] [PubMed]
- 41.Hood L., Tian Q. Systems approaches to biology and disease enable translational systems medicine. Genomics Proteomics Bioinformatics. 2012;10:181–185. doi: 10.1016/j.gpb.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hood L, Tian Q. Systems approaches to biology and disease enable translational systems medicine. Genomics Proteomics Bioinformatics 2012;10:181–5. [DOI] [PMC free article] [PubMed]
- 42.Eddington D.T., Puccinelli J.P., Beebe D.J. Thermal aging and reduced hydrophobic recovery of polydimethylsiloxane. Sens Actuators B Chem. 2006;114:170–172. [Google Scholar]; Eddington DT, Puccinelli JP, Beebe DJ. Thermal aging and reduced hydrophobic recovery of polydimethylsiloxane. Sens Actuators B Chem 2006;114:170–2.
- 43.Rossi J., Paczkowski P., Shen Y.W., Morse K., Flynn B., Kaiser A. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood. 2018;132:804–814. doi: 10.1182/blood-2018-01-828343. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rossi J, Paczkowski P, Shen YW, Morse K, Flynn B, Kaiser A, et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 2018;132:804–14. [DOI] [PMC free article] [PubMed]
- 44.Kleppe M., Kwak M., Koppikar P., Riester M., Keller M., Bastian L. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov. 2015;5:316–331. doi: 10.1158/2159-8290.CD-14-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov 2015;5:316–31. [DOI] [PMC free article] [PubMed]
- 45.Amir E.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]; Amir ED, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–52. [DOI] [PMC free article] [PubMed]
- 46.Wang N., Zheng J., Chen Z., Liu Y., Dura B., Kwak M. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat Commun. 2019;10:95. doi: 10.1038/s41467-018-07981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat Commun 2019;10:95. [DOI] [PMC free article] [PubMed]
- 47.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–14. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cytotoxic potential and cytokine secretion from CAR-T cells, Raji cells, and co-cultures A. Fluorescent imaging of cytolytic activity. The target Raji cells were prestained in red lipophilic dye. SYTOX Green dye was added in the co-culture and accumulated in Raji cells (yellow) as they were killed and lysed by the effector cells (CAR-T cells). B. Population-level assay of cytokine secretion using a pin-spotted antibody microarray, showing the cytokine secretion from CAR-T cells only (green), Raji cells only (red), and their co-culture (orange).
Quality assessment of scRNA-seq data from unstimulated and antigen-stimulated CAR-T cells A. Scatter violin plots showing total number of genes, transcripts (UMI), and mitochondrial genes detected in single cells of unstimulated CAR-T cells (control) and antigen-stimulated CAR-T cells (activated). Cells are filtered based on the percentage of mitochondrial genes (less than 10%) and gene counts to exclude cell multiplets and low quality cells. B. Rank ordered list of genes contributing to PC1 and PC2 after performing PCA of all single cell transcriptomes combined both activated (CMB) and control groups. C. Visualization of the expression of specific genes of interest in t-SNE plots. These genes include pan-T cell marker CD3D (encoding CD3), T cell checkpoint PDCD1 (encoding PD-1), housekeeping gene ACTB (encoding beta actin), as well as genes encoding effector proteins/cytokines GZMB (encoding granzyme B), PRF1 (encoding perforin), and IFNG (encoding IFNγ). Color (blue) intensity correlates with the level of gene expression. UMI, unique molecular identifier.
Computational identification of pure CAR-T cell data and removal of residual tumor cell data from scRNA-seq A. t-SNE plot of stimulated CAR-T cells following magnetic bead-based removal of target Raji cells. A subpopulation emerged (cluster 5, circled), distinct from the larger cell cluster. B. Expression of B cell markers (IGHM encoding Ig Mu and CD19) and T cell markers (CD3D and GZMB) to verify the identity of each cluster. Cluster 5 was identified as residual B cells with markedly increased expression of B cell markers and removed computationally before further analysis.
Heatmap of top-ranked DEGs between the unstimulated and antigen-stimulated CAR-T cells A range of activation-specific genes encoding cytokines and proteins appeared with various effector, stimulatory, and inflammatory functions such as PRF1 (encoding perforin), GZMB (encoding granzyme B), IL5, IL13, and CSF2 (encoding GM-CSF), as well as IL8 and IFNG (encoding IFNγ), as indicated by arrows. This analysis was performed in Seurat (https://satijalab.org/seurat/) using single-cell gene expression data.
Multiplexed measurement of cytokine secretion from single CAR-T cells and CAR-T:Raji cell interactions CAR-T cells and the target Raji cells were co-cultured and multiplexed measurement of cytokine secretion was performed with a microchamber array chip integrated with high-density antibody barcodes. A. Scanned microchamber images of CAR-T cells (no staining) and Raji cells (labeled with red fluorescent membrane dye) co-loaded to sub-nanoliter microchambers. B. Fluorescent signal detected corresponding to cytokine secretion in individual microchambers. C. Overlaid optical and fluorescent images. The optical image provides information on cell number, cell type, and cell integrity (tracked with SYTOX) that are correlated to fluorescent signals corresponding to cytokine secretion.
GM-CSF expression in activated CAR-T cells A. At the single-cell protein secretion level, 87% of CD4+ subset cells (blue) and 78.5% of CD8+ subset (red) secreted GM-CSF. Similarly, at the transcriptional expression level, 81% of CD4+ cells and 76.9% of CD8+ CAR-T cells expressed CSF2 (encoding GM-CSF). B. Gene expression level of CSF2 compared to that of the genes encoding signature cytokines including IFNG (IFNγ) in TH1, IL5 in TH2, as well as IL10 and TGFB1 (TGFβ) in Treg cells. C. Gene expression level of CSF2 compared to that of the genes encoding TFs, including TBX21 (T-bet) in TH1, GATA3 in Th3, and FOXP3 in Treg cells, as well as STAT5. Expression of CSF2 has a high correlation with that of cytokine genes in TH1 (71.7 % for IFNG) and TH2 (54.8% for IL5) cells, as well as TF genes in TH2 cells (53.8% for GATA3). D. Gene expression of STAT1, STAT3, STAT4, STAT5, and STAT6 genes was examined and the normalized expression values are plotted for CD4+ cells (blue) and CD8+ (red) CAR-T cells after activation. E. The percentage of CD4+ cells expressing different STAT genes alone and together with STAT5.
Gene set enrichment analysis of antigen-stimulated CAR-T cells vs. unstimulated CAR-T cells A ranked gene list is created based on genes differentially expressed between antigen-stimulated CAR-T cells and unstimulated CAR-T cells. A positive ES indicates that the expressed gene is at the top of the ranked gene list, whereas a negative ES indicates that the gene is at the bottom of the ranked gene list. A. Top ranked gene sets or pathways. B. Graphical view of enrichment scores for different gene sets. GS, gene set; ES, enrichment score; NES, normalized enrichment score; FWER, family-wise error rate.
Correlating CTLA4 gene expression to cytokine gene expression in single cells A. Top 10% and bottom 10% of CTLA4-expressing cells with the highest and lowest expression levels were classified as high expression and low expression groups and separated to evaluate the relationships between cytokine functions and CTLA4 expression. Normalized expression of TH1 (IFNG and TNF), TH2 (IL13 and IL5), and Treg (IL10 and TGFB1) signature cytokine genes in CD 4+ T cells with high (gray) and low (purple) CTLA4 expression. B. Normalized expression of TH1 (IFNG and TNF), TH2 (IL13 and IL5), and Treg (IL10 and TGFB1) signature cytokine genes in CD 8+ T cells with high (gray) and low (purple) CTLA4 expression. Elevated IL13 (TH2) and IL10 (Treg) expression was correlated with the increase in CTLA4 expression in CD4+ cells. Elevated TGFB1 (Treg) expression was correlated with the increase in CTLA4 expression in CD8+ cells. t-test was performed for statistical analyses. #P ≥ 0.1; *P < 0.1; **P < 0.05; ****P < 0.01.
List of antibodies used for cytokine assays
Live cell tracking of single CAR-T cell killing a target tumor cell using the SYTOX Green uptake assay
Live cell tracking of multiple CAR-T cells attacking a target tumor cell using the SYTOX Green uptake assay
Data Availability Statement
The single-cell RNA-seq data are available in the Gene Expression Omnibus (GEO) as GEO: GSE129007.