Abstract
How insects combat RNA virus infection is a subject of intensive research owing to its importance in insect health, virus evolution, and disease transmission. In recent years, a pair of potentially linked phenomena have come to light as a result of this work—first, the pervasive production of viral DNA from exogenous nonretroviral RNA in infected individuals, and second, the widespread distribution of nonretroviral integrated RNA virus sequences (NIRVs) in the genomes of diverse eukaryotes. The evolutionary consequences of NIRVs for viruses are unclear and the field would benefit from studies of natural virus infections co-occurring with recent integrations, an exceedingly rare circumstance in the literature. Here, we provide evidence that a novel insect-infecting phasmavirus (Order Bunyavirales) has been persisting in a phantom midge host, Chaoborus americanus, for millions of years. Interestingly, the infection persists despite the host’s acquisition (during the Pliocene), fixation, and expression of the viral nucleoprotein gene. We show that virus prevalence and geographic distribution are high and broad, comparable to the host-specific infections reported in other phantom midges. Short-read mapping analyses identified a lower abundance of the nucleoprotein-encoding genome segment in this virus relative to related viruses. Finally, the novel virus has facilitated the first substitution rate estimation for insect-infecting phasmaviruses. Over a period of approximately 16 million years, we find rates of (0.6 − 1.6) × 10−7 substitutions per site per year in protein coding genes, extraordinarily low for negative-sense RNA viruses, but consistent with the few estimates produced over comparable evolutionary timescales.
Keywords: EVEs, NIRVs, paleovirology, substitution rates, insect immunity
1. Introduction
Nonretroviral integrated RNA virus sequences (NIRVs) persist in DNA form within the genomes of myriad eukaryotic organisms from fungi to humans (Crochu et al. 2004; Taylor and Bruenn 2009; Belyi, Levine, and Skalka 2010; Katzourakis and Gifford 2010; Taylor, Leach, and Bruenn 2010). Yet, despite several interesting case studies demonstrating antiviral or immune-related roles for NIRV proteins (Fujino et al. 2014; Edwards et al. 2018; Warner et al. 2018), continued work is needed to better understand their importance during infections, and especially their consequences for virus evolution (Aswad and Katzourakis 2012). Interestingly, function at the protein level is not the only mechanism by which NIRVs may interact with cognate viruses. NIRVs in a wide range of arthropod genomes are integrated in Piwi-interacting RNA (piRNA) clusters and expressed as piRNAs (ter Horst et al. 2019). So, NIRVs may confer antiviral benefits from various gene products and become fixed in a natural population. Still, little is known of the interactions of host NIRVs and their cognate viruses in nature. NIRVs are generally ancient—few closely related (i.e. phylogenetic sisters at the nucleotide level) virus-NIRV sister genes have been identified. Perhaps the only known ‘young’ virus-NIRV pair is in the Flaviviridae (isolated from wild Aedes aegypti, Aedes albopictus, and Culex mosquitoes) where the nucleotide sequence similarity rises to 65 per cent but the viral prevalence is low at 12 per cent (Cook et al. 2006). As NIRV formation is common there should be detectable ‘young’ NIRV-virus systems in nature that would provide insights into a missing evolutionary window. Here we report on the discovery of a closely related virus (Phasmaviridae)-NIRV system that was initially detected while searching for RNA viruses in wild phantom midges.
Phasmaviridae is a family of insect-infecting viruses in the order Bunyavirales. Like all bunyaviruses, phasmaviruses encode segmented, negative-sense RNA genomes and do not produce a DNA intermediate during infection. Those in the genus Orthophasmavirus are commonly associated with diverse insect hosts, including model organisms, agricultural pests, and disease vectors. Orthophasmavirus-derived NIRVs are widespread in the genomes of these and other insects (Ballinger et al. 2014). Phantom midges have a worldwide distribution and are the non-blood-feeding sister taxon to mosquitos. As larvae, they are planktonic invertebrate predators that can reach very high population densities (Xie, Iwakuma, and Fujii 1998). Two divergent phasmavirids have been described infecting phantom midges, Kigluaik phantom orthophasmavirus (KIGV) and Nome phantom orthophasmavirus (NOMV) each strictly associated with its host, despite overlapping distributions (Ballinger et al. 2014, 2017). Prevalence screens revealed that KIGV occurs at high frequency (median 75%) in Chaoborus trivittatus populations across subarctic North America and phylogenetic relationships among geographically isolated C. trivittatus populations are mirrored by their KIGV strains, indicating a long-term association that predates the post-glaciation recolonization of North America (Ballinger et al. 2014). This degree of evolutionary persistence and fidelity in insect-microbe associations is a trademark of vertically-transmitted bacterial endosymbionts, but in insect viruses its occurrence is not well documented or studied (though see Longdon and Jiggins 2012). While the transmission route of Chaoborus-infecting phasmaviruses has not been demonstrated in a laboratory setting, several observations made from natural populations suggest vertical transmission, including significant association between mitochondrial and virus haplotypes and consistently high infection frequency in early (larval) life stages (Ballinger et al. 2017). The extent to which host-specific phasmaviruses are distributed among other phantom midges is unknown.
Here we report and examine a novel phasmavirus of Chaoborus americanus, Niukluk phantom orthophasmavirus (NUKV). Chaoborus americanus is a temperate Nearctic phantom midge common in fishless ponds (Borkent 1981). We find a high frequency of infection and little genetic variation for NUKV populations, consistent with the host’s recent invasion of the study region, a 4,000 sq. mi. hydrologically-disturbed tundra area in subarctic western Alaska, USA (Taylor et al. 2016). We also report an infection in southwestern British Columbia, Canada, and a virus-free population in NY, USA. Using the novel NUKV sequences together with previously described phasmaviruses, we estimate the long-term evolutionary rate for members of the family Phasmaviridae at (0.6 − 1.6) × 10−7 substitutions per site per year. Furthermore, we identify a NUKV-derived NIRV, which we name orthophasmavirus-derived integrated nucleoprotein (odin), in the genome of C. americanus from all three locales. Molecular evolutionary analyses suggest that NUKV has been persisting in odin-expressing hosts for approximately 4 million years (Pliocene), favoring the hypothesis that antiviral roles for NIRVs may be related to reducing virulence rather than clearing infection. While our analyses do not yet implicate odin as an antiviral factor, we do identify a major disparity in abundance between the NUKV nucleoprotein-encoding genomic segment and those of related phasmaviruses. Overall, our findings contribute to studies of long-term virus evolution and bring the field closer to understanding the function and evolutionary consequences of NIRVs for viruses and their hosts.
2. Materials and methods
2.1 Field collections and sample preservation
Chaoborus americanus larvae were collected from freshwater ponds in northwest Alaska in late July or early August of 2011–2014, in British Columbia in June of 2017, and in New York in July and August of 2014 and 2018 by multiple oblique tows using a 200–250 μm throw or dip net (Wildco Scientific) and stored in 100 per cent ethanol at −20 °C immediately following identifications. Species determinations were made based on morphological characteristics of the larval mandible and labral blade. Collection site global positioning system (GPS) coordinates are listed in Supplementary Table S1.
2.2 Short-read sequencing and assembly
An RNA-Seq library was produced for C. americanus from total RNA extracted from four pooled larvae with the RNeasy Mini Kit (Qiagen) and subsequently treated with RQ1 DNase I (Promega). Ribosomal RNA was reduced with the RiboZero Gold rRNA removal kit (Epicentre). The library was generated with the RNA ScriptSeq v2 RNA-Seq library preparation kit (Epicentre) and quantified with the Agilent 2100 Bioanalyzer RNA 6000 Pico Chip. RNA sequencing was carried out at the University at Buffalo Next Generation sequencing facility using Rapid 150-cycle paired-end sequencing on a single Illumina HiSeq 2000 lane. Due to technical error, only single end reads were obtained (approximately 33 million 150 bp reads). CLC Genomics Workbench (http://www.qiagenbioinformatics.com) was used for de novo sequence assembly with default parameters, which allowed the software to automatically select kmer size, bubble size, and paired distances. Assembled contigs were grouped into a custom sequence database in Geneious R7.1 (Biomatters, http://www.geneious.com) (Kearse et al. 2012) and queried with phasmavirus amino acid sequences using the Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) algorithm tBLASTn and an expect value cutoff of 10−5.
2.3 Reverse transcription polymerase chain reaction for RNA virus validation and prevalence screening
For virus screens, the number of larvae tested and NUKV prevalence results per population is provided in Supplementary Table S1. In total, 116 and 20 individual larvae were screened from Alaska and NY, respectively, and one pooled sample of 22 larvae from BC was screened. For individual extractions, second to fourth instar larvae were cut in half using a fresh microscope slide coverslip, the posterior tissues were dried briefly to evaporate the ethanol prior to RNA extraction and the anterior tissues were stored in 100 per cent ethanol at −20 °C. Dried tissues were individually ground in 1.5 ml microfuge tubes with QuickExtract (Epicentre) using sterilized pestles and incubated at 62 °C while shaking at 200 rpm for 30 min, or bead-beaten in PrepMan Ultra (Applied Biosystems) and incubated at 95 °C for 10 min. cDNA was generated using GoScript reverse transcriptase (Promega) or Superscript III Reverse Transcriptase (Invitrogen), and GoTaq master mix (Promega) or Taq polymerase (Fisher Bioreagents) was used for PCR amplification. To distinguish between possible nucleic acid sources of the two co-occurring NUKV nucleoprotein-like sequences, we performed combinations of nuclease-treatments, reverse transcription, and PCR, summarized in Supplementary Fig. S1. To test for the presence of a DNA template corresponding to each putative virus gene, i.e. L, Gn, N and the alternate N (odin), we targeted these loci in Taq-only PCR reactions, i.e. no reverse transcription step was performed following total nucleic acid extractions. To test for the presence of RNA copies for each, we treated with RQ1 DNase I (Promega) prior to cDNA synthesis. Appropriate controls were included and loaded alongside test samples for ensure each treatment was successful (Supplementary Fig. S1). This was performed for samples collected at all three geographic regions and amplified products were Sanger sequenced to confirm primer specificity and perform sequence analysis. PCR primer sequences and thermal cycling programs are listed in Supplementary Table S2. Circular and linear DNAs were differentiated via exonuclease V (New England Biolabs) digestion according to manufacturer’s protocols prior to PCR. All PCR products were assessed for amplification success and correct size on a 1 per cent agarose gel stained with ethidium bromide.
Unpurified PCR products amplified by NUKV primers, odin primers, and host mtDNA primers were sent to the high throughput DNA sequencing facility at the University of Washington or to Sequetech (CA, USA) for Sanger sequencing in both directions. Sequence chromatograms were assembled and examined in Geneious R7. Primer regions were trimmed and consensus sequences were generated based on highest quality. Unique haplotypes were supported in both sequencing directions. Sequence alignments were created in Geneious using the MAFFT algorithm version 7.388 (Katoh and Standley 2013) plugin. Alignment lengths and the number of sequences in each are listed in Supplementary Table S3.
2.4 Haplotype analysis and phylogenetic inference
Median joining haplotype networks were inferred and visualized using the software PopArt 1.6 (Leigh and Bryant 2015). Host mitochondrial gene sequences were concatenated and treated as a single locus for all analyses. Population differentiation statistics Snn (Hudson 2000) and the diversity estimate γST (Nei 1982) reported in Supplementary Table S4 were estimated using DNAsp v5 with 1,000 permutations (Librado and Rozas 2009). γST is similar to the traditional FST (Wright 1951), but substitutes nucleotide diversity (π) for heterozygosity. Unlike Hudson’s modified FST (Hudson, Slatkin, and Maddison 1992), γST accommodates identical sequences in the computation. Snn summarizes the extent to which the most closely related sequences are present in the same population. Maximum likelihood phylogenetic trees were inferred and SH-like approximate likelihood ratio test branch support scores were computed using PhyML version 3.1 (Guindon et al. 2010) implemented by SeaView v4.5.4 (Gouy, Guindon, and Gascuel 2010) and visualized with FigTree v1.4. Trees built from amino acid alignments used the LG + I + G + F substitution model.
2.5 Virus abundance estimates
The total RNA Illumina HiSeq dataset used to identify NUKV originally was used to estimate the abundance of NUKV genomes, antigenomes, and transcripts. First, we used bbmap (Bushnell 2014) to map short reads from the NUKV dataset to the full-length transcripts of the L, GnGc, and N genes. This approach did not differentiate between antigenomes and transcripts as both are positive sense. We also mapped short reads from published datasets generated from related phasmaviruses to their respective full genome sequences. These were KIGV (GenBank accessions NC_034462, NC_034463, NC_034469) (Ballinger et al. 2014) and Culex orthophasmavirus (CLX; GenBank accessions MF176242, MF176243, MF176244) (Shi et al. 2017). Reads per kilobase per million reads (RPKM) calculations were performed by bbmap version 37.93. Raw reads are available on GenBank under BioProject PRJNA509576.
2.6 Estimation of divergence times and rates
Pairwise nucleotide sequence alignments of the mitochondrial gene cytochrome oxidase subunit I (COI) were generated using MAFFT and nucleotide divergence was used to calculate divergence times given a rate of 3.54 per cent My−1 (Papadopoulou, Anastasiou, and Vogler 2010). BEAUti 2.5.1 (Drummond et al. 2012) and BEAST 2.5.1 (Bouckaert et al. 2014) were used to apply this calibration to codon alignments of virus and host genes L, Gn, N, and COI. There were five sequences per virus alignment, NUKV AK (Alaska), NUKV BC (British Columbia), KIGV AK, KIGV BC, and KIGV NU (Nunavut). For L and Gn, all positions were present for all taxa and for N, a full-length gene was present for AK strains of NUKV and KIGV, while 150 positions of NUKV BC overlapped with the KIGV N sequences from BC and NU. We calibrated the nodes corresponding to the ancestor of North American NUKV and KIGV strains to 0.7 and 2.67 My, respectively, and used the strict molecular clock parameters in BEAST to estimate host and virus divergence time and the odin-NUKV N split. Tracer 1.7.1 was used to view and export posterior distributions plots.
3. Results
3.1 A novel bunyavirus of C. americanus
Using a total RNA sequencing approach, we identified a novel virus in C. americanus larvae collected in subarctic Alaska. Molecular phylogenetic analysis of the complete polymerase protein revealed it is a member of the family Phasmaviridae in the order Bunyavirales. The novel virus is sister to KIGV, which is associated with a different phantom midge host (C. trivittatus), sharing 41.2 per cent amino acid sequence identity with KIGV across the full polymerase protein sequence when aligned with all viruses in the family Phasmaviridae. The GnGc and N protein sequences share 33 and 35 per cent amino acid identity with those of KIGV, respectively. We assembled the complete virus genome and found that each segment exhibits the typical coding architecture and non-coding elements of phasmavirids, shown in Fig. 1A. These include a large genomic segment (L) encoding the RNA-dependent RNA polymerase (L protein), the M segment which encodes the viral glycoprotein precursor, and exhibits the expected sequence signals consistent with its translocation and cleavage into the Gn and Gc proteins, and the S segment, which encodes a small putative nonstructural protein (NSs), the nucleoprotein (N), and a conserved third open reading frame (ORF3). For this virus, we propose the name NUKV, as the species was first collected in the Niukluk River valley near Council, AK.
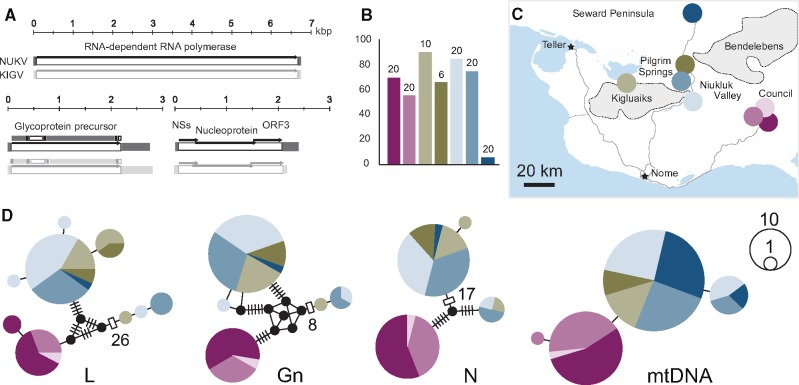
Figure 1.
Genome structure, prevalence, and genetic diversity of Niukluk phantom orthophasmavirus. (A) NUKV exhibits standard phasmavirus genome organization. The structure of each of the three genomic segments is shown above a reference segment from the related virus, KIGV. Untranslated regions are shown as filled rectangles and ORFs are shown as arrows. Above the M, predicted external (filled) and internal (hollow) regions of the viral glycoprotein are mapped. (B) NUKV infects Alaskan populations of Chaoborus americanus at high frequency. Bar graphs display prevalence in larvae collected from each of seven freshwater ponds. Numbers above plots refer to larvae screened per population. One fewer location is shown here relative to (C), as the eighth location in that panel is the source of the C. americanus RNA library. (C) Chaoborus americanus were collected from seven recently-colonized populations on the Seward Peninsula, Alaska. (D) Networks display structure of NUKV and C. americanus genetic diversity in the recently invaded study region. Pies represent unique haplotypes and bars indicate substitutions. Both organisms display limited genetic diversity and weak structure between populations, but divergence between haplotypes is greater for NUKV owing to elevated evolutionary rates.
3.2 NUKV is a common member of the C. americanus virome
To better understand the relationship between NUKV and C. americanus, we determined the infection frequency and characterized genetic variation in the Seward Peninsula populations. We screened 116 C. americanus larvae collected from subarctic freshwater ponds between 2012 and 2014 for NUKV. In total, 71 of those screened were positive for NUKV RNA (61%; Supplementary Table S1). Infection prevalence ranged from 5 to 90 per cent with a median of 70 per cent (Fig. 1B), though for two of seven populations we were able to screen only ten or fewer larvae, which could result in biased prevalence determinations for those populations. Excluding these yields a prevalence range from 5 to 85 per cent with a median of 70 per cent. To assess levels of virus genetic diversity in the study region (Fig. 1C), we sequenced cDNA products from each of the three genomic segments. These included fragments of the L, Gn, and N genes. We also sequenced two host mitochondrial loci, ND4 and COI. We find just four mitochondrial haplotypes, consistent with previous findings of the recent tundra invasion (Taylor et al. 2016; Ballinger et al. 2017). Likewise, recent genetic bottlenecks are evident in the virus loci, with nine, five, and four haplotypes found for the L, Gn, and N loci, respectively. Alignment lengths and number of polymorphic positions are provided in Supplementary Table S3. As expected due to its elevated mutation rate, the genetic distance between haplotypes is much greater for the virus loci than the host (Fig. 1D). Haplotype networks for virus and host show a weak genetic structure within the Peninsula interior, but strong structure between the interior populations and those on the opposite end of the Niukluk River valley (colored pink, Fig. 1C and D;Supplementary Table S4). Low haplotype diversity together with strong connectivity at both virus and host markers is consistent with a recent and rapid colonization of the Peninsula interior, which was also seen across eight seasons of temporal taxon sampling in the 2000–2014 interval (Taylor et al. 2016).
3.3 The viral nucleoprotein gene has been captured by the host
In addition to the NUKV S segment described above, we identified a second putative phasmavirus N transcript amongst our assembled contigs. BLAST search results and nucleotide alignments suggested it is closely related to NUKV N, but it does not cover the entirety of the S segment, only the N coding sequence is represented. We thought it could be either an incompletely-assembled fragment of a defective NUKV genomic segment or a copy of the N gene that had been integrated into and transcribed from the host genome. We carried out a series of nucleic acid digests and reverse transcriptions to determine whether the template genetic material was RNA or DNA (Supplementary Fig. S1). Two primer sets, one targeting the full-length NUKV S contig (i.e. the putative exogenous virus) and one targeting the alternate N-like gene (i.e. the potential host integration or defective viral genome segment), each produced amplicons of the intended size and did not cross-amplify. The primers targeting exogenous NUKV amplified a product in two of six Alaskan samples and did not amplify in Taq-only reactions, indicating DNA templates were absent, signaling that only those two individuals were infected with NUKV. The primer set for the N-like gene amplified from all samples and all digest treatments except the negative control, signaling its presence as both RNA and DNA in every individual tested.
3.4 The endogenous nucleoprotein gene is fixed in North American hosts
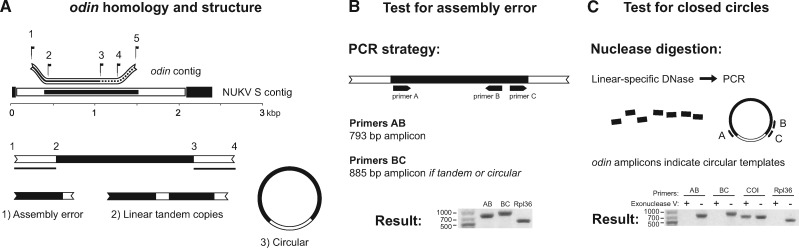
We collected C. americanus from freshwater ponds in British Columbia, Canada, and New York, USA. Using the same primer sets and nuclease treatment approach described above, followed by Sanger sequencing, we confirmed that the BC population was infected with NUKV while the NY population lacked detectable infection. Yet, both populations encode the NUKV N-like sequence in DNA form, indicating that it is endogenous to the host, fixed in North American populations, and at least as old as their common ancestor. Hereafter we refer to this C. americanus gene as odin. The odin mRNA in our transcriptome from Alaska is just 905 bp. We attempted to extend its boundaries by reviewing the flanking regions of reads mapping to its ends, and these suggested the contig ends were identical to each other, i.e. multiple adjacent copies are encoded in tandem and transcribed together. Other possibilities for the matching ends include cytoplasmic reverse transcription and circularization of the odin transcript (Fig. 2A), as has been reported for virally-derived DNA (vDNA) produced from defective RNA viral genomes during infections in other insects (Poirier et al. 2018), or technical errors in the reads or assembly. We ruled out technical error via PCR using primers designed to amplify both inward and outward from the coding region toward the putative second copy (or in the opposite direction around the circle, if circular; Fig. 2B). Amplification using the outward-facing primer confirmed that the repeating sequence in the assembly is real. Subsequently, we carried out nuclease digestions with a DNase enzyme specific to linear molecules, after which no odin templates could be amplified (Fig. 2C), but a mitochondrially-encoded locus could. Together, these results demonstrate that odin is present in tandem copy in the genome of C. americanus.
Figure 2.
The captured viral nucleoprotein, odin, is encoded in tandem copies in the genome of Chaoborus americanus. (A) The odin-encoding contig aligned to the NUKV S genomic segment. Homologous regions are limited to within the nucleoprotein coding region, shown as a solid black bar in the NUKV S segment. In odin, all sequences are derived from NUKV but only region 2–4 is one contiguous copy of the nucleoprotein gene. At position 3, perfect nucleotide identity with position 1 begins. Possible explanations for odin’s contig structure are (1) assembly error, (2) tandem copies, and (3) circular structure. (B) PCR primers directed inward and outward were used to confirm the accuracy of the repeat structure in our assembly. (C) Exonuclease V does not digest nicked or supercoiled circular DNA but does digest linear single- and double-stranded DNA; incubation with exonuclease five prior to PCR removed templates for odin and a nuclear gene, rpl36, but not a mitochondrial target, COI.
3.5 Unusual genome segment abundance of NUKV in C. americanus
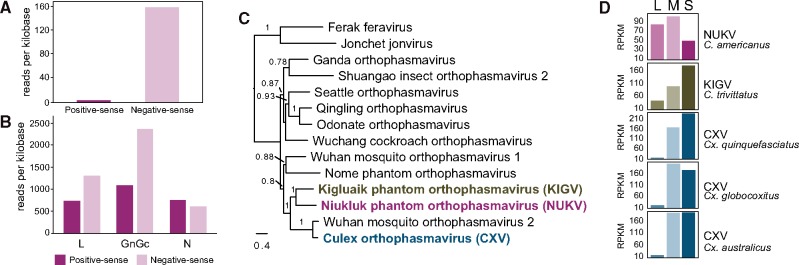
We found that odin is only transcribed in antisense (Fig. 3A), therefore it is unlikely to function at the protein level unless sense transcripts are produced under different circumstances. Unfortunately, fresh material needed to perform small RNA analysis is not currently available to us. Alternatively, we reasoned that an RNA-related role such as RNAi priming activity would result in reduced detectability by short-read mapping of the targeted genome segment, S, relative to the others. We calculated the abundance of positive and negative-sense reads in the C. americanus short-read dataset as a proxy for genome segment abundance (negative-sense reads) and transcript abundance (positive-sense reads) of NUKV. We found S to be the least abundant genome segment by mapped reads per kilobase, and whereas the L and M segments outnumbered L and Gn transcripts and antigenomes, S segments were slightly outnumbered by N transcripts and antigenomes (Fig. 3B). Both of these results were unexpected.
Figure 3.
Unusual genome segment abundance of NUKV in Chaoborus americanus. (A) Short-read mapping to the odin sequence shows that the gene is transcribed in antisense. (B) Short-reads mapping to the NUKV L and GnGc transcripts and genomes (negative sense) are more abundant than those of N—genome sequences appear to be disproportionately reduced. (C) A maximum likelihood phylogram of full-length L protein amino acid sequences shows evolutionary relationships between viruses in the family Phasmaviridae. KIGV and CXV are emphasized in bold typeface. Branches are labeled with SH-like approximate likelihood ratio test scores greater than 0.75. (D) Short-read mapping to full genome segment sequences reveals S segments are highly abundant in phasmaviruses related to NUKV (Ballinger et al. 2014; Shi et al. 2017).
To determine whether the observed genome segment ratio of NUKV is typical of phasmaviruses, we analyzed short-read datasets generated from closely related phasmaviruses (Fig. 3C) and their respective hosts. These additional datasets were for KIGV, which infects C. trivittatus (one short-read dataset) (Ballinger et al. 2014), and CLX, which infects Culex mosquitos (three datasets) (Shi et al. 2017). Segment abundance, inferred from mapped RPKM, exhibited a similar and unambiguous pattern in all three Culex hosts and in C. trivittatus. That is, M and S sequences are much more abundant than L sequences (Fig. 3D). We quantified these patterns as per cent contribution of S segment reads to total phasmavirus RNA. In the KIGV and CLX datasets, the S segment makes up 43–57 per cent of total phasmavirus RNA abundance (N = 4, M = 50.8%, SD = 6.5%), while in NUKV, S segment reads account for just 21 per cent of the total phasmavirus RNA. These data suggest that the relative S segment abundance of NUKV is highly unusual, possibly due to differences in production or stability of the viral RNAs in C. americanus.
3.6 Timescale and evolutionary rate of phasmaviruses in Chaoborus
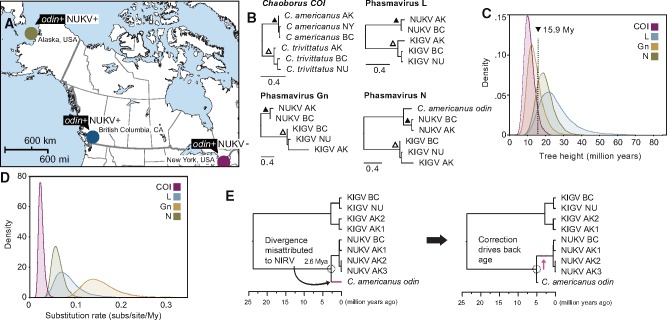
To investigate the evolution of Chaoborus-infecting phasmaviruses, we combined geographically-isolated NUKV and C. americanus sequences (Fig. 4A), which we know shared an infected ancestor due to the presence of odin in each, with host and virus sequences published previously for KIGV in Alaska, USA, British Columbia and Nunavut, Canada. These virus and host loci are shown in phylogenetic context with NUKV and C. americanus in Fig. 4B. We first asked whether Chaoborus divergence times are an appropriate calibration for phasmavirus evolution. Despite the anticipated differences in evolutionary rate between virus and host sequences, COI, L, Gn, and N gene trees calibrated at the most recent common ancestor for each species based on COI nucleotide divergence and a rate of 3.54 per cent My−1 (Papadopoulou, Anastasiou, and Vogler 2010), all yielded similar root ages (Fig. 4C). This observation is consistent with phasmavirus-host codivergence since these hosts split, approximately 16 Mya based on overlapping distributions of tree height estimates (dashed vertical line, Fig. 4C). Individual gene tree estimates were: COI: 11.2 My, 95 per cent highest posterior density (HPD) [6.2, 17.0]; L: 26.7 My [10.2–47.5]; Gn: 14.5 My [7.2–23.1]; N: 22.8 My [10.2–38.5]. Confidence intervals are wide at some loci, notably L, but there is general agreement among virus and host genes, thus the wide intervals may be due to limited sampling of modern phasmavirus diversity. Substitution rates generated during this analysis were 8.64 × 10−8 subs/site/year in L gene, 1.57 × 10−7 subs/site/year in Gn, and 6.19 × 10−8 subs/site/year in N (Fig. 4D). We note that the estimated rate of COI evolution in this analysis was 2.55 × 10−8 subs/site/year, making virus and host rates much more similar than expected.
Figure 4.
Molecular evolution of Chaoborus phasmaviruses and odin. (A) A map of North America shows collection sites and NUKV infection status of Chaoborus americanus. (B) Maximum likelihood phylograms for host and phasmavirus genes. Nodes that were calibrated to 0.7 and 2.67 My based on COI sequence divergence are marked with filled and hollow triangles, respectively. Note that odin was not present in the N alignment used for virus divergence and rate estimation. Scale bars display nucleotide distance. (C) Marginal density plots of tree heights for each gene tree in panel B following BEAST divergence analysis. Distributions for all loci overlap between 10 and 20 My, indicating virus and host have been diverging for similar timescales. (D) Marginal density plots of substitution rates estimated during BEAST divergence analysis show rates 3–10 times faster than the host mitochondrial gene, indicating a relatively slow long-term evolutionary rate for phasmaviruses. (E) The Bayesian consensus tree following BEAST divergence time analysis of phasmavirus N sequences and the odin gene displays an integration estimate of ∼2.6 My. Because this analysis did not account for the slowed evolutionary rate of odin after integration, the second tree illustrates that the effect of correcting for equal rates is to drive the true odin acquisition date further into the past.
3.7 NUKV has persisted with odin for millions of years
How old is the odin element detected in C. americanus? The gene is phylogenetically nested within Chaoborus-infecting phasmaviruses (with mosquito viruses being basal) and shares 75 per cent nucleotide identity with the C. americanus associated virus, NUKV N (Fig. 4E). The close relatedness of an extant RNA virus and a paleovirus is unusual and suggests a relatively recent integration. We found just one polymorphic nucleotide position within nearly full-length odin sequences from populations in AK, BC, and NY, supporting a recent divergence of the three. Meanwhile, the exogenous NUKV N gene shows 6 per cent sequence divergence between AK and BC strains (L gene: 5.9%, Gn: 8.3%; additional comparisons in Table 1). A second nuclear gene, ribosomal protein L36, shows one intronic polymorphism and one exonic polymorphism across 510 positions and a mitochondrion-encoded gene, COI, is 96.6–100 per cent identical, suggesting a common ancestor existed around one million years (My) ago for these populations based on an estimated divergence rate of 3.54 per cent My−1 (Papadopoulou, Anastasiou, and Vogler 2010). This serves as a minimum age estimation for odin, though it appears to fall considerably short of the time interval since integration. Attempting to extend beyond this point using host information alone would not be fruitful due to the limited sequence divergence within odin. However, by considering phasmavirus sequences as well, we were able to estimate an approximate time since integration to be 2.65 My (95% HPD 1.69–3.63; Fig. 4E). This analysis assumes that the substitution rate of odin and NUKV N is similar, which is not a reasonable assumption due to the considerable disparity between virus and host evolutionary rates. Therefore, 2.65 My is a conservative minimum age and odin may be up to twice as old (Fig. 4E). Applying the evolutionary rate of N estimated previously and attributing all differences to the virus yields an age of 4.02–4.08 Mya for comparisons with NUKV AK and NUKV BC, respectively.
Table 1.
Nucleotide divergences of Chaoborus and phasmaviruses.
| Pairing |
Nucleotide divergence (%) |
||||
|---|---|---|---|---|---|
| Sequence A | Sequence B | COI | L | Gn | N |
| AMER AK | AMER BC | 2.16 | 5.89 | 8.25 | 6.04 |
| AMER AK | TRIV AK | 16.70 | 32.38 | 57.80 | 50.00 |
| AMER AK | TRIV NU | 19.07 | 32.72 | 61.25 | 48.98 |
| AMER AK | TRIV BC | 18.39 | 32.47 | 53.62 | 47.51 |
| AMER BC | TRIV AK | 16.59 | 32.33 | 59.06 | 51.03 |
| TRIV AK | TRIV NU | 9.44 | 13.70 | 34.97 | 19.54 |
| TRIV AK | TRIV BC | 8.76 | 13.54 | 33.42 | 20.38 |
Letters indicate genes from each of three virus genome segments. L, the L protein (polymerase); Gn, glycoprotein; N, nucleoprotein. Nucleotide divergence is between representative haplotypes where AMER is Chaoborus americanus and TRIV is Chaoborus trivittatus.
4. Discussion
Our results provide insight into a rarely-glimpsed window of early NIRV-virus interactions in natural populations. Much is unknown about the function of NIRVs, but this discovery suggests that RNA viruses may be more resilient to these elements than has been hypothesized. The only comparable case of NIRV-virus co-occurrence we are aware of comes from Ae. aegypti and Ae. albopictus mosquitos that harbor endogenous flavivirus-like elements (Crochu et al. 2004; Suzuki et al. 2017). A virus closely related to these NIRVs (up to 65% nucleotide identity) was isolated from wild Ae. aegypti, Ae. albopictus, and Culex mosquitos collected in Puerto Rico and Culebra in 2006, albeit at a much lower prevalence (12% of over 500 pools) (Cook et al. 2006).
Exploring the diversity of insect-pathogen interactions in the wild can meaningfully inform our understanding of infections in ways that laboratory models alone cannot, but it also presents challenges. Infectious NUKV, additional library-quality RNA samples, and a laboratory-reared host colony are currently out of our reach, so our analyses draw from resources collected prior to our discovery of odin. Any discussion of odin’s function as an antiviral factor is speculative at this point, but our short-read mapping analysis strongly suggests NUKV abundance ratios are atypical, particularly in regard to the S segment, which encodes the N gene. In other bunyaviruses, segment ratios consistently find that M and S are much more abundant than L, for example one estimate from Rift Valley fever virus found a ratio of 1:2:2 (L:M:S) (Gauliard et al. 2006) and one from Uukuniemi virus found a ratio of 1:4:2 (Pettersson et al. 1977). Still, it is important to emphasize at this point that these results do not implicate odin or any other specific host factor in mediating these changes. Given that odin is only expressed in antisense, one possibility is that it dampens NUKV titer through an endogenous siRNA-like pathway, hybridizing with sense strand NUKV N transcripts, exposing them to detection by dicer and facilitating the loading of N-derived viral siRNAs onto an argonaute protein, which would result in downstream targeting of NUKV genomes. Effective silencing of N transcription could reduce N protein production with obvious downstream repercussions for replication and genome packaging as formation of the ribonucleoprotein complex is required for both processes.
Our discovery demonstrates that host co-option and fixation of viral elements can occur without qualitative changes in susceptibility to infection as the driving force. We estimate that NUKV has persisted in C. americanus alongside odin for millions of years. While this is just a short interval from a gene evolution perspective, it is a deep glimpse into specific and ongoing virus-host interactions. Some authors have proposed that the production and maintenance of vDNAs leads to viral persistence (at the level of the individual), which may alleviate negative health effects on the host without loss of infection (Poirier et al. 2018). The spread and maintenance of odin suggests that this degree of protection may be of significant biological importance in the wild. As previous studies have shown lifetime persistence of vDNA in insects, an alternative interpretation of our results could be that odin is present as unintegrated, linear DNA. From this perspective, it would follow that a chromosome-level assembly or, at the least further flanking sequence information, is required to conclude odin is integrated. However, there are several reasons why an unintegrated vDNA explanation is highly unlikely. First, there is no evidence that unintegrated vDNA is heritable, certainly not at the generational scale required for the continental (>5000 km) host distribution we report, yet we find temporal and spatial stability in odin presence and expression as RNA at an early host life stage across North American populations. Second, if not inherited, vDNA must be generated from a viral template within the lifetime of the host, and our screens find no evidence of any additional phasmavirus genes in eastern North American populations—only odin is present. Third, we find very little sequence variation within odin, suggesting it evolves slower even than host mitochondria. If it were renewed as vDNA within each generation from exogenous virus templates, we expect differing vDNA gene segments and evidence of a substitution rate consistent with that hypothesis. Therefore, integration is the only favorable interpretation of our results, not only because there is clear precedent of these integrated elements within diverse eukaryotic genomes, but because the only alternative explanation—long-term stable inheritance of an extrachromosomal linear DNA molecule that expresses RNA—is unknown in nature and appears unlikely to occur.
We also report here that NUKV and KIGV are descendant strains of a phasmavirus that infected the ancestor of C. americanus and C. trivittatus. Our analysis suggests that the host species and their viruses split about 16 Mya, which falls within the range of two previous host divergence estimates of 8 and 25 Mya (Berendonk, Barraclough, and Barraclough 2003). We note that calibration of recent virus nodes with host-derived divergence time estimates in order to examine evidence of deeper virus-host codivergence is in itself a circular methodology; however, odin’s presence attests to the positive NUKV infection status of hosts at those nodes, validating the early divergence time as a true dual calibration point. We recovered similar divergence timings from mitochondrial and viral markers when calibrating the younger nodes according to the mitochondrial divergence rate, suggesting that the evolutionary rate of some RNA viruses over extended time in the absence of host shifts can behave in a surprisingly clock-like manner. Additionally, these rates are lower than any negative-sense RNA virus reported. At just (0.6 − 1.6) × 10−7 subs/site/year, they are comparable to long-term evolutionary rates reported for hantaviruses, (2.4 − 2.7) × 10−7 subs/site/year (Hughes and Friedman 2000). Our findings here add to recent surprises and changing perceptions about the substitution rates of RNA viruses. For example, the age of the common ancestor for Ebolaviruses and Marburgviruses based on paleoviral evidence is orders of magnitude older than expected from clocks based on rates from historical outbreaks (Taylor et al. 2014). Our present results are consistent with analyses showing a strong influence of temporal window on observed evolutionary rates of viruses (Aiewsakun and Katzourakis 2016). Why viruses evolve slowly over deep relative to shallower timescales is unclear. One possibility is that long-term persistent infections may also be characterized by reduced replication rates (Holmes 2003), which limits the generation of genetic variation tied to the activity of error-prone RNA polymerases. Another hypothesis is that evolutionary rates are rapid following host shifts, but slow as the virus reaches its optima in the novel host niche, proceeding onward from that point at a pace set by the host’s adaptive responses (Simmonds, Aiewsakun, and Katzourakis 2018). From the perspective of our results, these ideas are not mutually-exclusive. Continued investigation of long-term infections will help clarify the contribution of these and other factors to virus evolution.
Supplementary Material
Acknowledgements
We are grateful to Alexey Kotov and Andrew Medeiros for assistance in the field. This work was supported by National Science Foundation grant 1023334 (awarded to D.J.T.) and by the COBRE program at Mississippi State University with a grant from the National Institute of General Medical Sciences - NIGMS (PG20GM103646) from the National Institutes of Health.
Data availability
Raw reads have been deposited on GenBank under BioProject PRJNA509576.
Additional nucleotide sequences generated during this work have been deposited on GenBank.
Funding
This work was supported by a grant from the National Institutes of Health (PG20GM103646).
Conflict of interest: None declared.
References
- Aiewsakun P. Katzourakis A. (2016) ‘Time-Dependent Rate Phenomenon in Viruses’, Journal of Virology, 90: 7184–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. [DOI] [PubMed] [Google Scholar]
- Aswad A. Katzourakis A. (2012) ‘Paleovirology and Virally Derived Immunity’, Trends in Ecology & Evolution, 27: 627–36. [DOI] [PubMed] [Google Scholar]
- Ballinger M. J. et al. (2014) ‘Discovery and Evolution of Bunyavirids in Arctic Phantom Midges and Ancient Bunyavirid-Like Sequences in Insect Genomes’, Journal of Virology, 88: 8783–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger M. J. et al. (2017) ‘Unexpected Differences in the Population Genetics of Phasmavirids (Bunyavirales) from Subarctic Ponds’, Virus Evolution, 3: vex015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi V. A. Levine A. J. Skalka A. M. (2010) ‘Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes’, PLoS Pathogens, 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk T. U. Barraclough T. G. Barraclough J. C. (2003) ‘Phylogenetics of Pond and Lake Lifestyles in Chaoborus Midge Larvae’, Evolution, 57: 2173–78. [DOI] [PubMed] [Google Scholar]
- Borkent A. (1981) ‘The Distribution and Habitat Preferences of the Chaoboridae (Culicomorpha: Diptera) of the Holarctic Region’, Canadian Journal of Zoology, 59: 122–33. [Google Scholar]
- Bouckaert R. et al. (2014) ‘BEAST 2: A Software Platform for Bayesian Evolutionary Analysis’, PLoS Computational Biology, 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. (2014) BBMap Short Read Aligner <http://www.sourceforge.net/projects/bbmap/> accessed 23 May 2019.
- Cook S. et al. (2006) ‘Isolation of a New Strain of the Flavivirus Cell Fusing Agent Virus in a Natural Mosquito Population from Puerto Rico’, The Journal of General Virology, 87: 735–48. [DOI] [PubMed] [Google Scholar]
- Crochu S. et al. (2004) ‘Sequences of Flavivirus-Related RNA Viruses Persist in DNA Form Integrated in the Genome of Aedes spp. mosquitoes’, The Journal of General Virology, 85: 1971–80. [DOI] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M. R. et al. (2018) ‘Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes’, Cell Reports, 24: 861–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K. et al. (2014) ‘Inhibition of Borna Disease Virus Replication by an Endogenous Bornavirus-Like Element in the Ground Squirrel Genome’, Proceedings of the National Academy of Sciences of the United States of America, 111: 13175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard N. et al. (2006) ‘Rift Valley Fever Virus Noncoding Regions of L, M and S Segments Regulate RNA Synthesis’, Virology, 351: 170–9. [DOI] [PubMed] [Google Scholar]
- Gouy M. Guindon S. Gascuel O. (2010) ‘SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building’, Molecular Biology and Evolution, 27: 221–4. [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Holmes E. C. (2003) ‘Molecular Clocks and the Puzzle of RNA Virus Origins’, Journal of Virology, 77: 3893–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R. (2000) ‘A New Statistic for Detecting Genetic Differentiation’, Genetics, 155: 2011–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R. Slatkin M. Maddison W. P. (1992) ‘Estimation of Levels of Gene Flow from DNA Sequence Data’, Genetics, 132: 583–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L. Friedman R. (2000) ‘Evolutionary Diversification of Protein-Coding Genes of Hantaviruses’, Molecular Biology and Evolution, 17: 1558–68. [DOI] [PubMed] [Google Scholar]
- Katoh K. Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A. Gifford R. J. (2010) ‘Endogenous Viral Elements in Animal Genomes’, PLoS Genetics, 6: e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. (2012) ‘Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data', Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. W. Bryant D. (2015) ‘POPART: Full-Feature Software for Haplotype Network Construction’, Methods in Ecology and Evolution 6: 1110–16. [Google Scholar]
- Librado P. Rozas J. (2009) ‘DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data’, Bioinformatics, 25: 1451–2. [DOI] [PubMed] [Google Scholar]
- Longdon B., Jiggins F. M. (2012) ‘Vertically transmitted viral endosymbionts of insects: do sigma viruses walk alone?', Proceedings of the Royal Society B: Biological Sciences, 279: 3889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (1982) 'Evolution of Human Races at the Gene Level’, in Bohhe-Tamir, B., Cohen, P., and Goodman, R. N. (eds) Human Genetics, part A: The Unfolding Genome, New York: Alan R. Liss. [Google Scholar]
- Papadopoulou A. Anastasiou I. Vogler A. P. (2010) ‘Revisiting the Insect Mitochondrial Molecular Clock: The Mid-Aegean Trench Calibration’, Molecular Biology and Evolution, 27: 1659–72. [DOI] [PubMed] [Google Scholar]
- Pettersson R. F. et al. (1977) ‘The Genome of Uukuniemi Virus Consists of Three Unique RNA Segments’, Cell 11: 51–63. [DOI] [PubMed] [Google Scholar]
- Poirier E. Z. et al. (2018) ‘Dicer-2-Dependent Generation of Viral DNA from Defective Genomes of RNA Viruses Modulates Antiviral Immunity in Insects’, Cell Host & Microbe, 23: 353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M. et al. (2017) ‘High Resolution Meta-Transcriptomics Reveals the Ecological Dynamics of Mosquito-Associated RNA Viruses in Western Australia’, Journal of Virology, 91: e00680–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Aiewsakun P. Katzourakis A. (2018) ‘Prisoners of War—Host Adaptation and Its Constraints on Virus Evolution’, Nature Reviews Microbiology, 17: 321–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. et al. (2017) ‘Uncovering the Repertoire of Endogenous Flaviviral Elements in Aedes Mosquito Genomes’, Journal of Virology, 91: e00571–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J. et al. (2014) ‘Evidence That Ebolaviruses and Cuevaviruses Have Been Diverging from Marburgviruses since the Miocene’, PeerJ 2: e556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J. et al. (2016) ‘Climate-Associated Tundra Thaw Pond Formation and Range Expansion of Boreal Zooplankton Predators’, Ecography, 39: 43–53. [Google Scholar]
- Taylor D. J. Bruenn J. (2009) ‘The Evolution of Novel Fungal Genes from Non-Retroviral RNA Viruses’, BMC Biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J. Leach R. W. Bruenn J. (2010) ‘Filoviruses Are Ancient and Integrated into Mammalian Genomes’, BMC Evolutionary Biology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Horst A. M. et al. (2019) ‘Endogenous Viral Elements Are Widespread in Arthropod Genomes and Commonly Give Rise to piRNAs’, Journal of Virology, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner B. E. et al. (2018) ‘Cellular Production of a Counterfeit Viral Protein Confers Immunity to Infection by a Related Virus’, PeerJ, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. (1951) ‘The Genetical Structure of Populations’, Annals of Eugenics, 15: 323–54. [DOI] [PubMed] [Google Scholar]
- Xie P. Iwakuma T. Fujii K. (1998) ‘Studies on the Biology of Chaoborus flavicans (Meigen) (Diptera : Chaoboridae) in a Fish-Free Eutrophic Pond, Japan’, Hydrobiologia, 368: 83–90. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads have been deposited on GenBank under BioProject PRJNA509576.
Additional nucleotide sequences generated during this work have been deposited on GenBank.