Abstract
Background
In vivo quantification of spinal cord atrophy in neurological diseases using MRI has attracted increasing attention.
Purpose
To compare across different platforms the most promising imaging techniques to assess human spinal cord atrophy.
Study Type
Test/retest multiscanner study.
Subjects
Twelve healthy volunteers.
Field Strength/Sequence
Three different 3T scanner platforms (Siemens, Philips, and GE) / optimized phase sensitive inversion recovery (PSIR), T1‐weighted (T1‐w), and T2*‐weighted (T2*‐w) protocols.
Assessment
On all images acquired, two operators assessed contrast‐to‐noise ratio (CNR) between gray matter (GM) and white matter (WM), and between WM and cerebrospinal fluid (CSF); one experienced operator measured total cross‐sectional area (TCA) and GM area using JIM and the Spinal Cord Toolbox (SCT).
Statistical Tests
Coefficient of variation (COV); intraclass correlation coefficient (ICC); mixed effect models; analysis of variance (t‐tests).
Results
For all the scanners, GM/WM CNR was higher for PSIR than T2*‐w (P < 0.0001) and WM/CSF CNR for T1‐w was the highest (P < 0.0001). For TCA, using JIM, median COVs were smaller than 1.5% and ICC >0.95, while using SCT, median COVs were in the range 2.2–2.75% and ICC 0.79–0.95. For GM, despite some failures of the automatic segmentation, median COVs using SCT on T2*‐w were smaller than using JIM manual PSIR segmentations. In the mixed effect models, the subject was always the main contributor to the variance of area measurements and scanner often contributed to TCA variance (P < 0.05). Using JIM, TCA measurements on T2*‐w were different than on PSIR (P = 0.0021) and T1‐w (P = 0.0018), while using SCT, no notable differences were found between T1‐w and T2*‐w (P = 0.18). JIM and SCT‐derived TCA were not different on T1‐w (P = 0.66), while they were different for T2*‐w (P < 0.0001). GM area derived using SCT/T2*‐w versus JIM/PSIR were different (P < 0.0001).
Data Conclusion
The present work sets reference values for the magnitude of the contribution of different effects to cord area measurement intra‐ and interscanner variability.
Level of Evidence: 1
Technical Efficacy: Stage 4
J. Magn. Reson. Imaging 2019;49:1078–1090.
Keywords: spinal cord MRI, segmentation, total cross‐sectional area, gray matter area, intrascanner interscanner reliability
Quantifying spinal cord atrophy and the more recently described spinal cord gray matter atrophy in various neurologic conditions including trauma, inflammation, or neurodegeneration has gained increasing attention, particularly with the development of dedicated spinal cord imaging techniques.1, 2, 3, 4, 5, 6 Spinal cord dedicated volumetric 3D T1‐weigthed (T1‐w) protocols, similar to the ones widely used for brain volume estimation, are becoming a standard for total cross‐sectional area (TCA) measurements.7 However, on images acquired with this and other conventional T1‐w and T2‐weighted (T2‐w) protocols, the gray matter (GM) / white matter (WM) contrast is suboptimal to allow separate assessment of these two tissues.
The most promising imaging techniques used so far to measure GM area/volume in the spinal cord are based on T2*‐weighted (T2*‐w) 3D or 2D gradient echo (GRE)8, 9, 10 or 2D T1‐w phase sensitive inversion recovery (PSIR) protocols.11, 12 These protocols also have good WM/CSF (cerebrospinal fluid) contrast that allows assessment of the TCA. Direct comparison of the and PSIR techniques in terms of intrascanner and interscanner reliability of TCA and GM area measurements is fundamental for effect‐size and sample‐size estimates in studies quantifying spinal cord and cord gray matter tissues atrophy.
The goal of this study was to perform direct comparisons of three selected optimized protocols for spinal cord GM and TCA segmentation on the same group of 12 healthy controls, on three 3T scanners produced by the three main vendors of human magnetic resonance imaging (MRI) scanners: Siemens, Philips, and General Electric (GE). The chosen protocols used product sequences available on all systems.
Materials and Methods
Research Participants
Twelve healthy subjects (five males, seven females, mean age/standard deviation [SD]: 33.5/9.7 years) with no history of neurological disorder were enrolled in the study. The Committee on Human Research at our institution approved the study protocols. Written informed consent was obtained from all participants.
Image Acquisition
All participants were scanned twice with a 30‐minute MRI protocol, with repositioning in between the scans (test/retest, 1 hour of total scan time per scanner) on three different scanners: a Siemens 3T Skyra (Siemens Healthineers, Erlangen, Germany), a Philips 3T Ingenia (Royal Philips, Amsterdam, The Netherlands), and a GE 3T Discovery MR750 (General Electric Healthcare, Chicago, IL). Between the test and retest scans, participants were asked to get off the scanner table and have a little walk in the MR room. The Siemens, Philips, and GE scanners were equipped, respectively, with a 64‐channel head‐neck coil, a neurovascular (NV) coil, and a HNS CTL 123 coil, all providing good signal‐to‐noise ratio in the upper cervical cord region. The 1‐hour sessions on the three different scanners for each participant were performed within a month for all subjects (median, mean/SD: 20, 19.1/8.3 days). All study acquisitions were performed between October 5 2017 and November 27 2017.
The scanning protocol included a sagittal cervical cord localizer, an axial single‐slice 2D PSIR acquisition at the spinal cord disc level C2‐C3, a axial 2D MEDIC/M‐FFE/MERGE (nomenclature, respectively for Siemens, Philips, and GE) acquired covering the cervical portion of the spinal cord from vertebra C1 down to about vertebra C6, and a T1‐w sagittal 3D MPRAGE/T1‐TFE/BRAVO acquisition centered on the C3 vertebral body.
The PSIR protocol was optimized on the three scanners based on experience with the previously developed protocols for the Skyra Siemens scanner.11, 12, 13 The specific sequence/parameters on each scanner were optimized conditional on the specific software/hardware available. The driving optimization rationale was to achieve similar contrast‐to‐noise ratio (CNR) across the scanners and hardware configurations. Perfectly matching all the acquisition parameters and acquisition times was not considered a priority.
The and T1‐w protocols used in the study were optimized in a worldwide collaborative initiative.14 The protocols are freely available for download on the website https://osf.io/tt4z9/ and are periodically updated to follow modifications suggested by the participants to the initiative. For the GE scanner, minor further tuning of some parameters was necessary because of hardware limitations (no anterior neck coils were available, therefore parallel imaging was not possible).
Parameters for the 2D PSIR, 3D T1‐w, and 2D protocols are reported in Table 1. Note that the definitions of some of the parameters, in particular repetition time (TR), inversion time (TI), and echo time (TE), vary by vendor. For example, the TR is defined for Siemens as the time between inversion/preparation pulses, while for Philips and GE the TR is defined as time between excitation pulses.
Table 1.
2D Phase Sensitive Inversion Recovery (PSIR), 3D T1‐w, and 2D T2*‐w Protocol Parameters
| 2D PSIR | Siemens | Philips | GE |
|---|---|---|---|
| Sequence name | CV | T1‐TFE | PSMDE |
| Dimension | 2D | 2D | 2D |
| TR (msec) | 4000 | 9.5 | 8.00 |
| TE (msec) | 3.22 | 4.7 | 3.76 |
| TI (msec) | 400 | 300 | 400 |
| # averages | 3 | 5 | 20 |
| Shots | 9 | 10 | 52 |
| Segments | 26 | 24 | 4/5 |
| Flip angle (deg) | 10 | 15 | 25 |
| Voxel sixe (mm) | .78 × .78 × 5 | .78 × .78 × 5 | .78 × .78 × 5 |
| Field of view (mm) | 200 × 200 × 5 | 200 × 200 × 5 | 200 × 200 × 5 |
| BW(Hz/Px) | 250 | 151.7 | 113.6 (22.73 kHz tot) |
| Phase encoding dir. | R > > L | R > > L | A > > P |
| Parallel acc. factor | no | no | no |
| Acq. time (min:sec) | 1:52 | 2:30 | ∼3 (dep. on heart rate) |
| Cardiac gating | simulated | not needed | finger pulse |
| Orientation | axial | axial | axial |
| 3D T1‐w | Siemens | Philips | GE |
|---|---|---|---|
| Sequence name | MPRAGE | T1‐TFE | BRAVO |
| Dimension | 3D | 3D | 3D |
| TR (msec) | 2000 | 7.77 | 8.71 |
| TE (msec) | 3.72 | 3.56 | 3.66 |
| TI (msec) | 1000 | 1000 | 450 |
| # averages | 1 | 1 | 2 |
| Flip angle (deg) | 9 | 8 | 12 |
| Voxel sixe (mm) | 1 × 1 × 1 | 1 × 1 × 1 | 1 × 1 × 1 |
| Field of view (mm) | 320 × 260 × 192 | 256 × 256 × 192 | 256 × 256 × 96 |
| BW (Hz/Px) | 150 | 191.5 | 97.65 (25.00 kHz tot) |
| Phase encoding dir. | A > > P | A > > P | R > > L |
| Parallel acc. factor | 2 | 2 | no |
| Acq. time (min:sec) | 4:44 | 4:56 | 7:28 |
| Orientation | sagittal | sagittal | sagittal |
| 2D T2*‐w | Siemens | Philips | GE |
|---|---|---|---|
| Sequence name | MEDIC | M‐FFE | MERGE |
| Dimension | 2D | 2D | 2D |
| TR (msec) | 627 | 625 | 777 |
| TE (msec) | 15 | 2.52 | 13.77 |
| # echoes | 3 | 3 | 3 |
| # averages | 2 | 1 | 2 |
| Flip angle (deg) | 30 | 30 | 30 |
| Voxel sixe (mm) | 0.5 × .0.5 × 3 | 0.5 × .0.5 × 3 | 0.5 × .0.5 × 3 |
| Field of view (mm) | 160 × 160 × 45 | 160 × 160 × 45 | 128 × 128 × 45 |
| BW (Hz/Px) | 240 | 241.1 | 195.31 (25.00 kHz tot) |
| Phase encoding dir. | R > > L | R > > L | A > > P |
| Parallel acc. factor | 2 | no | no |
| Acq. time (min:sec) | 3:52 | 4:02 | 6:44 |
| Orientation | axial | axial | axial |
Qualitative Quality Assessment
Two operators (NP and RGH) visually assessed the quality of all the images and assigned a consensual score (0, bad; 0.5, average; 1, good) to each image for three image characteristics: overall quality, overall noise/excessive motion, and GM/WM delineation/contrast (when present). Values were summed to assign a total score for each scanner and protocol for each of the three characteristics (maximum score = 24).
Data Processing
All data processing and analyses were performed by a single operator (NP) with more than 12 years of experience in brain and spinal cord MRI acquisition/analysis methods.
CNR Evaluation
CNR between GM and WM (CNRGM/WM) and between WM and CSF (CNRWM/CSF) was calculated by two operators (NP and RGH) in regions of interest (ROIs) at the C2‐C3 disc level on the test images for each scanner/protocol/subject.
The two operators manually drew a GM ROI on the anterior part of the GM (an area spanning the anterior horns). Three WM ROIs were symmetrically drawn in the region of the lateral and posterior columns. Two CSF ROIs were symmetrically placed in the right and left spinal canal. The same group of ROIs was used for all the acquisitions of a subject. Examples of ROIs are reported in Figs. 1, 2 and 3
Figure 1.
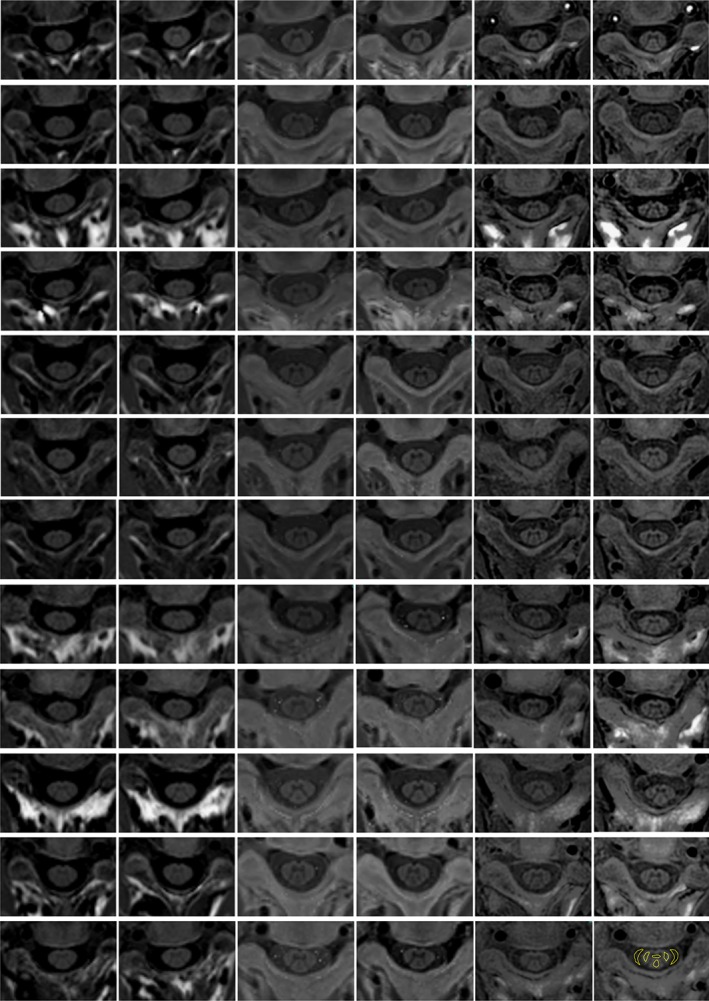
PSIR images acquired at the C2‐C3 disc level for the 12 subjects on the three different scanners. Each row is a subject and from left to right: Siemens scanner test acquisition, Siemens scanner retest acquisition, Philips scanner test acquisition, Philips scanner retest acquisition, GE scanner test acquisition, and GE scanner retest acquisition. In the bottom right image an example of ROIs used for the CNR evaluation is reported.
CNR between tissues 1 and 2 was computed for each subject, scanner, and protocol as previously defined15, 16, 17, 18:
| (1) |
where SI1, SI2, SD1, and SD2 respectively indicate the mean intensity value within the tissues 1 and 2 ROIs, and the corresponding standard deviations.
Average values and SDs between operators were computed and differences between scanners and protocols tested using two‐tailed t‐tests (P < 0.05).
Total Cross‐Sectional Area and GM Area Reliabilities
To estimate intra‐ and interscanner reliability of area measurements, TCA and GM were computed for all the available images acquired with the different protocols.
In order to assess the potential impact of the segmentation method on the spinal cord metrics, we selected the two most widely used approaches based on recent literature. The first approach is JIM v6 (Xinapse Systems, http://www.xinapse.com) that was previously used in different studies.1, 2, 3, 11, 12, 19, 20, 21, 22, 23, 24, 25 The second approach is the open source Spinal Cord Toolbox (SCT) (https://sourceforge.net/p/spinalcordtoolbox/wiki/Home/) that has seen recent utilization.14, 23, 26, 27, 28, 29, 30, 31, 32, 33
While the methods based on JIM have been previously optimized and tested for GM/TCA segmentation of single‐slice 2D PSIR images and for TCA extracted by 3D T1‐w and T2/ images, the SCT has been optimized and tested for TCA extraction from T1‐w/T2‐w acquisitions and for GM/TCA segmentation of acquisitions with multiple slices/volumetric coverage. Therefore, JIM was used to segment TCA on the T1‐w, PSIR, and images, and to segment GM on PSIR images. The automatic SCT was used to segment TCA on T1‐w and images, and to segment GM on images. We did not use the SCT on the single‐slice 2D PSIR images, because the SCT is optimized for multiple slices/volumetric coverage, and JIM for the GM segmentation of images, because we believe there are no exhaustive published data regarding the reliability of manual segmentations with this combination of software/contrast.
2D PSIR Images
TCA and GM areas for each participant and scanner were measured on the phase‐sensitive reconstructed images. TCA estimates were obtained in a semiautomated way using an active surface model34 available in JIM, with a method previously shown to have high intra‐ and inter‐rater reliability.1, 11 Briefly, this was done using the cord finder toolkit with fixed settings (nominal cord diameter 8 mm, number of shape coefficients 24, order of longitudinal variation 12). The marker requested by the toolkit was positioned by a single experienced operator (NP) on the mid‐sagittal WM, directly posterior to the gray commissure.
GM areas were manually measured using JIM with a segmentation technique that has been shown to be highly reliable. GM area was segmented three times using JIM by NP for each participant and scanner. The average GM area obtained from the three segmentations was finally calculated.1, 11
From previous experience1, 3, 11, 35, 36 the interoperator variability of the segmentations performed with the JIM methods are expected to have coefficient of variation (COV) <0.5% and intraclass correlation coefficient (ICC) >0.99 for the TCA semiautomated measurements, and COV in the range 3–5% and ICC ∼0.90 for the GM area manual measurements.
T1‐w and T2*‐W Images
Two methods were used to calculate TCA on T1‐w and acquisitions. The first method, semiautomated, was used in previous publications.3, 19, 20 TCA on T1‐w images was measured by reslicing the sagittal acquisitions and extracting five consecutive 1‐mm‐thick axial slices perpendicular to the long axis of the cord at the C2‐C3 disc level, and measuring the average area of the cord using the semiautomated cord finder toolkit of JIM with the same fixed settings used for PSIRs. The markers requested by the toolkit were placed at the center of the spinal cord in each of the five slices. For images a similar process was applied to a single axial slice at the C2‐C3 disc level without any reslicing.
The second method used the fully automatized SCT. Original images were preprocessed in the native space and then registered to the PAM50 spinal cord template.31 The TCA was extracted from T1‐w and scans following automatic cord segmentation (using in order the commands “sct_propseg,” “sct_label_vertebrae,” “sct_label_utils,” “sct_register_to_template,” and “sct_warp_template”33 with default parameters, following the documentation available at https://sourceforge.net/p/spinalcordtoolbox/wiki/tools/) and then averaged within the C3 vertebra automatically labeled by the software (command “sct_process_segmentation”). The GM area on images was extracted with the SCT following automatic gray matter segmentation (command “sct_segment_graymatter”)32 and extracting the averaged value within the C3 vertebra (command “sct_process_segmentation”).
Statistical Analysis
All statistical analyses were performed using JMP Pro 13 (SAS Institute, Cary, NC).
The coefficient of variation (COV = 100 × (absolute difference) / mean of measurements) for all the test/retest couples of TCA and GM area measured with the different segmentation methods/protocols was calculated and its median/mean (SD) on the group of subjects computed for each scanner.
The ICC was calculated between all the test/retest couples of TCA and GM area measured, for each different segmentation method/protocol, and for each scanner.
Bland–Altman plots were produced for each of the combinations segmentation method/protocol, representing each scanner with a different symbol. On the Bland–Altman plots, the difference of the retest and test measurements was reported on the y‐axis and their mean value on the x‐axis.
Mixed models with scanner as fixed effect, and test–retest and subject as nested random effects were used to estimate the contribution of subject, test–retest acquisition, and scanner to the variance of obtained measures.
To visualize interscanner differences in the calculated areas, the average values between test and retest acquisitions were computed and graphed for each of the combinations method/protocol.
To evaluate the effect of acquisition protocol when using the same segmentation method, analysis of variance (ANOVA) (t‐tests) were used between couple of measurements for TCA (PSIR, T1‐w, and images segmented with JIM and T1‐w and images segmented with SCT). ANOVA was used also to evaluate the effect of the segmentation method on the same protocol (T1‐w images and images segmented with JIM and SCT). Finally, ANOVA was used to see if there were statistically significant differences (P < 0.05) in the GM area values obtained with the two couples segmentation method/protocol (SCT/T2*‐w vs. JIM/PSIR).
Results
Qualitative Quality Assessment
All images acquired with PSIR and protocols at the C2‐C3 disc level for the 12 healthy controls on the three different scanners are reported in Figs. 1 and 2, demonstrating overall good quality of both the PSIR and images. Overall quality consensus scores for PSIR were 24 for all the scanners, while for images they were 23, 18, and 22, respectively for Siemens, Philips, and GE.
Figure 2.

T2*‐w images acquired at the C2‐C3 disc level for the 12 subjects on the three different scanners. Each row is a subject and from left to right: Siemens scanner test acquisition, Siemens scanner retest acquisition, Philips scanner test acquisition, Philips scanner retest acquisition, GE scanner test acquisition, and GE scanner retest acquisition. In the bottom right image an example of ROIs used for the CNR evaluation is reported.
Consensus visual qualitative assessment scores suggest that the PSIR images on the GE scanner appeared slightly more noisier/affected by motion (22) compared with the other two scanners (24), while according to this score, the images appeared to be of a slightly better quality on Siemens (23) and GE (24) scanners compared with Philips (22). With regard to the GM delineation/contrast, visual qualitative assessment indicated consistent good quality for the PSIR images (23, 24, 23 for Siemens, Philips, and GE), while the quality of some of the images (12 over 72, which means about 17%) was suboptimal (scores 24, 19, 23 for Siemens, Philips, and GE).
Illustrative images acquired on a single subject with the T1‐w protocol are shown in Fig. 3. The quality of T1‐w images was in general consistent with the reported example and consistent across the different scanners (overall quality scores and noise/motion were 24, 24, and 23, respectively, for Siemens, Philips, and GE). We excluded from the following analyses two T1‐w acquisitions because the subject clearly moved during the acquisition (both were test scans on the GE scanner).
Figure 3.
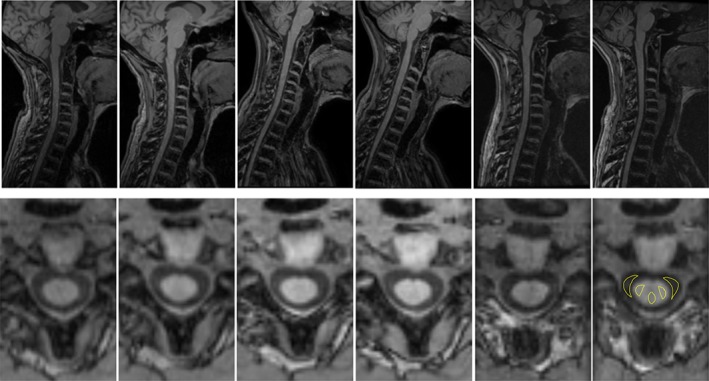
Illustrative example of T1‐w images acquired for a single healthy subject. Top row: sagittal original acquisition. Bottom row: axial reslicing at the C2‐C3 disc level. From left to right: Siemens scanner test acquisition, Siemens scanner retest acquisition, Philips scanner test acquisition, Philips scanner retest acquisition, GE scanner test acquisition, and GE scanner retest acquisition. In the bottom right image an example of ROIs used for the CNR evaluation is reported.
CNR Evaluation
In Table 2 the GM/WM and WM/CSF CNR measured at the C2‐C3 disc level for PSIR, , and T1‐w protocols is reported.
Table 2.
Between Operators Mean (Standard Deviation) Contrast‐to‐Noise Ratio (CNR) Between Gray Matter (GM) and White Matter (WM) Tissues, and Between WM and Cerebrospinal Fluid (CSF), for the Three Different Acquisition Protocols (PSIR, T2*‐w and T1‐w) on the Three Different Scanners
| CNRGM/WM | PSIR | T2*‐w | T1‐w |
|---|---|---|---|
| Siemens | 2.11 (0.15) | 1.56 (0.06) | — |
| Philips | 3.14 (0.16) | 1.09 (0.05) | — |
| GE | 2.39 (0.04) | 1.67 (0.02) | — |
| CNRWM/CSF | PSIR | T2*‐w | T1‐w |
|---|---|---|---|
| Siemens | 8.45 (0.22) | 3.54 (1.38) | 9.91 (0.14) |
| Philips | 3.06 (0.11) | 3.71 (1.05) | 9.08 (0.04) |
| GE | 3.40 (0.07) | 4.64 (2.16) | 7.26 (0.05) |
The GM/WM CNR for the PSIR protocol was higher compared with the protocol for all the scanners (P < 0.0001). GM/WM CNR for the protocol on the Philips scanner was lower if compared with Siemens (P < 0.0001) that was lower if compared with GE (P < 0.0001), confirming the visual qualitative impression of slightly worse quality on Philips. The visual impression that the PSIR images were noisier on GE, however, is not supported by the CNR evaluation.
The WM/CSF CNR for the T1‐w protocol was consistently higher for all the scanners compared with the PSIR and protocol images (P < 0.0001). The WM/CSF CNR for PSIR and protocols was comparable for GE (P = 0.06) and Philips (P = 0.042), while for Siemens for PSIR it was much higher (P < 0.0001).
TCA and GM Area Measurements
Test–retest COV and ICC for measured TCA and GM area for all the combinations of segmentation methods and protocols are reported in Table 3.
Table 3.
Test‐Retest COV (Median, Top Row, and Mean (SD), Middle Row) and ICC (Bottom Row) for TCA and GM Area Measurements on Images Acquired With the Three Protocols (PSIR, T1‐w, T2*‐w) and Using the Different Segmentation Methods (JIM, SCT) on the Group of 12 Controls
| JIM TCA T1‐w |
JIM TCA PSIR |
JIM TCA T2*‐w |
SCT TCA T1‐w |
SCT TCA T2*‐w |
SCT GM T2*‐w |
JIM GM PSIR |
|
|---|---|---|---|---|---|---|---|
| Siemens | 0.84 1.23(1.25) 0.9892 |
1.09 1.55(1.40) 0.9853 |
1.43 2.19(2.35) 0.9671 |
2.27 2.53(2.16) 0.9450 |
2.42 5.87(12.96) 0.8121 |
2.91 8.31(15.91) 0.6516 |
3.41 4.31(2.94) 0.8751 |
| Philips | 1.16 1.55(1.32) 0.9828 |
1.14 1.21(0.83) 0.9923 |
1.36 1.85(1.90) 0.9720 |
2.67 3.55(2.74) 0.8753 |
2.24 3.91(4.04) 0.8652 |
4.82 5.05(4.04) 0.8077 |
7.21 6.52(3.30) 0.6915 |
| GE | 1.02 1.34(0.96) 0.9893 |
2.38 2.56(1.53) 0.9515 |
0.82 1.15(0.96) 0.9903 |
2.36 3.37(3.75) 0.8770 |
2.76 5.60(7.14) 0.7937 |
3.64 8.30(10.46) 0.3423 |
6.52 6.25(4.64) 0.7443 |
Data for each scanner are reported.
For the TCA, with the JIM semiautomatic method, median COVs are very similar across the three protocols and smaller than 1.5% (with the only exception of PSIR on the GE scanner). ICC were always >0.95. The SCT performed very similarly when measuring TCA on T1‐w and protocols for all the scanners, with median COV in the range 2.2–2.75% and ICC in the range 0.79–0.95.
With regard to GM, median COV of measurements obtained with the SCT on images were consistently smaller compared to manual segmentation on PSIR images. Nevertheless, mean and SD of COV for the SCT/T2*‐w combination are in general larger than on JIM/PSIR, because of outlier values due to failures of the automatic segmentation algorithm (Bland–Altman plots reported in Fig. 4). The lowest ICC were 0.3423 for SCT/T2*‐w on GE and 0.6915 for JIM/PSIR on Philips.
Figure 4.
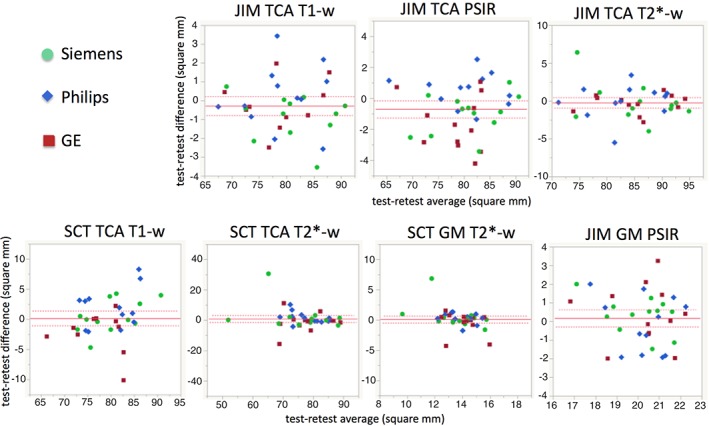
Bland–Altman plots reporting the difference between the TCA and GM area retest and test measurements (y‐axis) and their mean value (x‐axis), for all the combinations of software and protocol.
Furthermore, despite similar COV median values, mean/SD are bigger (and ICC smaller) for SCT on images than for T1‐w images, due to fewer automatic segmentation errors of the SCT on T1‐w images.
Statistical Analysis
In the mixed effect models, subject was always the main contributor to the variance of the area measurements (always significant, Wald P < 0.05). Among all the combinations of segmentation methods/protocols/tissues, session (test–retest) was statistically significant in explaining the variance of the area measurements only for TCA measured on T1‐w images with the SCT (P < 0.0001). Scanner (fixed effect) was instead often statistically significant in the mixed effect models for TCA measurements. The value of the estimated intercept (that can be read as values for Siemens for the way the variables were ordered in the model), the biases of the measurements that the model attributes to the other scanners and the related P‐values are reported in Table 4.
Table 4.
Mixed Effect Model Results
| JIM TCA T1‐w |
JIM TCA PSIR |
JIM TCA T2*‐w |
SCT TCA T1‐w |
SCT TCA T2*‐w |
SCT GM T2*‐w |
JIM GM PSIR |
|
|---|---|---|---|---|---|---|---|
| Siemens (intercept) |
79.94 | 80.63 | 84.62 | 78.81 | 77.49 | 13.90 | 20.28 |
| Philips | –0.62 P = 0.0012 |
0.37 P = 0.25 |
–1.54 P < 0.0001 |
0.82 P = 0.13 |
0.26 P = 0.74 |
0.04 P = 0.81 |
0.09 P = 0.48 |
| GE | –0.56 P = 0.0039 |
–1.42 P < 0.0001 |
0.64 P = 0.016 |
–1.59 P = 0.0047 |
0.84 P = 0.28 |
0.24 P = 0.15 |
0.08 P = 0.53 |
Estimated scanner contribution to biases: Estimated intercept (corresponding to Siemens) and the bias and related P value for the other scanners are reported (in bold when < 0.05). Values of areas are in square mm.
The mean value of the test and retest acquisitions for the 12 healthy subjects are reported in Fig. 5 for each segmentation method/protocol/area.
Figure 5.
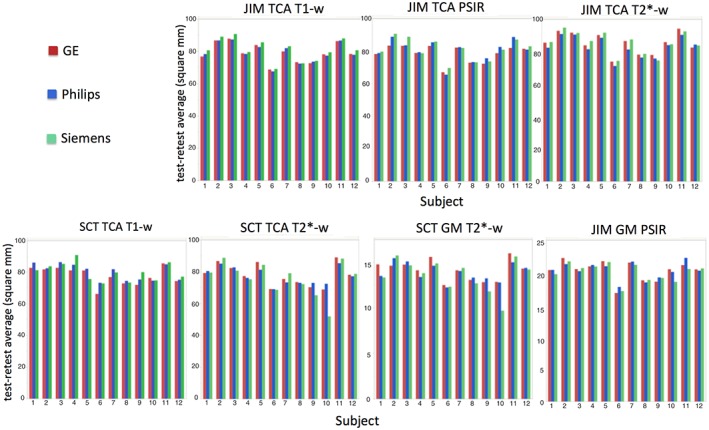
Plots reporting the mean of the values measured for the TCA and GM area in the test and retest acquisitions (y‐axis) for the 12 healthy subjects (x‐axis), for all the combinations of segmentation method and protocol (indicated above the plot).
According to the ANOVA, there was a statistically significant difference for TCA measurements on the protocol segmented with JIM, compared to both PSIR (P = 0.0021) and T1‐w (P = 0.0018). No difference was found between the T1‐w and protocols when the segmentation was performed using the SCT (P = 0.18).
Regarding the difference attributable to the segmentation method on the same protocol for TCA measurement, on T1‐w images JIM and SCT were not statistically different (P = 0.66), while there was difference for the contrast (P < 0.0001).
Finally, there were very significant differences in the GM area obtained using the SCT/T2*‐w vs. JIM/PSIR segmentation method/protocol combinations (P < 0.0001). However, no scanner‐related bias was detected for GM areas.
All results are graphed in Fig. 6, where P‐values for the comparisons between different combinations of segmentation method/protocol are also reported.
Figure 6.
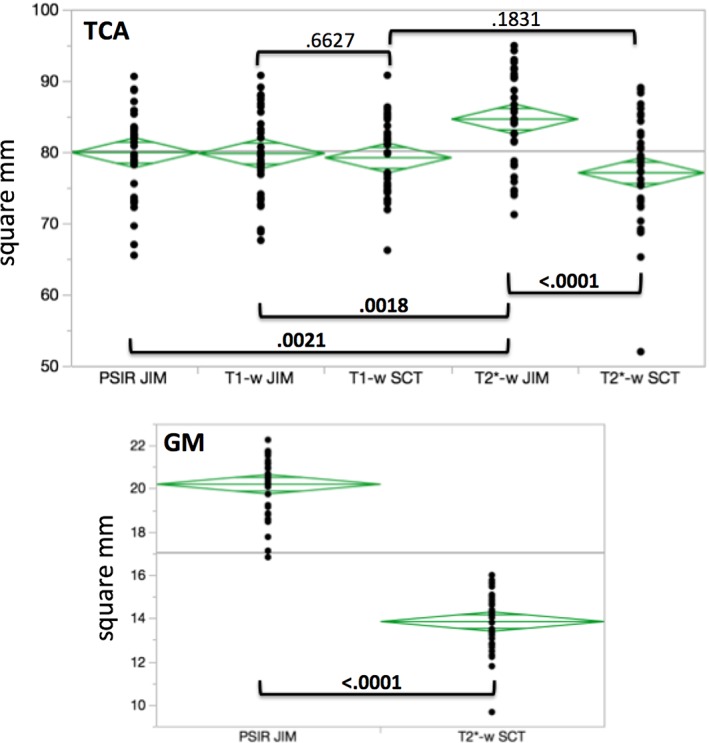
ANOVA for TCA and GM area measured with the different combinations segmentation method/protocol. P‐values for the different couples of comparisons are reported and highlighted in bold when differences were statistically significant (P < 0.05).
Discussion
In this work we present for the first time analysis of a rich MRI dataset, acquired on the same 12 healthy subjects on three 3T scanners produced by the main commercial brands with the most promising protocols for TCA and GM area assessments. A qualitative assessment and quantitative evaluation of CNR and intra‐ and interscanner reliability of area measurements at the C2‐C3 spinal cord level is presented.
For GM delineation, the quality of PSIR images was more consistent than images. These qualitative visual impressions were confirmed by CNR evaluations that showed that GM/WM CNR for PSIR images was higher than for images for all the scanners. The observed tendency for fuzzy appearance of GM on compared to PSIR images may be the result from higher sensitivity of the protocol to susceptibility artifacts and motion.
3D T1‐w images were consistently of good quality for all the scanners in terms of spinal cord/CSF delineation. This is not surprising, considering that the used protocols are optimizations of 3D inversion recovery spoiled gradient echo protocols that have become a standard for atrophy assessment on brain images over more than a decade.
The goal of having comparable CNR across different vendors was overall achieved with the chosen sequences/parameters/hardware, with in general 3D T1‐w protocols giving higher WM/CSF CNR than the other two protocols and PSIR higher GM/WM CNR than protocols. It has to be mentioned that, since there were different hardware configuration/protocol choices, we preferred not to correct CNR for acquisition times/coverage when evaluating CNR at the C2‐C3 level. It has also to be mentioned that different resolutions can affect the CNR.
For TCA estimates, all protocols performed very similarly in terms of intra‐ and interscanner reliability, for a given segmentation method. The semiautomatic method based on JIM showed better test–retest COV and ICC on both 3D T1‐w and protocols compared with the automatic SCT method.
We therefore think there is not an obvious choice of best protocol if the goal of a study is TCA evaluation. The choice has to be driven by a series of factors and considerations such as the acquisition time that can be spent on a protocol, the spinal cord levels to be covered, the specific hardware available, and the need of assessing TCA alone or GM as well. It is worth noting that protocols gave significant biases in comparison to 3D T1‐w and PSIR protocols for TCA estimates when JIM segmentation was used.
When using JIM, biases across scanners tended to be statistically more significant than when using the SCT; the test–retest COV and biases across scanners with JIM measurements were all bigger than the interoperator variability reported in the previous literature. The statistical sensitivity to scanner model for JIM could be explained considering the lower interoperator and intrascanner variability of JIM‐based segmentations, thereby providing statistical power to detect small biases across scanners.
GM segmentations were performed with the method/protocol couples JIM/PSIR and SCT/. The latter combination gave lower median COVs. There was a statistically significant large bias between values obtained with the two segmentation method/protocol combinations. derived GM areas were much smaller than PSIR derived ones. This could be due to the contrast difference or the higher resolution for the protocols. It has been shown previously that higher resolution PSIR gives smaller partial volume effects on the WM/GM edge and therefore smaller GM area estimates.37
Automatic methods have obvious advantages if compared to manual or semiautomatic methods, but if they often need corrections they are essentially semiautomatic methods. The SCT was not robust as an automatic method on these spinal cord data and had higher failure rates on images than T1‐w images. This explains the low median values, but bigger mean and standard deviations in some cases. In a few cases, the SCT repeated the same error for both test and retest acquisitions of a particular subject, in particular for images (for example, wrong vertebra assignment, or wrong total cord delineation and subsequently GM segmentation). These systematic errors gave a good intrascanner reproducibility of GM area, but it was evident with a visual check that the segmentations were not accurate in both the test and retest acquisitions. We also noticed that the SCT provided a GM segmentation result even when there was no WM/GM delineation (a suboptimal image, but even on a 3D T1‐w images with no GM visible). The SCT can be very useful also in these situations to create a probabilistic GM mask to be used to calculate metrics on other acquisitions/contrasts, but it could give misleading information if used to quantitatively assess the GM area.
These observations could explain why in a published work that tested the SCT GM segmentation method and other methods, the Dice Similarity Coefficients and Jaccard Index indicated moderate overlap of segmentations obtained with SCT with gold‐standard manual segmentations.15
Manually correcting SCT errors was beyond the scope of the present work, but there is clearly room for improving this very useful tool, maybe tuning it to the different specific acquisitions. Manual and semiautomatic methods have the disadvantage of being time‐consuming and can have high interrater variability. Nevertheless, the semiautomatic method based on JIM consistently gave better reliability for TCA estimates on all the tested protocols. For the GM manual segmentations performed with JIM, the observed test–retest COVs were of the same order of magnitude of the interoperator variability previously reported. The statistical power provided by the GM segmentation technique was therefore not sufficient to disentangle all the possible sources of variability in the measurement (scanner, positioning at acquisition, test–retest, interoperator variability of segmentations).
A limitation of this study is that software/hardware varied across scanners. For example, the available GE scanner did not have anterior neck coils, which prohibited the use of parallel imaging. For this reason, the implementation of some protocols on GE was a little different than on Siemens and Philips, where we managed to set more similar protocols. The T1‐w sequence on GE had a smaller field of view (FOV) compared to the other scanners, different bandwidth, but two averages were made to compensate for the CNR lost. Different FOVs can affect the quality of shimming and the CNR, and therefore affect the quality of segmentations. A different choice could have been made; for example, an IR‐SPGR sequence could have been used instead of BRAVO, or increasing the FOV instead of making two averages. Analogous differences due to the lack of parallel acceleration capability were present in the protocol. Also for PSIR, the way the different vendors implement the cardiac gating in the protocol forced differences in the settings on the different scanners.
Another limitation of this study is that we performed analyses with only two segmentation methods. We also decided not to perform segmentation for every protocol/segmentation method combination but constrained our analyses to only those applications already shown to be appropriate for the given segmentation method. Further developments in segmentation methods may help to reduce the variability in the area estimates. These data could be used for testing (and possibly improving) other algorithms.
Other limitations are the absence of a T2‐w protocol, the limited number of subjects, and the fact that only data on healthy subjects were acquired (all choices forced by the very demanding protocol that required an hour of scan on three different scanners in a very short time frame).
While there may be further room for optimization, we believe that our efforts reflect the expected biases and variability due to the choice of scanners, protocols, and segmentation methods.
The present work suggests that multiscanner/multicenter studies for TCA/GM segmentation are feasible with all the techniques explored in the study.
This study may set reference values for the magnitude of the contribution of different effects (scanner, protocol, segmentation method) to TCA/GM area measurement intra‐ and interscanner variability.
The data and results reported in the present study may help in making informed decisions when planning a specific study, depending on the different acquisition settings and study goals.
Further optimization of protocols and segmentation algorithms is warranted and this study can help in determining what are the directions in which the spinal cord MRI community should move along.
Acknowledgments
First and foremost, the authors would like to thank the twelve subjects who participated in this study. Repeating the MRI acquisitions on three different scanners in such a short time frame required significant time and effort. The authors would also like to thank Julien Cohen‐Adad of Polytechnique Montréal for his great work in leading the international effort that defined most of the T1‐w and protocols used in the study, as well as Antonella Castellano, Letterio Salvatore Politi and Andrea Falini of San Raffaele Institute (Milan, Italy), who were fundamental in optimizing the PSIR protocol for Philips scanners. Additionally, the authors would like to express their gratitude for the indispensable support received from Siemens (Gerhard Laub, Sinyeob Ahn, Kevin J. Johnson), Philips (Marcello Cadioli) and General Electric (Suchandrima Banerjee, Patrick D. Koon). Finally, the authors would like to thank William A. Stern, and all the MRI technologists and staff members who with their professionalism, kindness and availability made performing this study possible. This project was supported by the Research Evaluation & Allocation Committee (REAC), School of Medicine, University of California, San Francisco.
References
- 1. Schlaeger R, Papinutto N, Panara V, et al. Spinal cord gray matter atrophy correlates with multiple sclerosis disability. Ann Neurol 2014;76:568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schlaeger R, Papinutto N, Zhu AH, et al. Association between thoracic spinal cord gray matter atrophy and disability in multiple sclerosis. JAMA Neurol 2015;72:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castellano A, Papinutto N, Cadioli M, et al. Quantitative MRI of the spinal cord and brain in adrenomyeloneuropathy: in vivo assessment of structural changes. Brain 2016;139(Pt 6):1735–1746. [DOI] [PubMed] [Google Scholar]
- 4. Freund P, Curt A, Friston K, Thompson A. Tracking changes following spinal cord injury: insights from neuroimaging. Neuroscientist 2013;19:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen‐Adad J, El Mendili MM, Morizot‐Koutlidis R, et al. Involvement of spinal sensory pathway in ALS and specificity of cord atrophy to lower motor neuron degeneration. Amyotroph Lateral Scler Frontotemporal Degener 2013;14:30–38. [DOI] [PubMed] [Google Scholar]
- 6. El Mendili MM, Cohen‐Adad J, Pelegrini‐Issac M, et al. Multi‐parametric spinal cord MRI as potential progression marker in amyotrophic lateral sclerosis. PLoS One 2014;9:e95516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rocca MA, Comi G, Filippi M. The role of T1‐weighted derived measures of neurodegeneration for assessing disability progression in multiple sclerosis. Front Neurol 2017;8:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Leener B, Taso M, Cohen‐Adad J, Callot V. Segmentation of the human spinal cord. Magma 2016;29:125–153. [DOI] [PubMed] [Google Scholar]
- 9. Held P, Dorenbeck U, Seitz J, Frund R, Albrich H. MRI of the abnormal cervical spinal cord using 2D spoiled gradient echo multiecho sequence (MEDIC) with magnetization transfer saturation pulse. A T2* weighted feasibility study. J Neuroradiol 2003;30:83–90. [PubMed] [Google Scholar]
- 10. Yiannakas MC, Kearney H, Samson RS, et al. Feasibility of grey matter and white matter segmentation of the upper cervical cord in vivo: a pilot study with application to magnetisation transfer measurements. NeuroImage 2012;63:1054–1059. [DOI] [PubMed] [Google Scholar]
- 11. Papinutto N, Schlaeger R, Panara V, et al. 2D phase‐sensitive inversion recovery imaging to measure in vivo spinal cord gray and white matter areas in clinically feasible acquisition times. J Magn Reson Imaging 2015;42:698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Papinutto N, Schlaeger R, Panara V, et al. Age, gender and normalization covariates for spinal cord gray matter and total cross‐sectional areas at cervical and thoracic levels: A 2D phase sensitive inversion recovery imaging study. PLoS One 2015;10:e0118576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papinutto N, Datta E, Zhu AH, et al. Multisite feasibility study of spinal cord gray matter and total cord areas measurements on 2D phase sensitive inversion recovery images. In: Proc 24th Annual Meeting ISMRM, Singapore; 2016.
- 14. Alley S, Gilbert G, Gandini Wheeler‐Kingshott CAM, et al. Consensus acquisition protocol for quantitative MRI of the cervical spinal cord at 3T. In: Proc 26th Annual Meeting ISMRM, Paris; 2018.
- 15. Prados F, Ashburner J, Blaiotta C, et al. Spinal cord grey matter segmentation challenge. NeuroImage 2017;152:312–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grussu F, Schneider T, Zhang H, et al. Neurite orientation dispersion and density imaging of the healthy cervical spinal cord in vivo. NeuroImage 2015;111:590–601. [DOI] [PubMed] [Google Scholar]
- 17. Yiannakas MC, Grussu F, Louka P, et al. Reduced field‐of‐view diffusion‐weighted imaging of the lumbosacral enlargement: a pilot in vivo study of the healthy spinal cord at 3T. PLoS One 2016;11:e0164890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gringel T, Schulz‐Schaeffer W, Elolf E, et al. Optimized high‐resolution mapping of magnetization transfer (MT) at 3 Tesla for direct visualization of substructures of the human thalamus in clinically feasible measurement time. J Magn Reson Imaging 2009;29:1285–1292. [DOI] [PubMed] [Google Scholar]
- 19. Papinutto N, Bakshi R, Bischof A, et al. Gradient nonlinearity effects on upper cervical spinal cord area measurement from 3D T1‐weighted brain MRI acquisitions. Magn Reson Med 2018;79:1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z, Yaldizli O, Pardini M, et al. Cervical cord area measurement using volumetric brain magnetic resonance imaging in multiple sclerosis. Mult Scler Relat Disord 2015;4:52–57. [DOI] [PubMed] [Google Scholar]
- 21. Aymerich FX, Auger C, Alonso J, et al. Cervical cord atrophy and long‐term disease progression in patients with primary‐progressive multiple sclerosis. AJNR Am J Neuroradiol 2018;39:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singhal T, Tauhid S, Hurwitz S, Neema M, Bakshi R. The effect of glatiramer acetate on spinal cord volume in relapsing‐remitting multiple sclerosis. J Neuroimaging 2017;27:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yiannakas MC, Mustafa AM, De Leener B, et al. Fully automated segmentation of the cervical cord from T1‐weighted MRI using PropSeg: Application to multiple sclerosis. NeuroImage Clin 2016;10:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim G, Khalid F, Oommen VV, et al. T1‐ vs. T2‐based MRI measures of spinal cord volume in healthy subjects and patients with multiple sclerosis. BMC Neurol 2015;15:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valsasina P, Rocca MA, Horsfield MA, et al. Regional cervical cord atrophy and disability in multiple sclerosis: a voxel‐based analysis. Radiology 2013;266:853–861. [DOI] [PubMed] [Google Scholar]
- 26. Martin AR, De Leener B, Cohen‐Adad J, et al. Monitoring for myelopathic progression with multiparametric quantitative MRI. PLoS One 2018;13:e0195733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin AR, De Leener B, Cohen‐Adad J, et al. Can microstructural MRI detect subclinical tissue injury in subjects with asymptomatic cervical spinal cord compression?. A prospective cohort study. BMJ Open 2018;8:e019809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paquin ME, El Mendili MM, Gros C, et al. Spinal cord gray matter atrophy in amyotrophic lateral sclerosis. AJNR Am J Neuroradiol 2018;39:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasoanandrianina H, Grapperon AM, Taso M, et al. Region‐specific impairment of the cervical spinal cord (SC) in amyotrophic lateral sclerosis: A preliminary study using SC templates and quantitative MRI (diffusion tensor imaging/inhomogeneous magnetization transfer). NMR Biomed 2017;30. [DOI] [PubMed] [Google Scholar]
- 30. Martin AR, De Leener B, Cohen‐Adad J, et al. Clinically feasible microstructural MRI to quantify cervical spinal cord tissue injury using DTI, MT, and T2*‐weighted imaging: assessment of normative data and reliability. AJNR Am J Neuroradiol 2017;38:1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Leener B, Levy S, Dupont SM, et al. SCT: Spinal Cord Toolbox, an open‐source software for processing spinal cord MRI data. NeuroImage 2017;145(Pt A):24–43. [DOI] [PubMed] [Google Scholar]
- 32. Dupont SM, De Leener B, Taso M, et al. Fully‐integrated framework for the segmentation and registration of the spinal cord white and gray matter. NeuroImage 2017;150:358–372. [DOI] [PubMed] [Google Scholar]
- 33. De Leener B, Kadoury S, Cohen‐Adad J. Robust, accurate and fast automatic segmentation of the spinal cord. NeuroImage 2014;98:528–536. [DOI] [PubMed] [Google Scholar]
- 34. Horsfield MA, Sala S, Neema M, et al. Rapid semi‐automatic segmentation of the spinal cord from magnetic resonance images: application in multiple sclerosis. NeuroImage 2010;50:446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kearney H, Yiannakas MC, Abdel‐Aziz K, et al. Improved MRI quantification of spinal cord atrophy in multiple sclerosis. J Magn Reson Imaging 2014;39:617–623. [DOI] [PubMed] [Google Scholar]
- 36. Bischof A, Olney NT, Rosen HJ, et al. Phase Sensitive Inversion Recovery (PSIR) spinal cord imaging as a potential biomarker for Motor Neuron Disease. In: Proc 26th Annual Meeting ISMRM, Paris; 2018. [DOI] [PMC free article] [PubMed]
- 37. Datta E, Papinutto N, Schlaeger R, et al. Gray matter segmentation of the spinal cord with active contours in MR images. NeuroImage 2017;147:788–799. [DOI] [PubMed] [Google Scholar]


