SUMMARY
Abscission is the terminal step of mitosis that physically separates two daughter cells [1, 2]. Abscission requires the endocytic sorting complex required for transport (ESCRT), a molecular machinery of multiple subcomplexes (ESCRT-I/II/III) that promotes membrane remodeling and scission [3–5]. Recruitment of ESCRT-I/II complexes to the midbody of telophase cells initiates ESCRT-III assembly into two rings, which subsequently expand into helices and spirals that narrow down to the incipient site of abscission [6–8]. ESCRT-III assembly is highly dynamic and spatiotemporally ordered, but the underlying mechanisms are poorly understood. Here, we report that after cleavage furrow closure, septins form a membrane-bound double ring that controls the organization and function of ESCRT-III. The septin double ring demarcates the sites of ESCRT-III assembly into rings and disassembles before ESCRT-III rings expand into helices/spirals. We show that septin 9 (SEPT9) depletion, which abrogates abscission, impairs recruitment of VPS25 (ESCRT-II) and CHMP6 (ESCRT-III). Strikingly, ESCRT-III subunits (CHMP4B, CHMP2A/B) accumulate to the midbody, but they are highly disorganized failing to form symmetric rings and to expand laterally into the cone-shaped helices/spirals of abscission. We found that SEPT9 interacts directly with the ubiquitin E2 variant (UEV) domain of ESCRT-I protein TSG101 through two N-terminal PTAP motifs, which are required for the recruitment of VPS25 and CHMP6, and the spatial organization of ESCRT-III (CHMP4B, CHMP2B) into functional rings. These results reveal that septins function in the ESCRT-I-ESCRT-II-CHMP6 pathway of ESCRT-III assembly and provide a framework for the spatiotemporal control of the ESCRT machinery of cytokinetic abscission.
Keywords: abscission, cytokinesis, septins, ESCRT, TSG101, midbody
Graphical Abstract
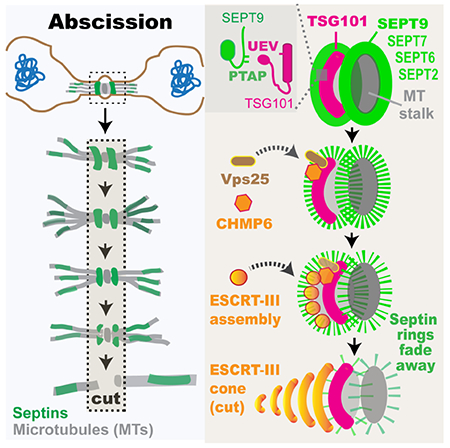
eTOC Blurb
Abscission, the last step of cell division, involves the ESCRT machinery of membrane scission. Karasmanis et al. show that a septin double ring is required for ESCRT-III assembly into functional rings and cones/spirals. Surprisingly, SEPT9 interacts directly and synergizes with TSG101 for proper recruitment and assembly of ESCRT-II/-III subunits.
RESULTS
Septins form a membrane-bound double ring on the midbody of telophase cells
During telophase in the budding yeast Saccharomyces cerevisiae, septins form a filamentous double ring, which is critical for the spatial coordination of factors involved in cytokinesis [9–11]. To examine whether septins have conserved this organization and function in mammalian cells, we imaged the spatiotemporal dynamics of septins in Madin-Darby canine kidney (MDCK) epithelial cells. In stable MDCK cell lines that express fluorescent SEPT9 or SEPT2 [12–14], septins accumulated to the ingressing cleavage furrow. Upon furrow closure, septins reorganized from an hourglass-like structure to a double bar, which was oriented parallel to the plane of cell division, flanking the center of the developing midbody (Figure 1A–B; arrowheads). Additionally, septins decorated microtubule (MT) segments distal to the midbody center (Figure 1B, Videos S1 and S2). Strikingly, the septin double bar became gradually dimmer and disassembled before abscission (Figure 1B and S1A, arrowheads; Videos S1 and S2).
Figure 1. A septin double ring flanks the midbody between cleavage furrow closure and abscission.
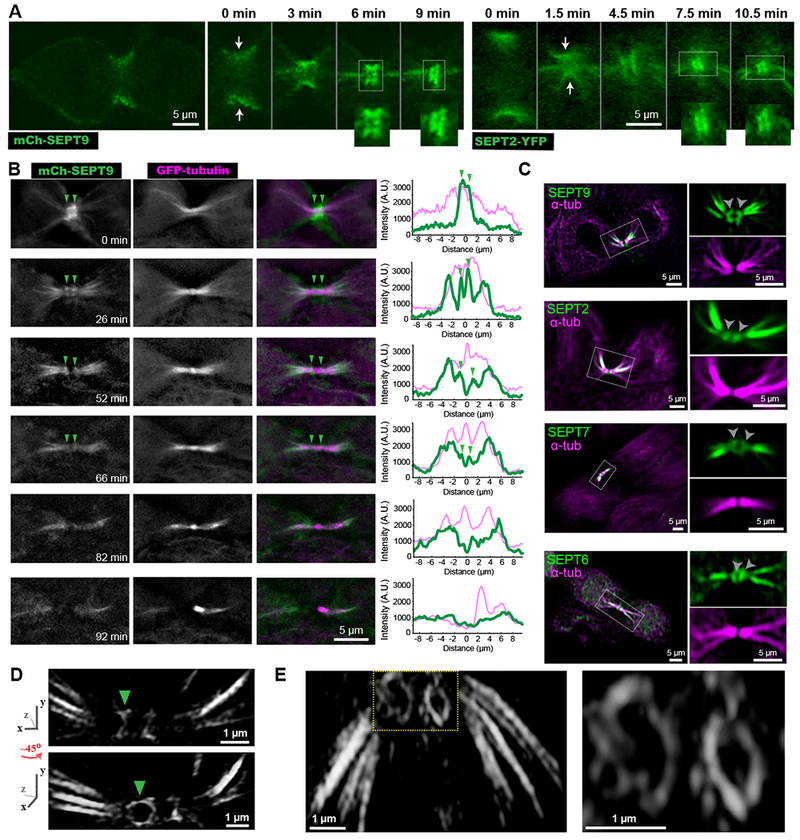
(A-B) Time-lapse images of MDCKs expressing mCherry-SEPT9 (A) and SEPT2-YFP (B) show septins transitioning from an hourglass-like shape (arrows) to a double bar (insets).
(B) Time-lapse frames of MDCK-mCherry-SEPT9 cells expressing GFP-α-tubulin. Arrowheads point to a double septin bar, which becomes progressively dimmer. Plots show a fluorescence line scan along nine microns left and right of the ICB center.
(C) Confocal images of endogenous α-tubulin and SEPT9, SEPT2, SEPT7, SEPT6. Outlined ICBs and double septin bars (arrowheads) are shown in higher magnification.
(D-E) Structured illumination super-resolution images of mCherry-SEPT9 (D) and endogenous SEPT2 (E) were 3D volume rendered and rotated from various angles. A ~45 degree rotation of the ICB around the y-axis (D) reveals that the septin bar (arrowhead) flanking the midbody is a ring. See also Figures S1 and S4, and Videos S1 and S2.
Endogenous septins (SEPT2/6/7/9) showed similar distributions and colocalized with one another as previously observed in interphase cells (Figures 1C and S1B) [15]. Extraction with Triton X-100 prior to fixation resulted in loss of the septin double bar, indicating a membrane-bound structure, while septin localization to MTs was not affected (Figure S1C–D). By combining structured illumination super-resolution microscopy with 3D image rendering, we resolved that the 3D organization of the septin double bar is a dual ring (Figure 1D–E). The septin rings often had an elliptical shape with long and short diameters of 1.1 ± 0.04 μm and 0.6 ± 0.07 μm (n = 8), respectively, which resemble the dimensions of the intercellular bridge (ICB). Hence, mammalian septins form a transient membrane-bound double ring, which flanks the center of the midbody, and marks the stage of telophase between cleavage furrow closure and abscission.
The septin double ring demarcates the recruitment and assembly of ESCRT-II/III proteins
The septin double ring coincides temporally with a stage of telophase, during which the ESCRT machinery of abscission is assembled [16, 17]. Initiated by a double ring of ESCRT-I proteins, which localize to the midbody, ESCRT assembly involves the recruitment of ESCRT-II and ESCRT-III, which assemble into rings that expand laterally to form helices and spirals - also appearing and referred to as cones - that narrow down to the sites of ICB severing [3, 4, 18–21]. ESCRT-III subunits were recently shown to undergo a continuous turnover and remodeling, which is enabled by the ATPase VPS4, raising the question of how ESCRT assembly is spatiotemporally controlled [22, 23].
To investigate whether septins have a role in ESCRT assembly, we first analyzed the spatiotemporal distribution and dynamics of septins with respect to ESCRT-I (TSG101) and ESCRT-II (VPS25), which localize to the center of the midbody as two nearly indistinguishable rings separated by a distance near the resolution limit of light microscopy, and ESCRT-III subunits (CHMP6, CHMP4B), which are further apart forming a discernable dual ring [4]. Strikingly, all ESCRTs overlapped partially with the septin ring structures (Figure 2A). Time-lapse microscopy of cells that stably express GFP-CHMP4B showed that the septin double ring demarcates the sites of ESCRT-III recruitment (Figures 2B–C and S2A, Video S3 and S4). A spatial overlap was observed between the fluorescence intensity curves of mCherry-SEPT9 and GFP-CHMP4B as well as between mCherry-SEPT2 and GFP-CHMP4B/CHMP2A (Figure 2C–D and S2A, asterisks). While initially septins demarcated the recruitment of ESCRT-III subunits, septin ring fluorescence decreased gradually showing an inverse correlation with ESCRT-III, which increased (Figure 2E and S2B). Concomitant with the diminution of septin rings, one of the two ESCRT-III rings generated a cone-like structure (Figure 2C–D and S2B; green arrow). Quantification of septin fluorescence showed a reduction to 18 ± 3% (n = 12) of maximum intensity prior to ESCRT-III cone formation, which marked the site of ICB severing by a single cut (Figure 2F). Notably, disassembly of the dual septin ring always preceded ESCRT-III cone formation, occurring 17 ± 8 and 11 ± 7 minutes (n = 12), respectively, prior to severing (Figure 2G–H). Further analysis indicated that the double septin ring forms synchronously with the arrival of TSG101 after cleavage furrow closure and is present during the recruitment of ESCRT-II/III subunits (Figure S2C). These data indicate that the septin double ring is spatiotemporally coupled to the assembly of the ESCRT machinery prior to forming the cone (helices/spirals), which promotes membrane constriction and fission.
Figure 2. Assembly of the ESCRT machinery of abscission is spatiotemporally coupled to the septin double ring.
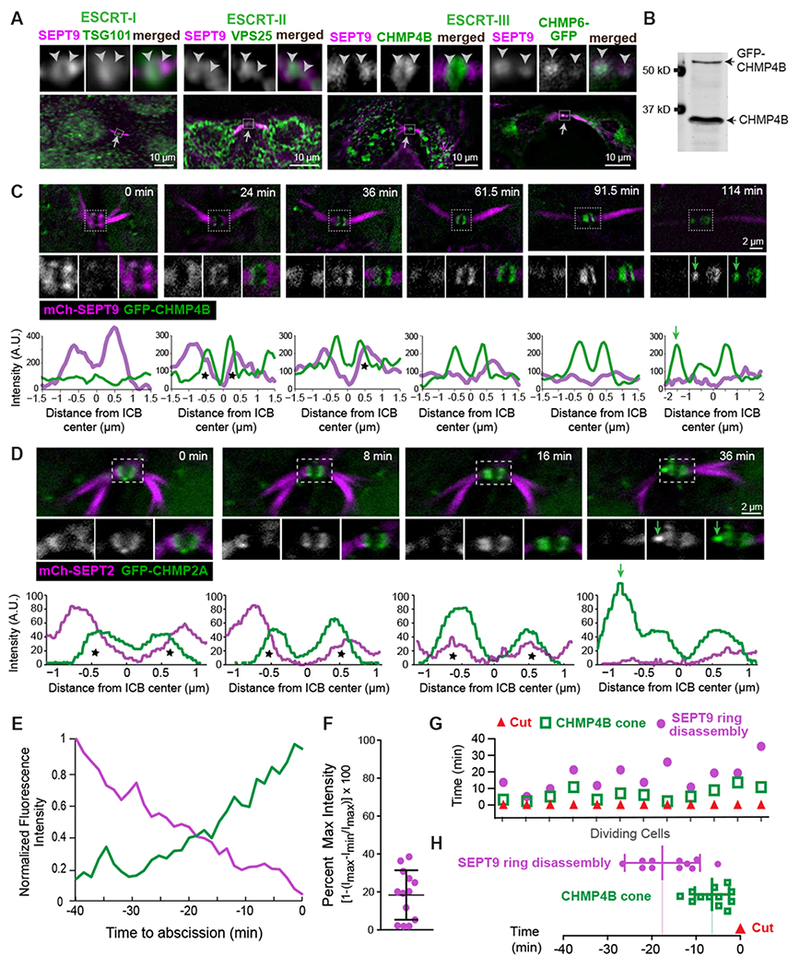
(A) SEPT9 localization with respect to endogenous TSG101, VPS25, CHMP4B and GFP-CHMP6 in the outlined ICB areas (arrows) of MDCK cells. Arrowheads are fixed in position and point to SEPT9-ESCRT overlap.
(B-D) Western blot (B) of MDCK-GFP-CHMP4B cell lysate and time-lapse images of GFP-CHMP4B with SEPT9-mCherry (C) and mCherry-SEPT2 with GFP-CHMP2A (D). Plots show the mean pixel intensity of septins (magenta) and GFP-CHMP2A/CHMP4B (green) across the width of the midbody. Asterisks highlight fluorescence overlap and green arrows point to the formation of ESCRT-III cones.
(E) Mean fluorescence intensity of GFP-CHMP4B and SEPT9-mCherry rings after normalization to a scale of 0 (min) to 1 (max) prior to abscission (t=0; n = 3 cells).
(F-H) Quantification of the fluorescence intensity of SEPT9-mCherry (percent of maximum) in the frame preceding the emergence a GFP-CHMP4B cone (F), and timing of SEPT9-mCherry ring disassembly and GFP-CHMP4B cone formation with respect to the ICB cut (t = 0) in individual cells (G) and as an average ± SD (H; n = 12 cells). See also Figure S2 and Videos S3 and S4.
Septins are required for the functional organization and activation of the ESCRT machinery of membrane fission
To test whether septins function in the assembly and activation of the ESCRT-III machinery, we knocked down the expression of SEPT9 (Figure 3A–B), which has been previously shown to impact abscission without affecting the ingression and closure of the cleavage furrow [24]. In contrast to cells treated with control shRNAs, which completed abscission under 2 h, SEPT9-depleted cells required longer times (2-8 h) and a significant fraction did not undergo abscission after 10 h or died (Figure 3C).
Figure 3. Septin 9 depletion abrogates the recruitment of VPS25 and CHMP6, and impairs proper ESCRT-III assembly.
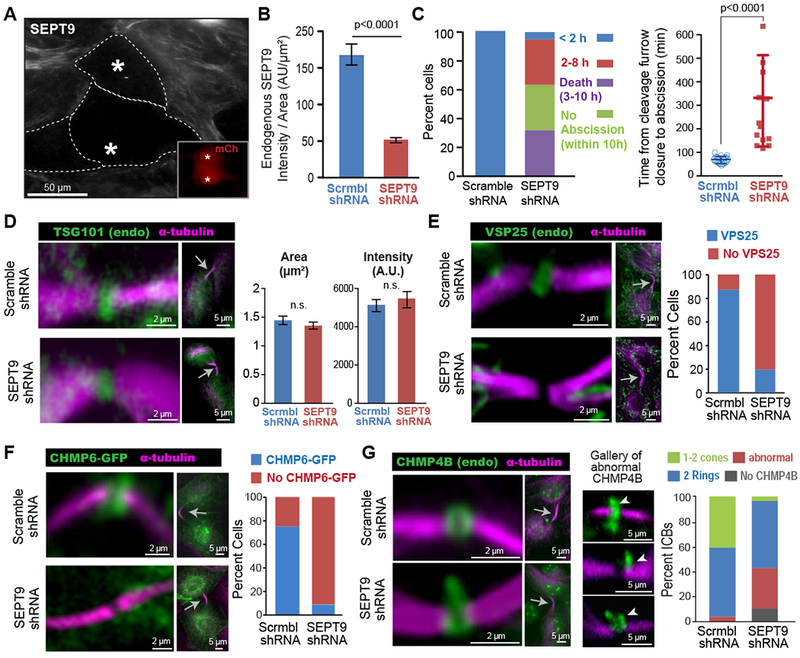
(A-B) Image (A) of SEPT9 in MDCK cells transfected with shRNAs/mCherry (insets; asterisks). Quantification (B) of mean SEPT9 fluorescence intensity per area (± SEM) in cells treated with shRNAs (n = 30-33).
(C) Percentage of total cells that completed abscission under 2 h or within 2-8 h after cleavage furrow closure, and percent of cells that failed to undergo abscission or died within 10 h (n = 19). Plot shows the mean duration (± SD) of completed abscissions in control (n = 19) and SEPT9-depleted cells (n = 13).
(D) TSG101 and α-tubulin in the ICBs (arrows) of shRNA-treated MDCKs. Quantification of the mean area and fluorescence intensity (± SEM) of TSG101 in the center of the ICB (n = 32-34).
(E-F) Images show the localization of a-tubulin, VPS25 (E) and CHMP6-GFP (F) in the ICBs of MDCKs treated with shRNAs. Graph shows the percentage of ICBs with or with no VPS25 (n = 50) and CHMP6-GFP (n = 25-26).
(G) Localization of α-tubulin and CHMP4B in ICBs (arrow) of cells transfected with shRNAs. A gallery of defective CHMP4B phenotypes is shown and arrowheads point to distended accumulations of CHMP4B. Quantification shows percent ICBs without CHMP4B (absent) and with abnormally localized CHMP4B or two rings and 1-2 cones of CHMP4B in control (n = 64) or SEPT9-depleted (n = 86) cells. See also Figure S3.
We asked if the abscission defects in SEPT9-depleted cells were due to mislocalization of ESCRT proteins. The ESCRT-I protein TSG101 was normally recruited and localized to the midbody of SEPT9-depleted cells (Figure 3D). Similarly, the ESCRT-III adaptor ALIX, which initiates a pathway of ESCRT-III assembly parallel to TSG101 [18], was not affected (Figure S3A). In contrast, the ESCRT-II subunit VPS25 was markedly absent from the midbody (Figure 3E). This defect was specific to VPS25 as recruitment of the ESCRT-II proteins VPS22 and GFP-VPS36 was not affected (Figure S3B–C).
Next, we examined the localization of ESCRT-III components. SEPT9 depletion abrogated the recruitment of CHMP6, which is critical for the assembly of ESCRT-III subunits such as CHMP4B [20, 25]. CHMP6-GFP was absent from the ICBs of >90% of SEPT9-depleted cells (Figure 3F). This phenotype was consistent with lack of VPS25 recruitment as VPS25 is posited to recruit CHMP6, providing a physical link between ESCRT-I and ESCRT-III [20, 26, 27]. Despite the lack of CHMP6 recruitment, only a small percentage (<10%) of cells lacked the ESCRT-III proteins CHMP4B, CHMP2B and GFP-CHMP2A (Figure 3G and S3D–F). However, in a significant fraction of ICBs (>20%), CHMP4B lacked the symmetric doublet that flanks the midbody center. Instead, CHMP4B was highly disorganized and distended with bulging accumulations (Figure 3G); CHMP2B and GFP-CHMP2A were similarly disorganized (Figure S3D–F). This abnormal organization of ESCRT-III was also accompanied by a dramatic reduction in the formation of ESCRT-III cones. ICBs with at least one cone of endogenous CHMP4B or CHMP2B were reduced by 70-95% and GFP-CHMP2A was also diminished by >50% (Figure 3G and S3D–F). Despite these defects, the ATPase VPS4, which drives the turnover and remodeling of ESCRT-III [22], was normally recruited and formed a symmetric double ring, but failed to localize in cones given the absence of ESCRT-III cones (Figure S3G–H). These data indicate that the defects in ESCRT-III organization and remodeling were primarily due to lack of VPS25 and CHMP6 recruitment. Therefore, septins are involved in the ESCRT-I-ESCRT-II-CHMP6 pathway of ESCRT-III activation, and they are critical for the functional assembly of the ESCRT-III machinery of cytokinetic abscission.
SEPT9 interacts with TSG101 through two PTAP motifs, which are required for the ESCRT-I-ESCRT-II-CHMP6 pathway of ESCRT-III assembly
Because SEPT9 is required for the proper assembly and function of the ESCRT machinery, we asked whether SEPT9 interacts directly with ESCRT proteins. A prominent ESCRT-interacting and recruiting motif is the sequence Pro-Thr/Ser-Ala-Pro (PTAP), which is present in viral proteins that hijack the ESCRT machinery for budding from the plasma membrane [28–30]. Intriguingly, the N-terminal proline-rich domain of SEPT9 contains two PTAP motifs (aa 176-179 and 199-202), which are not present in any other septins (Figure 4A). Because viral PTAPs are known to interact directly with the ubiquitin E2 variant (UEV) domain of TSG101, we performed in vitro binding assays with recombinant SEPT9 and the UEV of TSG101. SEPT9 bound the UEV and PTAP deletion disrupted the interaction (Figure 4B). Co-immunoprecipitation experiments in MDCK cell lysates showed that SEPT9 interacts with TSG101 in a PTAP motif-dependent manner (Figure 4C–D). This interaction appeared specific to TSG101 as neither VPS25 or CHMP6 co-immunoprecipitated with SEPT9 (Figure S4A).
Figure 4. Assembly of ESCRT-III requires a direct interaction between the UEV domain of TSG101 and PTAP motifs of SEPT9.
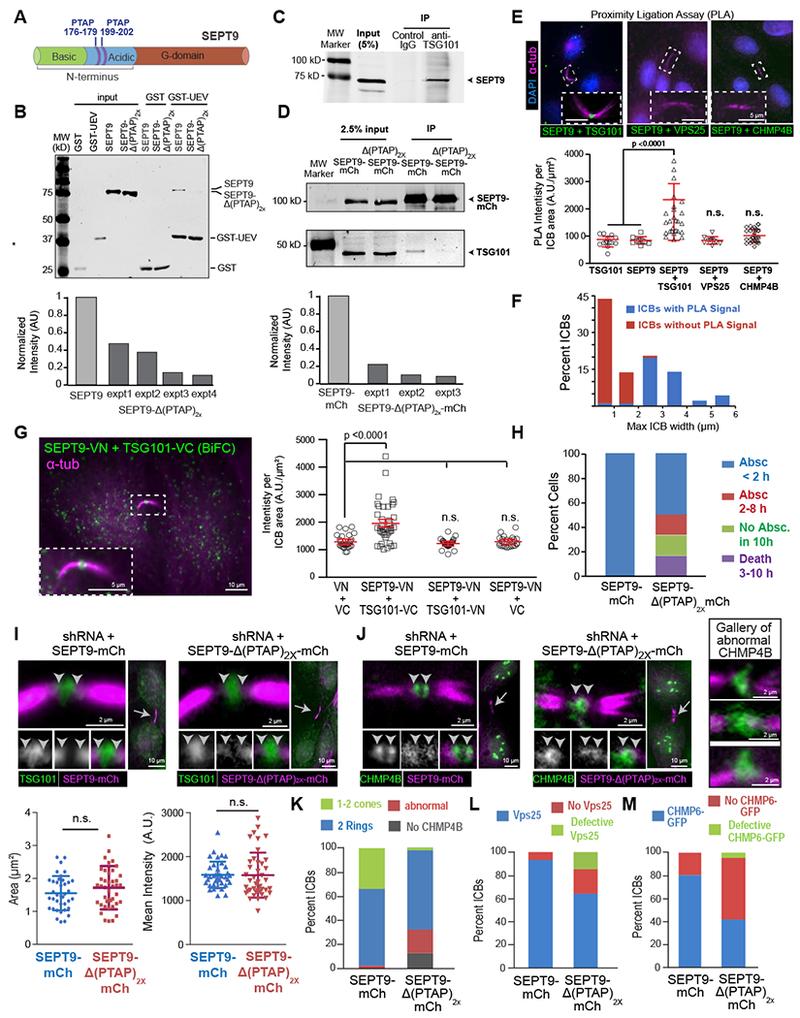
(A) Schematic of SEPT9 (isoform 1) domains and location of PTAP motifs.
(B) Western blots (anti-His/-GST) of protein binding assays with GST or GST-UEV (TSG101 aa1-145) and His-SEPT9 or His-SEPT9-Δ(PTAP)2x. Quantification shows the normalized intensity of His-SEPT9-Δ(PTAP)2x relative to His-SEPT9 (set to 1) in four independent experiments (expt).
(C-D) Western blots (anti-SEPT9) of immunoprecipitations (IPs) with control and anti-TSG101 IgGs (C). SEPT9-mCherry or SEPT9-Δ(PTAP)2x-mCherry were immunoprecipitated with RFP_Trap_A and blotted for mCherry and TSG101 (D). Graph shows normalized intensity of TSG101 in IP of SEPT9-Δ(PTAP)2x-mCherry relative to SEPT9-mCherry (set to 1).
(E) Images show PLA signal in the outlined ICBs of cells labeled with antibodies against SEPT9, TSG101, VPS25 and CHMP4B. PLA signal (mean ± SD) quantification for TSG101 alone (n = 14), SEPT9 alone (n = 10), SEPT9 plus TSG101 (n = 45), SEPT9 plus VPS25 (n = 10), and SEPT9 plus CHMP4B (n = 32).
(F) Distribution of the percentage of ICBs with and without PLA signal (SEPT9-TSG101) based on maximum diameter of their MT stalk (n = 45).
(G) BiFC signal between SEPT9-VN and TSG101-VC and mean fluorescence intensity (± SEM) for MDCKs expressing VN plus VC (n = 29), SEPT9-VN plus TSG101-VC (n = 39), SEPT9-VN plus TSG101-VN (n = 29), and SEPT9-VN plus VC (n = 26).
(H) Quantification of cells that completed abscission <2 h or within 2-8 h and failed to undergo abscission or died within 10 h in SEPT9-depletion rescue experiments with SEPT9-mCherry (n = 16) or SEPT9-Δ(PTAP)2x-mCherry (n = 12).
(I-M) MDCK cells were transfected with plasmids co-expressing SEPT9 shRNA and shRNA-resistant SEPT9-mCherry or SEPT9-Δ(PTAP)2x-mCherry. Cells were stained for endogenous TSG101 (I), CHMP4B (J) and VPS25 (L), or imaged for CHMP6-GFP expression (M). Images show the localization of TSG101 and CHMP4B in the outlined ICBs (arrows). Arrowheads point to SEPT9-ESCRT overlap. Quantifications show the mean surface area and fluorescence intensity (± SEM) of TSG101 (I; n = 38 - 41), and percent of ICBs without or with ESCRTs of normal and abnormal organization in cells stained for CHMP4B (n = 30-37), VPS25 (n = 40-46) and CHMP6-GFP (n = 55-78). See also Figure S4.
To further test whether SEPT9 associated with TSG101 in the ICB of cytokinetic cells, we performed a proximity ligation assay (PLA) and bimolecular fluorescence complementation (BiFC), which report respectively on the nano-proximity (< 30-40 nm) and interaction of two proteins [31, 32]. PLA for endogenous SEPT9 and TSG101 yielded fluorescence signal at the ICB center (Figure 4E). Quantification of the ICBs with PLA signal as a function of the maximum diameter of their MT stalk, which gradually compacts approaching abscission, showed that the preponderance of ICBs without PLA signal were under 2 μm-wide (Figure 4F). These data indicate that SEPT9 and TSG101 are within an interacting proximity in earlier stages of abscission. BiFC further confirmed that SEPT9-TSG101 interact in the ICB (Figure 4G).
To test if this novel TSG101-SEPT9 interaction was functionally important for the assembly of the ESCRT-III machinery, we expressed the deletion mutant SEPT9-Δ(PTAP)2x-mCherry and SEPT9-mCherry (control) while simultaneously depleting the endogenous SEPT9, which was done to circumvent epistasis from the native SEPT9. The abscission defects were fully rescued only by SEPT9-mCherry, indicating that SEPT9-Δ(PTAP)2x-mCherry was functionally impaired (Figure 4H and S4B). SEPT9-Δ(PTAP)2x-mCherry did not impact TSG101 localization (Figure 4I), but disrupted ESCRT-III (CHMP4B, CHMP2B) assembly into double rings and their expansion to cones (Figure 4J and S4C–D). These effects were accompanied by a marked increase in ICBs that lacked VPS25 and CHMP6-GFP (Figure 4L–M). Therefore, a direct interaction between the PTAP motifs of SEPT9 and the UEV domain of TSG101 is critical for the functional assembly of the ESCRT-III machinery of abscission.
DISCUSSION
Our results indicate that SEPT9 interacts with TSG101 in the early stages of abscission and is required for the recruitment of the ESCRT-II protein VPS25, which in turn binds CHMP6, a nucleator of ESCRT-III polymerization. Given that TSG101 contains an auto-inhibitory PSAP motif [33], it is plausible that SEPT9 may relieve the TSG101 autoinhibition enabling the recruitment of VPS25 and in turn CHMP6. However, SEPT9 does not appear to associate with VPS25 or CHMP6, suggesting that SEPT9 may function transiently and/or allosterically. Alternatively, SEPT9 may disengage from TSG101 upon docking of the ESCRT-II proteins VPS22 and VPS36, which interact with VPS25 [26, 34]; despite forming a complex, differences in the spatiotemporal organization of these ESCRT-II proteins have been reported [20]. Interestingly, SEPT9 does not impact the recruitment of VPS22/36 or ESCRT-III subunits such as CHMP4B and CHMP2A/B, which might be mediated by the ALIX pathway of ESCRT recruitment [18].
Surprisingly, SEPT9 is critical for ESCRT-III assembly into symmetric rings and their expansion into cones (helices/spirals), which poses the question of how septins control the higher-order organization of ESCRT-III subunits. Septin rings have been shown to consist of circumferential single- and double-stranded filaments (10-20 nm wide), which resemble ESCRT-III filaments in their arrangement and dimensions [3, 9]. As the septin rings precede the recruitment and assembly of ESCRT-III subunits, septin filaments may provide a physical barrier and/or template during ESCRT-III assembly. Hence, in the absence of septins, ESCRT-III polymerization could expand disorderly into larger membrane areas as indicated by the distended phenotypes, which in part might be due to ESCRT-III in membranes shed from the midbody. If the septin double ring controls physically the assembly of ESCRT-III into rings, it would be advantageous if this function were coupled to CHMP6, which promotes the clustering of CHMP4B into nucleation seeds and catalyzes ESCRT-III polymerization [25, 35]. SEPT9-dependent recruitment of CHMP6 suggests that ESCRT-III polymerization commences alongside the septins rings, which may provide a physical framework for the higher-order assembly of ESCRT-III.
Lateral expansion of ESCRT-III into the cones of abscission occurs concomitantly with the disassembly of septin rings, raising the possibility that septin rings may impede precocious ESCRT-III cone formation and abscission. However, despite the disassembly of the septin double ring, a single cone develops first in most cells, leading initially to a single cut. Moreover, SEPT9-depletion does not result in premature cone formation or ICB constriction/severing. Hence, septin ring disassembly alone cannot determine the timing of ESCRT-III cone emergence.
Previous studies have suggested that septins function in abscission by synergizing with anillin to promote ICB formation and elongation [36, 37], regulating the localization of the exocyst complex, which mediates vesicle docking, and destabilizing the MTs of the midbody [24, 38]. Consistent with a previous study [36], SEPT9 knock-down did not affect ICB formation and elongation. Interestingly, this study suggested that constriction of the ICB by anillin-septin filaments played a role in the recruitment of ESCRT-III (CHMP4B) to the abscission site [36]. However, our data show that septins function upstream of the abscission site during the initial assembly of ESCRT-III into rings. Although we cannot rule out that septins function in the vesicular delivery of ESCRT proteins to the ICB membrane via the exocyst complex, the interaction of SEPT9 with TSG101 is indicative of a direct role in the assembly of membrane-bound ESCRT complexes. Of note, septin knock-down has been shown to disrupt TSG101 association with endosomes and the biogenesis of multivesicular bodies, whose inward vesiculation is driven by membrane-associated ESCRTs [39]. Regarding the destabilization of midbody MTs, septins colocalized with MT segments that are distal to the midbody of MDCK cells and were absent from the sites of MT severing (Figure S4E). Lastly, SEPT9 depletion did not appear to affect the clearance of F-actin from the ICB (Figure S4F–G), which affects ESCRT-III recruitment and abscission [40, 41].
In summary, our results have uncovered septins as a key partner of the ESCRT machinery, shedding new light on how ESCRT assembly is spatiotemporally controlled. This new function of septins stems from their organization into a double membrane-bound ring, which is evolutionarily conserved between the telophase of yeast and mammalian cells. Although ESCRT complexes do not have a critical role in yeast cytokinesis, our findings highlight an evolutionary adaptation of the septin double ring for the ESCRT mechanisms of mammalian abscission.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elias T. Spiliotis (ets33@drexel.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Tissue culture and transfections
The MDCK type II/G cell strain was generated from the kidney of an adult female cocker spaniel. Authenticated low-passage stocks of the original II/G strain were passed on to our lab by Dr. W. James Nelson (Stanford University); MDCK-II cells are only available by the European Collection of Authenticated Cell Culture (ECACC 00062107), but the exact strain is unclear. MDCK-II/G cells were used to generate the stable MDCK-SEPT2-YFP [12], MDCK-mch-SEPT9 [42], MDCK-mCherry-SEPT2 [13] , MDCK-GFP-SEPT6 and MDCK-GFP-CHMP4B cell lines, which were maintained in low glucose DMEM with 1 g/liter NaHCO3 and 10% FBS, at 37°C in a 5% CO2 incubator. Cell lines were routinely tested for mycoplasma, maintained at sub-confluent densities and in low passage numbers, and monitored for proper morphology and expression of fluorescent chimeras. Typically, 80 X 103 cells were seeded on 22 mm glass coverslips (VWR) or 35 mm MatTek glass dishes coated with 30 μg/mL type I bovine collagen (Advanced Biomatrix). Cells were transfected with 0.5-1 μg plasmid DNA using Lipofectamine 2000 (Invitrogen) for 24-48 h prior to imaging. For live-cell imaging, cells were switched into phenol red-free media supplemented with 30 mM Hepes (Invitrogen) and the dishes were sealed with parafilm.
METHOD DETAILS
Plasmids and cloning
Plasmids encoding for fluorescently tagged GFP-TSG101 and GFP-CHMP4B were a gift from Dr. Paul Bieniasz (The Rockefeller University, NY) and were cloned into the pCR3.1 vector. Fluorescently labeled GFP-CHMP2A and CHMP6-GFP were purchased from addgene (CHMP2A_GFP_N_term; #31805 and CHMP6_GFP; #31806, respectively, deposited by the Daniel Gerlich lab) [3]. GFP-VPS25, GFP-VPS36 and GFP-VPS22 were a gift from Dr. Natalie Elia (Ben Gurion University of Negev, Israel) and were cloned into the pEGFP-C1 vector [20]. pEGFP-C1-VPS4 was a kind gift from Dr. Wes Sundquist (University of Utah School of Medicine, UT). pEGFP-C1-α-tubulin (GFP-α-tubulin) was a gift from J. Nelson (Stanford University, CA) [13].
The plasmids for the bimolecular fluorescence complementation (BifC) assay TSG101-VC and SEPT9_VN were made as follows: human TSG101 (GenBank accession number: NM_006292, derived from pCR3.1-GFP-TSG101; see above) was sub-cloned into the pBiFC-VN173 (addgene #22010; deposited by the Chang-Deng Hu lab) and pBiFC-VC155 (addgene #22011; deposited byt the Chang-Deng Hu lab) vectors by PCR amplification, with KAPA HiFi HotStart DNA polymerase (KK2502; KAPA BIOSYSTEMS) and the primers 5’- AAAAAAGAATTCAATGGCGGTGTCG-3’, 5’-TTTTTTGTCGACGTAGAGGTCACT-3’ and 5’- AAAAAAGAATTCAAATGGCGGTGTCG-3’, 5’-TTTTTTCTCGAGTGTAGAGGTCACT-3’, respectively. The amplified PCR fragments were cloned into the EcoRI-SalI and EcoRI-XhoI sites of pBiFC-VN173 and pBiFC-VC155, respectively. Human SEPT9 (GenBank accession number NM_001113491) was sub-cloned to pBiFC-VN173 vector by PCR amplification, with KAPA HiFi HotStart DNA polymerase (KK2502; KAPA BIOSYSTEMS) and the primers 5’- AAAAAAAAGCTTATGAAGAAGTCT-3’ and 5’- TTTTTTGTCGACCATCTCCGGGGC-3’. The amplified PCR fragment was cloned into the HindIII and SalI sites of pBiFC-VN173.
The plasmids used to create the stably expressing MDCK mcherry-SEPT2, mCherry SEPT9, SEPT2-YFP and GFP-SEPT6 have been described previously and are as follows: pmCherry-C1-SEPT2 [13], pmCherry-C1-SEPT9 [42]), pYFP-N1-(mouse) SEPT2 [12], and pEGFP-C3-SEPT6 [43]. Canine SEPT9 (GenBank accession number XM_022423896.1) was targeted using pSuper-mCherry shRNAs described previously [42] (see also the rescue construct below).
The plasmid for rescue of SEPT9 expression was generated by a three-step molecular cloning process. First, the canine SEPT9 shRNA oligos 5’-GATCCCCGACCGGCTGGTGAACGAGAAGTTTTCAAGAGAAACTTCTCGTTCACCAGCCGTCTTTTTA -3’ and 5’-AGCTTAAAAAGACCGGCTGGTGAACGAGAAGTTTCTCTTGAAAACTTCTCGTTCACCAGCCGGTCGGG -3’ were annealed into the BamHI and HindIII sites of pGFP-V-RS vector. Next, the human SEPT9 (GenBank accession number NM_001113491) mcherry sequence was cloned into the BgIII and NotI sites of the same vector and finally, the SEPT9 sequence was mutated at five different sites to create silent mutations that would not be recognized by the shRNA. Mutations were done using the QuikChange II Site-Directed mutagenesis kit (Agilent Technologies) using the primers 5’-GATGAGGACTCGGAGGACCGACTAGTAAATGAAAAGTTCCGGGAGATGATCCC -3’ and 5’-GGGATCATCTCCCGGAACTTTTCATTTACTAGTCGGTCCTCCGAGTCCTCATC -3’. The plasmid for rescue of SEPT9 lacking the two PTAP motifs (aa 176-179/199-202), SEPT9-Δ(PTAP)2x -mCherry, was generated using a two-step KAPA Biosystems Site-Directed mutagenesis protocol. The first PTAP motif (176-179 aa) was deleted using KAPA HiFi HotStart DNA polymerase (KK2502; KAPA BIOSYSTEMS) and the primers 5’-GTCCCCGAGGTGGCCACCGACGCA-3’ and 5’-TGCGTCGGTGGCCACCTCGGGGAC-3’. The amplified PCR products were treated with DpnI (R0176L; New England Biolabs) for 1h at 37°C before the reaction was heat inactivated at 80°C for 20 min and subsequently transformed in E.coli DH5a cells. The construct with the first PTAP sequence deletion (aa 176-179) was used as a template to delete the second PTAP motif (aa 199-202) employing the protocol described above with the primers 5’-CCTGCTGAGGCGAGCCCAGCCCAG-3’ and 5’-CTGGGCTGGGCTCGCCTCAGCAGG-3’.
pET28a- hSEPT9, which encodes for 6x His-SEPT9 (GenBank accession number NM_001113491) has been described previously [44].
His- SEPT9-Δ(PTAP)2x was produced by using the previously described His-SEPT9 [44] (GeneBank accession number NM_001113491) as a template and the identical steps and primers as described for Rescue SEPT9-Δ(PTAP)2x–mCherry.
GST- TSG101 UEV was produced by PCR amplifying aa 1-145 from GFP-TSG101 (see above), with KAPA HiFi HotStart DNA polymerase (KK2502; KAPA BIOSYSTEMS) and the primers 5’-AAAAAAAAGCTTATGGCGGTGTCGGAGAG-3’ and 5’-TTTTTTCTCGAGTCAAGGACGAGAGAAGAC-3’. The amplified PCR fragment was cloned into the HindIII and XhoI sites of pGEX-KT-ext.
Stable cell lines
MDCK-GFP-SEPT6 and MDCK-GFP-CHMP4B stable cell lines were created similarly to the previously described MDCK-SEPT2-YFP [12], MDCK-mCherry-SEPT9 [42] and MDCK-mCherry-SEPT2 [13] cell lines. In brief, MDCK cells were transfected with pEGFP-C3-SEPT6 or GFP-CHMP4B (see above) with Lipofectamine 2000 and selected for G418 resistance by continuous culturing in DMEM media supplemented with 1mg/mL geneticin (Invitrogen). After two weeks, cells were sorted based on expression levels and clones were selected.
Protein expression and purification
Recombinant GST, GST-TSG101-UEV, His-SEPT9 and His-SEPT9 Δ(PTAP)2X were transformed into E.coli Bl21 (DE3) (Invitrogen). Bacterial cultures were grown to OD600 of 0.5 and induced with 0.5 mM IPTG for 5 h at 22°C. Cultures were centrifuged at 4,000 rpm for 20 min at 4°C.
Cell pellets for His-SEPT9 and His-SEPT9 Δ(PTAP)2x were resuspended in lysis buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM PMSF, 1 mg/ml lysozyme and 10 mM imidazole and lysed by sonication (10 sets of 15 pulses on ice with 30 s interval between each set). Cell lysates were centrifuged at 13,000 rpm for 30 min at 4°C and passed through a 0.45 μm pore filter. Supernatants were loaded onto gravity flow columns with Ni-NTA agarose beads (745400.25; Macherey-Nagel), which were equilibrated with 10 ml lysis buffer. Columns were washed with 30 ml washing buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 10 mM imidazole). Proteins were eluted in elution buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol and 250 mM imidazole. Cell pellets for GST and GST-TSG101-UEV were resuspended in lysis buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT, 10% glycerol, 0.1% Triton X-100, 1 mM PMSF and 1 mg/ml lysozyme and lysed by sonication (10 sets of 15 pulses on ice with 30 s interval between each set). Cell lysates were centrifuged at 13,000 rpm for 30 min at 4oC and passed through a 0.45 μm pore filter. Supernatants were loaded onto gravity flow columns with Glutathione agarose beads (16100; Thermo Scientific) equilibrated with 10 ml lysis buffer. Subsequently, columns were washed with 30 ml washing buffer (50 mM Tris pH 8.0, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT, 10% glycerol) and proteins were eluted in elution buffer containing 50 mM Tris pH 8.0, 150 mM NaCI, 2 mM MgCl2, 5 mM DTT, 10% glycerol and 10 mM glutathione. All proteins were dialyzed overnight at 4°C in buffer containing 50 mM Tris pH 8.0, 150 mM NaCl and 10% glycerol.
GST-pull downs, immunoprecipitations and western blots
Pull down assays between recombinant purified GST- and His-tagged proteins were performed by incubating 10 μg of GST or GST-TSG101-UEV with 20 μl glutathione agarose beads (16100; Thermo Scientific) for 2 hours at 4°C. Subsequently, beads were washed twice with GST-pull down buffer (50 mM Hepes pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 1 mM PMSF, 5 mM DTT, 2 mM EGTA and 10% glycerol) and were incubated with 10 μg His-SEPT9 or His-SEPT9 Δ(PTAP)2x for 2 hours at 4°C. Beads were washed five times with GST-pull down buffer before they were resuspended with loading buffer and boiled. Samples were loaded onto 10% SDS-PAGE gels and transferred to a nitrocellulose membrane. Membranes were blocked with 5% non-fat dry milk and 1% BSA for 1 hour at room temperature. Membranes were washed with PBS-T (PBS1x/0.1% Tween 20) and incubated with mouse antibody against 6xHis tag (1:2,000; Qiagen) and rabbit antibody against GST tag (1:10,000; Santa Cruz) diluted in PBS-T over night at 4°C. Subsequently, membranes were incubated with anti-mouse or anti–rabbit secondary antibodies (LiCor) for 1 hour at room temperature, before scanning with an imaging system (Odyssey; LICOR).
Co-immunoprecipitations of SEPT9 and endogenous TSG101 were performed by lysing untransfected MDCK and MDCK-mCherry-SEPT9 cells (~ 8X106) for 1h at 4Ό in IP lysis buffer (25 mM HEPES, pH 7.6, 150 mM NaCl, 0.4 % NP40, 1 mM EDTA , 5% glycerol, 1 mM PMSF and 1X protease inhibitors) (EMD Millipore, Darmstadt, Germany). Cell debris was cleared by spinning at 16,000 x g for 20 min and supernatants were subsequently incubated with 20 μl of RFP-Trap®_A beads (ChromoTek) overnight at 4°C. Beads were washed 5X with 5 bead volume in lysis buffer, boiled in SDS loading buffer (2x bead volume) and run on a 10% SDS-PAGE gel. Gels were transferred to a nitrocellulose membrane, which was subsequently blocked with PBS-T (PBS1X and 0.1% Tween-20) containing 5% milk and 1% BSA for 1h RT. The same membrane was blotted sequentially with rabbit anti-TSG101 (1:3000; Proteintech), rabbit anti-CHMP6 (1:3000; Proteintech), rabbit anti-VPS25 (1:3000; Proteintech) and mouse anti-mCherry (1:10000; abcam).
Coimmunoprecipitations of endogenous TSG101 with endogenous SEPT9 was done by lysing untransfected MDCK cells in IP lysis buffer. The cell extracts were incubated overnight at 4°C with 1.5 μg of mouse anti-TSG101 antibody (Santa Cruz; Sc c-2) or control mouse IgG (Santa Cruz), followed by a 16h incubation at 4°C with 30 μl Protein A/G agarose beads (Santa Cruz). After washing with IP lysis buffer, the reactions were run on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane as described above. The membrane was blotted with rabbit anti-SEPT9 (1:3000).
Co-immunoprecipitations of rescue SEPT9-mcherry or SEPT9-Δ(PTAP)2x-mCh with endogenous TSG101 was performed by transfecting MDCK cells with the “rescue” constructs and lysing the cells 24 hours after transfection in IP lysis buffer. The cell extracts were incubated overnight at 4°C with 20 μl of RFP-Trap®_A beads (ChromoTek), run on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane as described above. The membrane was blotted first with rabbit anti-TSG101 (1:3000; Proteintech) and then mouse anti-mCherry (1:10000; abcam) antibodies. Subsequently, membranes were incubated with goat-anti rabbit IRDye® 680RD or goat anti-mouse IRDye® 800RD secondary antibodies (LICOR) for 1 hour at room temperature, before scanning with the Odyssey imaging system (LICOR).
To quantify the expression levels of the stable MDCK cells lines expressing GFP-CHMP4B, cells were lysed under stringent lysis conditions (1% SDS, 0.4% NP-40, 10mM Tris-HCl pH 7.5, 2mM EDTA) and extracts were collected as described above. Samples were run on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane and blotted with either rabbit anti CHMP4B (1:3000, Santa Cruz).
Antibodies and reagents
MDCK cells were immunostained with the following antibodies: rabbit anti-SEPT9 (1:200; Proteintech Group), mouse anti-α-tubulin (DM1α, 1:500; SIGMA), rabbit anti-SEPT2 (a5N5 1:300; gift from Makoto Kinoshita, Nagoya University,Japan), rabbit anti-SEPT7 (1:300; IBL America), rabbit anti-SEPT6 (1:300; gift from Makoto Kinoshita, Nagoya University,Japan), mouse anti-SEPT9 (1:200; EMD Millipore), rabbit anti-CHMP4B (1:100; Proteintech Group), mouse anti-ALIX (1:100; Santa Cruz), mouse anti-TSG101 (sc C-2 1:100; Santa Cruz), rabbit anti-VPS25 (1:200; AbClonal), rabbit anti-VPS22 (1:200; AbClonal), rabbit anti-CHMP2B (1:200; AbClonal). Secondary antibodies (1:200) were purchased from Jackson ImmunoResearch Laboratories and included donkey anti-mouse, or -rabbit antibodies conjugated with AMCA, Alexa 488, Alexa 594 or Alexa 647. Actin was stained using Alexa Fluor 488 phalloidin (1:100; Invitrogen). Western blots were performed with rabbit anti-TSG101 (1:3000; Proteintech Group), rabbit anti-VPS25 (1:3000; Proteintech Group), rabbit anti-CHMP6 (1:3000; Proteintech Group), rabbit anti-CHMP4B (1:3000; Santa Cruz), mouse anti-mCherry (1:10000; abcam), mouse anti- His tag (1:2000; Qiagen) and rabbit anti-GST (1:10000; Santa Cruz).
Immunofluorescence
MDCK cells fixed in warm fixation buffer (4% PFA, 60 mM Pipes-KOH, pH 6.9, 25 mM Hepes, 10 mM EDTA, and 2 mM MgCl2) for 10 min, permeabilized with PBS containing 0.1% Triton X-100 for 10 min and blocked with blocking buffer (PBS 1X, 2% BSA) for an additional 30 min. Primary antibodies were diluted in blocking buffer and spun at 100,000 g for 10 min at 4°C before adding on cells. Primary antibodies were incubated for 3 h at room temperature or 4°C overnight. Secondary antibodies were similarly diluted in blocking buffer, spun before adding on cells and incubated for 1 h in room temperature. Samples were mounted in FluorSave hard mounting medium (EMD Millipore CALBIOCHEM).
For the extraction experiments in Fig S1B–C, MDCK -mCherry-SEPT2 or MDCK-mCherry-SEPT9 cells were incubated in pre-warmed PHEM (60 mM Pipes-KOH, pH 6.9, 25 mM Hepes, 10 mM EDTA, and 2 mM MgCl2) buffer with or without 0.1% Triton X-100 for 30 seconds prior to fixation. For endogenous ESCRT (TSG101, VPS25, VPS22, CHMP2B and CHMP4B) staining, cells were incubated with prewarmed PHEM buffer containing 0.1% digitonin for 60 seconds prior to fixation, resulting in a gentle membrane extraction. For cells transfected with plasmids co-expressing mCherry and shRNAs, staining for endogenous ESCRTs (Figure 3) was performed by fixation in PHEM buffer containing 4% PFA and 0.1% digitonin to gently extract the cells while maintaining mCherry fluorescence.
Co-stains of rabbit anti-SEPT9 and rabbit anti-VPS25, -CHMP4B and -CHMP2B (Figure 2A) was done using the Zenon™ Alexa Fluor™ 568 Rabbit IgG Labeling Kit (Invitrogen), according to the manufacturer’s instructions. In brief, cells were fixed, permeabilized, blocked and stained for SEPT9 with an anti-rabbit-Alexa488 secondary antibody (Jackson ImmunoResearch Laboratories) as described above. Before mounting the samples, 1 μg of rabbit anti VPS25, CHMP4B or CHMP2B antibody was sequentially incubated with 5 μl of Zenon® rabbit IgG labeling reagent and the Zenon® blocking reagent for 5 minutes at room temperature. The complexes were then applied to the samples and incubated for 1h at room temperature. After washing with 1X PBS, cells were briefly fixed with prewarmed 2% PFA in PBS for 5 minutres and mounted in Fluorsave hard mounting medium (EMD Millipore CALBIOCHEM).
Proximity ligation assay
The proximity ligation assay was performed using the Duolink® PLA fluorescence kit (Sigma), according to the manufacturer’s instructions. In brief, cells were fixed and permeabilized as described above. Next, samples were incubated with the Duolink® blocking solution for 1 h at 37°C. Primary antibody mixtures of rabbit anti-SEPT9 and mouse-TSG101 or mouse anti-SEPT9 with Rabbit anti-VPS25 or anti-CHMP4B were diluted in Duolink® blocking solution and added to the cell samples for an overnight incubation at 4°C. The next day, samples were incubated with a 1:5 dilution of PLA mouse-Plus and rabbit-Minus Probes for 1 hour in a preheated humidity chamber (37°C). Subsequently, the probes were ligated by adding Ligase, diluted 1:40 in 1X ligation buffer (sigma) and incubating the samples for 30 minutes in a preheated (37°C). humidity chamber. After washing, the signal from the ligated probes was amplified with a polymerization step, by adding 1:80 Polymerase enzyme diluted in 1X Duolink® Amplification buffer and incubating the samples at 37°C for 100 minutes. After extensive washes, cells were mounted in Duolink® PLA mounting media with DAPI and coverslips were sealed using nail polish.
Microscopy
Fixed samples were imaged on a Zeiss AxioObserver Z1 inverted microscope with a Zeiss 63x/1.4 NA oil objective, a Hamamatsu Orca-R2 CCD camera and the Slidebook 6.0 software. Alternatively, fixed or live samples were imaged (Figures 1A (mch-SEPT9), 1C, 2A , 2E–H, 3C, S4B) using spinning disk laser microscopy on an Olympus IX83 equipped with a 60X/1.3 NA silicone oil objective, an iXon Life 888 EMCCD camera, green and red laser lines (U-M7553 U-FBNA; BX3 BLUE NARROW and U-M7673 U-FGNA; BX3 GREEN NARROW), a temperature and CO2 controlled environmental chamber (IX3-ILL) and the Olympus CellSens software. Live-cell imaging of MDCK cells (Figures 1A (SEPT2-YFP), 1B, 2C, 2D, S1A, S2A-C, S4E) was also performed at 37°C on a wide-field Olympus IX-81 equipped with a motorized stage (ProScanII; Prior), a Hamamatsu Orca-Flash4.0 (sCMOS) camera, an Olympus 60X/1.40 NA objective, a custom built stage-top chamber with a temperature controller (Air-Therm ATX; World Precision Instruments) and the SlideBook 6.0 software. Super resolution 3D structured illumination microscopy (SIM) imaging was performed with the OMX V4 microscope (GE Healthcare) using a 60X/1.42 NA objective, a z-step size of 0.125 μm and immersion oil with refractive index 1.514 (GE Healthcare).
Experimental Design
Replication and reproducibility were ensured through repetition of experiments by the same author or multiple authors independently of one another. Key results were reproduced through the use of independent and alternative approaches (e.g., staining for endogenous proteins to corroborate results with fluorescent chimeras, use of BiFC to corroborate results of proximity ligation assay, colocalization and protein-binding data). Multiple ESCRT-III subunits (CHP2A/B, CHMP4B) were also used to independently verify phenotypes of ESCRPT-III assembly. Characterization of spatiotemporal dynamics of septins with respect to ESCRTs was reproduced independently by three authors of the paper (E.P.K., D.H., J.R.B.), who performed time-lapse imaging experiments at separate times of the project. Imaging for phenotypes from knock-down experiments was performed in replicates of two or three, and repeated two to four times on different days using cells from different passages. The SEPT9-TSG101 interaction was biochemically confirmed with in vitro GST pull downs of recombinant proteins (four replicates on different days), endogenous and stable cell-line immunoprecipitations (four different experiments), immunoprecipitation of expressed SEPT9 (four replicates), proximity ligation assays (3 replicates) and a BiFC assay (2 replicates). Sample sizes were of a minimum of 25 cells from at least two different replicates per each experiment. Key results with more than two replicates had a proportionally larger sample size (n = 50-70). Sample variation stemmed from the varying number of cytokinetic cells per each sample and the levels of expression of fluorescent chimeras, which led to exclusion of data from cells that over-expressed GFP-tagged ESCRT-III subunits. Owing to these variations, no sample randomization or blinding were performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image processing and fluorescence quantifications
In Figures 2A, 4I–J, S4C, fluorescence levels in zoomed-in crops of the ICB center were adjusted uniformly but separately from the corresponding lower magnification images of whole cells, in which ESCRT fluorescence in the midbody was dimmer due to the autocontrast with the brighter intensities in the cell body compartments. In Figures 1B, S1A and 2C, intensity to distance plots were generated by using Fiji’s rectangular selection (Figure 2C) or straight-line tool (Figure 1B, S1A) and Plot Profile function. 3D SIM images in Figure 1D–E were imported into the Volocity software (PerkinELMER) and rotated to make the septin rings visible. Ring diameters were measured in Volocity using the Measure Distances function. In Figure 2D and S2A, ESCRT-III and septin average ring intensities across the width of the midbody were plotted using a custom-made MATLAB (MathWorks) code. The intracellular bridge (ICB) area containing the ring and MT-associated septins was isolated and the septin and ESCRT-III channels were converted into a matrix of intensity values. After background subtraction, the intensity values were averaged at every position across the ICB and plotted in MATLAB. For Figure 2E and Figure S2B, MDCK-GFP-CHMP4B cells transfected with SEPT9-mcherry and MDCK-mCherry-SEPT2 cells transfected with GFP-CHMP4B (n=3) were imaged live with 90 second or 2-minute intervals, respectively, until abscission. The mCherry-SEPT9 or mCherry-SEPT2 and GFP-CHMP4B rings in the ICB were masked by fluorescence thresholding for each timepoint and the mean intensities were measured using Fiji or Slidebook 6.0, respectively. After normalizing, the average intensities of the three cells for each timepoint was calculated and plotted in Excel. In Figure 2F–H, MDCK-GFP-CHMP4B cells expressing SEPT9-mcherry were imaged live with 90 second intervals until abscission (n=12). The septin ring on the side of the first cut was masked using the threshold function in Fiji and mean intensity of the septin ring was measured right after cleavage furrow ingression when septin rings are formed (Imax), and one frame before the appearance of the CHMP4B cone (Imin), along with the background intensity for these frames (Ibckgmax and Ibckgmin). The % reduced septin ring intensity was calculated using the formula {1-[( Imax - Ibckgmax) − (Imin - Ibckgmin)/(Imax - Ibckgmax)]} × 100. In Figure S2C, time-lapse movies of MDCK-mCherry-SEPT2 or MDCK-mCherry-SEPT9 cells co-expressing fluorescently tagged ESCRTs, were analyzed and compiled to temporally map the recruitment and re-organization of each component relative to the time of abscission. Each point in the graph represents a single cell. In Figure 3C and S4B, MDCK cells transfected with scramble or SEPT9 shRNA (Figure 3C) or SEPT9shRNA and SEPT9-mCherry or SEPT9-Δ(PTAP)2x-mCherry (Figure S4B) were identified by their co-expression of mCherry and were imaged live in DIC for 10 hours and the time between cleavage furrow completion and abscission (membrane fission between two daughter cells; visible in DIC) was tracked. To measure fluoresce intensity of endogenous SEPT9 in Figure 3A–B, images were imported to Fiji and the mCherry channel was thresholded to outline the cells expressing scramble or SEPT9 shRNA. The outline of each cell was stored as a region of interest (ROI) and its sum intensity and area were measured. For Figures 3D, and 4I, TSG101 accumulations in the intracellular bridge (marked by tubulin) were outlined manually using Fiji’s freehand selection tool. Similarly, for Figure 4E and 4G the tubulin channel was used to mask the ICB, which was saved as an ROI and the intensity for the PLA (Figure 4E) or BifC (Figure 4G) signal was measured. To generate the plot in Figure 4F, the widest width of the ICB as marked by the tubulin stain was measured for both sides of the ICB. The average of the two widths was calculated for each cell and the presence of PLA signal was scored. In Figure S4G, actin intensity was measured by using the tubulin channel to outline the ICB. The outlined areas were stored as a ROI and their sum intensity and area were measured. Percent ICBs with or without ESCRT accumulation and percent ICBs with different ESCRT-III morphologies (Figures 3E–G, 4K–M, S3A–H) were assessed visually and recorded for each capture. In Figure S1C–D, septin intact double rings, partially or fully extracted were assessed only for ICBs with non-compacted microtubules, as the septin rings dissipate in later stages of cytokinesis, where microtubules are tightly compacted. Quantifications of GST-pull downs of SEPT9 or SEPT9-Δ(PTAP)2x with GST-TSG101-UEV (Figure 4B) were done by normalizing the SEPT9/SEPT9-Δ(PTAP)2x band intensities (ISEPT9) to their respective input (Iinput) and amount of GST-UEV pulled (IUEV), using the formula ISEPT9/Iinput/IUEV. For each experiment, SEPT9-Δ(PTAP)2x was calculated in relation to SEPT9-WT (control; intensity set to 1). Similarly, quantification of immunoprecipitations of endogenous TSG101 with SEPT9- or SEPT9-Δ(PTAP)2x-mCherry (Figure 4E) were done by measuring TSG101 (ITSG101) normalized to TSG101 input (IinputTSG101) and SEPT9-/ SEPT9-Δ(PTAP)2x-mCherry captured with the RFP-Trap®_A beads (ChromoTek) (ISEPT9-mCh), using the formula ITSG101/IinputTSG101/ISEPT9-mCh. For each experiment, the intensity of TSG101 pulled down by SEPT9-Δ(PTAP)2x was calculated in relation to that pulled with SEPT9-WT (control; intensity set as 1).
Statistical Analysis
Statistical analysis of data was done using GraphPad Prism 6.0 software. Data sets were tested for assumptions of normality using the D’Agostino and Pearson normality test. p values were derived using a student’s t-test for normally distributed data with equal variance and the Mann Whitney U test for non-normally distributed data. P values lower than 0.05 were considered statistically significant. Data graphs were plotted in GraphPad Prism using the bar graph plot function showing the standard error of the mean (SEM) or the scatter dot plot function showing the mean ± SEM or mean ± the standard deviation of the mean (SD). N values and p values for each experiment are denoted in the corresponding figure and/or figure legend.
Supplementary Material
Video S1. Time-lapse movie of an MDCK-mCherry-SEPT9 (green) cell transfected with GFP-α-tubulin (magenta). Scale bar, 5 μm. Related to Figure 1.
Video S2. Time-lapse movie of an MDCK-mCherry-SEPT2 (green) cell transfected with GFP-α-tubulin (magenta). Arrow points to ICB cut. Scale bar, 5 μm. Related to Figure 1.
Video S3. Time-lapse movie of an MDCK-GFP-CHMP4B cell transfected with SEPT9-mCherry. Scale bar, 5 μm. Related to Figure 2.
Video S4. Time-lapse movie of an MDCK-mCherry-SEPT2 cell transfected with GFP-CHMP2A. Scale bar, 5 μm. Related to Figure 2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-SEPT9 | EMD Millipore | Cat#MABE992 |
| Rabbit anti-septin 9 | PTG labs | Cat#10769 – IAP; RRID:AB_2185012 |
| Mouse anti-α-tubulin (DM1α) | Sigma | Cat#T6199; RRID:AB_477583 |
| Rabbit anti-Septin 2 (clone: a5N5) | Kind gift form M. Kinoshita (Nagoya University, Japan) | N/A |
| Rabbit anti-Septin 6 | Kind gift form M. Kinoshita (Nagoya University, Japan) | N/A |
| Rabbit anti-Septin 7 | IBL America | Cat#18991; RRID:AB_1630825 |
| Rabbit anti-CHMP4B | PTG labs | Cat# 15669-1-AP |
| Mouse anti-TSG101 (clone C-52) | Santa Cruz | Cat# sc-7964, RRID:AB_671392 |
| Mouse anti-ALIX | Santa Cruz | Cat# sc-53540, RRID:AB_673819 |
| Rabbit anti-VPS25 | Abclonal | Cat# A13132, RRID:AB_2759982 |
| Rabbit anti-VPS22 | Abclonal | Cat# A10583, RRID:AB_2758124 |
| Rabbit anti-CHMP2B | Abclonal | Cat# A13410, RRID:AB_2760272 |
| Rabbit anti-TSG101 | PTG labs | Cat# 14497-1-AP, RRID:AB_2208090 |
| Rabbit anti-CHMP6 | PTG labs | Cat# 16278-1-AP, RRID:AB_2079498 |
| Rabbit anti-VPS25 | PTG labs | Cat# 15669-1-AP, RRID:AB_2215019 |
| Mouse anti-mCherry | Abcam | Cat# |
| Mouse anti-His tag | Qiagen | Cat# Cat# 34440, RRID:AB_2714179 |
| Rabbit anti-CHMP4B | Santa Cruz | Cat# sc-134946, RRID:AB_10608470 |
| Rabbit anti-GST | Santa cruz | Cat# sc-459, RRID:AB_631586 |
| goat-anti rabbit IRDye® 680RD | LI-COR | Cat# 925-68071; RRID:AB_2721181 |
| goat anti-mouse IRDye® 800RD | LI-COR | Cat# 925-32210; RRID:AB_2687825 |
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) | Invitrogen | Cat#C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Purified 6XHis-hSEPT9 (GenBank accession number NP_001106963; isoform a ) protein | [44] | N/A |
| Purified 6XHis-hSEPT9 Δ(PTAP)2x | This paper | N/A |
| GST-TSG101-UEV | This paper | N/A |
| Ni-NTA agarose beads | Macherey-Nagel | Cat# 745400.25 |
| Glutathione agarose beads | Thermo Scientific | Cat# 16100 |
| RFP-Trap®_A beads | ChromoTek | Cat# rta-10 |
| Critical Commercial Assays | ||
| Quick Change II site-mutagenesis kit | Agilent Technologies | Cat#200523 |
| Duolink® PLA – Protein Detection Technology | Sigma - Aldrich | Cat# DUO92101-1KT |
| Zenon™Alexa Fluor™ 568 Rabbit IgG Labeling Kit |
ThermoFisher Scientific | Cat# Z-25306; RRID:AB_2736952 |
| Experimental Models: Cell Lines | ||
| Madin Darby kidney canine epithelia cells (MDCK) II/G | Kind gift from J. Nelson, Stanford University, CA | ECACC #00062107 |
| MDCK mCherry- SEPT9 | [42] | N/A |
| MDCK mCherry- SEPT2 | [13] | N/A |
| MDCK SEPT2-YFP | [12] | N/A |
| MDCK GFP-SEPT6 | This paper | N/A |
| MDCK GFP-CHMP4B | This paper | N/A |
| Oligonucleotides – see Table S1 for sequences of primers used for this study | ||
| Recombinant DNA | ||
| GFP-TSG101 | Kind gift from Dr. Paul Bieniasz (The Rockefeller University, NY) | N/A |
| GFP-CHMP4B | Kind gift from Dr. Paul Bieniasz, The Rockefeller University, NY | N/A |
| GFP-CHMP2A | Addgene | Cat# 31805 , deposited by the Daniel Gerlich Lab |
| CHMP6-GFP | Addgene | Cat #31806 , deposited by the Daniel Gerlich Lab |
| GFP-VPS25 |
Kind gift from Dr. Natalie Elia, Ben Gurion University of Negev, Israel [20] | N/A |
| GFP-VPS36 | Kind gift from Dr. Natalie Elia, Ben Gurion University of Negev, Israel [20] | N/A |
| GFP-VPS22 | Kind gift from Dr. Natalie Elia, Ben Gurion University of Negev, Israel [20] | N/A |
| pEGFP-C1-VPS4 |
Kind gift from Dr. Wes Sundquist, University of Utah School of Medicine, UT | N/A |
| pEGFP-C1-α-tubulin | Kind gift from J. Nelson, Stanford University, CA [13] | N/A |
| pBiFC-VN173 | Addgene | Cat#22010 deposited by the Chang-Deng Hu lab |
| pBiFC-VC155 | Addgene | Cat#22010 deposited by the Chang-Deng Hu lab |
| SEPT9_VN | This paper | N/A |
| TSG101_VC | This paper | N/A |
| TSG101_VN | This paper | N/A |
| pmCherry-C1-SEPT2 | [13] | N/A |
| pmCherry -C1-SEPT9 | [42] | N/A |
| pYFP-N1- (mouse) SEPT2 | [12] | N/A |
| pEGFP-C3-SEPT6 | [43] | N/A |
| pmCherry-N-hSEPT9 | [42] | N/A |
| pGFP-VR-S-canine S9 shRNA-rescuehSEPT9-FL-mCherry | This paper | N/A |
| pGFP-VR-S-canine S9 shRNA1-rescuehSEPT9-Δ(PTAP)2x -mCherry | This paper | N/A |
| GST-TSG101 UEV | This paper | N/A |
| His-hSEPT9 | [44] | N/A |
| His-hSEPT9- Δ(PTAP)2x | This paper | N/A |
| Software and Algorithms | ||
| Slidebook | 3i | https://www.intelligent-imaging.com/ ; RRID:SCR_014300 |
| Fiji (ImageJ) | NIH | https://imagej.nih.gov/ij/ ; RRID:SCR_002285 |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ ; RRID:SCR_002798 |
| MATLAB | MATLAB | http://www.mathworks.com/products/matlab/; RRID:SCR_001622 |
Highlights.
Septins form a membrane-bound double ring in the midbody of telophase cells
Septin rings demarcate the membrane sites of ESCRT-III assembly
SEPT9 is required for ESCRT-III ring formation and expansion to spirals/cones
SEPT9 interacts with TSG101 for recruitment and assembly of ESCRT-II/-III subunits
ACKNOWLEDGMENTS
We are grateful to Drs. Paul Bieniasz (The Rockefeller University/HHMI), Natalie Elia (Ben-Gurion University of the Negev, Israel), Makoto Kinoshita (Nagoya University, Japan) and Wes Sundquist (University of Utah Medical School) for their generous sharing of plasmids. This work was supported by NIGMS grant R01GM097664 to E.T.S. and NCI pre-doctoral fellowship F31CA213776 to E.P.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Guizetti J, and Gerlich DW (2010). Cytokinetic abscission in animal cells. Semin Cell Dev Biol 21, 909–916. [DOI] [PubMed] [Google Scholar]
- 2.Nahse V, Christ L, Stenmark H, and Campsteijn C (2017). The Abscission Checkpoint: Making It to the Final Cut. Trends Cell Biol 27, 1–11. [DOI] [PubMed] [Google Scholar]
- 3.Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, and Gerlich DW (2011). Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620. [DOI] [PubMed] [Google Scholar]
- 4.Elia N, Sougrat R, Spurlin TA, Hurley JH, and Lippincott-Schwartz J (2011). Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A 108, 4846–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoten CL, and Carlton JG (2018). ESCRT-dependent control of membrane remodelling during cell division. Semin Cell Dev Biol 74, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoneberg J, Lee IH, Iwasa JH, and Hurley JH (2017). Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol 18, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henne WM, Stenmark H, and Emr SD (2013). Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuh AL, and Audhya A (2014). The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol 49, 242–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong K, Wloka C, Okada S, Svitkina T, and Bi E (2014). Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun 5, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lippincott J, Shannon KB, Shou W, Deshaies RJ, and Li R (2001). The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci 114, 1379–1386. [DOI] [PubMed] [Google Scholar]
- 11.Dobbelaere J, and Barral Y (2004). Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305, 393–396. [DOI] [PubMed] [Google Scholar]
- 12.Spiliotis ET, Kinoshita M, and Nelson WJ (2005). A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science 307, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen JR, Hwang D, Bai X, Roy D, and Spiliotis ET (2011). Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J Cell Biol 194, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolat L, and Spiliotis ET (2016). Septins promote macropinosome maturation and traffic to the lysosome by facilitating membrane fusion. J Cell Biol 214, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolat L, Hu Q, and Spiliotis ET (2014). Septin functions in organ system physiology and pathology. Biol Chem 395, 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlton JG, and Martin-Serrano J (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912. [DOI] [PubMed] [Google Scholar]
- 17.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, and Sundquist WI (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26, 4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christ L, Wenzel EM, Liestol K, Raiborg C, Campsteijn C, and Stenmark H (2016). ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J Cell Biol 212, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thoresen SB, Campsteijn C, Vietri M, Schink KO, Liestol K, Andersen JS, Raiborg C, and Stenmark H (2014). ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat Cell Biol 16, 550–560. [DOI] [PubMed] [Google Scholar]
- 20.Goliand I, Nachmias D, Gershony O, and Elia N (2014). Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol Biol Cell 25, 3740–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goliand I, Adar-Levor S, Segal I, Nachmias D, Dadosh T, Kozlov MM, and Elia N (2018). Resolving ESCRT-III Spirals at the Intercellular Bridge of Dividing Cells Using 3 D STORM. Cell Rep 24, 1756–1764. [DOI] [PubMed] [Google Scholar]
- 22.Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, Larios J, Poser I, Muller-Reichert T, Scheuring S, Roux A, et al. (2017). Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 19, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mierzwa B, and Gerlich DW (2014). Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell 31, 525–538. [DOI] [PubMed] [Google Scholar]
- 24.Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, and Trimble WS (2010). Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J Cell Biol 191, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee IH, Kai H, Carlson LA, Groves JT, and Hurley JH (2015). Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A 112, 15892–15897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teo H, Perisic O, Gonzalez B, and Williams RL (2004). ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell 7, 559–569. [DOI] [PubMed] [Google Scholar]
- 27.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, and Maki M (2005). Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J 387, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Serrano J, Zang T, and Bieniasz PD (2003). Role of ESCRT-I in retroviral budding. J Virol 77, 4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano J, Zang T, and Bieniasz PD (2001). HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7, 1313–1319. [DOI] [PubMed] [Google Scholar]
- 30.Pornillos O, Alam SL, Davis DR, and Sundquist WI (2002). Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol 9, 812–817. [DOI] [PubMed] [Google Scholar]
- 31.Hu CD, and Kerppola TK (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. (2006). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3, 995–1000. [DOI] [PubMed] [Google Scholar]
- 33.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, and Sundquist WI (2003). HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol 162, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, and Hurley JH (2004). Structure of the ESCRT-II endosomal trafficking complex. Nature 431, 221–225. [DOI] [PubMed] [Google Scholar]
- 35.Fyfe I, Schuh AL, Edwardson JM, and Audhya A (2011). Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J Biol Chem 286, 34262–34270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renshaw MJ, Liu J, Lavoie BD, and Wilde A (2014). Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol 4, 130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Amine N, Kechad A, Jananji S, and Hickson GR (2013). Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J Cell Biol 203, 487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menon MB, Sawada A, Chaturvedi A, Mishra P, Schuster-Gossler K, Galla M, Schambach A, Gossler A, Forster R, Heuser M, et al. (2014). Genetic deletion of SEPT7 reveals a cell type-specific role of septins in microtubule destabilization for the completion of cytokinesis. PLoS Genet 10, e1004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traikov S, Stange C, Wassmer T, Paul-Gilloteaux P, Salamero J, Raposo G, and Hoflack B (2014). Septin6 and Septin7 GTP binding proteins regulate AP-3- and ESCRT-dependent multivesicular body biogenesis. PLoS One 9, e109372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, Formstecher E, Salomon R, Goud B, and Echard A (2011). Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat Cell Biol 13, 981–988. [DOI] [PubMed] [Google Scholar]
- 41.Fremont S, Hammich H, Bai J, Wioland H, Klinkert K, Rocancourt M, Kikuti C, Stroebel D, Romet-Lemonne G, Pylypenko O, et al. (2017). Oxidation of F-actin controls the terminal steps of cytokinesis. Nat Commun 8, 14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolat L, Hunyara JL, Bowen JR, Karasmanis EP, Elgawly M, Galkin VE, and Spiliotis ET (2014). Septins promote stress fiber-mediated maturation of focal adhesions and renal epithelial motility. J Cell Biol 207, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinoshita M, Field CM, Coughlin ML, Straight AF, and Mitchison TJ (2002). Self- and actin-templated assembly of Mammalian septins. Dev Cell 3, 791–802. [DOI] [PubMed] [Google Scholar]
- 44.Bai X, Bowen JR, Knox TK, Zhou K, Pendziwiat M, Kuhlenbaumer G, Sindelar CV, and Spiliotis ET (2013). Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. J Cell Biol 203, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Time-lapse movie of an MDCK-mCherry-SEPT9 (green) cell transfected with GFP-α-tubulin (magenta). Scale bar, 5 μm. Related to Figure 1.
Video S2. Time-lapse movie of an MDCK-mCherry-SEPT2 (green) cell transfected with GFP-α-tubulin (magenta). Arrow points to ICB cut. Scale bar, 5 μm. Related to Figure 1.
Video S3. Time-lapse movie of an MDCK-GFP-CHMP4B cell transfected with SEPT9-mCherry. Scale bar, 5 μm. Related to Figure 2.
Video S4. Time-lapse movie of an MDCK-mCherry-SEPT2 cell transfected with GFP-CHMP2A. Scale bar, 5 μm. Related to Figure 2.


